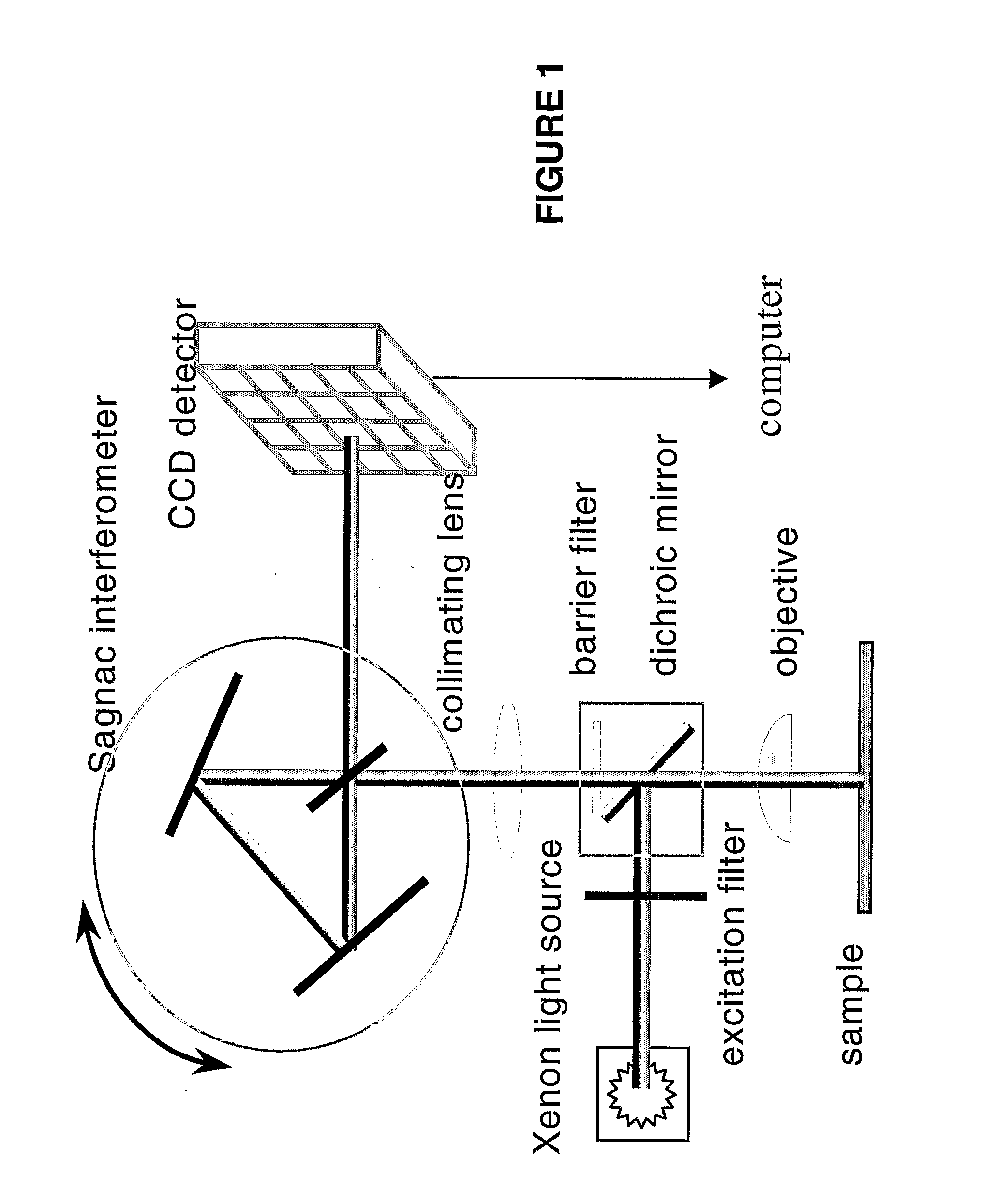
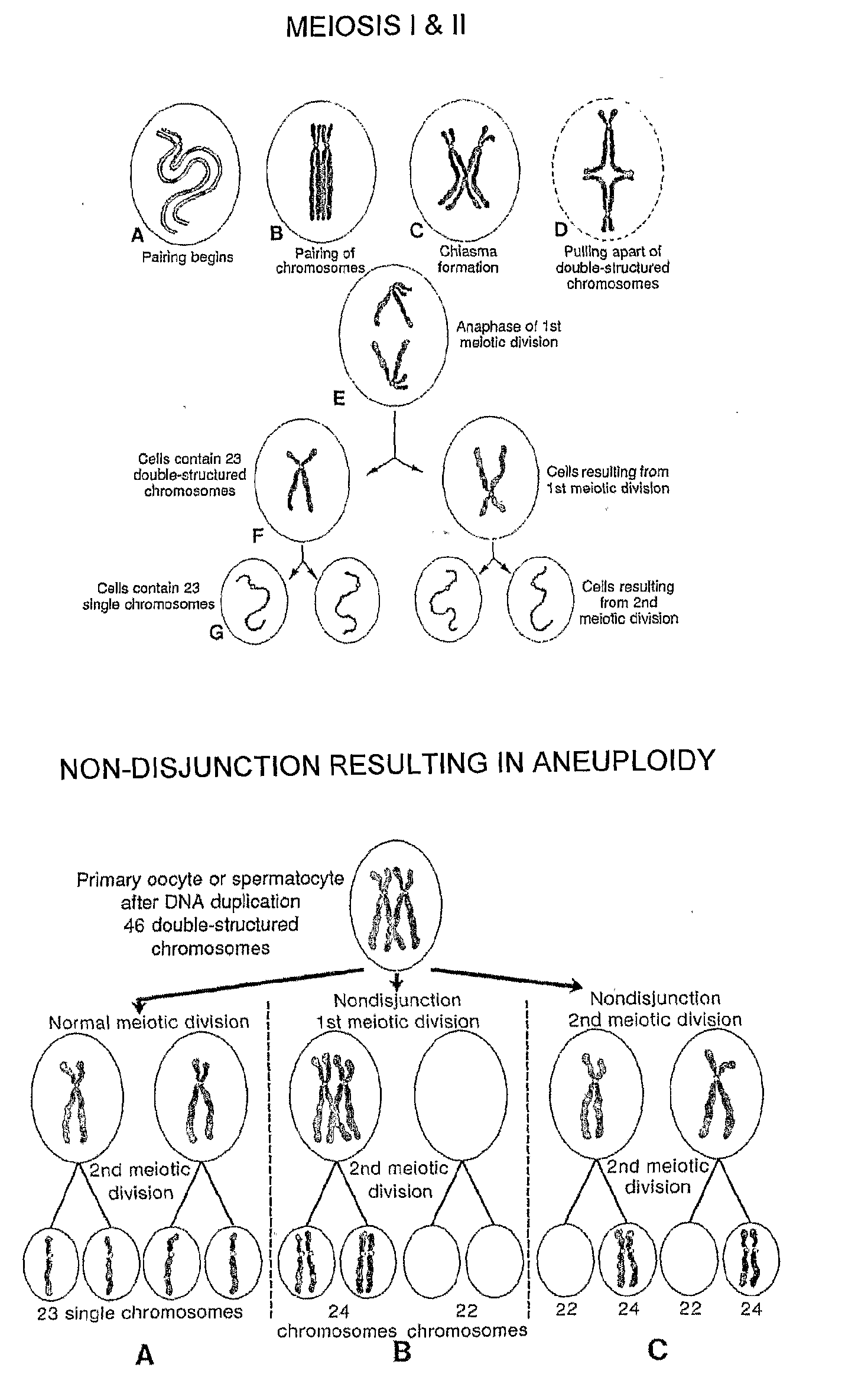
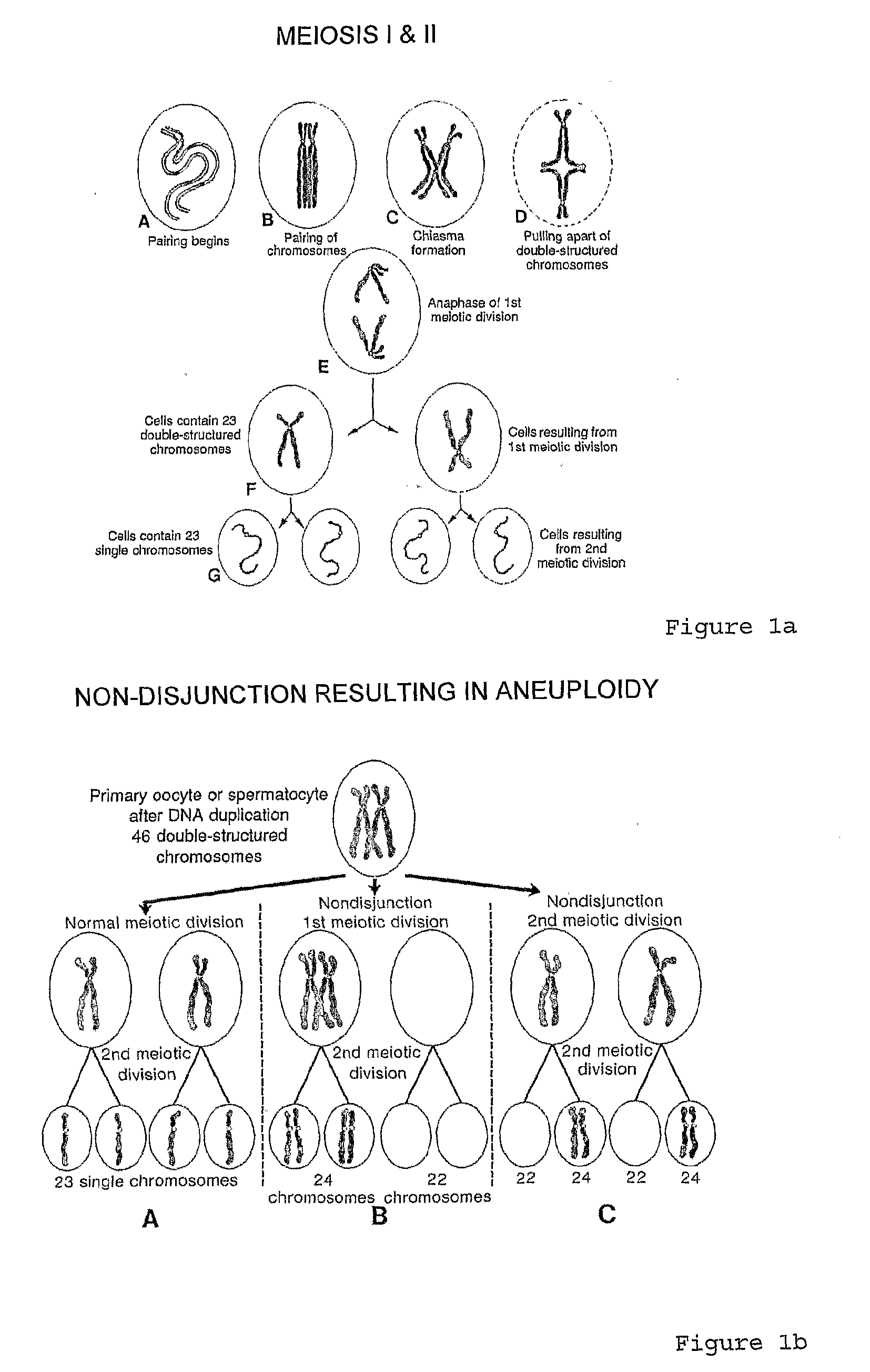
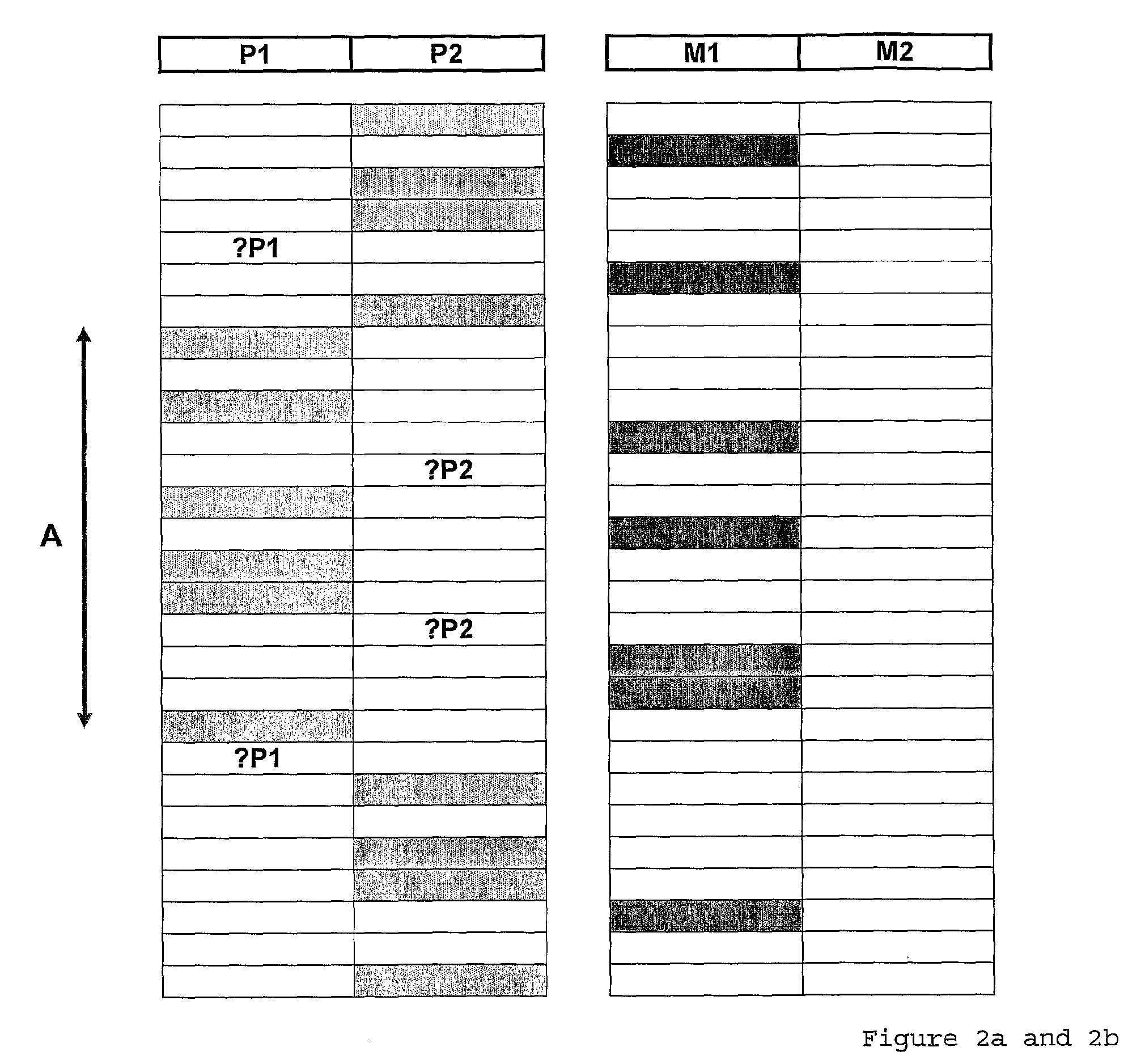
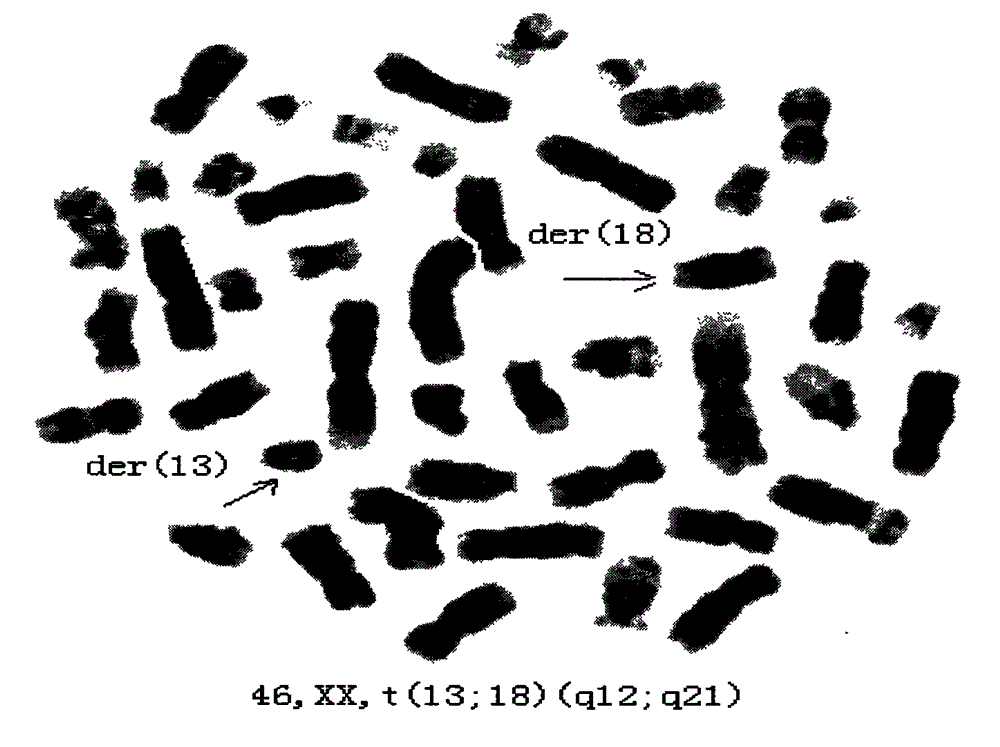
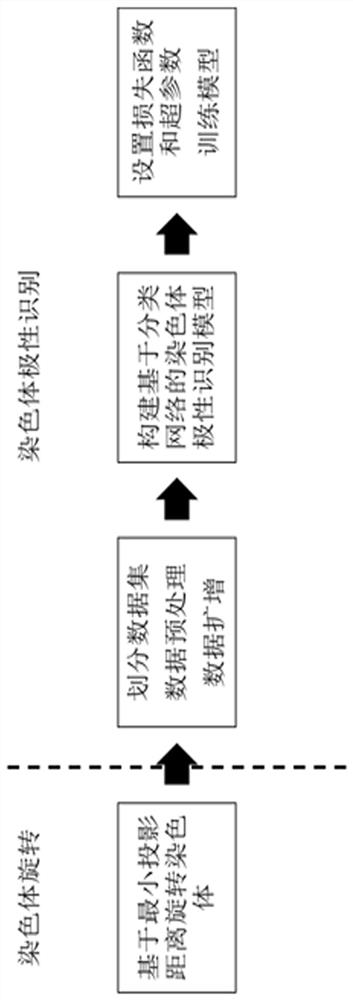
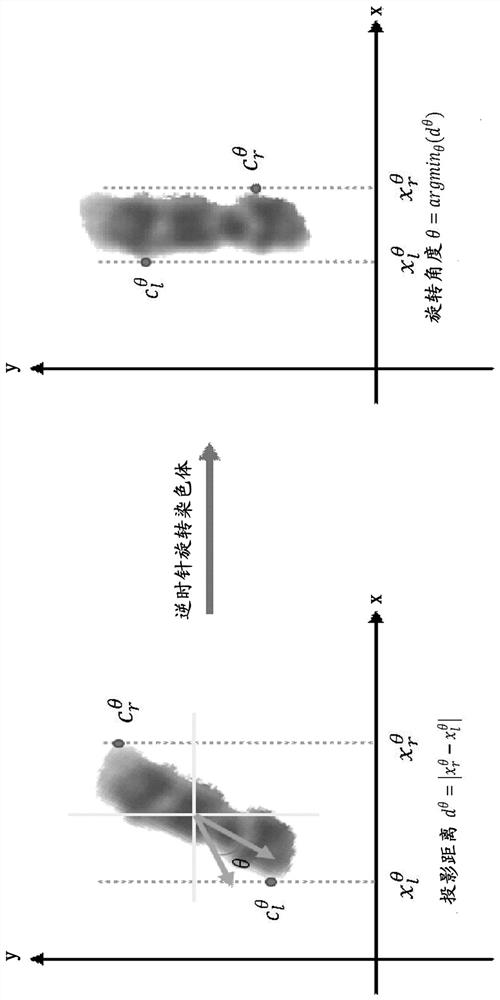
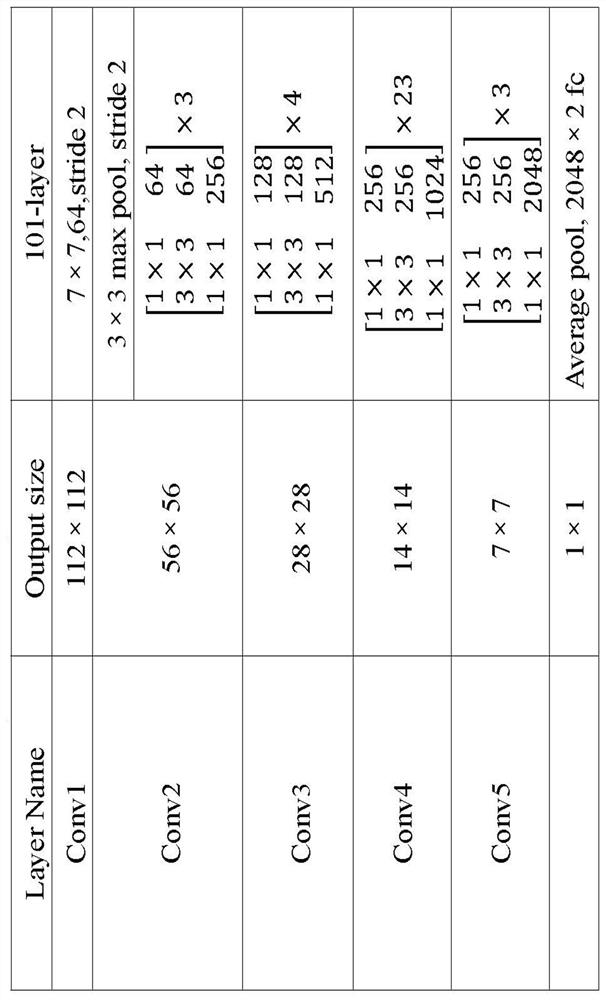
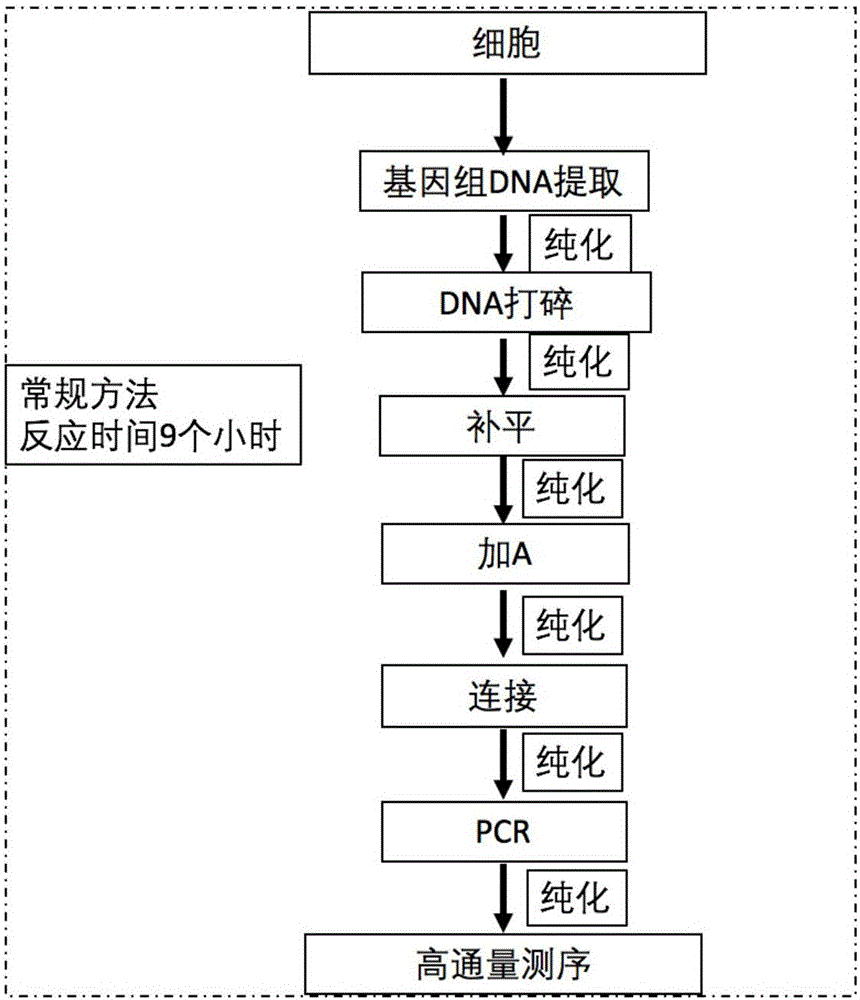
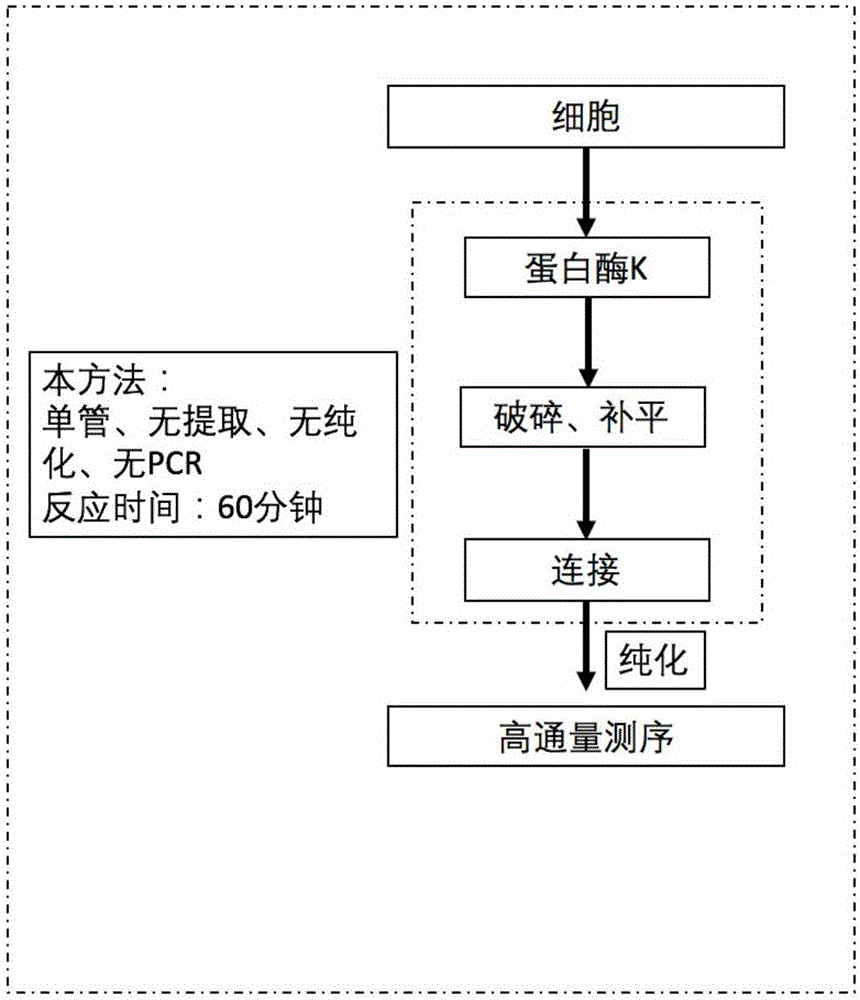
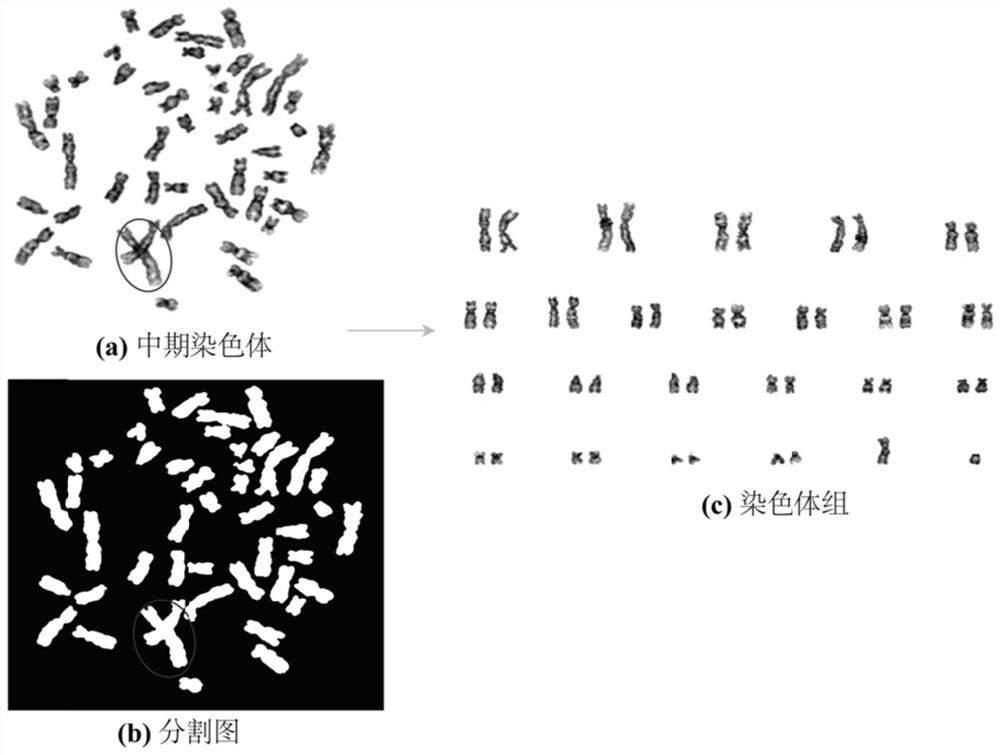
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
49 results about "Chromosome analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Chromosome analysis is the scientific examination of genetic material. Comprised of proteins and DNA, human chromosomes provide extensive information about the genetic makeup of individuals.
Full Karyotype Single Cell Chromosome Analysis
InactiveUS20090098534A1Improve accessibilityReduce nonspecific bindingSugar derivativesMicrobiological testing/measurementCell stainingTumor cells
A full set of 24 chromosome-specific probes to analyze single cells or cell organelles to test for abnormalities is described. When used in an assay based on sequential hybridization, the full set is comprised of three subsets of chromosome-specific probes with each set comprised of 8 different probes. Also described are assays using a set of probes to analyze single cells and cellular organelles to accurately determine the number and type of targeted human chromosomes in various types of cells and cell organelles, such as tumor cells, interphase cells and first polar bodies biopsied from non-inseminated oocytes. Methods of selection or generation of suitable probes and hybridization protocols are described, as are preferred probes for frill set of 24 chromosome-specific probes to target all 24 human chromosomes are described in the Tables.
Owner:REPROGENETICS +1
Chromosomal Analysis By Molecular Karyotyping
ActiveUS20080318235A1Highly accurate linkage analysisAccurate detectionBioreactor/fermenter combinationsBiological substance pretreatmentsY haplotypeChromosomal analysis
The invention provides a method of karyotyping (for example for the detection of trisomy) a target cell to detect chromosomal imbalance therein, the method comprising: (a) interrogating closely adjacent biallelic SNPs across the chromosome of the target cell (b) comparing the result at (a) with the SNP haplotype of paternal and maternal chromosomes to assemble a notional haplotype of target cell chromosomes of paternal origin and of maternal origin (c) assessing the notional SNP haplotype of target cell chromosomes of paternal origin and of maternal origin to detect aneuploidy of the chromosome in the target cell. Also provided are related computer-implemented embodiments and systems.
Owner:BLUEGNOME
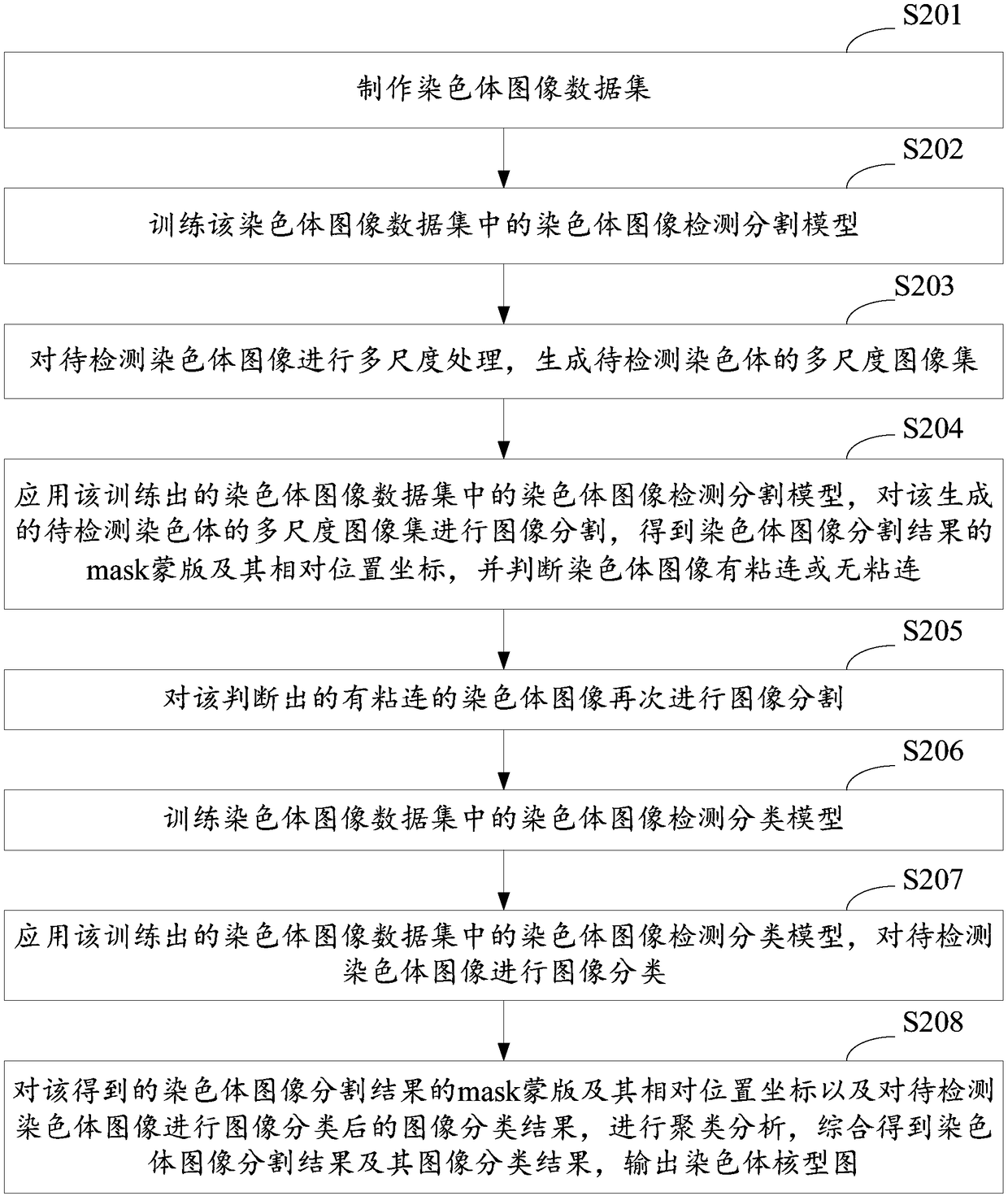
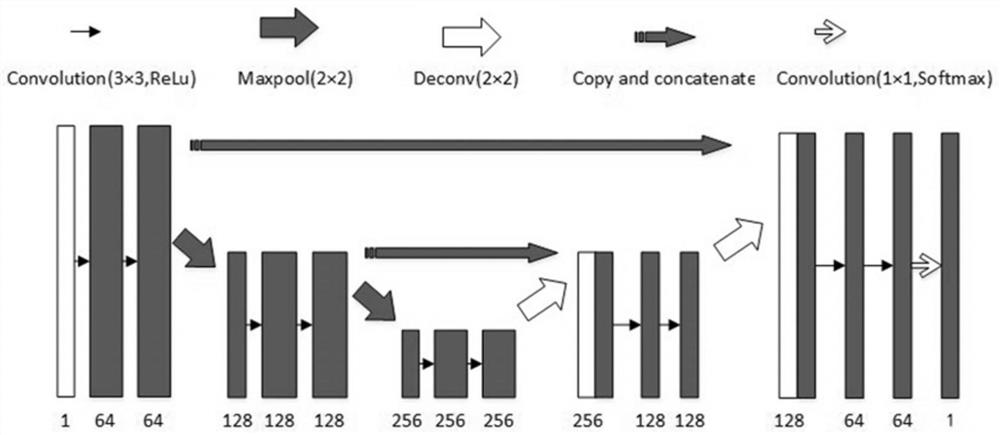
A method and system for automatic chromosome analysis based on depth learning
ActiveCN109344874ATaking into account the overall shapeTaking into account featuresImage enhancementImage analysisPattern recognitionChromosome localisation
The invention discloses a chromosome automatic analysis method and a chromosome automatic analysis system based on depth learning, which can adopt multi-level processing, stratify the independent formand overlapping form of chromosomes, and perform cluster analysis on chromosome position coordinates, classification labels and classification confidence to output a karyotype map. In this manner, the method of chromosome segmentation based on depth learning can be adopted, independent of specific chromosomal morphological patterns, The method has high generalization ability, can adopt chromosomeclassification method based on depth learning, take into account the global morphology and banding characteristics of chromosomes, improve classification accuracy, can adopt multi-scale processing, more fully utilize the detected images, and effectively improve the segmentation effect in the case of chromosome overlap and adhesion.
Owner:HUAQIAO UNIVERSITY
Polymer microsphere containing inorganic nano microparticles, and its preparing method and use
The invention relates to a method for connecting nanoparticles and polymer spheres with chemical bonding in the polymer microspheres and its preparation and biomedical applications, which recombines the water-soluble inorganic nano-particles into hydrophobic polymers with reactive surfactant through emulsion polymerization. In this way, the multi-functional polymer microspheres can be obtained through the recombination of different types of nanoparticles on the same microspheres. The positive charge on the microsphere surface can couple the biological molecules onto it by the electrostatic interaction to get a microsphere fluorescent probe used in immunodetection, chromosome analysis of multiple genes, protein array, DNA sequencing and simultaneous detection in different regions of gene chips and cell or biological tissues.
Owner:INST OF CHEM CHINESE ACAD OF SCI
A chromosome recognition method based on depth learning
ActiveCN109300111AAutomatic IdentificationAccurate identificationImage enhancementImage analysisStudy methodsWorkload
The invention discloses a chromosome identification method based on depth learning, belonging to the technical field of chromosome identification. At present, the method of chromosome analysis is basically manual operation, and the examiner needs a lot of training time to master the knowledge of identifying each chromosome type, and the workload is heavier. Even experienced physicians analyze andidentify patients' chromosomes, the process typically takes more than two weeks and takes a longer period of time. And artificial recognition, subjectivity is very strong, easy to be affected by the external environment, the accuracy is not high. As that depth learn method is adopted, the chromosome type can be accurately carry out, Compared with the existing identification technology, the high-efficient identification technology can effectively improve the efficiency of chromosome karyotype analysis, shorten the identification sorting time, complete the automatic classification and sorting ofchromosomes with high accuracy, at the same time, can effectively reduce the work burden of doctors, free from external interference, and the process is simple, reasonable, and can be applied to large-scale outward promotion.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
Grey scale characteristic graph-based automatic separation method for conglutinated chromosomes
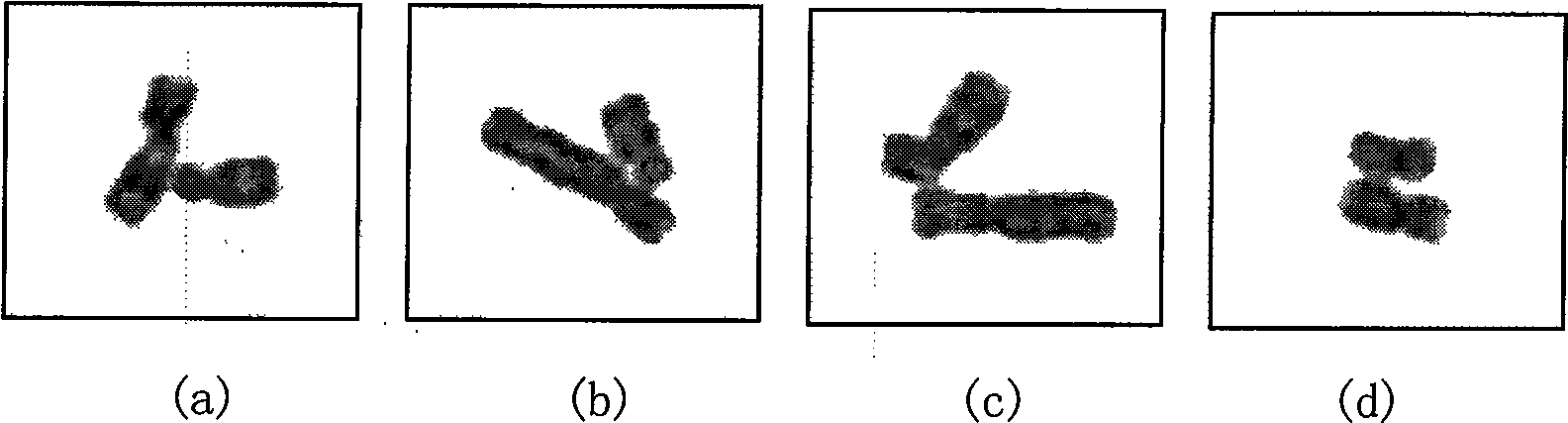
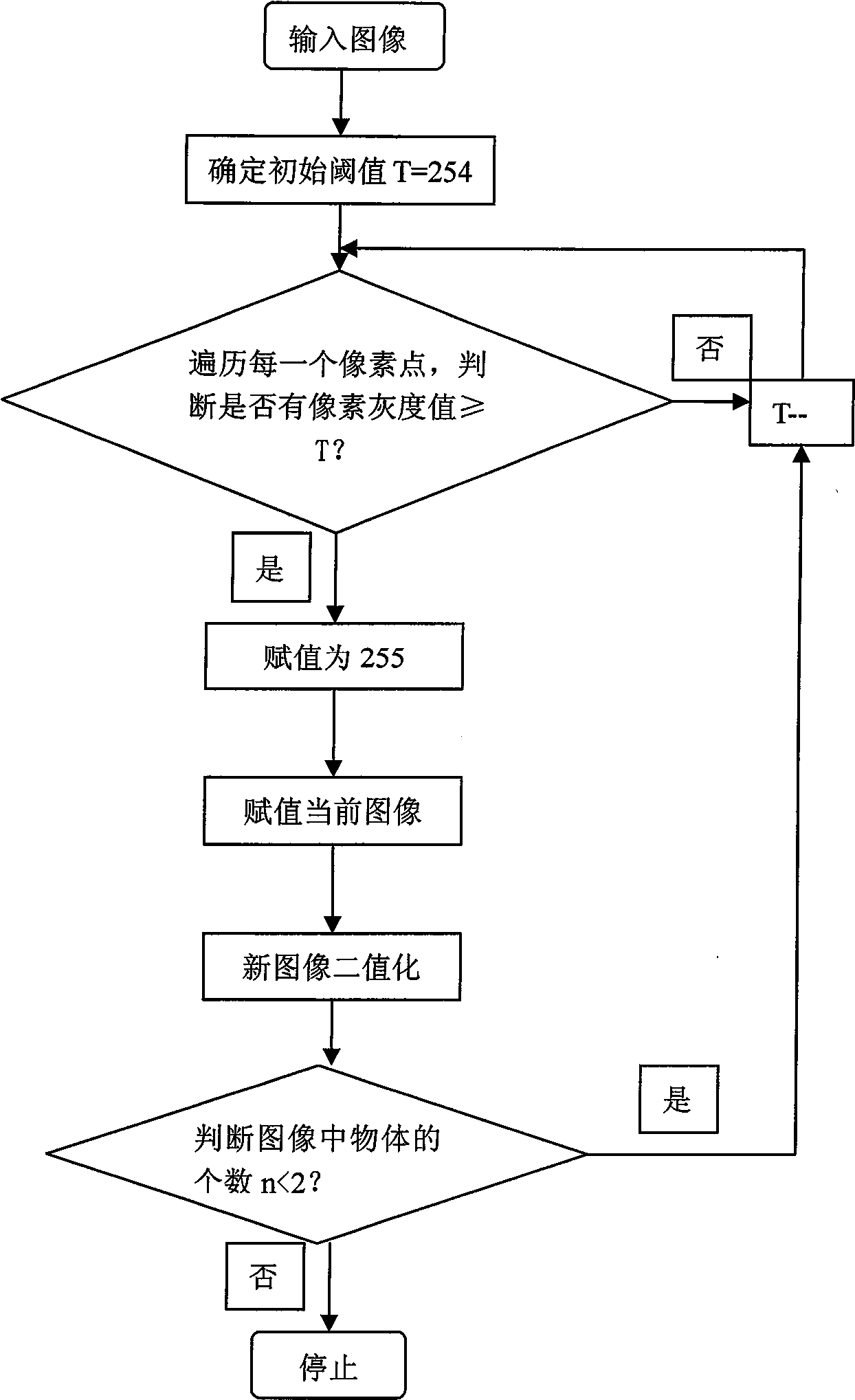
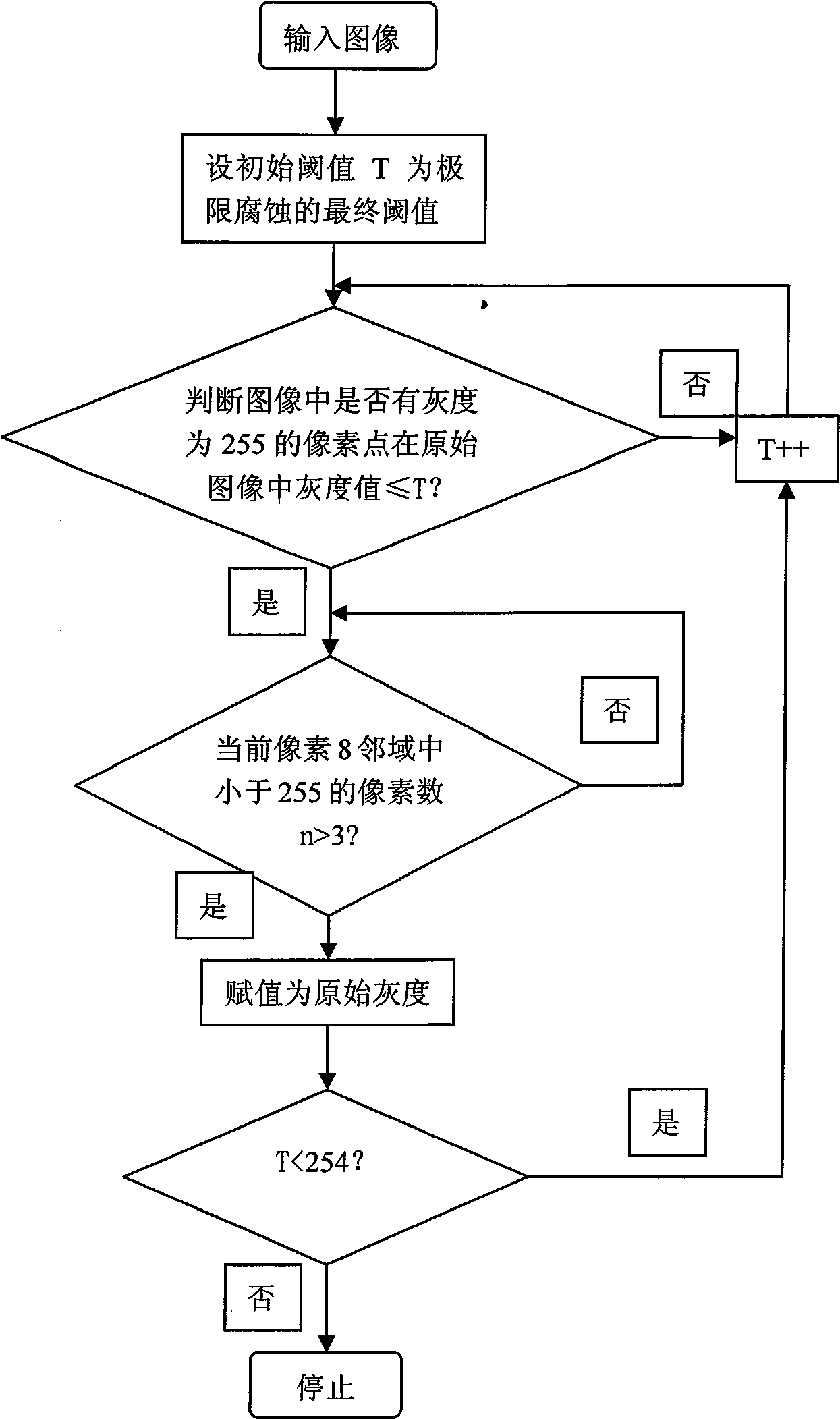
InactiveCN101520890AHigh degree of automationGood Continuity Boundary EffectImage analysisState of artBoundary effects
The invention provides an automatic separation method for conglutinated chromosomes. The method firstly carries out immersion and corrosion processing operation based on a grey scale characteristic graph of chromosomes, and then carries out conditioned expansion recovery processing operation. The invention designs a grey scale characteristic graph-based morphological processing method to separate the chromosomes which are seriously conglutinated but are neither sheltered nor overlapped. The method solves the technical problems and fills up the technical vacancy of separating the chromosomes which are seriously conglutinated but are neither sheltered nor overlapped in the prior art, meets the requirements of accurately separating the conglutinated chromosomes and detecting the number of the chromosomes in medical fields of chromosome analysis, cell research, pathological analysis, and the like, and further improves the automaticity in a chromosome analysis system. The method provides individual chromosomes with good continuous boundary effect and independence for the automatic separation processing in the chromosome analysis system, and particularly keeps concave and convex details of original chromosome image boundary completely and clearly so as to improve the separation automaticity of the conglutinated chromosomes and the accuracy of the chromosome number counting.
Owner:GUANGDONG VTRON TECH CO LTD
Construction method and ultralow temperature freezing and storing method of sinocyclocheilus grahami saccus olfactorius cell line
ActiveCN103305456AEasy to operateActiveAnimal cellsMicroorganism based processesFiberFresh water organism
The invention relates to a construction method and ultralow temperature freezing and storing method of a sinocyclocheilus grahami saccus olfactorius cell line, and belongs to the technical field of cell culture and ultralow temperature freezing and storing method of the biological cell of freshwater aquatic life. A sinocyclocheilus grahami saccus olfactorius tissue is used as a raw material to be cultured in an L-15 culture solution at the pH (Potential of Hydrogen) value of 7.0-7.2 and comprising fetal calf serum by means of a tissue explant and subculture is carried out by means of trypsin digestion. The method specifically comprises the preparation of a cell culture solution, primary culture and subculture. The construction method disclosed by the invention has the following beneficial effects: (1) primary culture takes short time, and cell mass is large and can be applied to chromosome analysis; (2) the constructed sinocyclocheilus grahami saccus olfactorius cell line is in fiber like morphology, can be subcultured continuously and directly applied to a research on biological characteristics, and satisfies the demands of the storage, the theoretical research and the application of sinocyclocheilus grahami germplasm resources; (3) the construction method is also applicable to the construction of saccus olfactorius cell lines of other fishes.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
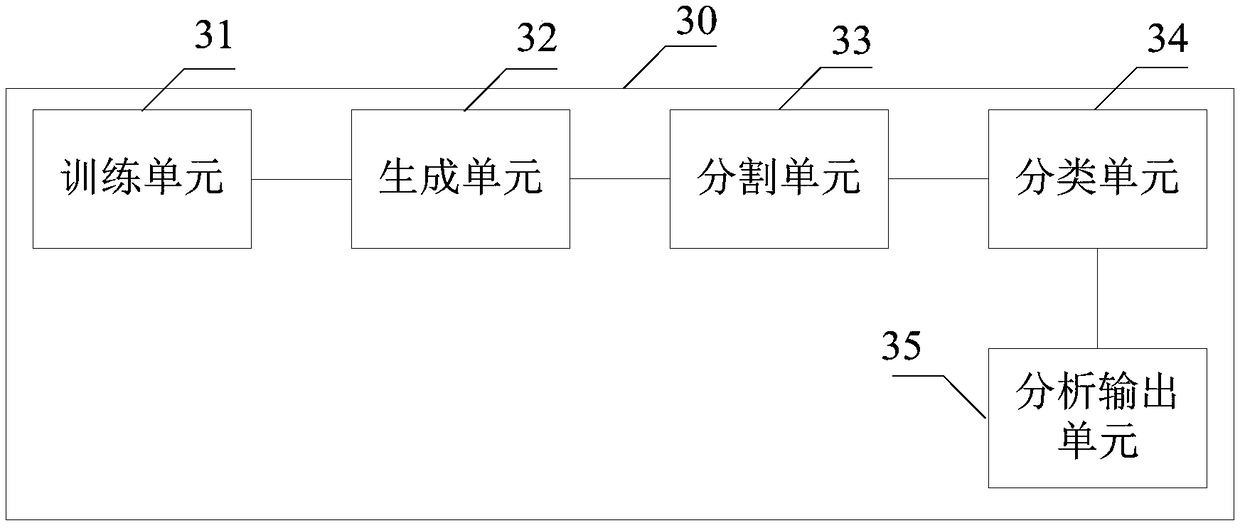
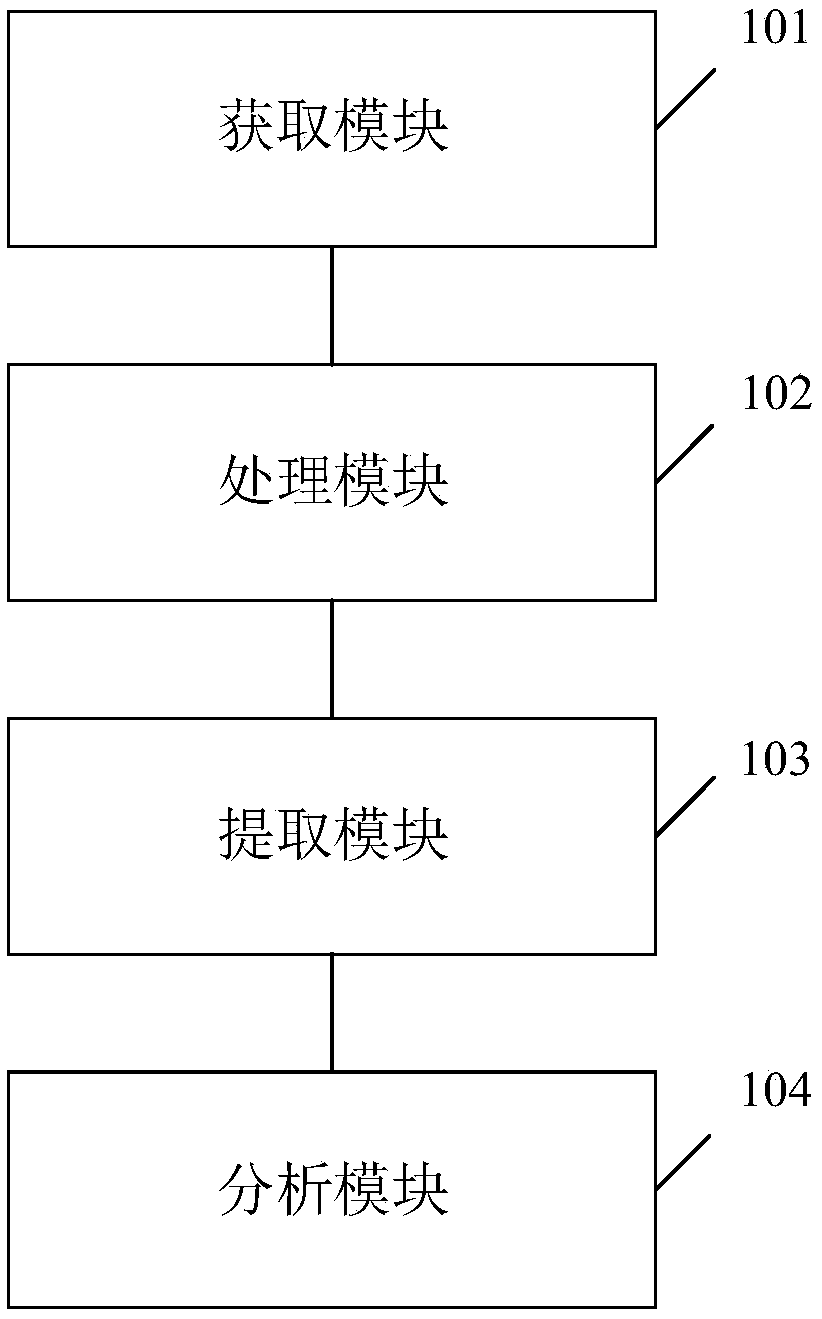
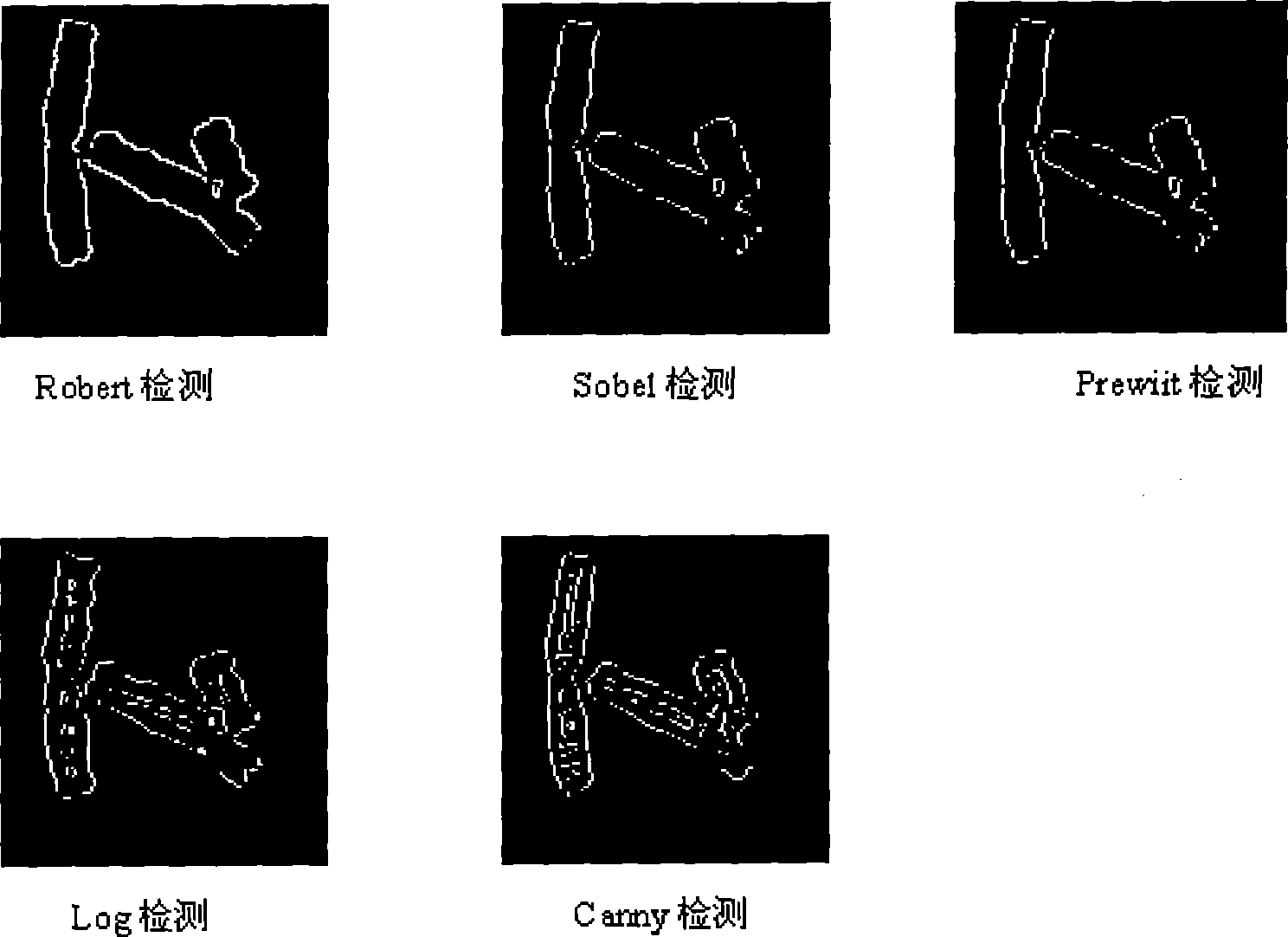
Human chromosome analysis device based on image recognition , apparatus and storage medium
InactiveCN109242842AImprove analysis efficiencyImprove reliabilityImage enhancementImage analysisFeature extractionDependability
The invention discloses a human chromosome analysis device based on image recognition. First, an image corresponding to the chromosome is acquired by an acquisition module, then the image is processedby the processing module to obtain the target image, And the chromosomes in the target image are segmented to obtain a plurality of independent chromosomes. The extraction module extracts the features of each independent chromosome to obtain the corresponding chromosome features. Finally, the analysis module classifies the independent chromosomes by using the trained classifier to analyze the independent chromosomes. Therefore, the present application can analyze chromosomes in an intelligent manner, which not only saves a large amount of time but also is not limited by technical experience compared with the manual method in the prior art, and greatly improves the analysis efficiency and reliability of chromosomes. In addition, the invention also discloses an analysis device and a storagemedium for chromosomes, and the effect is as above.
Owner:郑州金域临床检验中心有限公司
Method for detecting and extracting chromosome contour based on directional searching
InactiveCN101414358AMeet the needs of accurate detection of object contoursImprove accuracyCharacter and pattern recognitionRadiologyOperand
The invention discloses a chromosome contour detecting and extracting method based on directed search, comprising: giving binaryzation treatment to collected images of chromosome; making a point on the contour of the chromosome as a starting point in the image after the binaryzation treatment; searching for and extracting border information continuously along the edge of the chromosome contour in a clockwise direction till reaching the detecting and extracting starting point again, so as to detect the contour of the chromosome wholly. The detecting and extracting method disclosed by the invention satisfies the requirements that contours of objects should be accurately detected in medical treatment fields such as chromosome analysis, cell research, pathological analysis, etc. The method of the invention improves automation level of segregating and incising overlapped and adherent chromosomes according to the contour, perfects the shortcomings of traditional computing methods that the contour fractures, the contour is incontinuous and unevenness details of the contour are ignored, and improves the accuracy of detecting results, furthermore operand is greatly reduced.
Owner:GUANGDONG VTRON TECH CO LTD
Partition method for crossing-overlapped chromosome
InactiveCN101499165ARealize automatic segmentationImprove accuracyImage enhancementImaging analysisSlope ratio
The invention provides a method for partition cross overlapping chromosome which relates to the medicine microimage image analysis field of digital medical treatment. The method includes steps as follows: firstly, describing line segment in a central region of the cross overlapping chromosome; secondly, calculating slope ratio of the line segment and normal slope ratio of the line segment and obtaining the calculated result; thirdly, measuring the line segment length according with the calculated result and confirming a partition point according with the measured result; fourthly, partition the cross overlap chromosome according with the partition point. The method provided by the invention judges the partition point based on the method for measuring the line segment length and processes chromosome partition according to this which can reduce difficulty and working amount of manpower partition, increase automatization degree for analyzing chromosome, and has simple arithmetic.
Owner:GUANGDONG VTRON TECH CO LTD
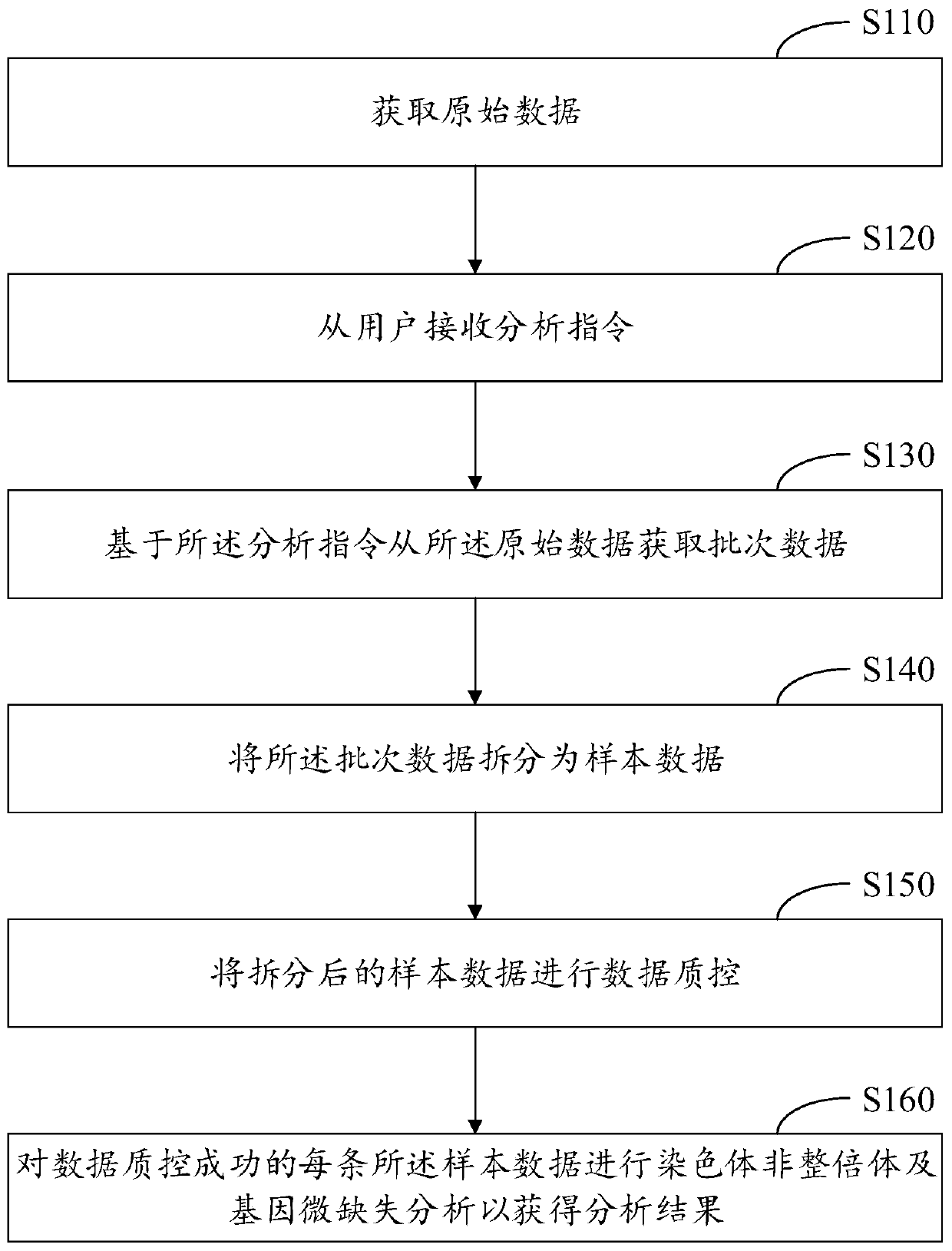
Sequencing data automated analysis method and device, and electronic device
The invention discloses a sequencing data automated analysis method and device, and an electronic device. The sequencing data automated analysis method includes the steps: acquiring raw data; receiving an analysis instruction from a user; acquiring batch data from the raw data based on the analysis instruction; splitting the batch data into sample data; and performing data quality control on the split sample data; and performing chromosomal aneuploidy and gene microdeletion analysis on each of the sample data being successful in data quality control, so as to obtain an analysis result. In thisway, chromosomal aneuploidy and gene microdeletion analysis results can be obtained directly from the raw data based on user instructions, so that the labor and time cost of chromosome analysis can be reduced and ease of use can be improved.
Owner:ZHEJIANG ANNOROAD BIO TECH CO LTD +2
Chromosome preparation method, as well as required culture medium and preparation method thereof
InactiveCN102443623AQuality improvementEasy to check and analyzeMicrobiological testing/measurementPhytohemagglutininsPrenatal diagnosis
The invention discloses a chromosome preparation method, as well as a required culture medium and a preparation method thereof, and is used for solving karyotype analysis problem of chromosome. The culture medium consists of RPMI (Roswell Park Memorial Institute) 1640, heparin sodium, HEPES (2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid), L-glutamine, NaHCO3, benzylpenicillin potassium, streptomycin sulphate, bovine serum and phytohemagglutinin (PHA). The detection method comprises the following steps: implanting 0.3 to 0.4ml of human peripheral blood into the culture medium; adding colchicinamide in 2-4 hours before culture is terminated to realize that the cell is terminated in anaphase; culturing the cell after 68 to 72 hours to harvest the cell; performing hypotonicity for 40 minutes, three times of fixation, banding, dyeing and other treatments; and performing chromosome analysis under a microscope to determine whether the peripheral blood supplier has a phenomenon of chromosome abnormality. The culture medium disclosed by the invention has convenience for use, simpleness in operation, low cost and low patient detection fee, and is suitable for genetic diagnosis, infertility and prenatal diagnosis in each level of hospitals.
Owner:苏州苏大赛尔免疫生物技术有限公司
Construction and ultralow temperature freezing preservation method of fin cell line of schizothorax grahami
ActiveCN103436490AEasy to operateActiveDead animal preservationVertebrate cellsSerial passageGermplasm
The invention relates to construction and ultralow temperature freezing preservation method of a fin cell line of schizothorax grahami, and belongs to the technical field of culture and ultralow temperature ultralow temperature freezing preservation of cells of fresh water aquatic organisms. The method comprises the following steps of: culturing in a DMEM(Dulbecco Modified Eagle Medium) / F12 culture solution which contains fetal calf serum and cell growth factors and has the pH value of 7.0-7.2 by taking the ventral fin tissue of the schizothorax grahami as a material and adopting a tissue explant method; carrying out subculture by adopting a trypsin digestion method. The method particularly comprises the steps of cell culture solution preparation, primary culture and subculture. The construction method disclosed by the invention is short in primary culture time consumption and large in cell quantity and can be used for chromosome analysis; the constructed fin cell line of the schizothorax grahami is in a fiber-like form, can be subjected to serial passage and directly applied to biological characteristic research, meets the requirements for germplasm resource conservation and theoretical research and application of the schizothorax grahami and is suitable for constructing the fin cell lines of other fishes.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
Adhesion chromosome separation method and device based on skeleton segmentation and key point detection
ActiveCN112200809AEffective for contact adhesion problemsContact adhesion is no longer limitedImage enhancementImage analysisGeneticsComputational biology
The invention belongs to the technical field of chromosome analysis, and particularly relates to an adhesion chromosome separation method and device based on skeleton segmentation and key point detection. The adhesion chromosome separation method based on skeleton segmentation and key point detection comprises the following steps: S1, scaling and normalizing an adhesion chromosome image to be separated to obtain an adhesion chromosome input image; and S2, inputting the adhesion chromosome input image obtained in the step S1 into a chromosome skeleton segmentation model for segmentation to obtain a chromosome skeleton image. The method provided by the invention does not depend on a specific morphological mode of chromosomes, has relatively high generalization ability, starts from adhesion points among the adhesion chromosomes, and utilizes the adhesion points to separate the adhesion chromosomes. The adhesion chromosome separation method and device based on skeleton segmentation and keypoint detection can effectively solve the chromosome separation problem during adhesion of multiple chromosomes.
Owner:SHANDONG IND TECH RES INST OF ZHEJIANG UNIV
Isolated culture method of hair follicle stem cells of Yangtze River delta white goat
InactiveCN109355255AOptimize the extraction processAvoid timeCell dissociation methodsDead animal preservationPhysiologyStem cell line
The invention provides an isolated culture method of hair follicle stem cells of Yangtze River delta white goat. By adoption of the method, an isolated cell line has a good growth and division state,and relatively pure hair follicle stem cells can be obtained and are of a typical paving stone shape; primary cells grow fast, so that aggregated growth of cells can be observed on the third day, morecell aggregates in aggregated growing can be observed on the fourth day, the cells can overgrow 60 to 70 percent of a culture dish on the fifth day, and the positive rate of hair follicle stem cell markers is up to 90 percent or above. Results of cell chromosome analysis, hair follicle stem cell marker detection, cell activity detection and the like show that a built hair follicle stem cell lineof the Yangtze River delta white goat is high in activity, and the hair follicle stem cell markers are high in positive rate; and a good practical model can be provided for researching gene expressionand passageway regulation of skin cells and growth and a mechanisms of skin appendages, in particular for researching a mutual action mechanism among main cells of the skin.
Owner:YANGZHOU UNIV
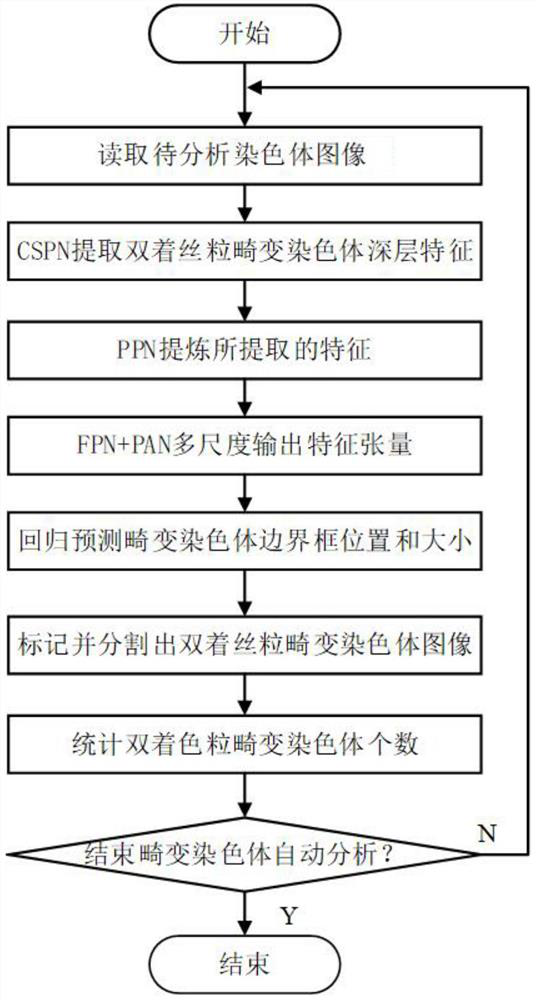
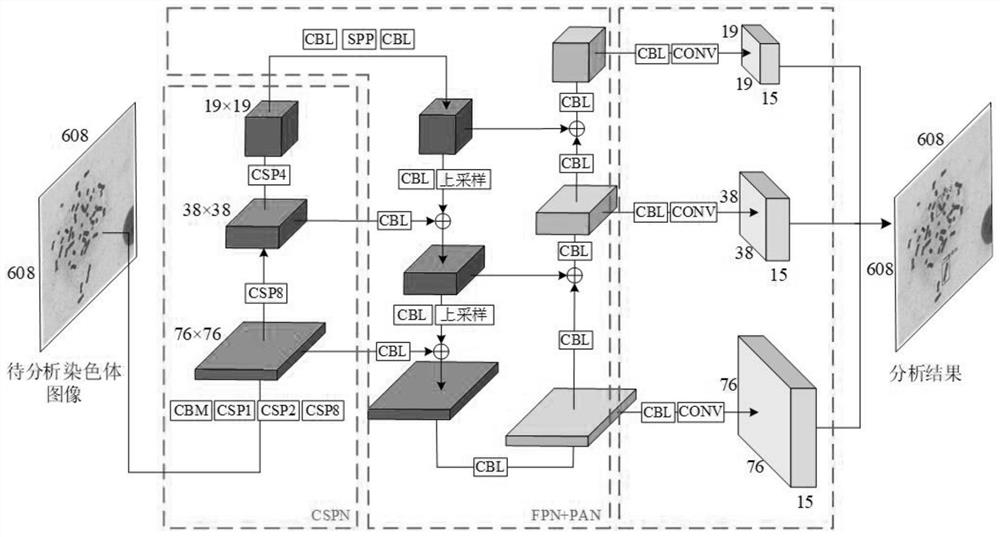
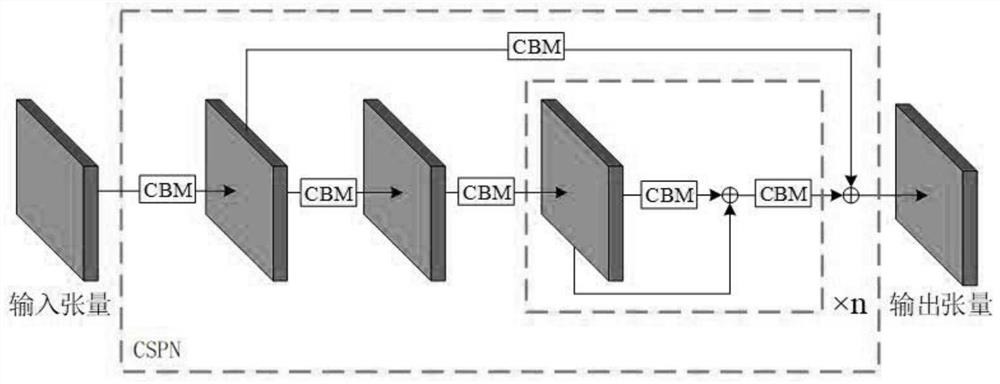
Double centromere distorted chromosome analysis and prediction method based on multi-scale fusion method
PendingCN112381806AFast and accurate automatic detection and analysisImprove analysis efficiencyImage enhancementImage analysisAlgorithmNetwork model
The invention relates to a chromosome analysis technology, and discloses a double centromere distorted chromosome analysis and prediction method based on a multi-scale fusion method, and the method comprises the steps: enabling a trained neural network model to extract the deep features of an inputted to-be-analyzed chromosome image through CSPN; carrying out Drop Block operation and spatial pyramid pooling operation to extract key features; respectively outputting three feature tensors on three scales by adopting a feature fusion strategy combining a feature pyramid network and a path aggregation network; after Drop Block operation is carried out, outputting three prediction tensors; adopting a DIOU NMS algorithm to screen the predicted bounding box; and analyzing the double centromere distorted chromosome. The trained neural network model can quickly mark the distorted chromosomes on the distorted chromosome image and count the number of the distorted chromosomes, the detection accuracy is higher, the robustness is higher, and a doctor is assisted in completing biological dose estimation.
Owner:SHANGHAI BEION MEDICAL TECH CO LTD
Chromosome image processing method and system
ActiveCN114240883AImprove generalizationImprove general performanceImage enhancementImage analysisImaging processingGenetics
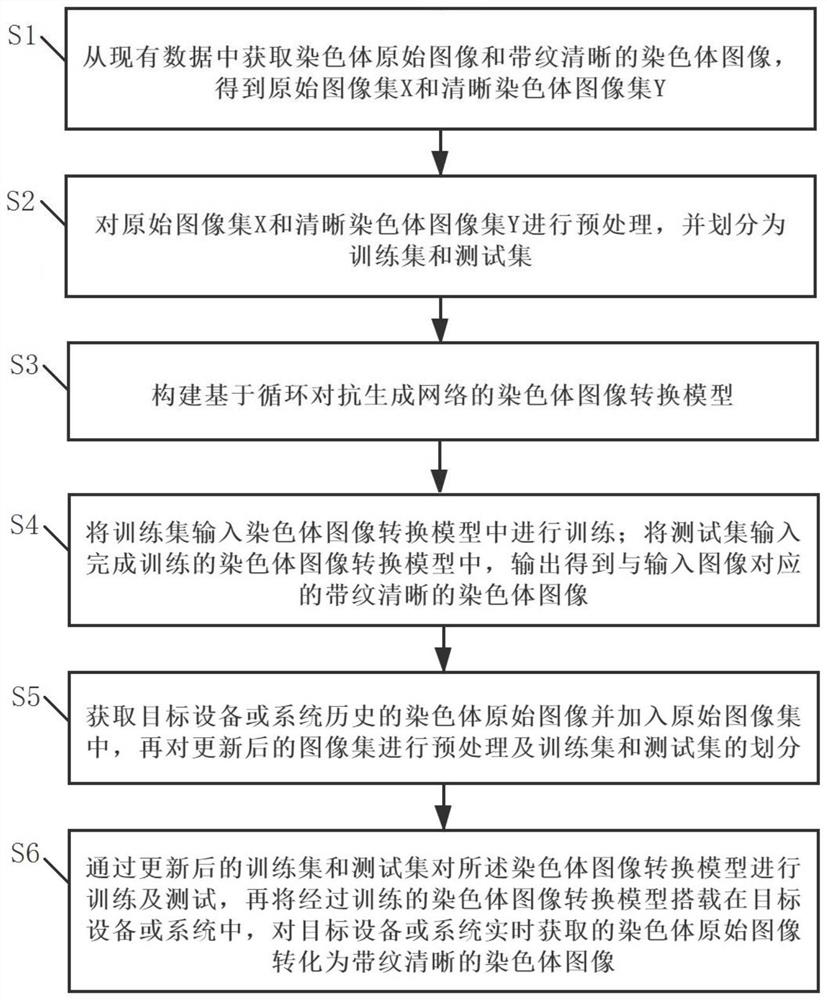
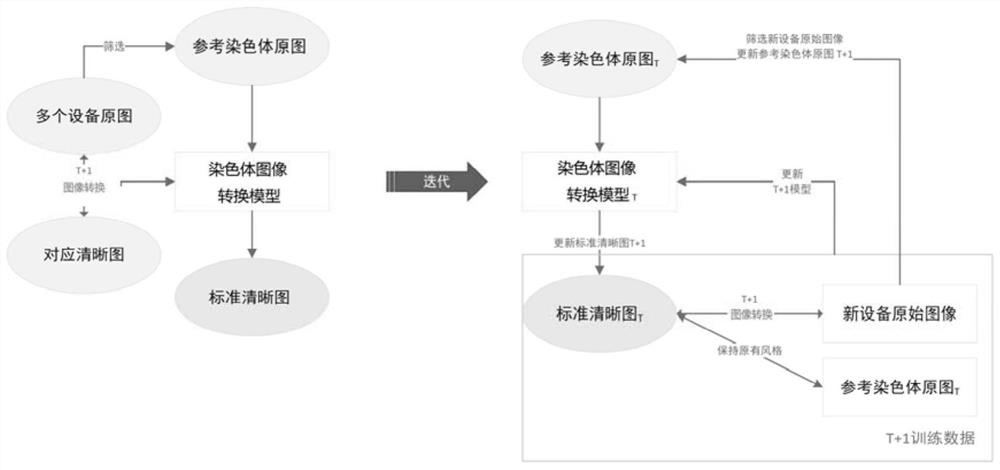
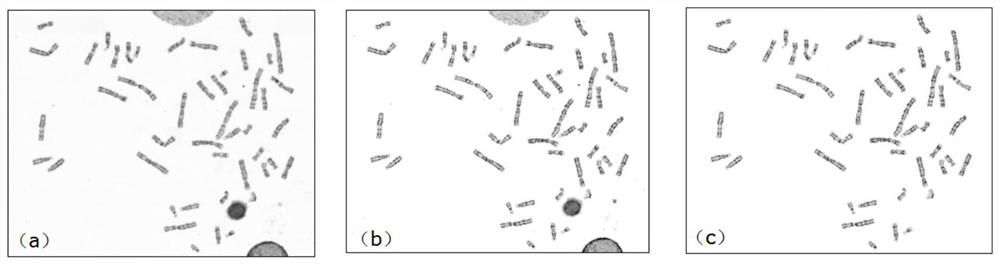
The invention relates to the technical field of chromosome analysis, and provides a chromosome image processing method and system, and the method comprises the following steps: obtaining an original chromosome image and a clear chromosome image with stripes, and obtaining an original image set and a clear chromosome image set; preprocessing the image set, and dividing the image set into a training set and a test set; constructing a chromosome image conversion model based on a cyclic generative adversarial network; respectively inputting the training set and the test set into a chromosome image conversion model for training and testing; acquiring a historical chromosome original image of a target device or system, adding the historical chromosome original image into an original image set, preprocessing the updated image set, and dividing a training set and a test set; and training and testing the chromosome image conversion model through the updated training set and test set, carrying the trained chromosome image conversion model in a target device or system, and converting an original chromosome image obtained by the target device or system in real time into a chromosome image with clear stripes.
Owner:易构智能科技(广州)有限公司
Abnormal karyotype image library and construction method for same
ActiveCN104064108AIncrease the difficultyIncreased complexityMaps/plans/chartsDiseaseExternal quality assessment
The invention relates to an abnormal karyotype image library used in the medical field and a construction method for the same. Remainder adherent growth living cells used for chromosome disease diagnosis due to a diagnosis need and already confirmed to be provided with complicated chromosome abnormality are taken, and subjected to repeated generation amplification, cells are corrected, lowly-permeated, fixed and stored at minus 20 DEG C, then one type or more types of cell suspensions are taken and mixed according to the needed ratio, thus only one abnormal karyotype contained in each part of the cell suspensions is converted to various abnormal karyotypes with different ratios and easy to carry out misdiagnose, the image library is prepared through chromosome preparation and image capture, the difficulty and complexity of differential diagnosis and chimera diagnosis are improved by the prepared images, and more chromosome abnormal combined types of karyotype images can be prepared as needed; specimens are easy to get and waste redundant cells are used for teaching, technical examination, external quality assessment and resource sharing for chromosome analysis, and a remarkable effect is acted on timely discovery for diagnosis quality problems and improvement for a diagnosis level.
Owner:ATTACHED OBSTETRICS & GYNECOLOGY OSPITAL MEDICALCOLLEGE ZHEJIANG UNIV
Chromosome polarity recognition method and system based on deep learning
ActiveCN113408505AHigh degree of analysis automationSimple processCharacter and pattern recognitionNeural architecturesData setAlgorithm
The invention provides a chromosome polarity recognition method based on deep learning. The method comprises the steps of (1) collecting a data set, (2) constructing a training set and a test set, (3) performing learning training on a chromosome polarity recognition model based on the training set, and (4) inputting the test set into the chromosome polarity recognition model for testing, and outputting a polarity result of a chromosome to be predicted. The invention further provides a chromosome polarity recognition system based on deep learning. The method and the system provided by the invention can accurately judge the current chromosome polarity category based on a deep learning classification algorithm, and finish chromosome polarity adjustment according to the current chromosome polarity category, so that all chromosomes are kept in a state that short arms face upwards. According to the method and the system, the chromosome polarity recognition accuracy reaches 96.36%, the data source is simple, the automation degree of chromosome analysis is high, the process is more concise, and the method and the system have wide industrial practicability.
Owner:中科伊和智能医疗科技(佛山)有限公司
Cell chromosome analysis rapid library building method and kit
InactiveCN106755337AEasy to keep addingReduce pollutionMicrobiological testing/measurementLibrary creationMicro cellA-DNA
The invention discloses a cell chromosome analysis rapid library building method. The method comprises the following steps: (1) directly adding proteinase K into cells for cracking the cells; (2) directly adding DNA cracking enzyme and a DNA end filling reagent into the reaction system of the step 1; and (3) directly adding a DNA ligation reagent into the reaction system of the step 2, so that the target to-be-detected DNA is in joint connection with the DNA. The sequencing library provided by the invention can be used for detection of a high-throughput sequencing instrument, the single-tube test can be realized, the genome DNA extraction, the PCR amplification and the purification among the steps are not needed, and thus the chromosome sequencing detection based on micro cells is enabled to be rapid, simple and convenient. The invention further discloses a cell chromosome analysis rapid library building kit.
Owner:HANGZHOU GENE META MEDICAL DEVICE CO LTD
Small-molecule fluorescent probe and application thereof in chromosome analysis and cytogenetics
ActiveCN111303145AOrganic chemistryMicrobiological testing/measurementHomologous chromosomeMolecular fluorescence
The invention discloses a small-molecule fluorescent probe and an application thereof in chromosome analysis and cytogenetics, and the small-molecule fluorescent probe can be used as a universal toolfor chromosome image segmentation and is used for identifying and distinguishing overlapped, aggregated or contacted chromosomes. The small-molecule fluorescent probe is used as a reference marker forpositioning centromere positions, is used for classifying homologous chromosomes, distinguishing different forms of chromosomes and estimating chromosome arm lengths, is used in cooperation with karyotype analysis for chromosome deletion and polyploid, is used in cooperation with fluorescence in situ hybridization (FISH), and is used for positioning designated genes and judging chromosome abnormalities. The invention provides a rapid and reliable method for auxiliary chromosome analysis, and has great potential in conventional clinical practice.
Owner:陈斯杰 +2
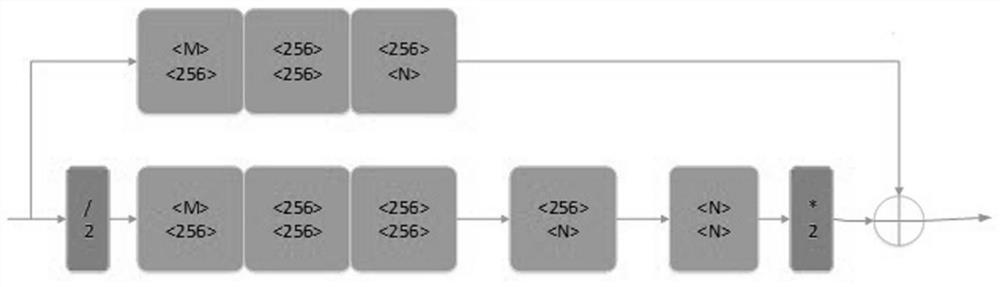
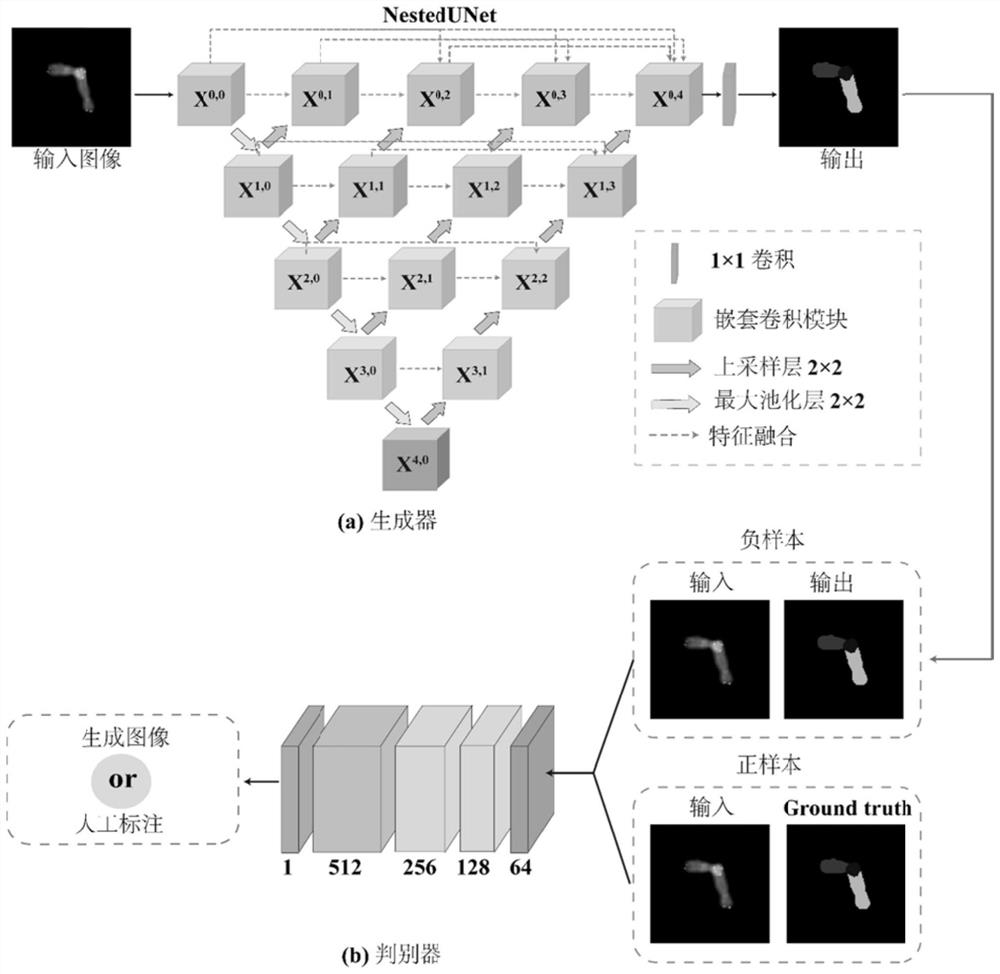
Automatic overlapping chromosome segmentation method based on adversarial learning multi-scale features
ActiveCN112215847AEasy accessImprove Segmentation AccuracyImage enhancementImage analysisFeature learningGenerative adversarial network
The invention discloses an automatic overlapping chromosome segmentation method based on adversarial learning multi-scale features. Challenges of human chromosome analysis are overlapping of automaticchromosome segmentation, which hinders medical diagnosis and biomedical research. Therefore, the invention provides an adversarial multi-scale feature learning framework, adopts a nested U-shaped network as a generator, and aims to explore 'optimal' representation of chromosome images by using multi-scale features. Conditional generative adversarial network (cGAN) adversarial learning are used topromote output distribution to be closer to a gold standard image; a least square GAN target is adopted to improve the training stability of the framework; and the continuity optimization is carriedout by using the Lovasz-Softmax loss, so that better performance is obtained. Experimental results show that the method is superior to other traditional algorithms in subjective visual effect and objective evaluation standard.
Owner:WUHAN UNIV
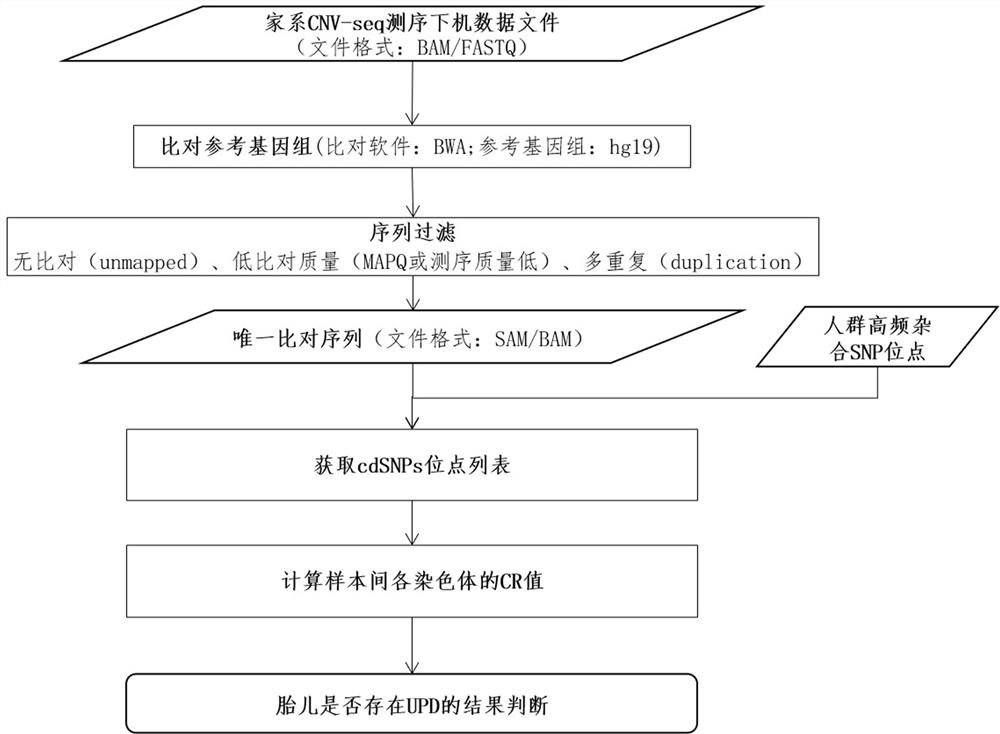
Family-based low-depth sequencing method for detecting chromosome monoparental disomes
The invention discloses a family-based low-depth sequencing method for detecting chromosome monoparental disomes by utilizing the characteristic that a large number of cdSNPs exist in chromosomes between samples of a fetus and parents, and the method comprises the following steps: S1, sequence comparison: comparing three original data files of family CNV-seq to obtain three files with comparison information; S2, performing sequence filtering; S3, selecting a high-frequency heterozygous SNP site data set of the crowd; S4, obtaining a cdSNP site list; S5, counting the total CR value of the mother-child and father-child samples and the CR value of each chromosome; and S6, performing fetal chromosome UPD analysis according to the CR value to obtain a conclusion. According to the method, the CNV-seq detection process and the detection cost do not need to be changed, analysis and comparison are carried out only by utilizing the CNV-seq original data file of the family, whether UPD exists in the chromosome of the fetus or not can be identified, and the method has the advantages of low detection cost and high detection accuracy.
Owner:广东博奥医学检验所有限公司
Method for acquiring Y chromosome sequence of pseudobagrus ussuriensis
ActiveCN114058713AShorten the timeEasy to operateMicrobiological testing/measurementClimate change adaptationGenomic sequencingAnimal science
The invention relates to the technical field of neutral chromosome analysis in aquaculture, and discloses a method for acquiring a Y chromosome sequence of pseudobagrus ussuriensis, which comprises the following steps: step 1, constructing an interspecific hybrid population which is obtained from male pseudobagrus ussuriensis and female Pseudobagrus tenuis through artificial insemination; step 2, identifying male hybrid individuals, wherein the male hybrid individuals in the step 2 are identified through a gonad characteristic observation method and a molecular marker method; step 3, sequencing and assembling the third-generation genomes of the male hybrid individuals, wherein the sequencing and assembling of the third-generation genomes in the step 3 are implemented by adopting HiFi and Hi-C technologies; and step 4, conducting comparing with a female genome chromosome to obtain a Y chromosome sequence. According to the method for obtaining the Y chromosome sequence of the pseudobagrus ussuriensis, the time required for obtaining the Y chromosome sequence under the condition of the prior art can be greatly shortened, the labor intensity is effectively reduced, the working efficiency is improved, and the sex control breeding process of the pseudobagrus ussuriensis is powerfully promoted.
Owner:HUAIYIN TEACHERS COLLEGE
Novel method for screening mutagenic polyploids of jujube test-tube seedlings
InactiveCN103004607AImprove selection rateShorten the timeHorticulture methodsPlant tissue cultureShootZiziphus jujuba
The invention discloses a novel method for screening mutagenic polyploids of jujube test-tube seedlings. The novel method comprises the following steps of: (1) basic procedures of jujube test-tube seedling mutation; (2) carrying out first screening on the mutated plants according to the morphological characteristics; (3) carrying out differentiation culture and carrying out second screening on the differentiated multiple shoots; (4) carrying out chromosome analysis on the screened target multiple shoots; and (5) culturing the determined polyploids. The method has the beneficial effects that the mutagenic polyploids of jujube test-tube seedlings are screened by adopting the way of proceeding from the exterior to the interior in combination with continuous differentiation culture, so the selection rate of the tetraploids of the jujube test-tube seedlings can be greatly increased and the time can be shortened; Continuous three-time tests carried out on the jujubes by applying the method help discover that more than 35.0% of tetraploids can be bred within 110-150 days.
Owner:YANAN UNIV
Swordtail fish larva fish cell line and construction method and application thereof
ActiveCN110527661AGood growthSensitiveCell dissociation methodsMicrobiological testing/measurementAnimal virusIn vitro toxicology
The invention discloses a swordtail fish larva fish cell line. The swordtail fish larva fish cell line is constructed by primary culture and subculturing. The invention further provides an applicationof the swordtail fish larva fish cell line to separation of aquatic product animal viruses and environmental pollutant monitoring. Swordtail fish larva fish is subjected to cell culture, the cell culture condition is primarily determined, chromosome analysis is performed, the sensibility of various aquatic animal viruses by cells is detected, and the inductive effect of antibiotic environmental pollutants namely rifampicin on cells is researched. The cell line establishes foundation for research of later stage aquatic animal viruses and application of the cell line as an in vitro toxicology cell model to toxicology correlational research of an environmental pollutant.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
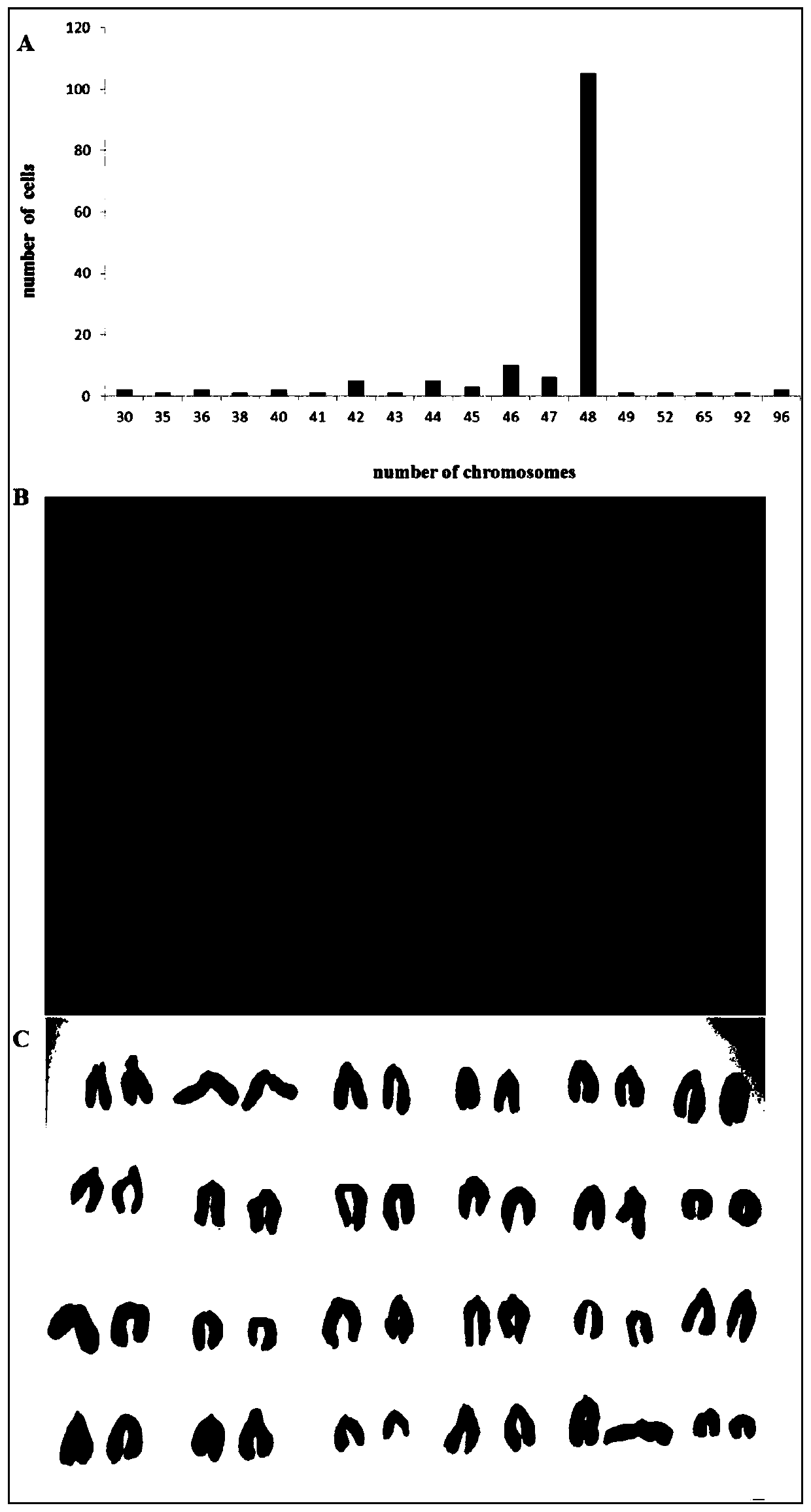
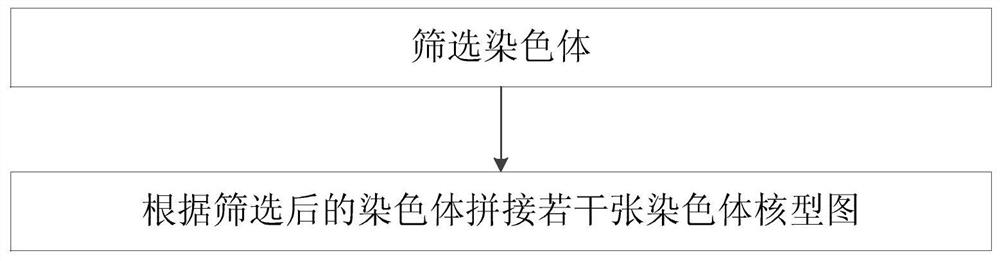
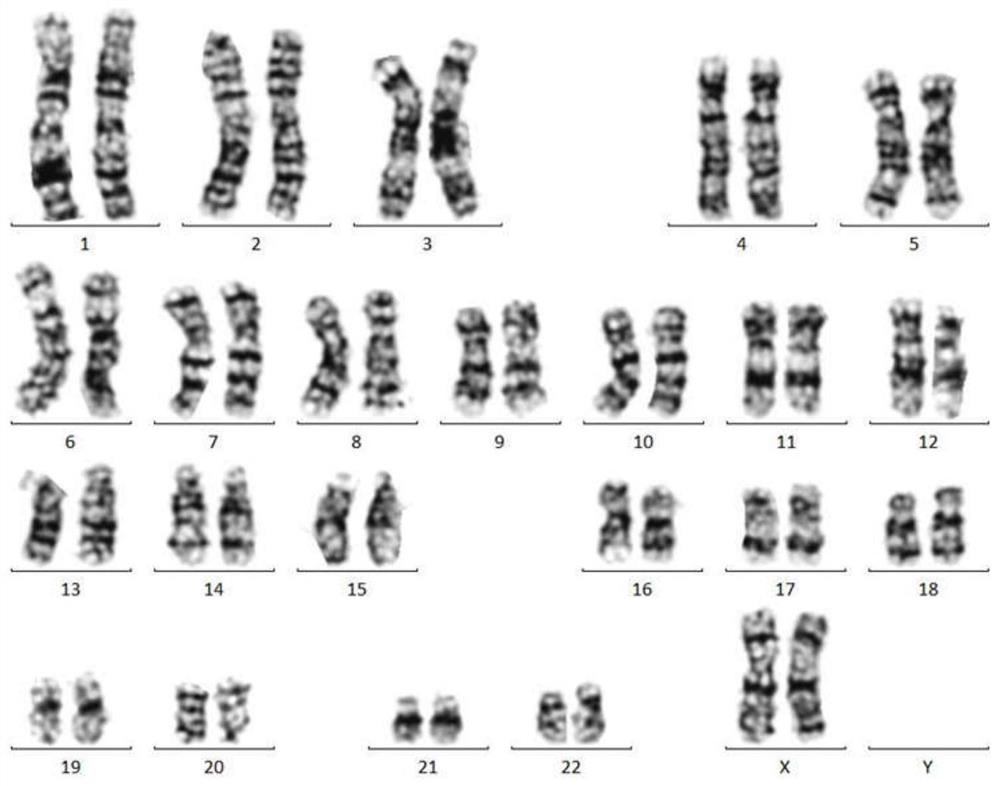
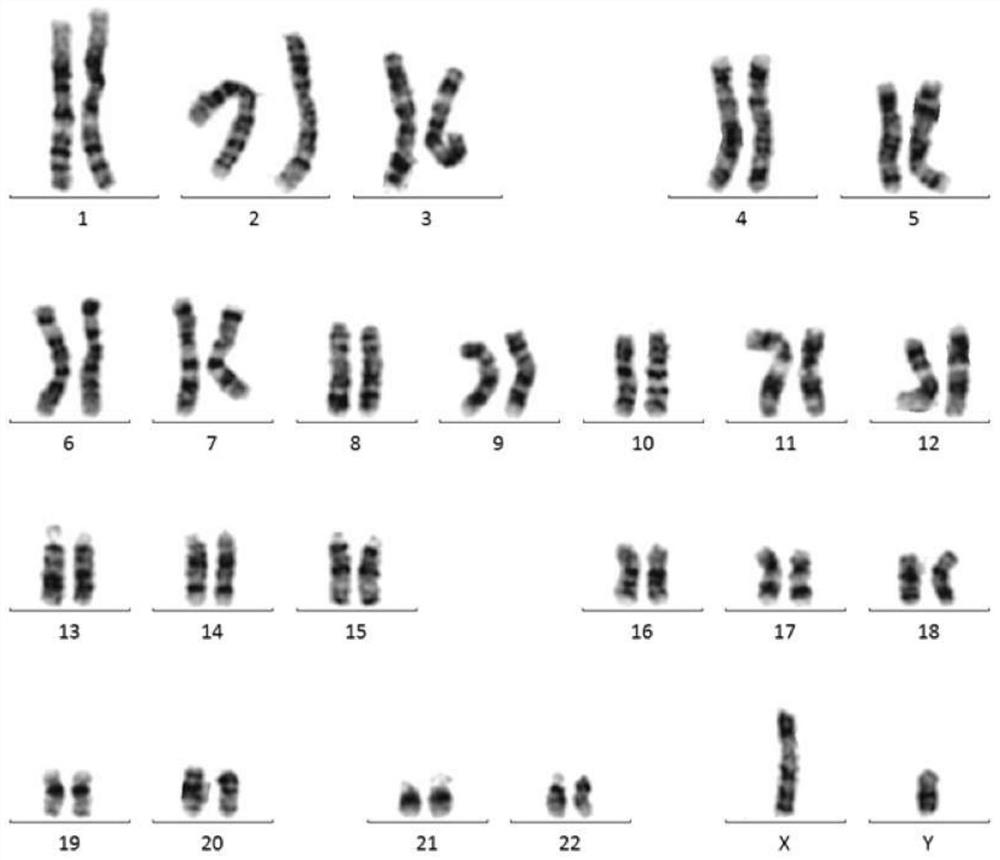
Optimal chromosome karyotype map splicing method, optimal chromosome karyotype map splicing system and chromosome karyotype analysis method
PendingCN111986081AAccurate analysisAvoid missed diagnosisGeometric image transformationProteomicsGeneticsMissed diagnosis
The invention belongs to the technical field of chromosome analysis, and particularly relates to a chromosome karyotype map optimal splicing method, a chromosome karyotype map optimal splicing systemand a chromosome karyotype analysis method. The chromosome karyotype map optimal splicing method comprises the steps that chromosomes are screened; and according to the screened chromosomes, a plurality of chromosome karyotype graphs are spliced to obtain a plurality of clear and complete chromosome karyotype graphs through optimal splicing in the metaphase chromosome division phase images, so that later chromosome karyotype analysis is more accurate, and missed diagnosis and misdiagnosis of illness conditions are avoided.
Owner:江苏医像信息技术有限公司
Improved fluorescence in-situ hybridization method and application thereof
InactiveCN106480204AClear clinical valueMicrobiological testing/measurementStainingSpecimen Handling
The invention provides an improved fluorescence in-situ hybridization method and an application thereof. The improved fluorescence in-situ hybridization method comprises the following steps: processing a specimen, conducting denaturation, processing a probe, conducting hybridization and conducting counter-staining. According to the improved fluorescence in-situ hybridization method provided by the invention, by conducting observation by virtue of a fluorescence microscope and calculating fluorescence hybridization signals of cells, conducting precise chromosome analysis on 81 patients with lymphoproliferative diseases and judging gene deletions and molecular genetic abnormalities of the patients, the following conclusions are obtained: molecular genetic analysis is conducive to diagnosis and differential diagnosis of lymphoid leukemia, and moreover, the molecular genetic analysis also serves as an important index for monitoring disease remission and recurrence and for judging a curative effect in the clinical field. On the basis of chromosomal karyotype analysis, the proper fluorescence in-situ hybridization probe and method are selected to conduct precise chromosome analysis on most lymphoid leukemia patients, and the various molecular genetic abnormalities are related to prognosis of the patients.
Owner:AFFILIATED HOSPITAL OF NANTONG UNIV
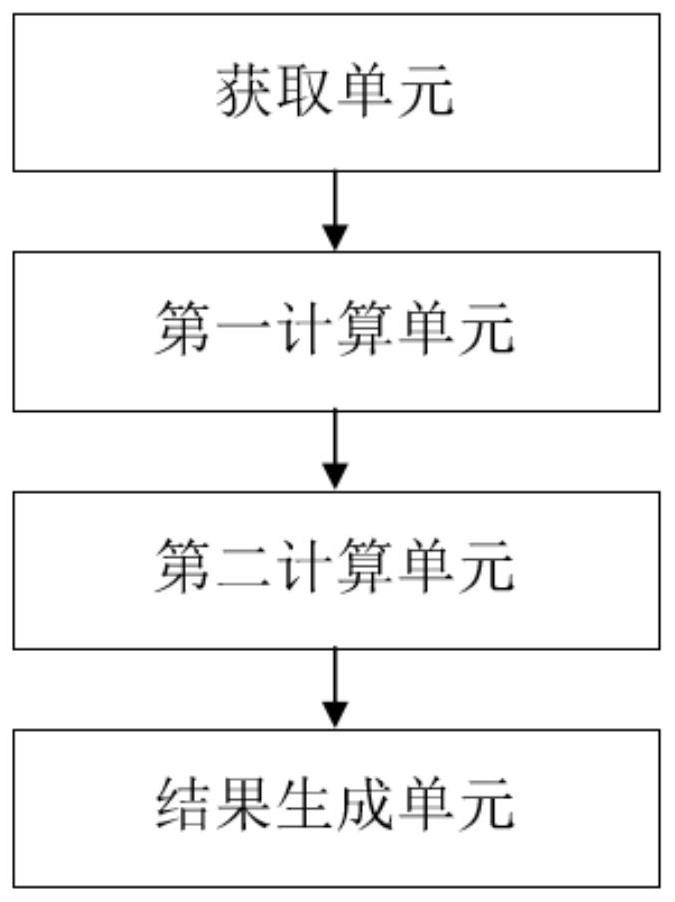
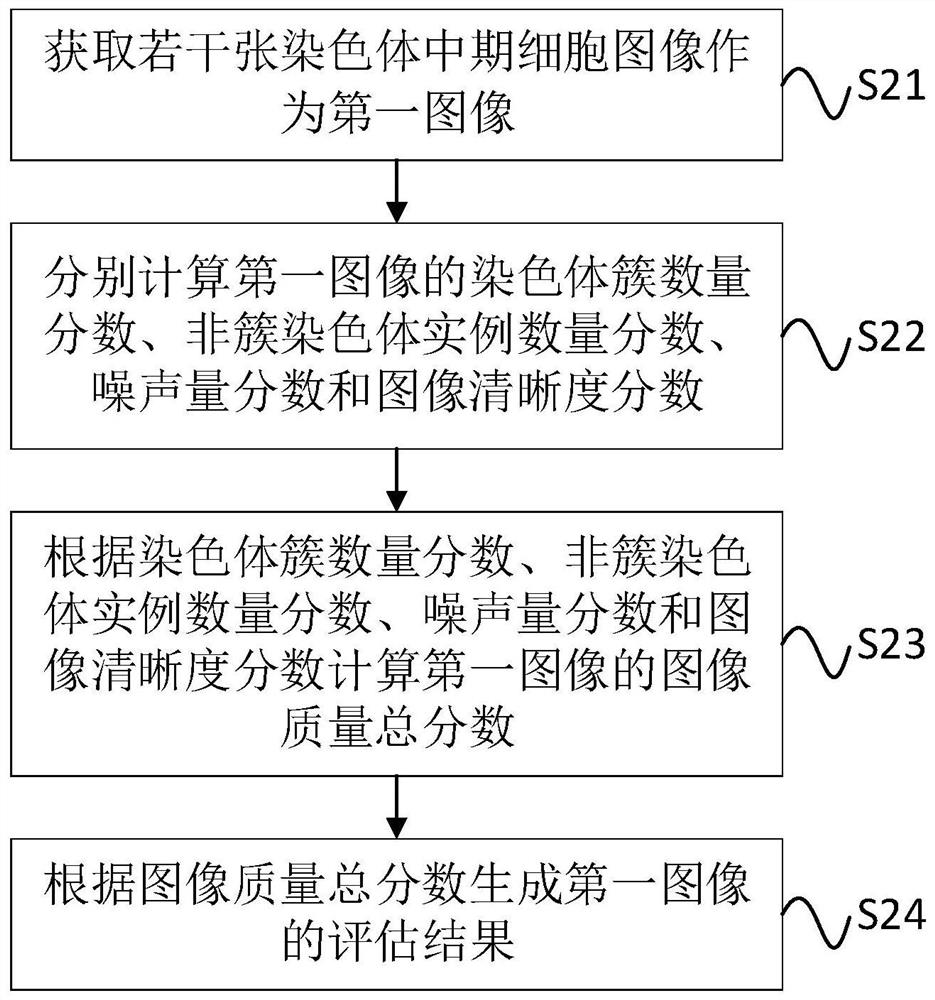
Chromosome metaphase cell image quality evaluation system and method
PendingCN112489007AReduce difficultyImprove accuracyImage enhancementImage analysisEvaluation resultImaging quality
The invention discloses a chromosome metaphase cell image quality evaluation system and method. The system comprises an obtaining unit which is used for obtaining a plurality of chromosome metaphase cell images as a first image; a first calculation unit which is used for respectively calculating a chromosome cluster quantity score, a non-cluster chromosome instance quantity score, a noise quantityscore and an image definition score of the first image; a second calculation unit which is used for calculating an image quality total score of the first image according to the chromosome cluster number score, the non-cluster chromosome instance number score, the noise amount score and the image definition score; and a result generation unit which is used for generating an evaluation result of the first image according to the image quality total score. The evaluation result skill generated by the result generation unit effectively reduces the difficulty of subsequent chromosome karyotype analysis and improves the analysis accuracy. The system and method can be widely applied to the technical field of chromosome analysis.
Owner:SOUTH CHINA NORMAL UNIVERSITY
Flounder tetraplont fry batch induction method
InactiveCN101595849BImprove induction efficiencyPloidy neatMicrobiological testing/measurementClimate change adaptationAgricultural scienceStart time
The invention discloses a flounder tetraplont fry batch induction method comprising an induction method and an identification method of tetraplont fries; wherein, the induction method comprises collection and insemination of unfertilized ovum, and determination of oosperm chromosome doubling starting time, hydrostatic pressure shock pressure and processing time, and the identification method comprises ploidy determinator identification and chromosome analysis identification. The method firstly builds a method of inducing the tetraplont fries of flounder, such as paralichthysolivaceus and the like by hydrostatic pressure, screens hydrostatic pressure processing starting time and hydrostatic pressure shock conditions for inhibiting first merogenesis and carrying out chromosome doubling, andbuilds a tetraplont fry batch induction method. Tetraplont fries obtained with the method have high induction rate of 90%; the method of the invention has the characteristics of advancing, high efficiency, accuracy and reliability, and has important application value in fish polyploidy induction and sterile offspring seed production, and has wide promotion and application prospect in fish cultivation and breeding.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com