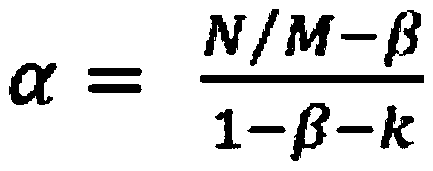Polyploid organism genome diploidization degree quantitative evaluation method
A quantitative evaluation and genomics technology, applied in genomics, bioinformatics, proteomics, etc., can solve problems such as quantitative evaluation of the degree of diploidization of polyploid genomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1: material preparation
[0028] In this experiment, Schizothorax, a unique fish in the Tibetan Plateau, was used as the research object. Female Schizothorax adults caught in the wild from Yajiang, Tibet. Based on previous studies, Schizothorax is a polyploid fish with a tetraploid karyotype. After recording the basic traits and indicators, take the fins and freeze them with liquid nitrogen for one hour, and store them in a -80 degree refrigerator.
Embodiment 2
[0029] Example 2: Schizothorax DNA extraction and high-pass sequencing
[0030] Frozen Schizothorax is expected to be ground in a single weight until ground to a powder. Afterwards, add 1 mL of digestion buffer so that the buffer fully contacts the ground sample and submerges. Place the entire sample in a 1.5 mL centrifuge tube and let stand at 37 °C for 5 h. Afterwards, cool the digestion solution to room temperature, take 0.5 mL of equilibrated phenol, mix gently and centrifuge at 3000-5000×g for 10 min, and suck out the aqueous phase. Add 0.5 mL of a mixture of phenol: chloroform: isoamyl alcohol (25:24:1 by volume) and extract twice again, extract the aqueous phase and add NaCl to a concentration of 0.3M. Add 3 times the volume of ethanol to make the solvents mix with each other, centrifuge at 3000×g for 10 min, rinse twice with 70% ethanol, suck out the supernatant, and dry in the air. The obtained sample was suspended in TE, NaCl was added to 100 mM, RNase A was added...
Embodiment 3
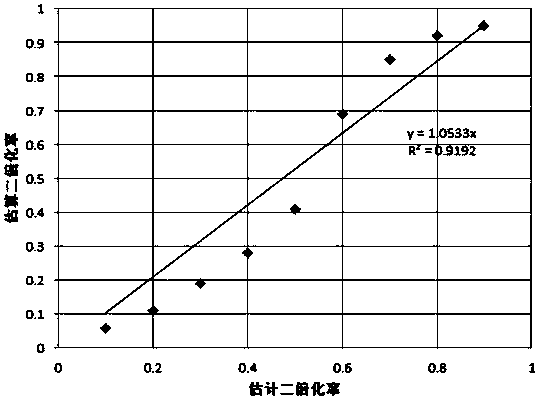
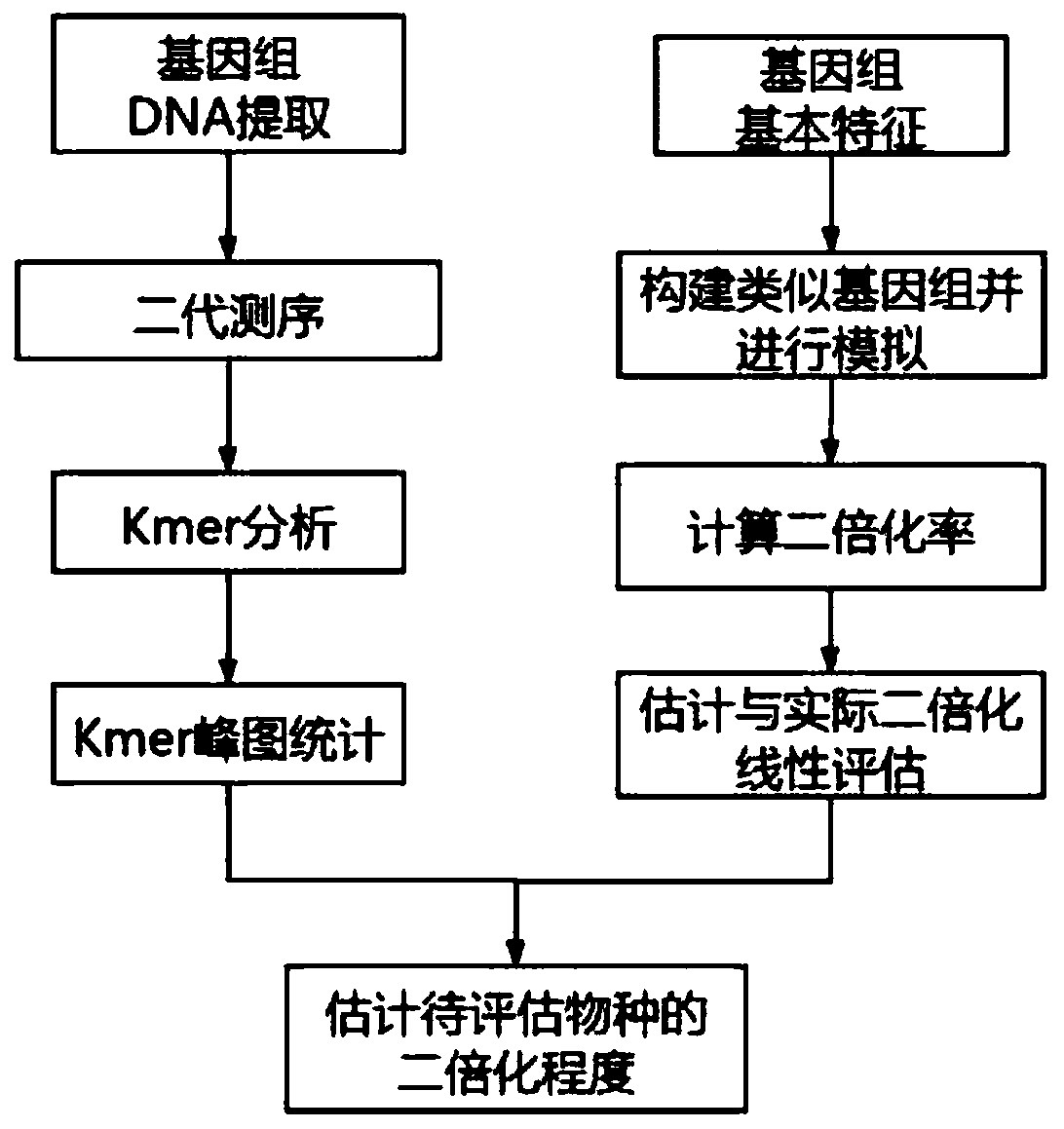
[0032] Example 3: Estimate the heterozygosity of the genome based on the results of genome resequencing
[0033] According to the genome resequencing results, after quality control and filtering by Fastqc and HTQC software, the number of Kmers of each type was calculated according to the Kmers with a read length of 17 bp, and the distribution of different Kmers was constructed. Using the method of Kmer distribution, the genomic characteristics information such as genome size and heterozygosity of Schizothorax was obtained. In this project, the calculated Schizothorax genome size is 2.2Gb, and the heterozygosity is 0.71%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com