Nucleic acid sequencing process based on micro array chip
A microarray chip and nucleic acid sequencing technology, which is applied in the field of biomedical nucleic acid array chip applications, can solve the problems of high nucleic acid sequencing cost, low preparation cost, and high sequencing throughput, and achieve the effects of simple implementation, high throughput, and mature technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
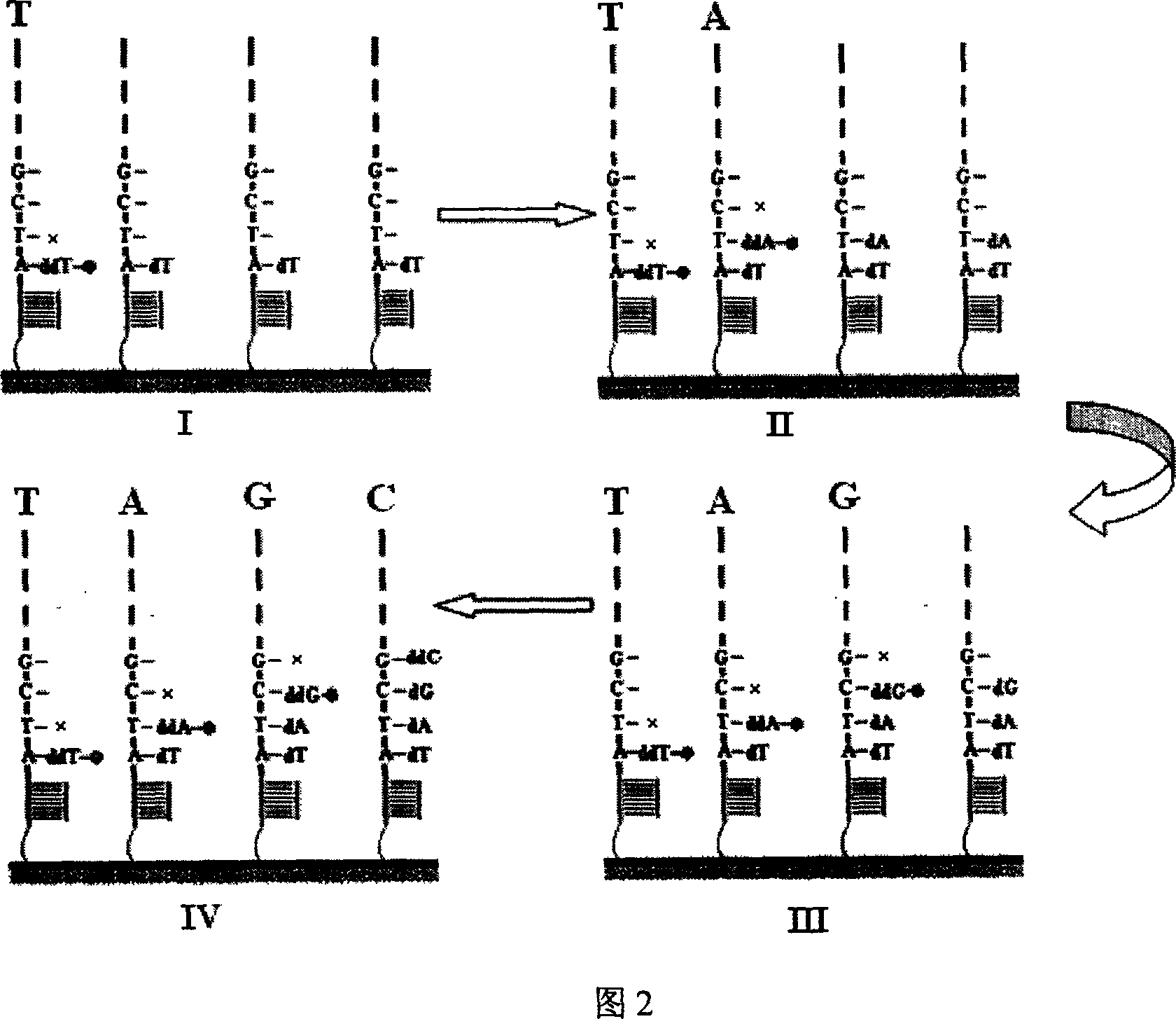
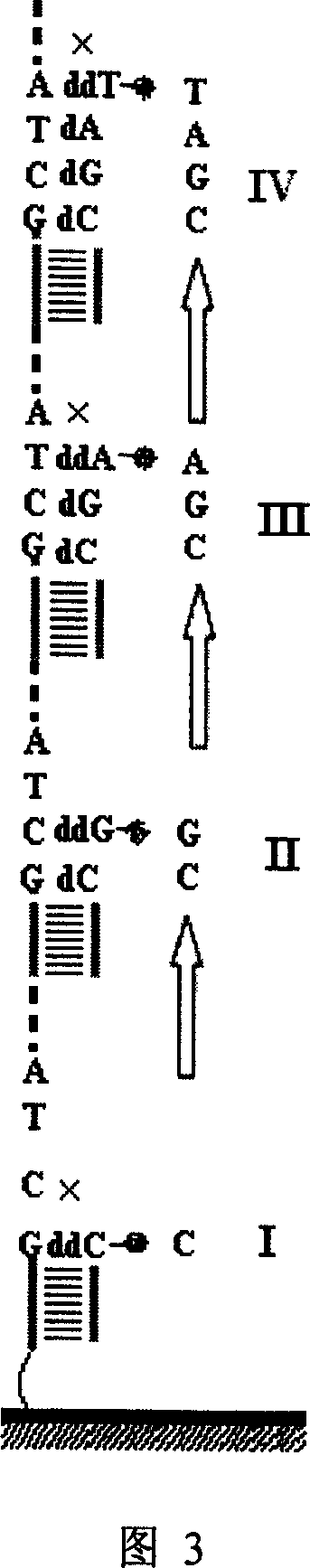
[0035] Example 1. Nucleic acid microarray chip prepared from immobilized PCR products for sequencing
[0036] 1. Preparation of Universal Primer PCR Products
[0037] Taking amplification as an example, to measure the nucleic acid sequence of the hepatitis B virus DNA fragment 78-257, the specific method is:
[0038] 5'...GGCATGGACATTGATCCTTATAAAGAATTTGGAGCTACTGTGGAGTTACTCTCCTTTTTG
[0039] CCTTCTGACTTCTTTCCGTCGGTACGAGATCTTAGATACCGCCTCAGCTCTATATCGGGAAGCCTTAGAGTCTCCTGAACATTGCTCACCTCACCATACTGCACTCAAGCAAGCAATT...3', design a pair of specific amplification primers, P1:
[0040] 5’...NH 2 -TTTTTTTTTTCCGTACCTGTAACTAG...3',
[0041] P2: 3'...CTCAAGCAAGCAATT GCATGGCCACGGGCGGAC ...5'. The italic part is the specific primer sequence, and the bold part is the general sequence, that is, the 5' end of all PCR downstream primers has this fragment. The HBV 78-257 fragment is amplified in a conventional PCR amplification system by using the pair of primers.
[0042] 2. Preparation of ge...
Embodiment 2
[0047] Example 2. Nucleic acid microarray chip prepared by fixed rolling circle amplification technology for sequencing
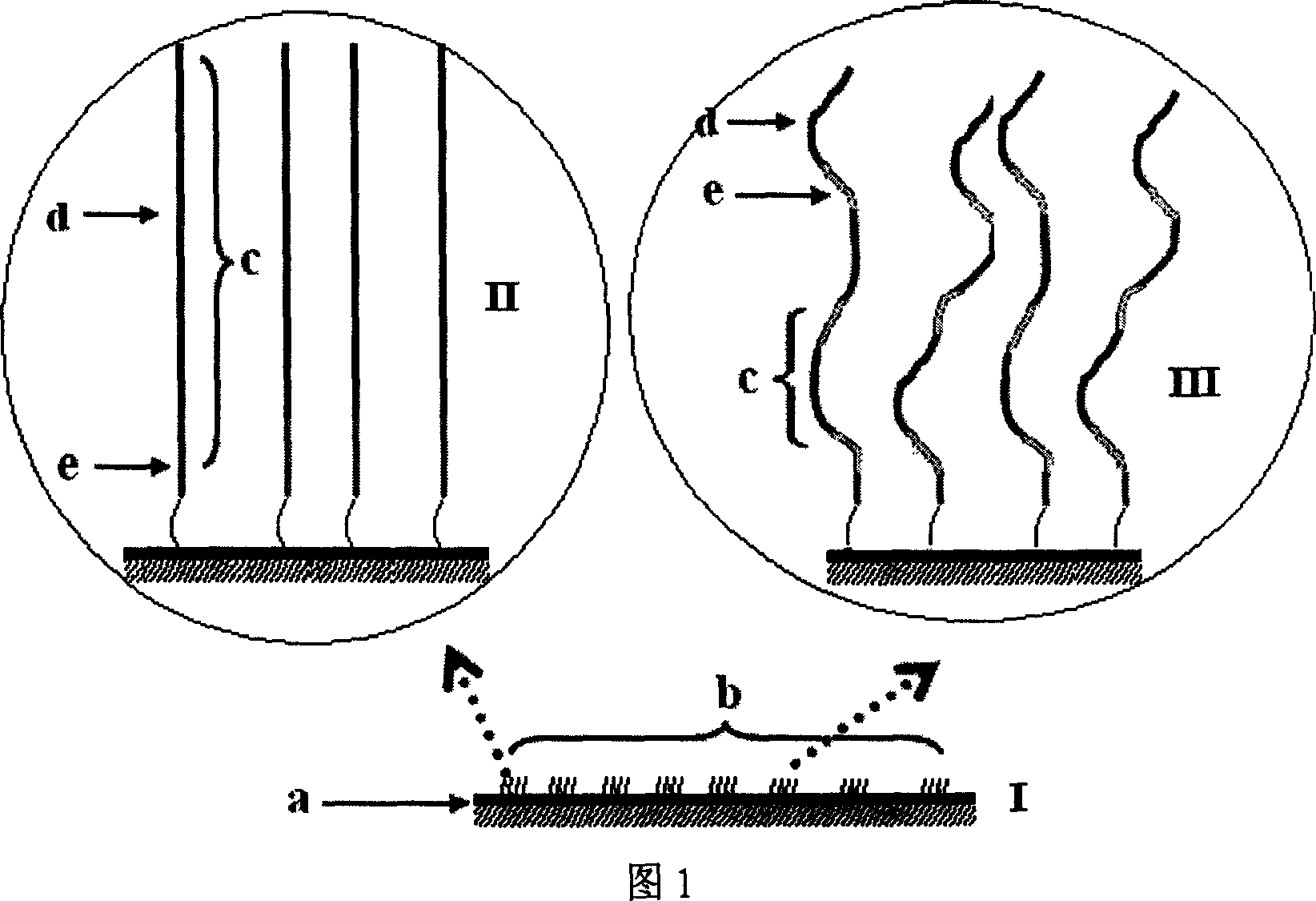
[0048] 1. Apply the rolling circle amplification technique to prepare microarray chips of single-molecule multi-copy DNA fragments of the whole genome (as shown in Figure 1-III).
[0049]Fragmentation of genomic DNA: as shown in Figure 6, ①Use an ultrasonic instrument to break the genomic DNA into fragments of 100-500 bp at a certain intensity, and take 1 μl of the product to gel in 1.5% agarose Gel electrophoresis for 30 minutes to detect the effect of ultrasonic fragmentation of genomic DNA; ②Take a certain amount of ultrasonic product, add an appropriate amount of mung bean nuclease system, and react at 30°C for 30-60 minutes, because this enzyme can remove single-stranded 3′ or 5′ overhangs blunt end of all DNA fragments, and then inactivate mung bean nuclease at 80°C for 15 minutes; ③Add an appropriate amount of Lambada DNA nuclease system to the react...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com