Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
405 results about "Protease K" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Reagent for extracting and purifying DNA
ActiveCN101613696AMeet the extraction requirementsEasy to operateSugar derivativesDNA preparationMicrosphereMagnetic bead
The invention discloses a reagent for extracting and purifying DNA, comprising the following substances: preprocessing lysis solution with pH of 7.4-8.5: water serves as solvent, and solute comprises the substances with the following final concentration: 10-100mM of buffer salt, 0.5%-5% of surface active agent, 1-100mM of chelating agent, 50-150mM of soluble salt, and 0.2-4mg / ml of protease K; pyrolysis combined liquid with pH of 6.4-7.4: water serves as solvent, and solute comprises the substances with the following final concentration: 20-150mM of buffer salt, 3-8 of MChaotropic salt, 1-10% of surface active agent, 0.5-4% of amphoteric ion detergent, 10-100mM of chelating agent and 15-30% of alcohol; cleaning solution I: water serves as solvent, and solute comprises the substances with the following final concentration: 4-6 of MChaotropic salt, 25-50% of alcohol and 10-100mM of chelating agent; cleaning solution II is 75% of ethanol water solution; spent regenerant is TE (10mM Tris-HCl, 1mMEDTA, and pH is 8.0); magnetic bead suspension is nanoscale silicone coated magnetic microsphere (50-100mg / ml).The DNA extracting efficiency is as high as 93.67% by adopting the reagent.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Magnetic bead process nucleic acid extraction and transformation kit, and using method thereof
InactiveCN109022417AAvoid wastingHigh purityMicrobiological testing/measurementDNA preparationLysisMagnetic bead
The invention provides a magnetic bead process nucleic acid extraction and transformation kit, and a using method thereof. The kit includes a lysate, a protease K, a magnetic bead solution, a transforamtion reagent, a nucleic acid protection agent, a binding solution, a washing solution I, a washing solution II and an eluent. The using method of the kit includes the following steps: (1) lysis; (2)transformation; (3) layering; (4) binding; (5) primary washing; (6) secondary washing; (7) drying; and 8) elution. The kit and the using method thereby can complete the transformation while achievingautomatic extraction of nucleic acid from a sample. The sample is lysed, releases the nucleic acid, and then begins to transform, the nucleic acid is separated from proteins by the binding solution after the transformation is completed, the methylated nucleic acid is purified by a magnetic bead process, and a pre-denaturation step in subsequent qPCR detection of the sample is used to replace a previously omitted sulfo group removal step, so a purification process in the original nucleic acid extraction step is omitted in the entire extraction and transformation process.
Owner:SUREXAM BIO TECH
Kit for detecting methylation of colorectal cancer related gene multi-sites and application of kit
ActiveCN106893777AMeet the requirements of early screeningHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationMulti siteA-DNA
The invention discloses a kit for methylation of colorectal cancer related gene multi-sites and application of the kit. A primer combination and a probe can specifically detect the methylation degrees of a plurality of colorectal cancer related gene sites. The invention further provides application and a method of the primer combination and the probe for detecting the methylation in colorectal cancer and precancerous polyps screening. The method comprises the steps that a cell lysis solution is added to cell sediment and mixed uniformly, then PVPP is added for crude extraction, a cell sediment sample after primary treatment contains a little protein and RNA, protease K and RNA enzyme are added for completely removing the protein and the RNA, cell lysate is added to a centrifugal column with an adsorption film, after impurities are washed by a DNA rising solution, a DNA eluent is added to elute the DNA in the absorption film, and finally a DNA solution is collected through centrifugation. Efficient pipe collection detection is performed aiming at the plurality of gene sites, the flexibility and the specificity are improved, and the tumor early screening requirement can be improved.
Owner:WUHAN AIMISEN LIFE TECH CO LTD
Direct PCR (Polymerase Chain Reaction) method without extracting animal tissue DNA (Deoxyribonucleic Acid)
InactiveCN102344919ASimple methodFast wayMicrobiological testing/measurementDNA preparationBiotechnologyEthylene diamine
The invention relates to a direct PCR (Polymerase Chain Reaction) method without extracting animal tissue DNA (Deoxyribonucleic Acid). The method comprises the following steps of: digesting animal tissues with tissue lysis solution; and directly performing PCR amplification by using digestion solution as a template, wherein the used tissue lysis solution consists of Tris-C1, EDTA (Ethylene Diamine Tetraacetic Acid), protease K and NaCl. Compared with the conventional common method, the method has the advantages that: the method can be used for simultaneously treating a large number of animal tissue samples; high-flux molecular operation based on PCR amplification is realized; operation steps are quickly simplified; and cost is saved.
Owner:CHINA AGRI UNIV
Composite for preservation of human saliva and preparation method there of
InactiveCN102919218APollution suppressionAvoid pollutionDead animal preservationN-Acetyl-5-methoxytryptamineGenomic DNA
The invention relates to a composite for preservation of human saliva and a preparation method of the composite. The composite comprises the following components: Tris-HCl with pH being 6-8.5 (5-20mmol / L), EDTA (Ethylene Diamine Tetraacetic Acid) (0.5-2mmol / L), NaOAc (2.5-3.5mol / L), cane sugar (0.1-0.4mol / L), N-acetyl-5-methoxytryptamine (1-3mmol / L), propylparaben (1-4mg / mL), diazonium imidazolidinyl urea (1-3mg / mL), protease K(10mu g / mL) and a system with pH being 7.5-8.5. When used for preservation of human saliva, the composite is capable of inhibiting nuclease activity and preventing nuclease from being contaminated by external microbes or being oxidized by itself, thereby guaranteeing the integrality and the purity of genomic DNA (deoxyribonucleic acid). The composite is long in preservation period, wide in preservation temperature range and applicable to gene detection and related scientific researches.
Owner:湖北维达健基因技术有限公司
Kit for extracting DNA/RNA of virus through magnetic bead method and using method
The invention discloses a kit for extracting DNA / RNA of a virus by utilizing magnetic beads and a using method. The kit comprises the following eight components: an extracting solution I (cracking), an extracting solution II (binding), an extracting solution III (scrubbing), an extracting solution IV (scrubbing), an extracting solution V (eluting), magnetic bead suspension, a protease K working solution and a nucleic acid settling agent. The kit extraction method comprises the following steps: cracking, binding, scrubbing and eluting. The kit and the extraction method can be used for extracting the DNA / RNA of the viruses of different types of samples, the impurities such as proteins and lipids in the samples are effectively removed, the extracted nucleic acid is high in purity and complete in fragments, and the extraction process is safe and non-toxic.
Owner:宝瑞源生物技术(北京)有限公司
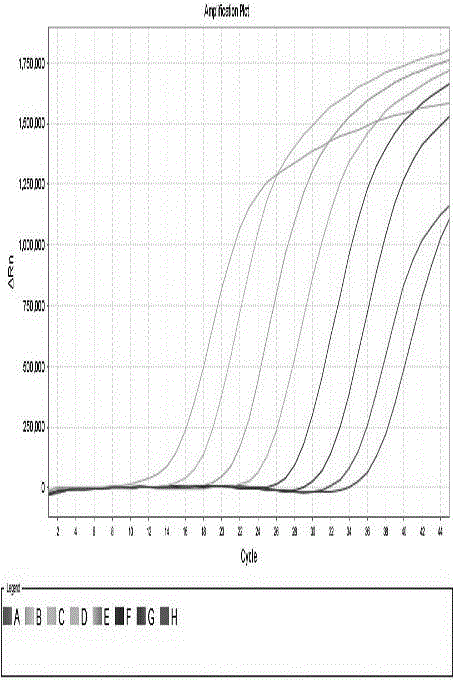
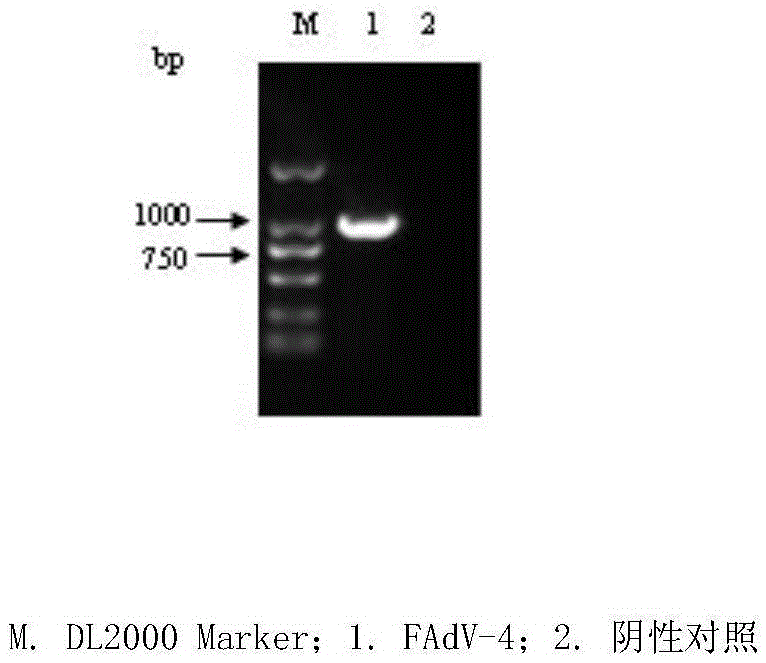
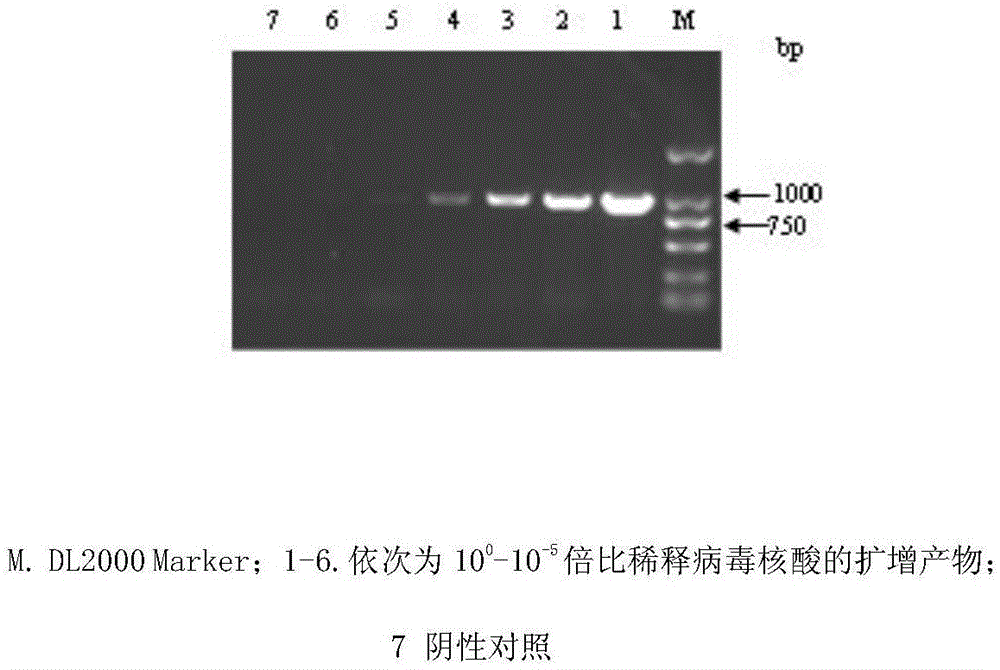
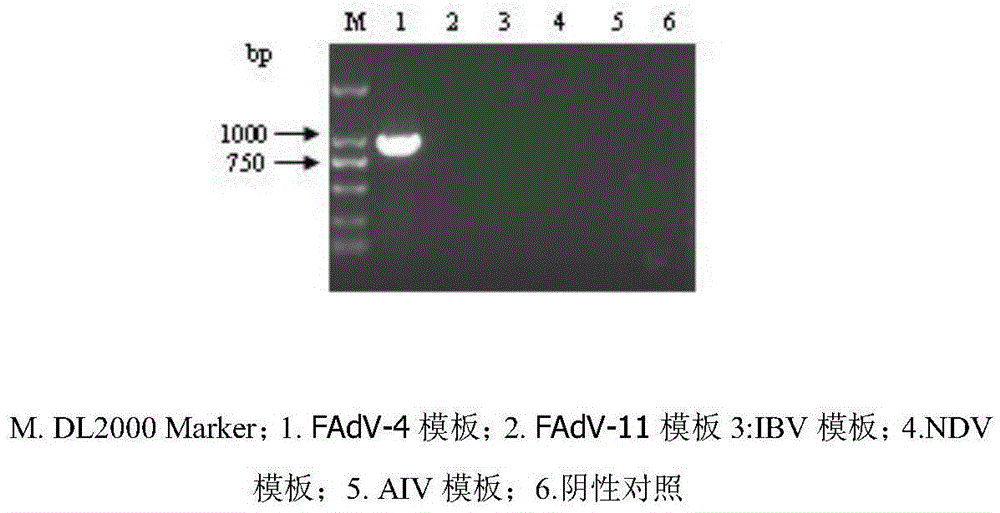
FAdV-4 PCR detection kit and detection method
ActiveCN105483292ASpecific detectionQuick checkMicrobiological testing/measurementSodium acetateDistilled water
The invention discloses an FAdV-4 PCR detection kit and detection method. The kit comprises protease K, a lysing solution, sodium acetate, saturated phenol, chloroform, isoamylol, anhydrous ethanol and sterile distilled water, and is characterized by comprising 2*TaqPCR Mix, a sense primer and a reverse primer, wherein the sequence of the sense primer is shown as SEQ ID No.1, and the sequence of the reverse primer is shown as SEQ ID No.2. The kit can be used for detecting FAdV-4. The method for detecting FAdV-4 by adopting the kit is also provided, and FAdV-4 can be detected quickly and specifically.
Owner:SHIJIAZHUANG SHIMU ANIMAL HUSBANDRY PHARMA CO LTD
Nucleic acid extraction method by virtue of paramagnetic particle method
The invention discloses a nucleic acid extraction method by virtue of a paramagnetic particle method. The nucleic acid extraction method comprises the following steps: sub-packaging 'N' [mu]M (M refers to mol / L) of sample ('N' is less than or equal to 300 and greater than or equal to 50), 'N' [mu]M of lysate and '0.1N' [mu]M of protease K in a No.1 nucleic acid extraction plate, conducting a cracking reaction at 70 DEG C for 10-60min, and then adding '1.1N' [mu]M of magnetic particle-binding buffer; sub-packaging '2.5N' [mu]M of washing solution A, '2.5N' [mu]M of washing solution B and '0.25-0.5N' [mu]M of TE solution in No.2, No.3 and No.4 nucleic acid extraction plates; and under the action of magnetic force, adsorbing magnetic particles in the No.1 nucleic acid extraction plate by virtue of a magnetic bar, conducting transferring, releasing, rapid mixing and magnetic absorption and sequentially processing the magnetic particles by virtue of the No.2, No.3 and No.4 nucleic acid extraction plates, so as to obtain extracted nucleic acid. With the application of the method disclosed by the invention, the influence of impurities on nucleic acid extraction from saliva and other biological samples is effectively reduced, so that nucleic acid extraction efficiency and nucleic acid purity are improved.
Owner:广东国盛医学科技有限公司
Method for separating and extracting exosomes in lamination centrifugal-filtration mode
The invention discloses a method for separating and extracting exosomes in a lamination centrifugal-filtration mode. The method is used for the technical field of molecular biology and clinical examinations. A device comprises a lamination centrifugal-filtration device for separating and extracting the exosomes and an exosome separating and extracting kit composed of an incubating buffering solution and protease K. To-be-measured samples are incubated through the incubating buffering solution and the proper amount of protease K in the kit at the room temperature, then centrifugation is carried out through the lamination centrifugal-filtration device in a matched centrifugal machine, the product is mixed to be even, trapped liquid in an ultrafiltration pipe is collected, and the exosomes can be obtained. According to the method, in addition to the centrifugal machine, large experimental devices are not required, cost is low, operation is convenient and fast, time is short, a large amount of sample operation can be parallelly carried out, and the obtained high-purity exosomes can meet the requirements of clinical large-scale application and popularization.
Owner:东莞微量精准检测研究院有限公司
Pinewood nematoda detection reagent-box and detection method
InactiveCN1529164AImprove practicalityImprove detection efficiencyMicrobiological testing/measurementWood testingNematodePinewood nematode
Two pairs of primer are designed: Pl / P3 is an idiocratic primer for pine wood nematode; P2 / P3 an idiocratic primer for quasi pine wood nematode. The kit includes 1.25ml solution I,1ml solution II, 100U tag enzyme, and 50ul for each contrapositive DNA of wireworm. The solution I is as following constitutes: Worm Lysis Buffer, 500mM KCl, 100mMTris-ClpH 8.0, 15mM MgCl2, 10mMDTT, 4.5% Tween20, 0.1%gelatin, 0.2ug / ul protease K. The solution II is as following constitutes: 2xPCR reaction Buffer, 4.0mM MgCl2, 0.2mM dNTP, 0.8 mu MP1, P2, P3. The invented kit possesses high specificity, sensitiveness and stability, providing fast, sensitive and specific technical method.
Owner:NANJING AGRICULTURAL UNIVERSITY
Human feces DNA extraction method
ActiveCN102864138AQuality improvementHigh repeat stabilityDNA preparationTE bufferBovine serum albumin
The invention discloses a human feces DNA extraction method. The prior art is complicated and time-consuming; and the trace DNAs extracted by the previous sampling method is too little to perform molecular diagnosis. The extraction method uses selective sample collection PSC equipment for feces samples and adopts a low temperature preservation method. The extraction method comprises the following steps: picking a sample by using a sterilized gun head or sterilized blade and directly placing the picked sample on ice; and adding a feces lysis solution, fully cracking and evenly mixing, adding bovine serum albumin, centrifuging to remove the supernatant, adding protease K to perform a digestion reaction at 58 DEG C, then adding a protein precipitation solution to precipitate the protein, centrifuging to obtain the supernatant, adding a silica gel membrane binding solution to fully bind DNAs, centrifuging to obtain precipitates, adding a silica gel membrane washing solution to wash twice, centrifuging, and adding a TE (Tris + EDTA) buffer solution to finally obtain a preservation solution with DNAs. The human feces DNA extraction method has the advantages of high repeatability and stability, simpleness and short time.
Owner:ZHEJIANG CENTURY CONDOR MEDICAL SCI & TECHNOLOTY CORP
Nucleic acid fast extraction kit and application method thereof
The application discloses a nucleic acid fast extraction kit, which includes a mixed enzyme working solution, a cracking adsorption liquid, a detergent liquid, a rinsing liquid and an eluate liquid that are individually prepared. The mixed enzyme working solution includes protease K and lysozyme. The cracking adsorption liquid includes Gu-HCl, Tris-HCl, EDTA, isopropanol, SDS, Triton-100, beta-mercaptoethanol and sodium citrate. The detergent liquid includes LiCl, NaCl, MOPS and ethanol. The rinsing liquid is ethanol. The eluate liquid is EDTA and Tris-HCl. The kit is suitable for extraction of genomic DNA of whole blood, culture cell, tissues, saliva, G<-> bacteria and G<+> bacteria. The purity of extracted DNA is high, and the DNA is suitable for downstream digestion and PCR biological operation. The kit is stale in reagent property, can meet common laboratory demands and can overcome defects of toxicity, long time consumption and tedious operation of a traditional method.
Owner:ZUNYI MEDICAL UNIVERSITY
Rapid no-heating nucleic acid extraction kit based on magnetic bead method
PendingCN109750030ASimple stepsReduce pollutionMicroorganism lysisDNA preparationMagnetic beadPhenol
The invention provides a rapid no-heating nucleic acid extraction kit based on a magnetic bead method. The kit comprises a solution containing magnetic beads, protease K, a polyA solution, deproteinizing liquid, washing liquid, eluant and lysate. The invention also provides a method for extracting nucleic acid by using the kit. The method includes the steps that the lysate and the magnetic beads are added to a sample and fully shaken for uniform mixing, and when lytic cells release nucleic acid, the nucleic acid is captured with the magnetic beads; then, the specific deproteinizing liquid andwashing liquid are adopted for washing respectively to remove protein, salt ions and other impurities on the surfaces of the magnetic beads; finally, the specific eluant is used for rapidly eluting the nucleic acid at room temperature. Compared with a conventional nucleic acid extraction kit based on a magnetic bead method, the kit has the advantages that the steps are simple and rapid, and the whole process only takes about 15 min; besides, heating is not needed, so that the probability of sample room contamination possibly caused by aerosol is reduced; no chloroform phenol or other toxic reagents are used, so that the kit is safe and free of pollution.
Owner:SHANGHAI KEHUA BIO ENG
Whole blood genomic DNA high-flux plate type extracting kit and extracting method
InactiveCN106636064AImprove throughputHigh-throughput Whole Blood DNA ExtractionDNA preparationMagnetic beadChelex 100
Owner:LUOYANG G N T BIOLOGICAL TECH CO LTD
Method for determination of form of selenium in selenium-rich plant material
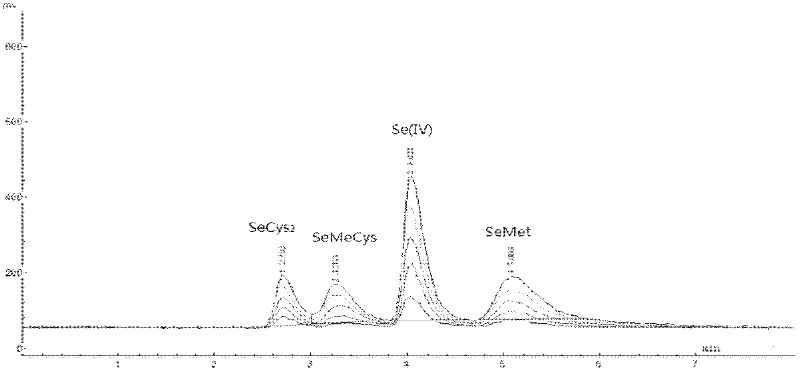
InactiveCN102520104AStable formImprove extraction efficiencyComponent separationAnalysis by material excitationSpectroscopyAtomic fluorescence spectrometry
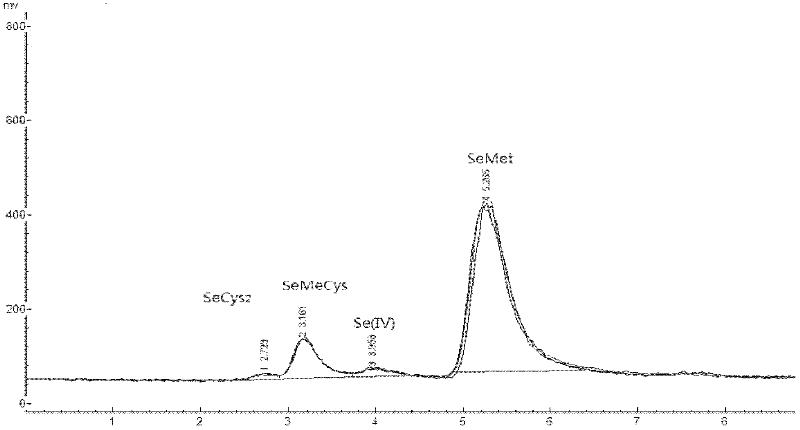
The invention discloses a method for determination of the form of selenium in a selenium-rich plant material. The method provided by the invention is characterized by comprising the following steps of 1, a selenium-rich plant material pretreatment process comprising carrying out extraction of a crushed selenium-rich plant material sample by an extraction buffer Tris-HCl, after the extraction, adding cellulase into the extract, carrying out wall breaking of cells of the selenium-rich plant material sample, orderly adding effective amounts of protease K and protease XIV into the extract, and carrying out hydrolysis extraction to obtain a solution of a sample needing to be detected, and 2, carrying out determination processes respectively on a solution of selenium having a standard form and the solution of the sample needing to be detected by the combination of the hydride generation-atomic fluorescence spectroscopy technology and the high performance liquid chromatography. The method provided by the invention is simple, can be realized easily, adopts nontoxic reagents and does not produce pollution.
Owner:SUZHOU SETEK
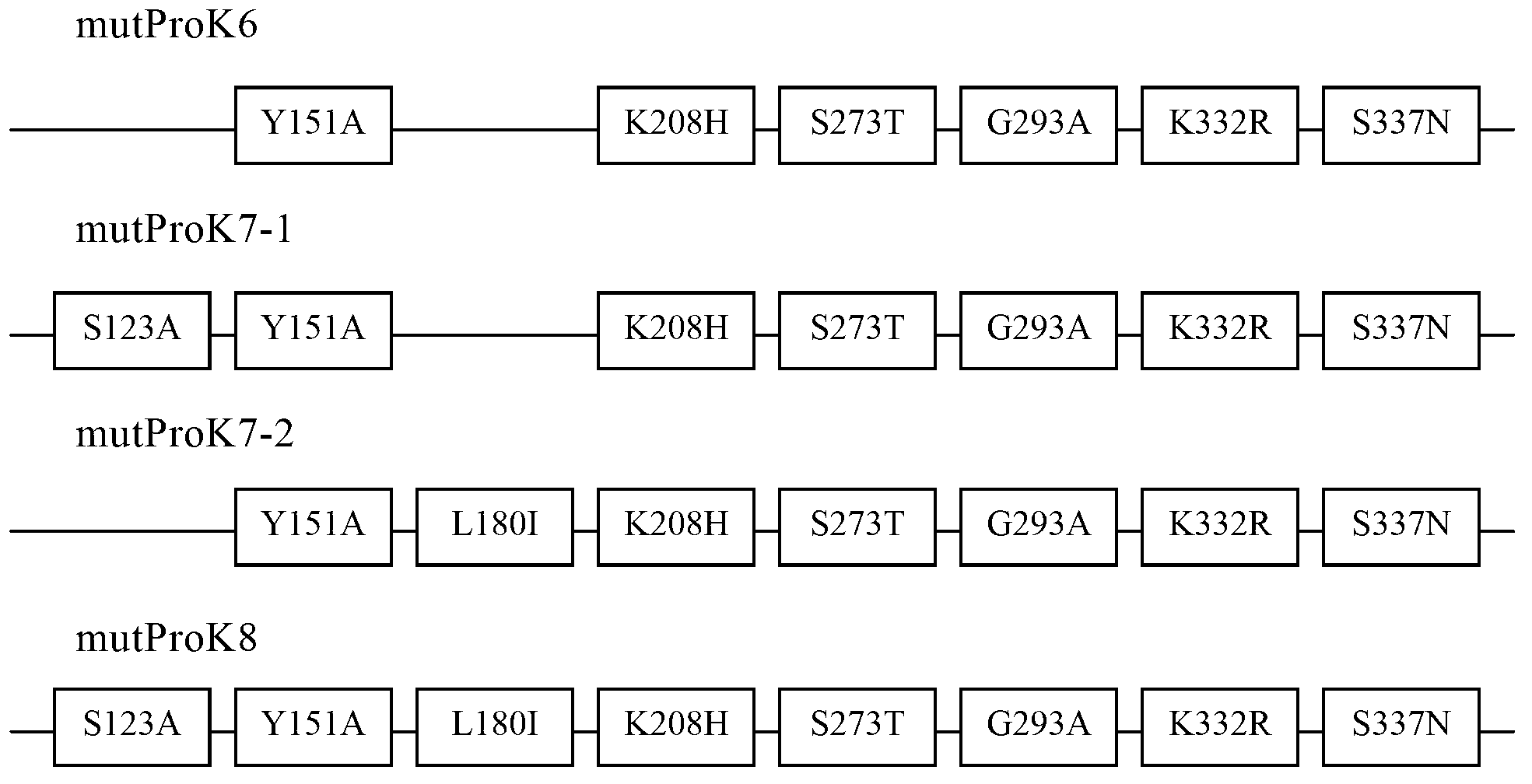
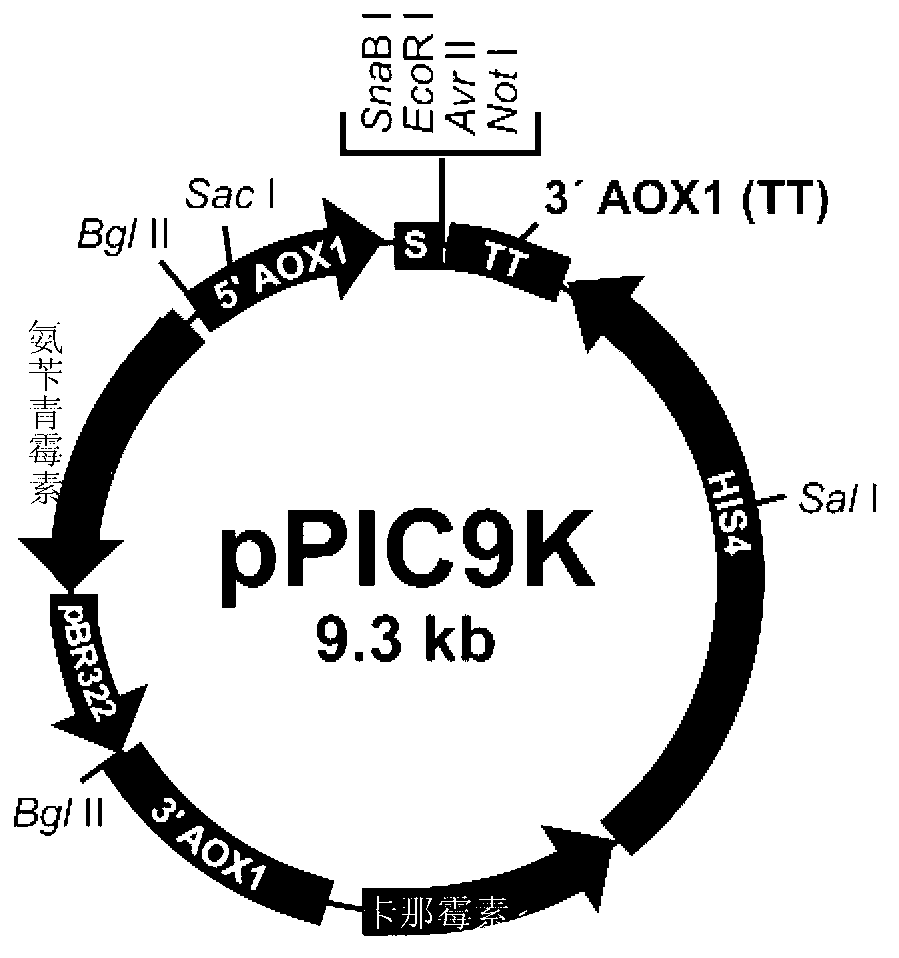
Gene mutation type recombined protease K and industrialized production method thereof
The invention relates to a gene mutation type recombined protease K and an industrialized production method thereof. Specifically, the invention relates to the gene mutation type recombined protease K which is obtained by performing gene reformation and protein engineering and has an enzymatic activity being more than two times of the enzymatic activity of the same natural protein, and further relates to the industrialized production method and a technical process for utilizing yeast cells to large-scale culture expressing foreign proteins and prepare the gene mutation type recombined protease K.
Owner:GPROAN BIOTECH (SUZHOU) INC
Kit and method for quickly extracting DNA (deoxyribonucleic acid) from colla corii asini
ActiveCN104046616AIncrease concentrationHigh purityMicrobiological testing/measurementDNA preparationChelex 100A-DNA
The invention discloses a kit and a method for quickly extracting DNA (deoxyribonucleic acid) from colla corii asini. A combined technology of an SDS (sodium dodecyl sulfate)-protease K and Chelex-100 combined process DNA extraction technology and a Glassmilk DNA purification technology is applied to DNA extraction of the colla corii asini. The dosage of needed colla corii asini paste is controlled to be 0.3ml to 0.5ml, the weight of colla corii asini blocks can be controlled to be 0.2mg to 0.6mg, and extracted DNA is high in concentration, excellent in purity, stable and difficult to degrade; due to extracted DNA, sequence fragments of 100bp-500bp can be amplified, and subsequent molecular biology experiments of gene cloning, sequencing and the like can be carried out. According to the method, the operation is simple, the dosage is low, and the consumed time is short; the method is especially suitable for DNA extraction of various types of colla corii asini products with low nucleic acid content and high degradation degree caused by deep processing; a foundation is provided for realizing colla corii asini source identification by adopting a DNA molecular biology identification technology.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
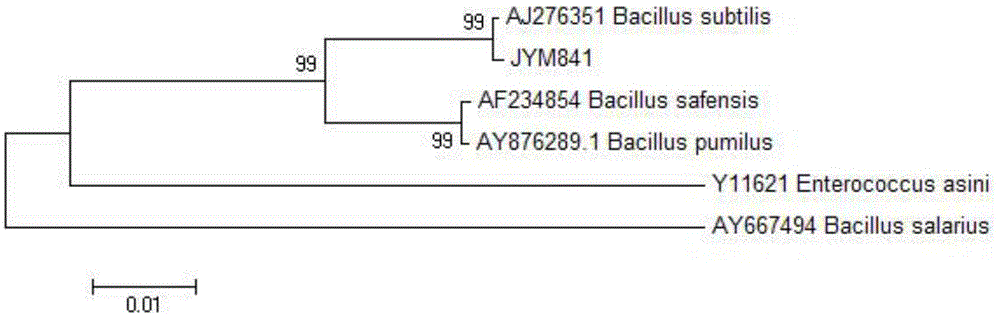
Bacillus subtilis with antibacterial activity and application of bacillus subtilis
ActiveCN104450590AStrong antibacterialBroad antibacterial spectrumAntibacterial agentsAntimycoticsStaphylococcus capraeCandida sanitii
The invention discloses bacillus subtilis JYM841 with antibacterial activity. The bacillus subtilis is preserved in a China center for type culture collection in January 10, 2014, and the preservation number is CCTCC NO: M 2014012. The bacillus subtilis JYM841 has a strong inhibitory effect on staphylococcus aureus and candida albicans, is high in antibacterial activity, wide in antibacterial spectrum, high in enzyme production, sensitive to bacterial antibiotic, drug-resistant to fungal antibiotic, high in heat-resisting ability and tolerant to acid and alkaline, has high antibacterial activity after treated at the temperature of 100 DEG C for 20min, and has tolerance to various enzymes such as trypsin, papain and protease K. An acid and alkali resistant pH (potential of hydrogen) value ranges from 6 to 10, and metabolite is stable, so that the bacillus subtilis can be applied to preparation of medicines for treating genital tract infection.
Owner:山东宝来利来生物工程股份有限公司
Efficient extraction method for whole blood genome DNA (deoxyribonucleic acid)
The invention relates to a method for quickly and efficiently extracting whole blood genome DNA (deoxyribonucleic acid), and belongs to the field of nucleic acid purification. The method comprises the following steps: adding a red cell lysis solution into whole blood, eddying until red cells are completely split, removing supernatant, and sucking all residual liquid; adding a white cell lysis solution and protease K, and splitting white cells in a water bath of 37 DEG C until the white cells are deposited and dissolved; adding sodium chloride and chloroform, carrying out centrifugal separation on DNA, and transferring the supernatant to another clean EP (epoxy) pipe; adding ammonium acetate and absolute ethanol for depositing DNA, washing by 75% of ethanol, naturally drying, and adding TE (Tris-EDTA(ethylene diamine tetraacetic acid)) for dissolving to obtain DNA. The method has the characteristics of high speed, high efficiency and non-toxicity. The extracted DNA can be used for various subsequent molecular biology studies such as polymerase chain reaction (PCR), methylation detection and gene cloning.
Owner:NORTH CHINA UNIVERSITY OF SCIENCE AND TECHNOLOGY
Method for separating free nucleic acid from blood serum or blood plasma sample by using magnetic bead
InactiveCN103952397AGuaranteed purityAvoid the inconvenience caused by high temperature incubationDNA preparationMagnetic beadBlood plasma
The invention relates to a method for separating free nucleic acid from a blood serum or blood plasma sample by using a magnetic bead, and belongs to the field of nucleic acid purification. The method comprises the following steps: firstly adding the blood serum / blood plasma sample, a lysis buffer, protease K and magnetic bead suspension liquid into a nuclease-free centrifugal tube, fully and uniformly mixing, then performing pyrolysis, adsorbing nucleic acid on the surface of the magnetic bead, performing purification under the effect of an external magnetic field, removing impurities such as protein, polysaccharide, salt and the like through a rinsing step, and finally eluting by adding nuclease-free water to obtain a free nucleic acid solution. The method disclosed by the invention has the characteristics of simplicity, high speed, stability, high efficiency and cheap price; the purified free nucleic acid is complete and high in purity, and can be directly used for subsequent experiments.
Owner:TIANGEN BIOTECH BEIJING
Kit for extracting viral genome nucleic acid and use method thereof
InactiveCN104805073ANo pollution in the processLarge nucleic acid adsorption capacityDNA preparationHigh concentrationMagnetic bead
The invention relates to a kit for extracting viral genome nucleic acid and a use method thereof. The kit is characterized by comprising protease K, silica-based modified magnetic beads, an extraction buffer solution, a rinsing liquid I, a rinsing liquid II, RNase-free water and the like, and further comprising a magnetic frame in a matched manner. The use method is characterized by comprising the following steps: adding protease K and the silica-based modified magnetic beads into the extraction buffer solution for one-time digestion pyrolysis of cells and adsorption pyrolysis of released virus nucleic acid; extracting guanidinium thiocyanate in the extraction buffer solution with the pH value of 5.5, so that the silica-based modified magnetic beads can adsorb more nucleic acid, that is, each gram of the silica-based modified magnetic beads can adsorb 10 mg or more of nucleic acid; utilizing hexadecyl trimethyl ammonium bromide contained in the extraction buffer solution for removing protein, polyose, phenols and other impurities, and utilizing polyvinylpyrrolidone K25 for removing pigments in the extraction buffer solution; conducting further purification on the silica-based modified magnetic beads adsorbed on the magnetic frame to obtain high-concentration genome nucleic acid. When the kit is utilized for extracting genome nucleic acid, independent digestion pyrolysis, washing and re-extraction on a source sample are not required in advance, so that the clinical test time is shortened, and the cost is greatly reduced.
Owner:EBIOS BEIJING BIOTECH
Extraction process of black corn silk polysaccharide
InactiveCN102504039AHigh recovery rateHigh purityMetabolism disorderAntineoplastic agentsCelluloseCombined method
The invention discloses an extraction process of black corn silk polysaccharide, which comprises a raw refining process, a protein removing process, a decolorization process and a grading and purifying process, wherein in the raw refining process, protease K water digestion and ultrasound extraction combined method is adopted in the raw refining process, the protein removing process is that sevage method and TDA (trichloroacetic acid) method are alternatively used for deproteinization, an ethanol decolorizing and peroxide decolorizing combined method is adopted in the decolorization process, and DEAE- cellulose (chlorine type) column chromatography method is adopted in the grading and purifying process. Compared with the prior art, the invention has the characteristics of high recovery ratio, excellent decolorization effect, high purity of polysaccharide and the like. The prepared black corn silk polysaccharide has better anticancer and hypolipemic effects.
Owner:ANHUI NORMAL UNIV
Simple method for efficiently extracting DNA (deoxyribonucleic acid) from soil samples
ActiveCN103993007APracticalHigh degree of reliabilityMicroorganism based processesDNA preparationFreeze thawingHigh concentration
The invention provides a simple method for efficiently extracting DNA (deoxyribonucleic acid) from soil samples. According to the simple method, the DNA in environmental samples is fully released and exposed by using combination of methods such as glass bead grinding, liquid nitrogen repetitive freeze-thawing, SDS (sodium-dodecyl sulphate), lysozyme and a protease-K method for the different soil samples, the DNA purification is carried out through multiple phenol imitation extraction processes, and the determination of the DNA extraction efficiency is carried out by adding an internal standard bacterial strain Xcc8004. The simple method has simple requirements on sample pretreatment; the quantity of the required environmental samples is small; the obtained DNA has relatively high concentration and purity; the extraction efficiency is relatively high; the analysis and detection of antibiotic resistance gene pollution in the complex environmental samples can be met; and the method is simple and reliable and has practical application value.
Owner:NANKAI UNIV
Method of simultaneously extracting animal DNA (Deoxyribonucleic Acid) virus and RNA (Ribonucleic Acid) virus nucleic acid in blood serum and double swabs
The invention discloses a method of simultaneously extracting animal DNA (Deoxyribonucleic Acid) virus and RNA (Ribonucleic Acid) virus nucleic acid in blood serum and double swabs. The method comprises the following steps of: carrying out lysis on a to-be-extracted substance by using guanidinium isothiocyanate lysate; adsorbing RNA by a silica gel membrane; removing impure protein by washing liquor I; removing impurities by washing liquor II; carrying out DEPC (Diethylpyrocarbonate) water-washing to remove nucleic acid, wherein the guanidinium isothiocyanate lysate comprises 3M-7M guanidinium isothiocyanate, 0.6%-1.0% TriTon-100, 30mM-50mM Tris-Cl, 5mM-15mM DTT (DL-Dithiothreitol), 60 mu g / mL-90 mu g / mL protease K, 10mM-30mM EDTA (Ethylene Diamine Tetraacetic Acid), and PH of the guanidinium isothiocyanate lysate is 4.3-4.6; the washing liquor I comprises 5M-6M guanidine hydrochloride, 53%-59% absolute ethyl alcohol, 70-90 mu g / mL protease K, and PH of the washing liquor I is 6.4-6.6; the washing liquor II comprises 70%-80% alcohol. The method disclosed by the invention has the advantages of being simple in extracting process, short in period, low in cost, and capable of simultaneously extracting RNA virus and DNA virus nucleic acid in an animal blood serum sample and a double-swab sample.
Owner:安徽华卫集团禽业有限公司
LAMP (loop-mediated isothermal amplification) rapid detection kit and detection method for salmonella
InactiveCN102199665AResolution cycleResolve SensitivityMicrobiological testing/measurementAgainst vector-borne diseasesRibonucleosideFluorescence
The invention belongs to the field of biological products, and relates to a detection kit and a detection method for rapidly detecting salmonella by utilizing an LAMP (loop-mediated isothermal amplification) technique. The kit is composed of a salmonella genome extracting reagent and an LAMP reaction reagent, wherein the salmonella genome extracting reagent comprises the following components: SDS (sodium dodecyl sulfate), protease K, CTAB (cetyltrimethyl ammonium bromide) / NaCl solution and NaAC; and the LAMP reaction reagent is composed of a reaction liquid A and a reaction liquid B, the reaction liquid A comprises the following components: 10*Lampbuffer, Bst DNA (deoxyribonucleic acid) polymerase, dNTP (deoxy-ribonucleoside triphosphate), an interprimer 1, an interprimer 2, an outer primer 1, an outer primer 2, lycine and ultrapure water, and the reaction liquid B is a fluorescent dye. The invention can be used to carry out LAMP detection on salmonella, has the characteristics of rapid detection, high accuracy, high sensitivity and convenient field application, and can be widely applied in the fields of veterinarian, food and exit-entry quarantine, etc.
Owner:镇江出入境检验检疫局检验检疫综合技术中心
Extraction kit and method for genome DNA of evergreen woody plants
ActiveCN107326023APrevent combined browningPrevent browningDNA preparationSecondary metaboliteGenetic engineering
The invention relates to an extraction kit and method for genome DNA of evergreen woody plants and belongs to the field of genetic engineering. The extraction kit comprises a rinsing liquor, a lysate, a leaching liquor A, a leaching liquor B, a binding buffer liquor, a column buffer liquor, an elution buffer liquor and an adsorption column, and further comprises a grinding assisting agent, wherein the grinding assisting agent comprises insoluble PVPP, quartz sand and ascorbic acid; the rinsing liquor comprises Tris-HCl, EDTA-Na2, NaCl, KAc, glucose, PVP, ascorbic acid and PEG; and the lysate comprises CTAB, Tris-HCl, EDTA-Na2, NaCl, sorbitol, borax, PVP, ascorbic acid and protease K. The extraction method is flexible and quick in operation, low in cost and wide in application range, and can radically remove secondary metabolites and other interfering substances in cells, and the extracted genome is good in quality, high in purity and less in protein pollution.
Owner:HENAN UNIV OF SCI & TECH
Extraction method of total DNA of microorganisms from soil sample
InactiveCN104651350ALow costImprove efficiencyMicrobiological testing/measurementDNA preparationWater bathsFreeze thawing
The invention relates to an extraction method of total DNA of microorganisms from a soil sample. The extraction method particularly includes following steps: (1) adding a DNA extraction buffer solution to the soil sample and performing repetitive freeze-thawing in liquid nitrogen and hot water bath; (2) adding a protease K, performing a water bath treatment process at 37 DEG C for 30 min; (3) adding a sodium dodecyl solution, performing a water bath treatment process at 65 DEG C for 30 min; (4) performing centrifugation to remove precipitations and collect a supernate; (5) performing extraction with a mixed solution containing phenol, chloroform and isoamylol and performing centrifugation to collect a supernate; (6) performing extraction with a mixed solution containing chloroform and isoamylol and performing centrifugation to collect a supernate; (7) adding isopropanol to the supernate in the step (6) to performing precipitation and collecting a precipitation after centrifugation; (8) adding ethanol for washing the precipitation and drying the precipitation at the room temperature; (9) adding a TE buffer solution for dissolving the precipitation to obtain a DNA crude extraction; and (10) purifying the DNA crude extraction through an adsorption column, packaging the purified DNA crude extraction and storing the purified DNA crude extraction at -20 DEG C. The extraction method is especially effective when being used in samples from deep soil and dry sand which are less in biomass and in a clayed soil sample. The DNA extraction can be directly used in subsequent molecular biological detection.
Owner:CHINA PETROLEUM & CHEM CORP +1
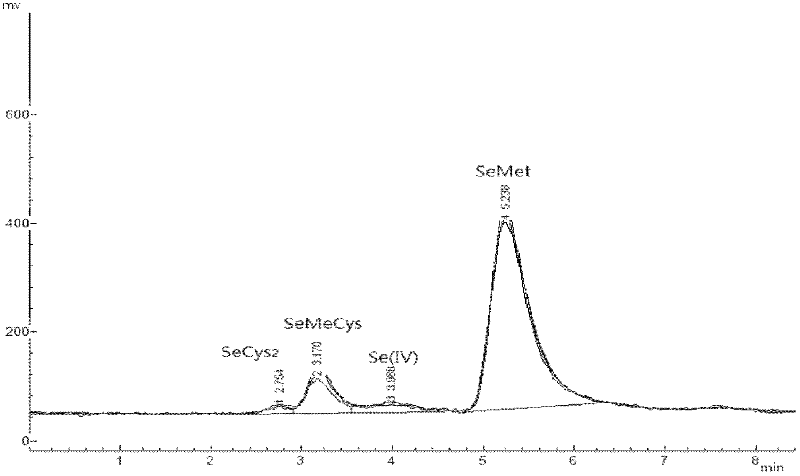
Method for determining selenium form in selenium-rich egg
The invention discloses a method for determining a selenium form in a selenium-rich egg, which is characterized by comprising the following steps: 1) pretreating the selenium-rich egg, using an extraction buffer Tris-HCl for extracting a sample of the selenium-rich egg, adding lipase for extracting if the selected selenium-rich sample is a yolk sample, then successively adding protease K and protease XIV with an effective amount for hydrolysis extraction to obtain a sample solution to be measured; directly and successively adding protease K and protease XIV with an effective amount hydrolysis extraction to obtain the sample solution to be measured if the selected selenium-rich egg sample is a egg white sample; 2) using combination of hydride generation-atomic fluorescence spectroscopy and high performance liquid chromatography for respectively determining a solution with a standard selenium form and the sample solution to be measured. The method of the invention is simple and easy to operate, the repeatability is good, and the used reagent has the advantages of no toxicity and no pollution.
Owner:SUZHOU SETEK
Salt-tolerant ethanol-tolerant protease-tolerant surfactant-tolerant exoinulinase, gene thereof, vector and strain
InactiveCN103981161APromote hydrolysisGood Salt Tolerance Ethanol Tolerance Tolerance To ProteaseBacteriaMicroorganism based processesExoinulinase activityBiofuel
The invention discloses a salt-tolerant ethanol-tolerant protease-tolerant surfactant-tolerant exoinulinase, a gene thereof, a vector and a strain. The exoinulinase InuAJB13 possesses the following properties: the optimum pH is 5.5, 50% or more of enzymatic activity is maintained in the pH scope of 4.0-7.0; the remnant enzyme activity reaches 90% or more after exoinulinase is processed by a buffer with a concentration of 0.1 M and pH of 4.0-7.0 for 1 h; the optimum temperature is 55 DEG C, and exoinulinase has the enzyme activity at 10-70 DEG C; a NaCl solution with a concentration of 0.6-4.5 M is capable of improving the enzyme activity by 0.2-0.6 times; 100% of the enzyme activity can be kept after exoinulinase is processed by a NaCl solution with a concentration of 0.2-4.5 M at 37 DEG C for 60 min; exoinulinase keeps the activity in 10% (V / V) of ethanol; exoinulinase keeps 88% or more of the activity after being processed in 3.0-15.0% (V / V) ethanol at 37 DEG C for 60 min; exoinulinase activity is not influenced or slightly influenced by trypsin, protease K, surfactants, most of metal ions, and commercial liquid laundry detergents; and exoinulinase is capable of hydrolyzing inulin, cane sugar, raffinose, stachyose, beta-2,6-fructan (levan) and soluble starch. The exoinulinase disclosed by the invention is applicable to industries such as feed, foodstuff, washing and biofuels.
Owner:YUNNAN NORMAL UNIV
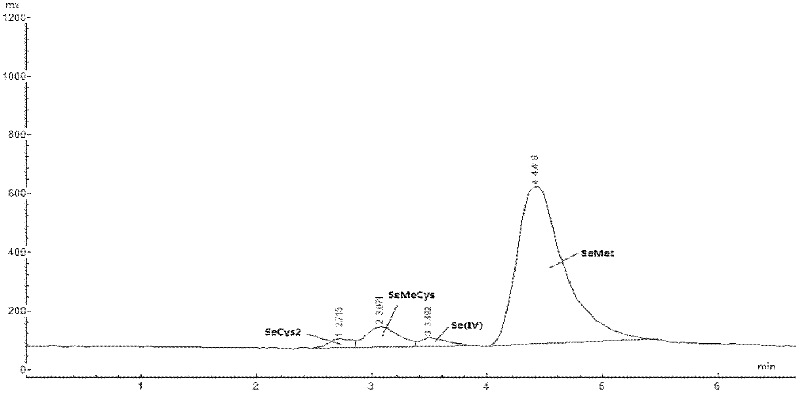
Method for determining form of selenium in selenium-enriched edible fungi
InactiveCN102519933AReduce dosageNo pollution in the processComponent separationFluorescence/phosphorescenceAtomic fluorescence spectrometryHydrolysis
The invention discloses a method for determining the form of selenium in selenium-enriched edible fungi. The method is characterized by comprising the following steps of: (1) pretreating selenium-enriched edible fungi: using deionized water as an extracting solution to extract a crushed selenium-enriched edible fungus sample, then adding effective amounts of protease K and protease XIV for hydrolysis so as to extract a sample solution to be detected; (2) conducting hydride generation: combining atomic fluorescence spectrometry with high performance liquid chromatography to determine a standard selenium form solution and the sample solution to be detected respectively. The method of the invention has the advantages of simplicity and easy operation, good repeatability, and employment of non-toxic and pollution-free reagents.
Owner:SUZHOU SETEK
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com