High-activity transposase and use thereof
A transposase, high activity technology, applied in the fields of molecular biology and biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] The acquisition of embodiment 1 highly active bz-hyPBase mutant
[0053] Based on the original sequence (amino acid sequence shown in SEQ ID NO: 1) of the existing highly active piggybac transposase (hyPBase for short), we made the following changes to obtain the protected baize piggyBac transposase (bz-hyPBase for short) sequence information:
[0054] (1) Based on human codon usage preferences, we optimized the codons of the existing highly active piggybac transposase to obtain the nucleotide sequence shown in SEQ ID NO:4 to improve the expression level of the transposase;
[0055] (2) The human c-myc nuclear localization signal is added after the start codon to improve the integration efficiency of foreign genes in host cells;
[0056] (3) The following method was used to randomly mutate the nucleotide sequence shown in SEQ ID NO:4 to obtain a mutant whose transposition efficiency was significantly better than that of the existing highly active piggybac transposase, ...
Embodiment 2
[0084] Example 2 bz-hyPBase has higher transposition efficiency in yeast
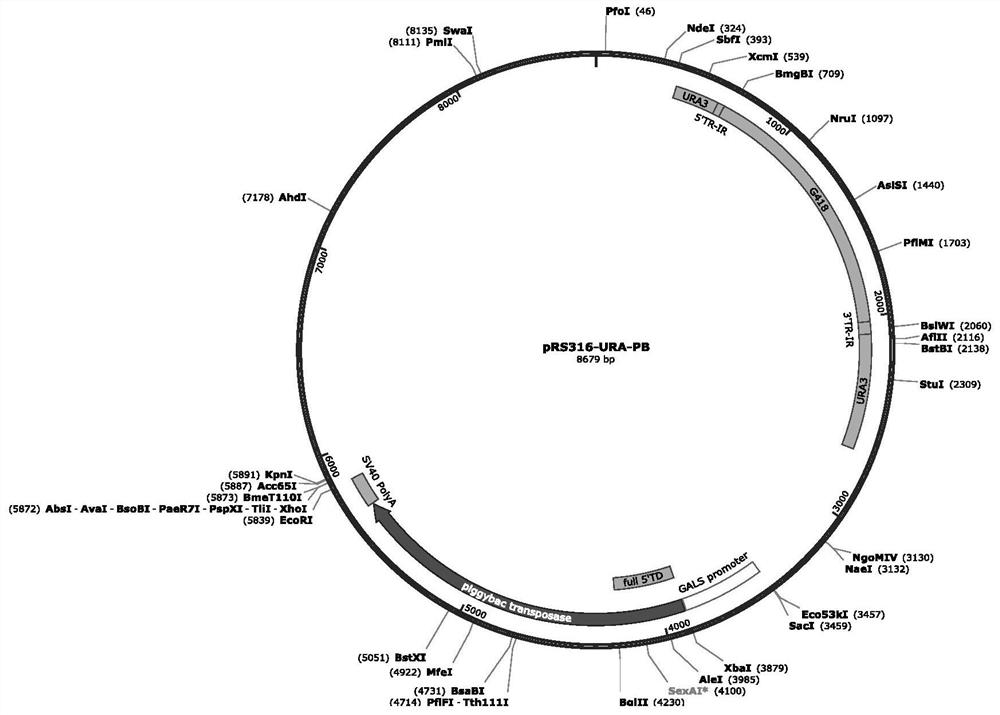
[0085] We inserted the transposon with the G418 resistance gene into the URA3 gene in the yeast plasmid PRS316 to disrupt the expression of the URA gene, and cloned the transposase with an inducible promoter into PRS316 to generate the plasmid PRS316-URA- For Pbase, plasmids carrying different transposases WT PBase, hyPBase, optimized hyPBase, and bz-hyPBase were prepared in parallel. The plasmid was transformed into ura-deficient Saccharomyces cerevisiae BJ2168, which could not survive in ura-deficient medium. The expression of transposase is turned on under the regulation of the inducer galactose, which promotes the transposition of the transposon, the transposition of the transposon, the normal expression of the URA gene, and the normal growth of the transposed clone in the ura-deficient medium . The transposition efficiency of the transposase in Saccharomyces cerevisiae can be calculated by counti...
Embodiment 3
[0087] Example 3 bz-hyPBase has higher gene editing efficiency in CHO cells
[0088] We cloned optimized hyPBase and bz-hyPBase into mammalian cell expression vectors to generate plasmid ploxP-optimized hyPBase (structure the same as Figure 5 , only the Figure 5 The medium transposase was replaced by bz-hyPBase with optimized hyPBase) and ploxP-bz-HyPB( Figure 5 ) to express the transposase. The promoters of optimized hyPBase and bz-hyPBase are connected with human c-myc nuclear localization signal. The transposon with the EGFP gene was cloned into the vector pSAD-EGFP ( Image 6 ) to express green fluorescent protein. The two plasmids expressing transposase and transposon were co-electrotransformed into CHO cells, and the transposon with EGFP would be inserted into the genome under the action of transposase to stably express green fluorescent protein. After two subcultures Afterwards, on the 7th and 14th day, the cells expressing the green fluorescent protein were cou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com