Accurate and efficient editing method of upland cotton genome
A genome and upland cotton technology, applied in the field of genetic engineering, can solve the problems of complex and huge genome, and no reports of allotetraploid cotton, and achieve highly specific effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
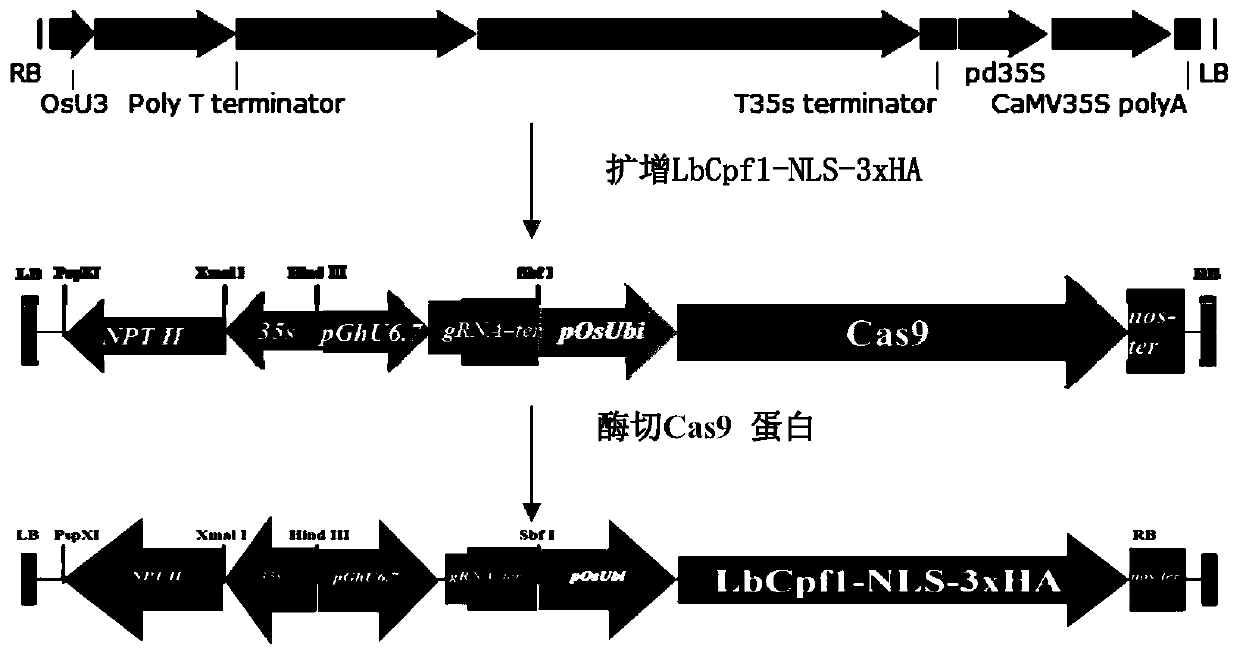
[0036] Example 1: Amplification of LbCpf1-NLS-3xHA target sequence
[0037] Using pHUN611 vector (sequence list as SEQ ID NO: 2) as a template for PCR amplification, the target sequence LbCpf1-NLS-3xHA (sequence list as SEQ ID NO: 4) was obtained. The primers for amplification are:
[0038] LbCpf / F:AAAAAGCAGGCTTCGAAATGTCCAAGCTGGAGAAGTTTACA,
[0039] And LbCpf / R: GAAAGCTGGGTCTAGATTAGGCATAGTCGGGGACATC;
[0040] The PCR reaction system is shown in Table 1.
[0041] Table 1 PCR reaction system of LbCpf1-NLS-3xHA sequence
[0042]
Embodiment 2
[0043] Example 2: Construction of transformation vector GhRBE3
[0044] Double-enzyme digestion of pRGEB32-GhU6.7-NPTⅡ (see SEQ ID NO: 1 for the sequence list). The digestion system is shown in Table 2. The digestion product is digested at 37°C for 5 hours, and the digested product is observed by gel electrophoresis. Add 4 μL of BstbI, digest the enzyme at 65°C for 20 minutes, observe whether the digested band is correct by gel electrophoresis, and then use a gel recovery kit (purchased from OMEGA, catalog number D2500-02) to purify the digested product. The enzyme digestion system is shown in Table 2.
[0045] Table 2 Restriction digestion system of pRGEB32-GhU6.7-NPTⅡ
[0046]
[0047] Connect the digested pRGEB32-GhU6.7-NPTⅡ vector and the LbCpf1-NLS-3xHA fusion protein fragment through the ClonExpress II One Step Cloning Kit (Vazyme C112-02), transform it into E. coli competent, and pick the positive clones. After sequencing, the plasmid with the correct sequence was named GhRB...
Embodiment 3
[0051] Example 3: Construction of GhRBE3-crRNA vector
[0052] 1. The crRNA design of GhCLA gene
[0053] The Gh_A10G2292 gene of upland cotton 1-deoxyxylulose-5-phosphate synthase (Cloroplasto alterados, CLA) was selected as the verification gene. The bioinformatics software sgRNAcas9_3.0.5 (Xie et al 2014) was used to design a crRNA target sequence with the PAM sequence "-TTTV" in the gene exon region, and one crRNA was selected to construct a plant expression vector for the gene editing system. The sequence of the crRNA is shown in Table 4.
[0054] Table 4 Sequence of crRNA
[0055]
[0056] 2. Connection of crRNA and GhRBE3 vector
[0057] The target inserted into the GhRBE3 vector sequence is a repetitive sequence of tRNA-crRNA-gRNA. The target sequence is obtained by gene synthesis, and then amplified by PCR and connected to the GhRBE3 vector. Now take crRNA as an example. The PCR primers are as follows: pRGEB32-7 / S:aagcatcagatgggcaAACAAAGCACCAGTGGTC, CLA1 / AS:TCTAGCTCTAAAACTG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com