Micro-array chip detection method for 10 on-melon diseases and used chip probe
A microarray chip and detection method technology, applied in biochemical equipment and methods, microbial measurement/testing, chemical library, etc., can solve problems such as low sensitivity, long separation cycle, affecting detection results, etc., and shorten the identification process , Accelerate the speed of port inspection and release, and the effect of accurate inspection and quarantine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0247] 2.3 Preparation of oligonucleotide chips
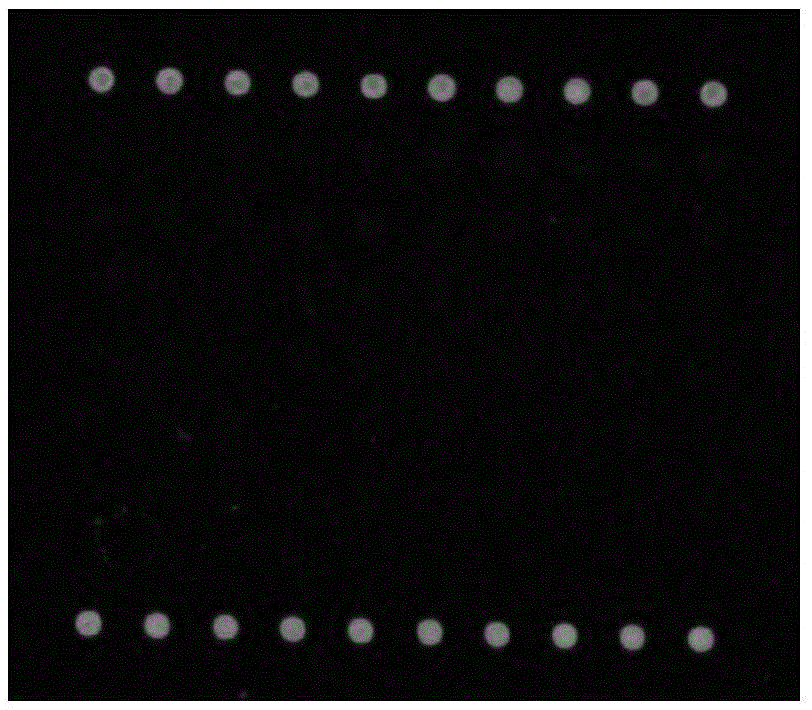
[0248] According to the chip array arrangement (as shown in Table 6), various detection and quality control probes were spotted on the aldehyde-based substrate, fixed for 24 hours, washed, and scanned. Pre-scan results such as figure 2 , the positive localization control sample spot is clear and uniform, the background of the chip is low, and no fluorescent signal appears at other probe sites, which is suitable for the subsequent hybridization of the experiment.
[0249] Table 6. Arrangement of gene chip probes
[0250]
[0251]
[0252] Note: The array is 9 rows and 10 columns, the upper and lower rows are DW positive positioning controls, and the other probes are repeated 5 times.
[0253] 2.4 Optimization of hybridization reaction system
[0254] 2.4.1 Hybridization time
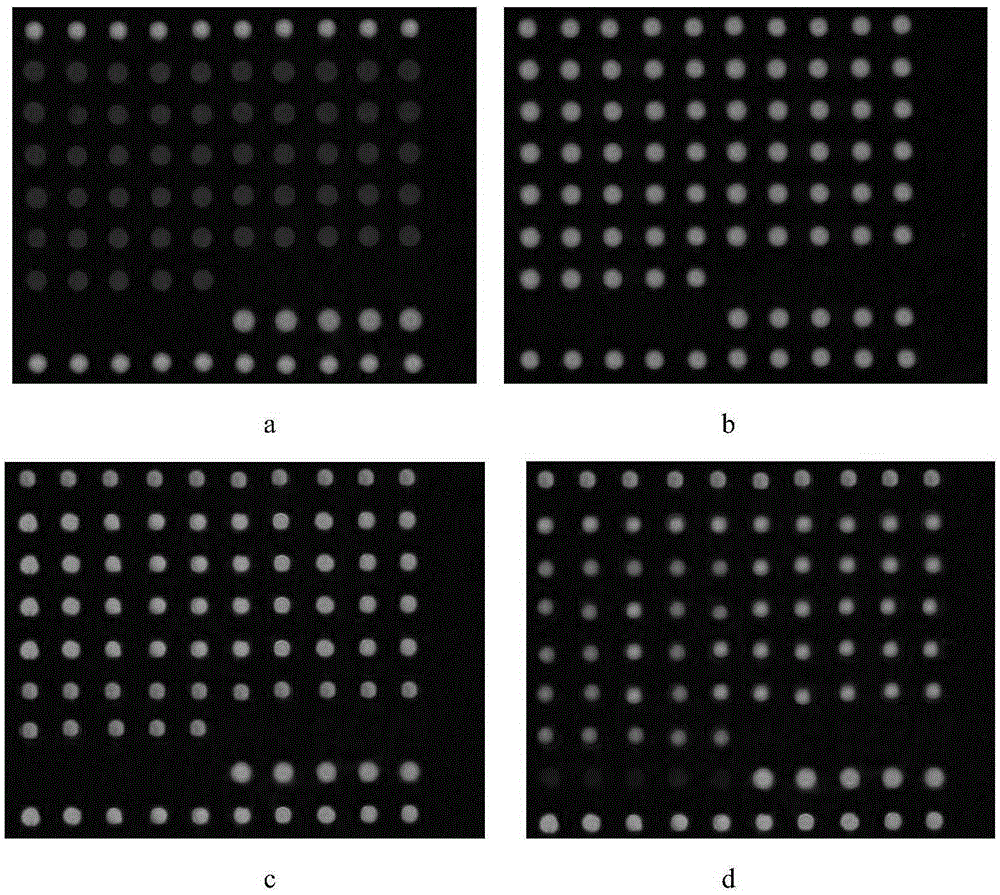
[0255] Four hybridization times were set for optimization, namely 30min, 60min, 90min, and 120min. Hybridization results such as image 3 . ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com