Medicine and bacterium resistant detection chip, method for preparation and application thereof
A technology for detecting chips and drug-resistant bacteria, applied in the field of microbial testing, can solve problems such as difficulty in popularization and application, high operational requirements, and poor practicability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0110] Embodiment 1 Probe Design and Chip Fabrication
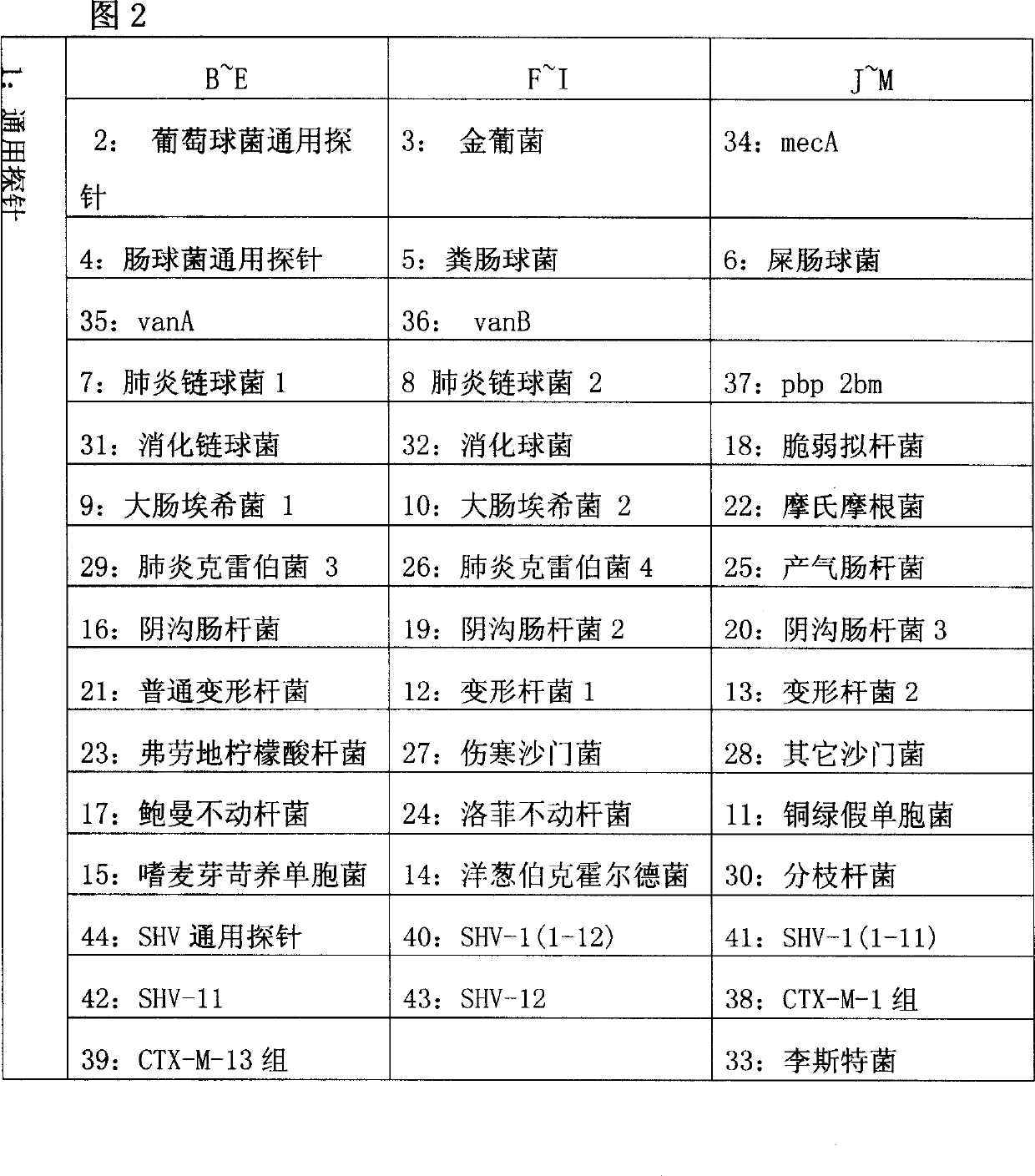
[0111] According to the known 23S rRNA gene sequences and drug resistance gene sequences of different bacteria, design probes with similar length, about 20mer. As shown in Table 1 and Table 2.
[0112] The synthesized oligonucleotide probes were dissolved in ionized water and mixed with equal volumes of the spotting solution so that the final concentration was 75 pmol / μl; The surface of the glass slide modified by aldehyde groups; placed in a relative humidity of 70%, fixed at room temperature for 48 to 72 hours; then, at room temperature, immerse the glass slide in 0.2% SDS and shake for several minutes, then immerse in double distilled water Shake for a few minutes, then immerse in 100°C double distilled water for 30 seconds, dry and set aside.
Embodiment 2
[0113] Example 2 Fluorescent labeling of target DNA fragments in specimens
[0114] Design two pairs of primers, and label the 5' end of each pair of primers with Cy3 or Cy5, and use PCR to amplify the target fragment of fluorescently labeled sample DNA, where the template is bacterial genomic DNA, 23s RNA gene primer sequence and drug resistance gene The primer sequences are listed in Table 3. The amplification system is as follows: Mix 2 μl of sample DNA solution with 23 μl of PCR amplification system (the amplification system includes 2.5 μl of 10×PCR reaction buffer, 5 nmol dATP, 5 nmol dGTP, 5 nmol dGTP, 5 nmol dCTP, and 15 pmol upstream and downstream primers each) . The system was kept at 95°C for 5min, then cycled 40 times at 95°C for 30S, 55°C for 30S, and 72°C for 30S, and finally kept at 72°C for 5min.
Embodiment 3
[0115] Example 3 Hybridization of the labeled fragment of interest with the chip
[0116] Take 2 μl of the target DNA fragment sample containing the fluorescently labeled specimen, mix it with 13 μl of Easy Hyb hybridization solution (Roche), denature at 99°C for 5 minutes, take 15 μl and drop it on the surface of the chip after ice bathing, cover the cover glass, and place it in a 40°C humid chamber Hybridization in medium for 60 min, then rinsed in washing solution I (1×SSC, 0.1% SDS) at room temperature in the dark for 10 min, and dried in the air.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com