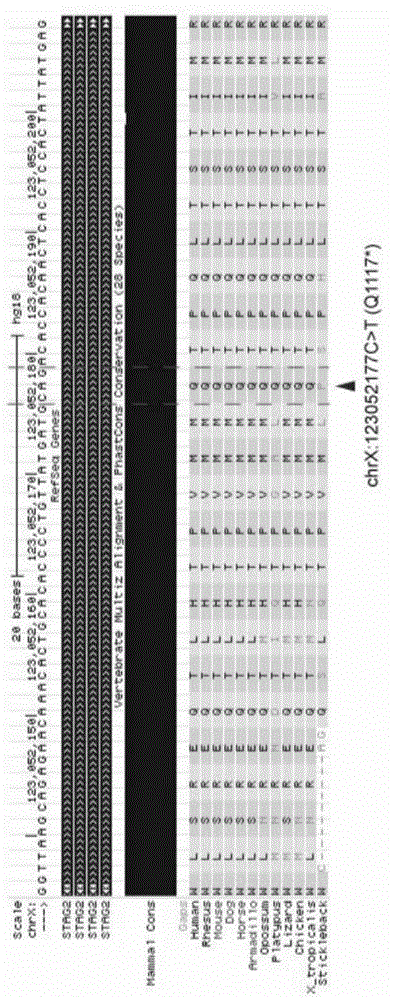
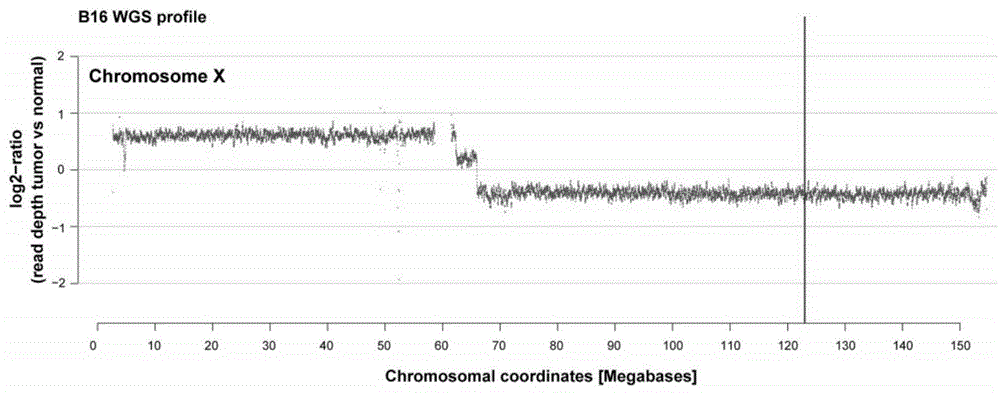
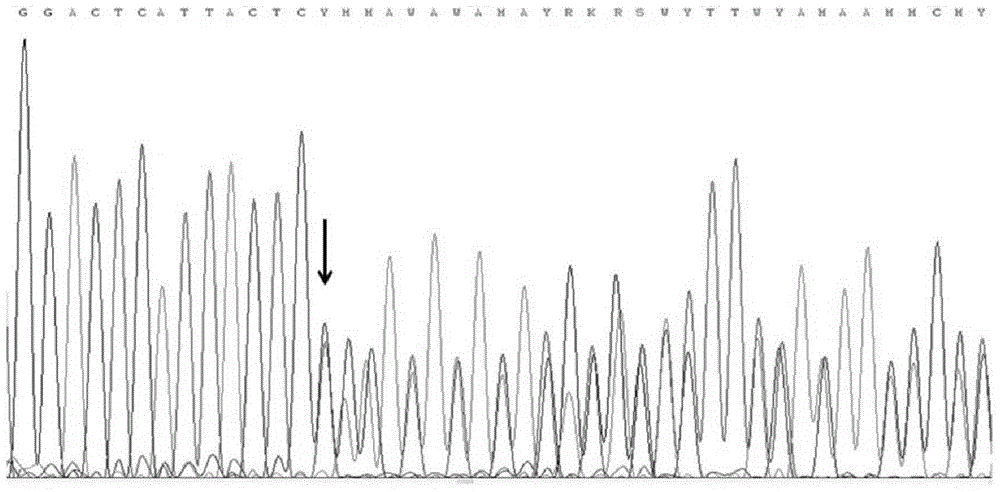
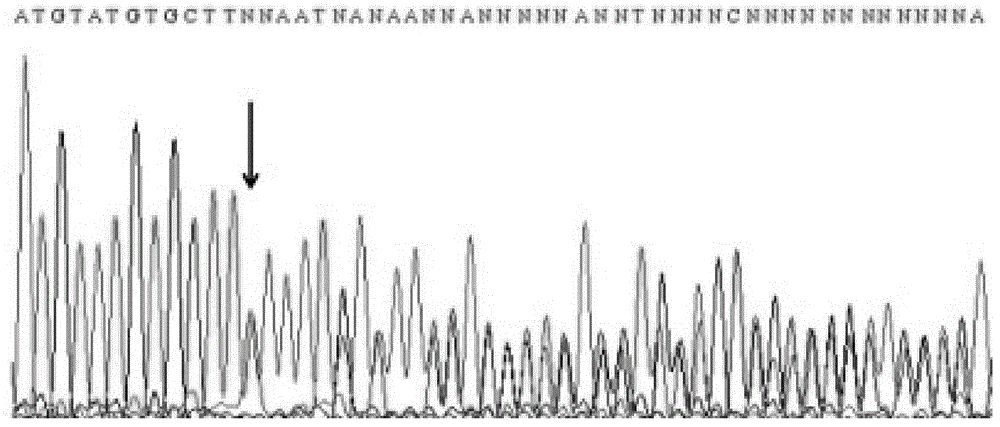
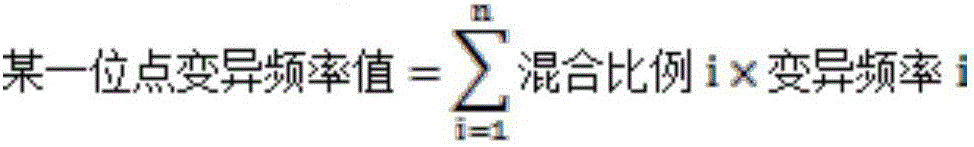Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
368 results about "Sanger sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Sanger sequencing is a method of DNA sequencing first commercialized by Applied Biosystems, based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. Developed by Frederick Sanger and colleagues in 1977, it was the most widely used sequencing method for approximately 40 years. More recently, higher volume Sanger sequencing has been replaced by "Next-Gen" sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use, for smaller-scale projects, and for validation of Next-Gen results. It still has the advantage over short-read sequencing technologies (like Illumina) that it can produce DNA sequence reads of > 500 nucleotides.
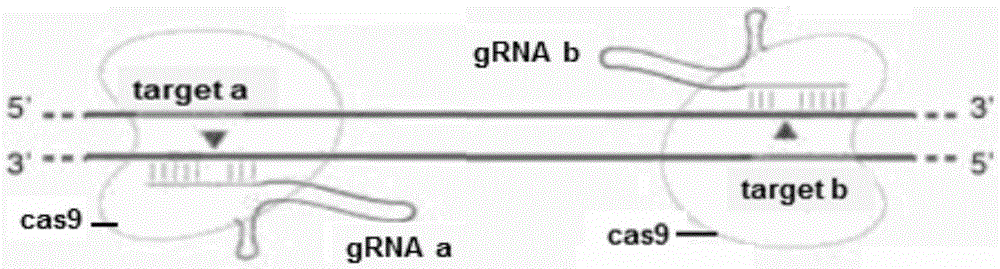
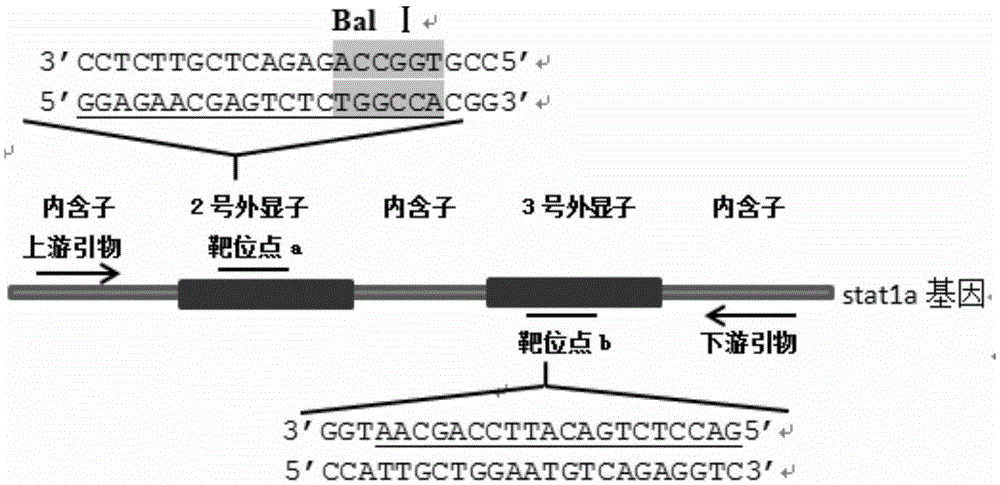
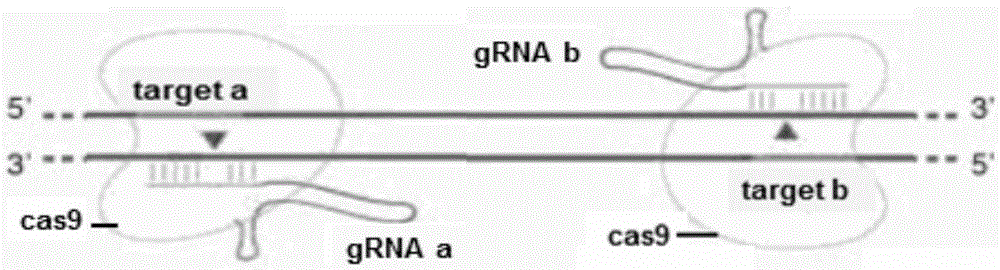
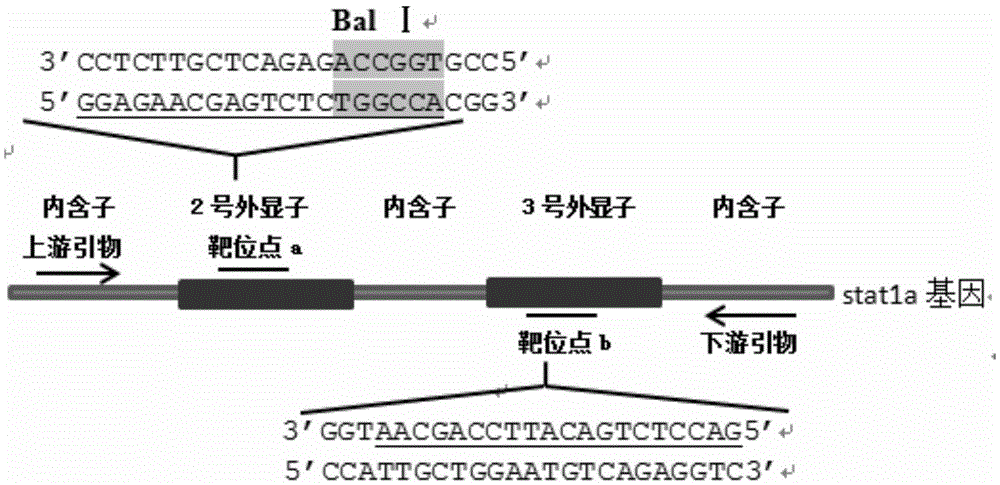
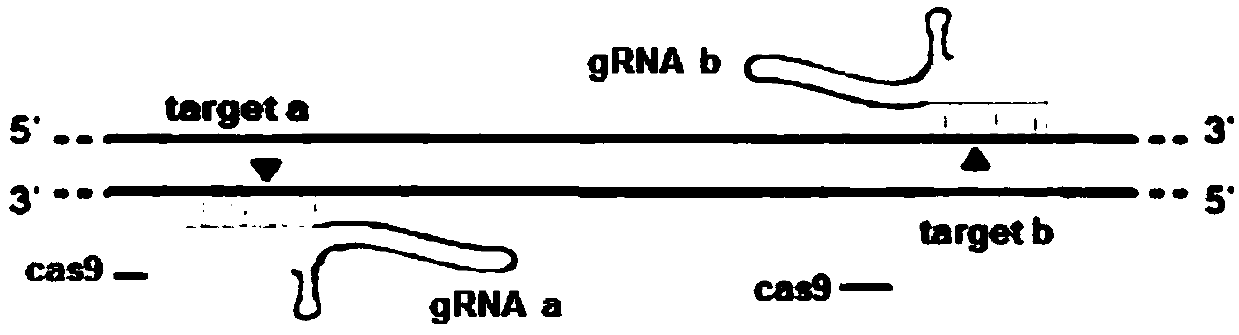
Method for breeding stat1a (signal transducer and activator of transcription 1) gene-deleted zebra fish through gene knockout
InactiveCN105647969AInefficient shooting techniqueLow costMicrobiological testing/measurementPeptidesFish embryoEmbryo
A method for breeding stat1a (signal transducer and activator of transcription 1) gene-deleted zebra fish through gene knockout comprises steps as follows: design of a CRISPR / Cas9 gene knockout target site: a gRNA expression carrier is established and gRNA is synthesized in vitro; micro-injection of a zebra fish embryo; detection of effectiveness of the target site with a T7E1 method through Sanger sequencing; tail cutting identification according to the identification steps after two months of injection; TA cloning of a target sequence; Sanger sequencing of plasmids; obtaining of heritable F1 generation of a zebra fish mutant; obtaining of F2 generation homozygote of the zebra fish mutant, F3 generation pure line inheritance of the gene-deleted zebra fish with the above method, and obtaining of a new zebra fish strain.
Owner:HUNAN NORMAL UNIVERSITY
Statla gene deletion type zebra fish
InactiveCN105594664AInefficient shooting techniqueLow costMicrobiological testing/measurementPeptidesFish embryoEmbryo
The invention provides statla gene deletion type zebra fish. After an experiment design region is knocked out, the sequence is represented as SEQ ID No.1; an experiment comprises the following steps: designing a CRISPR / Cas9 gene knockout target point: constructing a gRNA expression vector and synthesizing gRNA in vitro; carrying out micro-injection of zebra fish embryos; detecting the effectiveness of the target point by a T7E1 method and Sanger sequencing; two months after injection, carrying out tail shearing and identification and carrying out identification steps above; carrying out TA cloning of a target sequence; carrying out Sanger sequencing of plasmids; obtaining an F1 generation of descendible zebra fish mutants; obtaining F2 generation homozygotes of the zebra fish mutants; and carrying out F3 generation homozygous heredity of the gene deletion type zebra fish according to the method above to obtain a new zebra fish line.
Owner:HUNAN NORMAL UNIVERSITY
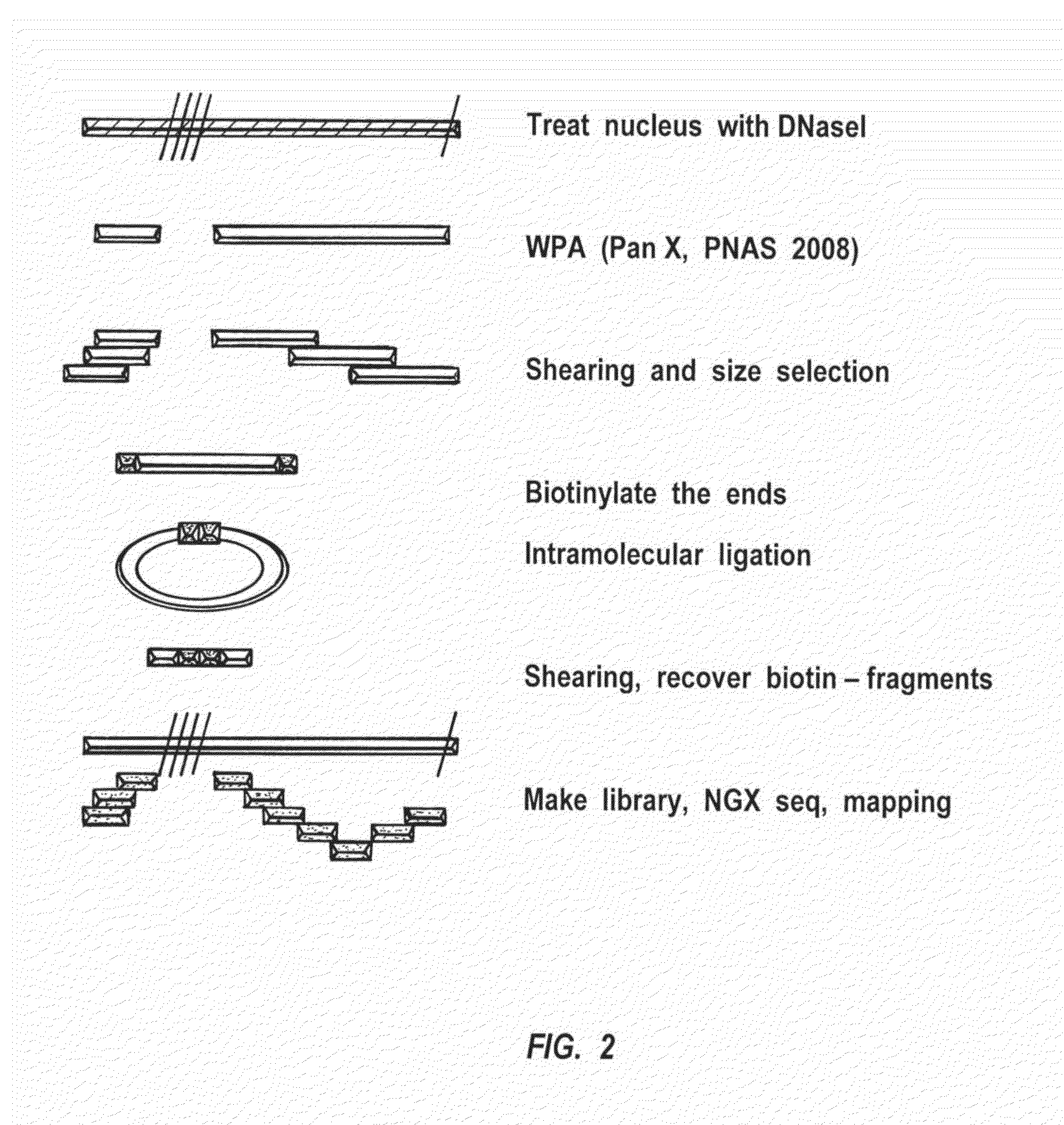
Methods for closed chromatin mapping and DNA methylation analysis for single cells
ActiveUS20150368694A1Reduce and minimize and prevent chanceLess efficiently ligatedMicrobiological testing/measurementLibrary screeningDNA methylationGenomic DNA
Methods of identifying DNase I Hyper-Resistant Sites (DHRS), or in board sense, highly compact chromatin and characterizing the DNA methylation status of DMRs such as CpG islands and CpG island shores are provided. The methods are particularly useful for analysis of genomic DNA from low quantities of cells, for example, less than 1,000 cells, less than 100 cells, less than 10 cells, or even one cell, and can be used to generate chromatin and methylation profiles. The downstream analyses include in parallel massive sequencing, microarray, PCR and Sanger sequencing, hybridization and other platforms. These methods can be used to generate chromatin and DNA methylation profiles in drug development, diagnostics, and therapeutic applications are also provided.
Owner:YALE UNIV
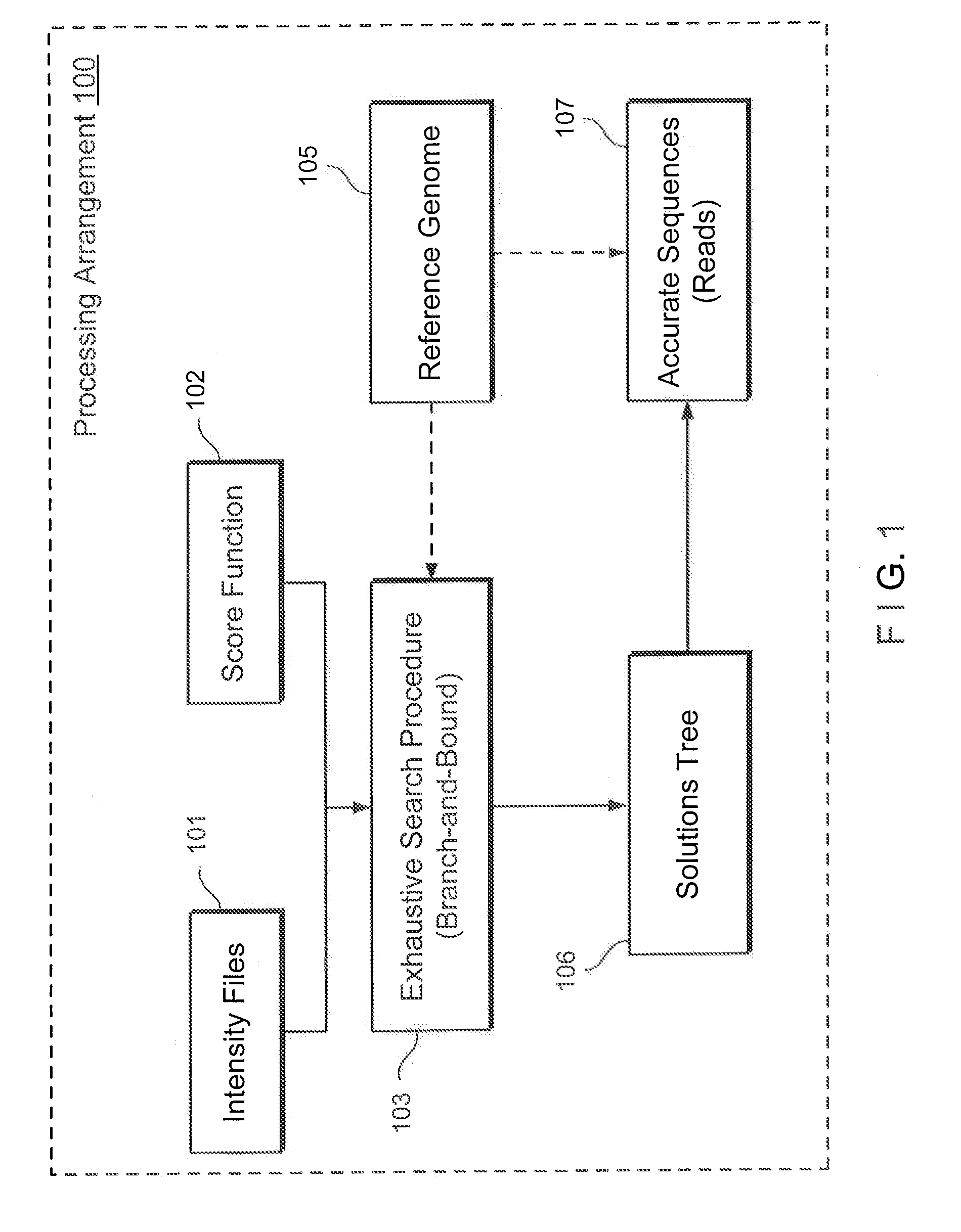
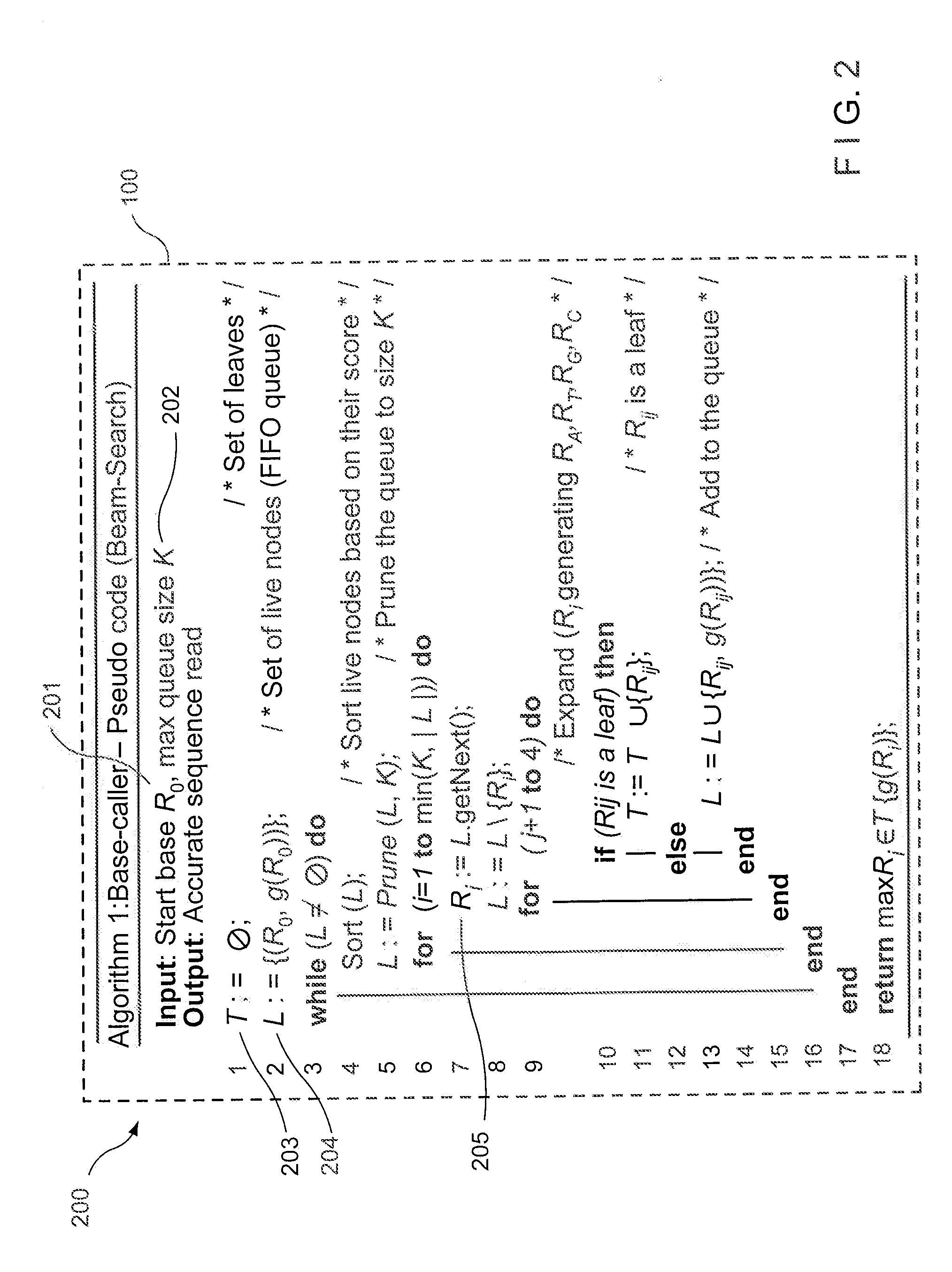
Method, computer-accessible medium and system for base-calling and alignment
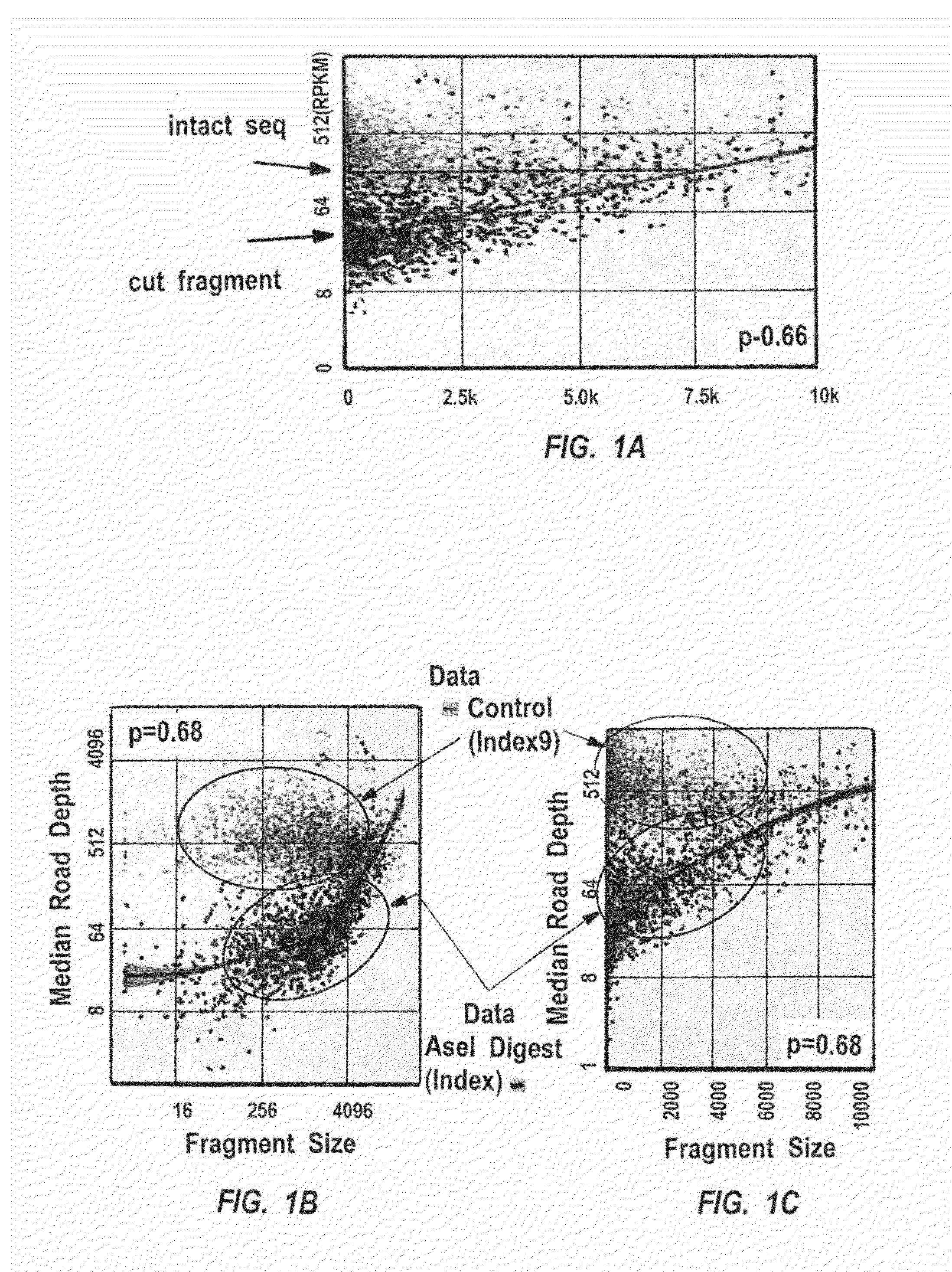
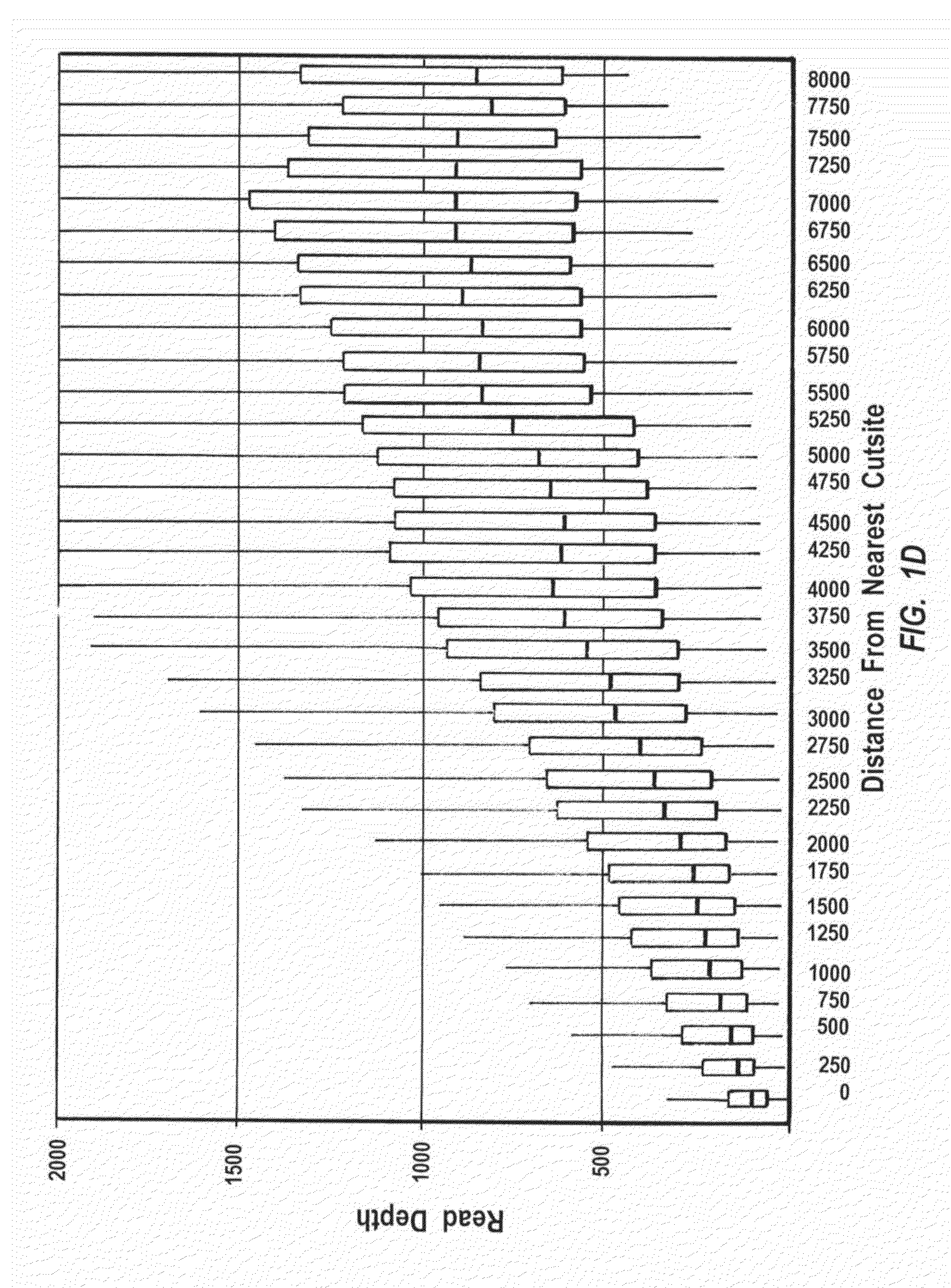
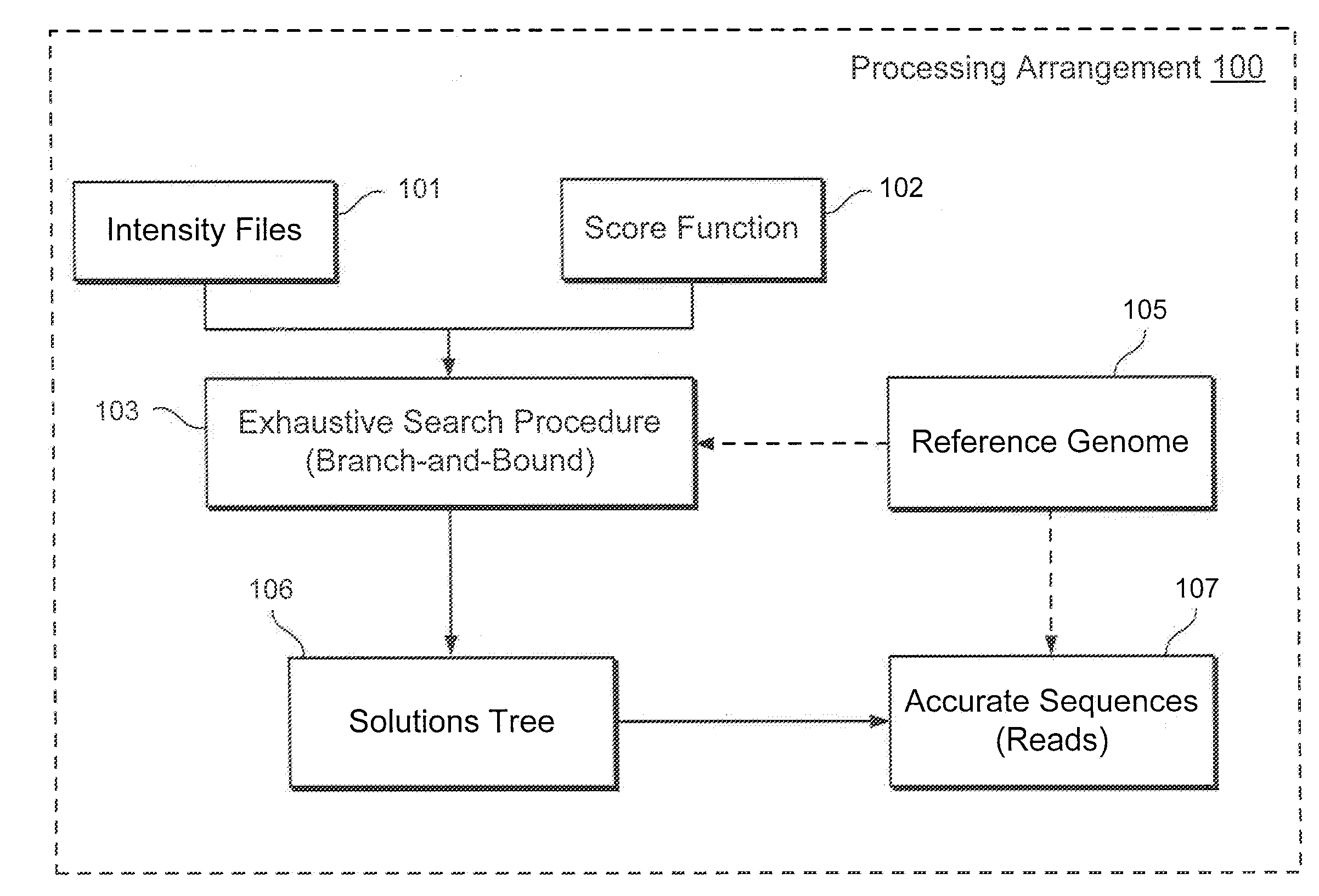
Exemplary methods, procedures, computer-accessible medium, and systems for base-calling, aligning and polymorphism detection and analysis using raw output from a sequencing platform can be provided. A set of raw outputs can be used to detect polymorphisms in an individual by obtaining a plurality of sequence read data from one or more technologies (e.g., using sequencing-by-synthesis, sequencing-by-ligation, sequencing-by-hybridization, Sanger sequencing, etc.). For example, provided herein are exemplary methods, procedures, computer-accessible medium and systems, which can include and / or be configured for obtaining raw output from a sequencing platform configured to be used for reading fragment(s) of genomes, obtaining reference sequences for the genomes obtained independently from the raw output, and generating a base-call interpretation and / or alignment using the raw output and the reference sequences. For example, a score function can be determined based on information associated with the sequencing platform that can be used to analyze polymorphisms based on the base-call interpretation and / or alignment.
Owner:NEW YORK UNIV
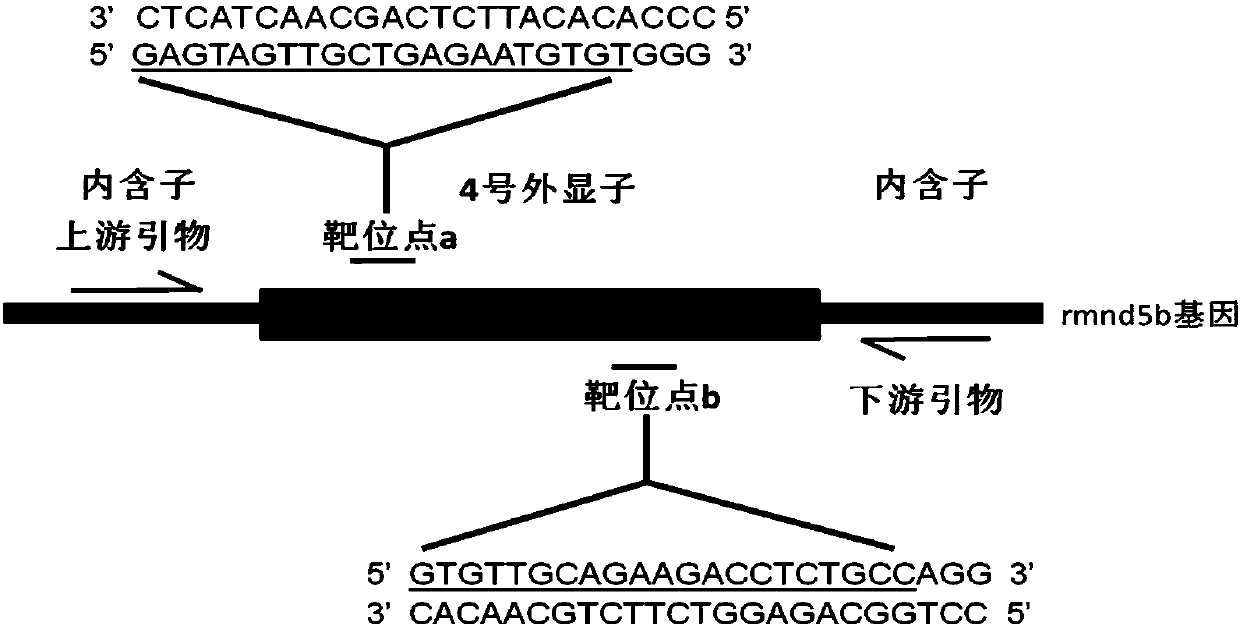
Method for breeding rmnd5b (required for meiotic nuclear division 5homolog B) gene deletion type zebra fish through gene knockout
InactiveCN108018316AEasy to makeLow miss rateHydrolasesStable introduction of DNAMeiotic nuclear divisionEmbryo
The invention discloses a method for breeding rmnd5b (required for meiotic nuclear division 5homolog B) gene deletion type zebra fish through gene knockout and belongs to the field of gene knockout. According to the method, construction of a gRNA expression vector and gRNA in-vitro synthesis are performed through design of a CRISPR / Cas9 (clustered regularly interspaced short palindromic repeats / CRISPR-associated 9) gene knockout target site, micro-injection is performed on an embryo of the zebra fish, the effectiveness of the target site is detected, tail cutting identification is performed, TA cloning is performed on a target sequence, plasmids are subjected to Sanger sequencing, an F1 generation of heritable zebra fish mutants is obtained, the same mutant female fish and male fish are picked from mutants of the F1 generation, hybridization is performed, an F2 generation of the zebra fish mutants is obtained, F2 generation homozygote is picked from the F2 generation of the zebra fishmutants, F3 generation pure-line inheritance is performed, and an rmnd5b gene deletion type zebra fish strain is obtained. The method is lower in off-target rate and has good medical research value inresearch of the correlation between rmnd5b gene deletion and development of other organs.
Owner:HUNAN NORMAL UNIVERSITY
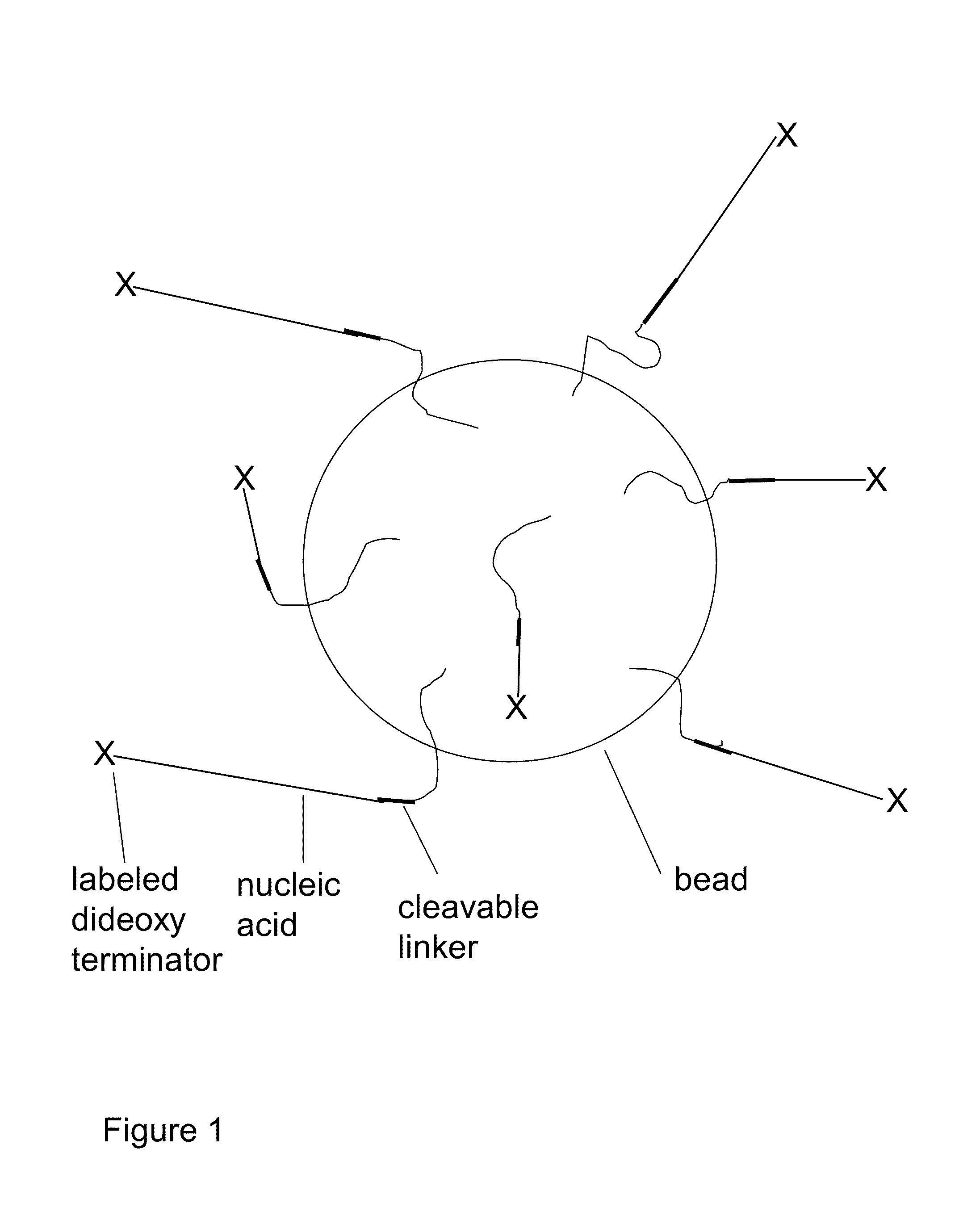
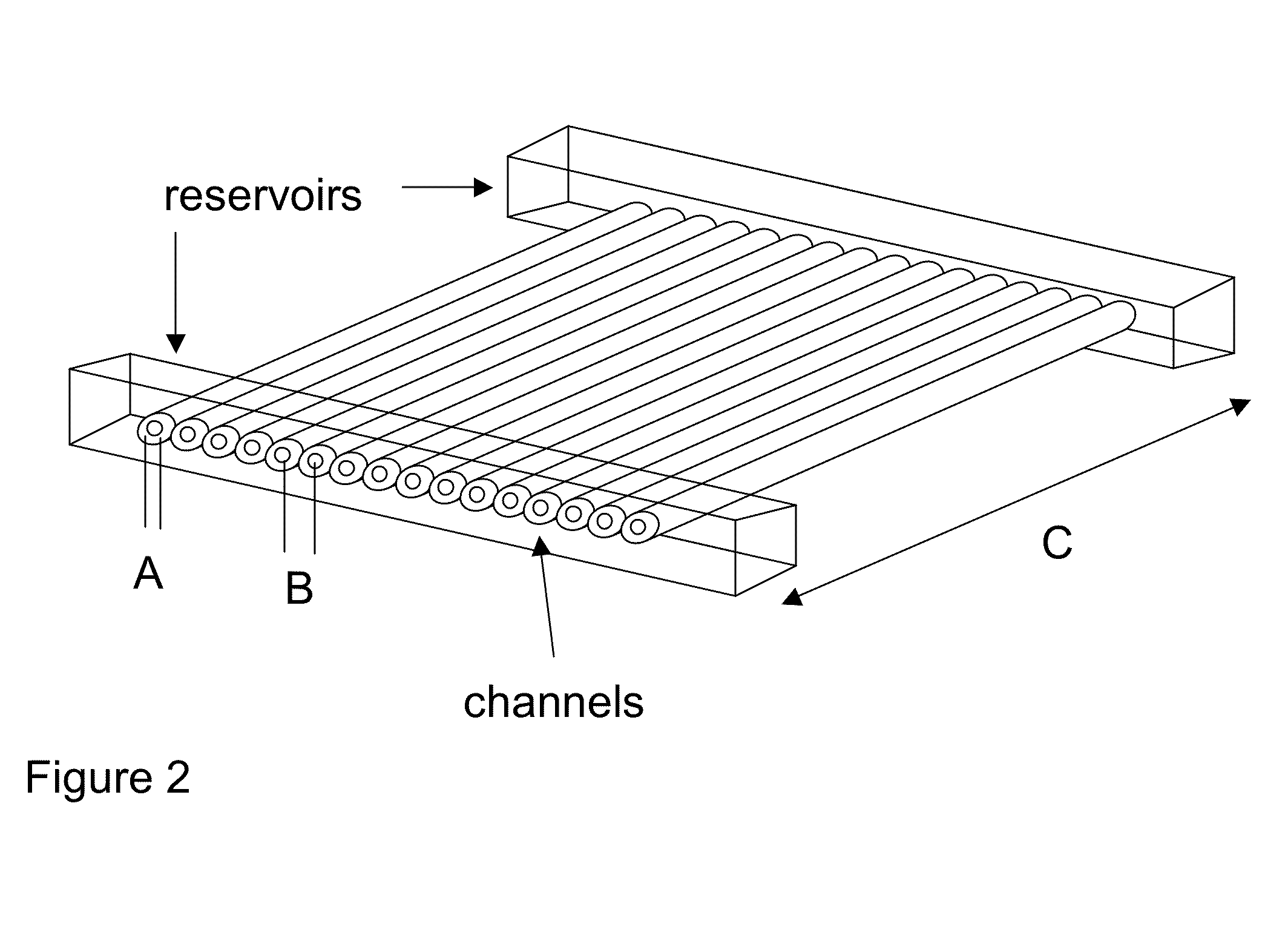
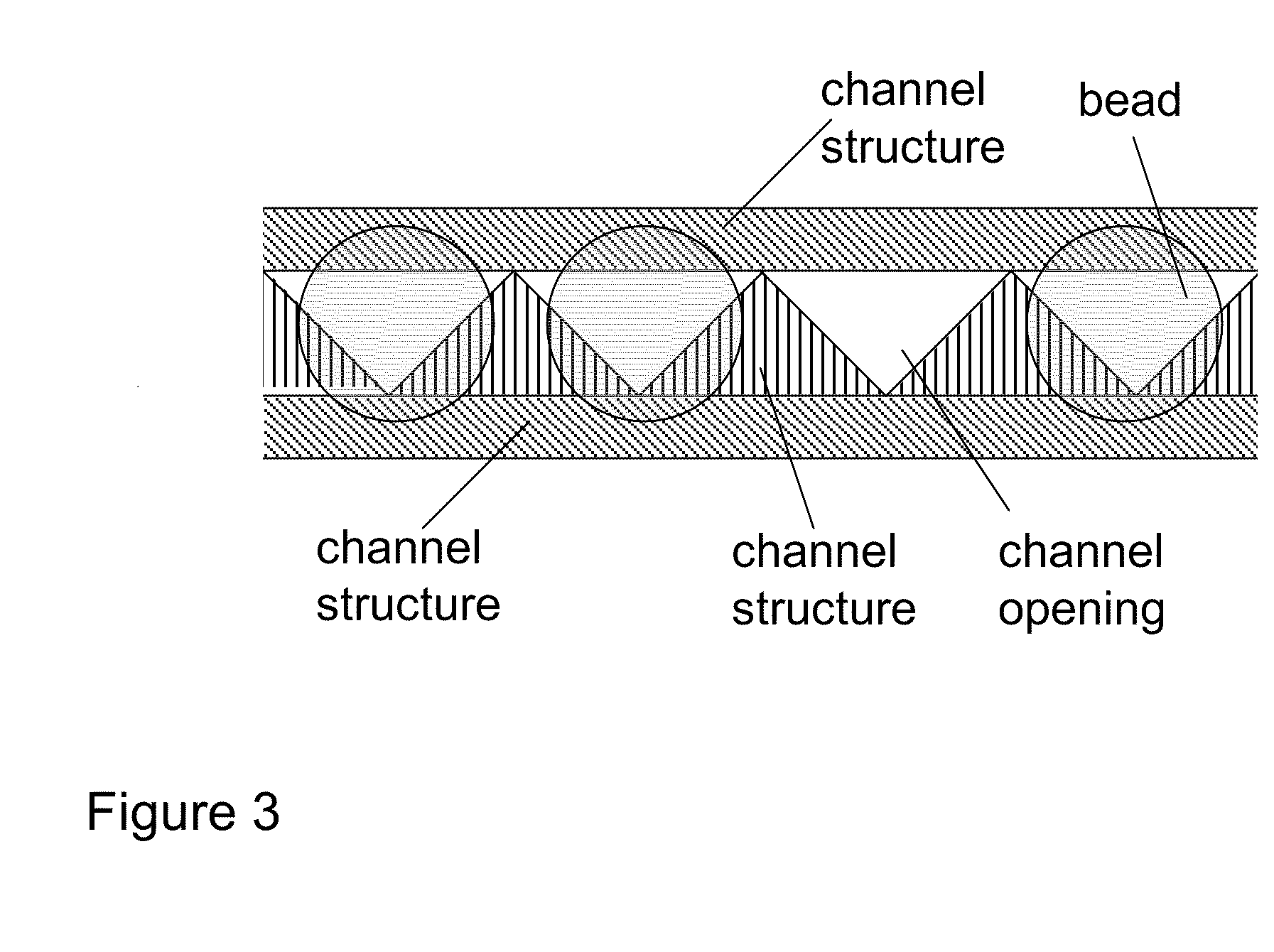
Methods for sanger sequencing using particle associated clonal amplicons and highly parallel electrophoretic size-based separation
InactiveUS20100112588A1Low accuracyLarge amount of sampleMicrobiological testing/measurementSanger sequencingComputational biology
Methods for highly parallel Sanger sequencing are discussed. In particular, provided herein are methods using particles to clonally amplify templates and to introduce the amplified nucleic acids into many parallel channels with a single template per channel. Once in the channels, the nucleic acids are separated by size using electrophoresis to produce long read length sequencing information. Methods involving optical detection of the size-separated nucleic acids and analysis of the resulting electropherograms to yield the sequences are disclosed.
Owner:CAERUS MOLECULAR DIAGNOSTICS
Method and primer for detecting mutation sites of all exon sequences of human BRCA1 and BRCA2 genes
ActiveCN104531862AGood amplification efficiencyImprove risk assessmentMicrobiological testing/measurementDNA/RNA fragmentationGeneticsBrca1 gene
The invention relates to the technical field of gene detection, and provides a method and a primer for detecting mutation sites of all exon sequences of human BRCA1 and BRCA2 genes. The specific primers include forward and backward primers for amplifying all 24 exons covering the BRCA1 gene, and base sequences of the primers are as shown in SEQ ID NO: 001-062, as well as forward and backward primers for amplifying all 27 exons covering the BRCA2 gene, and base sequences of the primers are as shown in SEQ ID NO: 063-142. By virtue of a Sanger sequencing method, the method and the primer disclosed by the invention can be used for detecting the mutation of all exons of the BRCA1 gene and the BRCA2 gene and can be used for extending all exons of the entire BRCA1 and BRCA2 genes, covering all to-be-detected mutation sites. The method and the primer disclosed by the invention are quite high in specificity and accuracy, simple to operate and low in cost, and can be used for greatly enhancing the risk assessment of hereditary breast cancer and effectively reducing the onset risk of the breast cancer.
Owner:上海润达榕嘉生物科技有限公司
Human idiopathic basal ganglia calcification pathogenic gene and detection method thereof
ActiveCN106834299AEfficient and comprehensive acquisitionAccurate acquisitionMicrobiological testing/measurementGenetic engineeringNervous systemInformation analysis
The invention relates to a human idiopathic basal ganglia calcification pathogenic gene and a detection method thereof. Human idiopathic basal ganglia calcification, namely IBGC is a neurodegenerative genetic disease. The invention provides seven mutation forms of four pathogenic genes including SLC20A2 (Sodium-dependent phosphate transporter 2), PDGFRB (Platelet-derived Growth Factor Receptor Beta), PDGFB (Platelet-derived Growth Factor Subunit B) and XPR1 (Xenotropic and Polytropic Retrovirus Receptor 1) and sequences of the seven mutation forms are shown as SEQ ID NO.1 to SEQ ID NO.7. The form of the pathogenic gene provided by the invention is not reported until now and can provide evidence and lay a foundation for analysis and medicine development of a pathogenic mechanism, pathogenic gene screening and detection, formulation of a therapeutic regimen and the like. Meanwhile, the invention constructs a pathogenic gene detection method; the pathogenic gene detection method comprises the following steps: firstly, capturing a pathogenic gene exon region by utilizing multi-PCR (Polymerase Chain Reaction); carrying out next generation sequencing on the pathogenic gene exon region and carrying out information analysis to find out mutation; finally, identifying the mutation by utilizing Sanger sequencing, wherein a PCR captured primer group comprises amplification primer sequences SEQ ID NO.12 to SEQ ID NO.23, and amplification primer sequences of a Sanger sequencing segment are shown as SEQ ID NO.24 to SEQ ID NO.35. The detection method provided by the invention covers all exons of the four pathogenic genes and can be used for efficiently, comprehensively, rapidly and accurately acquiring mutation information.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
SNP (single nucleotide polymorphism) marker combination and identification method of small Meishan pig and raw meat product
ActiveCN107699624AEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyRaw meat
The invention provides an SNP (single nucleotide polymorphism) marker combination and identification method of a small Meishan pig and a raw meat product. The method comprises the following steps: extracting a genome DNA of raw pork or a meat product, performing AGE (agarose gel electrophoresis) and Sanger sequencing after performing PCR (polymerase chain reaction) amplification on the genome DNA,and identifying the small Meishan pig and the meat product thereof according to an SNP genotype of a characteristic site of a sequencing result, wherein in the step of Sanger sequencing, peculiar mutation occurs at identified sites: SNP6, SNP12, SNP16, SNP71 and SNP79. According to the method, the problem that an identification method of the small Meishan pig and the meat product thereof does notexist in the prior art is solved.
Owner:SHANGHAI JIAO TONG UNIV
Somatic mutation site excavation method based on genomic sequencing
InactiveCN104946765AShorten the timeLow costMicrobiological testing/measurementGenomic sequencingRe sequencing
The invention discloses a somatic mutation site excavation method based on genomic sequencing. By means of depth re-sequencing on various tissue of a wild type shaddock and a mutation type shaddock, the obtained high-quality sequencing reads and the sweet orange genome are compared, and the single nucleotide polymorphism (SNP) of various sets of materials is obtained. Based on SNP sites distributed on some whole genomes, the case-control type correlation analysis is designed on the wild type shaddock and the mutation type shaddock, potential sites related to mutation are obtained and annotated, the chromosome area where the character correlation variation happens is speculated, finally, the potential sites are verified through a Sanger sequencing method, and the variation site of the mutation type shaddock is obtained. The method has the biggest advantages of being low in cost and short in period. The cost for finishing excavating of one mutant mutation site is about 20 thousand, the excavation can be finished in three months, and combining and innovation exploring of the genome advanced technology and the traditional citrus breeding study are expressed. The invention further discloses development and application of the method.
Owner:HUAZHONG AGRI UNIV
Quality control method for detecting human BRCA1/2 genovariation based on high-throughput sequencing and reagent kit
ActiveCN106381334AMeet quality control requirementsConducive to controlling the accuracy of analysisMicrobiological testing/measurementSanger sequencingNegative control
The invention relates to a quality control method for detecting human BRCA1 / 2 genovariation based on high-throughput sequencing, a reagent kit, and an application thereof. In the quality control method, a plurality of genome DNAs of a human tumor cell line with positive BRCA1 / 2 genovariation are extracted, and are determined by means of a Sanger sequencing method, wherein a variation positive locus is used as a positive comparison locus while a wild type locus is a negative comparison locus; the genome DNAs are mixed at a certain ratio to obtain a quality control product that can be used for detecting human BRCA1 / 2 genovariation through high-throughput sequencing. The reagent kit disclosed by the invention comprises the quality control product for detecting human BRCA1 / 2 genovariation.
Owner:上海思路迪医学检验所有限公司
High-flux detection method of circulating DNA liver cancer driver gene
PendingCN107723352AGuaranteed catchHigh sensitivityMicrobiological testing/measurementBlood plasmaExon
The invention discloses a high-flux detection method of a circulating DNA liver cancer driver gene. According to the method, a mutant gene which is the most common of the existing liver cancer is selected and combined with a second-generation high-flux sequencing technology to more comprehensively detect a gene mutant condition of the liver cancer. Since a blood plasma sample of a patient is adopted, the method can greatly reduce the detection injury of a patient; secondly, the whole-exome detection is performed on 75 liver cancer genes, the method is more comprehensive compared to the detection method based on real-time PCR and first-generation Sanger sequencing technology; and since only 75 genes are detected, the sample can be detected in a greater sequencing depth, so that the detection precision can reach 0.1 percent. Therefore, a reference can be provided for more and better individual precision therapeutic schemes of the patient.
Owner:JIAXING YUNYING MEDICAL INSPECTION CO LTD
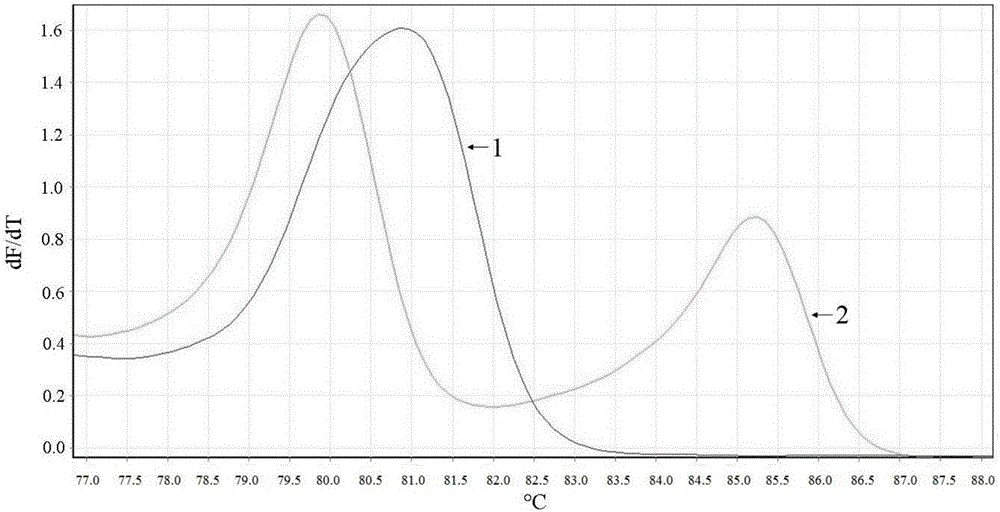
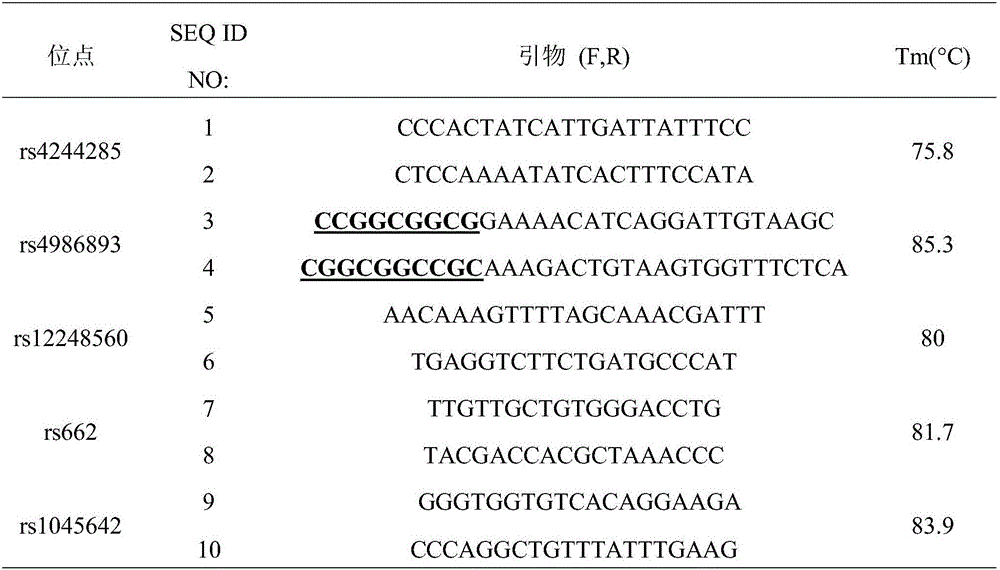
Clopidogrel drug-resistant gene detection method based on multiple HRM analysis
ActiveCN106636337AReduce dosageShorten detection timeMicrobiological testing/measurementDNA/RNA fragmentationResistant genesTyping
The invention relates to a clopidogrel drug-resistant gene detection method based on multiple HRM analysis. A primer in the method is shown as SEQ ID NO: 1-10. The invention further provides a primer combination related to HRM analysis and a kit. The clopidogrel drug-resistant gene detection method has the advantages that a method of HRM typing aiming at multiple SNP sites is established, three sites of rs4244285, rs4986893 and rs12248560 and / or two sites of rs1045642 and rs662 can be detected in a same reaction system, so that reagent consumption is reduced, and detection time is shortened. The method is used to detect samples of patients receiving percutaneous coronary interventional angiography, and a Sanger sequencing method is used for verification, so that accuracy is high.
Owner:SHANGHAI CHILDRENS MEDICAL CENT AFFILIATED TO SHANGHAI JIAOTONG UNIV SCHOOL OF MEDICINE
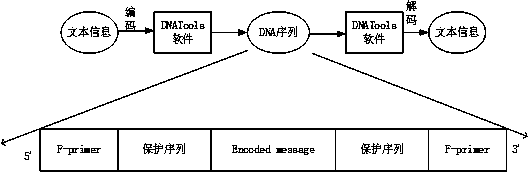
Method for taking DNA as text information efficient storage medium
InactiveCN110427786AIncrease synthesis costRaise the cost of sequencingDigital data protectionInternal/peripheral component protectionChinese charactersA-DNA
The invention provides a method for taking DNA as an efficient text information storage medium, which is used for storing text information and is beneficial to culture protection. The method for taking DNA as a text information efficient storage medium comprises a storage process and a decoding process. The storage process comprises the following steps: establishing an information sequence, dividing text content into a plurality of storage units, and converting each storage unit into a DNA sequence by utilizing DNA tool software, thereby converting the whole text into a plurality of DNA sequences; adding 20bp protection sequences to the two ends of the information sequence respectively; adding primer binding regions at two ends of the protection sequence. The decoding process comprises thefollowing steps: culturing bacteria, extracting plasmids, carrying out polymerase chain reaction to obtain an amplification product, carrying out agarose gel electrophoresis and imaging identification on the amplification product, sequencing the amplification product by using a Sanger sequencing method, and reducing the DNA sequence obtained by sequencing into a Chinese character text by using DNATools software.
Owner:西藏自治区人民政府驻成都办事处医院
Primer combination and reagent kit for complete mitochondrial genome detection
ActiveCN106755456ASimple and fast operationShort detection timeMicrobiological testing/measurementDNA/RNA fragmentationDirect sequencingMitochondrial disease
The invention relates to a primer combination and a reagent kit for complete mitochondrial genome detection. A PCR and Sanger sequencing technology is used for detecting a complete mitochondrial genome for auxiliary diagnosis of a mitochondrial disease caused by mtDNA (mitochondrial deoxyribonucleic acid) mutation. Six specific primer pairs are used for amplification of the whole mitochondrial genome first; each amplification product is 2700-3500bp in size; 200bp or more parts of adjacent amplification segments are overlapped; then, 21 sequencine primers are used for direct sequencing of the amplification products; the sequencing range covers the whole mitochondrial genome. With the adoption of the primer combination, the whole mitochondrial genome can be detected by 6 amplification reactions and 21 sequencing reactions; the detection workload is greatly reduced; moreover, the detection cost is also greatly lowered.
Owner:北京圣谷智汇医学检验所有限公司
Multi-factor model for predicting breast cancer onset risk and establishing method of multi-factor model
InactiveCN107201401AVerify accuracyAccurate Gene Mutation InformationMicrobiological testing/measurementDiagnostic recording/measuringFamilial breast cancerIntervention measures
The invention discloses a multi-factor model for predicting a breast cancer onset risk and an establishing method of the multi-factor model. The establishing method of the multi-factor model comprises screening gene detection, wherein the screening comprises the following steps: (1) collecting and treating a peripheral blood sample; (2) establishing library DNA of the sample, and performing computer sequencing on a Hiseq sequencing platform; (3) performing biological information analysis on computer data, and finding pathogenic mutation; (4) sequencing mutation sites by using a golden standard Sanger sequencing technique, thereby obtaining accurate gene mutation information. By adopting the multi-factor model which is disclosed by the invention and is used for predicting the breast cancer onset risk, a part of people can be screened and diagnosed at an early stage, active interference measures can be taken, and the multi-factor model is particularly applicable to prediction on high-risk people suffering from familial breast cancer.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
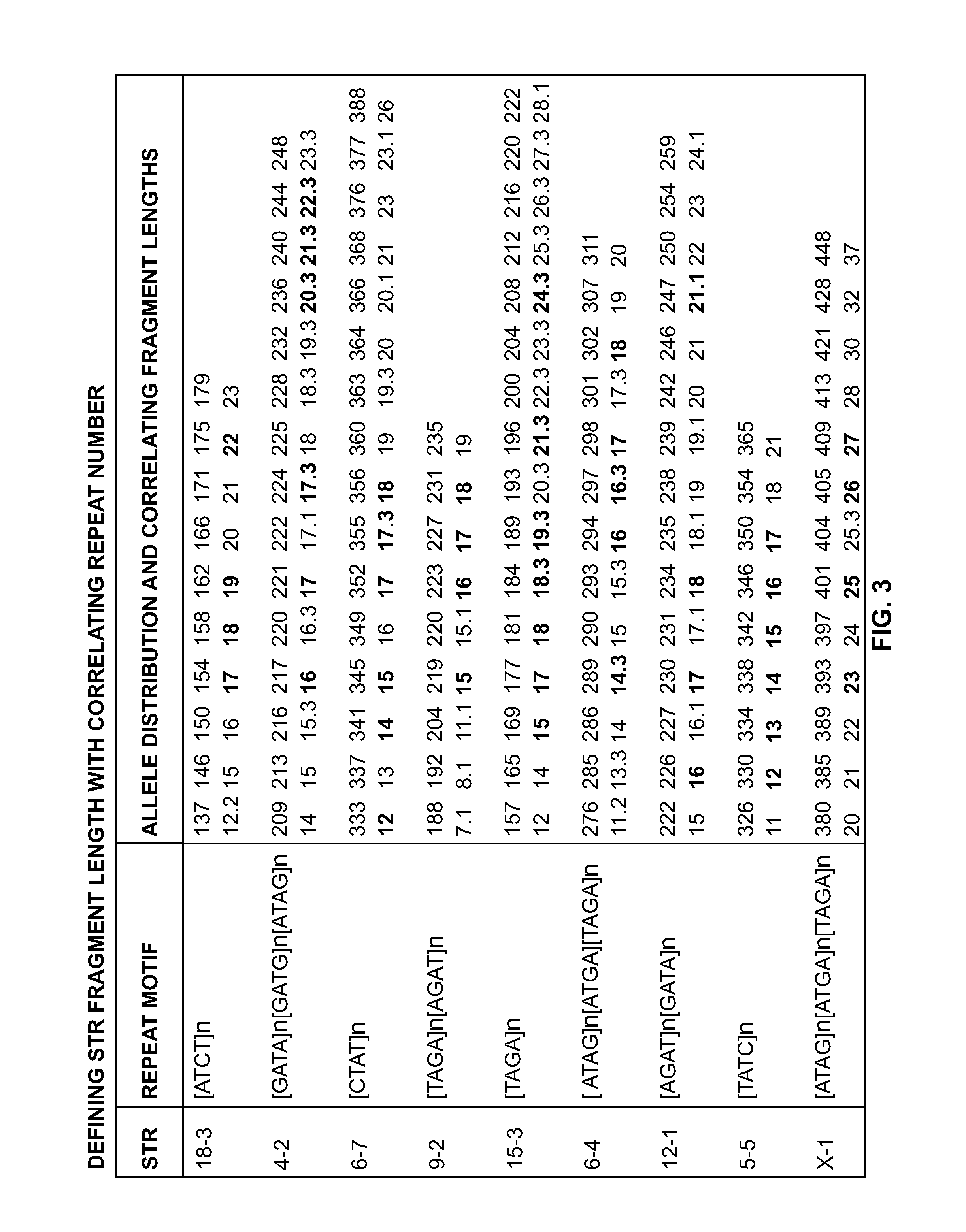
Mouse cell line authentication
ActiveUS20140066322A1High passage numberNucleotide librariesMicrobiological testing/measurementGenotypeTandem repeat
A multiplex polymerase chain reaction assay that targets nine tetranucleotide short tandem repeat (STR) markers in the mouse genome. Unique profiles were obtained from seventy-two mouse samples that were used to determine the allele distribution for each STR marker. Correlations between allele fragment length and repeat number were determined with DNA Sanger sequencing. Genotypes for L929 and NIH3T3 cell lines were shown to be stable with increasing passage numbers as there were no significant differences in fragment length with samples of low passage when compared to high passage samples. In order to detect cell line contaminants, primers for two human STR markers were incorporated into the multiplex assay to facilitate detection of human and African green monkey DNA. This multiplex assay is the first of its kind to provide a unique STR profile for each individual mouse sample and can be used to authenticate mouse cell lines.
Owner:GOVERNMENT OF THE UNITED STATES OF AMERICA AS REPRESENTED BY THE SEC OF COMMERCE THE NAT INST OF STANDARDS & TEHCNOLOGY
Targeted trapping and sequencing kit for lung cancer-related 285 genes and application of targeted trapping and sequencing kit
InactiveCN108148910AImprove detection efficiencyDetecting Gene FrontiersMicrobiological testing/measurementSmall sampleTrapping
The invention discloses a targeted trapping and sequencing kit for 285 lung cancer-related genes and application of the targeted trapping and sequencing kit. The kit is based on hybrid trapping of a probe, and can be used for preparing a high-throughput library of the lung cancer-related 285 genes and simultaneously detecting all variation loci on exons of the 285 genes, so that the disadvantagesof high cost, high time consumption, tedious operation and the like of conventional single-gene detection are overcome. Detection of a plurality of genes with a single small sample is implemented in the small sample for aspiration biopsy of lung cancer, and the clinical practice requirements of maximally using the small sample and obtaining a maximum amount of information are met. The kit is basedon an Ion Proton platform, has high accuracy and sensitivity, and has obvious advantages compared with a Sanger sequencing method, and a detection result in a residual DNA (deoxyribonucleic acid) sample of a paraffin sample is stable.
Owner:GUANGDONG GENERAL HOSPITAL
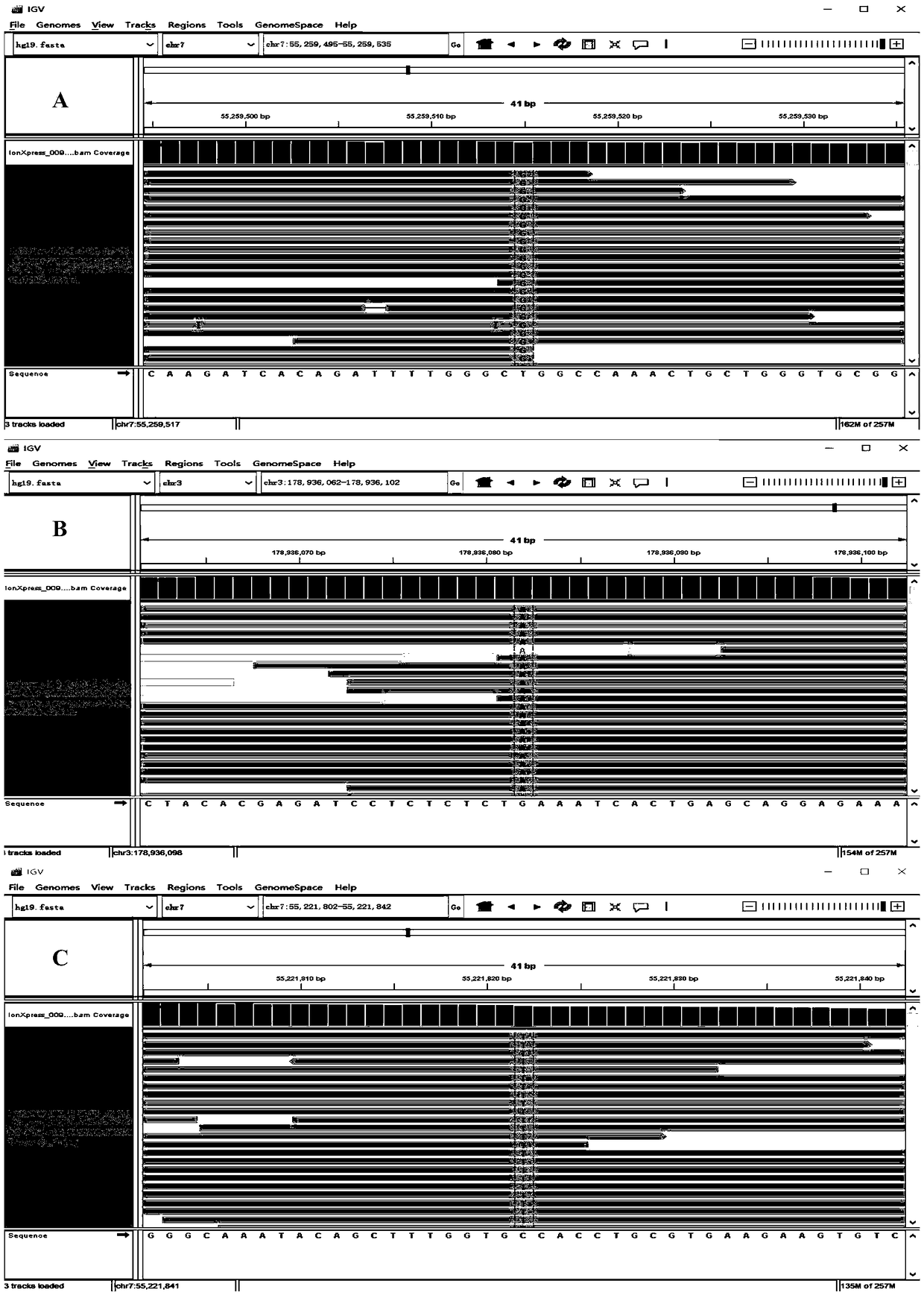
Primers and method for detecting polymorphic hotspot mutation condition of DOCK2 gene
InactiveCN105506107AGood amplification efficiencyThere will be no missed detectionMicrobiological testing/measurementDNA/RNA fragmentationSevere combined immunodeficiency diseaseMutation detection
The invention discloses primers and a method for detecting myelosis myelodysplastic syndrome and especially detecting DOCK2 gene mutation of a patient with myelosis myelodysplastic syndrome. The primers comprise (i) primers for amplifying 6th, 22nd, 33rd, 37th, 40th and 44th exon sequences of the DOCK2 gene. An Sanger sequencing technique and sequencing primers are adopted. The primers and method can be used for quickly detecting mutation of 6th, 22nd, 33rd, 37th, 40th and 44th exons of the DOCK2 gene in the body of a patient with myelosis myelodysplastic syndrome. The primers and method have the advantage of accurate detection result, can be used for assisted diagnosis of severe combined immunodeficiency disease, and have important reference meanings for early intervention and early therapy.
Owner:杭州艾迪康医学检验中心有限公司
Quality control method and kit for detecting human KRAS gene variation based on high-throughput sequencing
InactiveCN106498079AConducive to controlling the accuracy of analysisImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationHuman tumorKRAS
The invention relates to a quality control method and a kit for detecting human KRAS gene variation based on high-throughput sequencing, and application thereof. The quality control method comprises the following steps of extracting a plurality of genomic DNA (deoxyribonucleic acid) of a human tumor cell system with positive KRAS gene variation; measuring a positive variation site as a positive contrast site by a Sanger sequencing method; performing segmenting on the genomic DNA, and mixing according to certain ratio, so as to obtain a quality control product for detecting the human KRAS gene variation based on the high-throughput sequencing. The kit comprises the quality control product for detecting the human KRAS gene variation.
Owner:3D BIOMEDICINE SCI & TECH CO LTD
Primer combination, method and kit for constructing targeted library of multiple inherited metabolic liver diseases based on high-throughput sequencing
ActiveCN109468372ASave time and costEasy to operateMicrobiological testing/measurementLibrary creationExonLiver disease
The invention discloses a primer combination, method and kit for constructing a targeted library of multiple inherited metabolic liver diseases based on high-throughput sequencing. The base sequence of the primer combination is shown as SEQ ID No. 1 to SEQ ID No. 2245, and the primer combination covers total 703 exons and cutting regions of 43 genes such as SERPINA1, G6PC, SLC37A4, AGL and the like, and is capable of detecting 41 sub-type gene variation loci of totally 20 inherited metabolic liver diseases comprising alpha-1-antitrypsin deficiency, glycogen storage disease, citrullinemia, aminosuccinuria, hemochromatosis, porphyrinopathy, cystic fibrosis, bile acid synthesis defect, Gaucher disease, hereditary fructose intolerance, cholesterol ester storage disease, Gilbert syndrome, Dubinsyndrome, Rotor syndrome, progressive familial intrahepatic choleatasia and the like. The primer combination disclosed by the invention is simple in operation, high in accuracy and low in time consumption, and the time cost and manual cost used for detecting genes and fragments one by one during Sanger sequencing can be greatly saved.
Owner:THE FIRST AFFILIATED HOSPITAL OF ARMY MEDICAL UNIV
Method and primer for detecting whole exons of FIX gene
InactiveCN105506106AEasy to operateThe process is convenient and fastMicrobiological testing/measurementDNA/RNA fragmentationExonSanger sequencing
The invention discloses a method, primer and a kit for detecting whole exons of an FIX gene. Each of the primer and the kit includes seven pairs of forward and reverse primers which are expanded to cover the whole exon sequence of the FIX gene; a pair of universal primers is utilized as sequencing primers for detecting mutation conditions of the whole exon sequence of the FIX gene by virtue of a Sanger sequencing method. The primer and the method can be applied to the aspects of gene diagnosis, family female carrier detection, prenatal gene diagnosis and the like of patients with hemophilia B.
Owner:杭州艾迪康医学检验中心有限公司
InDel molecular marker related to cashmere traits of goat and application of InDel molecular marker
InactiveCN108410996ASpeed up the breeding processAn Accurate and Reliable Method for Detecting InDel Polymorphism of FGF5 Locus in Chinese Goat BreedsMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAllele
The invention relates to an InDel molecular marker related to cashmere traits of goat and an application of the InDel molecular marker. The marker is located between the 95454685bp and the 95455189bpof the No.6 chromosome of the goat; the polymorphism of the InDel molecular marker is - / insertion fragment; and a sequence of the insertion fragment is shown as SEQ ID NO.1. The invention also provides specific primers and a method for detecting the InDel marker; and sequences of the specific primer pair are shown as SEQ ID NO.2-3. Through the specific primer pair provided by the invention, on thebasis of an electrophoretic band obtained through PCR and agarose gel electrophoresis as well as analysis and statistics on Sanger sequencing and in accordance with cashmere goat allele and non-cashmere goat allele types on an InDel site, it can identify cashmere or non-cashmere traits of a goat variety. The InDel molecular marker provided by the invention is low in amount of detected samples inneed, simple and convenient in method and accurate in identifying result, and the InDel molecular marker has a good prospect of application and promotion.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
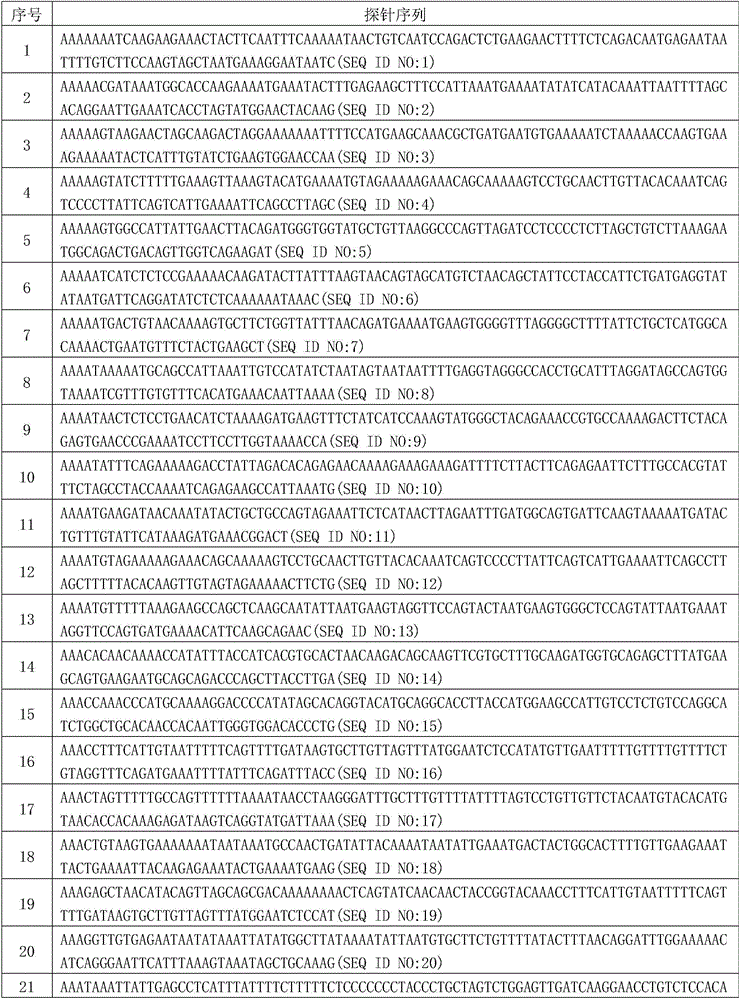
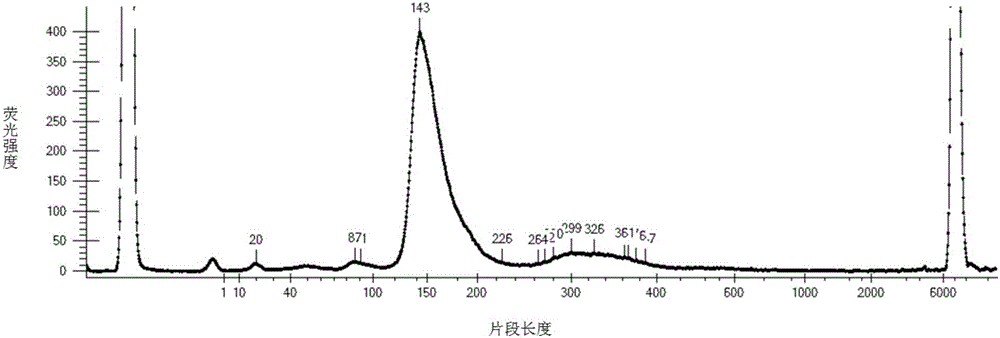
Method for identifying SNP in individual in Sanger sequencing oriented to PCR products of diploid
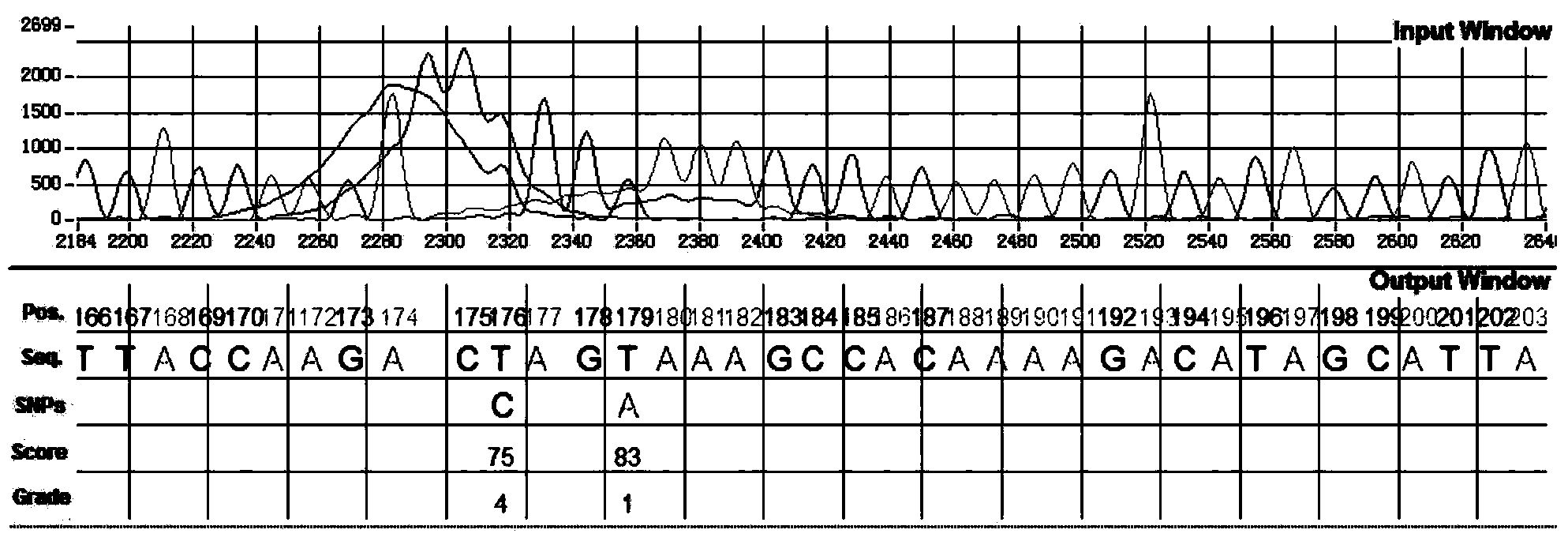
InactiveCN103593659ASolve the problem of automatic identification of SNP within an individualEfficient detectionBiological neural network modelsCharacter and pattern recognitionCytosineLevenberg–Marquardt algorithm
The invention discloses a method for identifying SNP in an individual in Sanger sequencing oriented to PCR products of diploid. According to the method, firstly, fluorescent data of four bases of adenine A, guanine G, cytosine C and thymine T contained in a chromatogram map are independently separated, filtering and noise reduction processing is carried out on the separated fluorescent data by adopting a small wave multiscale analysis method; the waveform characteristics of the fluorescent data of the four bases are further analyzed, a first peak and a second peak of the waveform are detected, and the peak distance, the height specific value and the fluctuation degree specific value of the waveform characteristics are selected as the elements for judging SNP loca, a BP nerve net with the structure of 3-10-1 is selected as a classifier for the detection of the SNP loca, and training is carried out on the BP nerve net by adopting a Levenberg Marquardt algorithm; output is mapped as SNP evaluation scores from 0 to 100 by adopting piecewise linear transformation, the classification of the SNP loca is defined from a 1 level to a 5 level according to the evaluation scores, and the SNP confidence coefficient of the loca is judged according to the classification. The method for identifying the SNP in the individual in the Sanger sequencing oriented to the PCR products of the diploid can effectively detect the SNP loca in the individuals in sequencing files.
Owner:SOUTH CHINA AGRI UNIV +1
Identification and application methods for MC1R gene haplotype
PendingCN110273010AImprove accuracyAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAnimal Genetics
The invention belongs to the technical field of animal husbandry or animal genetic breeding and discloses identification and application methods for an MC1R gene haplotype. The identification method for the MC1R gene haplotype comprises the steps that an MC1R gene is subjected to PCR amplification, and single-ended Sanger sequencing is adopted for obtaining a variation sequence; based on the established haplotype, the genotype and phenotype of the hair color of a breeding group are subjected to association analysis, and the haplotype for controlling a black feather and the black feather homozygous genotype are identified. According to the identification method, the haplotype for controlling the black feather and the black feather homozygous genotype can be accurately identified. According to the identification method, key SNPs loci are simultaneously identified in one sequencing reaction; but in the prior art, two sequencing primers are used for simultaneously identifying the SNPs loci. Compared with a PCR-RFLP identification method in the prior art, through Sanger sequencing, the identification accuracy is further improved.
Owner:SOUTHWEST UNIVERSITY
Accurate and efficient editing method of upland cotton genome
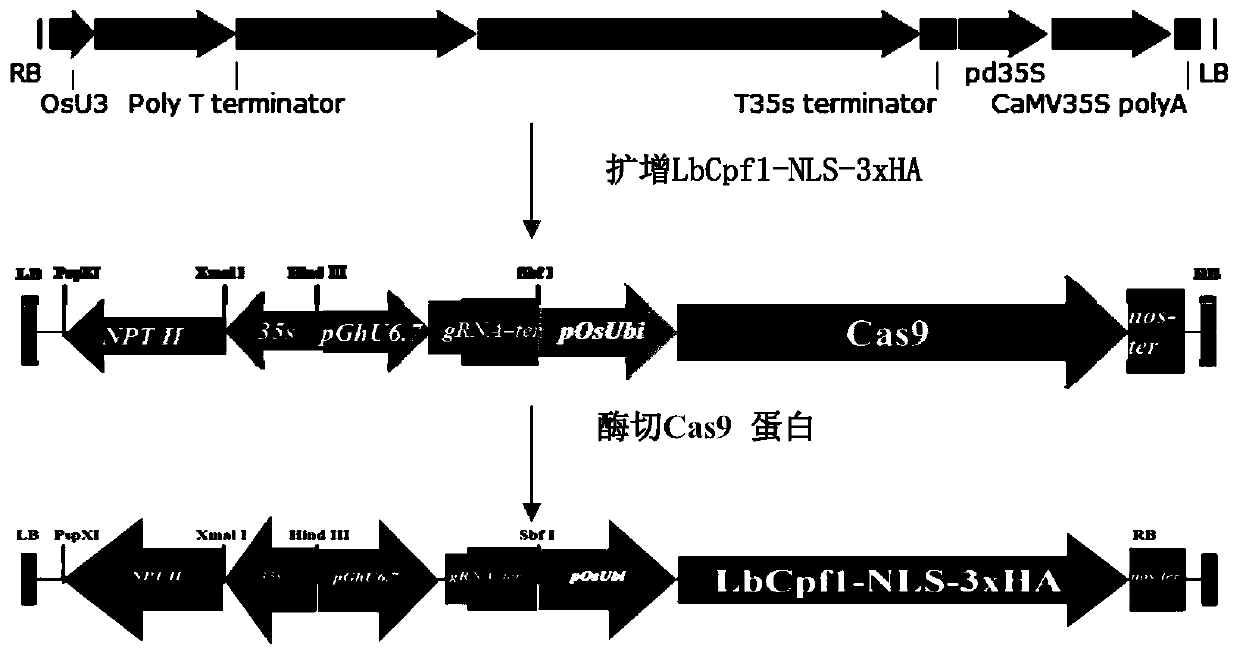
ActiveCN110283840AStrong specificityVectorsVector-based foreign material introductionHeterologousAgricultural science
The invention belongs to the technical field of plant gene engineering and particularly relates to an accurate and efficient editing method of an upland cotton genome. A pRGEB32-GhU6.7-NPT II vector containing a cotton endogenous promoter pGhU6-7 is modified, original Cas9 protein is replaced with LbCpf1 protein, and a vector GhRBE3 with editing capacity in cotton is constructed. GhCLA is selected as a target gene to verify application of GhRBE3 in cotton. One target is designed, a Cpf1 gene editing system is imported into the cotton genome by agrobacterium tumefaciens mediated genetic transformation, Sanger sequencing and high-throughput sequencing are performed on transgenic plants to detect the editing efficiency and the off-target effect of the method in the allotetraploid cotton genome. The method has good editing efficiency and specificity.
Owner:HUAZHONG AGRI UNIV
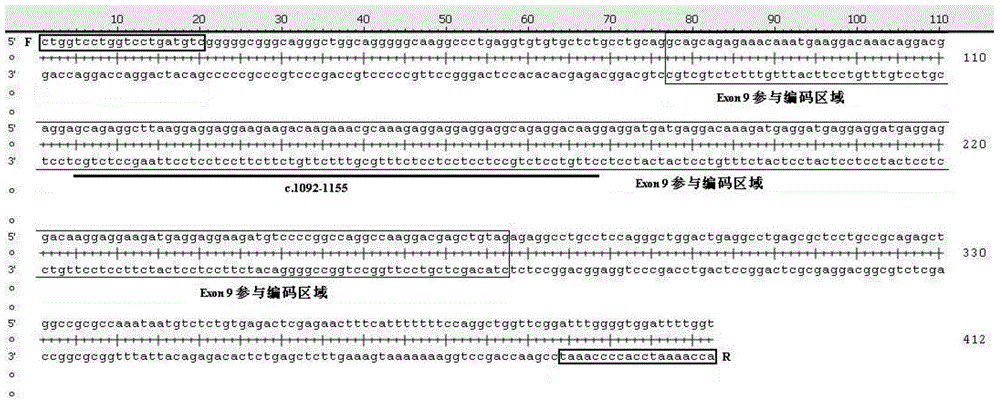
Reagent and method for detecting ninth exon mutation of CALR gene
The invention provides a reagent and a method for detecting the ninth exon mutation of a CALR gene by using a Sanger sequencing technology. The reagent and the method are high in detection specificity, and the mutation types of insertion and deficiency of the ninth exon of CALR can be specially analyzed, so that the false positive problem is eliminated, and missing inspection of rare mutation can also be avoided. The reagent and the method can be used for clinically screening genes of patients suffering from primary thrombocythemia and primary myelofibrosis.
Owner:南京艾迪康医学检验所有限公司
High-throughput assay method for leukaemia driver genes
PendingCN107723353AGuaranteed catchHigh sensitivityMicrobiological testing/measurementAssayIndividualized treatment
The invention discloses a high-throughput assay method for leukaemia driver genes. By choosing mutant genes and fusion genes commonly seen in leukaemia at present, in combination with the second-generation high-throughput sequencing technology, the method can more comprehensively assay the gene mutation and fusion conditions of leukaemia. Compared with the classical Sanger sequencing method mostlyadopted at present, the second-generation high-throughput sequencing technology has the advantages of high throughput, short sequencing time, high precision, low cost and rich information, and can accurately locate and analyze target gents in a short time. With the help of the high-throughput assay technology, the method can provide more detailed gene mutation and fusion spectra for clinical doctors to comprehensively judge the prognosis of leukaemia and guide individualized treatment.
Owner:JIAXING YUNYING MEDICAL INSPECTION CO LTD
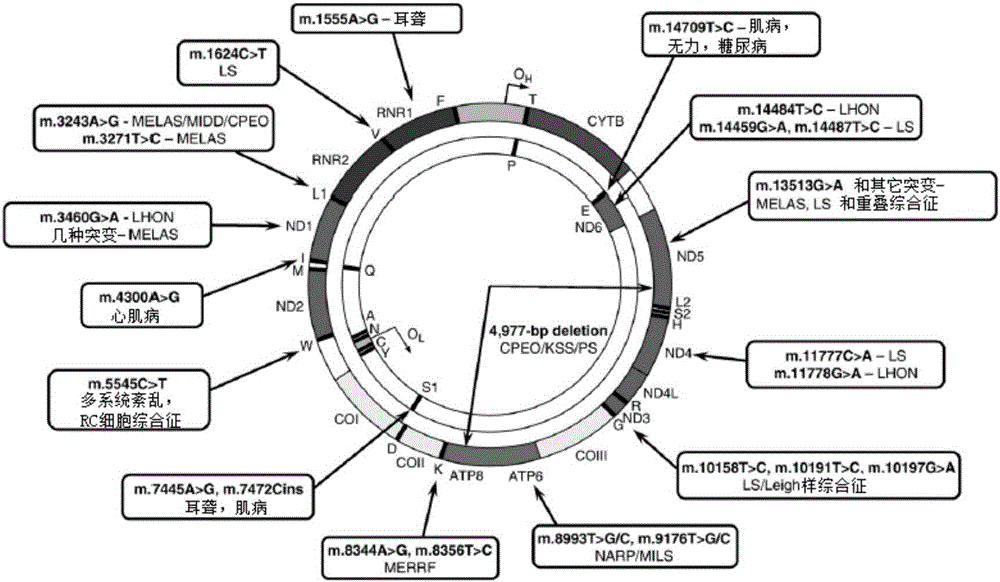
Detection primer, detection kit and detection method of Leber's hereditary optic neuropathy (LHON) mitochondrial DNA gene mutations
ActiveCN106755335AResolve SensitivitySolve the characteristicsMicrobiological testing/measurementDNA/RNA fragmentationGenetic dnaGene mutation
The invention relates to a detection primer, a detection kit and a detection method of Leber's hereditary optic neuropathy (LHON) mitochondrial DNA gene mutations. The invention, on the basis of Sanger sequencing principle, has the characteristics of being highly sensitive, stable and accurate; four most common pathogenic mutations of LHON mtDNA can be simultaneously detected, so that the problems of an existing LHON mitochondrial DNA mutation detection method, which is low in sensitivity, not strong enough in specificity, capable of causing pollution easily, high in expense and the like, can be solved; therefore, an accurate gene diagnosis method is provided for LHON; and the invention has important implications for genetic counseling and gene therapy of the disease (the LHON).
Owner:ZHONGSHAN OPHTHALMIC CENT SUN YAT SEN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com