Transgenic product low-density gene chip detecting method
A gene chip and detection method technology, applied in the field of detection of genetically modified products using low-density gene chips, can solve the problems of human and environmental hazards, lack of specificity and sensitivity, and achieve the effect of reducing the mismatch rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Selection of appropriate primers and probes is the key to successful DNA analysis and detection. Aiming at the sequences of the 9 most commonly used exogenous DNAs and control genes (such as 18S ribosomal RNA, soybean lectin endogenous gene and maize invertase endogenous gene) in transgenic products, various parameters are adjusted through repeated combinations, and the software Primer Express TM Search and analyze with OLIGO 4.0, and compare each oligonucleotide, and select primers and probes that meet the requirements. The selected primers and related information are shown in Table 1, and the probes used and related information are shown in Table 2.
[0039] Primer number
Primer sequence (5'-3')
target DNA
18S-1
TAA TAG AGC AAT GAA CAG TCG G
18S-2
CTT TCA ACA ACG GAT CTCTT G G
18S ribosomal RNA
lec-1
GTG CTA CTG ACC AGC AAG GCA AAC TCA GCG
lec-2
GAG GGT TTT GGG GTG CCG TTT TCG TCA AC
Soybean lec...
Embodiment 2
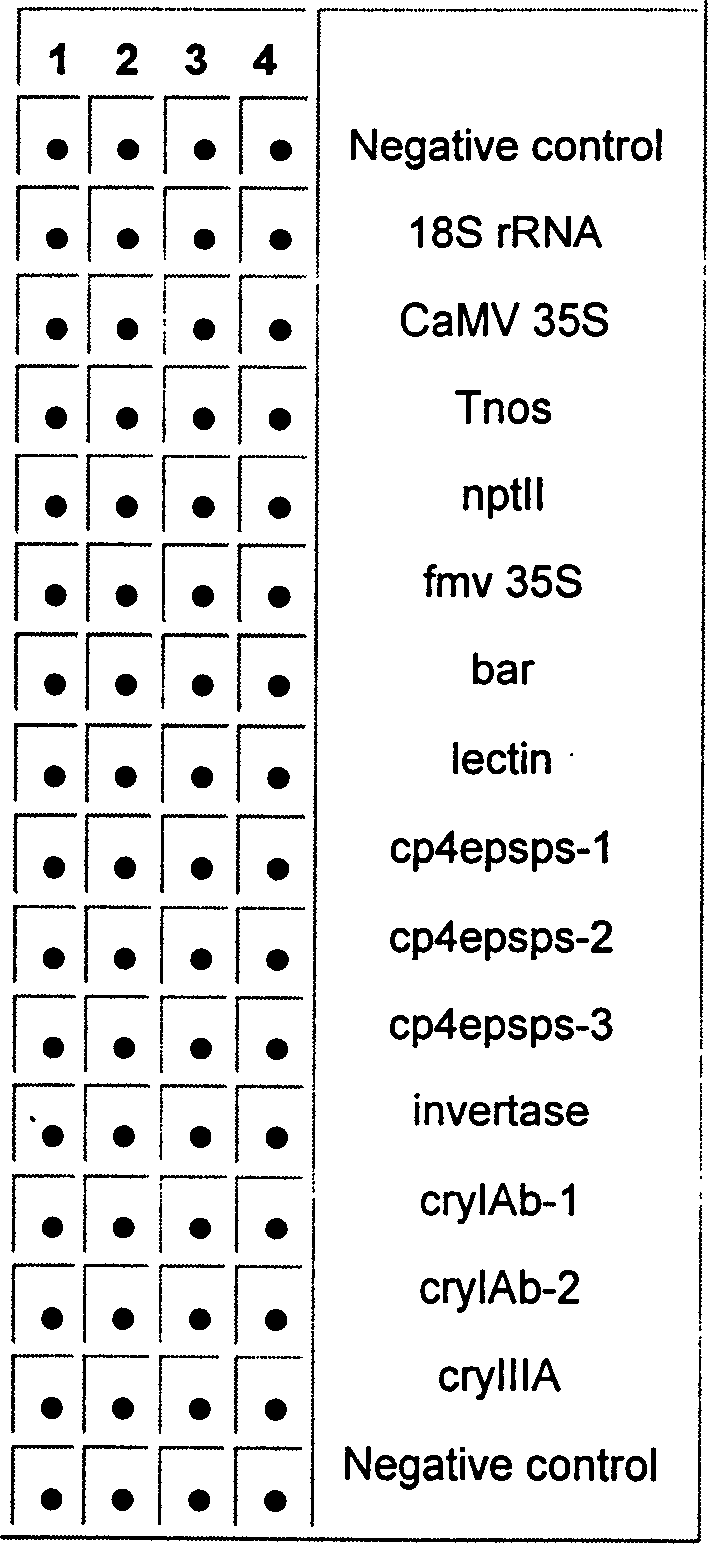
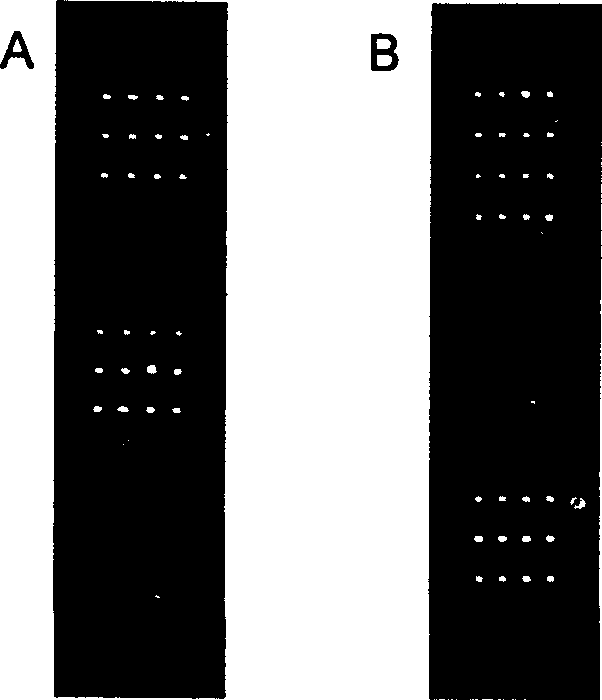
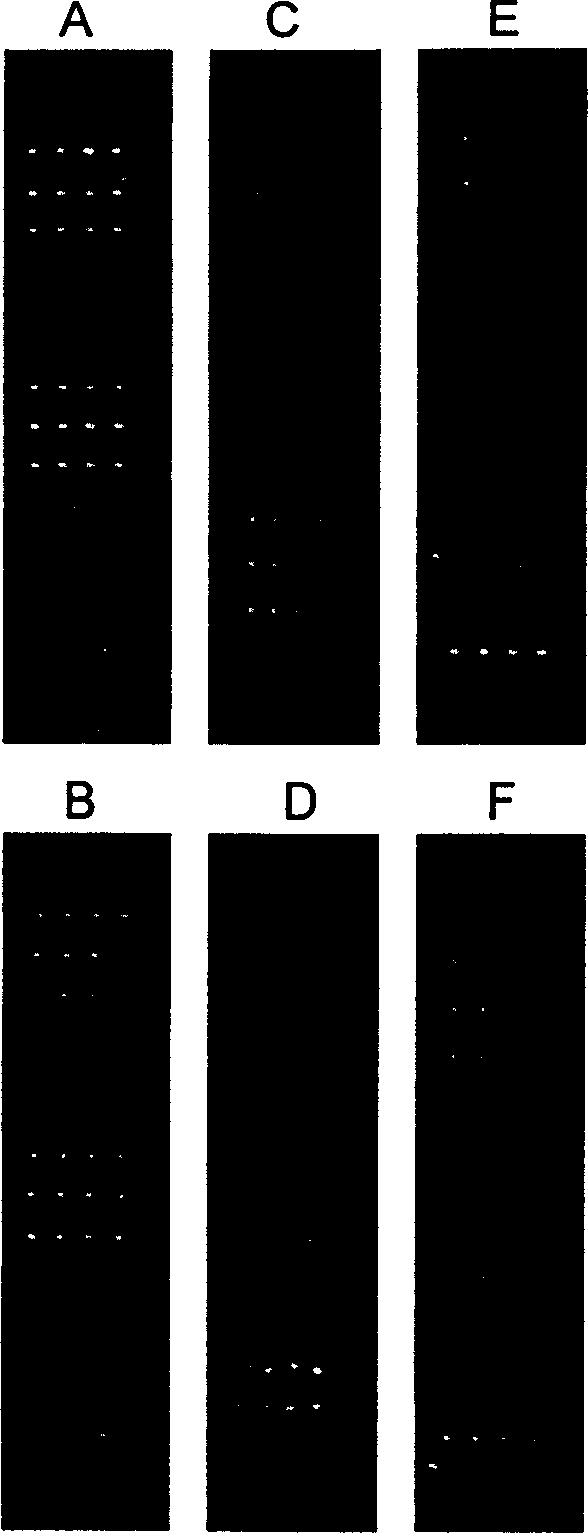
[0048] Using PixSys TM Model 5500 automatic spotting instrument, designed as a 16×4 array, contains 14 detection target probes, 2 negative controls, and 4 spots for each probe (see figure 1 ). A human gene that has almost no homology with the detection probe is set on the chip as a negative control to exclude the interference of non-specific hybridization during the hybridization process.
Embodiment 3
[0050] Chip hybridization refers to the process in which fluorescently labeled samples react with probes on the chip and generate a series of information. Factors that affect the formation of chip hybridization double strands include target concentration, hybridization solution components, and hybridization temperature. Selecting appropriate conditions can make most of the hybridization reactions in the best condition, that is to say, make as many correct pairs as possible. Missed, crosses with mismatches are minimized. The listed hybridization procedures (including reaction conditions such as probe modification, target concentration, hybridization temperature, hybridization time, and slide washing) have been optimized, such as amination of the 5' end of the probe and ligation of 10 thymines After the connecting arm composed of nucleosides (T), the immobilization efficiency of the probe on the slide can be significantly improved, and the hybridization effect can be improved; t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com