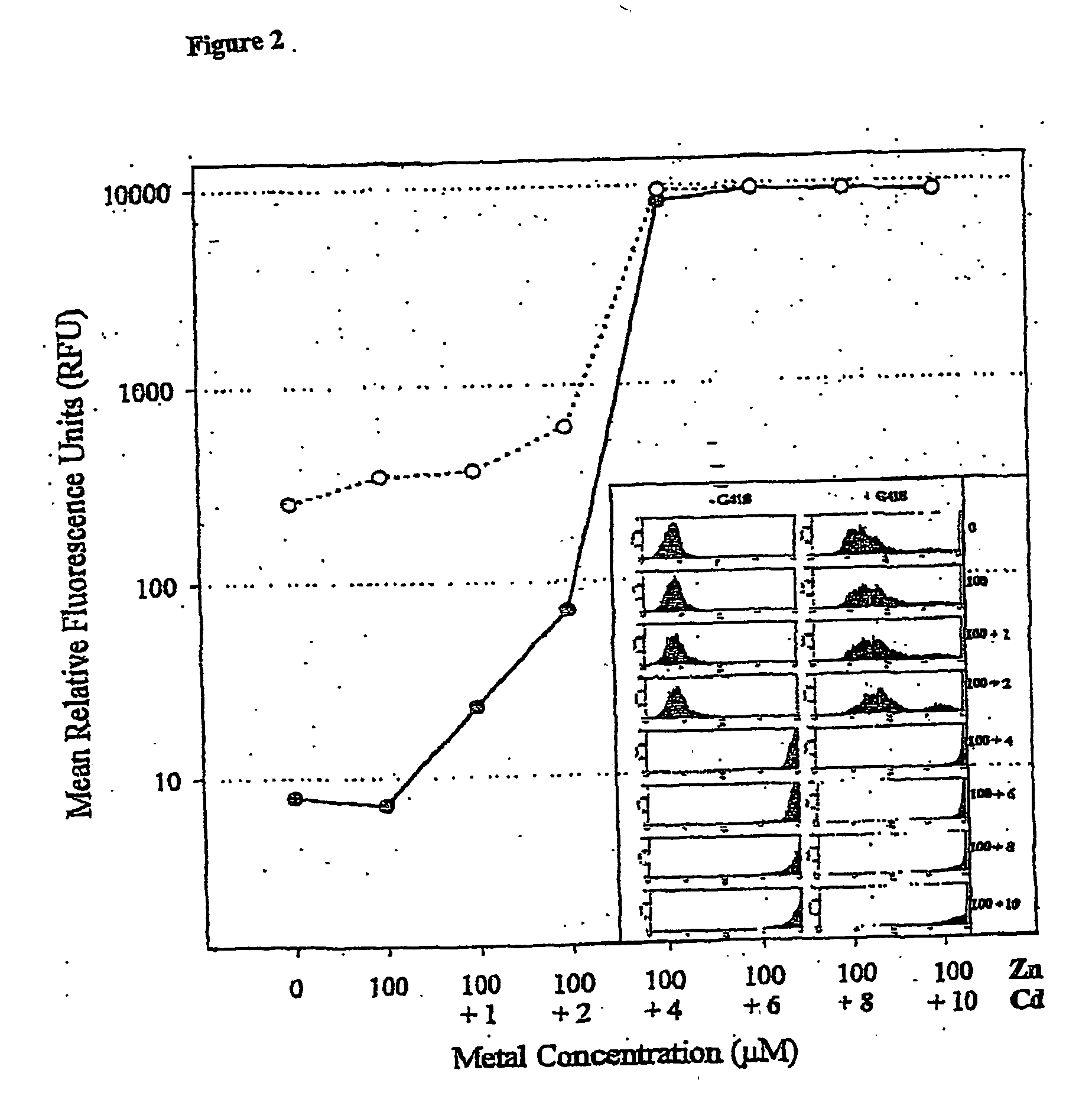
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
42 results about "Gene Organization" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Organization of Genes. Genes are organized into units known as chromosomes. Humans have 23 pairs of chromosomes (46 in total). One chromosome in each pair comes from the person’s mother and the other from their father.
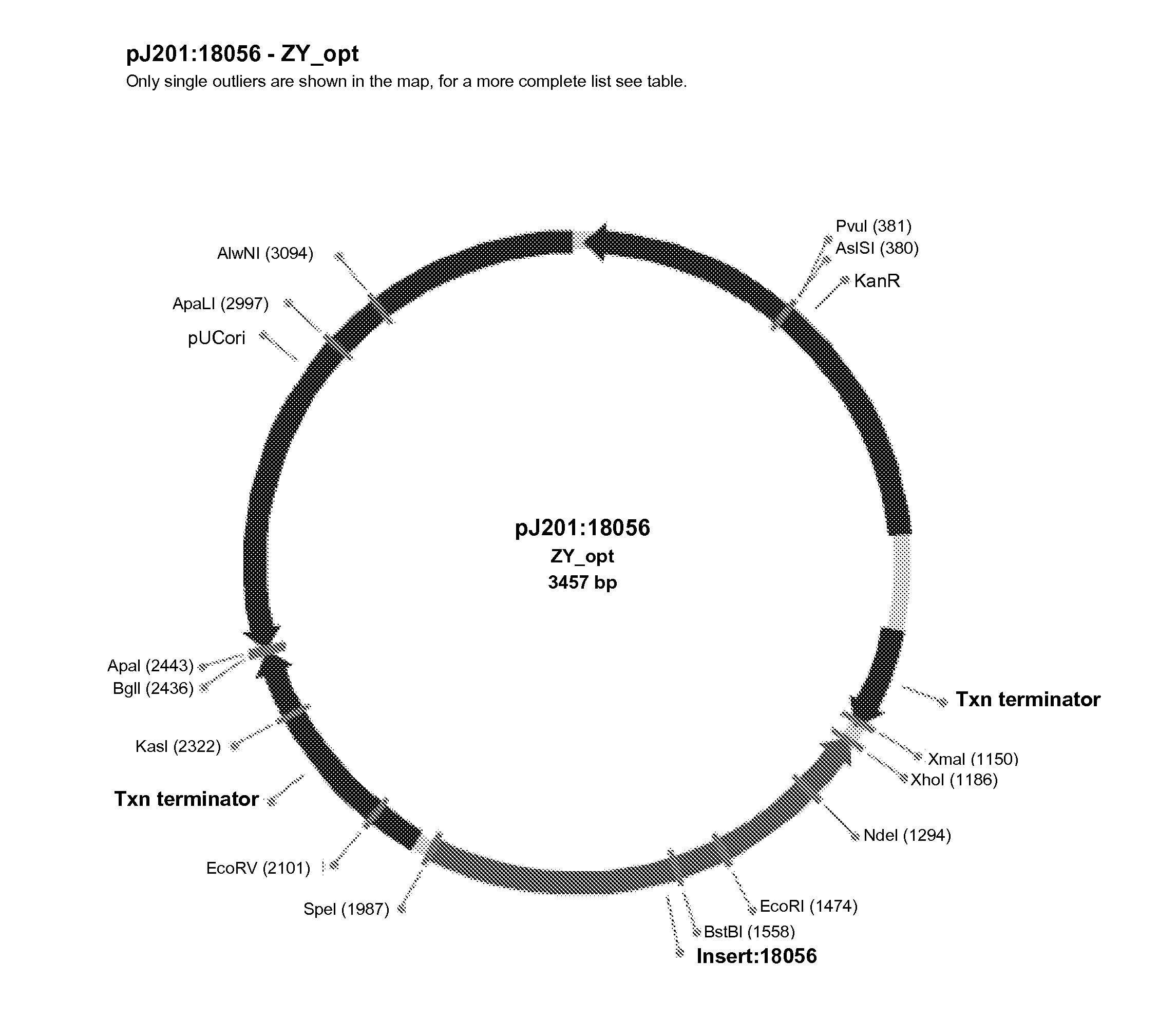
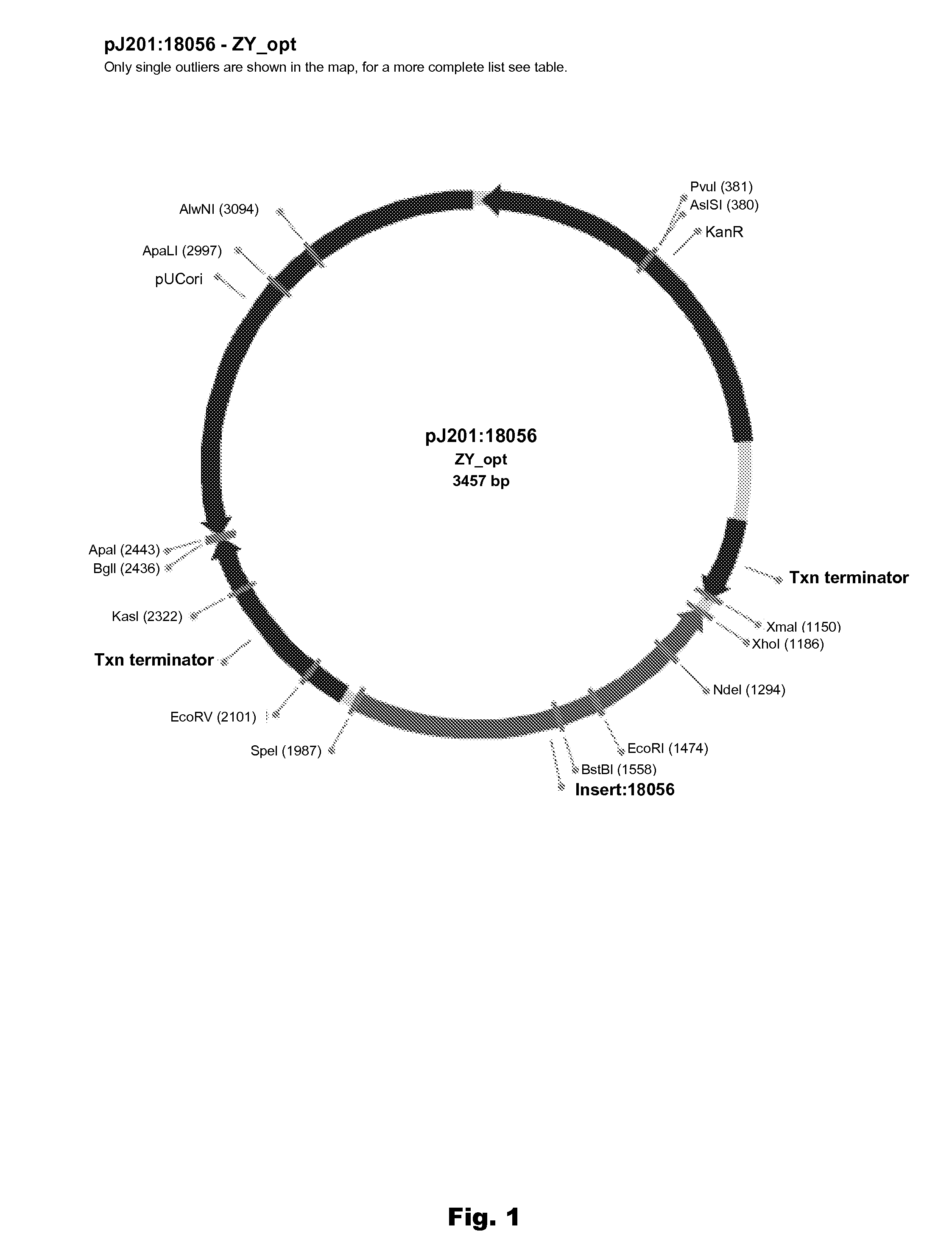
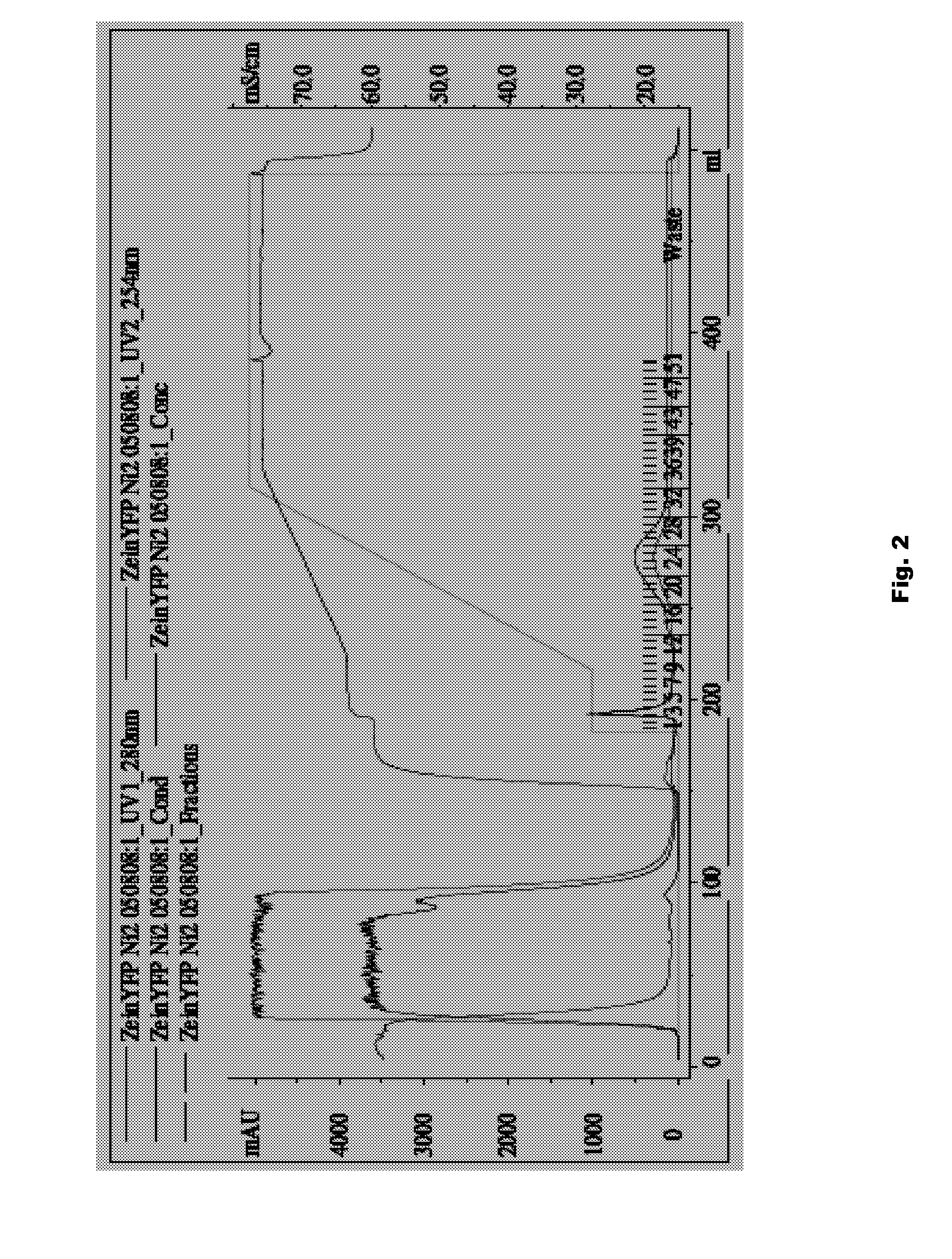
Plant peptide gamma-zein for delivery of biomolecules into plant cells
A method of introducing a molecule of interest into a plant cell having a cell wall includes interacting a gamma-zein peptide with a molecule of interest to form a gamma-zein linked structure. The gamma-zein linked structure is then placed in contact with the plant cell having a cell wall, and allowing uptake of the gamma-zein linked structure into the plant cell. Alternatively, a gene of interest can be expressed in a plant cell having an intact cell wall by interacting a gamma-zein peptide with the gene of interest to form a gamma-zein linked gene structure, allowing uptake of the gamma-zein linked gene structure into the plant cell, and expressing the gene of interest in the plant cell and its progeny.
Owner:CORTEVA AGRISCIENCE LLC
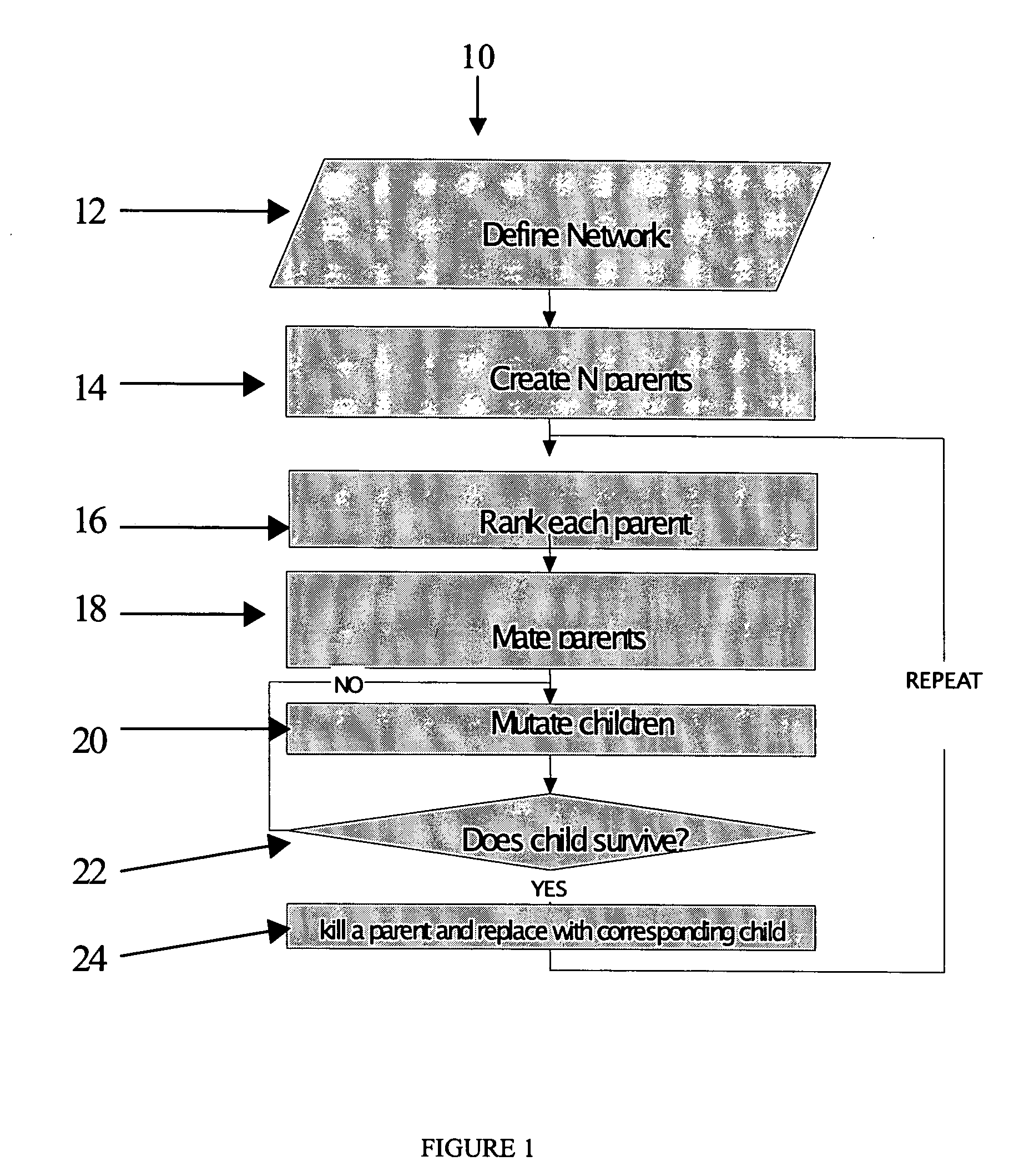
Method of determining at least one variable of a WDM optical network
ActiveUS20070025730A1Robust and cost-effectiveRobust and networkWavelength-division multiplex systemsTransmission monitoringWdm optical networksOptimal design
A method of optimizing a WDM optical ring network configuration using a genetic algorithm. Preferably, the method includes the steps of initially defining one or more parameters for a WDM optical network or portion of a WDM optical network and creating a plurality of parents each having a gene structure corresponding to the one or more parameters of the WDM optical network or portion thereof. The parents are then ranked according to predetermined fitness criteria. The highest ranking parents are then mated to form one or more children. Preferably, the children are then optionally mutated before or after testing to ensure that they satisfy predetermined network demands. Suitable children replace lower ranking parents to form new parents. The process is then repeated by mating the highest ranking parent in an effort to further optimize the network. Various steps in the genetic algorithm can be repeated until an optimal design is achieved.
Owner:CIENA
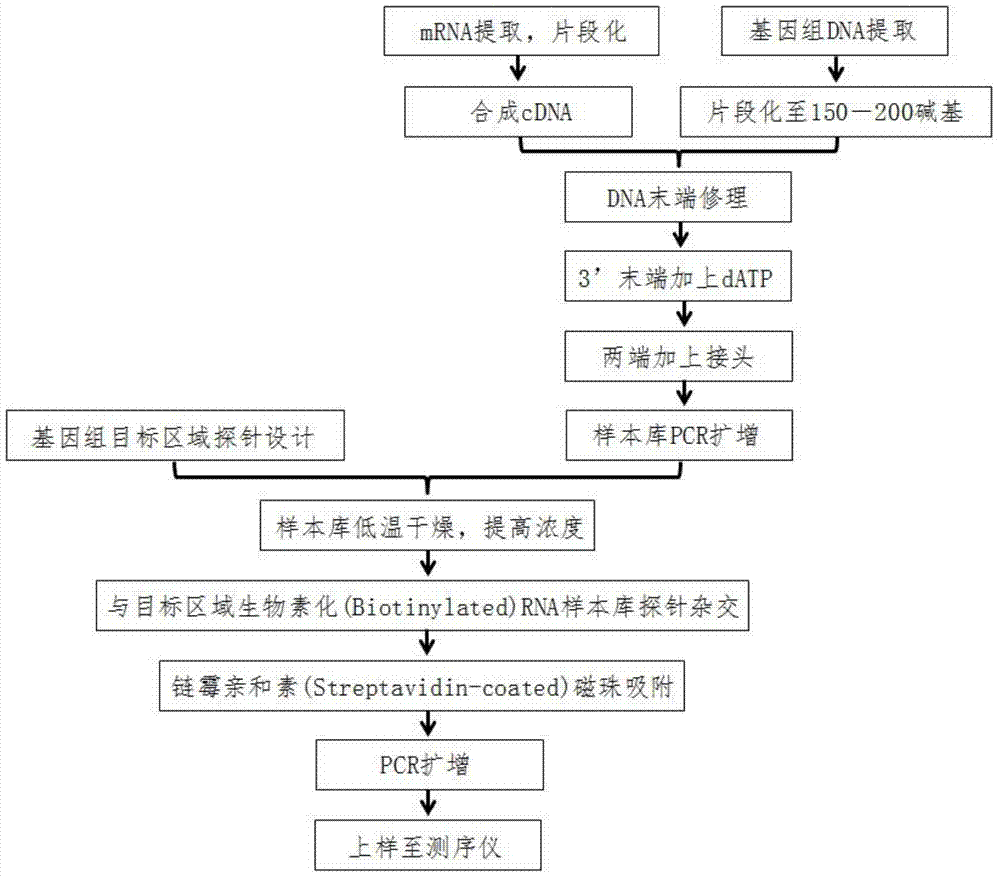
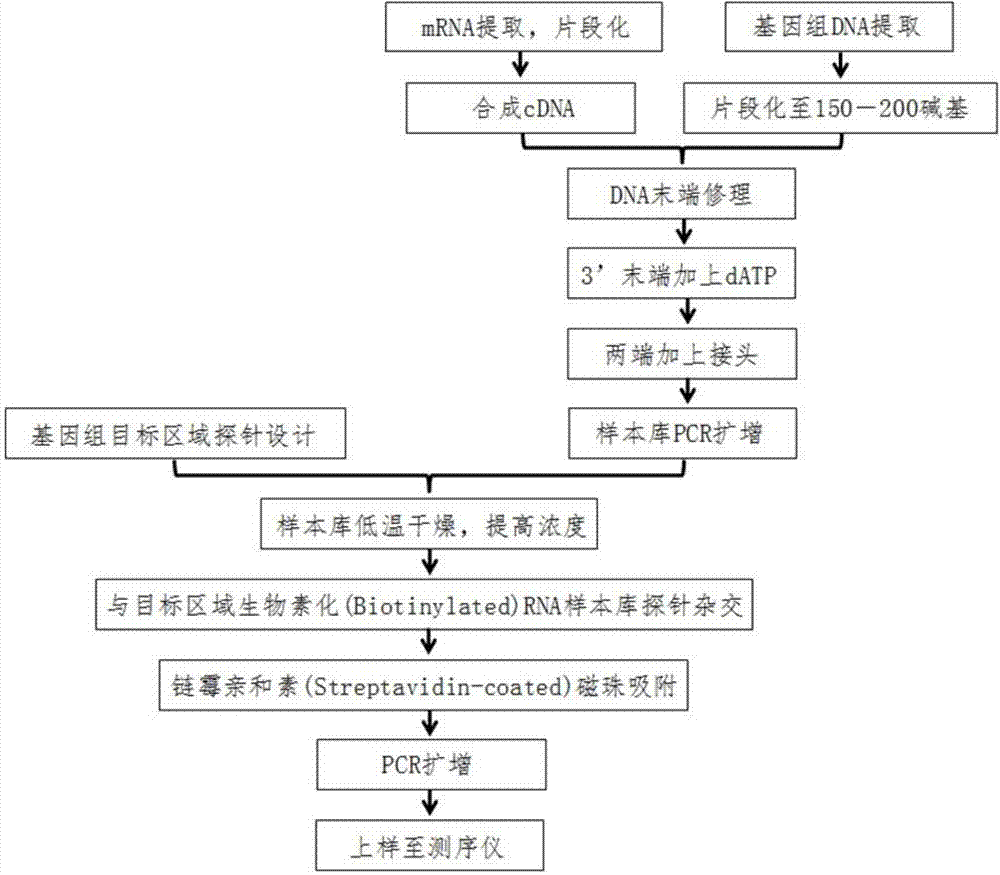
DNA (Deoxyribonucleic Acid) probe library used for hybridization with KRAS gene and method for enriching KRAS gene segments by adopting DNAprobe library
ActiveCN103667267AGuaranteed accuracyImprove accuracyNucleotide librariesMicrobiological testing/measurementBase JKRAS
The invention provides a DNA (Deoxyribonucleic Acid) probe library used for hybridization with a KRAS gene. The DNA probe library comprises one or more DNA probes which can be hybridized with the KRAS gene, wherein the DNA probes include the following sequences: SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO.6, SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.9 or SEQ ID NO. 10. The invention further provides a method for enriching the KRAS gene segments by adopting the DNA probe library. Based on this, the invention further provides a method for detecting gene structure mutations of the KRAS gene. The method disclosed by the invention can be adopted to obtain the KRAS gene segments by enriching in thousands of times, and the KRAS gene segments can be used for a next generation sequencing technology to detect the gene structure mutations including single base mutation, mRNA deletion or increase, mRNA structure transversion and mRNA splicing change.
Owner:GENESEEQ TECH INC
DNA (Deoxyribonucleic Acid) probe library hybridized with BRAF (v-Raf murine sarcoma viral oncogene homolog B1) gene, and method for enriching BRAF gene segments by adopting same
ActiveCN103667268AIncreased sensitivityGuaranteed accuracyNucleotide librariesMicrobiological testing/measurementViral OncogeneGenes mutation
The invention provides a DNA (Deoxyribonucleic Acid) probe library hybridized with BRAF (v-Raf murine sarcoma viral oncogene homolog B1) gene. The DNA probe library comprises one or more DNA probes which can be hybridized with the BRAF gene, each DNA probe comprises the following sequences: SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, OR SEQ ID NO.6. The invention also provides a method for enriching BRAF gene segments by adopting the same. Based on this, the invention further provides a method for detecting the gene mutation of the BRAF gene. The BRAF gene segments can be enriched by thousands of times through adopting the method, the BRAF gene segments can be used for next-generation sequencing technology for detecting gene structure mutation including single base mutation, mRNA deficiency or increase, mRNA structure transversion and mRNA splicing change.
Owner:GENESEEQ TECH INC
DNA (deoxyribonucleic acid) probe library for hybridization with EGFR (epidermal growth factor receptor) gene and method for enriching EGFR gene segments by use of same
ActiveCN103667270AGuaranteed accuracyImprove accuracyNucleotide librariesMicrobiological testing/measurementBase JA-DNA
The invention provides a DNA (deoxyribonucleic acid) probe library for hybridization with an EGFR (epidermal growth factor receptor) gene. The DNA probe library comprises one or more DNA probes which can be hybridized with the EGFR gene, and the DNA probe comprises the following sequence: SEQ ID No.1, SEQ ID No.2, SEQ ID No.3, SEQ ID No.4, SEQ ID No.5, SEQ ID No.6, SEQ ID No.7, SEQ ID No.8, SEQ ID No.9 or SEQ ID No.10. The invention also provides a method for enriching the EGFR gene segments by use of the DNA probe library. On the basis, the invention also provides a method for detecting the gene structure mutation of the EGFR gene. By adopting the method provided by the invention, the EGFR gene segments can be enriched by thousands of and even tens of thousands of times, and the EGFR gene segments can be applied to the next-generation sequencing technology to detect the gene structure mutation, including single base mutation, mRNA (messenger ribonucleic acid) deletion or increase, mRNA structure transversion and mRNA splicing change.
Owner:GENESEEQ TECH INC
Cloud management system and method for gene data analysis and processing
ActiveCN112185468AGood application effectData visualisationBiostatisticsWireless transmissionStructured data analysis
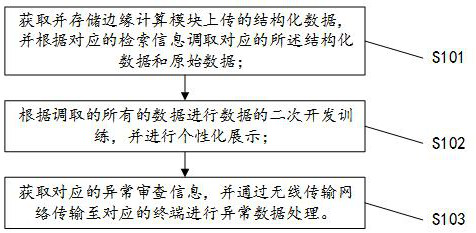
The invention discloses a cloud management system and method for gene data analysis and processing, and the method comprises the steps: firstly receiving and storing structural data transmitted from an edge calculation module in a 5G wireless transmission mode; calling the uploaded gene structured data and the original data of the fluorescence signals of the genes stored in the edge computing equipment according to the corresponding retrieval information, performing personalized display of different functions by utilizing the data, and performing analysis and mining on the gene data by utilizing an artificial intelligence related model; and finding out information with relatively high value for service requirements to carry out secondary development of corresponding functions. Besides, when there is abnormal information transmitted by the edge computing module and suspected abnormal samples obtained by analyzing the structural data by related applications in the system, an alarm is sent to the abnormal processing module in time, the abnormal processing module is reminded to take corresponding measures on the corresponding abnormal information, and the application effect of actual production is improved.
Owner:NANJING SUPERYEARS GENE TECH CO LTD
Hemangioma pathotype subgroup J avian leukosis virus gene and construction of infectious clone thereof
InactiveCN101857873AIncrease contentRich research methodsViruses/bacteriophagesFermentationStructure and functionAngiomatosis
The invention discloses a hemangioma pathotype subgroup J avian leukosis virus gene and the construction of infectious clone thereof. A full-length genome sequence of a subgroup J avian leukosis virus of the invention is shown as SEQ ID NO:1. Then, the hemangioma pathotype subgroup J avian leukosis virus gene shown as SEQ ID NO:1 is subjected to amplification by adopting a specific primer; and pBlueskript SK (I) is used as a vector and chick embryo fibroblast is used as a host cell to construct the infectious clone of the hemangioma pathotype subgroup J avian leukosis virus gene so as to lay the foundation for subsequent research on relation among pathogenic mechanism, a gene structure and functions, virus reproduction, genetic variation rules and the like of the hemangioma pathotype subgroup J avian leukosis virus ALV-J.
Owner:SOUTH CHINA AGRI UNIV
Living cell marking method of biotinylated Curli protein and application of living cell marking method
PendingCN112608876AStrong specificityAchieve biotinylated expressionBacteriaMicroorganism based processesEscherichia coliReceptor
The invention belongs to the technical field of gene engineering, and particularly relates to a living cell marking method of biotinylated Curli protein and application of the living cell marking method. The recombinant bacterium for expressing the biotinylated Curli protein is successfully constructed, escherichia coli BL21 (DE3) is used as a host strain, biotin receptor peptide (APtag) is fused into a gene structure unit csgA of escherichia coli BL21 (ED3) extracellular amyloid protein Curli, a co-expression plasmid pETDuet-1-CsgA-APtag-BirA is constructed, CsgA-APtag and biotin ligase BirA are highly expressed, and biotinylated expression of living cellCsgA is realized; under the induction of IPTG, the streptomycin probe coupled with the fluorophore can be specifically combined with biotinylated CsgA-APtag, and recombinant bacterial cells expressing Curli fibers can be subjected to fluorescence marking. The marking is high in specificity, and the potential influence of the marking on the cell expression function can be reduced to a certain extent.
Owner:JIANGSU UNIV
Method for changing feeding habits of bombyx mori and application of method
The invention relates to a method for changing the feeding habits of the bombyx mori and application of the method. The hestia gene of the bombyx mori is targeted at a fixed point, a gene structure ofthe bombyx mori is damaged to affect normal feeding behavior of the bombyx mori, and thus, the feeding habits of the bombyx mori are changed into omnivory from oligophagy. By the method, a breeding mode of feeding the bombyx mori with folium mori only in sericultural production is solved, and a new through is provided for large-scale bombyx mori feeding.
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI
Method of genetic screening using an amplifiable gene
InactiveUS20050142550A1Reduce productionHigh expressionVectorsMicrobiological testing/measurementHigh-Throughput Screening MethodsGene Organization
The present invention pertains to genetic screening methods and related cells and genetic constructs. In particular, the invention relates to a method of screening cells for an alteration, typically an amplification, in the copy number of a nucleic acid of interest using an amplifiable nucleic acid linked to a reporter nucleic acid; a method of screening cells for increased expression of a polypeptide of interest derived from the nucleic acid of interest; and related cells and genetic constructs. The method allows for high throughput screening of recombinant cells expressing a polypeptide or protein of interest and, in particular, for screening of cells expressing the polypeptide or protein of interest at elevated levels.
Owner:UNISEARCH LTD
A method for rapid cloning of physical quality regulation genes of elite winter sports athletes
The present invention proposes a method for rapid cloning of physical quality expression and regulation genes of elite winter sports athletes. Through extraction peripheral blood genomic DNA, and by using an improved overlap extension polymerase chain reaction (OE-PCR) technique to amplify physical quality related genes, products which are hard to get by using methods relying on restriction endonuclease digestion, can rapidly be obtained by using this technique. The method is time and effort saving, economical, accurate, efficient and simple for studying and exploring rapid cloning of physical quality expression and regulation genes of elite winter sports athletes, providing a most scientific method for the in-depth study of structures and functions of genes related to physical quality of winter sports athletes, and laying a solid theoretical and technical foundation for establishing a scientific system for early prediction of potential, evaluation and selection of winter sports athletes.
Owner:HARBIN INST OF PHYSICAL EDUCATION
Complex disease-related SNP site primer composition and use thereof
InactiveCN113403380AImprove the effect of prevention and controlAccurate acquisitionMicrobiological testing/measurementDNA/RNA fragmentationDisease riskMedicine
The invention provides a complex disease-related SNP site primer composition and use thereof, and belongs to the technical field of molecular biology. A group of SNP site amplification primers are designed and developed through an NGS method. A change of a gene structure related to complex disease in an individual body is detected through a bioinformatics analysis method. The complex disease-related SNP site primer composition predicts an individual disease risk and improves prevention and / or treatment effects of complex disease. The evaluation system and the detection method have low cost, are simple and convenient to operate, and have good sensitivity, high accuracy, good repeatability and great clinical application value.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Novel milchgoat multiparous and monotocous ovary differential expression associated gene, screening method thereof, identification method thereof and functional analysis method thereof
InactiveCN101967484AMicrobiological testing/measurementFermentationSubtractive Hybridization TechniquesOvarian tissue
The invention discloses a novel milchgoat multiparous and monotocous ovary differential expression gene, and a screening method thereof, an identification method thereof and a functional analysis method thereof. Xinong Saanen milchgoats are taken as experimental materials, the forward and reverse subtracted cDNA library of ovarian tissue in a milchgoat multiparous (3 goats) and monotocous anestrus is constructed by using suppression subtractive hybridization technology, differential expression gene fragments are screened, the differential expression gene is verified by real-time quantitative polymerase chain reaction (PCR) and the gene structure and potential function are analyzed by bioinformatics.
Owner:NORTHWEST A & F UNIV
Efficient Bt15A3 strain with excellent gene organization and its separation and application
A Bacillus thuringiesis 15A3 strain is disclosed, which has unique cry gene organization including crylAa, crylAc, crylc, crylD, coyII and cryV. It is obtained by screening with PCR method and bioactivity measuring method. Its wettable powder and liquid preparation has high effect on preventing and eliminating the pests, such as beet noctuid, cotton ballworm, etc..
Owner:NANKAI UNIV
Detection and analysis method of ultra-low frequency mutation sites based on duplex-seq
ActiveCN106599616BMeet the requirements of subsequent analysisIncrease diversityProteomicsGenomicsBarcodeClinical phenotype
A duplex-seq-based ultra-low frequency mutation site detection and analysis method disclosed in the present invention comprises the following steps: 1) evaluating the quality of original sequencing data, reducing data noise, and providing effective data for subsequent analysis; 2) random barcode Extract the title line of each sequence of the sequence file, which is convenient for subsequent quick retrieval of barcode and creation of consistent sequences; 3) Create consistent sequences based on family barcode and duplex barcode, and exclude mutations introduced during the library construction process or PCR process ; 4) Construct a double-stranded consensus sequence according to duplex-tag, and further exclude asymmetric mutation sites in the sequence; 5) Perform local quality correction on the compared data, and perform low-frequency mutation site detection; Three-level annotation of gene structure, function, and clinical phenotype; 6) Statistics of SSCS, DCS sequence numbers, comparison results, and variation site information, and output visual charts.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
A kind of Brassica napus nac47 transcription factor and its preparation method and application
ActiveCN110857317BPositive regulation of agingHigh activityPlant peptidesFermentationBiotechnologyRapeseed
The invention belongs to the technical field of biogenetic engineering, and in particular relates to a NAC47 transcription factor of Brassica napus and its preparation method and application. Specifically disclosed the gene structure of NAC47 transcription factor of Brassica napus, its preparation method and its application in positively regulating the senescence of rapeseed leaves; and disclosed that the expression of Brassica napus NAC47 gene can cause the necrosis of cells to cause the increase of electrical conductivity, The increase of hydrogen peroxide content, the decrease of chlorophyll content, the increase of anthocyanin content and the increase of malondialdehyde content; make the expression of RbohD, RbohF, YLS9, BFN1, CEP1 and two VPE genes better in Brassica napus controlled activation and expression.
Owner:NORTHWEST A & F UNIV
Circovirus 3 type double-copy full-length gene infectious clone plasmid as well as construction method and application thereof
The invention relates to the technical field of virology and biology, and relates to a circovirus 3 type (GyV3) double-copy full-length gene infectious clone using a reverse genetics technology as well as a construction method and application thereof. Specifically, the structural characteristics of a cyclic genome of GyV3 and enzyme cutting sites of the GyV3 are utilized, a GyV3 double-copy full-length infectious clone pcDNA3.1-2GyV3 is constructed by amplifying to two sections of virus whole genomes with different starting and ending points by using a high-fidelity enzyme and connecting to eukaryotic expression pcDNA3.1 (+) through two times of connection transformation, and virus rescue and pathogenicity research in an SPF (Specific Pathogen Free) chicken body are realized; and finally, a simple and rapid circle virus reverse genetics operating system is provided, a platform is provided for GyV3 pathogenicity, gene structure and function research and the like, and a foundation is laid for research of genetic engineering vaccines for GyV3.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
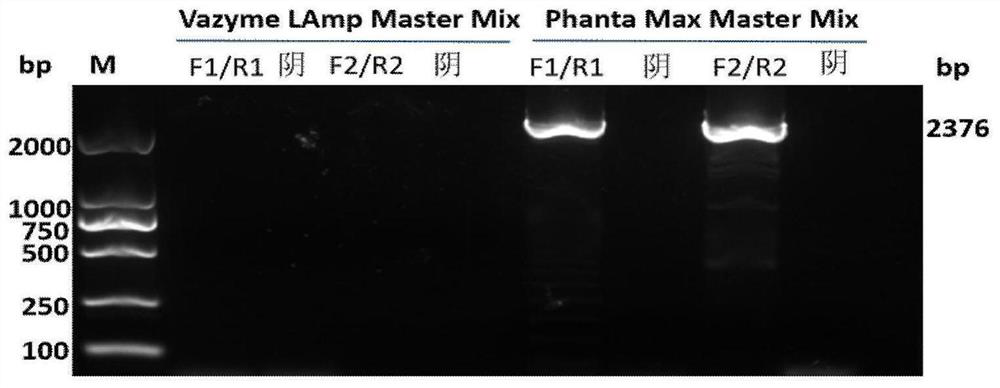
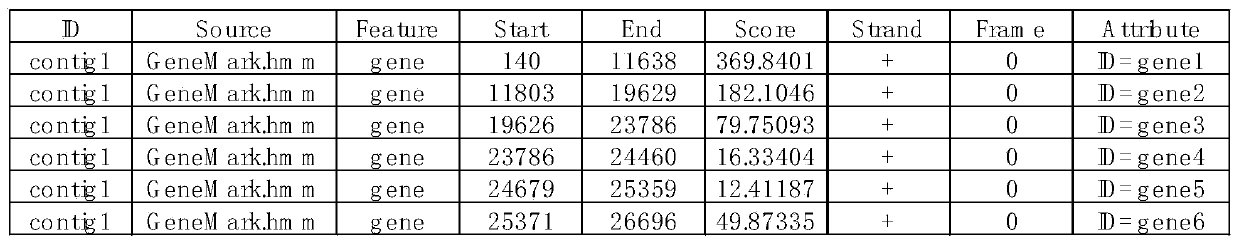
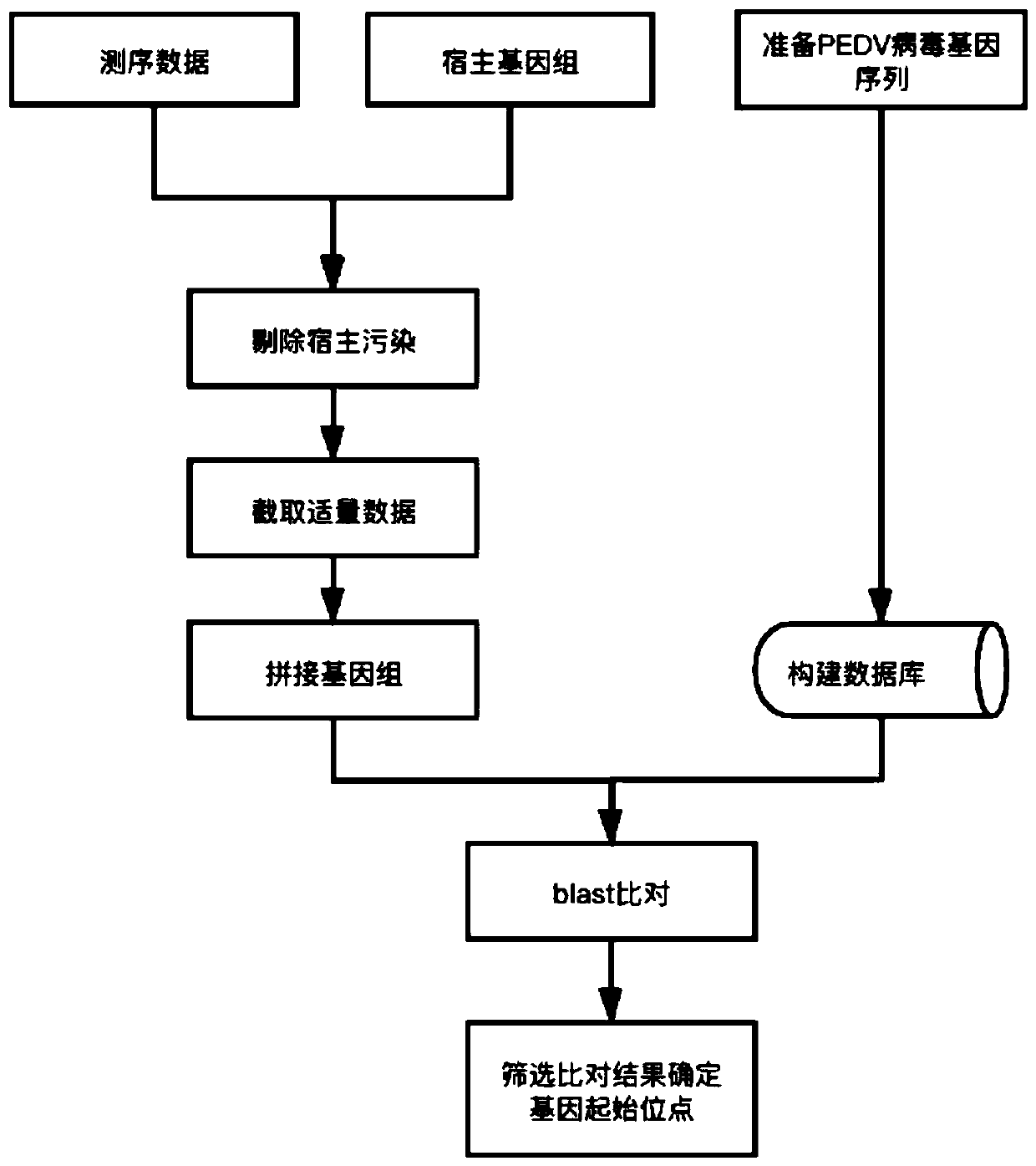
PEDV genome analysis method based on next-generation sequencing
The invention discloses a PEDV genome analysis method based on next-generation sequencing. The method is characterized by comprising the following steps: downloading a genome sequence and a transcriptsequence of a host pig as a host reference sequence; acquiring a sequencing sequence for removing host pollution; splicing the genome; carrying out homologous comparison on the genome; selecting a comparison result; and taking the compared starting site as a starting site of the gene. Compared with the prior art, the method has the advantages that gene structure can be accurately predicted, and omission is avoided.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Tumor-related SNP site primer composition and application
InactiveCN113416782AEasy to solveGood treatment effectMicrobiological testing/measurementDNA/RNA fragmentationMedicineTherapeutic effect
The invention provides a tumor-related SNP site primer composition and application thereof, and belongs to the technical field of molecular biology. According to the invention, a group of SNP site amplification primers are designed and developed through an NGS method, and through a bioinformatics analysis method, the change of a gene structure related to tumors in an individual body is detected, the disease risk of the individual body is predicted, and the prevention and / or treatment effect of the tumors is improved. The evaluation system and the detection method disclosed by the invention are low in cost, simple and convenient to operate, good in sensitivity, high in accuracy and good in repeatability, and have great clinical application value.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Method for Structural Annotation and Alignment Assessment of Full-Length Transcripts
The invention discloses a structure annotation and comparison result evaluation method of a full-length transcript. According to the disclosed comparison result evaluation and gene structure annotation method, matchAnnot software is used, an effect of a script is to modify an existing annotation gtf file and a sam file according to a format required by the matchAnnot software, the matchAnnot is used for structure annotation and comparison result evaluation, a presentation mode of a matchAnnot result is optimized, and counting is carried out.
Owner:南京派森诺基因科技有限公司
Cotton ghmate1 gene and its application in improving the color of cotton brown fiber
The invention relates to an improved cotton fiber color gene technology, and particularly relates to a cotton gene and a plant expression binary vector constructed by the gene and application to improvement of cotton brown fiber color. A nucleotide sequence of the cotton GhMATE1 gene is shown in SEQ ID: NO.1. An amino acid sequence of a protein coded by the gene is shown in SEQ ID: NO.2. On the basis of an early-cloned cotton GhTT12a gene (Chinese patent application number: 201310380270.5, and data of application: 2013-8-27), by using a bioinformatics analysis and electronic cloning method, a new gene containing an MATE (multidrug and toxic compound extrusion) conserved domain database is cloned, but a gene structure and a shearing mode of the cotton GhMATE1 gene are completely different from those of the GhTT12a gene, and the cotton GhMATE1 gene has 78 percent of sequence homology with a nucleotide sequence of an arabidopsis thaliana testa brown pigment synthetic TT12 (Transparent Testa) gene.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES +1
DNA probe library for hybridizing with kras gene and method for enriching kras gene fragments using the same
ActiveCN103667267BGuaranteed accuracyImprove accuracyNucleotide librariesMicrobiological testing/measurementKRASA-DNA
The invention provides a DNA (Deoxyribonucleic Acid) probe library used for hybridization with a KRAS gene. The DNA probe library comprises one or more DNA probes which can be hybridized with the KRAS gene, wherein the DNA probes include the following sequences: SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO.6, SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.9 or SEQ ID NO. 10. The invention further provides a method for enriching the KRAS gene segments by adopting the DNA probe library. Based on this, the invention further provides a method for detecting gene structure mutations of the KRAS gene. The method disclosed by the invention can be adopted to obtain the KRAS gene segments by enriching in thousands of times, and the KRAS gene segments can be used for a next generation sequencing technology to detect the gene structure mutations including single base mutation, mRNA deletion or increase, mRNA structure transversion and mRNA splicing change.
Owner:GENESEEQ TECH INC
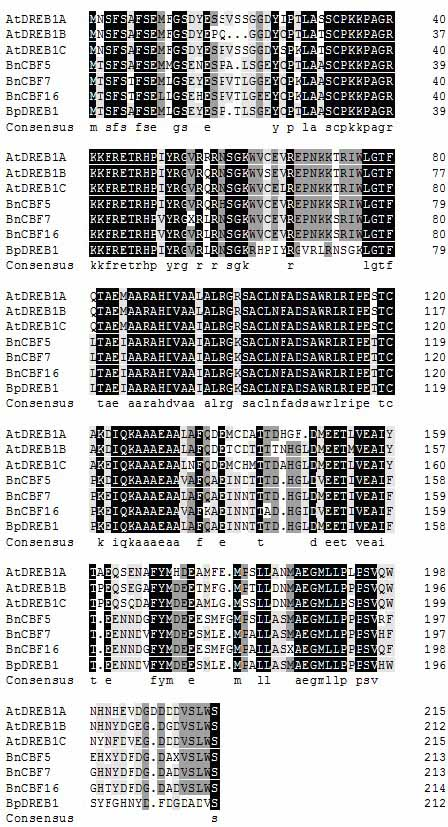
Gene sequence of Chinese cabbage dehydration transcription factor BpDREB1 and application thereof
The invention relates to a DREB transcription factor-BpDREB1 (EF219470). The overall length of a gene sequence is 647bp. A coding protein is predicted to contain 213 amino acids, the relative molecular weight of the protein is 23kDa and the theoretical isoelectric point of the protein is 5.11. The sequence homology of BpDREB1 and the transcription factors in the Chinese cabbage is 94%. An evolutionary tree shows that BpDREB1 belongs to the A1 subtribe in the DREB subfamily. The inducible expression mode analysis of the gene shows that BpDREB1 is subjected to intense and rapid inducible expression at low temperature, has response to drought stress to a certain extent but hardly has response to high salinity treatment. After transgenic arabidopsis thaliana over-expressing BpDREB1 is inducedat low temperature, the contents of soluble sugar and proline in arabidopsis thaliana are substantially increased. The above results show that the transcription factor BpDREB1 has the characteristicsof family member gene structures and plays an important role in a low-temperature and drought response pathway.
Owner:TIANJIN NORMAL UNIVERSITY
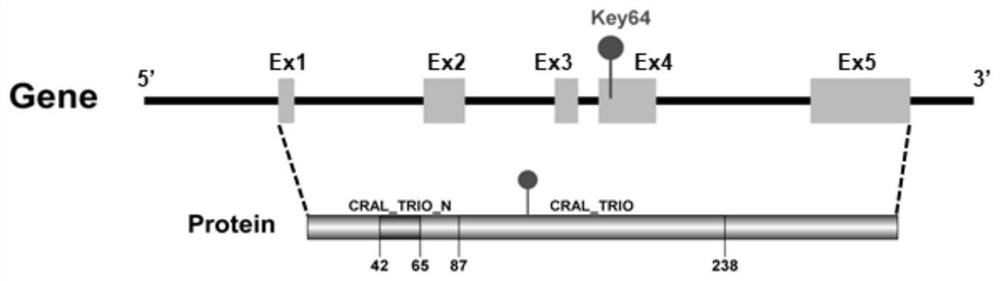
Application of Citrus citpitp1 Gene and Its Nucleotide Sequence in Transformation of Prokaryotic Vector Promoter
The invention discloses the application of a citrus CitPITP1 gene and its nucleotide sequence in prokaryotic vector promoter transformation. The nucleotide sequence of the CitPITP1 gene is shown in SEQ ID NO:1. The present invention screens the CitPITP1 gene from the genome of citrus, and finds from the CitPITP1 gene that a 64bp nucleotide sequence on exon 4 encoding the CRAL_TRIO domain of the CitPITP1 gene can drive gene expression in prokaryotes; therefore, the sequence containing CitPITP1_key64 The method can be applied to the transformation of a vector promoter in the field of genetic engineering.
Owner:HUAZHONG AGRI UNIV
A method for changing the feeding property of silkworm and its application
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI
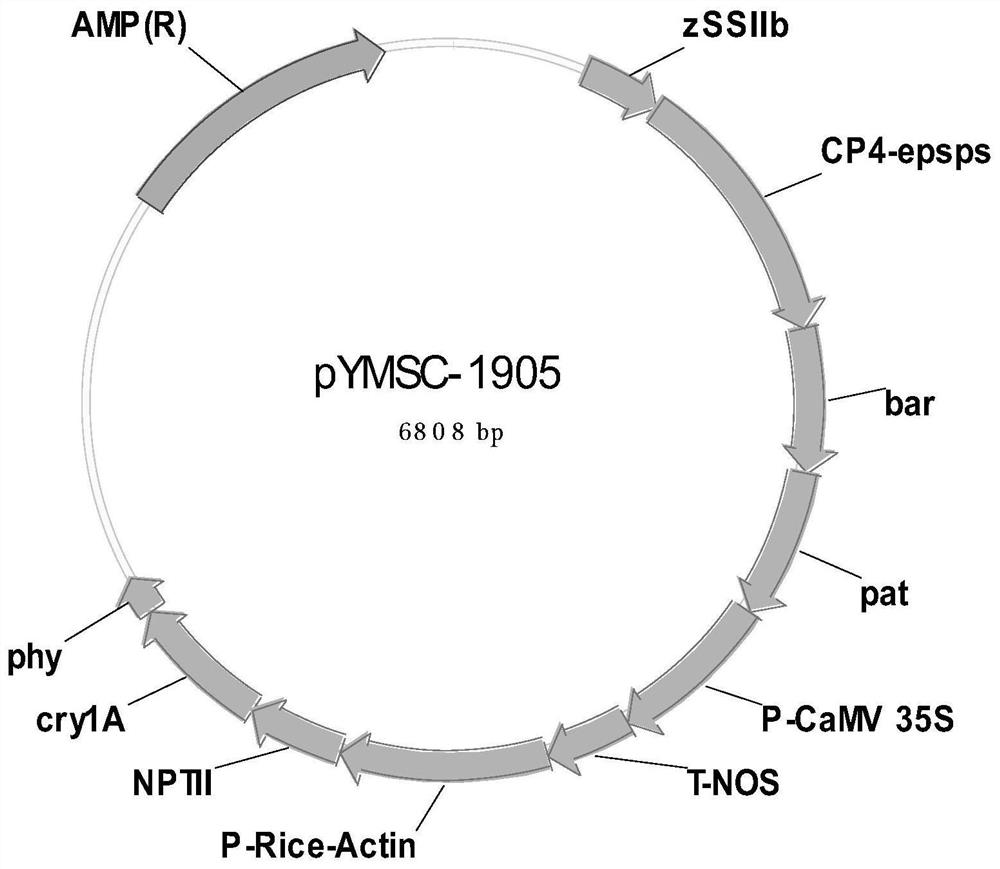
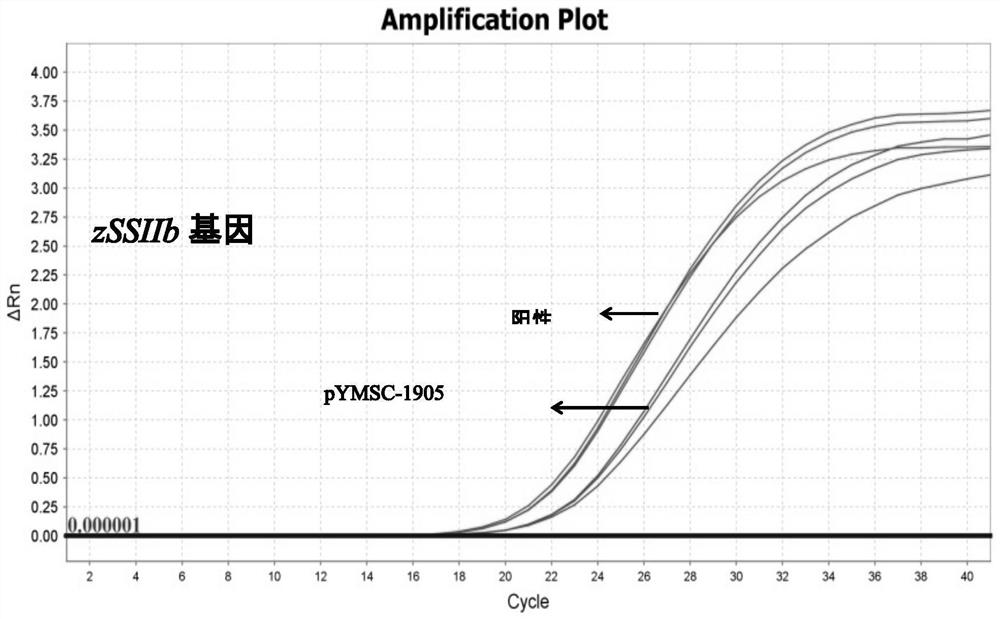
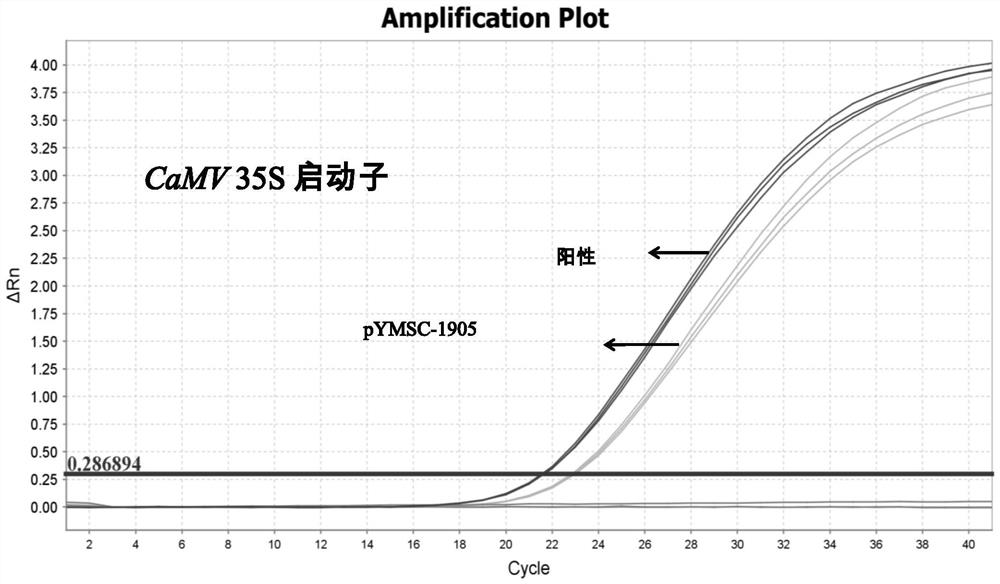
Positive plasmid molecule pYMSC-1905 for screening unauthorized transgenic maize
InactiveCN111826388AMicrobiological testing/measurementVector-based foreign material introductionPlasmid VectorFluorescent pcr
The invention discloses a positive plasmid molecule pYMSC-1905 for screening unauthorized transgenic maize. The plasmid molecule pYMSC-1905 is obtained by constructing structure specific fragments ofa CaMV 35S promoter, an NOS terminator, a bar gene, a pat gene, a CP4-EPSPS gene, an NPT II gene, a cry1A gene, a Rice-Actin promoter and a phy gene in the transgenic maize and a standard gene zSSIIbin the maize to a plasmid vector pUC18. A plasmid disclosed by the invention can screen out 261 kinds of transgenic maize including complex characters, and covers 91.6% of 285 kinds of maize transformants, and the plasmid molecule can be used as a positive control for common qualitative PCR detection and can also be used as a positive control for real-time fluorescent PCR detection.
Owner:黑龙江省农业科学院农产品质量安全研究所 +1
Cancer-targeted, virus-encoded, regulatable t (catvert) or nk cell (catvern) linkers
PendingCN113646334AOrganic active ingredientsSplicing alterationNucleotideNatural Killer Cell Inhibitory Receptors
Owner:RES INST AT NATIONWIDE CHILDRENS HOSPITAL
Glucose dehydrogenase gld gene of Leaperia bancroft and its amplification primer combination and cloning method
The invention provides the glucose dehydrogenase GLD gene of H. bancroft and its amplification primer combination and cloning method, belonging to the technical fields of molecular biology and gene cloning. The nucleotide sequence of the GLD gene is shown in SEQ ID No.6. The acquisition of this gene can further study the composition and gene structure of the gene in the cells of Leaperia bancrofti, and provide a theory for its function and functional region location in the reproductive physiology of Leaperia banecroft, as well as the difference from other species in accordance with.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Molecule marker of red peel gene of weedy rice
InactiveCN101338338BNot affectedEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationOryzaAgricultural science
The invention relates to a molecule marking method for a red peel gene of weed rice and belongs to the field of molecular genetics. A rice type of 02428(white peel) is utilized to be hybridized with Yangzhong weed rice (red peel) to confirm that the red peel gene of the Yangzhong weed rice is a dominance single gene which is decided by a maternal gene; a reciprocal cross result shows to have no cytoplasm effects. An InDel marker RID14 is designed according to the gene structure of the red peel gene Rc of the cloned common wild rice; the InDel marker is co-separated with the red peel gene of the weed rice. The marker can be utilized to accurately and effectively identify the red weed rice and the derived line thereof from the cultivated rice and can provide a molecule basis for the identifying of the weed rice.
Owner:JIANGSU ACAD OF AGRI SCI
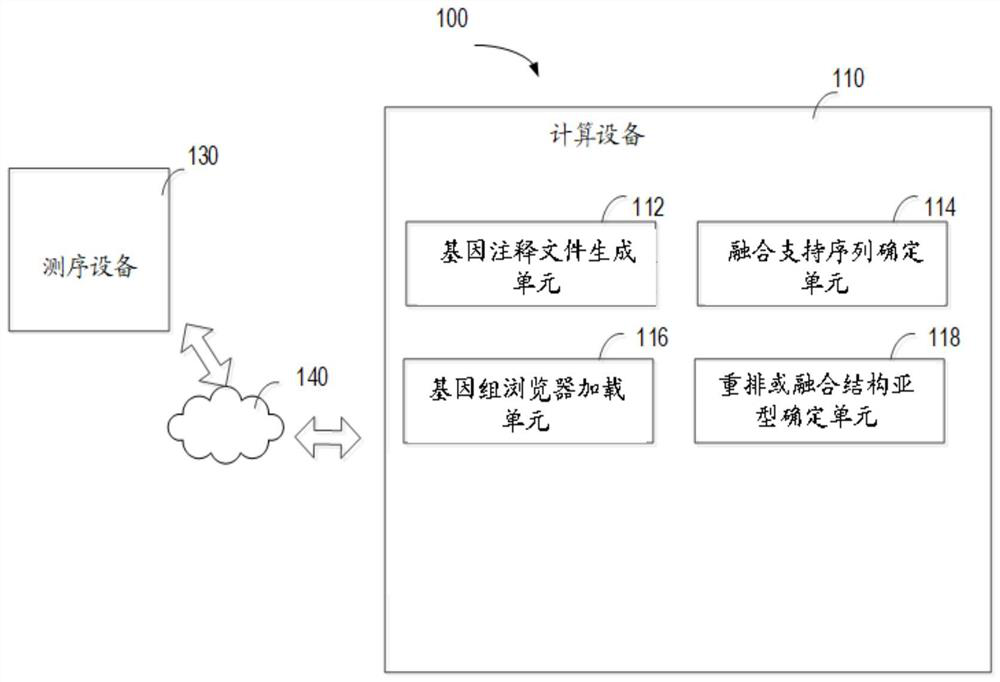
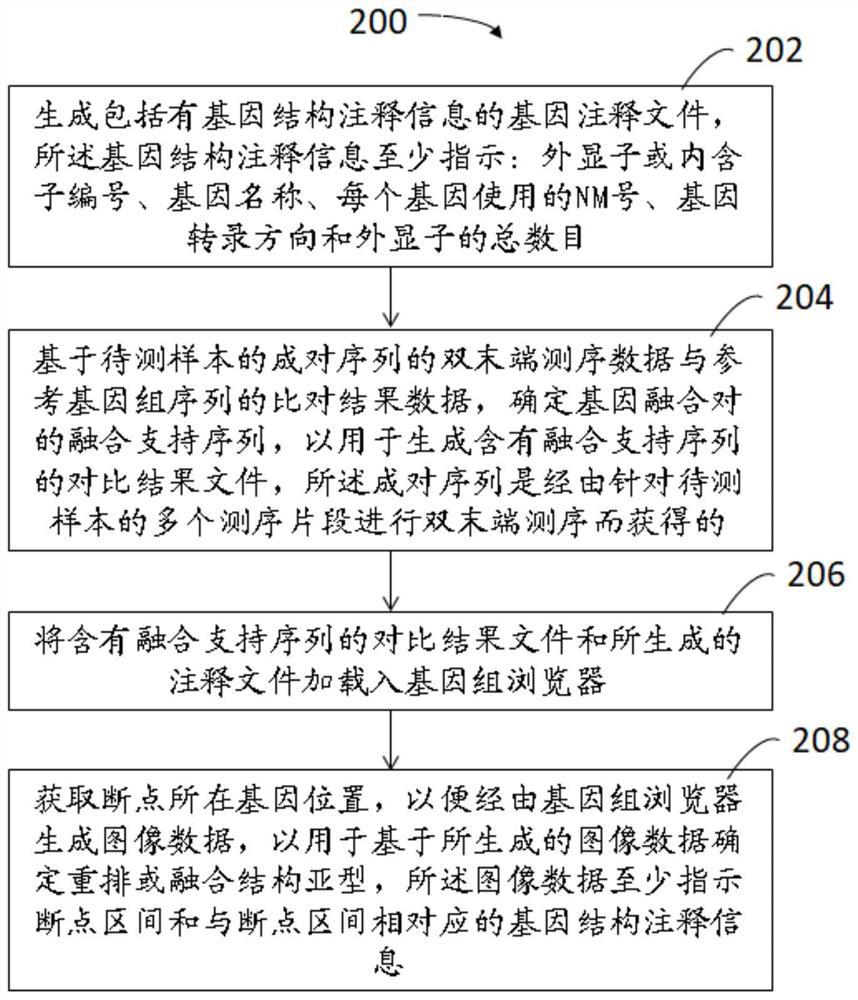
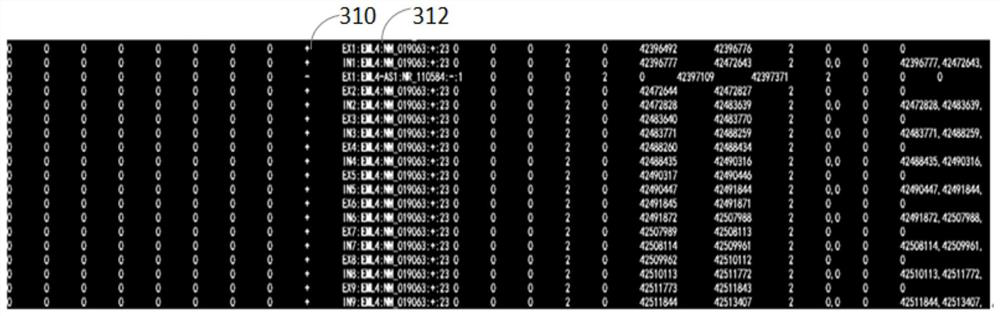
Methods, devices, and media for presenting rearranged or fused structural subtypes
The present invention relates to a method, apparatus and medium for presenting rearranged or fused structural subtypes. The method comprises the following steps: generating a gene annotation file comprising gene structure annotation information, wherein the gene structure annotation information at least indicates an exon or intron number, a gene name, an NM number used by each gene, a gene transcription direction and the total number of exons; determining a fusion support sequence of the gene fusion pair based on the double-end sequencing data of the paired sequence of the to-be-detected sample and the comparison result data of the reference genome sequence, so as to generate a comparison result file containing the fusion support sequence; loading the comparison result file and the generated annotation file into a genome browser; and obtaining a gene position where the breakpoint is located, so as to generate image data through a genome browser and determine a rearrangement or fusion structure subtype based on the generated image data. According to the method, the gene structure where the rearrangement breakpoint is located and the subtype structure formed after rearrangement can be read through a visualization method.
Owner:SHANGHAI ORIGIMED CO LTD +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com