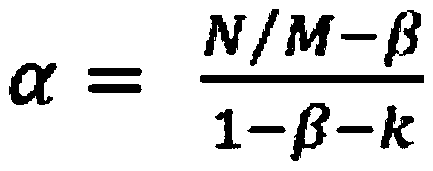Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
71 results about "Genome research" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
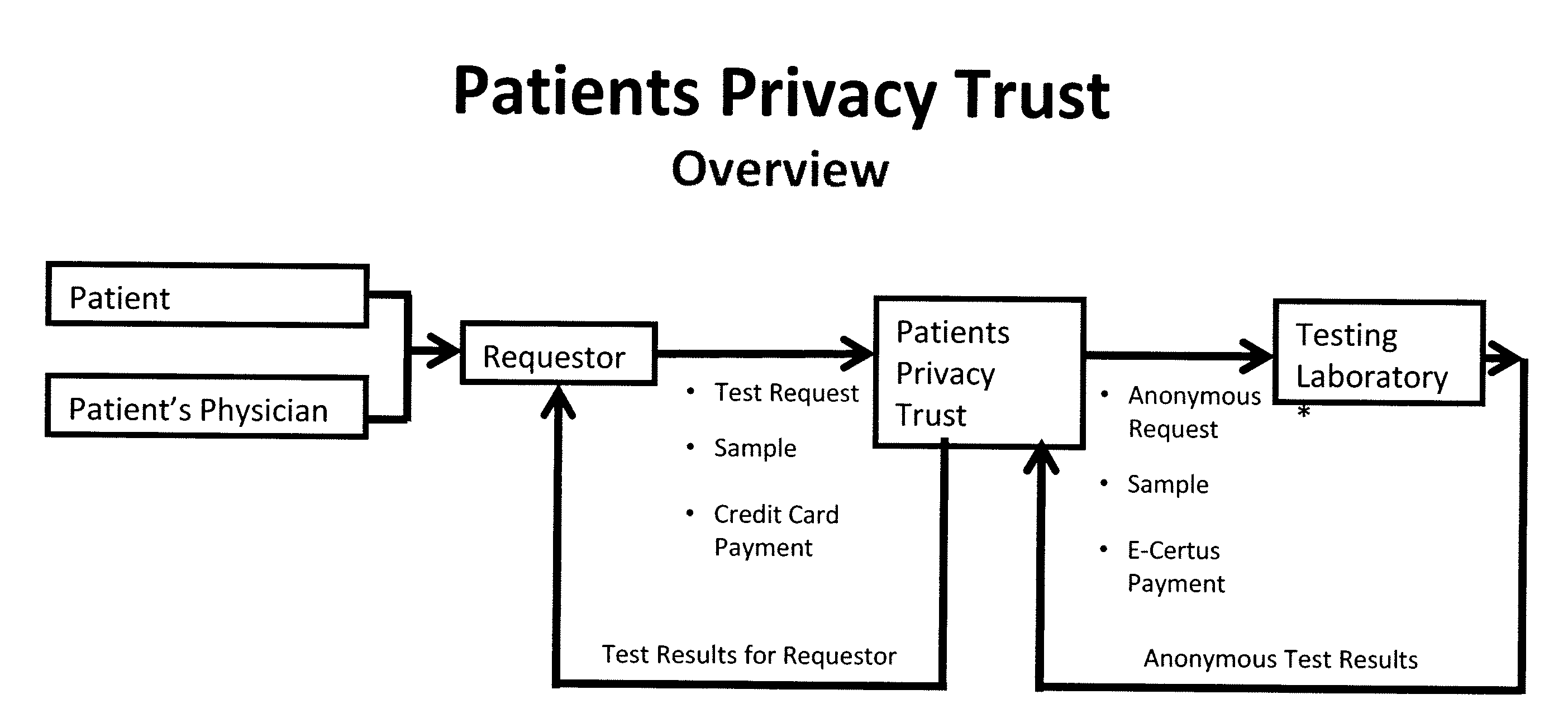
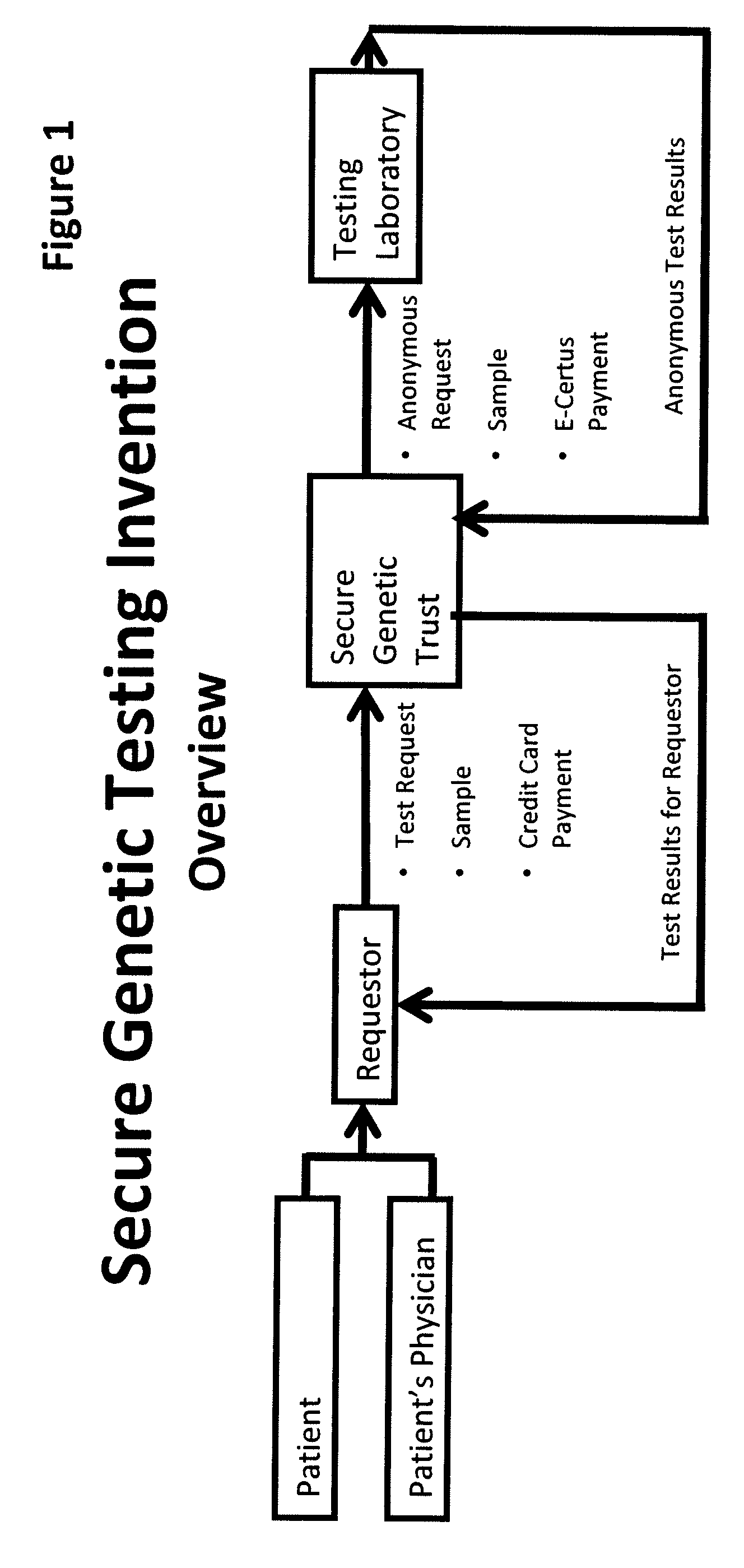
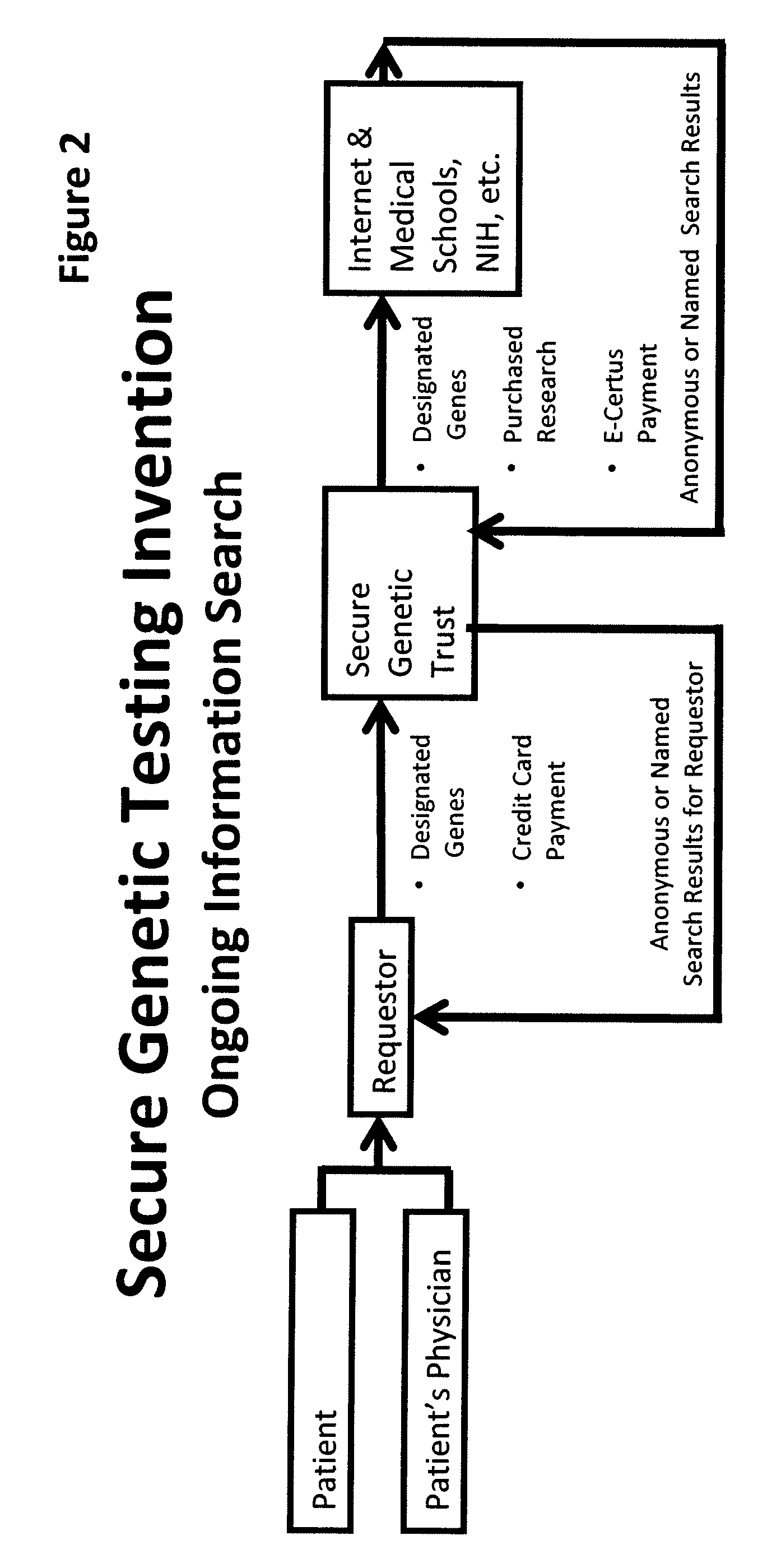
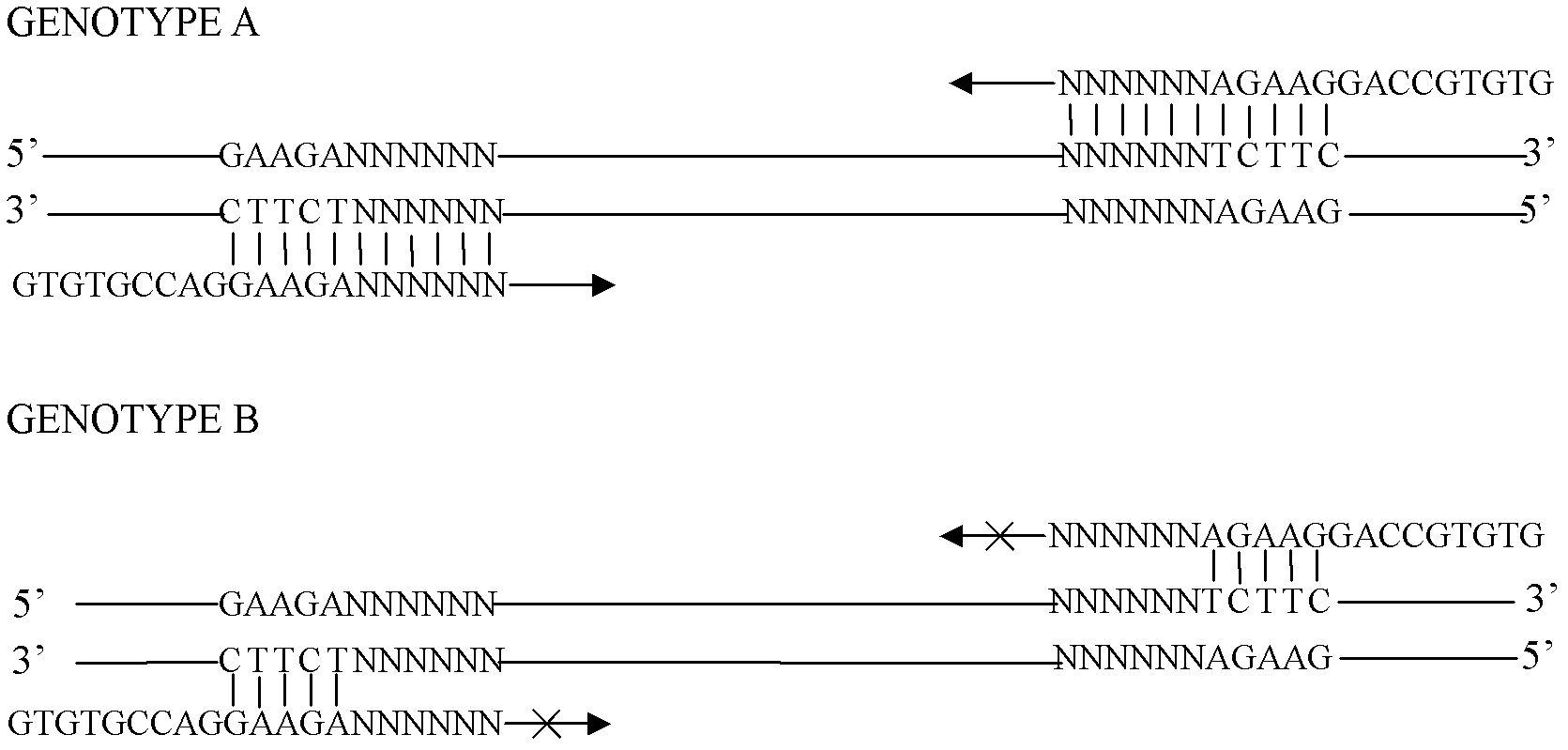
System for gene testing and gene research while ensuring privacy
InactiveUS20090198519A1Data processing applicationsSpecial data processing applicationsElectronic databaseGENETIC ABNORMALITY
A system, method and program product, the method comprising, in one embodiment, providing a secure testing service for patient's identification and payment data encrypted at the data level, non-identifiable method for a patient to have a genetic tests to identify variants or mutations of their genes or combinations of genes that predispose the patient to develop or have an identified disease, comprising: obtaining electronically genomic information for a patient comprising at least one of, (a) DNA information, (b) RNA information, (c) complementary DNA or RNA information, (d) transfer RNA (tRNA) information (e) messenger RNA (mRNA) information, and (f) Expressed Sequence Tags (EST) to identify an abnormal gene; searching by one or more computers electronic databases using the identified abnormal gene to obtain genetic sequencing and basic research, patient predispositions, and pharmacognetics that predict the response and reaction of patients with identified genetic abnormalities related to the identified abnormal gene and individual medications that may be prescribed relating to the identified abnormal gene or a relationship with said identified abnormal gene; performing an update search on at least a periodic basis to learn about subsequent genomic research developments and treatments for the identified abnormal gene, specific genes with variants or mutated genes identified in the genetic test; sending electronically via an Internet communication link data comprising or derived from the searching step and the update search to the patient or a third party; and with the sending step performed using a privacy component that prevents transmission to any third party unless predetermined permission clearance data is in the system.
Owner:MCNAMAR RICHARD TIMOTHY
Marine fish mitochondrion 12S rRNA (ribosomal ribonucleic acid) gene amplification primer and design and amplification method thereof
InactiveCN102912012AAmplify high efficiency and specificityImprove developmentMicrobiological testing/measurementDNA preparationBiotechnologyNucleotide
The invention belongs to the field of marine fish mitochondrial genome research, and particularly relates to a marine fish mitochondrion 12S rRNA (ribosomal ribonucleic acid) gene amplification primer. The primer is composed of two single-chain oligonucleotide chains, wherein a light chain primer is a nucleotide sequence shown as SEQ ID NO.1, and a heavy chain primer is a nucleotide sequence shown as SEQ ID NO.2. The invention also provides a design method of the amplification primer and a method for amplifying a marine fish DNA (deoxyribonucleic acid) solution by using the amplification primer. The invention can efficiently and specifically amplify multiple marine fish mitochondrion 12S rRNA genes and can be used for the analytical study on the system evolution of different taxonomic categories of fishes, thereby providing a powerful tool for fish species identification, germplasm resource survey and system evolution research.
Owner:ZHEJIANG OCEAN UNIV
Microdrop oscillatory silicone groove polymeraxe chair reaction biological chip
InactiveCN1477208ALow costReduce volumeMicrobiological testing/measurementFermentationMicroheaterEngineering
The present invention relates to a microdrop shaking silicon groove type PCR biological chip, including microgroove and thermostatic region positioned in the back surface of microgroove and separatedby heat-insulating groove, resistive microtemperature sensor positioned on the back surface of the microgroove and microheaters which are symmetrically placed at two sides of said temperature sensor.The biological chip has extensive application in the fields of biological genome research, forensic medicine, genetic analysis, therapeutic effect evaluation and medical diagnosis, etc.
Owner:TSINGHUA UNIV +1
Disease biomarker screening analysis method, disease biomarker screening analysis platform, server and disease biomarker screening analysis system
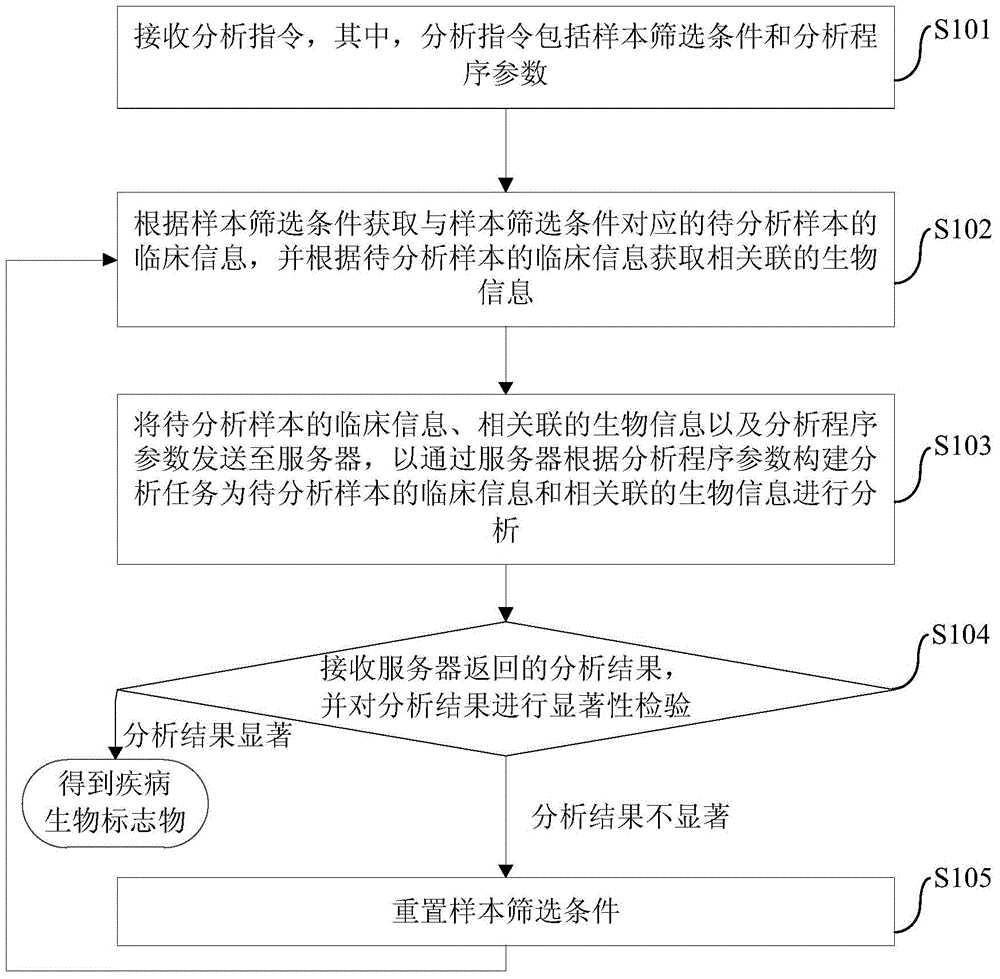
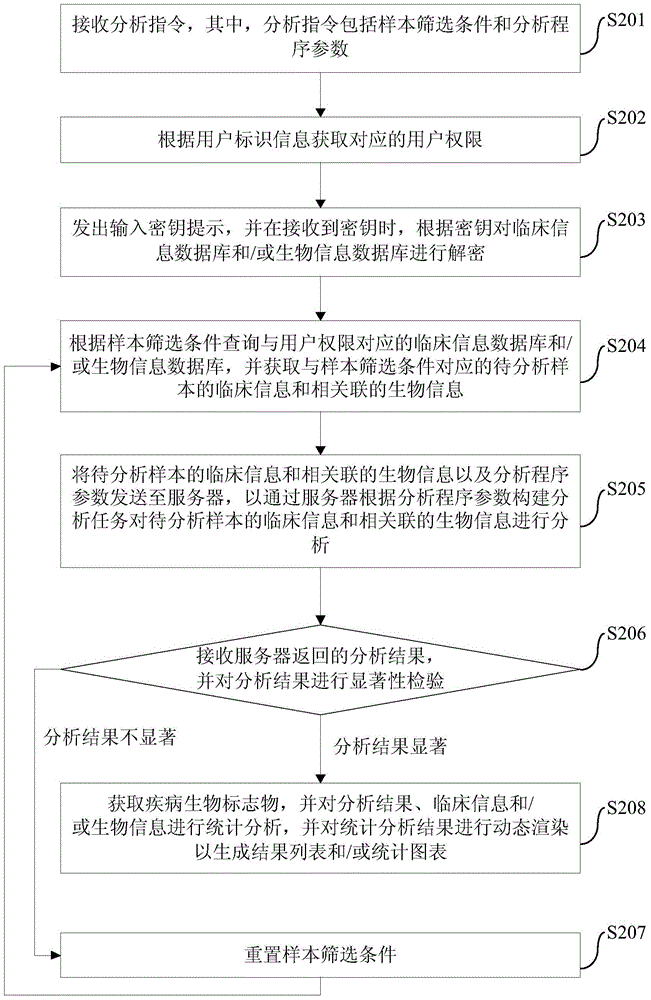
The invention provides a disease biomarker screening analysis method, a disease biomarker screening analysis platform, a server and a disease biomarker screening analysis system, wherein the disease biomarker screening analysis method comprises the following steps of: receiving an analysis instruction, wherein the analysis instruction includes sample screening conditions and analysis program parameters; obtaining clinical information of samples to be analyzed corresponding to the sample screening conditions according to the sample screening conditions, and obtaining relevant biological information according to the clinical information of the samples to be analyzed; sending the clinical information of the samples to be analyzed, the relevant biological information and the analysis program parameters to the server so as to analyze the clinical information of the samples to be analyzed and the relevant biological information; receiving an analysis result returned by the server, and performing significance inspection on the analysis result; and if the analysis result is non-significant, resetting screening conditions, and performing analysis again until the analysis result is significant. The method provides a platform associating the clinical information with genome information for the genome research aspect, and the great-sample-size gene verification experiment is facilitated.
Owner:GENERAL HOSPITAL OF PLA +1
Novel plant DNA virus carrier and its use
InactiveCN1425772AEasy accessClear functionMicrobiological testing/measurementGenetic engineeringViral FunctionForeign protein
The plant DNA virus carrier includes the infectious carrier of Chinese tomato yellowing curly leaf virus DNA-A and DNA-beta, derived plant gene silencing carrier and virus expressing carrier. The present invention also provides one new kind of satellite DNA molecule constructing plant DNA virus carrier and it has at least 72% homology with any one sequence of SEQ ID No.1-12. The present invention also provides the use and method of utilizing plant DNA virus carrier in virus functional genome research, plant functional genome research and foreign protein expression. The present invention provides one important platorm for researching virus genomee structure, function and expression and regulation mechanism and the interaction between virus and plant; provides one excellent tool for researching plant functional gene; and establishing one technological platform for using plant as bioreactor expressing foreign protein.
Owner:ZHEJIANG UNIV
Organization chip for researching functional genome as well as preparation method and application thereof
The invention relates to the biochips field, in particular to a tissue chip used for functional genome research and the preparation method as well as the application thereof. The tissue chip of the invention includes a substrate and a paraffin section arranged on the surface of the substrate, wherein, the paraffin section is provided with tissue sample points which are in different development stages and array according to the tissue dynamic development process. Because the tissue chip of the invention can be combined with a gene chip technology, the space-time expression of a difference expressive gene screened out by the chip technology in the tissue dynamic development process is validated and researched in the high-quality, economic and available way.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Method for cloning rice auxin induced protein gene
InactiveCN101492671AHigh homologySimple implementation stepsFermentationPlant genotype modificationMutantDNA Intercalation
The invention relates to a method for using rice auxin to induce protein genes to be cloned, which is characterized in that the implementation steps comprise: (1) the preparation of a rice transformation receptor; (2) the genetic transformation of the rice; (3) the screening of kanamycin-resistant callus tissue and the regeneration of plants; (4) the screening of the mutant of a T-DNA inserted progenies; (5) Tail-PCR; (6) the comparison and analysis of sequences on the Internet. In the invention, a rice mutant with a short plant height is obtained when carrying out rice functional genome research by using T-DNA label method; the lateral neighboring sequences of the mutant are researched by using TAIL-PCR technology; meanwhile, the position where the mutant T-DNA inserts the rice genome is arranged on the No. 4 chromosome of the rice by the comparison on the databases of NCBI and TIGR on the Internet; moreover, the rice BAC clone (OSJNBa0084K01) of the position is found out. The T-DNA is inserted between the two genes of the clone by analyzing the clone. Known functional genes with a very high homology with BAC cloning code amino acid sequence are forecasted by the sequence comparison on the Internet.
Owner:TIANJIN AGRICULTURE COLLEGE
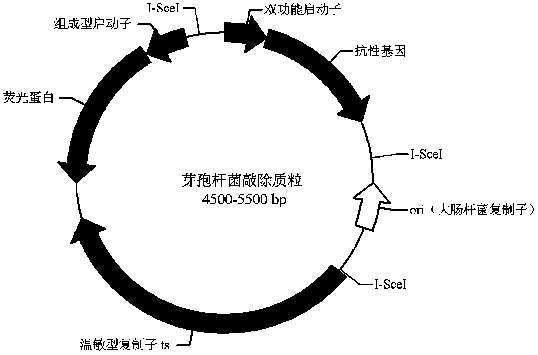
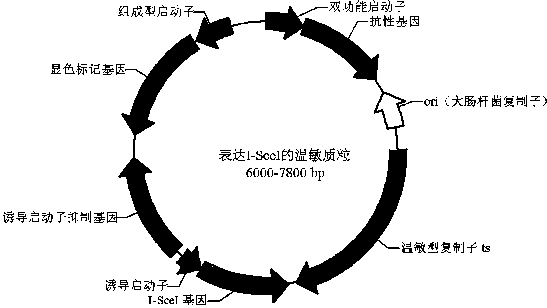
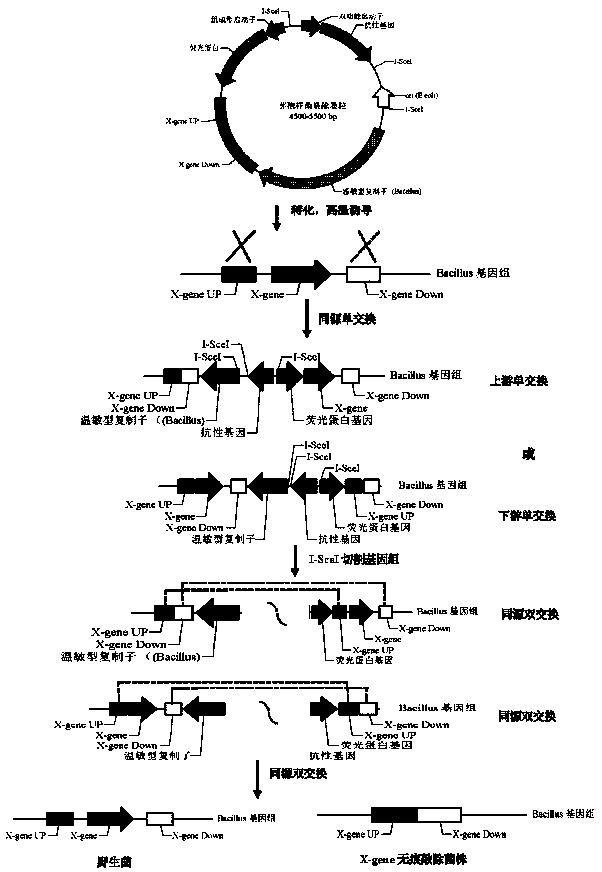
Bacillus gene traceless knockout/knockin plasmid and method, and kit
ActiveCN105505975AEasy to filterImprove efficiencyNucleic acid vectorVector-based foreign material introductionEscherichia coliPromoter
The invention discloses a bacillus gene traceless knockout / knockin plasmid and method, and a kit. The genome of the bacillus gene traceless knockout / knockin plasmid contains only one kind of resistance gene marked with a positive selection marker, a replicon capable of being duplicated in escherichia coli, a thermo-sensitive type replicon capable of being duplicated in bacillus, at least one I-Scel enzyme cutting site and only one kind of chromogenic protein gene, wherein promoters in front of the resistance gene and the chromogenic protein gene are both constitutive promoters in bacillus. The invention further provides a helper plasmid used for improving the knockout / knockin efficiency of the bacillus gene traceless knockout / knockin plasmid, and resistance gene and chromogenic protein gene in the genome of the helper plasmid are different from those of the knockout / knockin plasmid. By the adoption of the two plasmids, one or more target DNA sequences in the genome of bacillus can be modified continuously or iteratively, and the plasmids can be applied to multiple fields including bacillus genetic modification, metabolic process researching, functional genome researching and industrial application researching.
Owner:WUHAN KANGFUDE BIOTECH CO LTD
Method for developing marker through tetraploid potato high-throughput sequencing and application of method
ActiveCN105925680AEvenly distributedExcavation applicableMicrobiological testing/measurementDNA/RNA fragmentationHigh densityOrganism
The invention discloses a method for developing a marker through tetraploid potato high-throughput sequencing and an application of the method. The method for developing the marker through the tetraploid potato high-throughput sequencing disclosed by the invention can greatly reduce the complexity of a potato genome and can facilitate operation; and by virtue of the method, high-density SNP (single nucleotide polymorphism) sites can be rapidly identified, a chromosome label difference genetic interval for controlling target character can be obtained, and finally, the molecular marker can be developed on the basis of the difference genetic interval. With the application of the method disclosed by the invention, the application of an 2b-RAD sequencing technology in potato tetraploid material marker development is successfully achieved; the difficulty on developing the potato tetraploid material marker is solved, and meanwhile, a development cycle of the character closely linked marker is greatly shortened. In addition, the method disclosed by the invention also provides a reference for genetic interval mining, marker development and genome research of other tetraploid organisms having complex genomes.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Method for identifying indica rice and japonica rice varieties via metabolite content
The invention provides a method for identifying indica rice and japonica rice varieties via metabolite content, specifically identifying indica rice and japonica rice varieties via the weight percentage ratio of asparagine to alanine in rice seeds. The weight percentage ratio of asparagine to alanine in seeds of indica rice variety Guang-Lu-Ai No.4 is used as a reference value, in the same test conditions, if the weight percentage ratio of asparagine to alanine in seeds of to-be-tested variety is larger than the reference value, the to-be-tested variety is judged to be indica rice, and if the weight percentage ratio of asparagine to alanine in seeds of to-be-tested variety is smaller than the reference value, the to-be-tested variety is judged to be japonica rice. The method provided by the invention is the first proposal for identifying indica rice and japonica rice varieties via the weight percentage ratio of asparagine to alanine in rice seeds, plays an important guiding role in rice functional genome research and indica / japonica rice differentiation research, and can also promote the scientific research in indica / japonica rice cross breeding and environmental adaptation of varieties.
Owner:SHANGHAI JIAO TONG UNIV
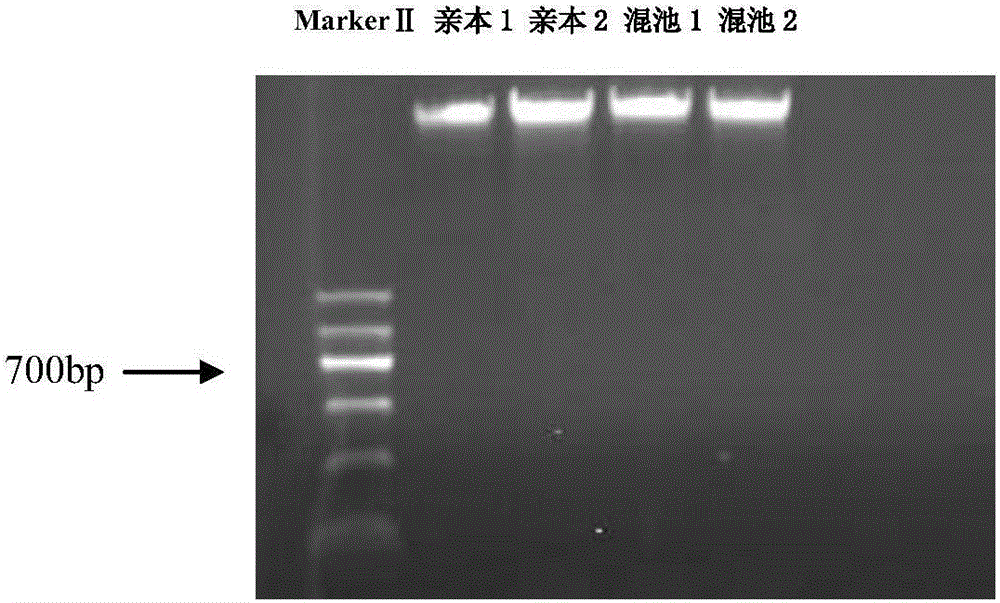
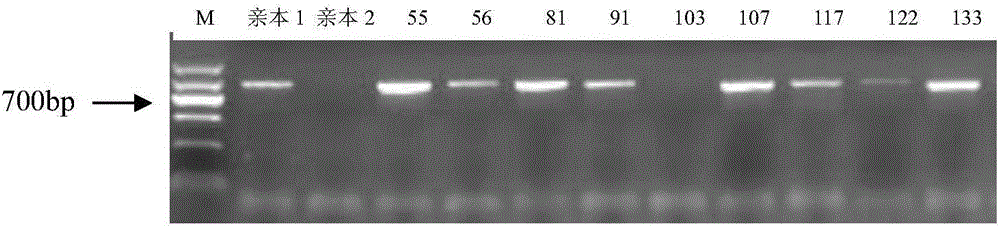
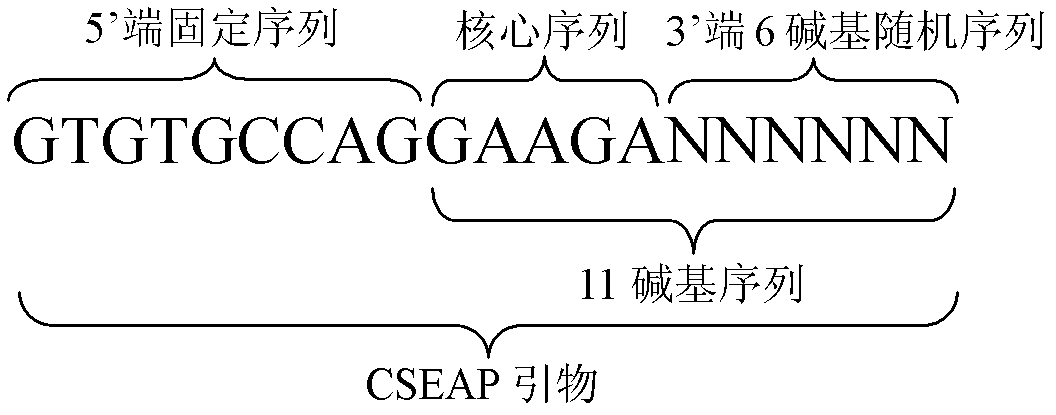
Exon conserved sequence amplified polymophic molecular marker and its analysis method
InactiveCN102443583AReduce design costLarge amount of amplified bandsMicrobiological testing/measurementDNA/RNA fragmentationConserved sequenceGenetic diversity
The invention discloses an exon conserved sequence amplified polymorphic molecular marker and its analysis method. According to the invention, mRNA sequence information of known species in public database can be utilized to find out five bases oligonucleotide conserved sequence of gene exon. The conserved sequence is taken as a core to link six bases random sequence to its 3' terminal to form an eleven bases sequence which makes up a primer of 20bp together with a fixed sequence of 5' terminal. Primers with higher occurrence frequency of the eleven bases sequence are preliminarily screened out by mRNA database. Then PCR is carried out for repeatability screening and the FCR based exon conserved sequence amplified polymorphic marker is obtained. The method provided by the invention has advantages of simple operation, high effectiveness, good repeatability, reduced time and cost of primer screening. It is a new effective molecular marking method for the genetic diversity analysis. The method can be widely applied in various fields of genetic map construction, gene or QTL location, marker-assisted breeding, genetic diversity analysis, genome research, mRNA differential expression analysis and the like and has a bright prospect.
Owner:GUANGXI UNIV
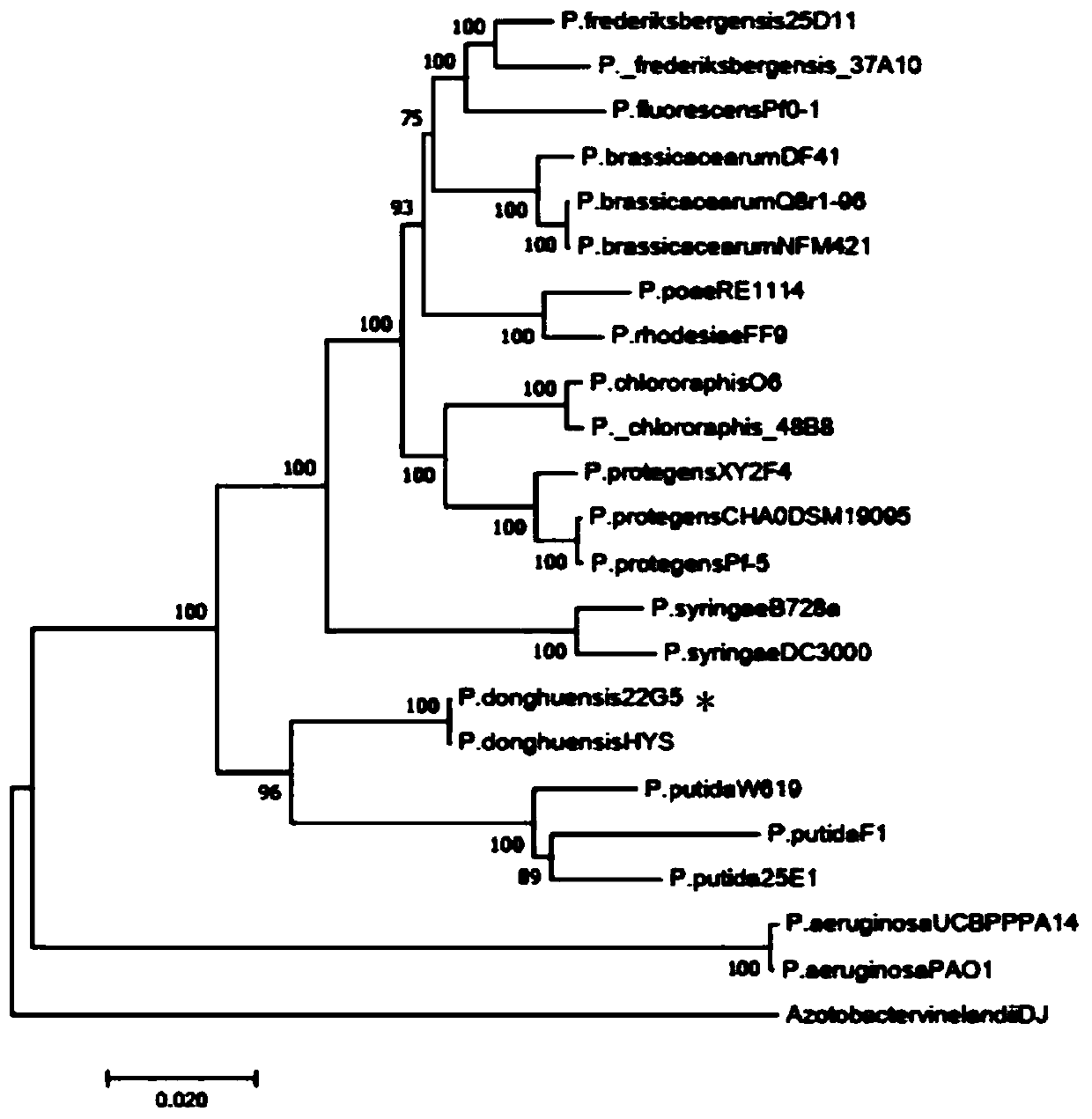
Pseudomonas donghuensis 22G5 and application thereof in prevention and treatment of crop verticillium wilt
InactiveCN110684686AAlleviate verticillium wiltGood inhibitory effectBiocideBacteriaBiotechnologyKetone
The invention discloses a pseudomonas donghuensis 22G5 and an application of the pseudomonas donghuensis 22G5 in prevention and treatment of crop verticillium wilt. The pseudomonas donghuensis 22G5 ispreserved in the China General Microbiological Culture Collection Center on July 08, 2019, and a preservation number of the pseudomonas donghuensis 22G5 is CGMCC No. 18084. The pseudomonas donghuensis 22G5 has no pathogenicity to plants, has a remarkable inhibition effect on cotton verticillium wilt caused by Verticillium dahliae, can remarkably reduce the occurrence of cotton verticillium wilt,and has a very good biological control effect. The genome studies show, the pseudomonas donghuensis 22G5 has a gene cluster for synthesizing gene cluster of siderophore 7-hydroxycycloheptyltrienol ketone, and the compound can be used as an iron ion chelating agent to inhibit the iron element necessary for the growth of verticillium dahliae, so that the growth of pathogenic bacteria is inhibited. The strain can be used for developing biopesticides for crop verticillium wilt, and provides a new thought and a new method for biological prevention and treatment of crop verticillium wilt.
Owner:ZHEJIANG UNIV
Peony genome DNA molecule marking method
InactiveCN103820546AEasy to operateLow costMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresesEnzyme digestion
A peony genome DNA molecule marking method is as follows: extracting a peony genomic DNA for enzyme digestion, connecting an enzyme digestion product with a joint DNA; performing PCR amplification of a connection product and a pre amplified primer to obtain a pre amplified product; taking the pre amplified product for PCR amplification with iPBS primer which is selectively amplified by use of an incision enzyme joint to obtain a PCR selectively-amplified product; then taking the PCR selectively-amplified product to add into a dyeing liquid, applying a sample for electrophoresis, after the electrophoresis, silver staining, observing, photographing and analyzing results. The peony genome DNA molecule marking method is a molecule marking technology based on the PCR and enzyme digestion, and has the advantages of simple operation, low cost and strong applicability; designed for the peony genome study technology, a new molecule marking system based on the retrotransposon is developed, and provides a new technological means for researches on genetic differentiation and evolution of germplasm resources of peony.
Owner:HENAN UNIV OF SCI & TECH
Method for extracting mitochondria and mitochondrial protein from cotton
ActiveCN107266521AHigh purityIncrease the number of extraction washesPeptide preparation methodsContamination rateMetabolism regulation
The invention discloses a method for extracting mitochondria and mitochondrial protein from cotton. The method has minimum mechanical damage to mitochondria, nucleus components and plasmid components except mitochondria as well as phenolic and polysaccharide substances can be removed effectively, the contamination rate is reduced, the purity and the quality of mitochondrial protein are improved, experimental technique requirements of cotton genome research, mitochondrial proteome research, metabolism regulation network research and the like are met, and related research work fields are broadened.
Owner:CHINA AGRI UNIV
SM gene for regulating and controlling cotton trichome, and application thereof
The invention relates to an SM gene for regulating and controlling cotton trichome, belonging to the technical field of genetic engineering. The cDNA sequence of the SM gene is shown by SEQ ID No: 1. The invention provides theoretical basis for the regulation and control of the cotton trichome and provides another way for improving the quality of cotton fiber; and the molecular marker obtained by the SM gene can be applied to multiple aspects such as building a genetic map, locating the gene or quantitative trait locus (QTL), breeding assisted by marking, analyzing genetic diversity, researching genome and the like.
Owner:ZHEJIANG FORESTRY UNIVERSITY
New method for capturing chromatin nucleosome vacancy district at high-throughput complete genome level and use therefor
InactiveCN102691111AMicrobiological testing/measurementLibrary creationTranscriptional regulationFunctional genomics
The invention relates to a new method for high-efficiently capturing a chromatin nucleosome vacancy district at a whole-cell level. The new method can locate the chromatin nucleosome vacancy district more accurately and sensitively in the range of a complete genome. The method for capturing the chromatin nucleosome vacancy district at a high-throughput complete genome level can be used to search the chromatin nucleosome vacancy district of the transcriptional regulation of eukaryotic cells, and is an effective means for functional genomics research.
Owner:CAPITAL UNIVERSITY OF MEDICAL SCIENCES
Method for identifying plant gene functions in total genome ranges
ActiveCN105734130AMicrobiological testing/measurementLibrary creationSingle nucleotide mutationNucleotide
The invention relates to a method for identifying plant gene functions in total genome ranges.The method includes 1), mutating single nucleotide of starting plants and establishing plant mutant libraries; 2), detecting phenotypes of mutants in the plant mutant libraries and establishing plant mutant phenotype databases; 3), detecting genotypes of the mutants in the plant mutant libraries and establishing plant mutant genotype databases; 4), determining functions of genes where SNP (single nucleotide polymorphism) sites caused by mutation of the single nucleotide in the starting plants by the aid of the plant mutant phenotype databases and the plant mutant genotype databases.The method has the advantages that the genes related to plant development procedures and biological procedures of stress-tolerant reaction or metabolic pathways or the like can be accurately and quickly found by the aid of the plant mutant phenotype databases and the plant mutant genotype databases which are established by the method, and novel ways can be created for plant functional genome research.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI +1
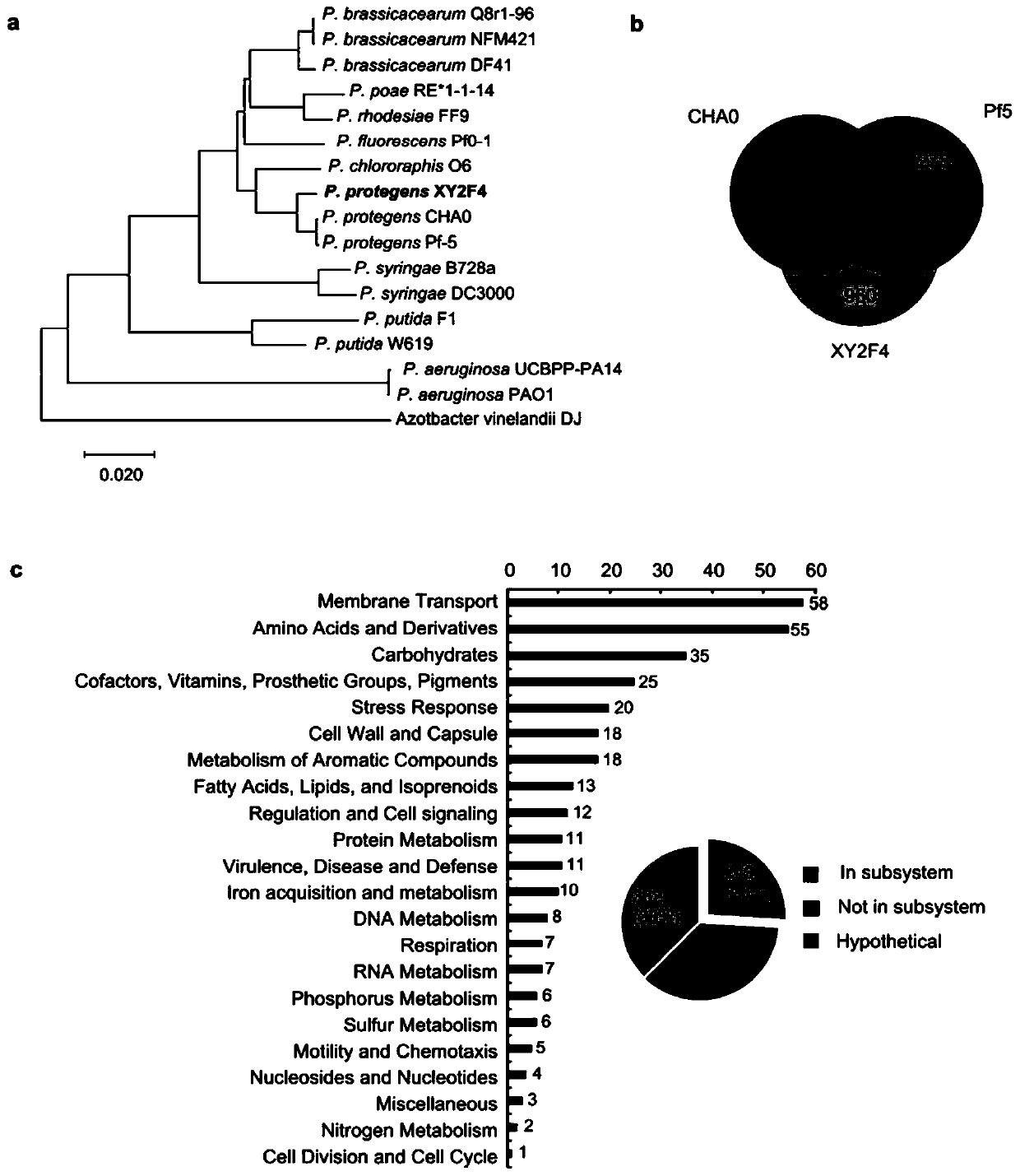
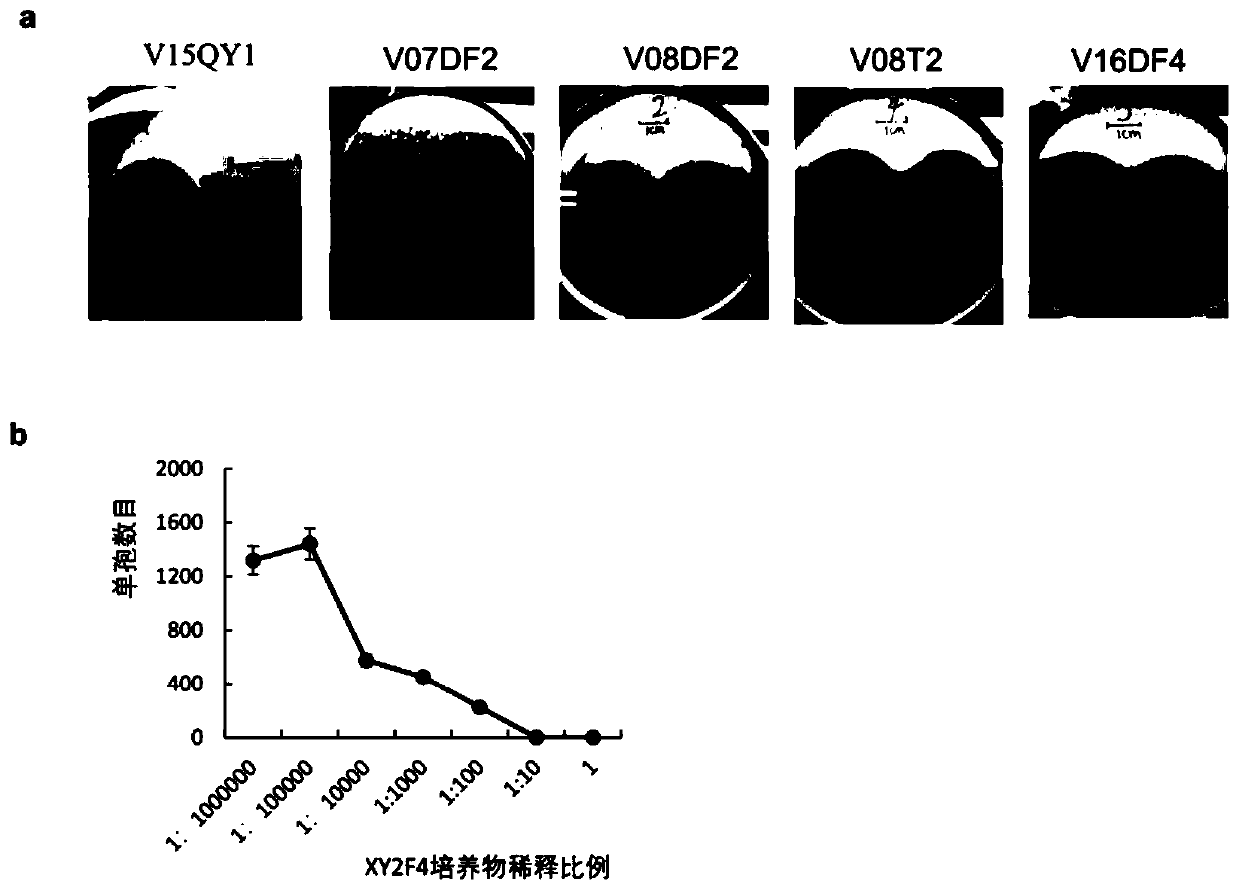
Pseudomonas protegens XY2F4 and application thereof in control of crop verticillium wilt
ActiveCN110643538AAvoid infectionReduce morbidityBiocideBacteriaBiotechnologyMicrobiological culture
The invention discloses Pseudomonas protegens XY2F4 and application thereof in control of crop verticillium wilt. The Pseudomonas protegens XY2F4 was collected in the China General Microbiological Culture Collection Center (CGMCC) of the China Committee for Culture Collection of Microorganisms (CCCCM) on June 24, 2019, with the collection number of CGMCC No.18017. The pseudomonas protegens XY2F4 has no virulence for plants, has a significant inhibition function on cotton verticillium wilt caused by verticillium dahliae, can remarkably relieve occurrence of cotton verticillium wilt and has goodbiological control effects. Genome research shows that the Pseudomonas protegens XY2F4 has a series of biological control related compound synthesis gene clusters, can be used for developing biological pesticides for crop verticillium wilt diseases, and provides a new thought and a new method for biological control of the crop verticillium wilt.
Owner:ZHEJIANG UNIV
Primer special for D-loop region complete sequence sequencing of culter erythropterus and application
InactiveCN107475408AGuarantee personal safetyRapid precipitationMicrobiological testing/measurementDNA preparationCulter erythropterusComplete sequence
The invention relates to the field of biological genome research and specifically discloses a primer special for D-loop region complete sequence sequencing of culter erythropterus and an application. According to the invention, a degenerate primer is adopted for performing PCR amplification and then a special primer is used for sequencing. A D-loop complete sequence sequencing method provided by the invention has the advantages that cost is low, mastering is easy and the complete sequence can be acquired by once positively and negatively sequencing, so that an efficient experimental technical support is supplied for geographical population identification of culter erythropterus, research on molecular genetics and molecule system evolutionism and early resource identification and the enlightenment is supplied for the other animal experiments.
Owner:INST OF AQUATIC LIFE ACAD SINICA
Portunidae mitochondrial COI gene universal primer, design and amplification method thereof
InactiveCN109234409AEfficient amplificationEfficient batch expansionMicrobiological testing/measurementDNA/RNA fragmentationForward primerDNA barcoding
The invention relates to the research field of cephalopod mitochondrial genome, and especially relates to a portunidae mitochondrial COI gene universal primer, and design and amplification methods thereof. The universal forward primer of mitochondrial COI gene of Portunidae is SEQ ID NO. 1: 5 '- TCNACAAAYCATAAAGAYATYGG-3 ', and that reverse prim is SEQ ID NO. 2: 5'-TANACYTCWGGRTGHCCRAARAAYCA- 3'.The present invention solves the problem that the prior art does not utilize the DNA barcode technology to identify the family Portunidae, and has the following advantages: 1) the universal primer ofmitochondrial COI gene of the family Portunidae provided by the present invention can amplify a plurality of families of Portunidae in batch at one time with high efficiency, and can provide great convenience for the species identification of the family Portunidae; 2) design universal primers in two conservative region of mitochondrial COI gene of family Portunidae to amplify an intermediate mutation sequence, that amplification length is about 600-700 bp, sufficient information sites are used for identification and differentiation of physical evidence, and a sequencing reaction can be complete, the operation is simple, and the cost is saved.
Owner:ZHEJIANG OCEAN UNIV
High-activity transposase and use thereof
The invention belongs to the field of molecular biology and biological medicine, and particularly relates to a high-activity transposase and use thereof. The high-activity transposase has an amino acid sequence as shown in SEQ ID NO: 2 and a coding nucleotide sequence of the amino acid sequence is shown as SEQ ID NO:3. When the high-activity transposase based on the amino acid sequence is applied to a transposon system, the gene transfer activity of transposons can be remarkably improved. The high-activity transposase and the nucleotide sequence for coding the high-activity transposase can be used for constructing a gene transfer system, and prepare or used as drugs, preparations or tools for genome research, gene therapy, cell therapy or multifunctional stem cell induction and / or differentiation.
Owner:SHANGHAI CELL THERAPY RES INST +1
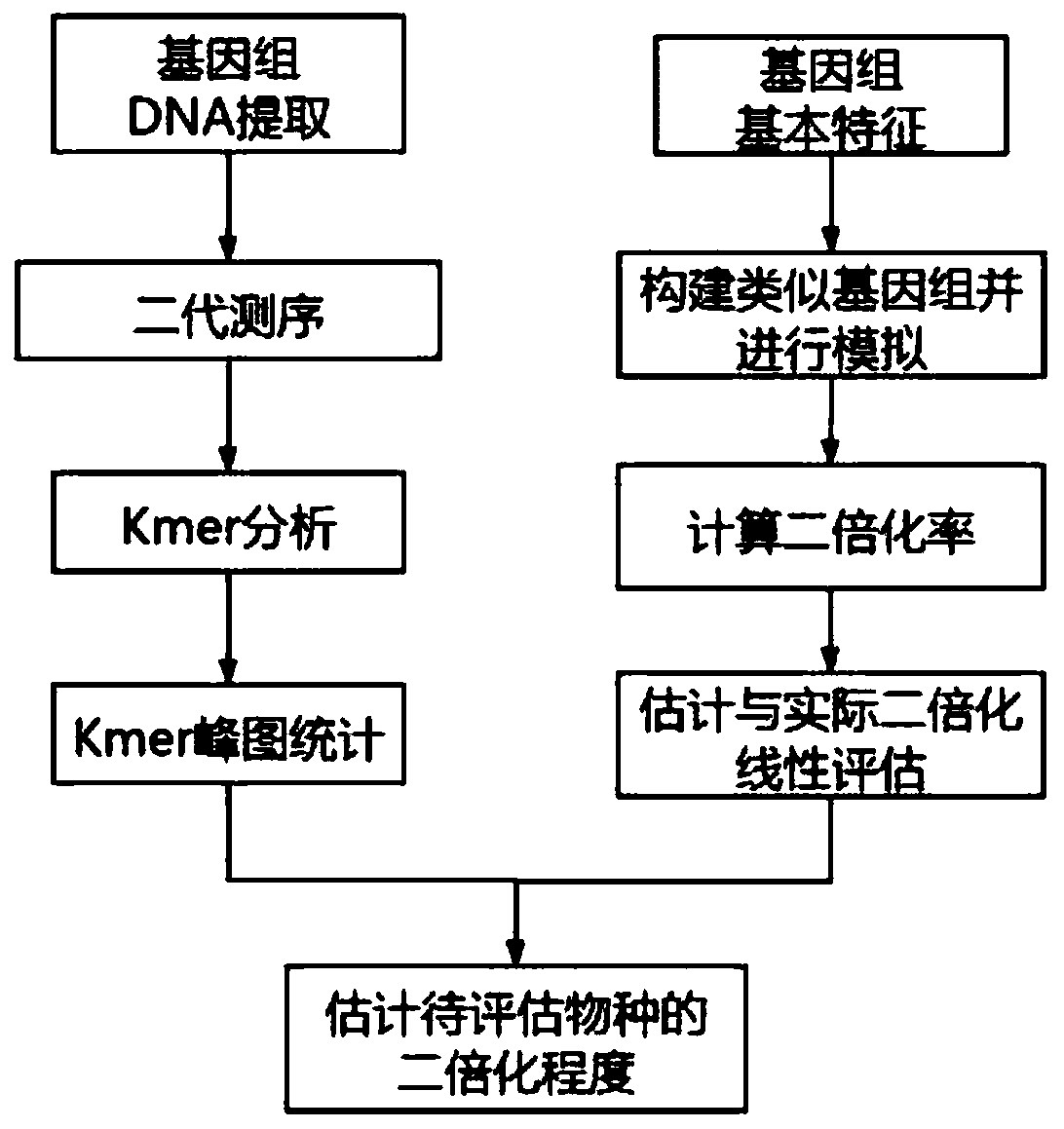
Polyploid organism genome diploidization degree quantitative evaluation method
The invention relates to a polyploid organism genome diploidization degree quantitative evaluation method, which comprises the following steps: 1) evaluating the heterozygosity of a genome according to the Kmer analysis result of the genome; 2) constructing a polyploid genome characteristic model according to the heterozygosity characteristics of the genome; 3) simulating polyploid genomes with different diploidization degrees on the basis of the polyploid genome characteristic model; and 4) evaluating the accuracy of polyploid diploidization rate estimation, and calculating to obtain the diploidization degree of the polyploid genome. According to the method, resequencing data is used for polyploid genome diploidization analysis for the first time, the cost is low, and the method is suitable for polyploid genome research of animals and plants.
Owner:西藏自治区农牧科学院水产科学研究所
Fish transgenosis breeding method
The invention relates to a fish transgenosis breeding method which belongs to the technical field of molecule biology fish breeding. The method mainly comprises the following steps: preprocessing the genome DNA of a fish and obtaining a DNA rearrangement fragment distinguished from the original genome DNA; guiding the DNA rearrangement fragment into an oocyte of an original fish by a transgenosis method to achieve the function with the genome of the original fish; forming a variable gene and generating individuals with mutational appearance; screening the individual with favorable production characteristics and carrying out further breeding. The invention judges whether the DNA rearrangement fragment enters the genome of the original fish or not by a TRAP method, obtains the variable gene by a two-step amplification method, provides a base for carrying out functional genome research later and obtains a unique molecular mark. The method is simple and can be widely applied to the breeding of fishes, other animals and plants, and the like and the research of relevant functional genomes.
Owner:FRESHWATER FISHERIES RES CENT OF CHINESE ACAD OF FISHERY SCI
Method for identifying plant introgression line with high tissue culture ability
The invention discloses a method for screening a Yuanjiang common wild rice introgression line material with high tissue culture ability. The method comprises the following steps of: 1) performing inducing culture on seeds of plant introgression lines to be identified to obtain calluses, and counting the inductivity; 2) sequentially subculturing the calluses obtained in step 1) twice to obtain first subcultured calluses and second subcultured calluses respectively, and counting the browning rate of the first subcultured calluses and the browning rate of the second subcultured calluses respectively; and 3) differentiating the calluses obtained in step 2), and counting the differentiation rate. The experiment proves that: Yuanjiang common wild rice and Teqing are utilized to construct an introgression line population, mature embryos of 80 introgression lines are cultured, and phenotype data of characters related to culture abilities of the introgression lines are investigated to screen the line with high culture ability by using the Teqing as control, so that a transgenic receptor with high culture ability can be provided for genetic improvement and functional genome research of rice.
Owner:CHINA AGRI UNIV
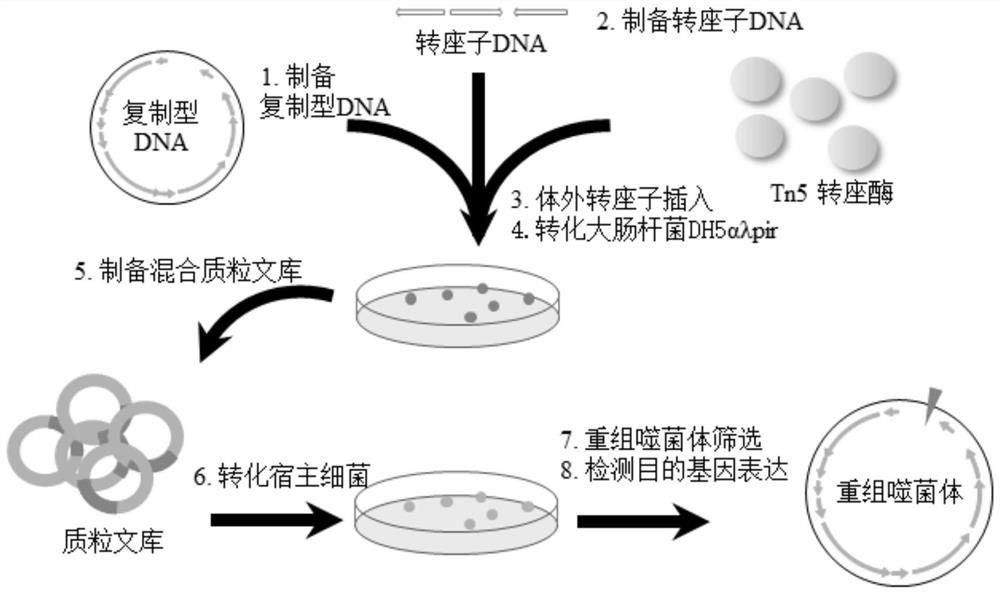
Construction method of recombinant bacteriophage
ActiveCN113136372AEasy to operateBacteriophagesVector-based foreign material introductionEscherichia coliEnterobacteriales
The invention relates to the technical field of molecular biology, in particular to a construction method of a recombinant bacteriophage. The construction method of the recombinant bacteriophage comprises the following steps: (1) extracting replicative DNA of the filamentous bacteriophage; (2) preparing a Tn5 transposon DNA with a target gene; (3) inserting the transposon DNA in vitro; (4) transforming escherichia coli DH5[alpha][lambda]pir; (5) preparing a mixed plasmid library; (6) transforming host cells; (7) screening recombinant bacteriophages; and (8) detecting the expression of the target gene. The construction method provided by the invention is simple to operate, can construct the recombinant filamentous phage without depending on the research on the functional genome of the target filamentous phage, and has important significance on the research on the application of the target recombinant filamentous phage.
Owner:GUANGXI UNIV
Changbai mount rhododendron chrysanthum pall genome DNA extraction method
InactiveCN103074326AHighlight substantiveSignificant progressDNA preparationMolecular levelDNA extraction
The invention belongs to the technical field of genetic engineering, and discloses an alpine plant Changbai mount rhododendron chrysanthum pall genome DNA extraction method. According to the method, a CTAB method and a SDS method are combined at the first time to extract and purify Changbai mountain rhododendron chrysanthum pall genome DNA; in the extraction method, a PVP use amount in a CTAB extraction buffer solution is 4% and is increased by 2% compared to a PVP use amount in the conventional method, a 2 beta-mercaptoethanol use amount is 4.3% and is increased by about 1 time compared with a 2-mercaptoethanol use amount in the conventional method, and a 65 DEG C warm bath time is prolonged to 4 h from 1 h of the conventional method; and the SDS method is combined to purify the extracted genome so as to successfully extract the high purity and good quality Changbai mountain rhododendron chrysanthum pall genome DNA. With the present invention, disadvantages of difficult extraction, low purity of the extracted genome and easy degradation of the existing alpine plant are overcome, and important significances are provided for subsequent molecular level researches and alpine plant genome researches.
Owner:JILIN NORMAL UNIV
A kind of amplification primer and screening method of mitochondrial cox3 gene of common northern razor clams
InactiveCN105200047BSingle stripeHigh amplification efficiencyDNA preparationDNA/RNA fragmentationGenetic diversityScreening method
The invention relates to an amplification primer and a screening method for a common northern razor clam mitochondrial cox3 gene. The amplification primer cox3CF is 5' TATCATTTAGTHGATATAAGACC 3'; the amplification primer cox3CR is 5' CCATGAAADCCAGTTATYACAA 3'; the preparation method of the amplification primer comprises Download sequence, sequence comparison, primer design and synthesis, DNA extraction, primer amplification five steps; the present invention screens out a pair of primers cox3CF and cox3CR for amplifying the cox3 sequence of the common northern razor clam mitochondria, and determines its reaction System and PCR amplification conditions, experiments have proved that the primer has the characteristics of a single band and high amplification efficiency, and can meet the needs of the research on the mitochondrial genome of razor clams and the subsequent research on genetic diversity and phylogenetics.
Owner:山东省海洋资源与环境研究院
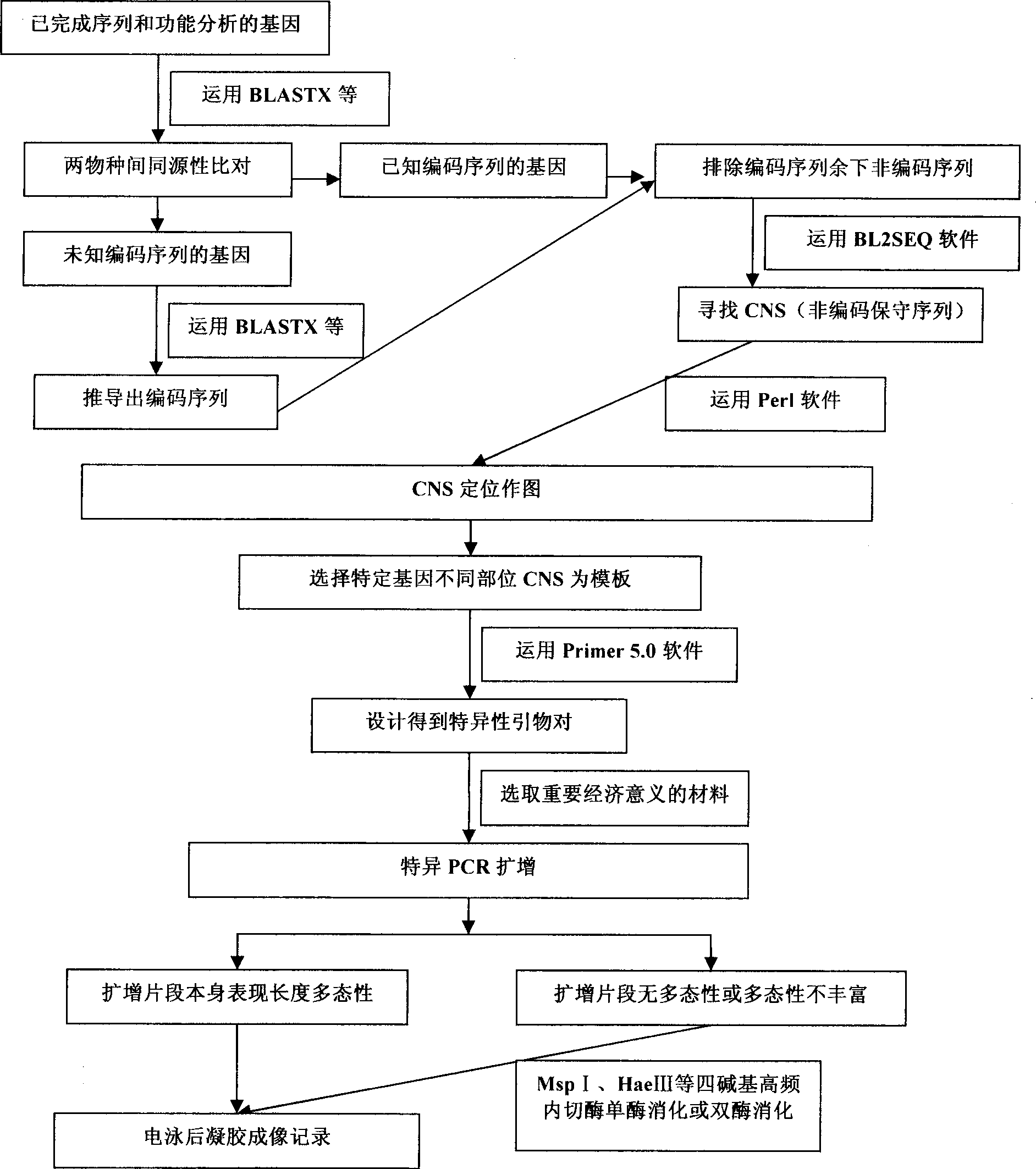
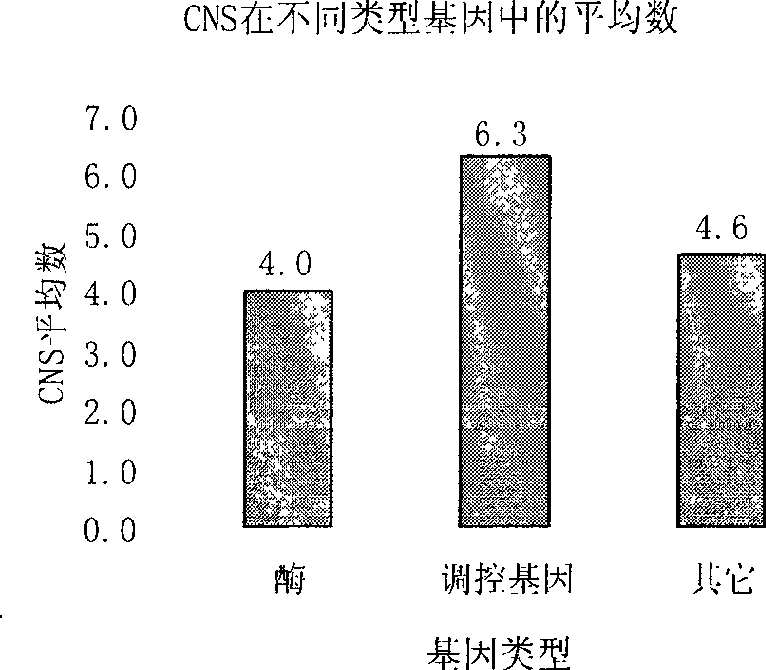
General use molecular marker CNS-AFLP for gramineae
InactiveCN101509043AOvercoming the problem of narrow application rangeGuaranteed FeaturesMicrobiological testing/measurementFermentationBiotechnologyGenomics
The invention discloses a method for building gramineae general molecular marker CNS-AFLP and developed primer sequence with good universality in gramineae crop, which is applicable to the technical fields of genomics, molecular biology and bioinformatics. Coding sequence or EST is taken as a template so as to design the primer; and conserved non-coding sequence (CNS) is taken as the template so as to design the primer. Detecting polymorphism is base sequence length and restriction enzyme cutting site variation of non-coding region between two CNSs and between CNS and Exon. The invention comprises steps of CNS developing, primer designing, PCR amplifying, detecting the polymorphism of amplified fragment, preparing and analyzing the polymorphism of restriction enzyme cutting fragment, etc. Compared with SSR mark, ILP mark and EST mark, the invention is characterized in that transspecies application range is wider and the designed primer is applicable to various species in the gramineae, thereby improving research efficiency. The method is a molecular marking method based on PCR technology, so that the designed primer has high universality and wide transspecies application range, thus effectively promoting the relatively laggard gramineae crop genome research and cereal crop comparative genetics.
Owner:GRAIN RES INST HEBEI ACAD OF AGRI & FORESTRY SCI +1
Goby mitochondrion COIII and ND3 gene amplimer, design and amplification method
The invention belongs to the fish mitochondrial genome research field, and concretely relates to a COIII and ND3 gene amplimer, the gene amplimer is composed of two strips of single chain oligonucleotide chains, wherein a light chain primer is a nucleotide sequence shown in SEQ ID NO.1, and a heavy chain primer is a nucleotide sequence shown in SEQ ID NO.2. The invention also provides a design and an amplification method of the amplimer. The invention can specifically amplify a plurality of goby mitochondrion COIII and ND3 gene sequences with high efficiency, and can be used in goby different classification category systems evolution and classification research.
Owner:ZHEJIANG OCEAN UNIV
Duck mitochondrion COI gene amplification primer and application thereof
PendingCN113481200ASingle stripeHigh amplification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversitySingle band
The invention relates to the technical field of duck biological mitochondrion gene acquisition, and particularly discloses a duck mitochondrion COI gene amplification primer and application thereof. The sequence of the primer is as follows: the sequence of an upstream primer COI-F: 5'-CCATCTTACCCGTGACCT-3', and the sequence of a downstream primer COI-R: 5'-TGGCTTGAAACCAGTGTA-3'. The duck mitochondrion COI gene amplification primer can increase the length of an obtained duck mitochondrion COI gene segment to about 1551bp, has the characteristics of single band and high amplification efficiency, and can better meet the requirements of subsequent duck mitochondrion genome research and duck population genetic diversity evaluation.
Owner:ANHUI AGRICULTURAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com