Method for preparing detection chip and method for detecting pathogen by using the same
A technology for detecting chips and pathogens, which is applied in the field of nucleic acid detection of pathogenic microorganisms, and can solve the problems of difficult preservation of primers, high price, and fluorescence quenching.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1, a kind of preparation method of detection chip, carries out following steps successively:
[0058] 1), design bacterial oligonucleotide probes:
[0059] The genome sequences of Escherichia coli, lactic acid bacteria, Salmonella, Shigella, Staphylococcus aureus, mold and yeast were obtained from the NCBI database, and then designed using Arraydesigner4.2 software. The resulting probe sequences are specifically shown in the table 1.
[0060] All the probes shown in Table 1 were used for each bacterium, namely, the four probes shown in SEQ ID NO: 1 to SEQ ID NO: 4 were used for Escherichia coli, and so on for other bacteria.
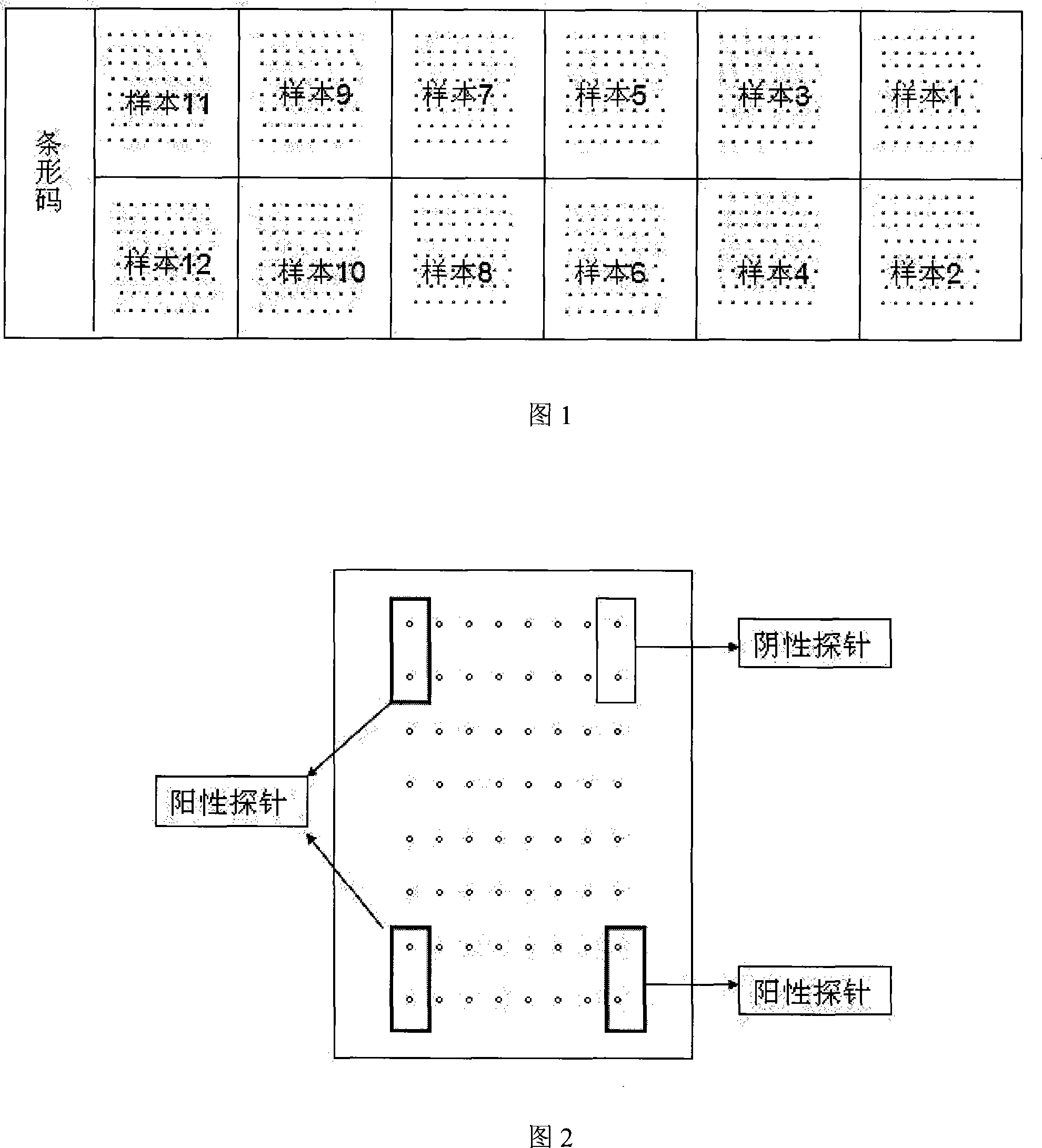
[0061] 2), chip preparation:
[0062] Select 28 probes shown in Table 1 for use, and 1 negative (sequence is:
[0063] 5-CTCAATCCTTTGGGTGTATGGGTCGTAGCGAACTGAGAAGGGCCGAGGTATTGTGGCA-3), 1 positive probe (GAPDH:
[0064] 5-GTCCAGTTAATTTCTGACCTTTTACTCCTGCCCTTTGAGTTTGATGATGCTGAGTGTAC-3), a total of 30 probes. Each probe was spotted in d...
Embodiment 2
[0066] Embodiment 2, a method for detecting pathogens in multiple food samples, select the detection chip obtained in embodiment 1, and perform the following steps in sequence:
[0067] 1), design primers:
[0068] Get the genome sequences of Escherichia coli, lactic acid bacteria, Salmonella, Shigella, Staphylococcus aureus, mold and yeast from the NCBI database, and then use the primer5.5 software to design the primers corresponding to each of the above bacteria The sequence is specifically shown in Table 2.
[0069] 2), the extraction of DNA in the sample to be tested, with 12 kinds of emulsion or liquid dairy products as the sample to be tested, each sample to be tested is followed in turn by the following steps:
[0070] ①. Take a 50ml sample, centrifuge at 1000-2000rpm at 4°C for 20min, and discard the supernatant;
[0071] ② Add 10ml of PBS, shake gently to resuspend the pellet, centrifuge at 1000-2000rpm at 4°C for 20min, discard the supernatant;
[0072] ③, repeat ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com