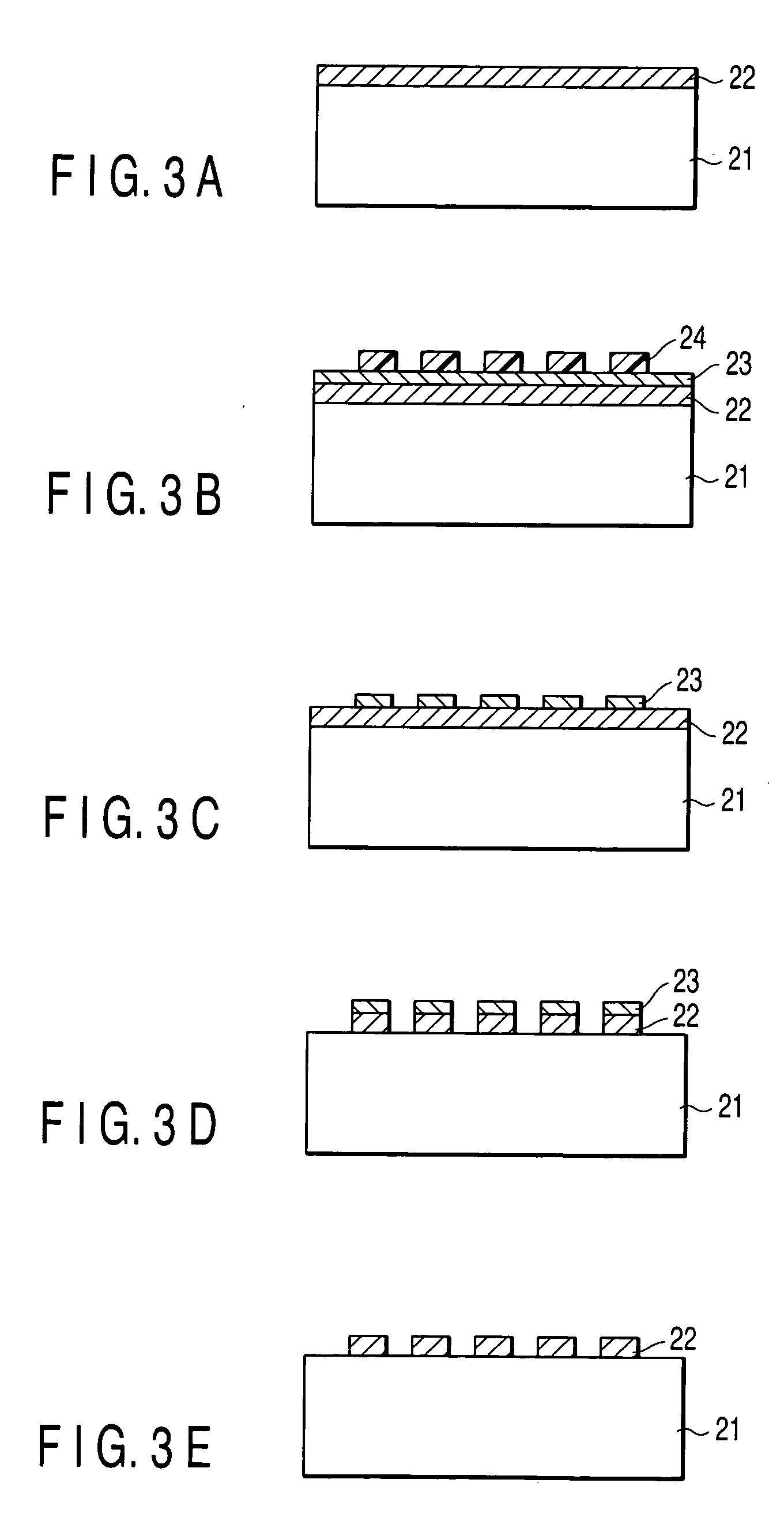
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
229 results about "Dna microchips" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
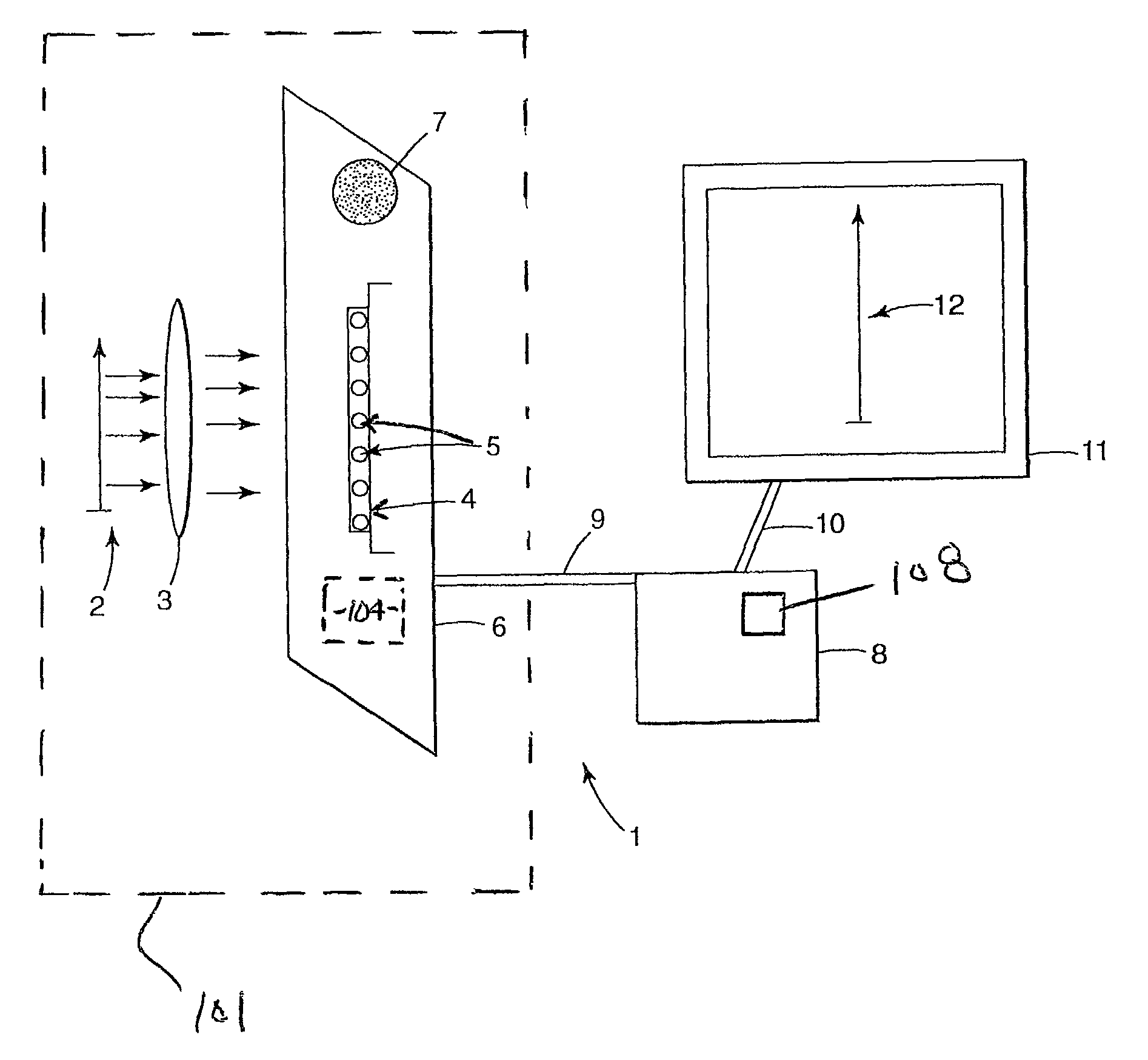
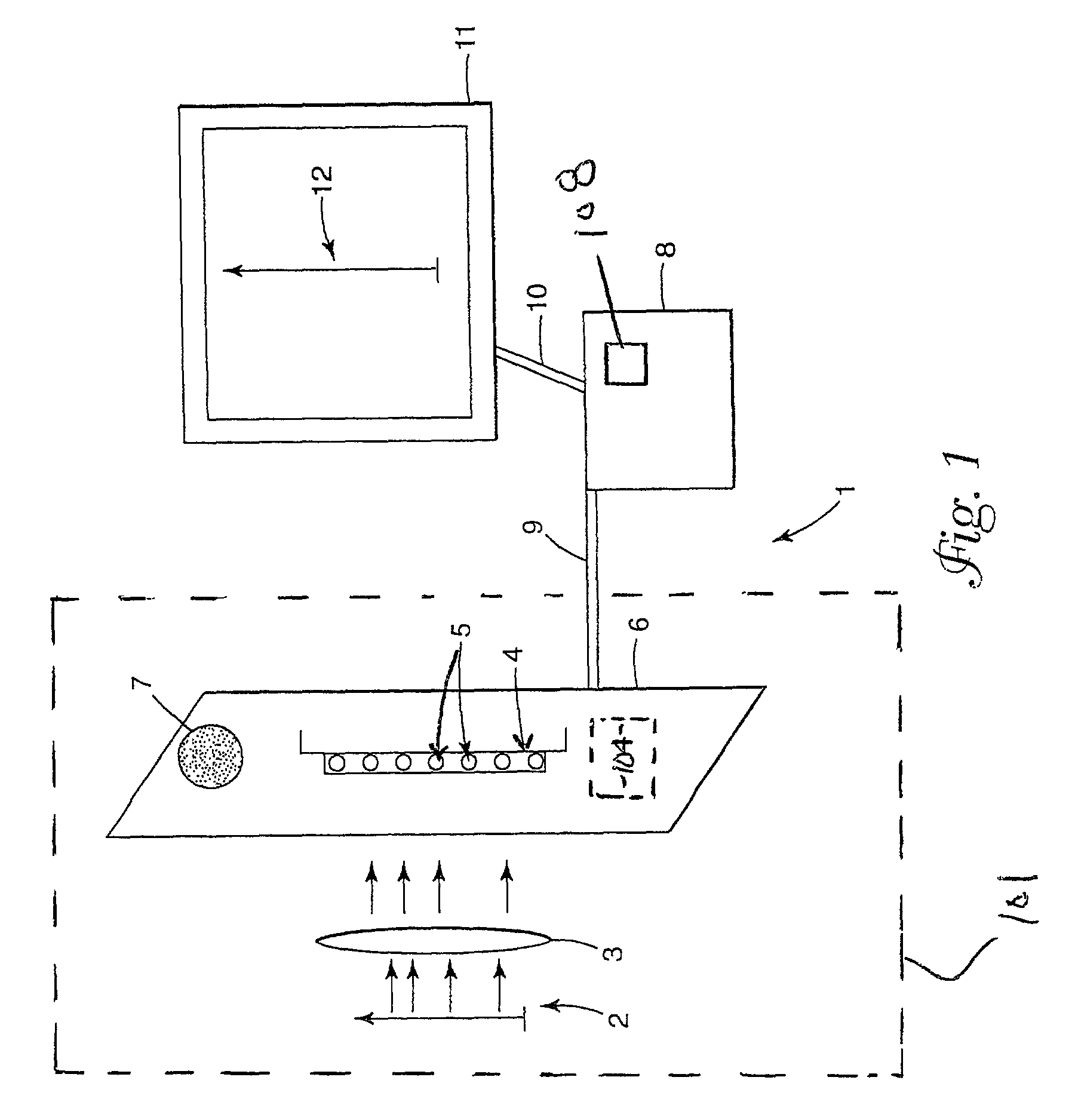
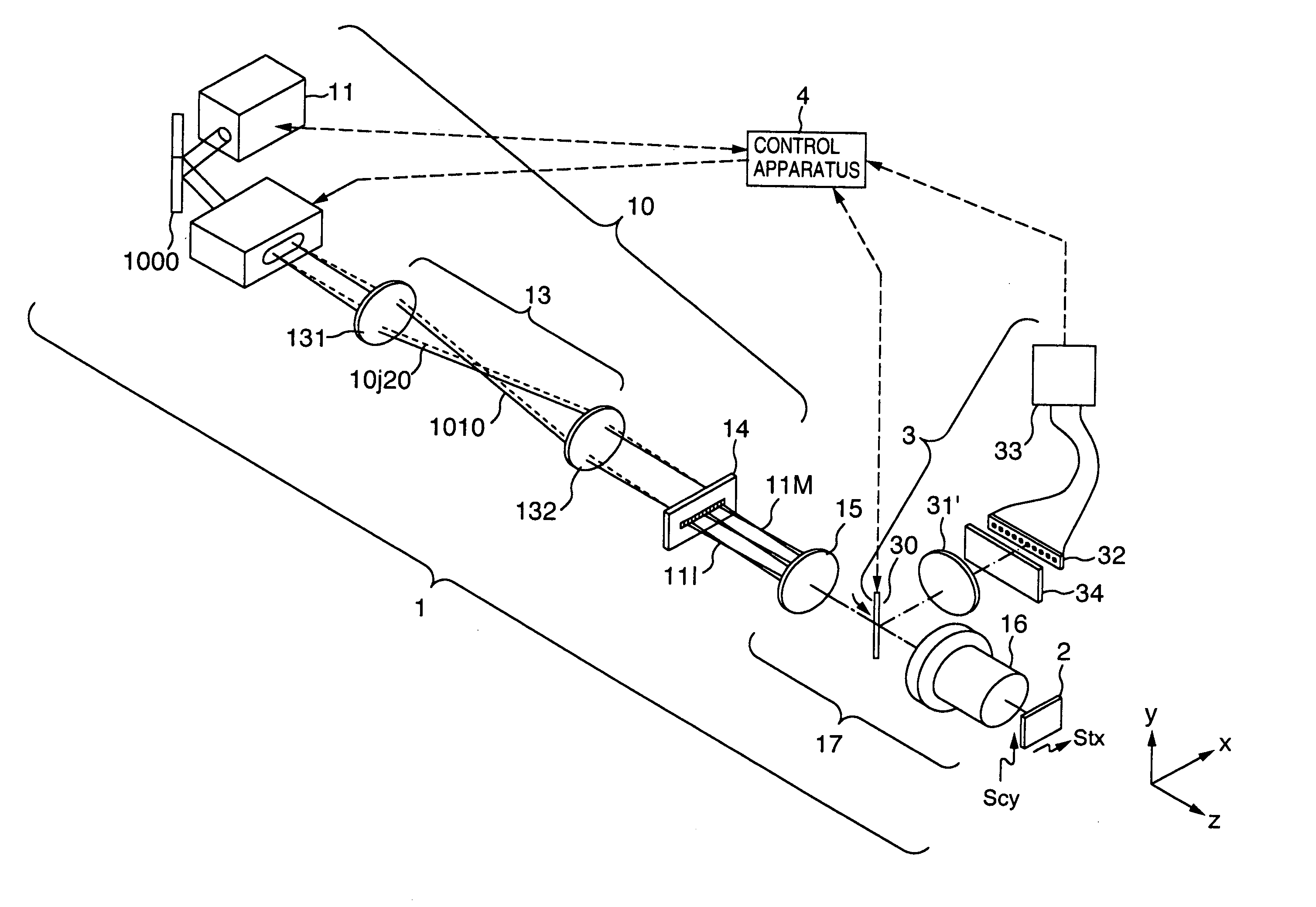
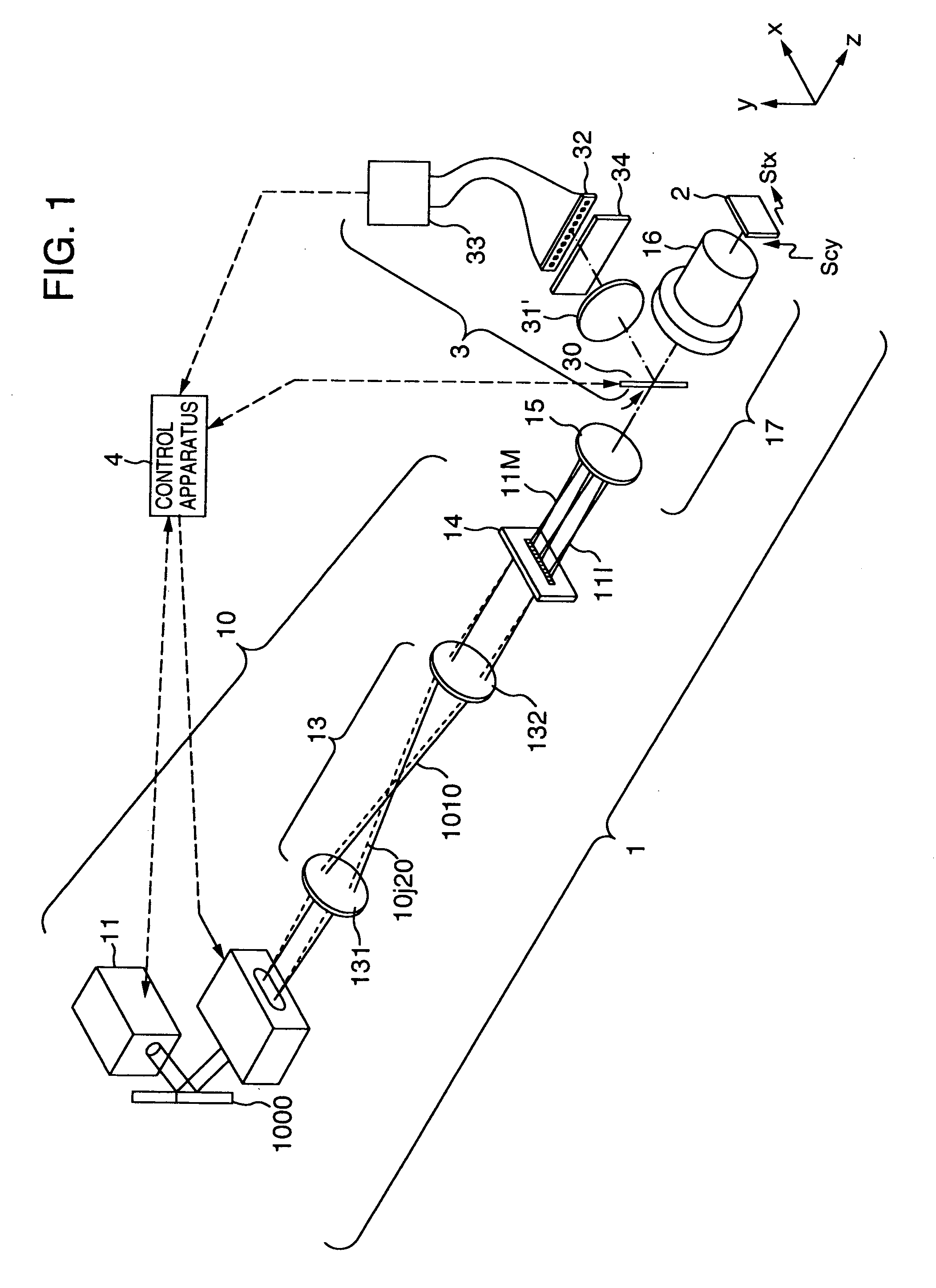
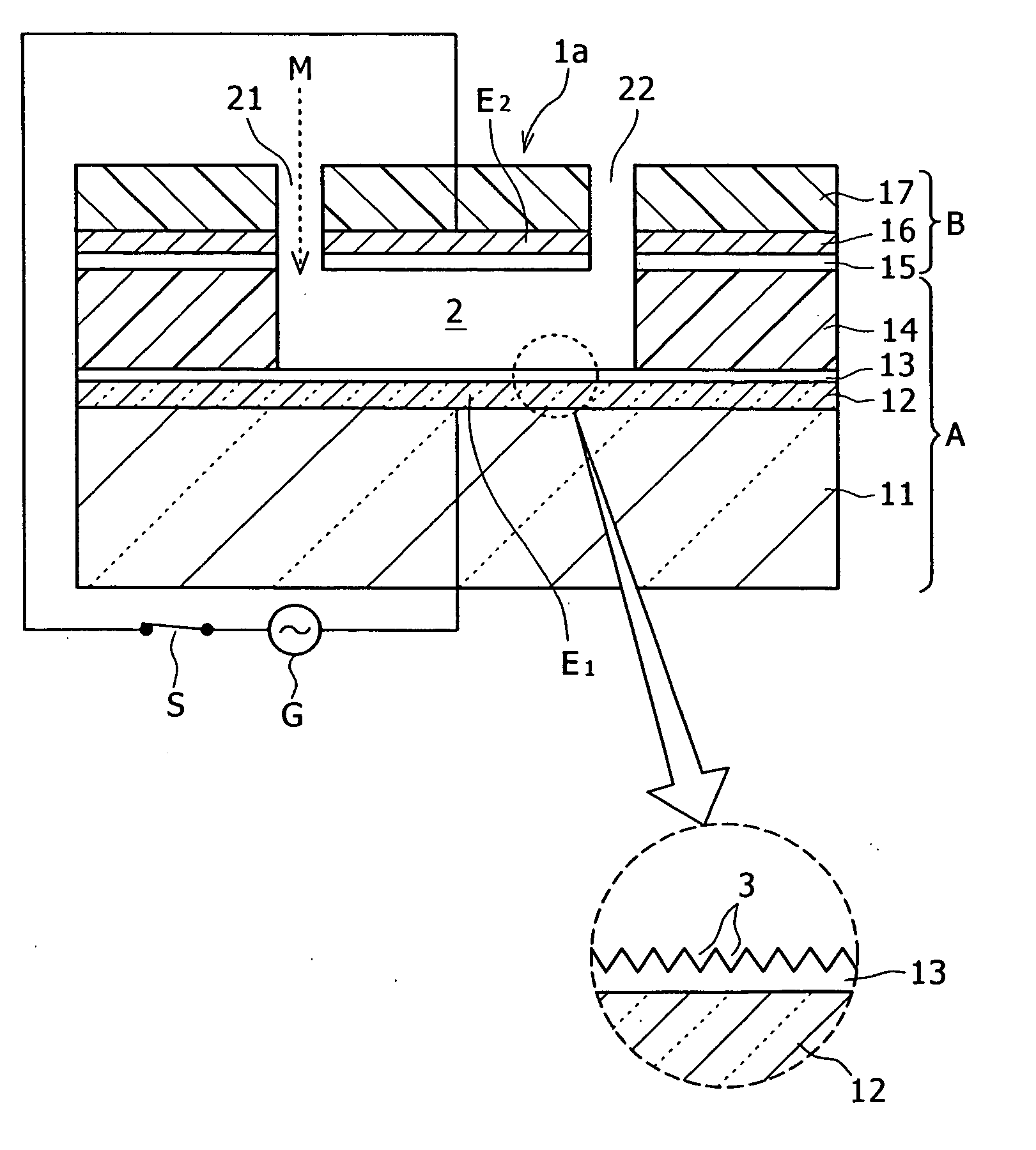
Imaging of biological samples using electronic light detector
A system and method using an electronic light detector array, e.g., a CCD or a CMOS-based detector array, is used to acquire a visual image of a biological sample that includes biological material associated with a biological material holding structure (e.g., a DNA spot array on a DNA chip, protein bands in a 2-D gel, etc.). For example, fluorescence from a biological sample may be detected.
Owner:RGT UNIV OF MINNESOTA
Systems and methods of conducting multiplexed experiments
A method for multiplexed detection and quantification of analytes by reacting them with probe molecules attached to specific and identifiable carriers. These carriers can be of different size, shape, color, and composition. Different probe molecules are attached to different types of carriers prior to analysis. After the reaction takes place, the carriers can be automatically analyzed. This invention obviates cumbersome instruments used for the deposition of probe molecules in geometrically defined arrays. In the present invention, the analytes are identified by their association with the defined carrier, and not (or not only) by their position. Moreover, the use of carriers provides a more homogenous and reproducible representation for probe molecules and reaction products than two-dimensional imprinted arrays or DNA chips.
Owner:VITRA BIOSCIENCES ASSIGNMENT FOR THE BENEFIT OF CREDITORS LLC +1
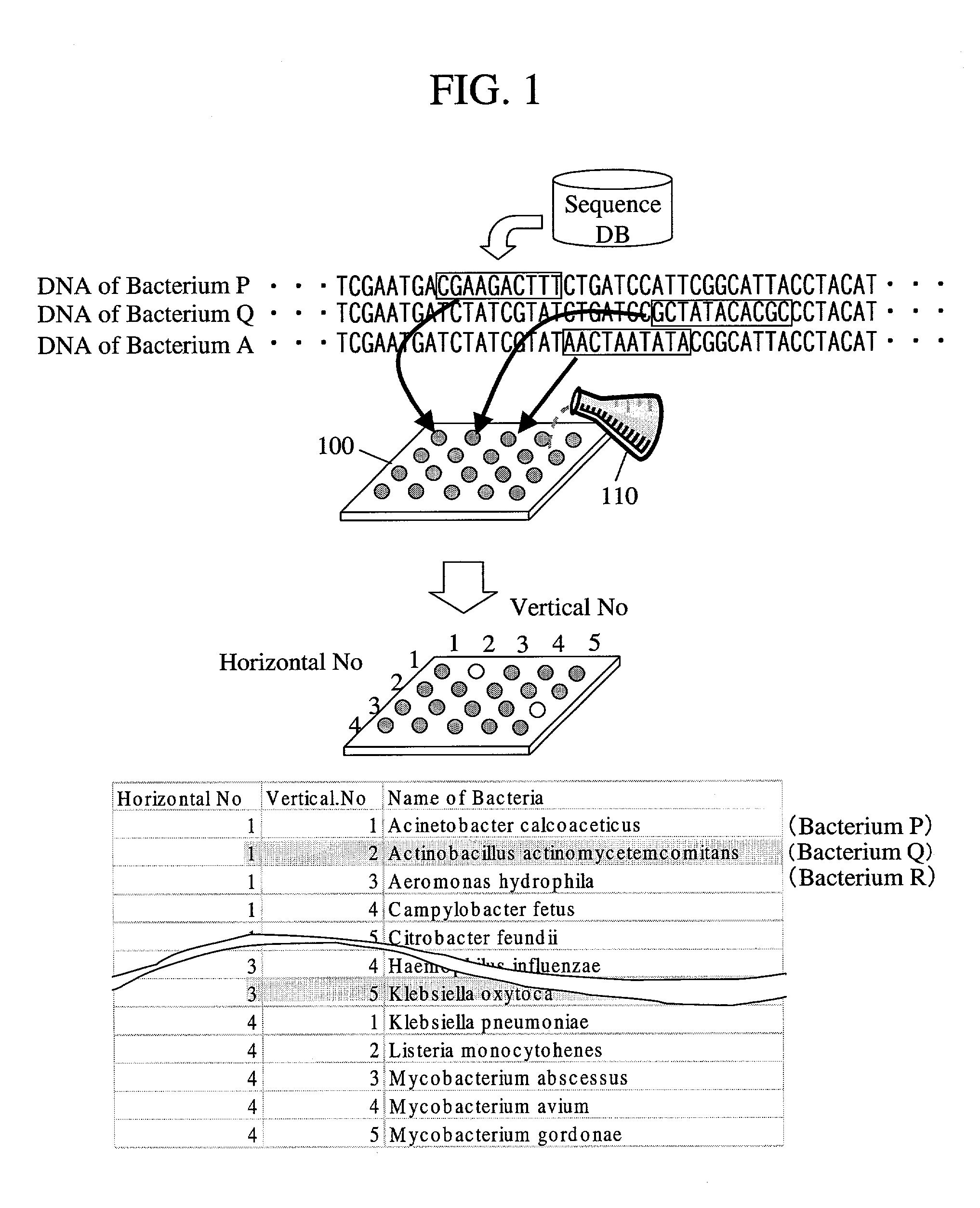
Probe designing apparatus and probe designing method
InactiveUS7035738B2Maximize precisionFlexible responseBioreactor/fermenter combinationsBiological substance pretreatmentsDesign methodsDNA
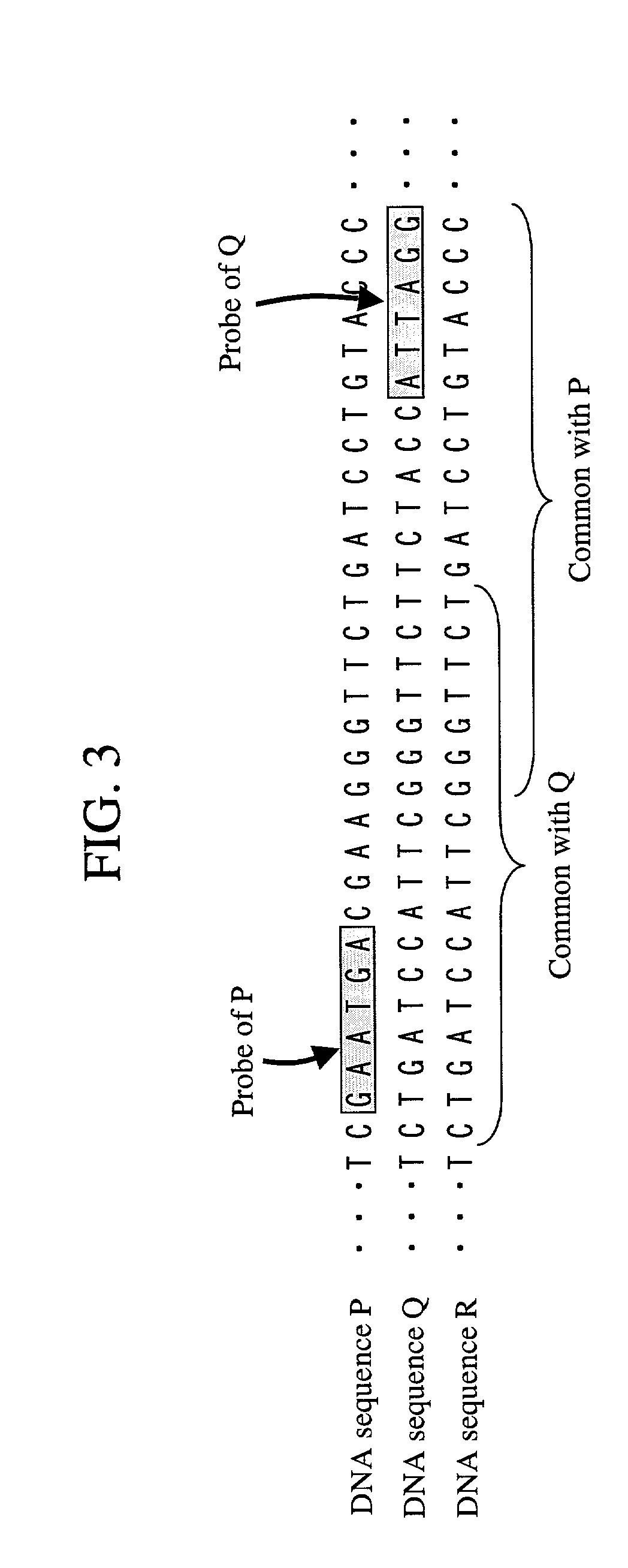
Where a DNA chip corresponding to a bacterium that is a subsequently added identification object is produced, temporal and pecuniary cost is reduced, and even where a probe unique to the DNA sequence of an identification object cannot be designed, precision in identifying DNA comprised in a sample is maximized. First, on designing a probe, a plurality of different probes are prepared to one kind of bacterium. Where some probes come to be not available by addition of bacteria that are new identification objects, identification is carried out using the remaining probes. Where a probe unique to an identification target bacterium cannot be designed, a probability of correct identification is increased by using multiple probes. Moreover, in consideration with a possibility that multiple kinds of bacteria simultaneously exist, a combination of probes that maximizes a probability of correct identification is selected.
Owner:HITACHI SOFTWARE ENG
Thin-film layered centrifuge device and analysis method using the same
ActiveUS20100297659A1Bioreactor/fermenter combinationsBiological substance pretreatmentsLab-on-a-chipEngineering
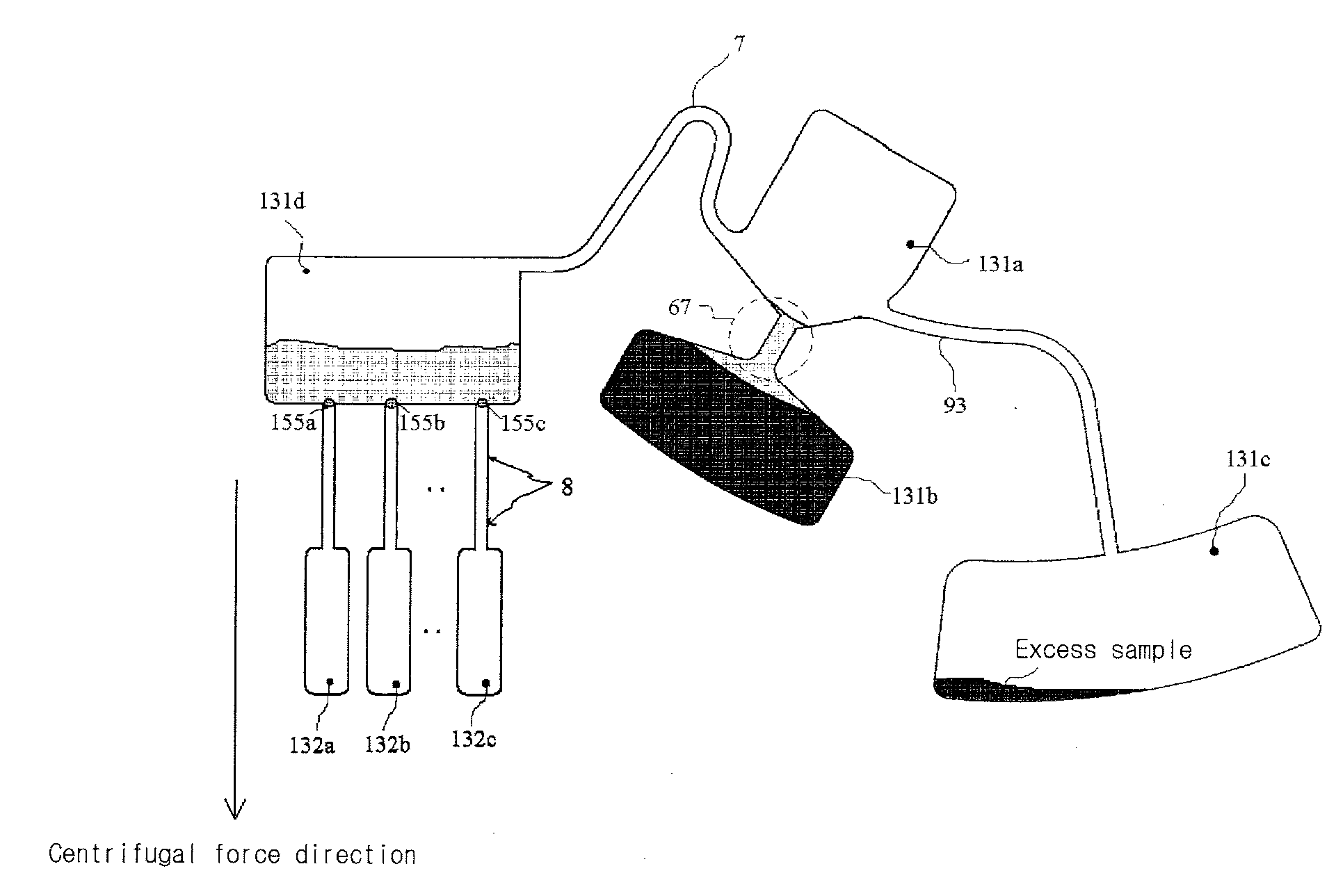
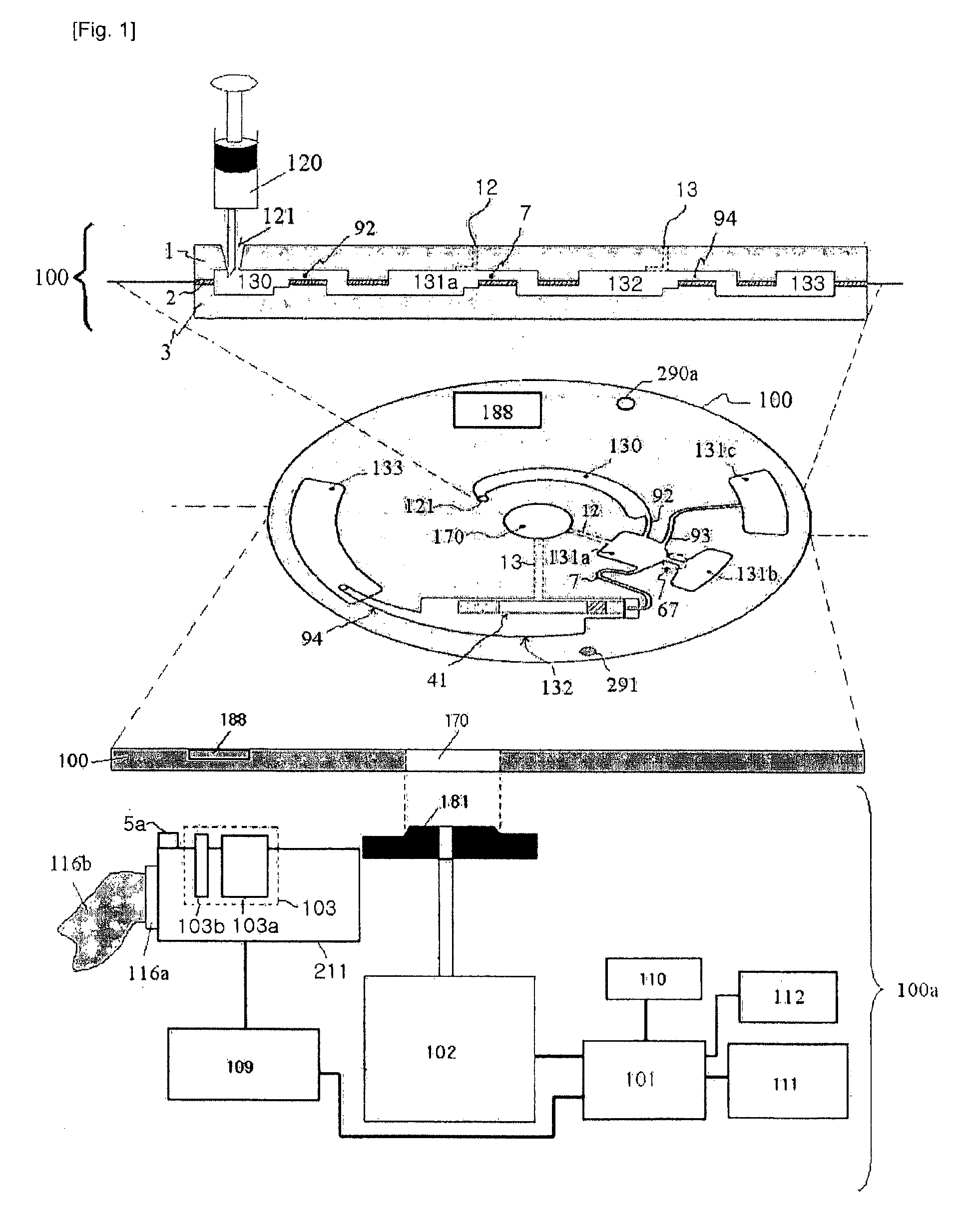
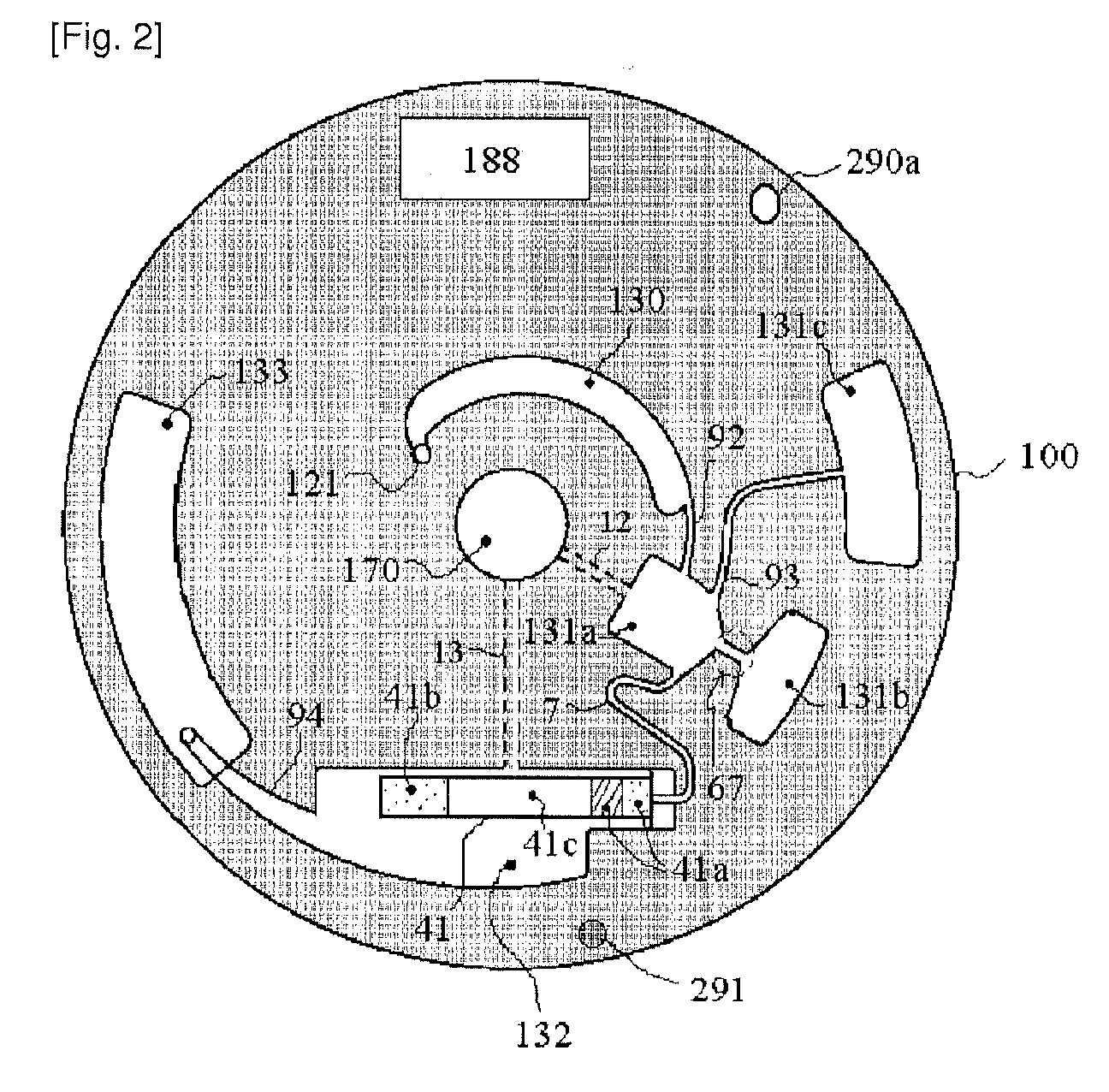
Disclosed herein is a thin-film layered centrifuge device and an analysis method using the same. One example of an embodiment of the present invention is a thin film layered centrifuge device where a device, such as a lab on a chip, a protein chip and a DNA chip, for diagnosing and detecting a small amount of material in a fluid is integrated into a rotatable thin-film layered body, and to an analysis method using the thin-film layered centrifuge device.
Owner:NEXUS DX
Biomolecule detecting element and method for analyzing nucleic acid using the same
ActiveCN1826525ADetection is accurate and stableBioreactor/fermenter combinationsBiological substance pretreatmentsField-effect transistorFloating electrode
An inexpensive DNA chip / DNA microarray system capable of highly accurate measurement with low running cost. A DNA probe 8 is immobilized on a floating electrode 7 connected to a gate electrode 5 of a field effect transistor. Hybridization with a target gene is carried out on the surface of the floating electrode, when a change in surface charge density is detected using a field effect.
Owner:NAT INST FOR MATERIALS SCI
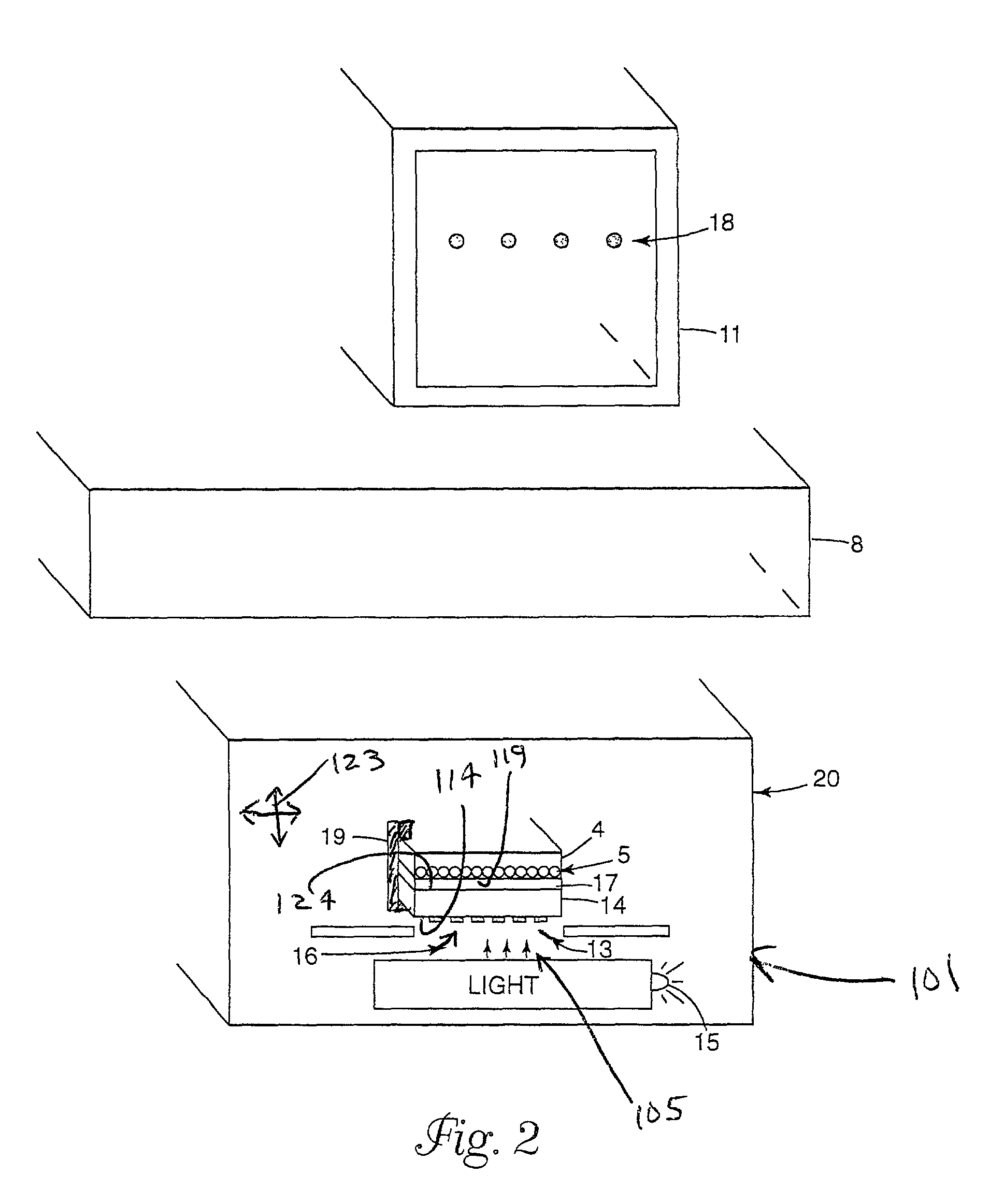
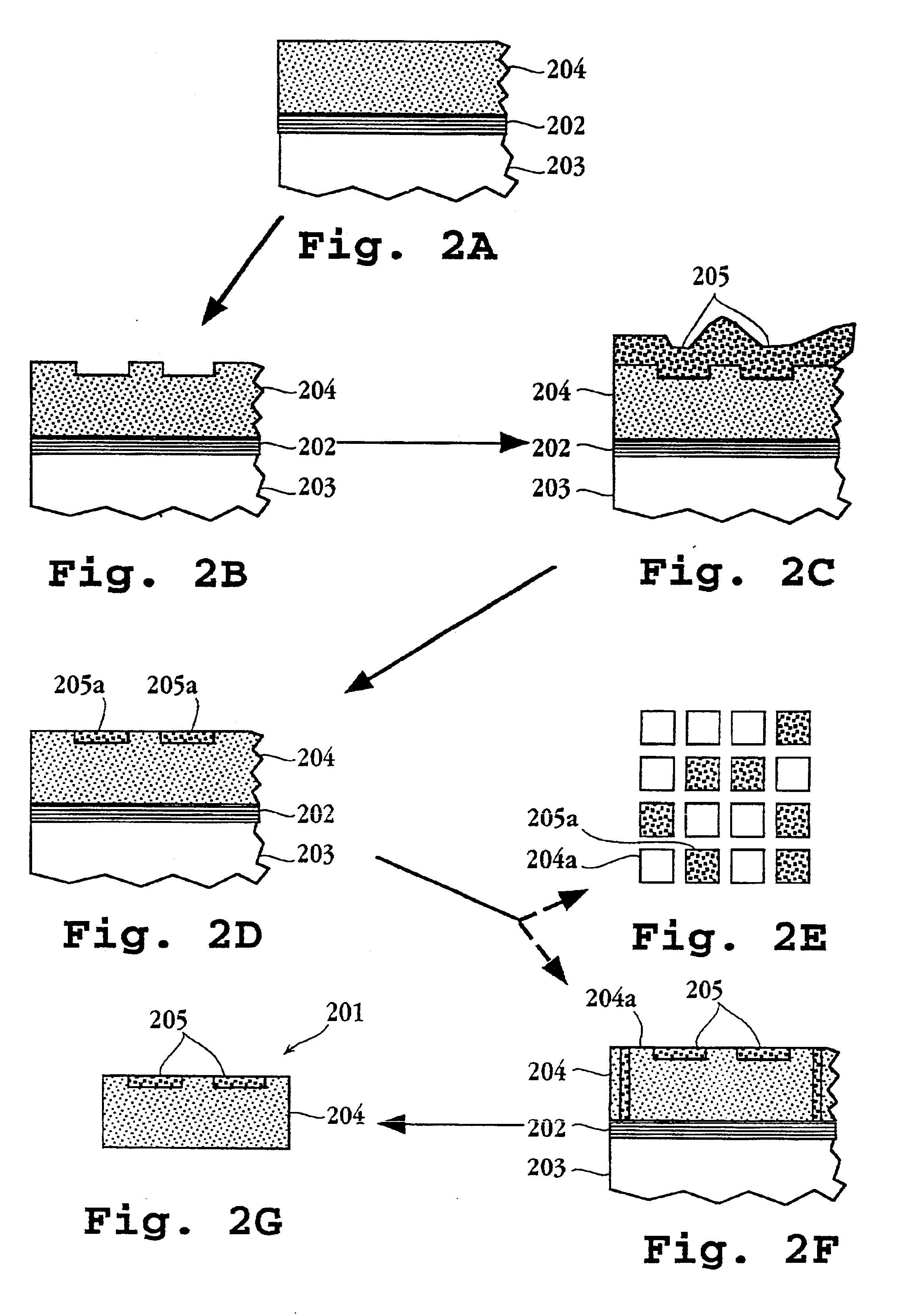
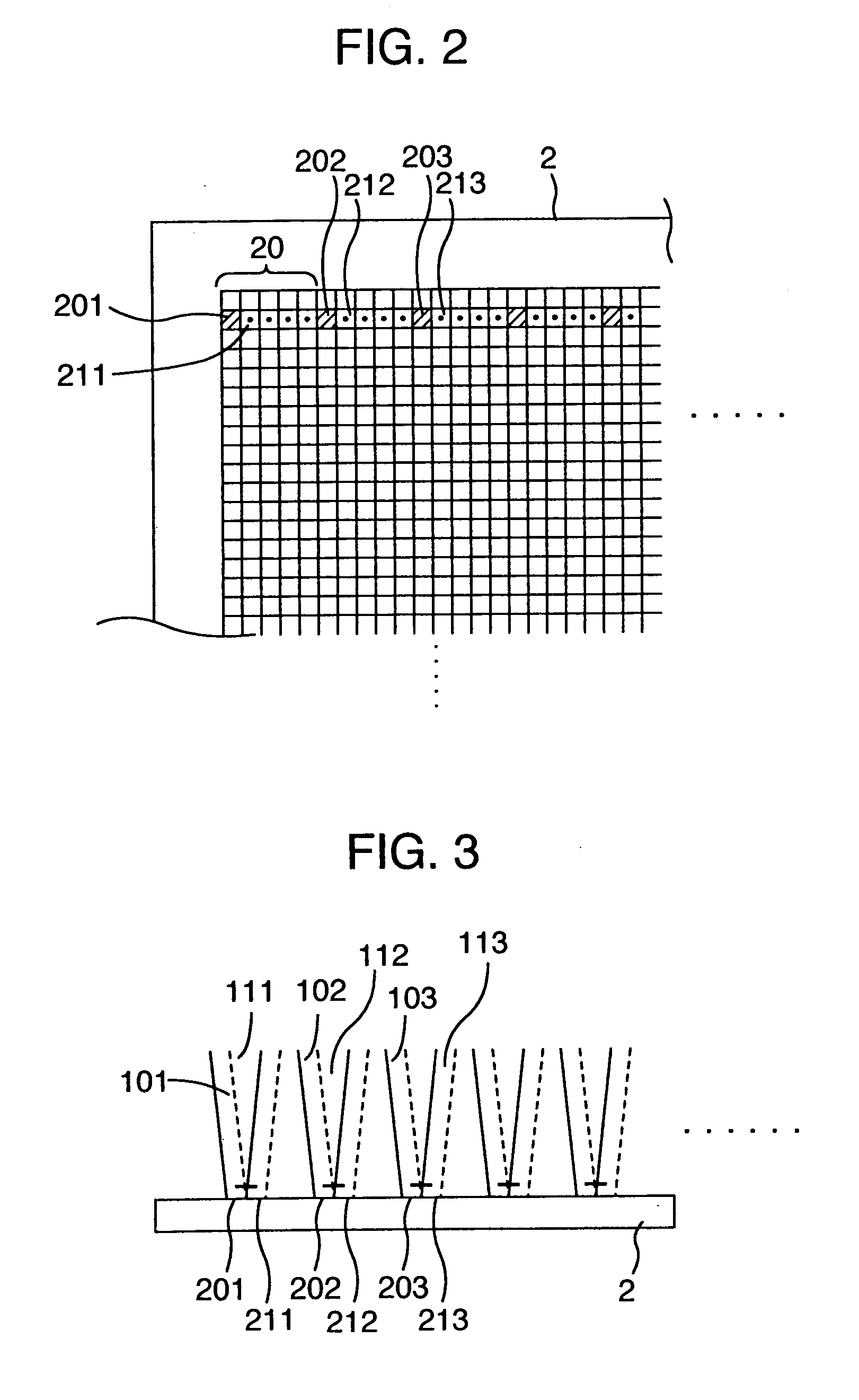
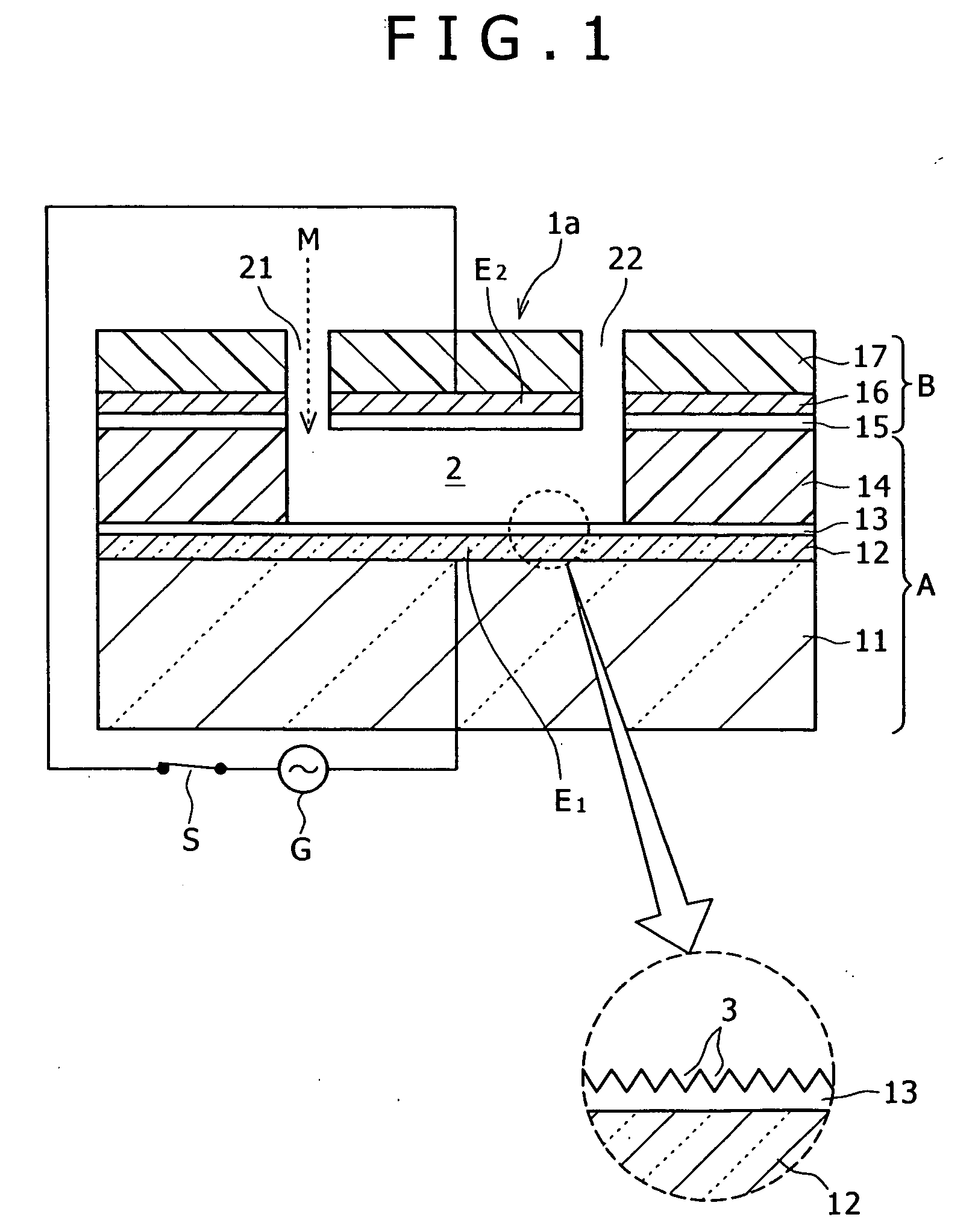
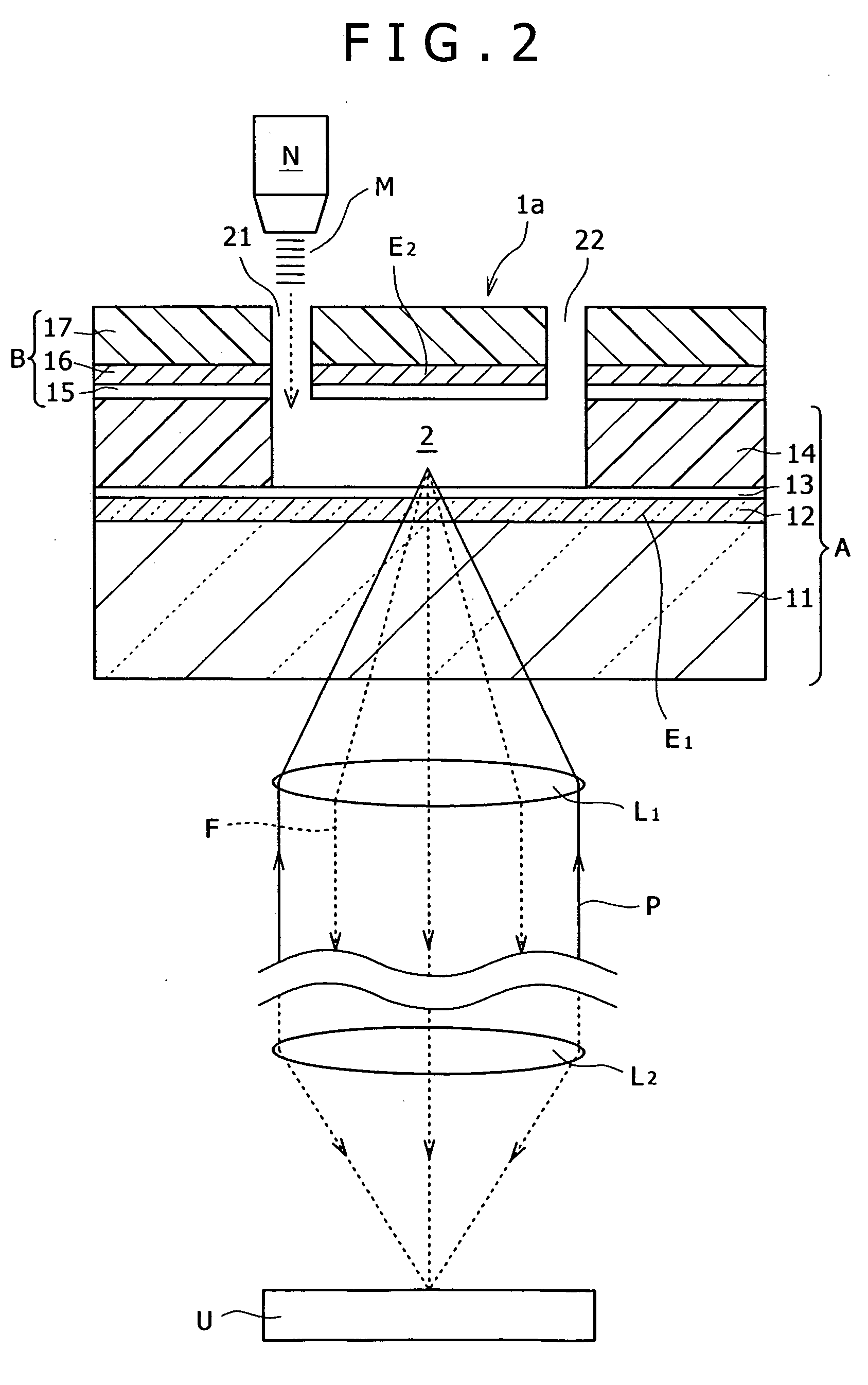
Method of inspecting a DNA chip
InactiveUS7217573B1Enhanced informationEasy to detectBioreactor/fermenter combinationsBiological substance pretreatmentsFluorescenceMulti detector
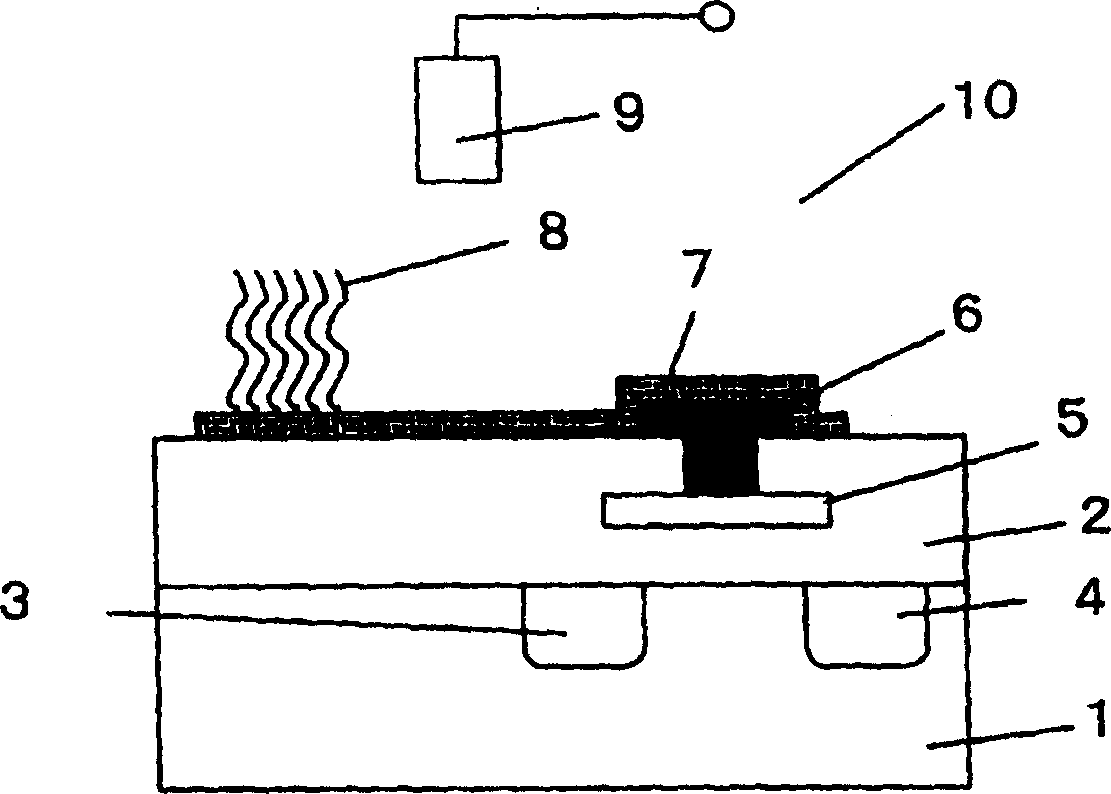
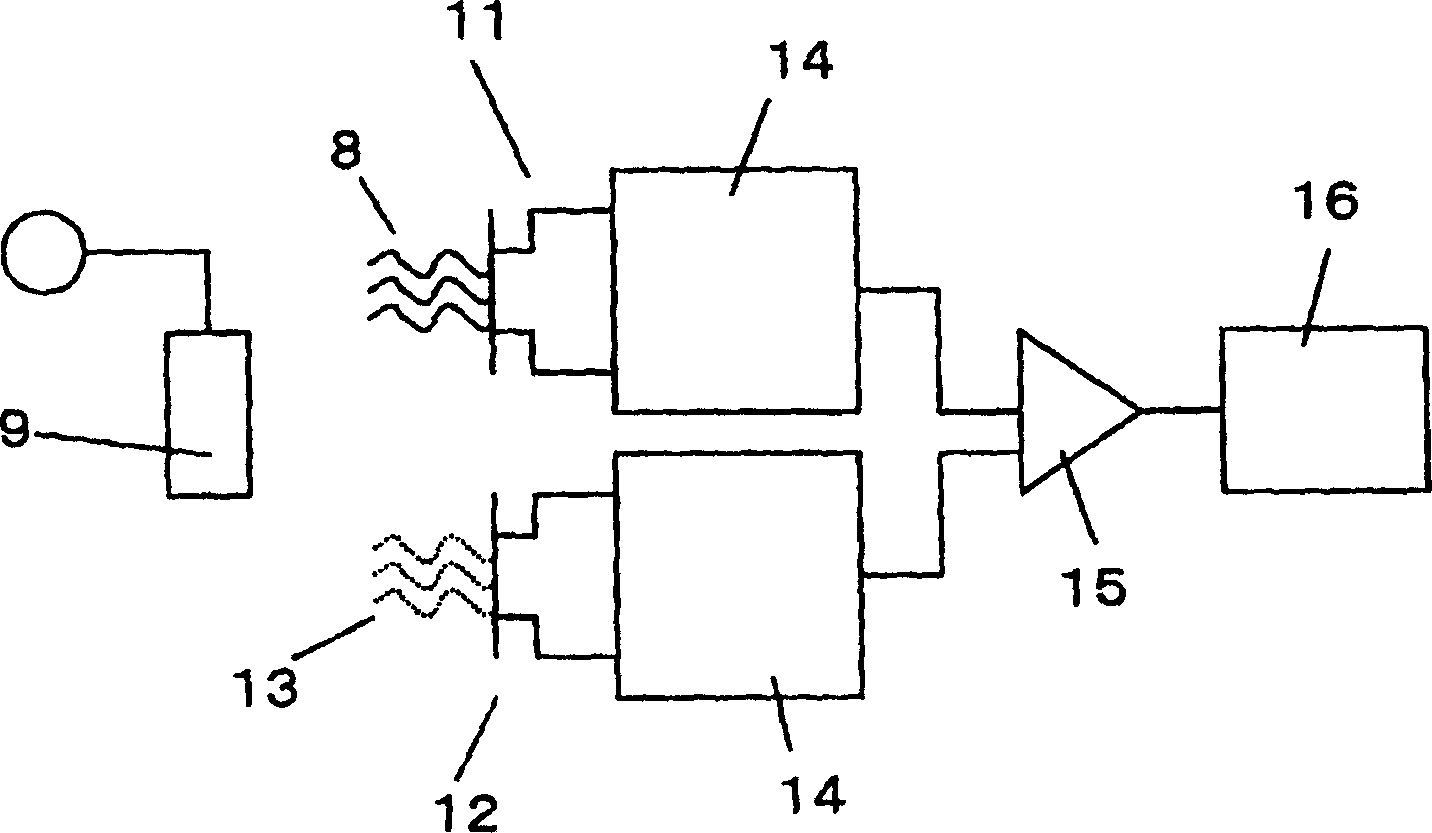
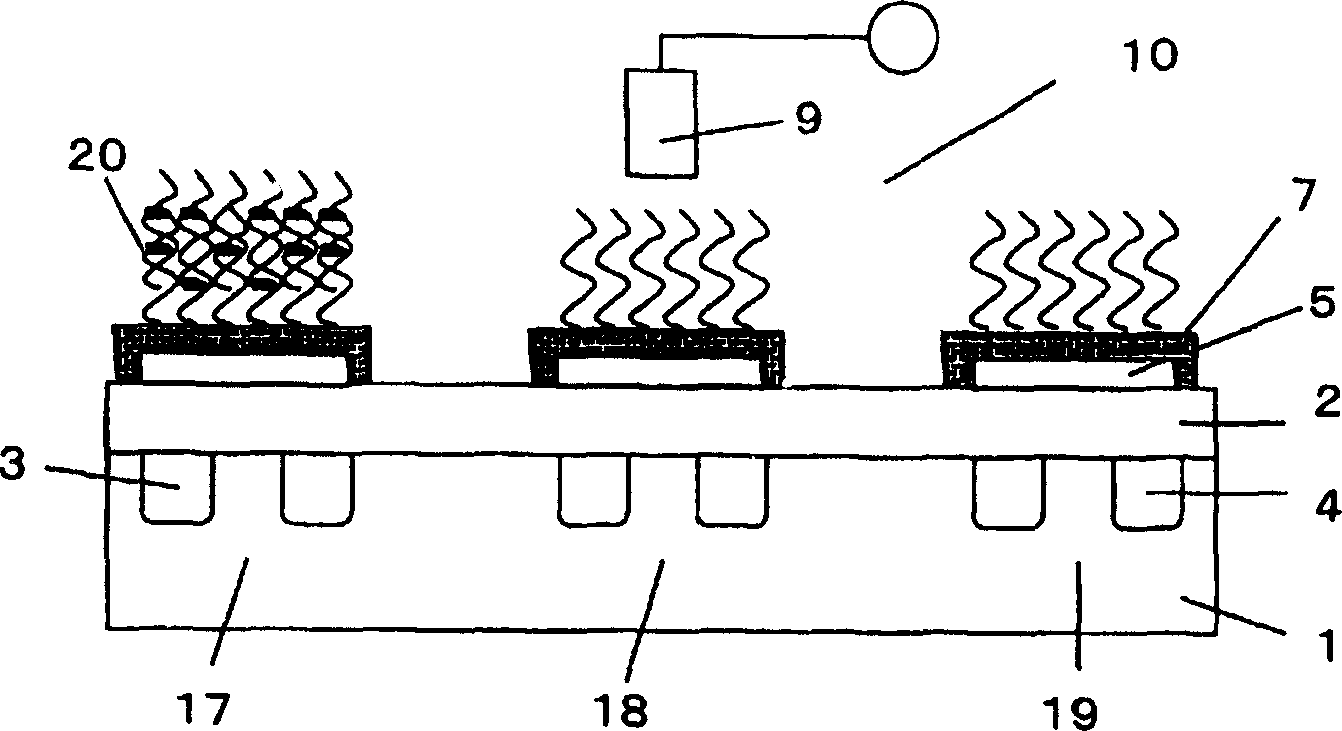
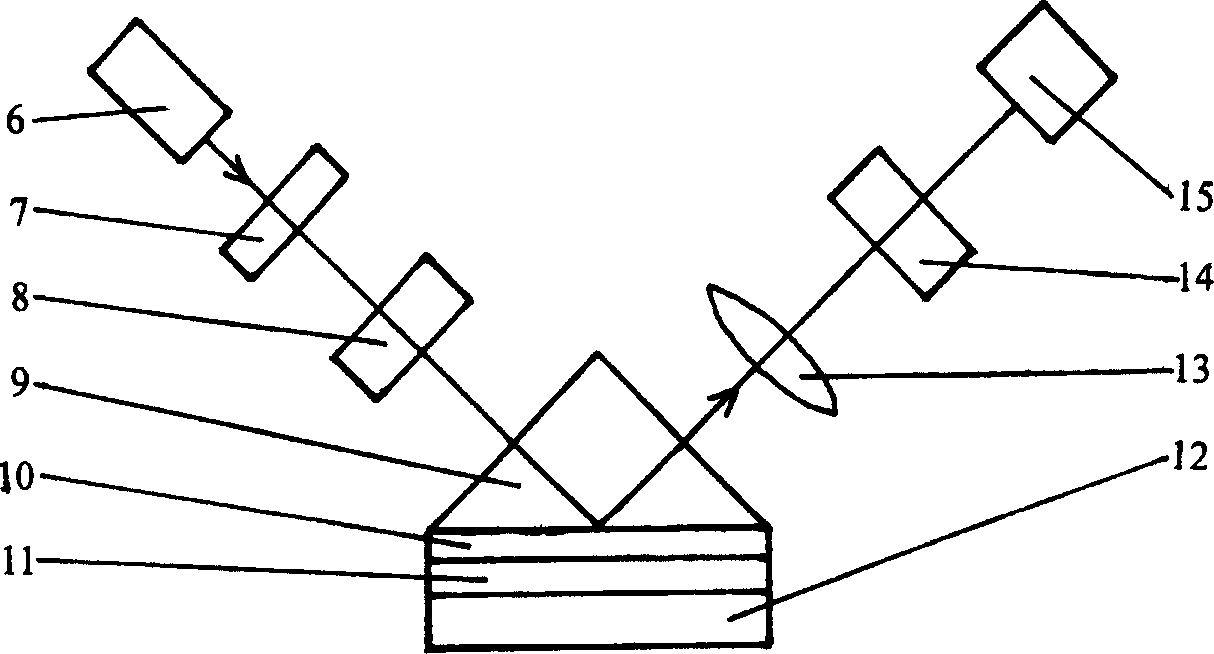
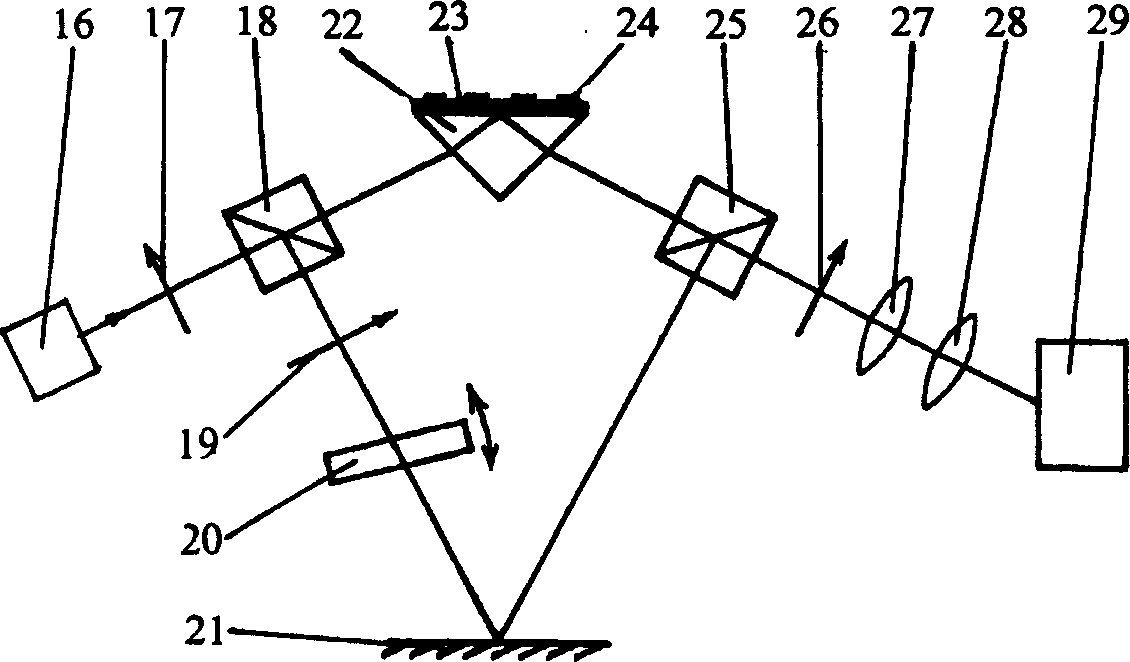
A method of inspecting a DNA chip and an apparatus therefor that allow a picture to be reconstructed in the following steps: A plurality of irradiation spots are formed on a DNA probe array mounted on a stage. Then, the stage is displaced in X, Y directions so as to execute a scanning, thereby irradiating substantially all the entire surface of the DNA probe array. Next, a plurality of emitted fluorescent lights, which are generated from the plurality of irradiation spot portions on the DNA probe array, are converged and are then detected simultaneously by multi detectors. Finally, a data processing apparatus processes the detected signals, thereby reconstructing the picture.
Owner:HITACHI LTD
Nano-crystal diamond film, manufacturing method thereof, and device using nano-crystal diamond film
A nano-crystal diamond film synthesized on a substrate and containing, as a major component, nano-crystal diamond having a grain diameter from 1 nm to less than 1000 nm. This nano-crystal diamond film can be formed on a substrate by means of a plasma CVD method using a raw material gas containing a hydrocarbon and hydrogen, allowing the formation of the nano-crystal diamond film to take place outside the plasma region. This nano-crystal diamond film is applicable to the manufacture of an electrochemical device, an electrochemical electrode, a DNA chip, an organic electroluminescent device, an organic photoelectric receiving device, an organic thin film transistor, a cold electron-emission device, a fuel cell and a catalyst.
Owner:TOPPAN PRINTING CO LTD +1
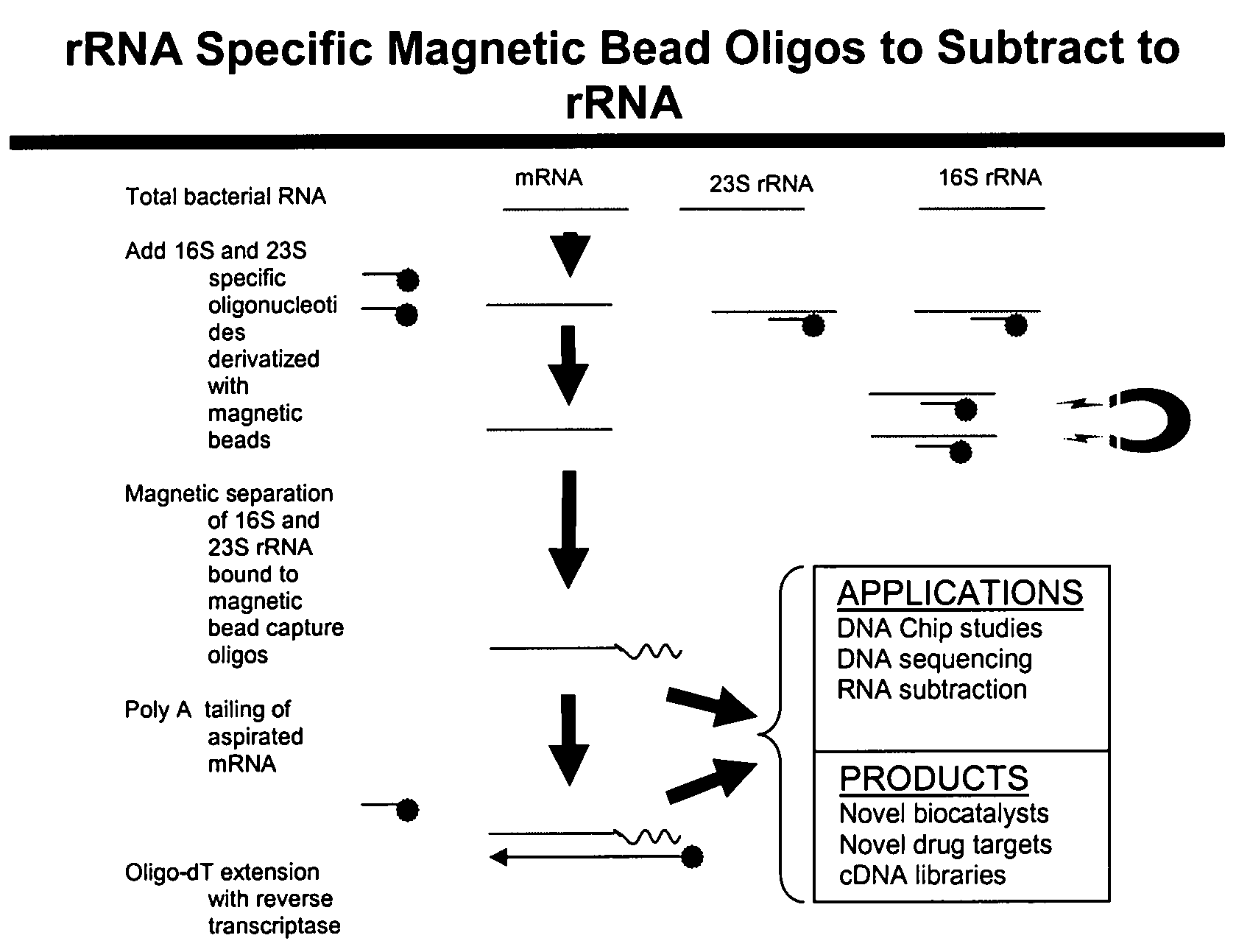
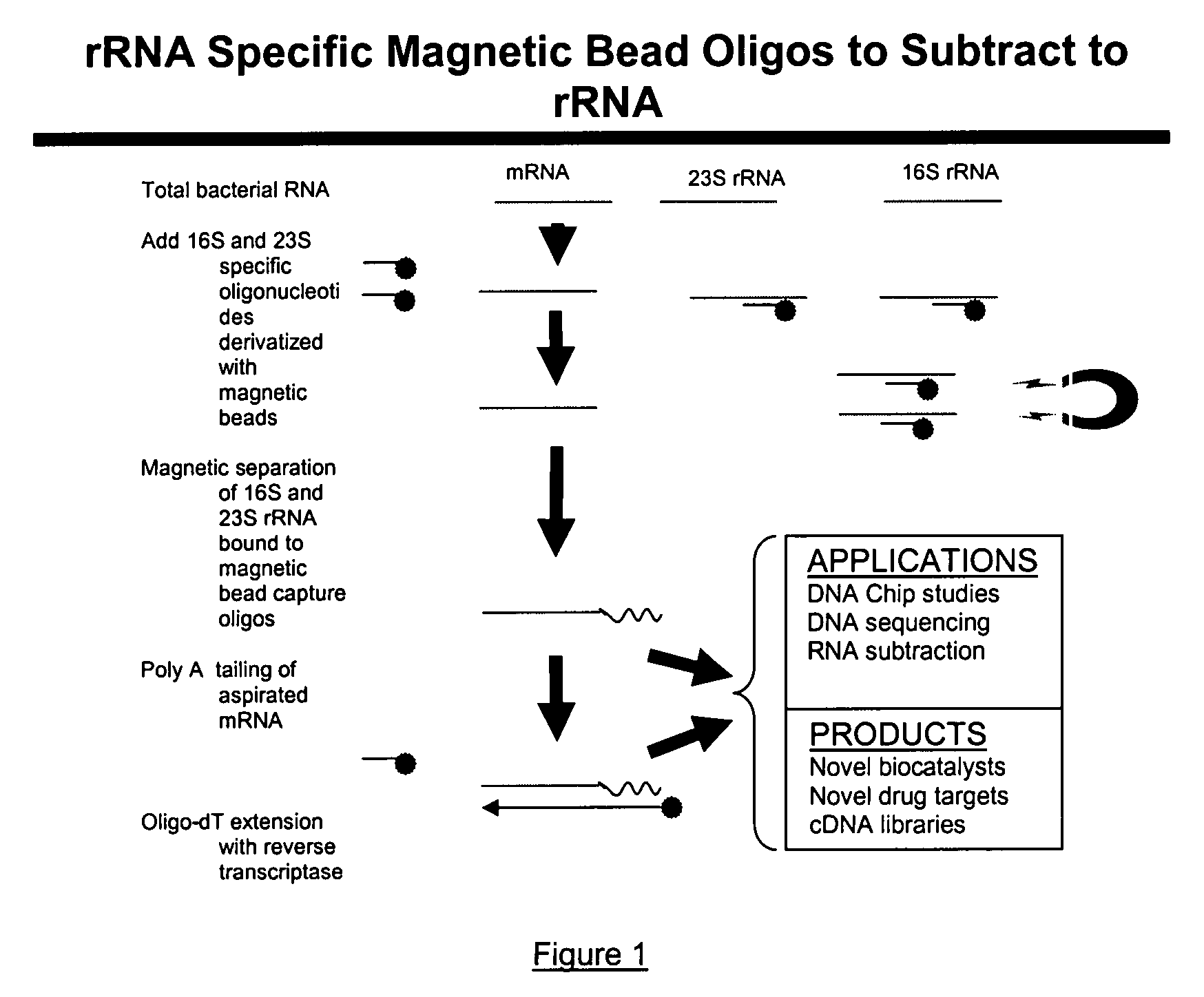
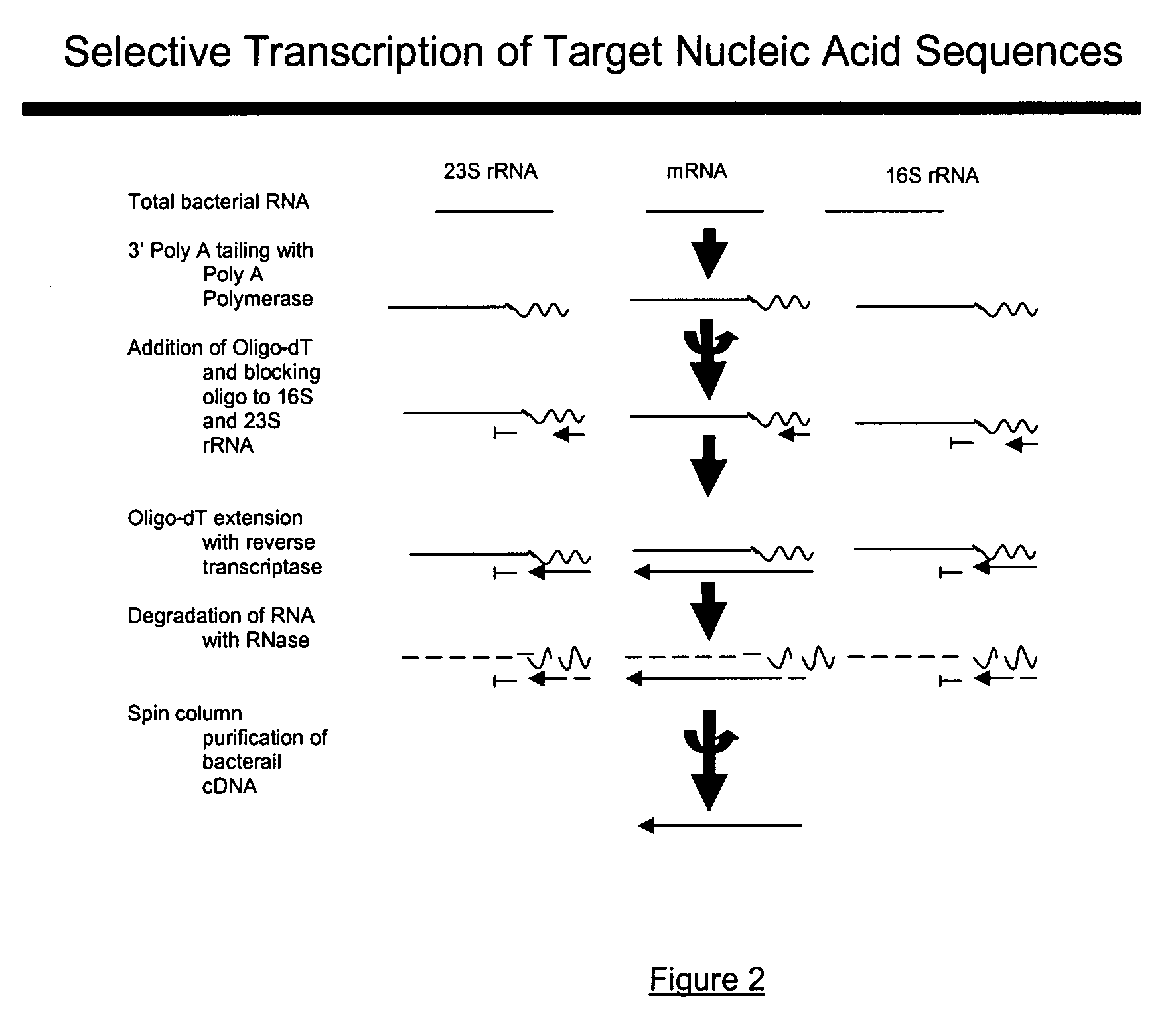
Methods and kits for negative selection of desired nucleic acid sequences
The present invention pertains to a method to isolate, separate, enrich or amplify a targeted nucleotide polymer such as mRNA through selective reverse transcription of the targeted polymer into cDNA from a sample comprising of chemically identical or similar polynucleotide polymers such as rRNA. The enrichment of the targeted nucleic acid such as mRNA is accomplished by blocking the reverse transcription of undesired rRNA while allowing unrestricted reverse transcription of the targeted polymer. The invention also embodies that the cleavage of the non-targeted nucleic acid such as rRNA bound to an oligonucleotide through enzymatic activity (RNase H). The invention further embodies methods and kits to accomplish the utility of the invention through the following steps 1) 3′ tailing of chemically identical or similar nucleotide polymers in a sample that includes bacterial mRNA 2) a 3′ tail capable of binding to a oligo-dN primer 3) at least one oligonucleotide capable of preventing the extension of oligo-dN bound to at least one non-targeted nucleotide polymers by a DNA polymerase such as a reverse transcriptase without restricting conversion of bacterial mRNA into cDNA 4) where the non-targeted molecule is prevented as a template for cDNA synthesis by enzymatic cleavage (RNase H) of template (rRNA)-oligonucleotide hybrid 5) where the reverse transcriptase is physically blocked by the oligonucleotide bound to the non-targeted nucleic acids such as rRNA 5) purification of the selectively transcribed cDNA. In further embodiments of the present invention, methods and composition to enable the study of bacterial transcriptomics-an analysis of genes expressed by a bacterial infection of a host, an isolated bacterial culture or a bacterial community, such as recovered from soil, intestine, mouth, biofilm, water etc are also included for use in DNA-chip or sequencing analyses.
Owner:SOWLAY MOHANKUMAR R
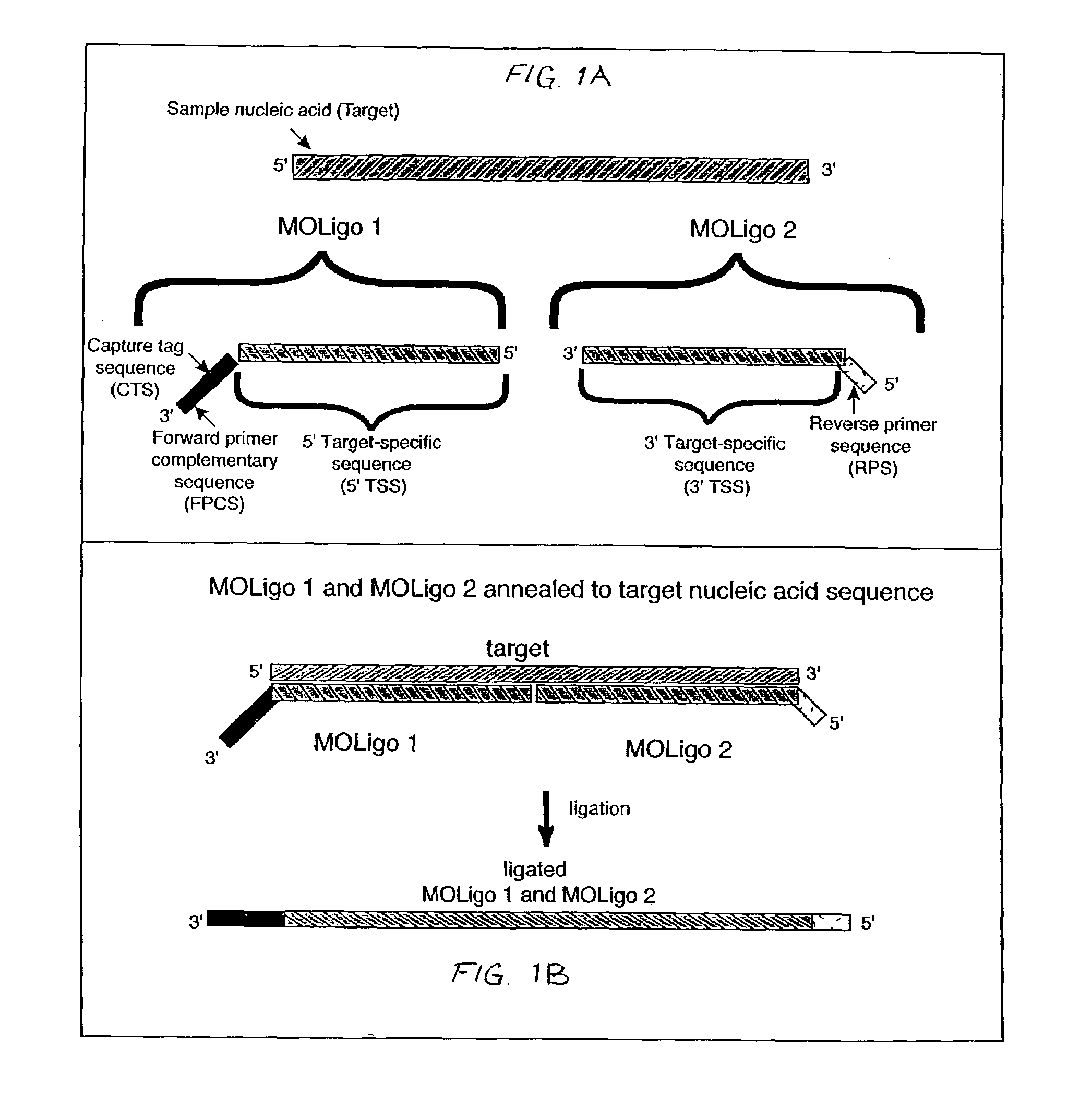
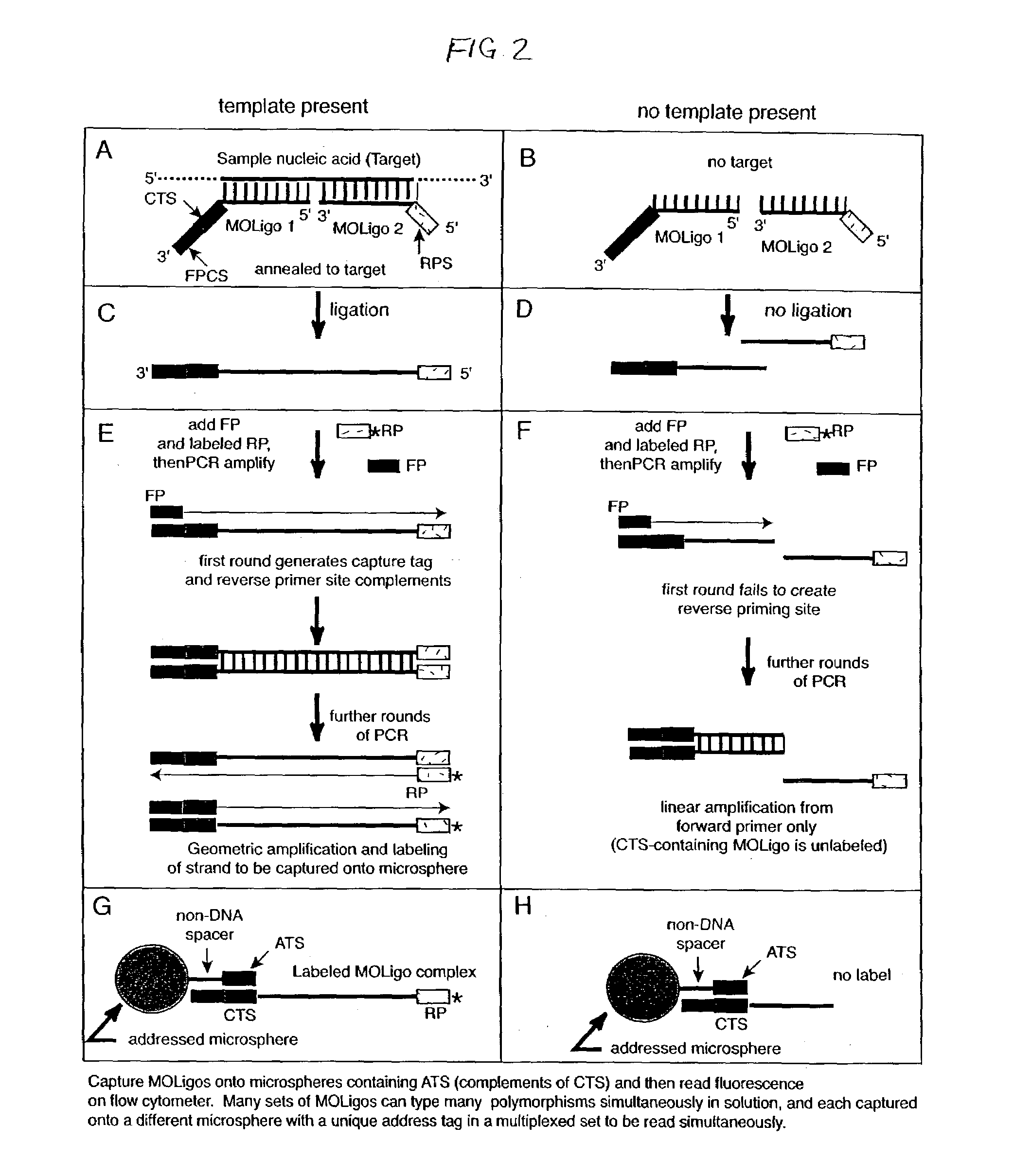
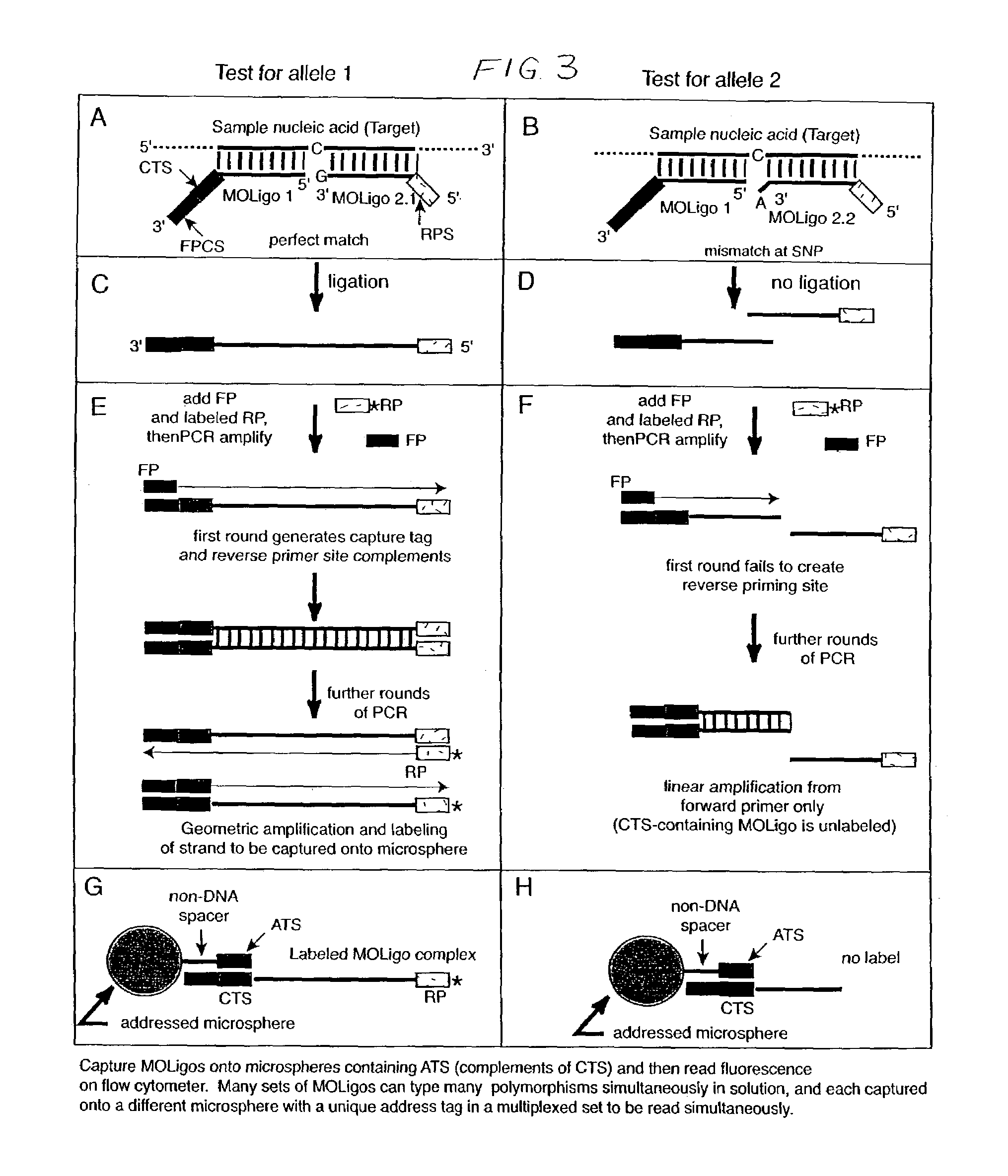
Nucleic acid sequence detection using multiplexed oligonucleotide PCR
InactiveUS7153656B2Provides redundancyRedundancy in allele discriminationSugar derivativesMicrobiological testing/measurementMicrosphereBiology
Methods for rapidly detecting single or multiple sequence alleles in a sample nucleic acid are described. Provided are all of the oligonucleotide pairs capable of annealing specifically to a target allele and discriminating among possible sequences thereof, and ligating to each other to form an oligonucleotide complex when a particular sequence feature is present (or, alternatively, absent) in the sample nucleic acid. The design of each oligonucleotide pair permits the subsequent high-level PCR amplification of a specific amplicon when the oligonucleotide complex is formed, but not when the oligonucleotide complex is not formed. The presence or absence of the specific amplicon is used to detect the allele. Detection of the specific amplicon may be achieved using a variety of methods well known in the art, including without limitation, oligonucleotide capture onto DNA chips or microarrays, oligonucleotide capture onto beads or microspheres, electrophoresis, and mass spectrometry. Various labels and address-capture tags may be employed in the amplicon detection step of multiplexed assays, as further described herein.
Owner:LOS ALAMOS NATIONAL SECURITY
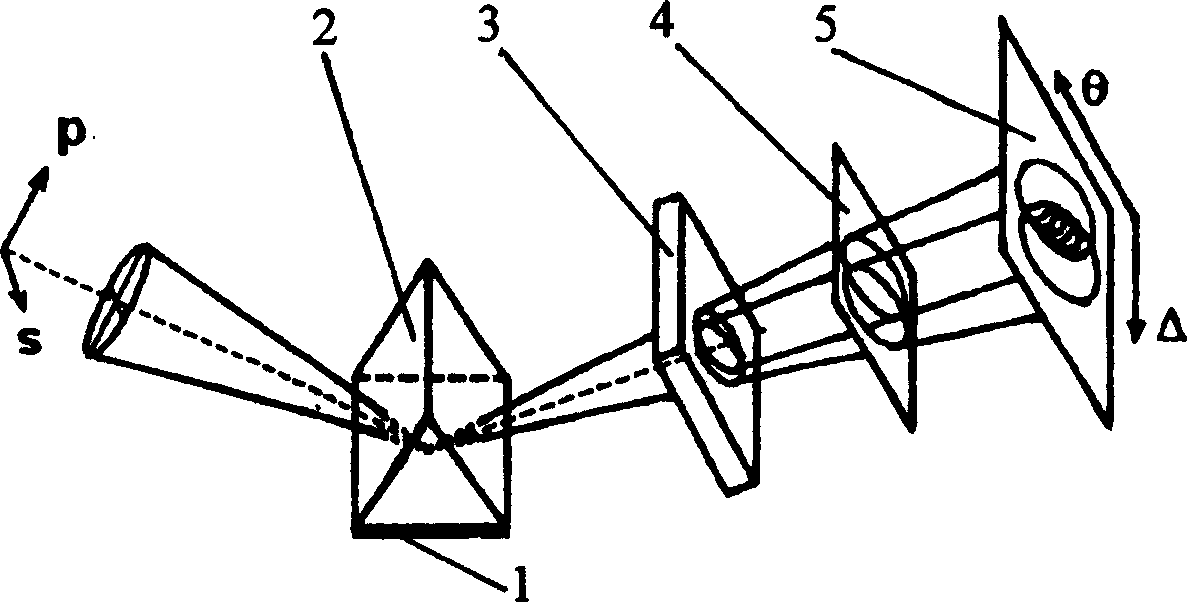
Method and system for detecting biological chip by space phase modulation interference array
InactiveCN1588064AHigh sensitivityImprove detection efficiencyPhase-affecting property measurementsColor/spectral properties measurementsPolarizerPrism
The invention relates to a method and system of adopting space phase modulation and interference array to detect the biochip, which belongs to biological technical field. The invention aims to overcome the deficiency of current technique and provides a new biochip detecting method and system, which can detect different volumes of DAN chips and protein chips dispense with mark and real-time achieve the information about the interaction of the biomolecules. The principle of the detection by space phase modulation and interference array is: the light reflected by the biosensor unit entering wallaston prism after combined by the one dimensional magnifying lens, the contained s ray and p ray which is polarizationdirection is divided into the two rays transmitting in different direction and realizing co-lightpath space phase modulation and interference in wallaston prism; the interference fringe being converted into electrical signal through the polarizer and imaging lens, the information of real-time phase change of the each unit on the array chip being acquired after processing and be offered to the biologist and medical scientists to parse.
Owner:TSINGHUA UNIV
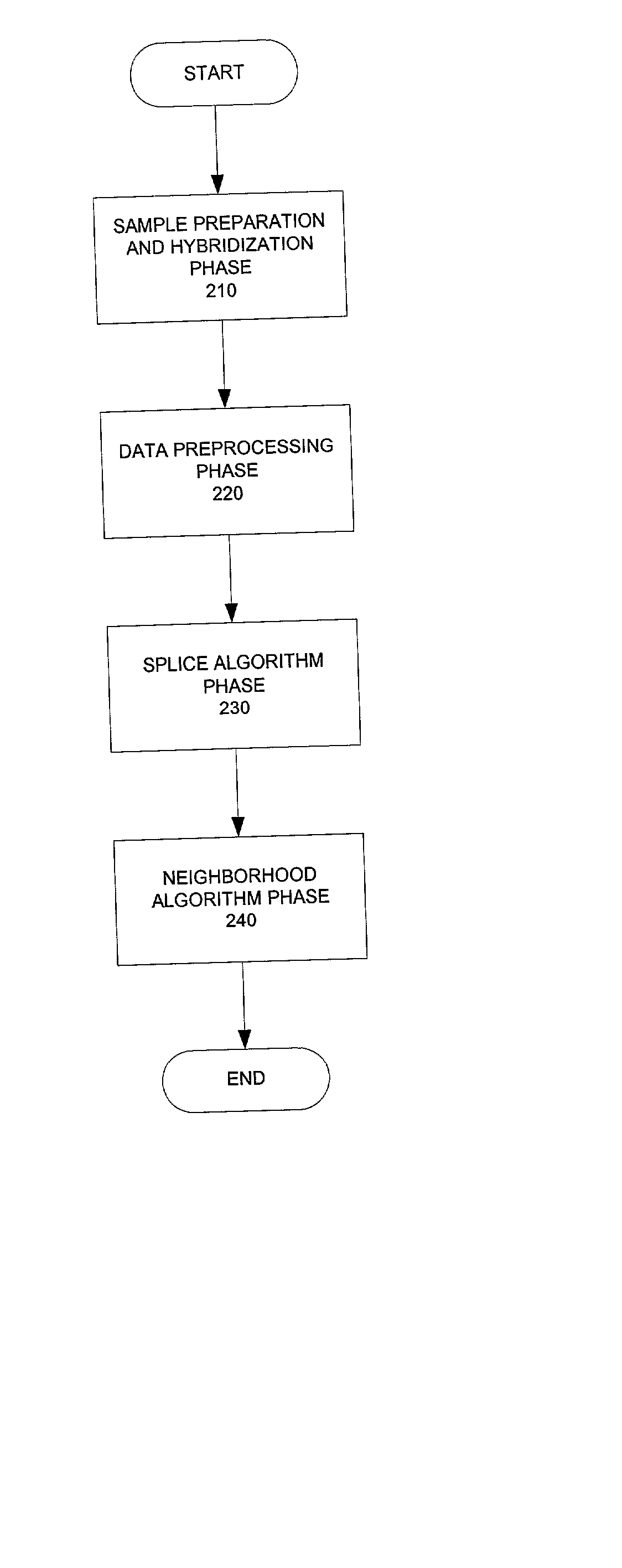
Method and system for predicting splice variant from DNA chip expression data
InactiveUS20020029113A1Microbiological testing/measurementBiological testingEukaryotic geneMessenger RNA
A system and method predict alternative splicing transcripts using DNA chip expression data as a primary data source. The system and method may perform prediction of alternative splicing of pre-messenger RNA that may be used, for example, for regulating eukaryotic gene expression.
Owner:WARNER-LAMBERT CO
Method of agitating solution
ActiveUS20070178603A1Stir wellAvoid damageImmobilised enzymesBioreactor/fermenter combinationsHigh signal intensityAnalyte
The method of stirring a solution according to the present invention is a method of stirring a solution comprising contacting a selective binding substance immobilized on the surface of a carrier with a solution containing an analyte substance reactive with the selective binding substance, and mixing the fine particles or air bubbles into the solution containing an analyte substance, and moving the fine particles or air bubbles without allowing contact thereof with the selective binding substance-immobilized surface. The present invention provides a stirring method that accelerates the reaction of a carrier-immobilized selective binding substance with an analyte substance and detects a trace amount of analyte at high signal intensity and high S / N ratio. The present invention enables diagnosis and examination in the clinical setting by using a selective binding substance-immobilized carrier such as DNA chip.
Owner:TORAY IND INC
Hybridization detecting unit including an electrode having depression parts and DNA chip including the detecting unit
InactiveUS20050274612A1Effective informationAvoid electrochemical reactionsImmobilised enzymesBioreactor/fermenter combinationsDNAElectric field
Disclosure herein is a hybridization detecting unit including, a reaction area for providing a field for hybridization between nucleic acid for detection and target nucleic acid, and opposed electrodes disposed so that an electric field can be applied to a medium stored or retained in the reaction area, wherein an electrode surface of at least one of the opposed electrodes has depression parts.
Owner:SONY CORP
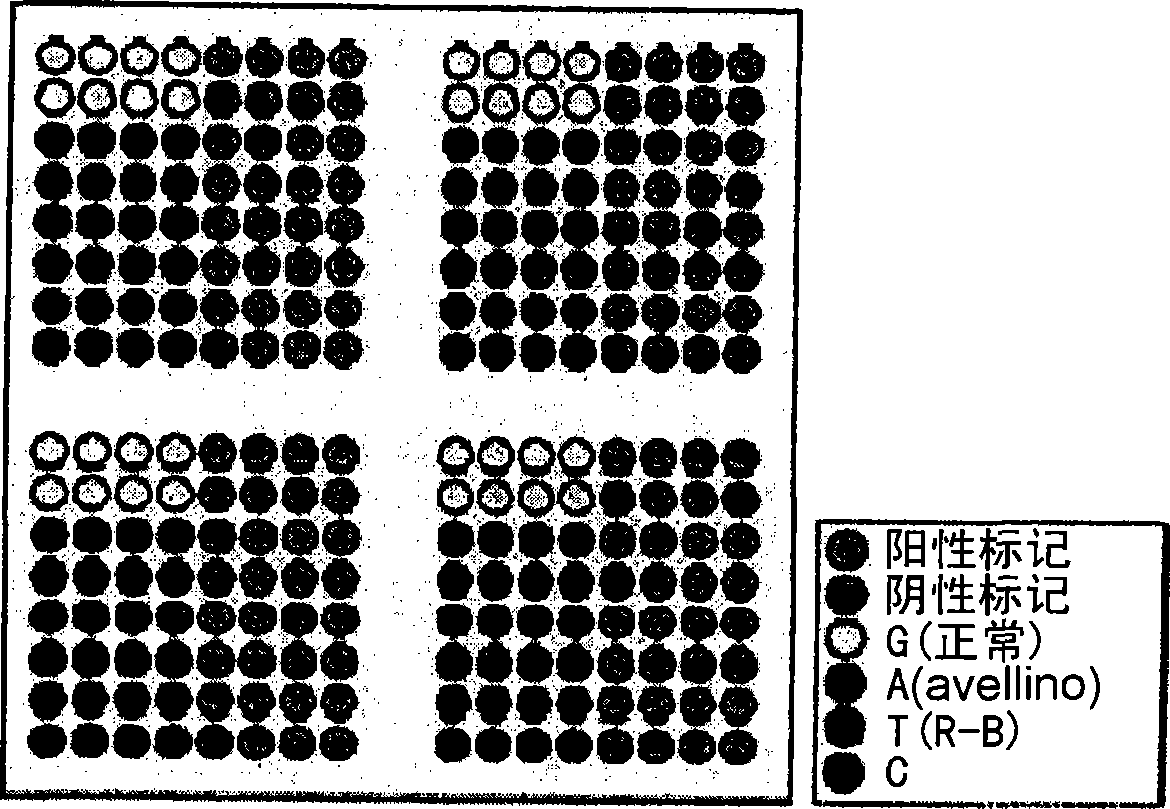
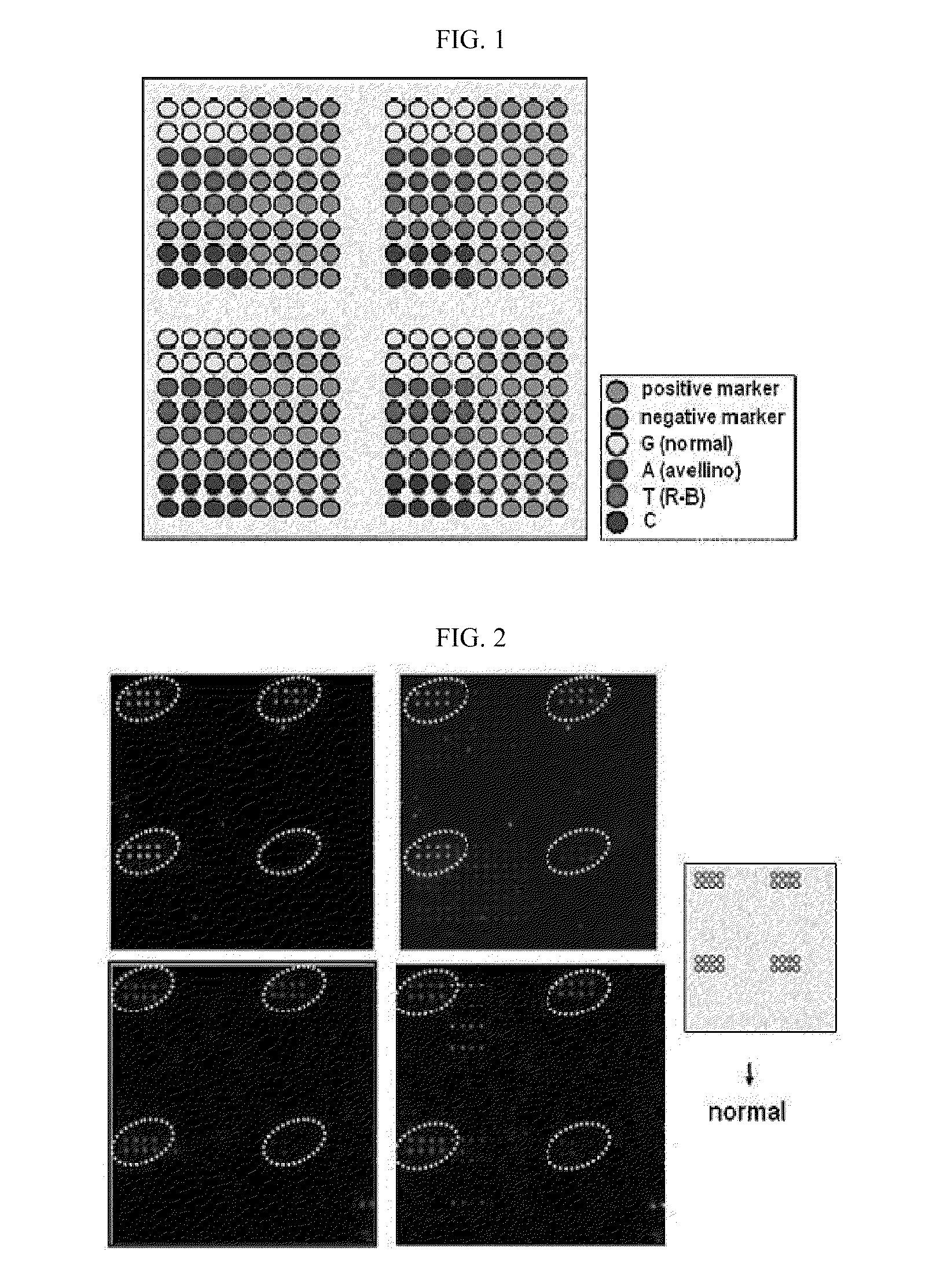
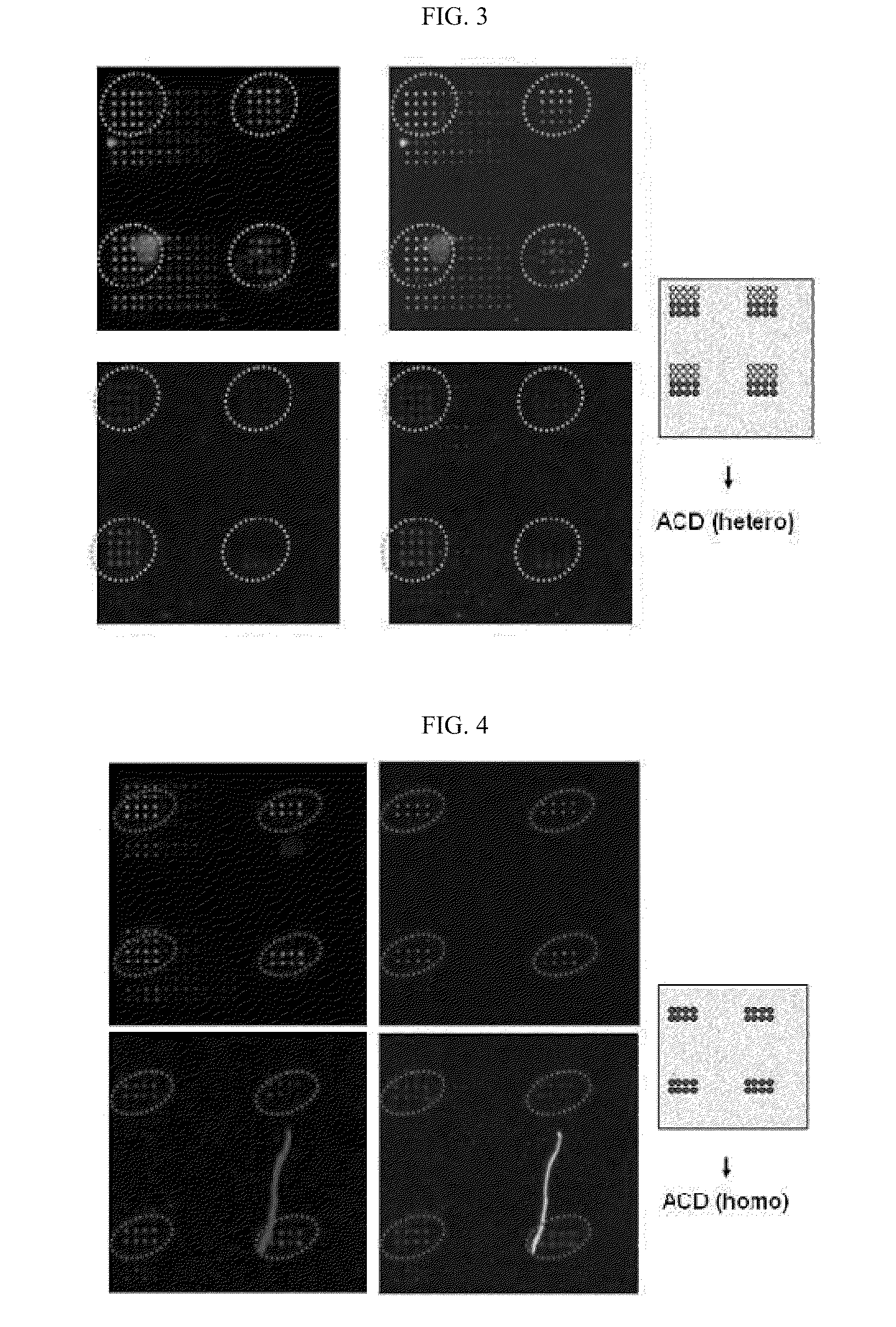
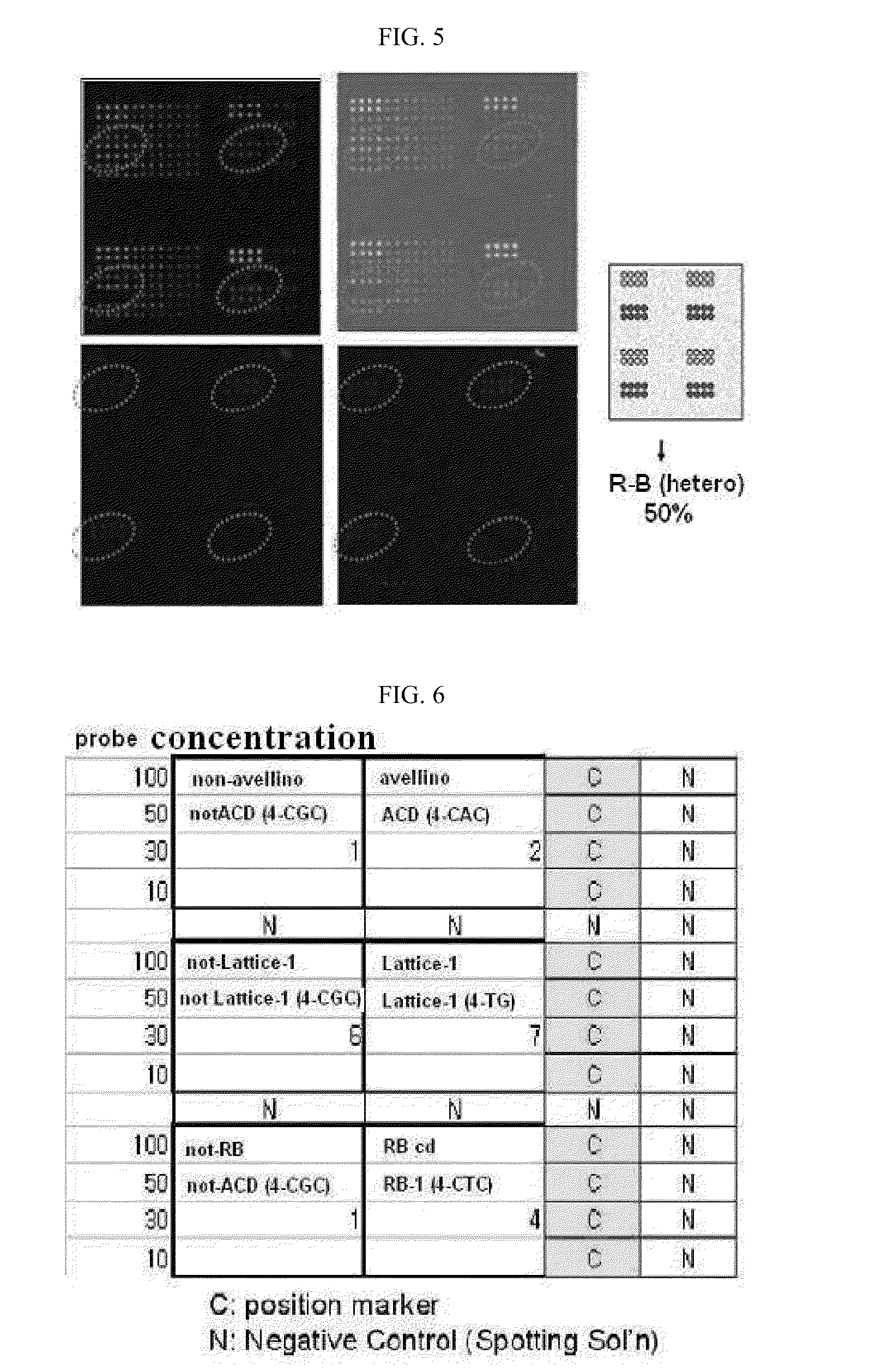
DNA chip for diagnosis of corneal dystrophy
InactiveCN101374850ASugar derivativesMicrobiological testing/measurementCornea dystrophySurgery procedure
The present invention relates to oligonucleotides for diagnosis of corneal dystrophy. More particularly, the present invention relates to oligonucleotides for detecting mutation of BIGH3 gene for diagnosis of corneal dystrophy including Avelllino corneal dystrophy, which must be precisely diagnosed before vision correction surgery, and a DNA chip for diagnosis of corneal dystrophy, which has the oligonucleotides fixed thereon. According to the present invention, conventional microscopic diagnosis of corneal dystrophy can be replaced with a precise genetic method, which prevents a patient with corneal dystrophy from losing eyesight by eyesight correction surgery after erroneous diagnosis.
Owner:MEDIGENES CO LTD
Thin film bio valve device and its controlling apparatus
ActiveUS20100288949A1Easy to usePlug valvesOperating means/releasing devices for valvesLab-on-a-chipProtein chip
Owner:PRECISIONBIOSENSOR INC
DNA chip and detection method and production method thereof
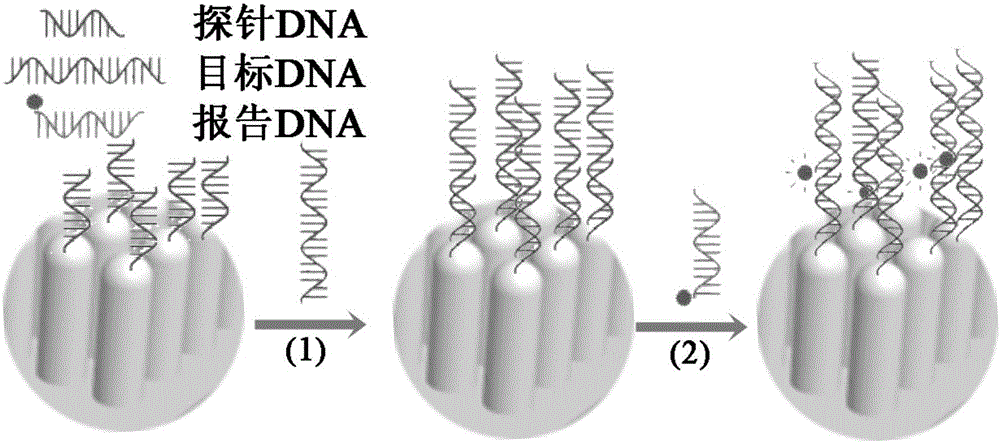
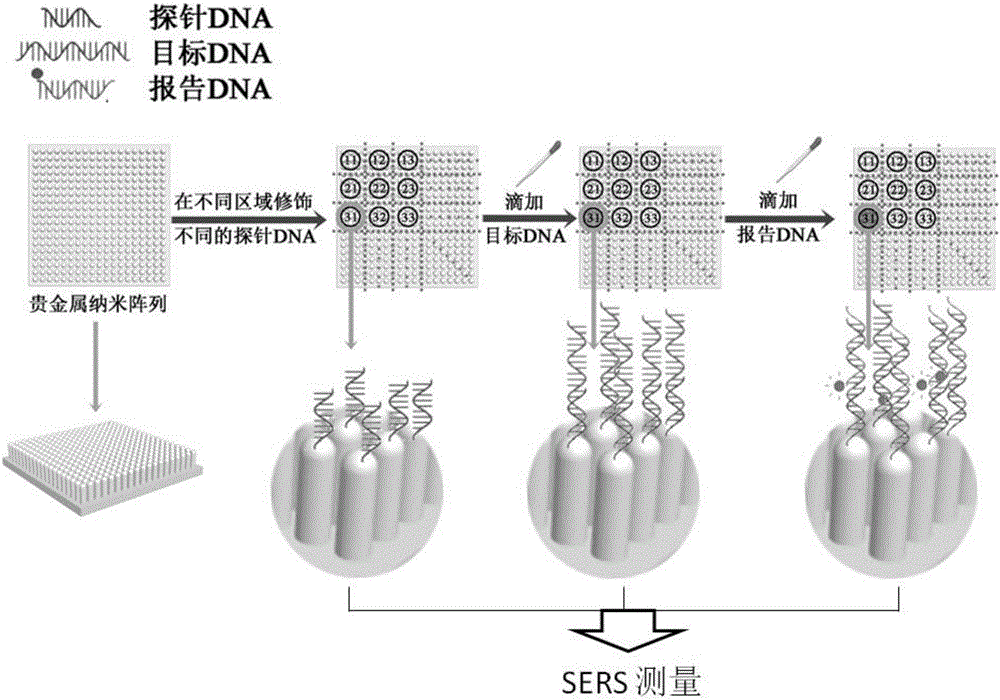
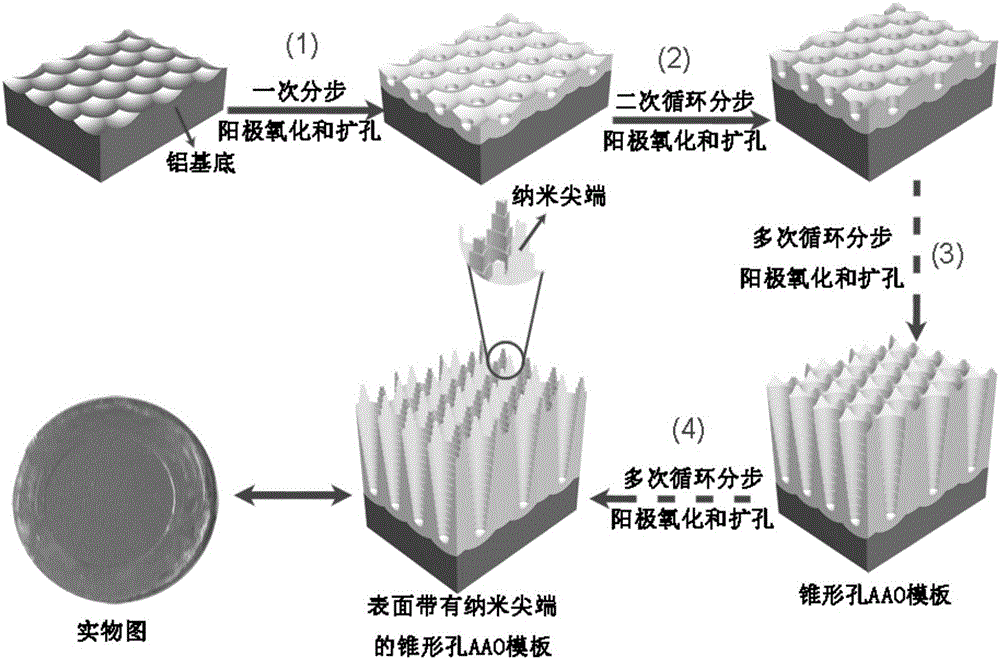
ActiveCN105019033AEasy to detectNucleotide librariesMicrobiological testing/measurementSurface-enhanced Raman spectroscopyGene
The present invention discloses a DNA chip and a detection method and a production method thereof. The substrate of the DNA chip is a SERS substrate taking a noble metal nano array as a substrate; the noble metal nano array is divided into a plurality of areas, the surface of each area is modified and connected with a plurality of different DNA probes, so that the DNA chip contains different probe DNA, the probe DNA is used to detect a target DNA to produce a SERS signal, and based on the frequency change and magnitude difference of the SERS signal, the identification and quantitative detection of the target DNA can be achieved. The DNA chip is designed by use of the noble metal nano array and surface enhanced raman scattering for achievement of detection and analysis of the target DNA and genes. The present invention also discloses a detection method and a production method of the DNA chip.
Owner:HEFEI INSTITUTES OF PHYSICAL SCIENCE - CHINESE ACAD OF SCI
DNA chip for diagnosis of corneal dystrophy
InactiveUS20090305394A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCornea dystrophyOligonucleotide
The present invention relates to oligonucleotides for diagnosis of corneal dystrophy. More particularly, the present invention relates to oligonucleotides for detecting mutation of BIGH3 gene for diagnosis or corneal dystrophy including Avellino corneal dystrophy, which must be precisely diagnosed before vision correction surgery, and a DNA chip for diagnosis of corneal dystrophy, which has the oligonucleotides fixed thereon. According to the present invention, conventional microscopic diagnosis of corneal dystrophy can be replaced with a precise genetic method, which prevents a patient with corneal dystrophy from losing eyesight by eyesight correction surgery after erroneous diagnosis.
Owner:MEDIGENES CO LTD
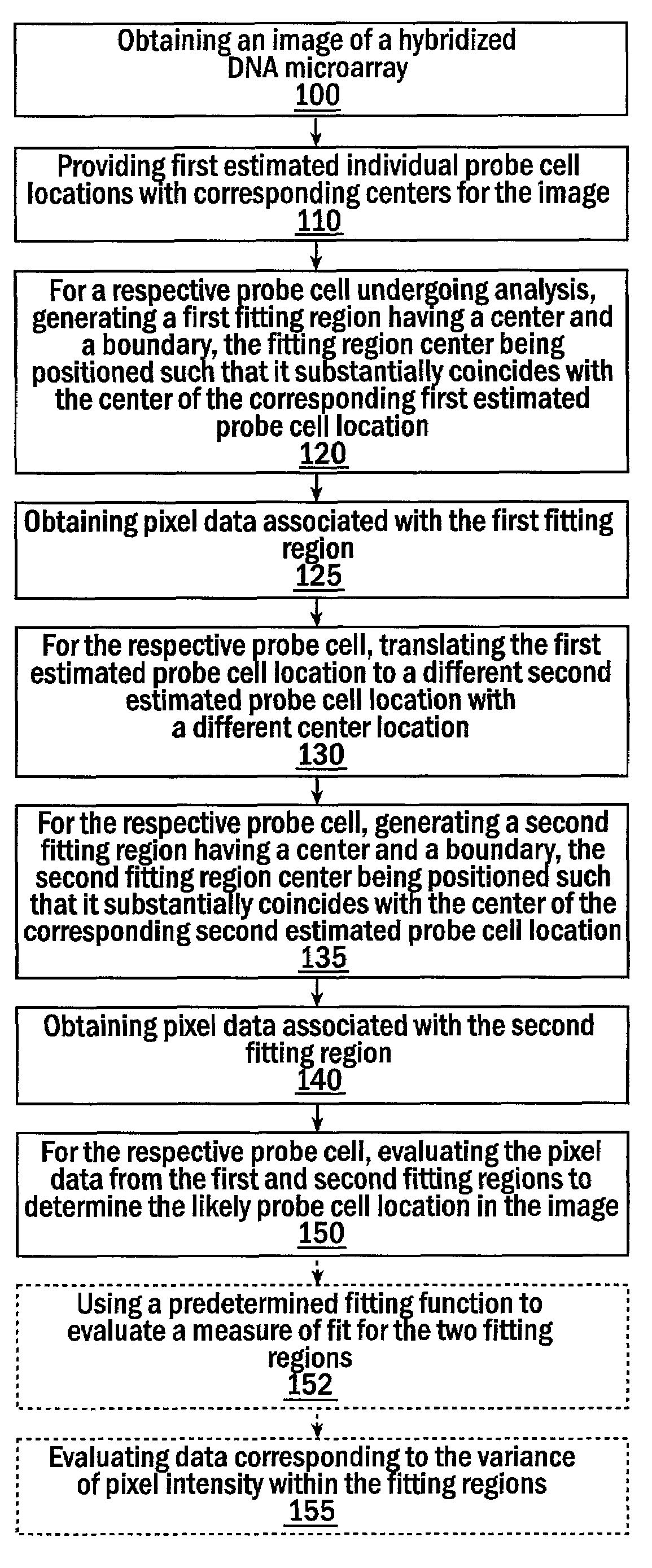
Methods for estimating probe cell locations in high-density synthetic DNA microarrays
InactiveUS6993173B2Easy to optimizeAccurate attributionImage enhancementImage analysisHigh-Density MicroarrayHigh density
Methods, systems, and computer program products for estimating the location of a probe cell in an image of a high-density microarray DNA chip interrogate a plurality of different closely spaced estimated locations to identify the most likely estimated location of the probe cell in the image.
Owner:DUKE UNIV
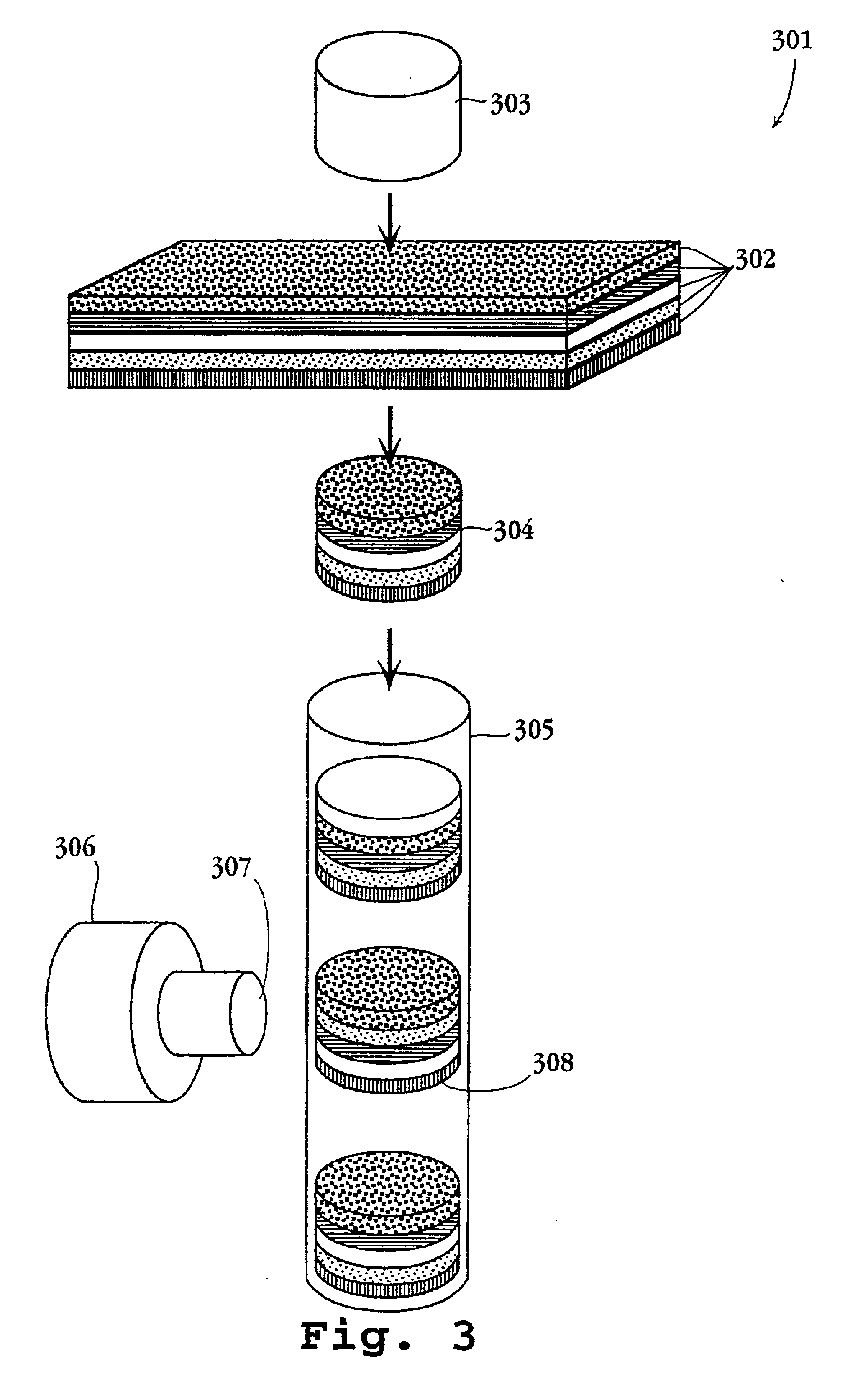
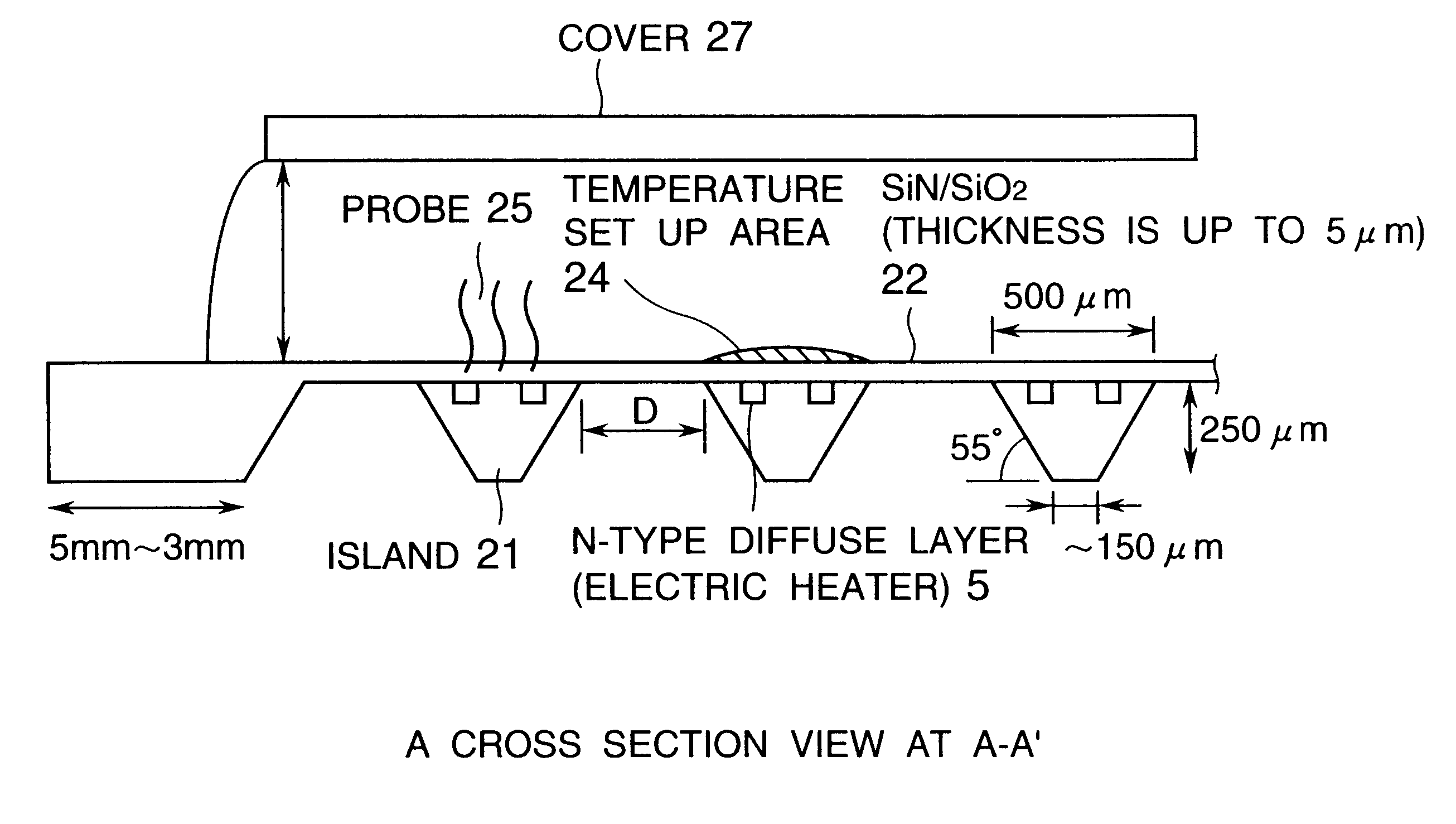
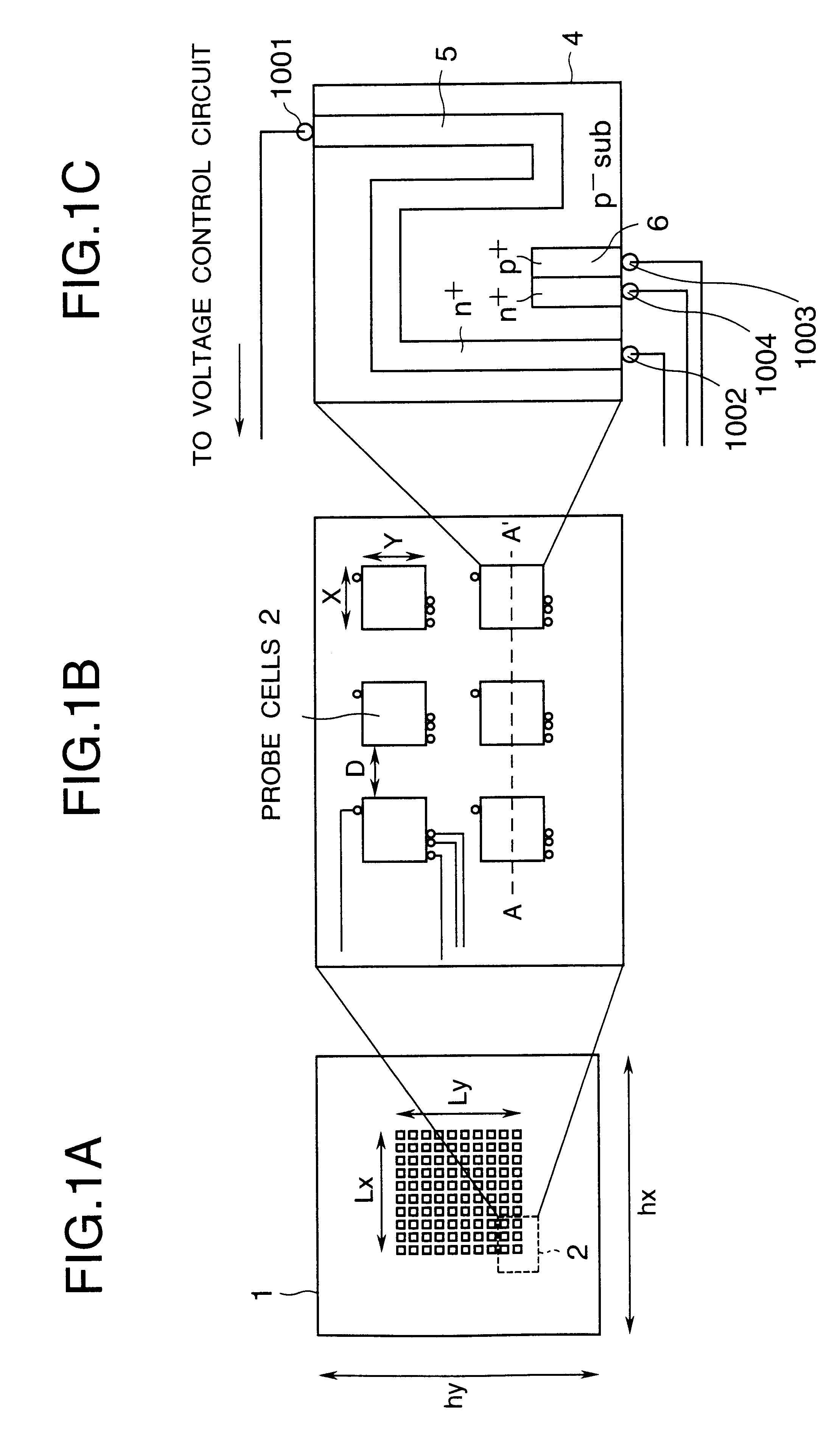
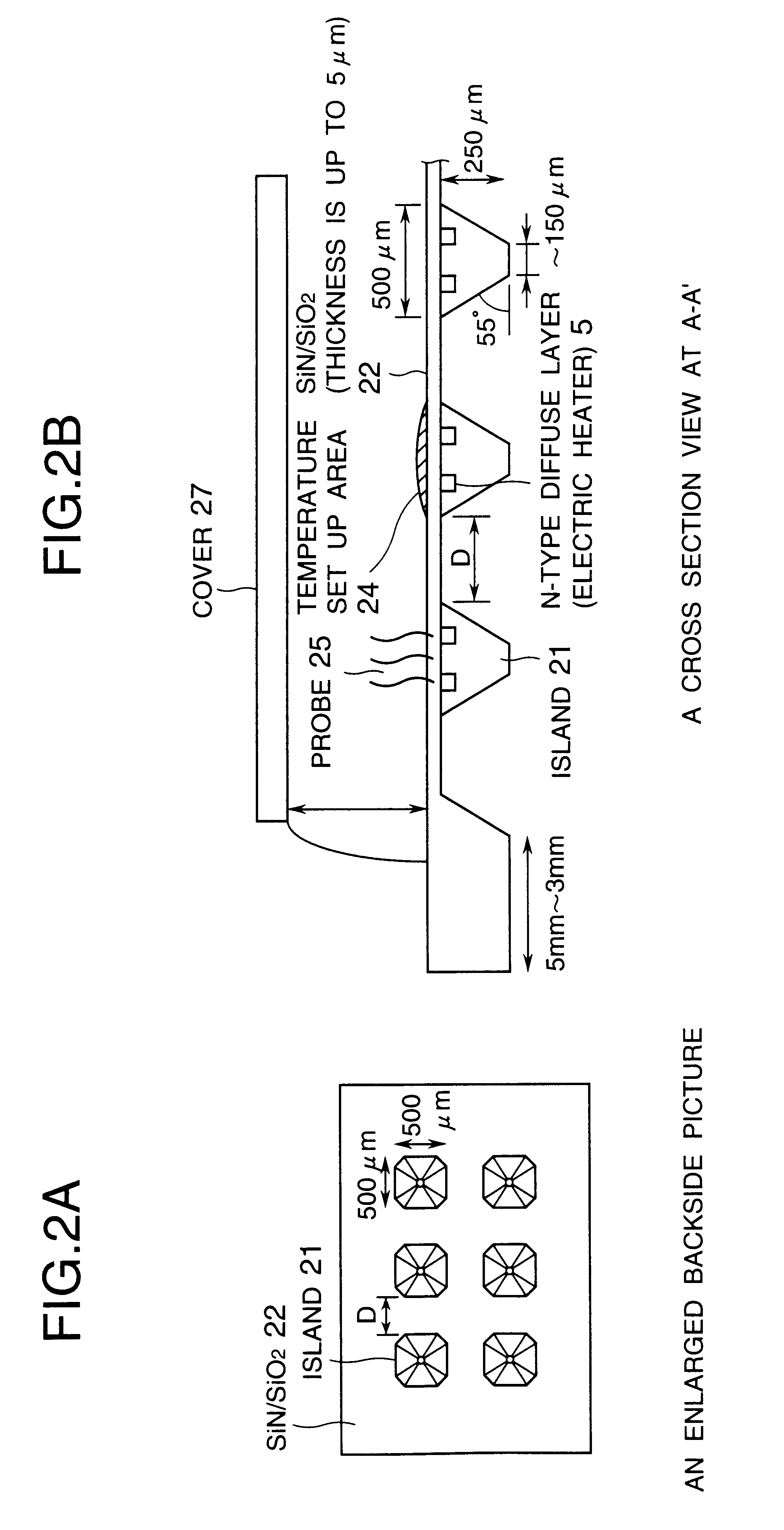
Advanced thermal gradient DNA chip (ATGC), the substrate for ATGC, method for manufacturing for ATGC, method and apparatus for biochemical reaction, and storage medium
A biochemical reaction detection chip capable of controlling the temperature for biochemical reactions including hybridizations and its substrate. The function of the chip is performed by comprising a plurality of islands of a heat conducting material on the membrane of the substrate, the islands being spaced from each other and individually provided with temperature controllers, and the probes immobilized on the substrate.
Owner:HITACHI LTD
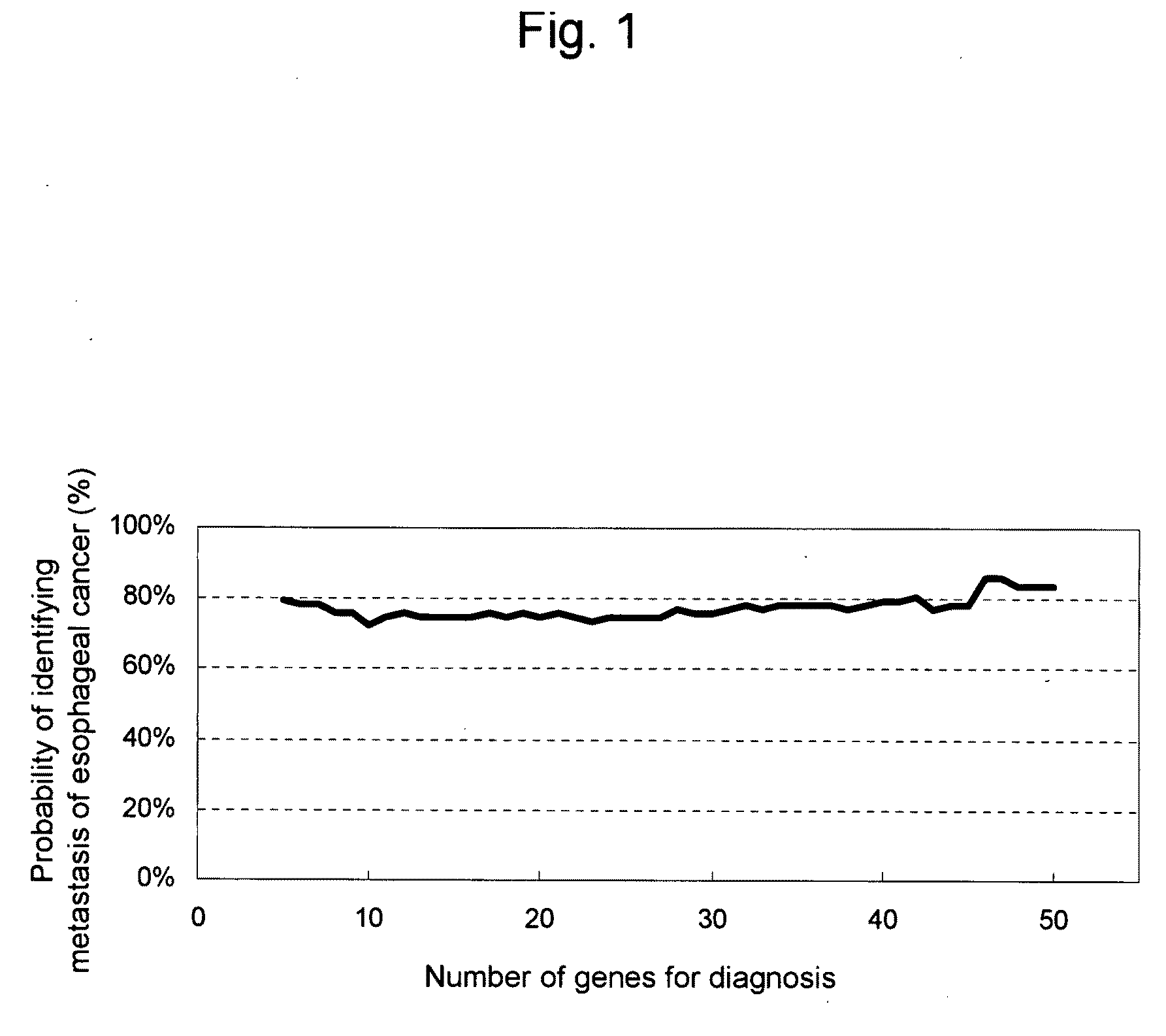
Composition and method for diagnosing esophageal cancer and metastasis of esophageal cancer
InactiveUS20090270267A1Simple and highly and and simple methodEasy to detectSugar derivativesMicrobiological testing/measurementLymphatic SpreadEsophageal cancer
This invention relates to a composition, kit, or DNA chip comprising polynucleotides and antibodies as probes for detecting, determining, or predicting the presence or metastasis of esophageal cancer, and to a method for detecting, determining, or predicting the presence or metastasis of esophageal cancer using the same.
Owner:TORAY IND INC +1
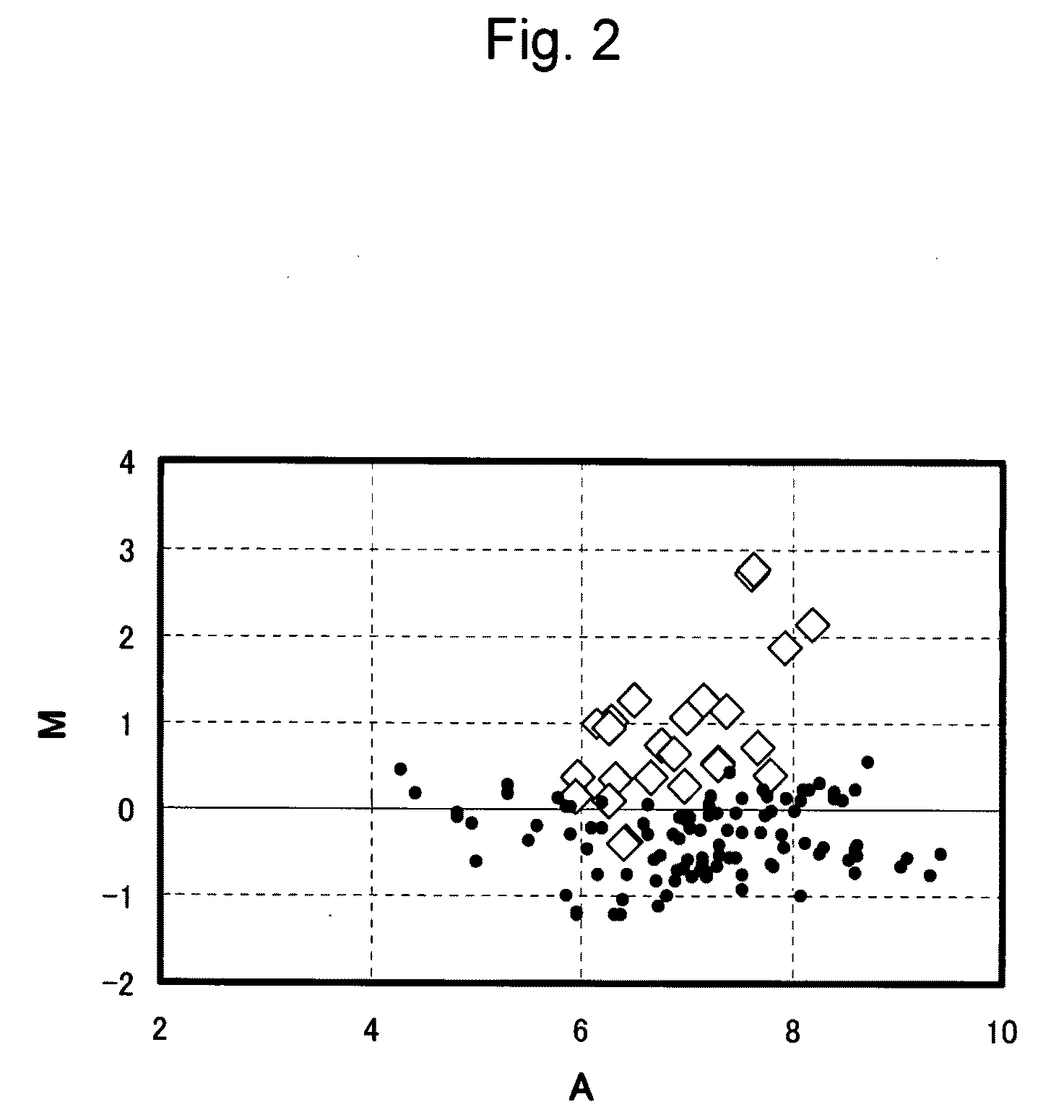
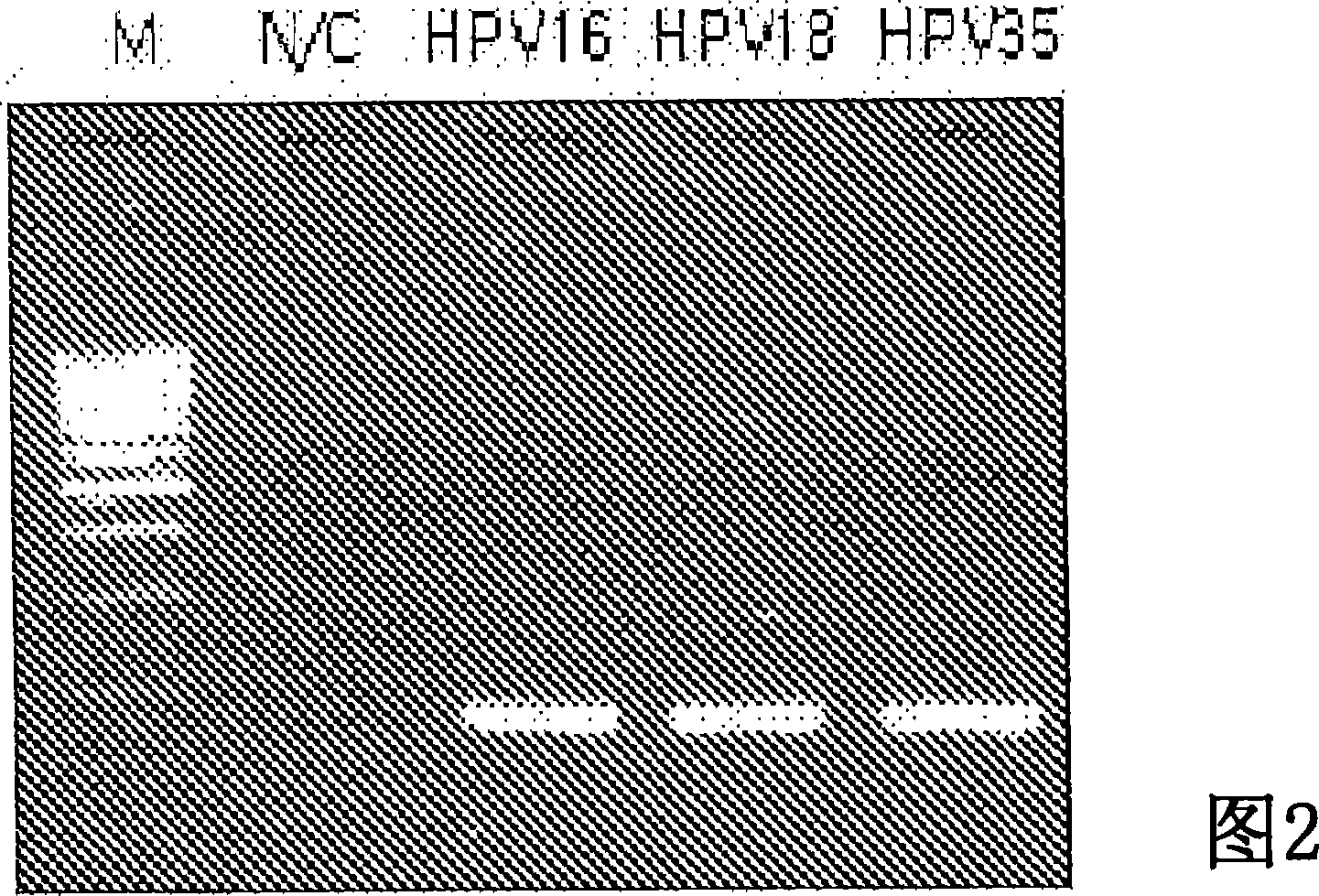
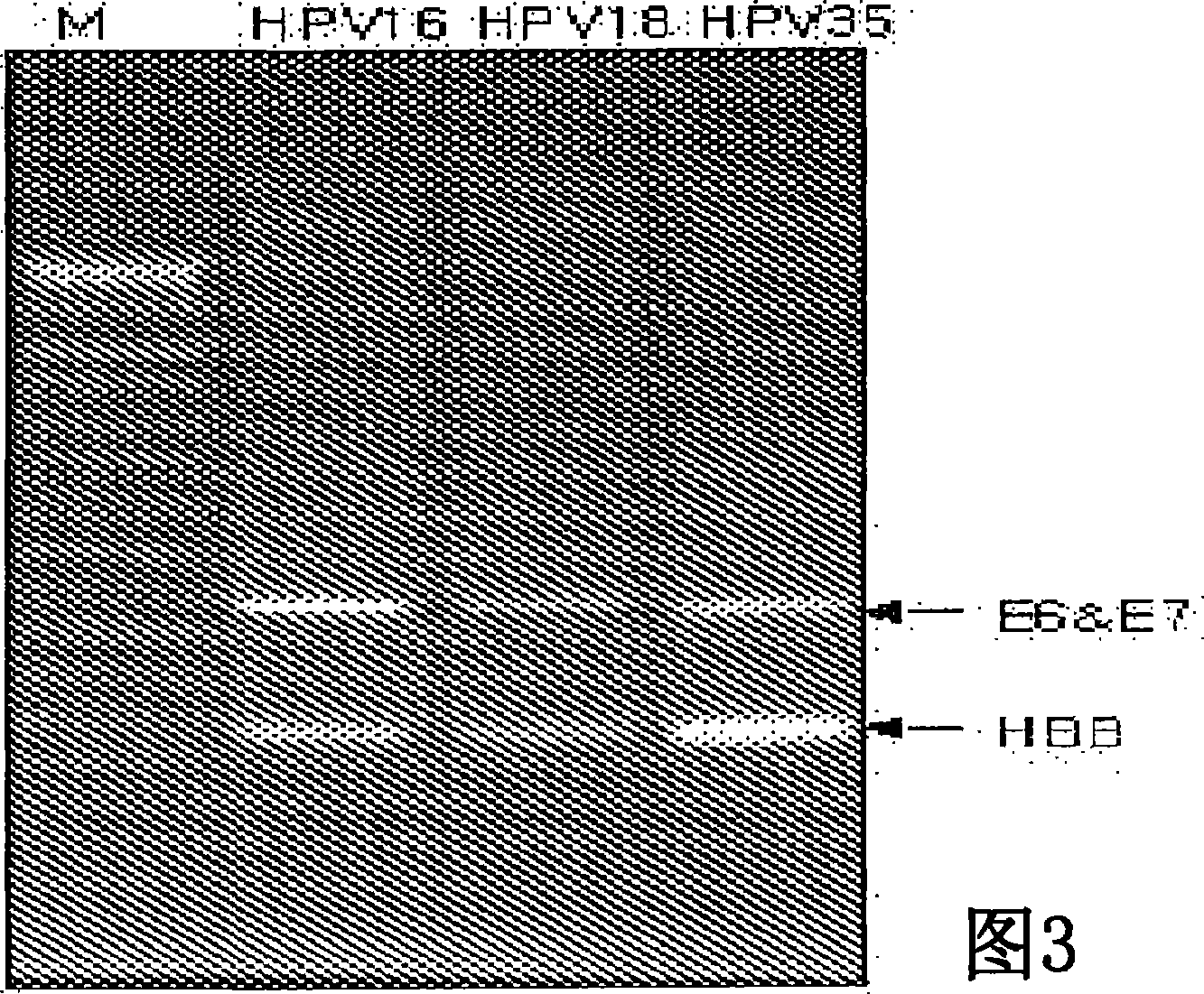
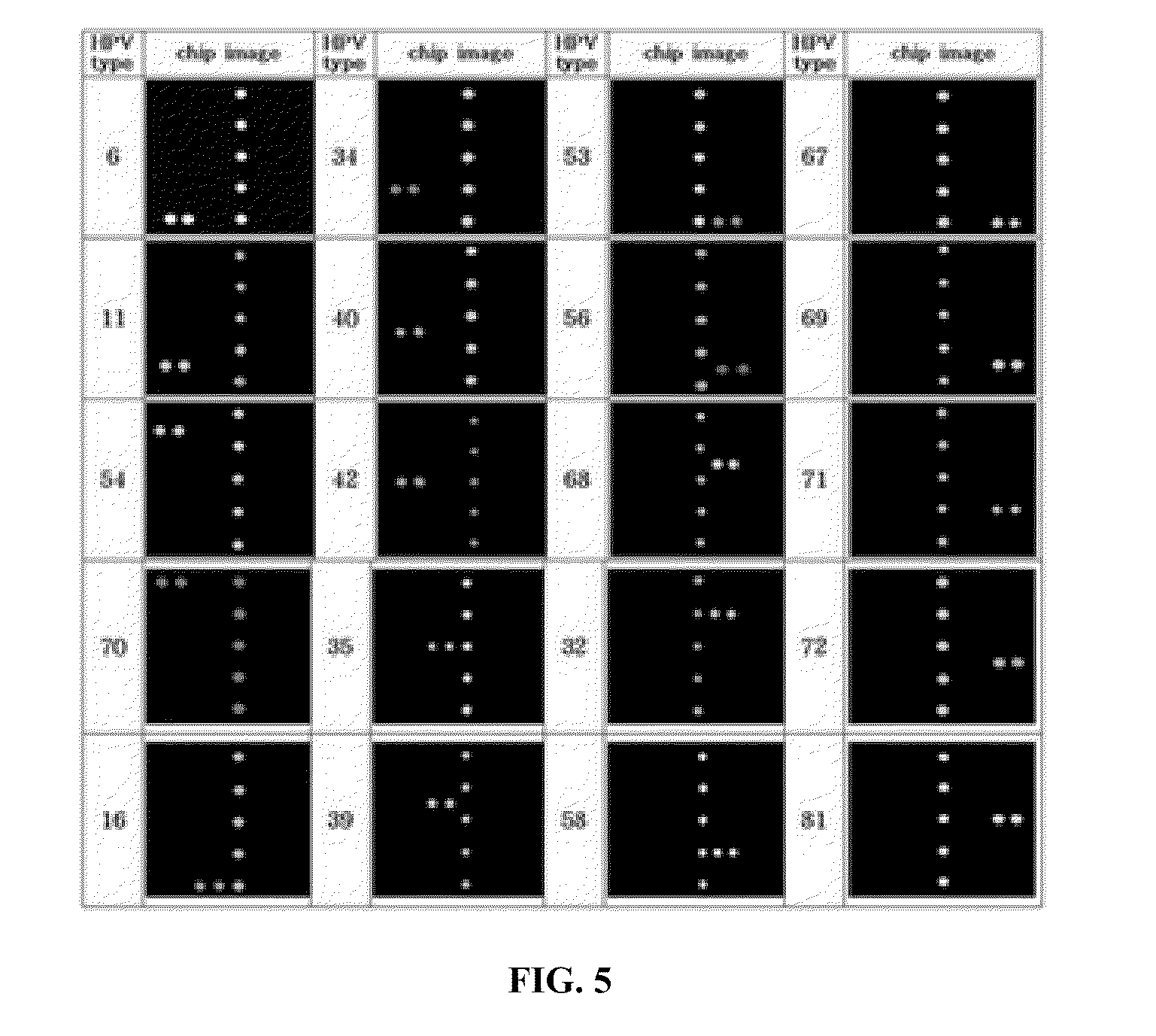
Probe of human papillomavirus and DNA chip comprising the same
ActiveUS7670774B2High selectivityHigh sensitivityBioreactor/fermenter combinationsBiological substance pretreatmentsHuman papillomavirusBiology
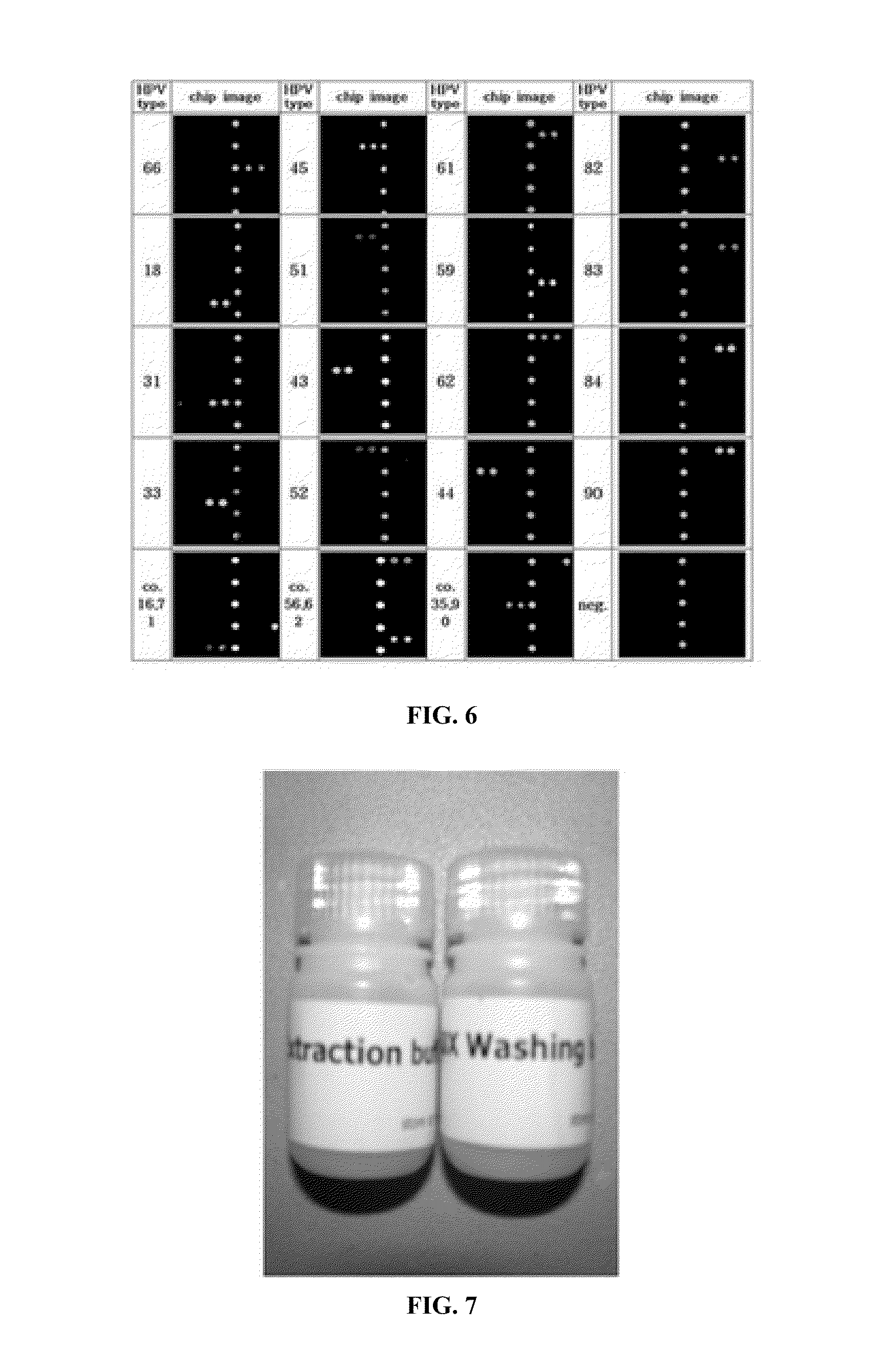
Oligonucleotide probes for analyzing 40 types of HPV were synthesized, and DNA chips were produced by using the oligonucleotide probes. The synthesis of the oligonucleotide probes is based on clones of L1 and E6 / E7 genes of 35 types of HPV obtained from cervical cell specimens from 4,898 Korean adult women and tissue specimens from 68 cervical cancer cases in addition to information based on American and European cases. The DNA chips can analyze the 40 types of HPV found in cervical, diagnose complex infection by at least one type of HPV, and have excellent diagnostic sensitivity and specificity on HPV genetic type up to 100% and reproducibility. Also, the DAN chips are superior to conventional analytic method, and very economical, since they can analyze numerous specimens in shortest time. Accordingly, the DNA chips are useful for predicting cervical cancer and precancerous lesion.
Owner:GOODGENE
New system and kit for screening and diagnosing phenylketonuria
ActiveCN105755109ALow costImprove throughputMicrobiological testing/measurementGenes mutationImage resolution
The invention discloses a new system for screening and diagnosing phenylketonuria.41 common phenylalanine hydroxylase gene mutation sites and 4 most common 6-pyruvoyl tetrahydrobiopterin synthetase (PTPS) gene (PTS) mutation sites are screened to form the gene mutation detection system of phenylketonuria.The system is low in cost and high in throughput.Compared with the prior art, the system has the advantages that most mutation sites can be detected in one system, and 45 mutation sites can be detected; the comprehensive cost is the lowest, and the total cost of the system is lower than built methods using the DNA chip technology, the 'molecular switch' technology, the high-resolution melting curve method and the SNaPShot technology to detect PAH gene mutation; most mutation sites can be detected, the number of the detected mutation sites is twice as the number of the detected PAH gene mutation sites of the high-resolution melting curve method built by Xiamen University, the number of the detected mutation sites is 4 times as the number of the detected mutation sites of the DNA chip technology and the SNaPShot technology, and the number of the detected mutation sites is 6 times as the number of the detected mutation sites of the 'molecular switch' technology.
Owner:SUZHOU MUNICIPAL HOSPITAL +1
Oligonucleotide sequences and DNA chip for identifying filamentous microorganisms and the identification method thereof
Owner:IND TECH RES INST
Liquid barcode and liquid barcode reader
InactiveUS20100213255A1Improve reliabilityIncrease in information amountMaterial nanotechnologyDiagnostics using lightCredit cardLab-on-a-chip
The inventive concept relates to a product authentication and identification device using a liquid barcode formed by liquid bars stored in liquid chambers. For example, according to the inventive concept liquid barcodes and a liquid barcode reader for reading liquid barcodes used for product authentication and identification of thin-film chemical analyzers such as a lab-on-a-disc and a bio disc in which bio chips such as a lab-on-a-chip, a protein chip, and a DNA chip for diagnosing and detecting a small amount of material in a fluid are integrated, a credit card, or other products can be provided.
Owner:SAMSUNG ELECTRONICS CO LTD
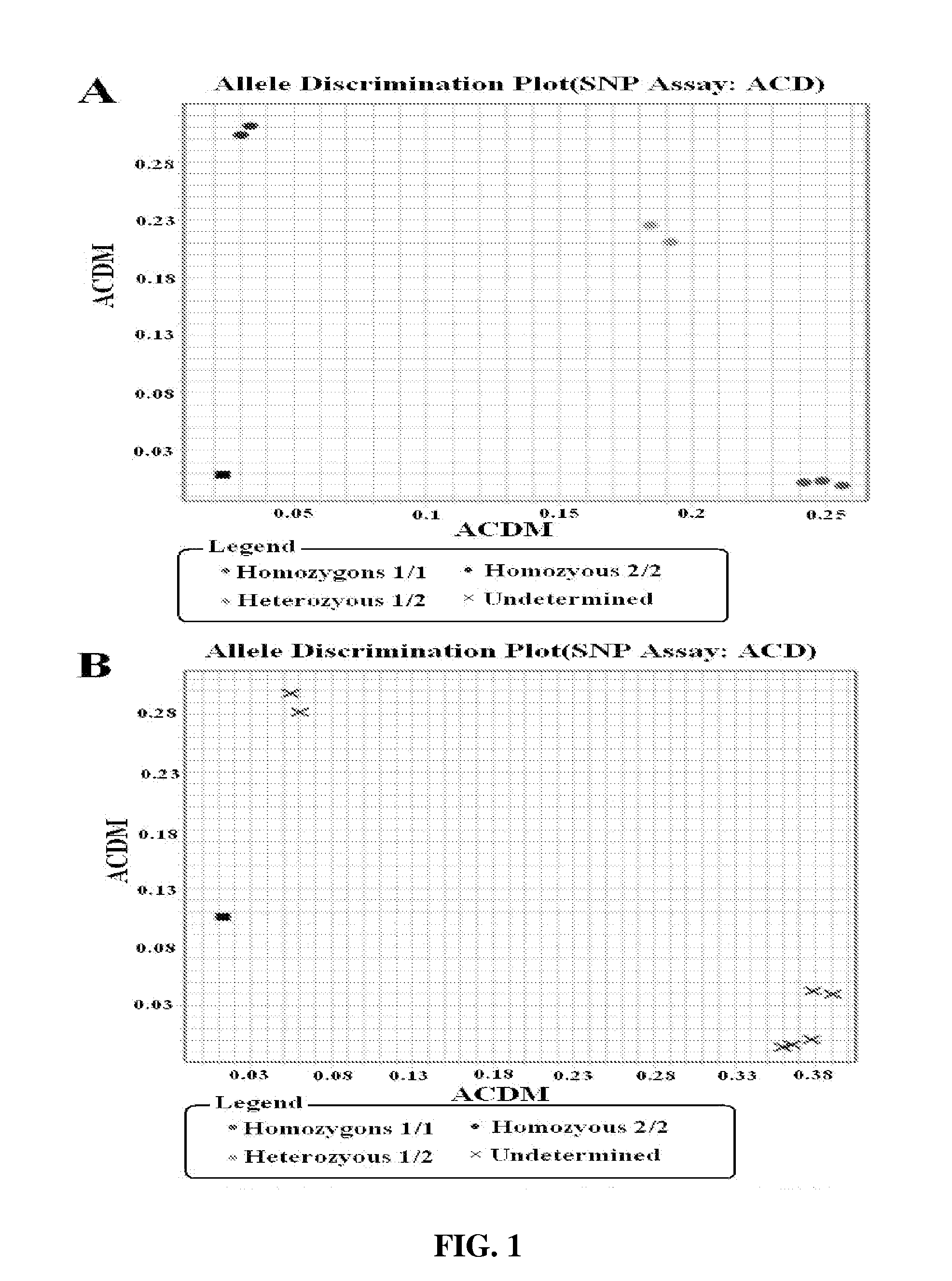
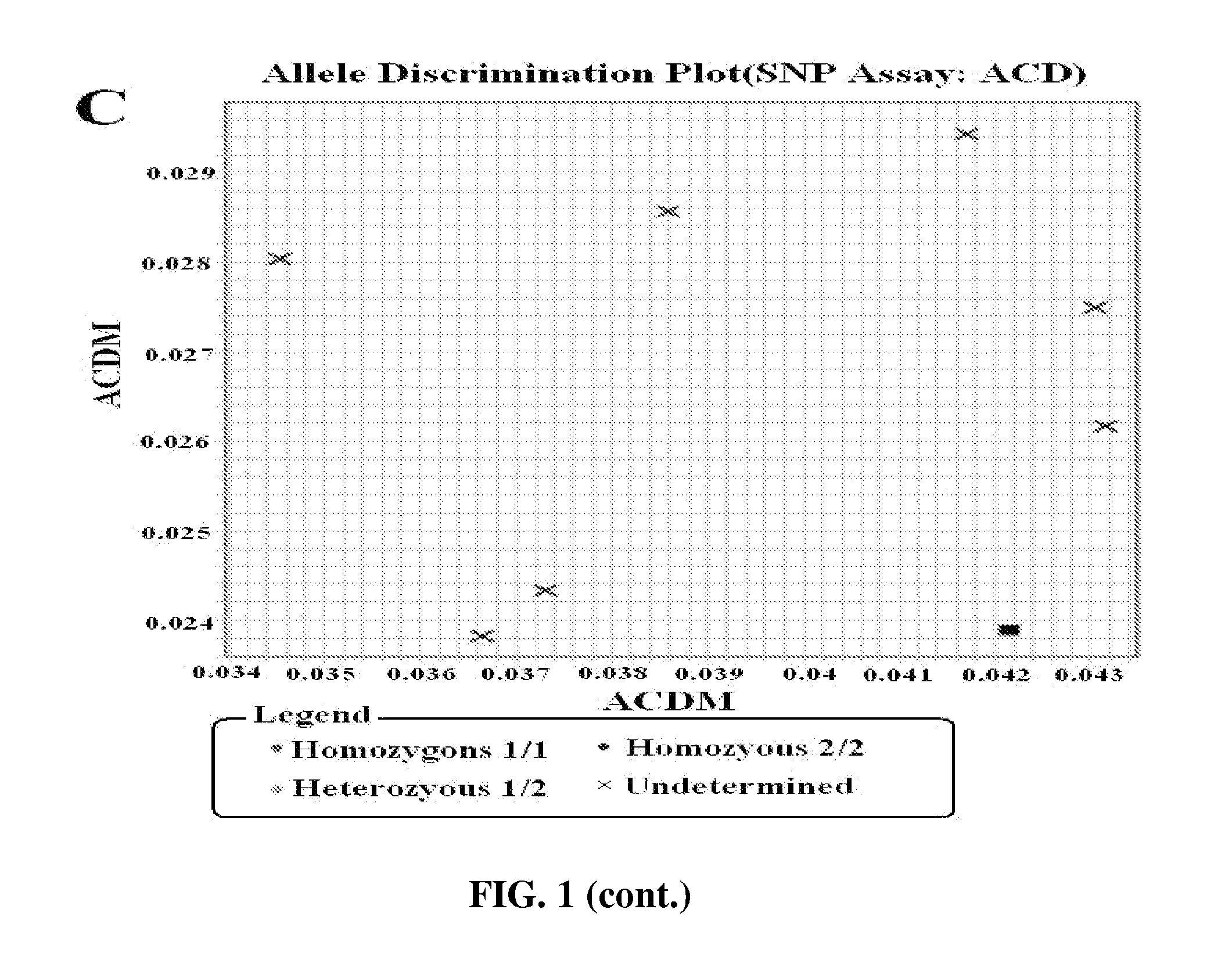
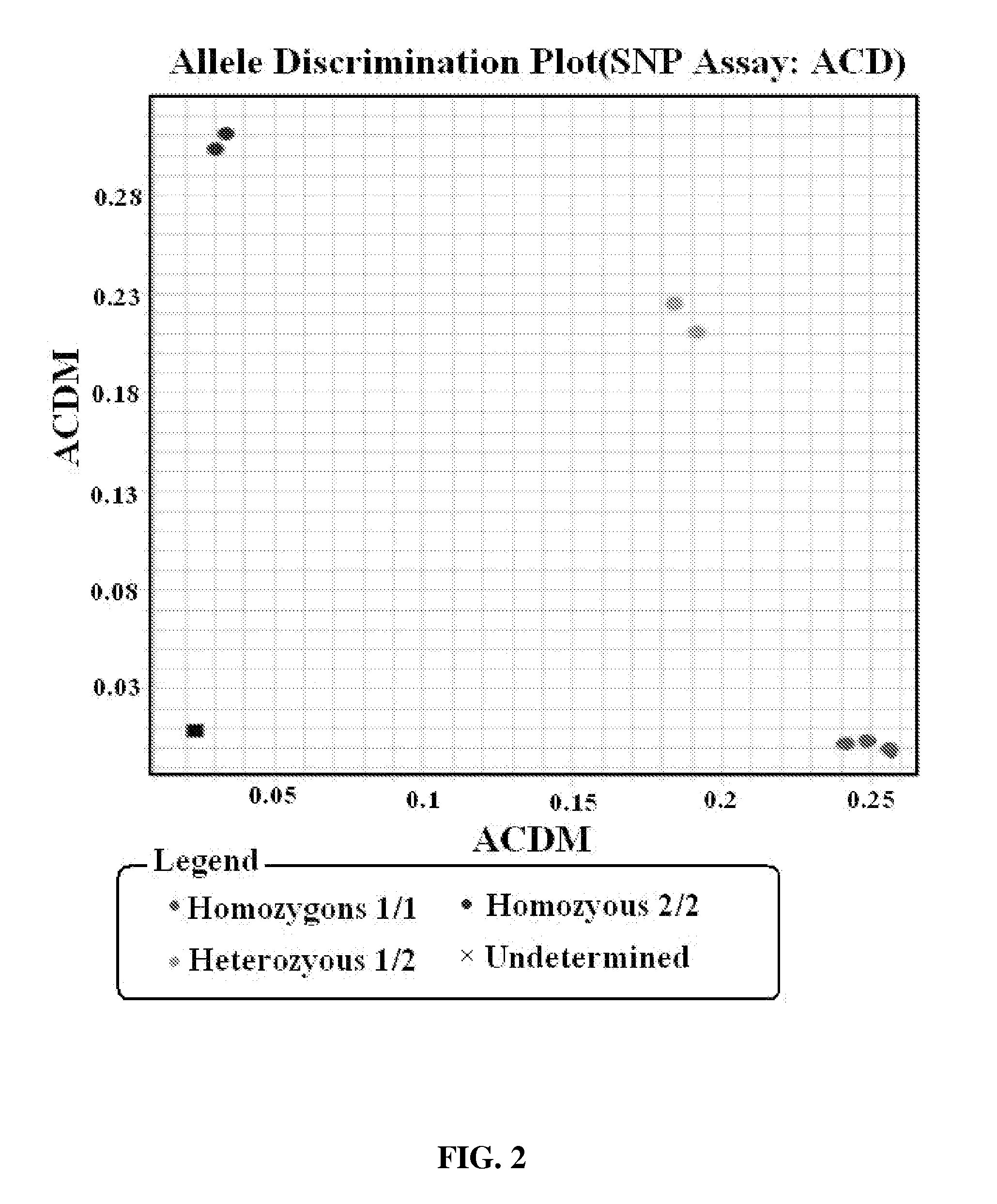
Methods for identification of alleles
ActiveUS20100041563A1Low homologyMicrobiological testing/measurementLibrary screeningPolymerase LGenomic DNA
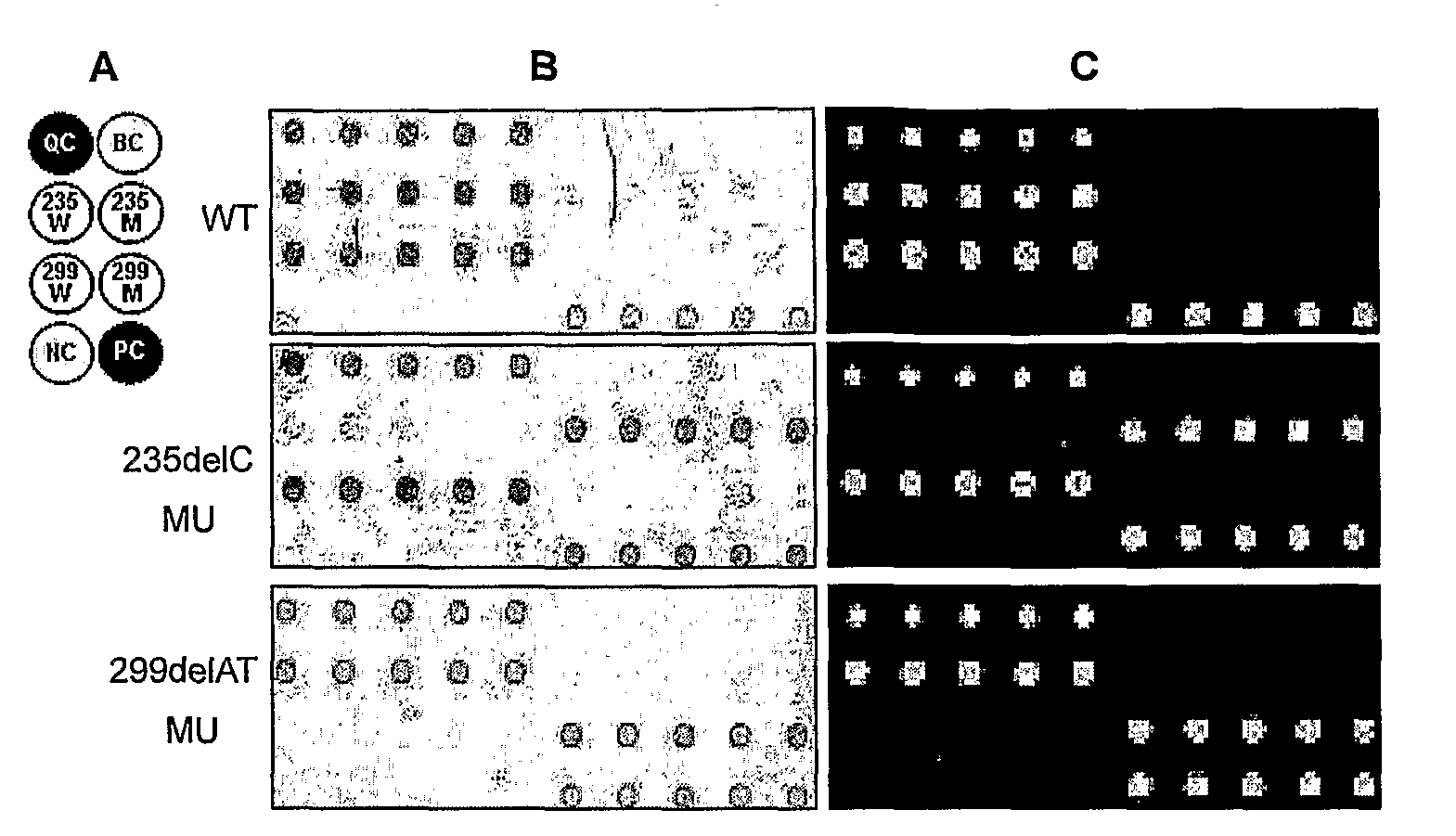
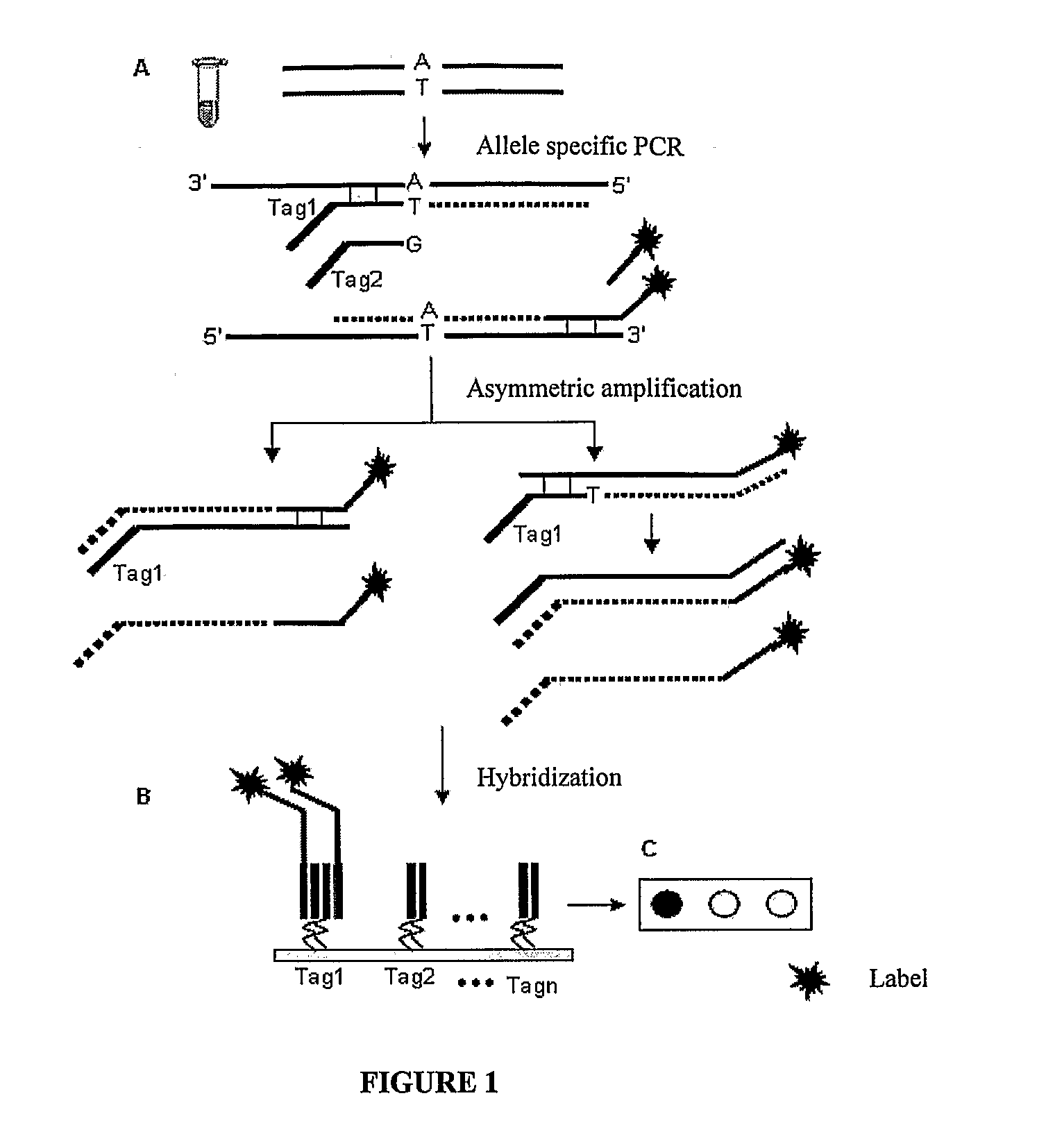
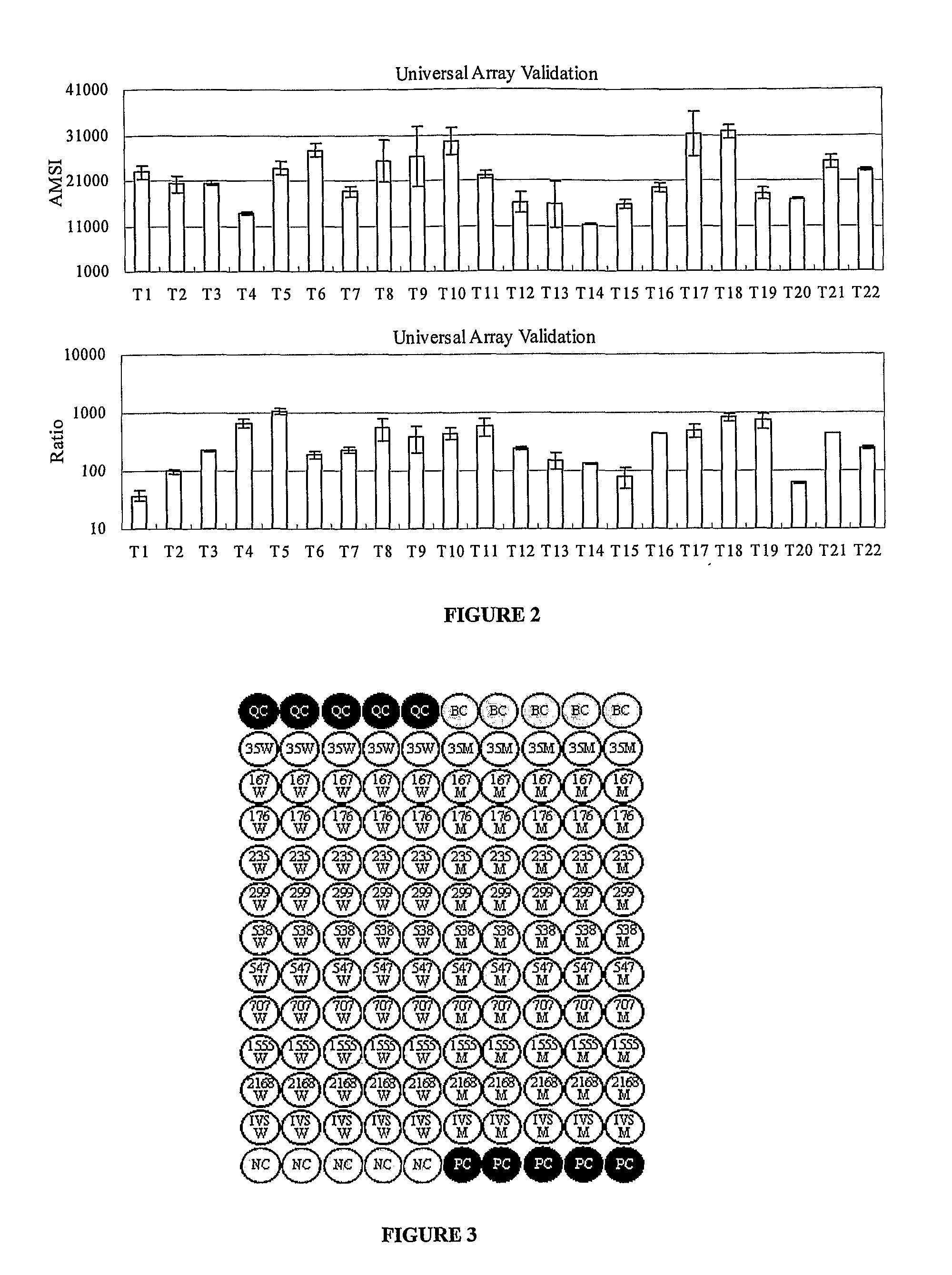
The invention provides a method for identification of alleles. In this method, genomic DNA is used as target. Multiple allele-specific PCR amplification are carried out with a group of primers comprising one or more allele-specific primers for a target gene, a universal primer, and a common primer; and a DNA polymerase without 5′ to 3′ exonuclease activity. The PCR products are hybridized with tag probes immobilized on a DNA chip. Results are determined based on the signal intensity and the position of the probe immobilized on the array. Each allele-specific primer comprises a unique tag sequence at the 5′ end. Each tag probe immobilized on the DNA chip comprises a sequence identical to its corresponding tag sequence; and each tag probe hybridizes only with the complementary sequence in the PCR amplification product.
Owner:CAPITALBIO CORP +1
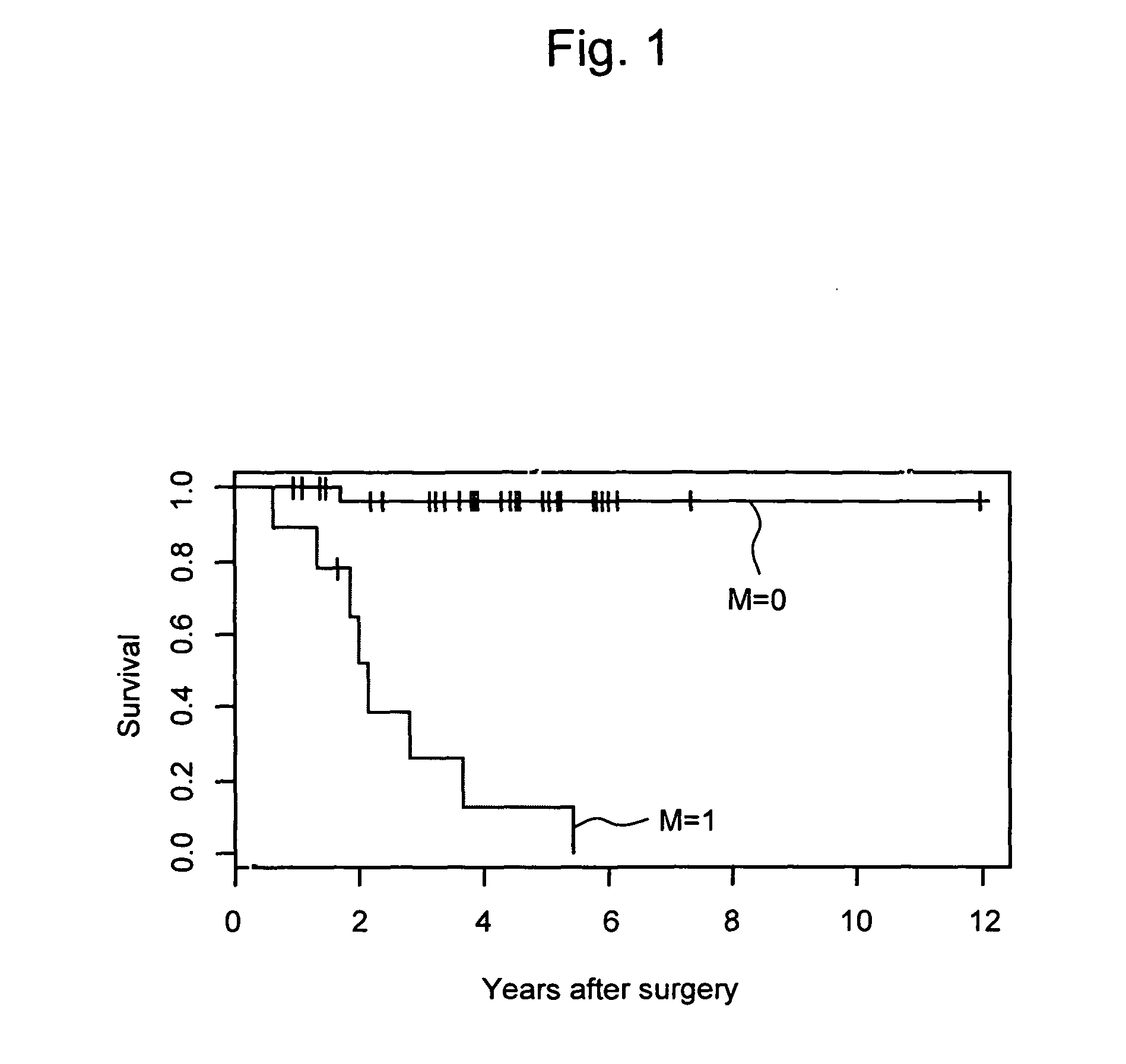
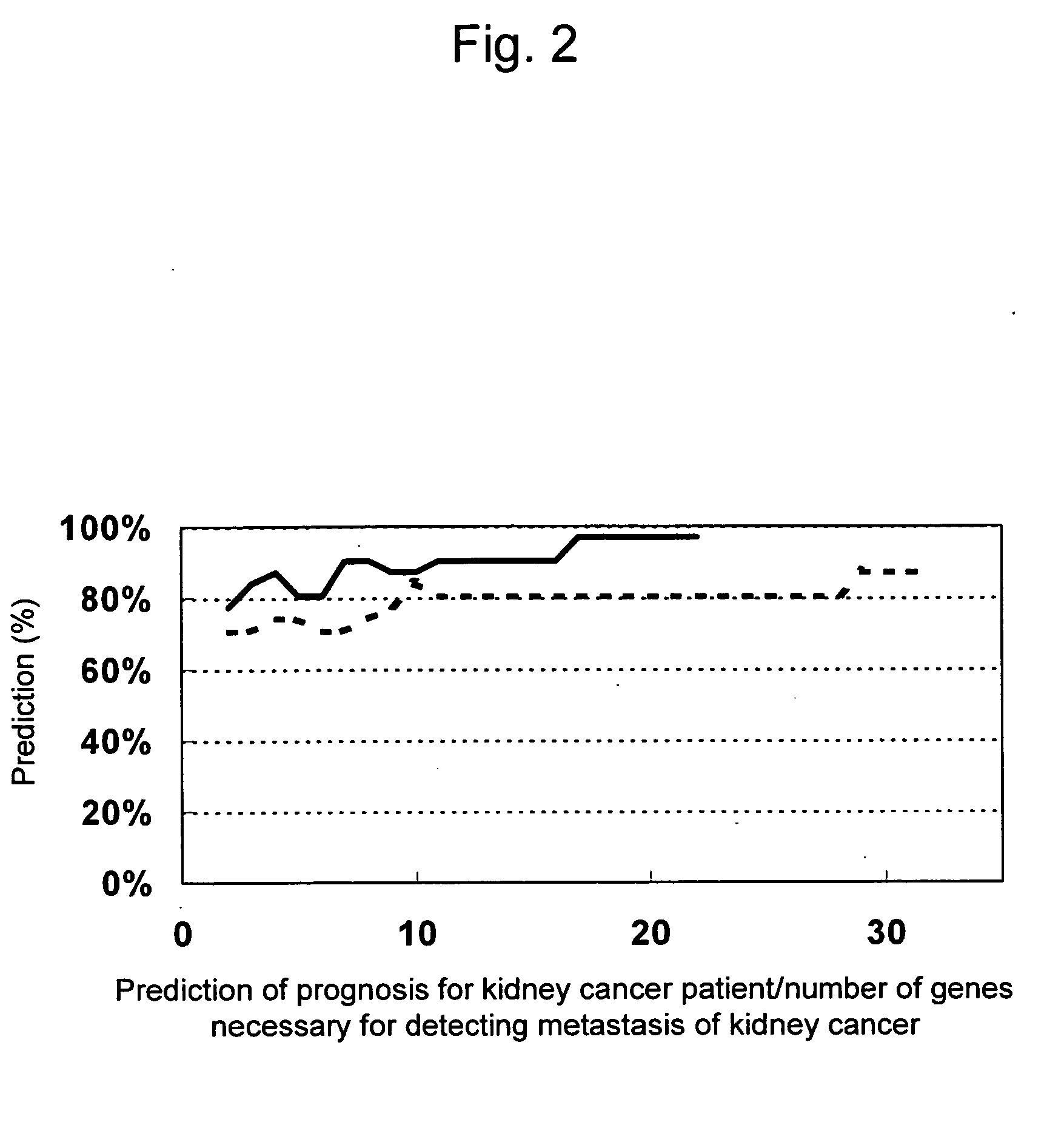
Composition and method for diagnosing kidney cancer and for predicting prognosis for kidney cancer patient
InactiveUS20100152055A1Strong specificitySimple and rapid methodNucleotide librariesMicrobiological testing/measurementLymphatic SpreadKidney cancer
This invention relates to a composition, kit, DNA chip, and use thereof for detecting, diagnosing, and predicting metastasis of kidney cancer and / or for predicting the prognosis for kidney cancer, comprising one or a plurality of polynucleotides selected from the group consisting of polynucleotides, mutants thereof or fragments thereof, the expression levels of which vary in kidney cancer cells from a patient with a poor prognosis when compared with that in kidney cancer cells from a patient with a good prognosis; or antibodies or fragments thereof that bind specifically to polypeptides, mutants thereof or fragments thereof, the expression levels of which vary in the similar manner.
Owner:TORAY IND INC +1
Primers for diagnosing avellino corneal dystrophy
InactiveUS20120077200A1Efficiently and accurately diagnosingSugar derivativesMicrobiological testing/measurementReal-Time PCRsCornea dystrophy
The present invention relates to a real-time PCR primer pair and probe for diagnosing Avellino corneal dystrophy, and more particularly to a real-time PCR primer pair and probe for diagnosing Avellino corneal dystrophy, which can accurately diagnose the presence or absence of a mutation in exon 4 of BIGH3 gene, which is responsible for Avellino corneal dystrophy. The use of the primer pair and probe according to the invention can diagnose Avellino corneal dystrophy in a more rapid and accurate manner than a conventional method that uses a DNA chip or PCR.
Owner:AVELLINO
Probe of human papillomavirus and DNA chip comprising the same
InactiveCN101035896AHigh selectivityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationCervical cellsHuman papillomavirus
Oligonucleotide probes for analyzing 40 types of HPV were synthesized, and DNA chips were produced by using the oligonucleotide probes. The synthesis of the oligonucleotide probes is based on clones of Ll and E6 / E7 genes of 35 types of HPV obtained from cervical cell specimens from 4,898 Korean adult women and tissue specimens from 68 cervical cancer cases in addition to information based on American and European cases. The DNA chips can analyze the 40 types of HPV found in cervical, diagnose complex infection by at least one type of HPV, and have excellent diagnostic sensitivity and specificity on HPV genetic type up to 100% and reproducibility. Also, the DAN chips are superior to conventional analytic method, and very economical, since they can analyze numerous specimens in shortest time. Accordingly, the DNA chips are useful for predicting cervical cancer and precancerous lesion.
Owner:文宇哲
Primer, probe, DNA chip containing the same and method for detecting human papillomavirus and detection kit thereof
InactiveUS20100285485A1Strong specificityShorten the timeSugar derivativesMicrobiological testing/measurementHpv testingTrue positive rate
The present invention discloses a primer, a probe, a DNA chip, and a test kit for diagnosis and genotyping of Human Papilloma Virus (HPV) as well as a method for testing HPV genotype. According to the present invention, it is possible to screen HPV genotypes with high sensitivity and specificity, and to diagnose infection by multiple HPV types which was not possible in other conventional HPV testing methods
Owner:JUNG WOON WON +1
Kit for detecting alpha-thalassemia genes
Owner:亚能生物技术(深圳)有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com