Living cell marking method of biotinylated Curli protein and application of living cell marking method
A technology of biotinylation and biotin, applied in the field of genetic engineering, to achieve the effect of reducing potential impact, low biological toxicity and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of Escherichia coli expressing biotinylated Curli protein
[0032] (1) wxya It is the gene unit of extracellular amyloid Curli fiber, and it is expressed in Escherichia coli BL21(DE3) wxya The gene is the target gene, the peptide domain GLNDIFEAQKIEWHE is translated into a nucleotide sequence, and the flexible sequence GS and wxya The C-terminal fusion connection of the gene was constructed to express the gene fragment CsgA-APtag containing the biotin acceptor peptide; wxyaThe gene is used as a template, and Primer Premier 5.0 software is used to design amplification primers. There are two upstream primers, which are shown in SEQ ID NO.3, namely: TAGCAGCAATTGCAGCAAT, and SEQ ID NO.4, namely: TATTTGCTCTAGAATGAAACTTTTAAAAGTAGCAGCAATTGCAGCAAT; there are three downstream primers , as shown in SEQ ID NO.5, namely: CGACCGCTCATCAGTAC, as shown in SEQ ID NO.6, namely: CGACCGCTCATCAGTACGGCAGTGGCCTGAACGATATCTTTGAGGCACAAAAAATCGAGTGGCA and as shown in SEQ ...
Embodiment 2
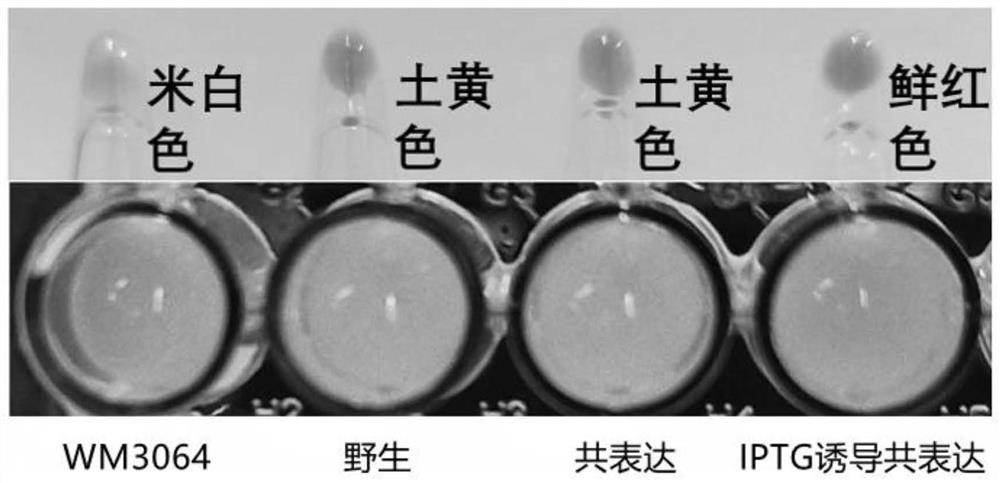
[0039] Example 2: Verification of expression of biotinylated Curli protein in co-expressed BL21 (DE3) strain
[0040] In this example, four groups of different strains were used to verify the expression performance of biotinylated Curli protein, namely: Escherichia coli WM3064, wild Escherichia coli BL21 (DE3) strain, co-expression BL21 (DE3) strain and IPTG-induced co-expression Express BL21(DE3) strain; put Escherichia coli WM3064, wild Escherichia coli BL21(DE3) strain and two groups of single colonies co-expressing BL21(DE3) strain in 37℃ environment for 12-16h, respectively, at 1% ratio Transfer to fresh LB (containing 80uM biotin) medium; the medium of Escherichia coli WM3064 must be supplemented with Dap, and the medium of the co-expressing BL21 (DE3) strain must be supplemented with 40-50 μg / mL ampicillin resistance (Amp). Detection of strain OD by UV spectrophotometer 600 When the value is 0.4~0.6, randomly select a group of co-expressed BL21 (DE3) and add 0.4~0.6mM ...
Embodiment 3
[0044] Example 3: Fluorescent labeling of biotinylated Curli protein in co-expressed BL21 (DE3) strain
[0045] The wild BL21 (DE3) bacterial solution in Example 2 and the IPTG-induced co-expression BL21 (DE3) bacterial solution were mixed with FITC-coupled streptomycin at a volume ratio of 4-6:1, and incubated in the dark for 10 min. Take 1-3 μL of bacterial liquid and drop it on a glass slide, cover it with a cover glass, wash with PBS to remove excess avidin protein, and perform fluorescence microscopy imaging.
[0046] Figure 5 FITC-Avidin-labeled fluorescence imaging comparison chart; in the figure, the left picture is the strain imaging picture under bright field, and the right picture is the corresponding fluorescence imaging picture under 488nm. like Figure 5 It can be seen that the wild BL21(DE3) has no corresponding fluorescently labeled strain, indicating that the expressed Curli does not specifically bind to the fluorescent probe, and no biotinylated Curli is e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com