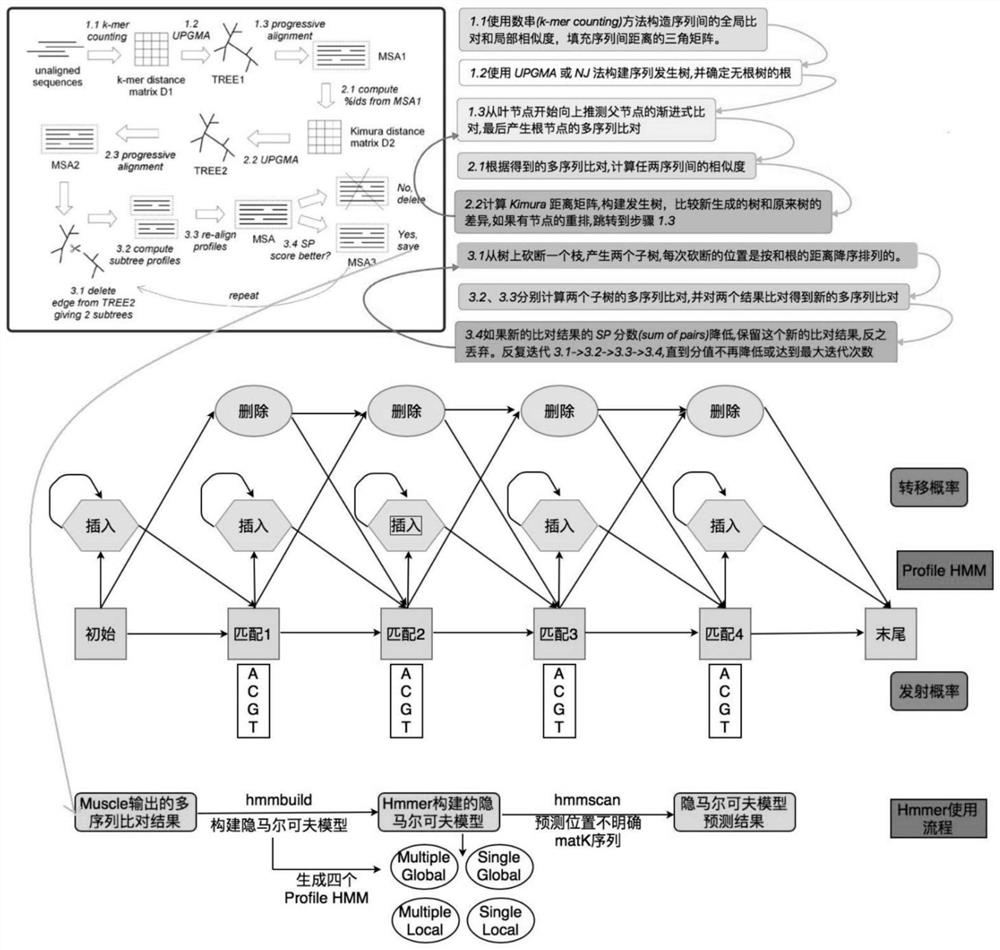
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
48 results about "Species level" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Species are as specific as you can get. It is the lowest and most strict level of classification of living things. The main criterion for an organism to be placed in a particular species is the ability to breed with other organisms of that same species. The species of an organism determines the second part of its two-part name.
Establishment method of gene library of fecal flora based on high-throughput gene sequencing
InactiveCN105937053AFast database buildingImprove throughputMicrobiological testing/measurementLibrary creationSequence databaseGenomic library
An establishment method of a gene library of fecal flora based on high-throughput gene sequencing employs a ''nested PCR'' method for enrichment amplification on 16S rDNA, so as to reduce the host and food residue genome pollution by the maximum; and V3 and V6 are combined for specific amplification and massive parallel sequencing, so as to obtain the a target gene sequence database of the flora. The library can increase the identification of the bacterial flora from Genus level to Species level. The method can maximally avoid the interference of host cell nucleic acid, and completely and efficiently detect 16SrDNA tag sequence of all bacteria varieties, so as to accurately determine the ecological structure of the intestinal flora.
Owner:GUANGZHOU SAGENE BIOTECH
Method of detecting microorganisms
InactiveUS7303870B2Broad specificityAccurately determineSugar derivativesMicrobiological testing/measurementSequence analysisMedical diagnosis
The present invention relates generally to a method for detecting, enumerating and / or identifying microorganisms in a sample. More particularly, the present invention provides a method for determining total microbial content in a sample by detecting the presence of nucleotide sequences associated with all or part of 16S rDNA or its corresponding 16S rRNA or its homologue, functional equivalent or derivative. The nucleotide sequences of the present invention may be used as an indicator of any microorganism and, hence, represents a universal target sequence which is indicative of total microbial content in a sample. The universal target sequence may also be varied to render same genus or species specific or the universal target used to trap microbial DNA or RNA which may be subsequently analyzed by sequence analysis or genetic probe technology. The universal target sequence is useful inter alia to design as universal primers and probes to amplify any microbial-derived genomic sequence, as a means to detect and enumerate total microorganisms and to identify microorganisms in a sample at the genus or species level. Such uses enable improved methods of enviroprotection, bioremediation, medical diagnosis and industrial microbiology. The present invention further relates to the universal target sequence in isolated form and / or primers or probes capable of hybridizing to same and kits for the detection of total microbial content in a sample.
Owner:THE UNIV OF SYDNEY +1
Method for designing species specific primer for detecting species with known genome information in microbial community and method for measuring bacterium content
InactiveCN106222249AAccurate understandingLower Sequencing CostsMicrobiological testing/measurementDNA/RNA fragmentationInformation analysisFermentation
The invention relates to a method for designing a species specific primer for detecting the species with known genome information in a microbial community and a method for measuring the bacterium content. The designing method is based on a species specific primer of some species with known genome information in a microbial community. The method comprises the following steps: cloning and sequencing a nearly full-length ribosomal gene library; carrying out biological information analysis (BLAST, etc.) to determine the species in a bacterial community and relative abundance of the species; inquiring the species with known genome sequences; based on a PRIMER-BLAST tool, designing a species specific primer, and detecting the specificity of the primer. The bacterium content is measured through a QPCR method: the provided species specific primer is used to quantitatively measure the content of corresponding species in a microbial community. The provided method for measuring the content of known microbes in a microbial community can trace and inspect the number change of some important microbial species in an important biological process (liquor fermentation, for example) in the species level. The provided method can measure the copy number of non-ribosomal gene sequence of species specificity in some microbial genome and cannot obtain the specific number of cells of species in a microbial community.
Owner:HARBIN INST OF TECH AT WEIHAI
Hybrid Chinese tuliptree rapid molecular detection specific primer and use method thereof
InactiveCN101348830AFast and reliable detectabilityFast and reliable identificationMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAgricultural science
The invention discloses primers for detecting hybrid trembling poplar molecule and a usage thereof, being specially used in quick molecular detection and identification of trembling poplar, liriodendron tulipifera and hybrid trembling poplar, and belonging to the molecular identification field for new plant variety. A pair of SSR primers, i.e. LT008L / R is designed, and can realize specific amplification of a 190bp product inside the trembling poplar, a 180bp product inside liriodendron tulipifera and a 180bp product, and a 190bp product inside hybrid trembling poplar. The primer adopts a pair of specific SSR primers to carry out once PCR amplification reaction, and carries out electrophoresis by use of 8 percent polyacrylamide gel; moreover, judgment can be carried out directly according to product fragment. The primers can be directly used for the species level identification of hybrid trembling poplar, trembling poplar and liriodendron tulipifera.
Owner:NANJING FORESTRY UNIV
High through-put detection of pathogenic yeasts in the genus trichosporon
InactiveUS20060216723A1Rapid and simple to performSugar derivativesMicrobiological testing/measurementSequence analysisPresent method
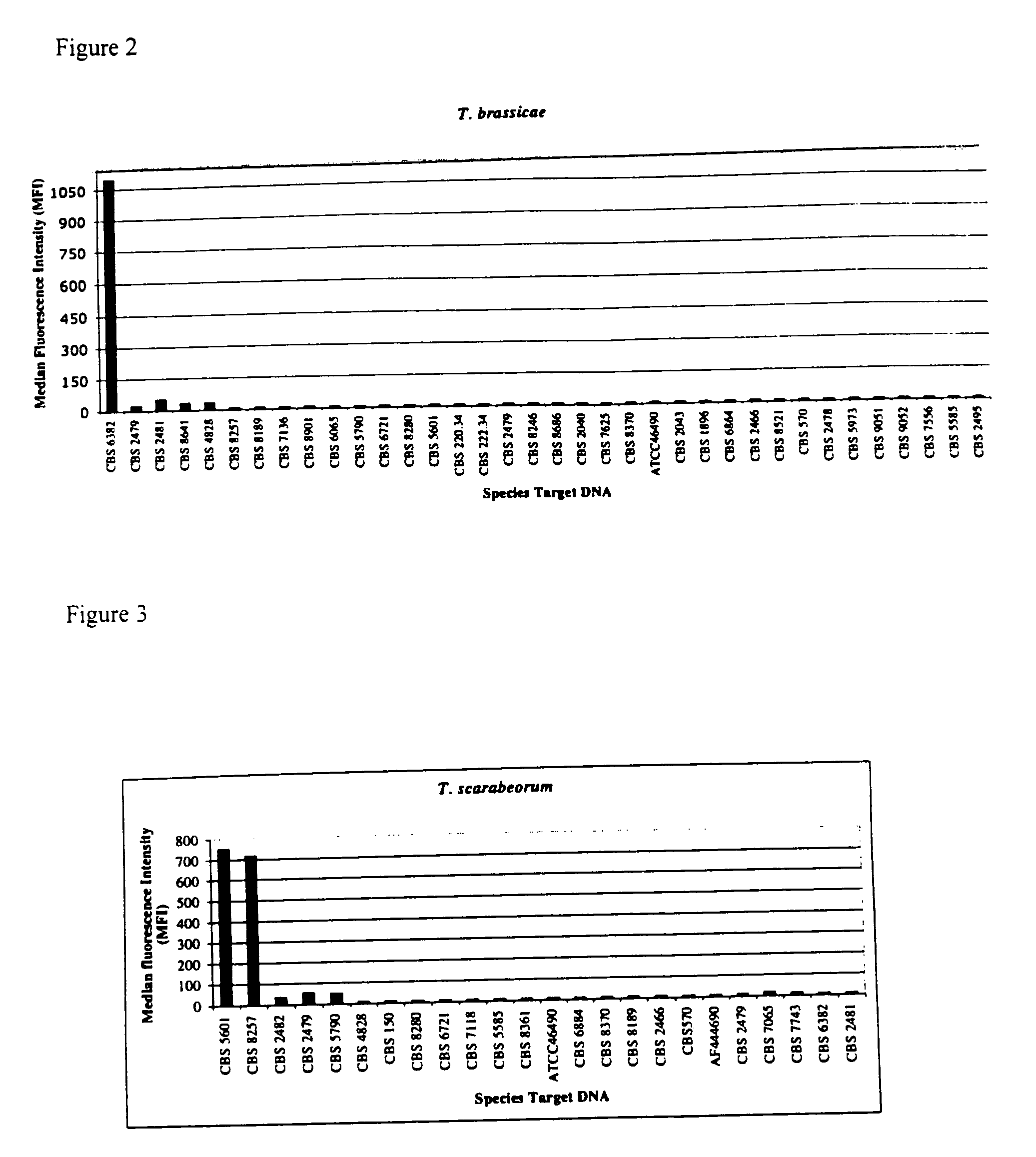
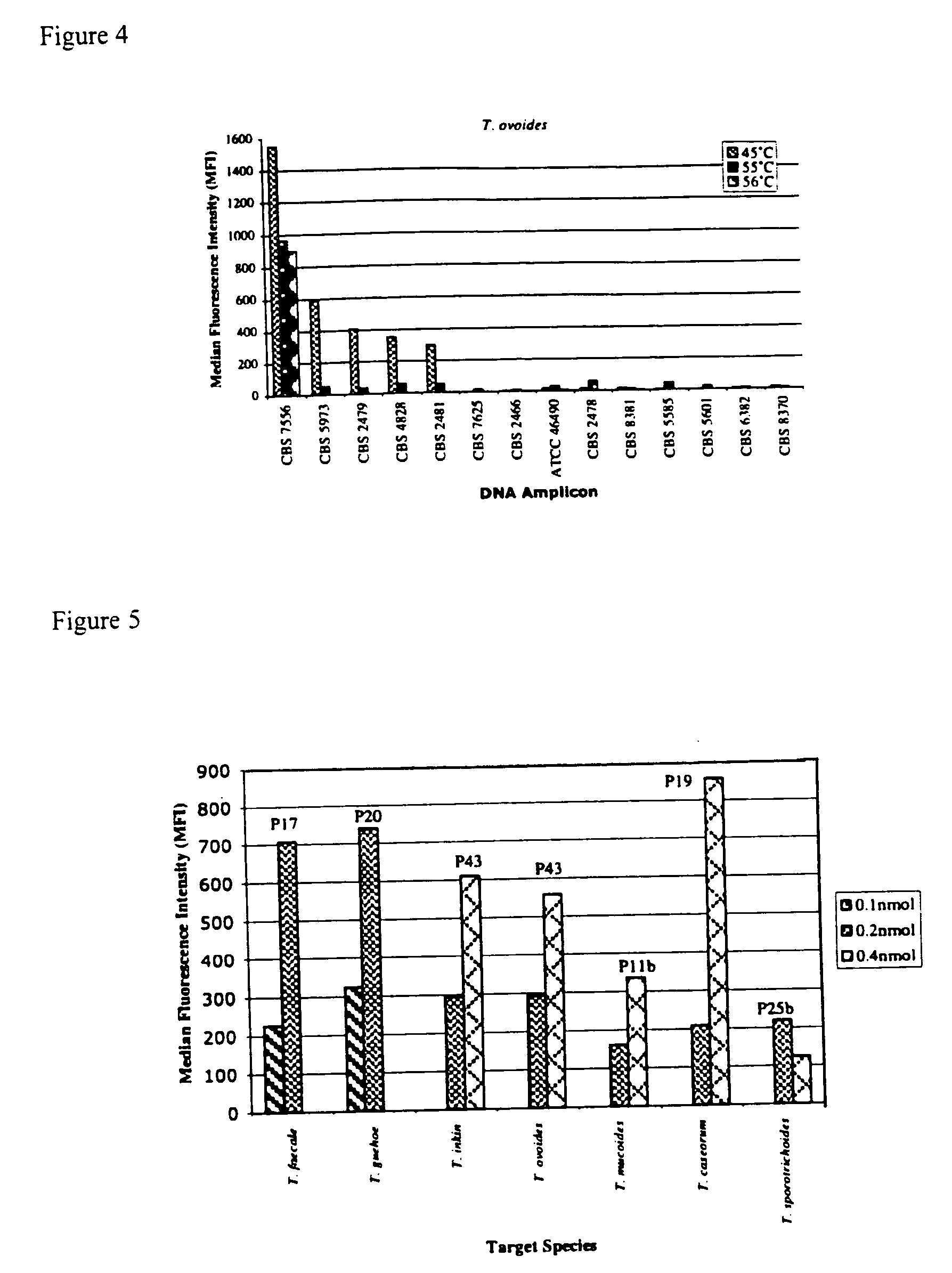
The emergence of opportunistic and antifungal resistant strains has given rise to an urgent need for a rapid and accurate method for the detection of fungal pathogens. In this application, we demonstrate the detection of medically important fungal pathogens at the species level. The present method, which is based on a nucleotide hybridization assay, consists of a combination of different sets of fluorescent beads covalently bound to species specific capture probes. Upon hybridization, the beads bearing the target amplicons are classified by their spectral addresses with a 635 nm laser. Quantitation of the hybridized biotinylated amplicon is based on the fluorescent detection with a 532 nm laser. Using this technology we designed and tested various multiplex formats, the performance of forty eight species specific and group specific capture probes designed from sequence analysis in the D1 / D2 region of ribosomal DNA, internal transcribed spacer regions (ITS), and intergenic spacer region (IGS). Species-specific biotinylated amplicons (>600 bp) were generated with three sets of primers to yield fragments from the three regions. The developed assay was specific and relatively fast, as it discriminated species differing by one nucleotide and required less than 50 min following amplification to process a 96 well plate with the capability to detect up to 100 species per well. The sensitivity of the assay allowed the detection as low as 102 genome molecules in PCR reactions and 107 to 108 molecules of biotinylated amplification product. This technology provided a rapid means of detection of Trichosporon species and had the flexibility to identify species in a multiplex format by combining different sets of beads. The assay can be expanded to include all known pathogenic fungal species.
Owner:MIAMI UNIVERISTY OF
System and method for productizing human capital labor employment positions/jobs
A method includes configuring a human-capital-management (HCM) master taxonomy and a HCM language library. The HCM master taxonomy includes a plurality of levels that range from more general to more specific, each level of the plurality of levels comprising a plurality of nodes. The plurality of levels include a job-species level and a job-family level, the job-species level including a level of greatest specificity in the plurality of levels, the job-family level including a level of specificity immediately above the job-species level. In addition, the method includes transforming human-capital information via the HCM language library. Further, the method includes classifying the transformed human-capital information into a job-family node selected from the plurality of nodes at the job-family level.
Owner:IQNAVIGATOR
PCR identification method for sorghum halepense and similar species and kit
InactiveCN101748191AAddressing fake sorghumSolve the problem of identification of its close speciesMicrobiological testing/measurementSorghum sudanenseMolecular identification
The invention relates to identification filed of importing quarantine weed species level, in particular to an identification method for sorghum halepense, black sorghum, and similar specicies and a kit, silk sorghum, sorghum propinquum, and sorghum sudanense these five species and the kit used by the method. The identification method comprises: 1, identifying PCR; 2, electrophoresis detection. The kit comprises 3 pairs of PCR reaction primers and a standard map. The advantages of the invention are as follows: the invention can solves the problem of identification of the sorghum halepense and the similar species of the sorghum halepense, and a kit which completes molecular identification of the sorghum halepense and the similar species of the sorghum halepense in 8 hours is provided.
Owner:INSPECTION & QUARANTINE TECH CENT OF FUJIAN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
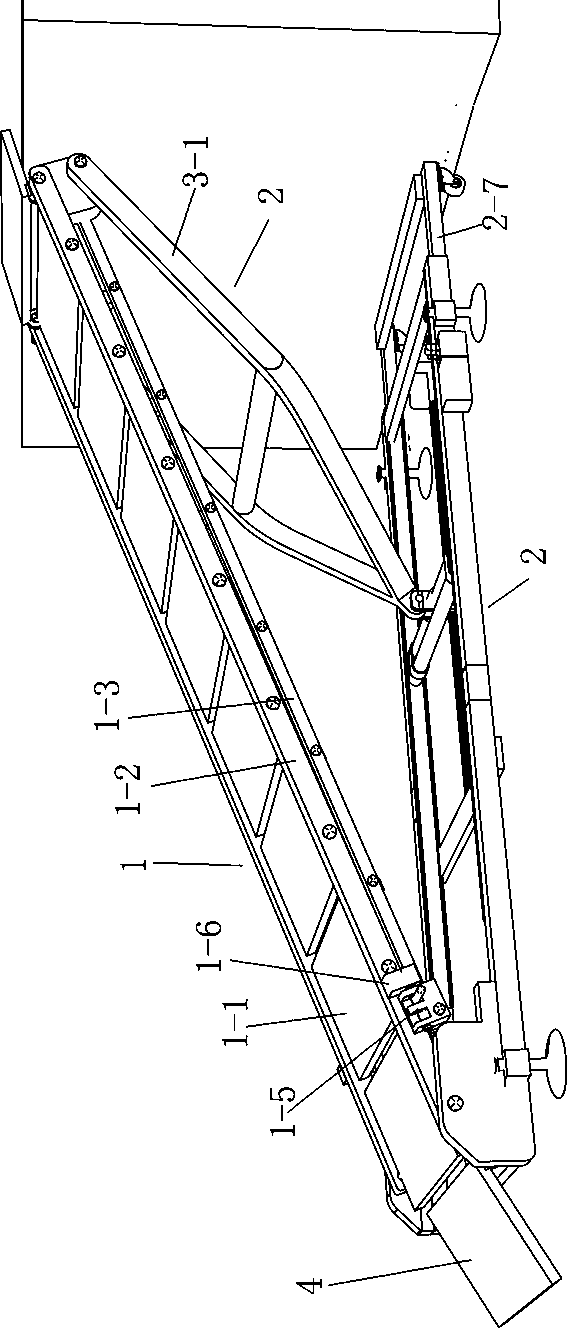
Emergency flattened folding ladder
Owner:唐晓腾
Method for detecting bartonella elizabethae with TaqMan real-time fluorescent quantitative PCR
InactiveCN104789657AGood repeatabilityGood linear relationshipMicrobiological testing/measurementDNA/RNA fragmentationDiseaseBartonella
The invention discloses a method for detecting bartonella elizabethae with TaqMan real-time fluorescent quantitative PCR. The method selects a Be specific gene sequence and obtains specific primers (SEQ ID No.2 and SEQ ID No.3) and a TaqMan probe (SEQ ID No.4) based on the specific gene sequence design. The method can detect the bartonella elizabethae under species level specificity and high sensitivity and provides effective means for researches such as early and rapid diagnosis and monitoring of a series of diseases caused by the bartonella and epidemiological investigation.
Owner:ICDC CHINA CDC
Method for evaluating dominance of species and application thereof in monitoring of endangered species and diagnosis and risk prediction of human flora-related diseases
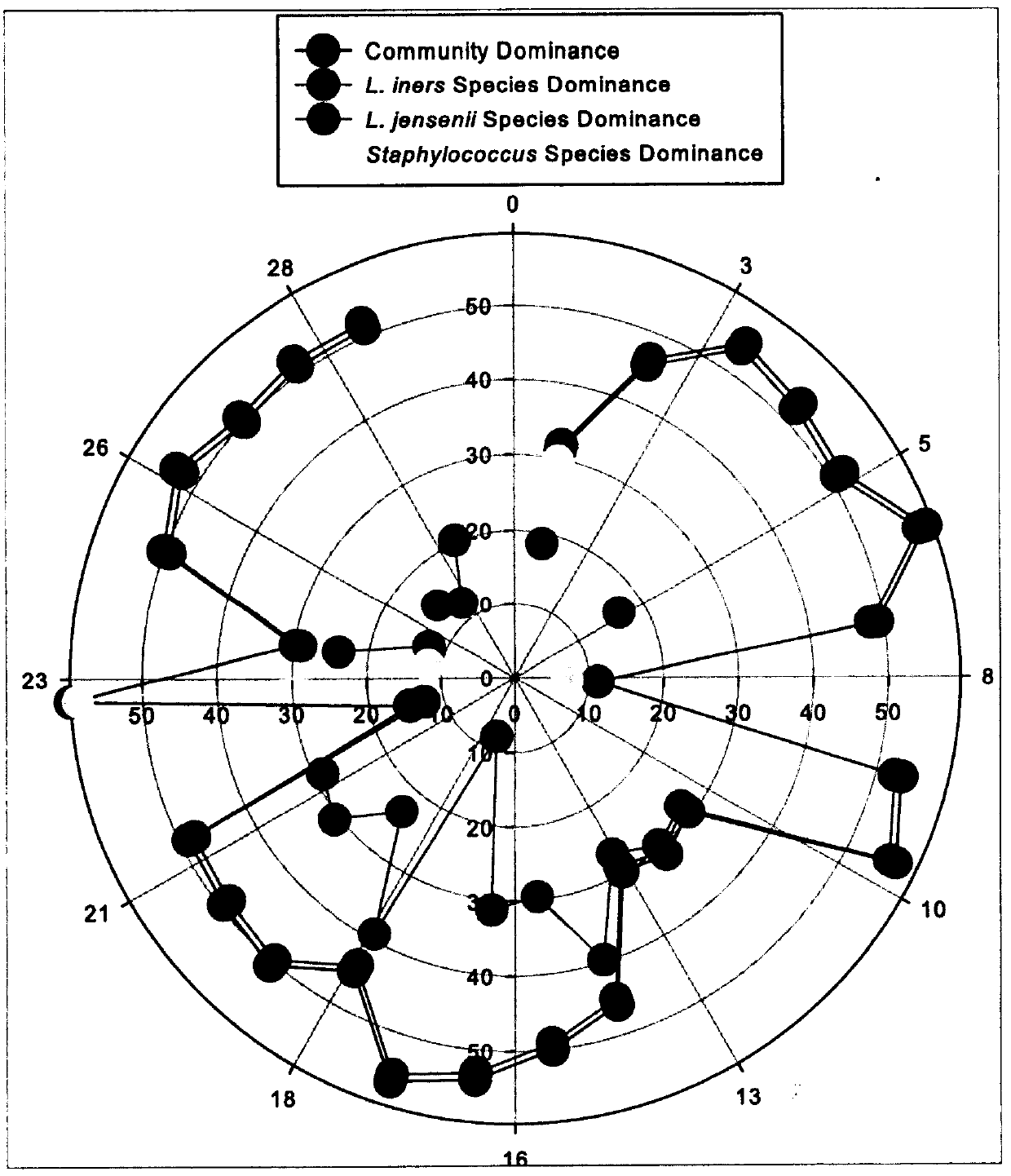
The invention relates to the dominance evaluation of ecological communities at the community level and species level, and mainly invents a concept and a method capable of comprehensively evaluating the dominance at the community level and species level. The concept and the method are used to monitor endangered species in macro-ecological communities and to diagnose and predict the risk of human flora-related diseases in microbial communities. The inventor defines a dominance index at the community level, a species dominance distance at the species level and a species dominance index. The threeindexes are defined by a unified mathematical concept and method, and can be used to evaluate the dominance of various biological / ecological communities (including macro-ecological communities (suchas tropical rainforests) and human microbial communities) in various time and / or space scales. In the specification, the microbial flora (community) in the female reproductive tract is taken as an example, and the dominance index is used to study the dominance change of the community in the time scale and the dominance of each species in the community and identify the difference in dominance of the same species in different communities (in the space scale).
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
Detection Of Bacteria Belonging to the Genus Campylobacter By Targeting Cytolethal Distending Toxin
ActiveUS20100248238A1Microbiological testing/measurementAgainst vector-borne diseasesEscherichia coliBacteroides
Multiplex PCR primers that can amplify the cdt genes of C. jejuni, C. coli, and C. fetus in a bacterial species-specific manner were prepared. Multiplex PCR with the primers was assessed using Campylobacter bacteria, other cdt gene-positive bacteria, and representative bacteria responsible for enteric infection. As a result, the present inventors' multiplex PCR using cdtB amplification primers was proven to enable simultaneous detection of different Campylobacter bacteria with high specificity. The methods of the present invention can identify Campylobacter at the bacterial species level in a single manipulation even when domestic animals or humans are infected with different bacterial species of Campylobacter.
Owner:FUSO PHARMA INDS +1
Chromogenic medium for the detection and identification of vancomycin resistant enterococci and method therefor
ActiveUS20080145881A1Facilitates direct identification and differentiationMicrobiological testing/measurementBiological material analysisMicroorganismChromogenic Substrates
A microbe-specific medium, containing specific chromogenic substrates, for the detection of vancomycin-resistant enterococci in a biological sample, whereby both the detection and identification of vancomycin-resistant enterococci at the species level is achieved utilizing one sample and one test.
Owner:BECTON DICKINSON & CO
Method and device to identify microbial species from sample
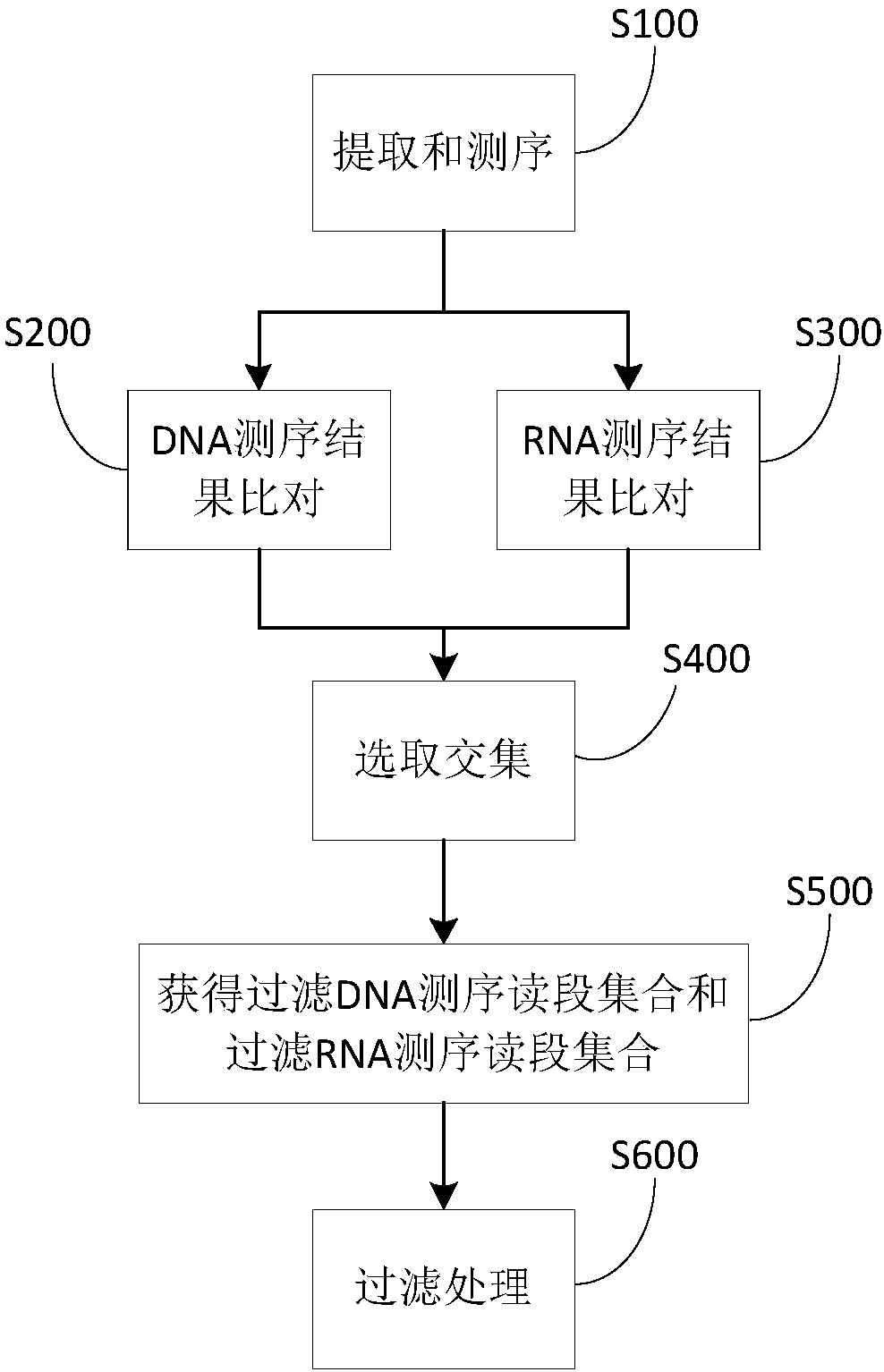
ActiveCN109082479AAccurate identificationAvoid interferenceBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismSpecies level
The invention provides a method and device to identify microbial species from a sample. The method includes: acquiring DNA and RNA sequencing results; comparing the DAN sequencing results to obtain afirst candidate microbe set and a specific DNA sequencing read set; comparing the RNA sequencing results to obtain a second candidate microbe set and a specific RNA sequencing read set; using at leastpart of an intersection of the first and second candidate microbe sets as a third candidate microbe set; selecting specific DNA and RAN sequencing reads of same quantity respectively from the specific DNA and RNA sequencing read sets so as to respectively obtain filtered DAN and RAN sequencing read sets; filtering the third candidate microbe set to obtain a fourth candidate microbe set, thereby forming microbial species in the sample. Therefore, the method and device allow species-level microbes to be accurately identified, can avoid interference from background microbes, are suitable for finding actively expressed pathogenic microbes, and are simple to operate.
Owner:SHENZHEN HUADA GENE INST
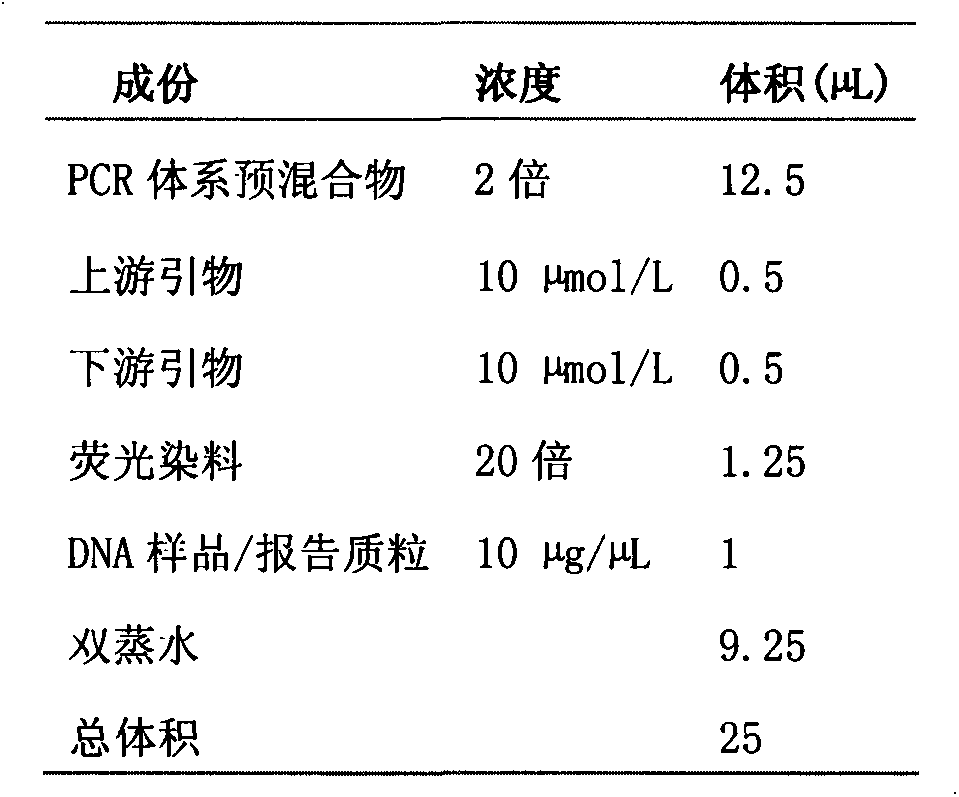
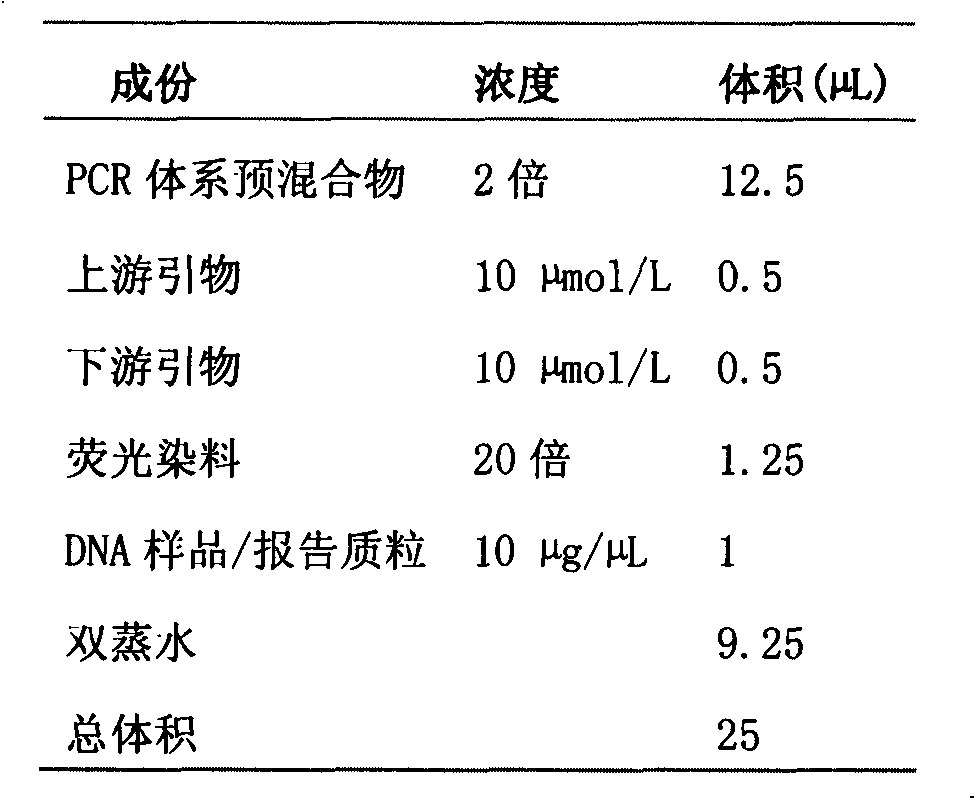
Fluorescent PCR (polymerase chain reaction) quick detection primer and kit for Xanthomonas campestris pv. campestris
InactiveCN103114136AGuaranteed reliabilityRealize integrated closure detectionMicrobiological testing/measurementFluorescence/phosphorescenceXanthomonas campestrisSpecies level
The invention relates to a real-time fluorescent PCR (polymerase chain reaction) quick detection primer and kit for Xanthomonas campestris pv. campestris. The primer comprises a forward primer of which the sequence is SEQ ID NO:1 and a reverse primer of which the sequence is SEQ ID NO:2. In the invention, a specific conserved sequence is obtained through molecular biological analysis according to a pthBXam and Tn3 gene sequence of Xanthomonas campestris pv. manihotis, and a specific amplification primer thereof is designed. The conserved gene sequence is shared by different Xanthomonas campestris pv. manihotis strains to ensure the reliability in detection of Xanthomonas campestris pv. manihotis from different sources on a species level. Besides, the primer provided by the invention adopts fluorescence labeling; and compared with the common PCR technology, observation does not need to be performed through a gel electrophoresis method, thus realizing the integrated sealed detection in the detection process.
Owner:郑媛
Library-building sequencing method for detecting full length of bacterial 16S rDNA
ActiveCN110452974ASuitable for structure detectionAccurate ecological structureMicrobiological testing/measurementSequence analysisMicroorganismSpecies level
The invention discloses a library-building sequencing method for detecting full length of bacterial 16S rDNA. By adding a molecular tag UMI to each sample, a ligation library and a splicing library are separately amplified, the two libraries are sequenced, the data is extracted by recognizing the UMI combination, and the full length sequence of 16S rDNA was assembled, and the species type is determined by comparing the databases. The library-building method of the invention is suitable for the detection of the flora structure of all types of samples, and can upgrade the traditional flora structure identification from a genus level to a species level, compared with the genus level, the species level has the advantage that an ecological structure of environmental microorganisms can be more accurately determined, which facilitates deep research, and method realizes the detection of specific bacterial species in samples.
Owner:北京群峰纳源健康科技有限公司
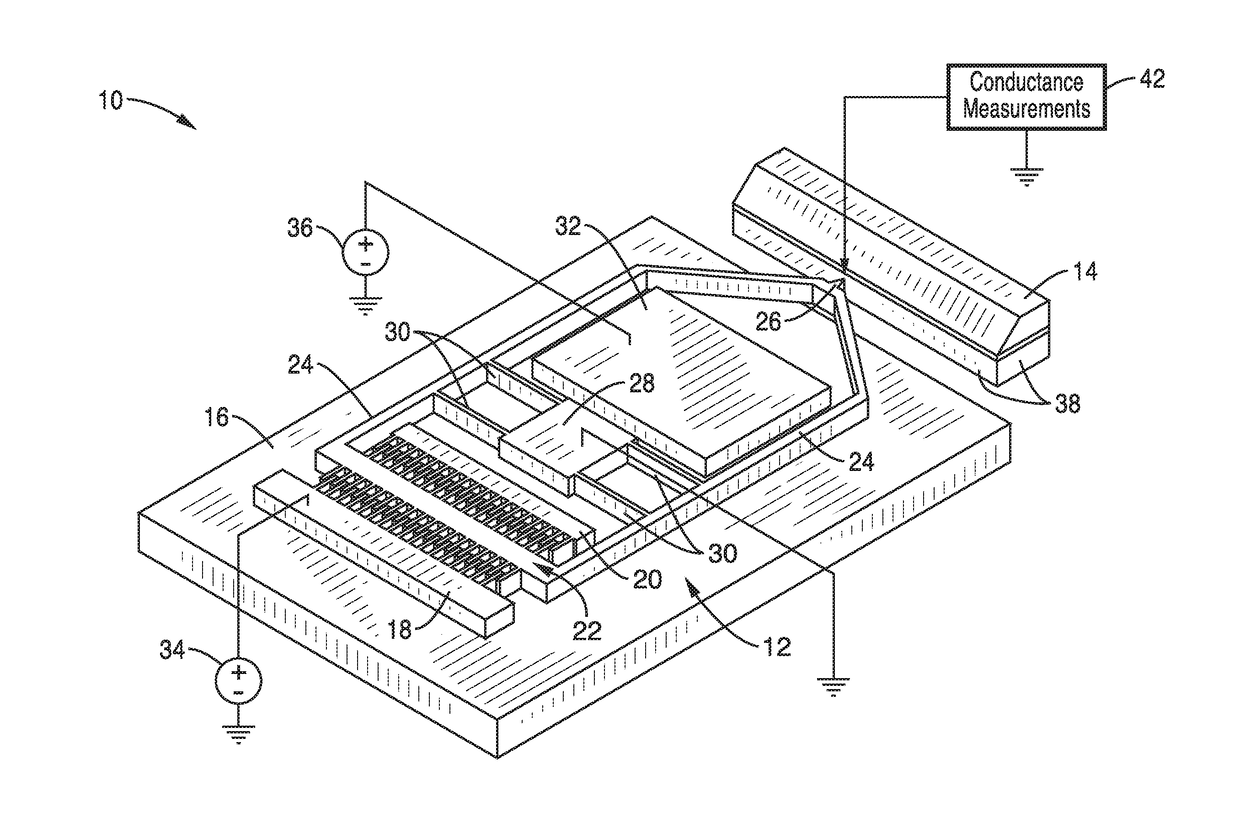
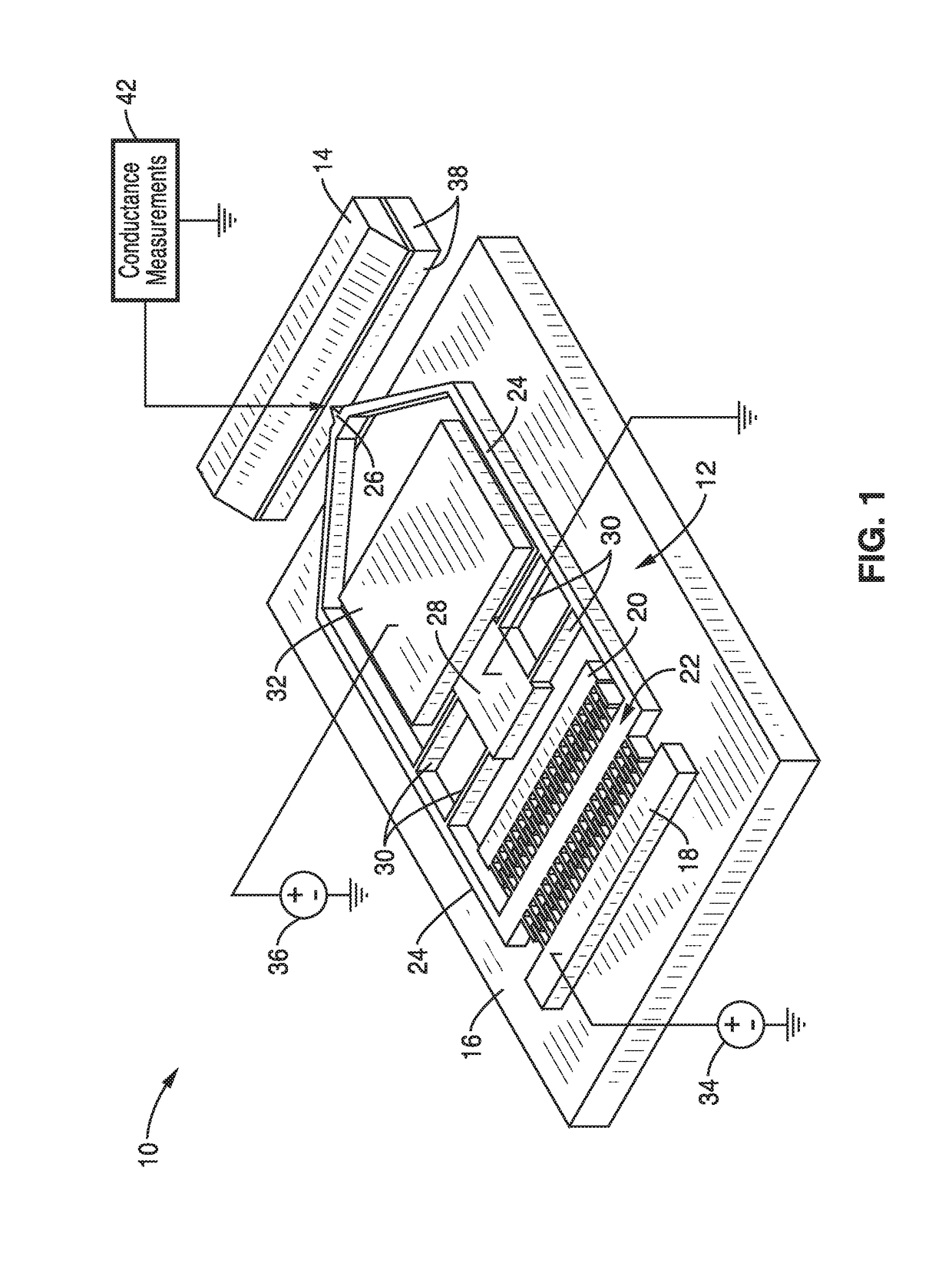
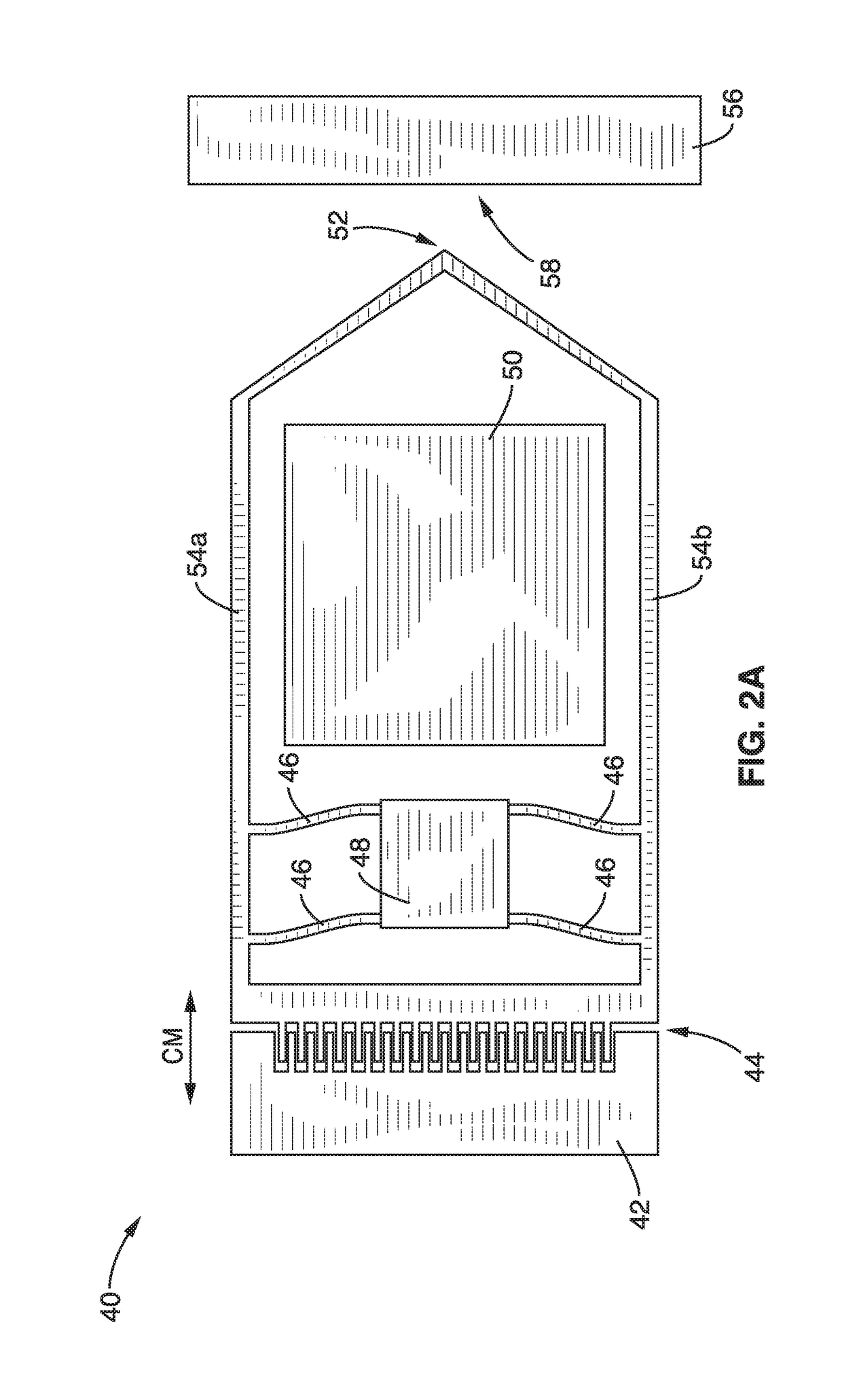
On-chip platform for single-molecule electrical conductance measurements
ActiveUS20170343531A1Easy to integrateAmenable to multiplexingMicrobiological testing/measurementMaterial analysisIn planeStatistical analysis
A micro-electromechanical platform and array system and methods for identifying microbial species with single molecule electrical conductance measurements are provided. The electromechanical platform has a two-tier actuation mechanism with a long stroke provided by a comb drive and a fine stroke provided by an in-plane flexural actuator. The platform is capable of making contact with a single-molecule, applying a bias, measuring the current, and performing a large number of measurements for statistical analysis. The system is capable of detecting any microbial species without requiring enzymatic amplification by detecting specific RNA sequences, for example. With oligonucleotide target molecules, the conductance is extremely sensitive to the sequence so even single-nucleotide polymorphisms can be identified. The system can also discern between subspecies using the same DNA probe. The system provides reliable, efficient, and inexpensive detection and species-level identification of microorganisms in complex detecting environments.
Owner:RGT UNIV OF CALIFORNIA
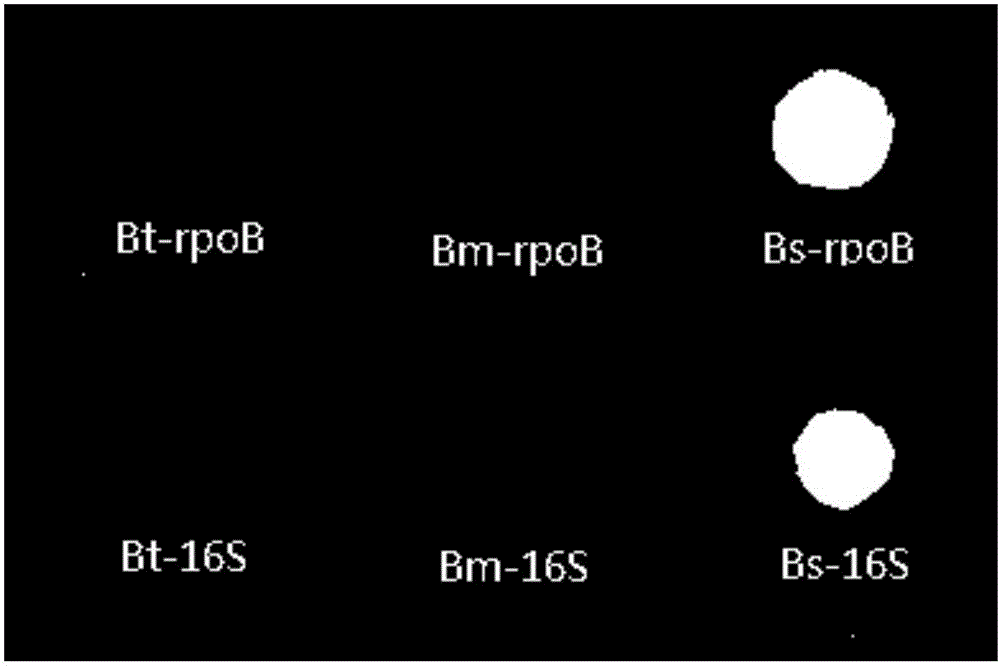
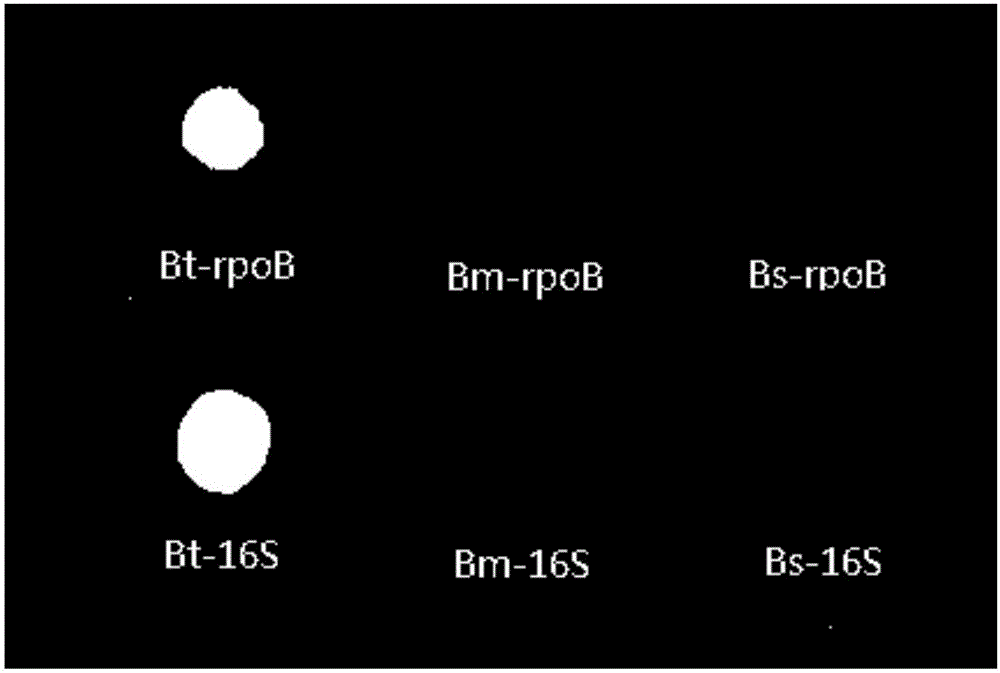
Gene chip for performing specific detection on bacillus subtilis
InactiveCN106191078ARealize classification and identificationReliable resultsMicrobiological testing/measurementFermentationFertilizerSpecies level
The invention discloses a gene chip for performing specific detection on bacillus subtilis, and belongs to soil microbe gene detection technology. According to a reported bacillus subtilis 16SDNA sequence and a reported rpoB gene sequence, a bacillus subtilis rpoB-specific gene fragment and a 16S-specific gene fragment are selected as detected target sequences, and corresponding multiple PCR amplification primer pairs, probes and detection chips are designed. Through the gene detection chip, the bacillus subtilis can be qualitatively and quantitatively detected within a short time, and classification and identification of the bacillus subtilis in the species level are achieved. The gene chip provides a powerful tool for colonization and propagation of the bacillus subtilis in fertilizer or soil and the like.
Owner:SHANXI UNIV
Method for evaluating temperature and humidity states of growing environment of nursery pig individual based on relative abundance of nasal cavity prokaryotic microorganisms
The invention discloses a method for evaluating temperature and humidity states of a growing environment of a nursery pig individual based on relative abundance of nasal cavity prokaryotic microorganisms. The method comprises the following steps: quantitatively detecting the relative abundance of the nasal cavity internal door, outline, order, family, genus and species level prokaryotic microorganisms of the nursery pig relative to the nasal cavity total prokaryotic microorganisms, and evaluating the temperature and humidity states of the growing environment of the nursery pig individual by using the relative abundance combination.
Owner:INST OF SUBTROPICAL AGRI CHINESE ACAD OF SCI
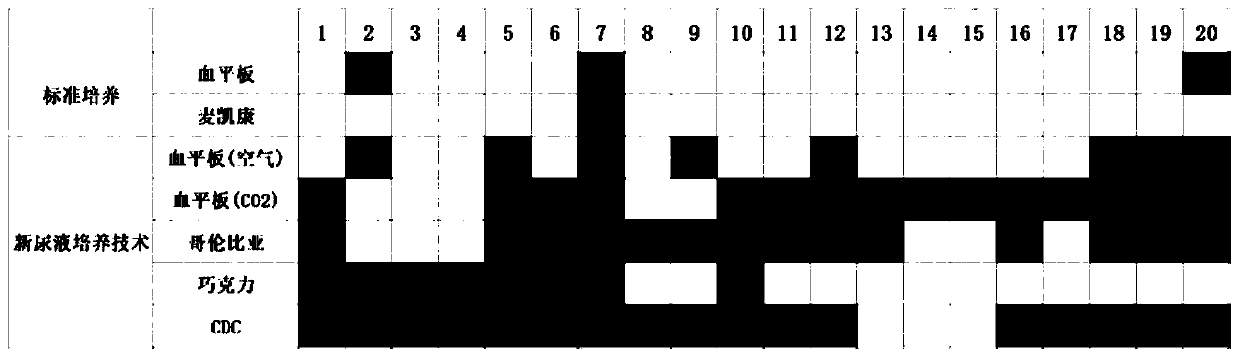
Novel detection method of urine bacteria
PendingCN110628867AExtend incubation timeLarge amount of specimenMicrobiological testing/measurementAnaerobic bacteriaCell culture media
The invention discloses a novel detection method of urine bacteria. The bacteria in urine are identified by increasing the amount of culture specimens, changing different growth media and gas conditions, and prolonging the culture time of bacteria. The method comprises the following steps: collection of urine; preservation and transportation; preliminary subculture; sub-subculture; and identification of bacteria. According to the present invention, the kind of bacteria in urine can be determined at the species level, and the further biochemical characteristics and metabolic characteristics ofthe cultured bacteria are analyzed and studied; and missing bacteria by standard culture can be identified by the method, anaerobic bacteria, aerobic bacteria and microaerophilic bacteria that survivein urine can be cultured and identified, and the method of the invention can more fully reflect the situation of all bacteria in urine compared with the standard culture. The time required by the method of the present invention is significantly shorter than the time of 16S rDNA sequencing, and the operation is simpler; the requirements for laboratory and experimental equipment are relatively low,which can be realized by most hospitals and laboratories; and the method has high operability and popularization.
Owner:NANFANG HOSPITAL OF SOUTHERN MEDICAL UNIV
Fluorescent PCR (polymerase chain reaction) quick detection primer and kit for pear fire blight pathogenic bacteria
InactiveCN103114137AGuaranteed reliabilityRealize integrated closure detectionMicrobiological testing/measurementFluorescence/phosphorescenceForward primerConserved sequence
The invention relates to a real-time fluorescent PCR (polymerase chain reaction) quick detection primer and kit for pear fire blight pathogenic bacteria. The primer comprises a forward primer of which the sequence is SEQ ID NO:1 and a reverse primer of which the sequence is SEQ ID NO:2. In the invention, a specific conserved sequence is obtained through molecular biological analysis according to an ITS gene sequence of pear fire blight pathogenic bacteria, and a specific amplification primer thereof is designed. The conserved gene sequence is shared by different pear fire blight pathogenic bacterium strains to ensure the reliability in detection of pear fire blight pathogenic bacteria from different sources on a species level. Besides, the primer provided by the invention adopts fluorescence labeling; and compared with the common PCR technology, observation does not need to be performed through a gel electrophoresis method, thus realizing the integrated sealed detection in the detection process.
Owner:郑媛
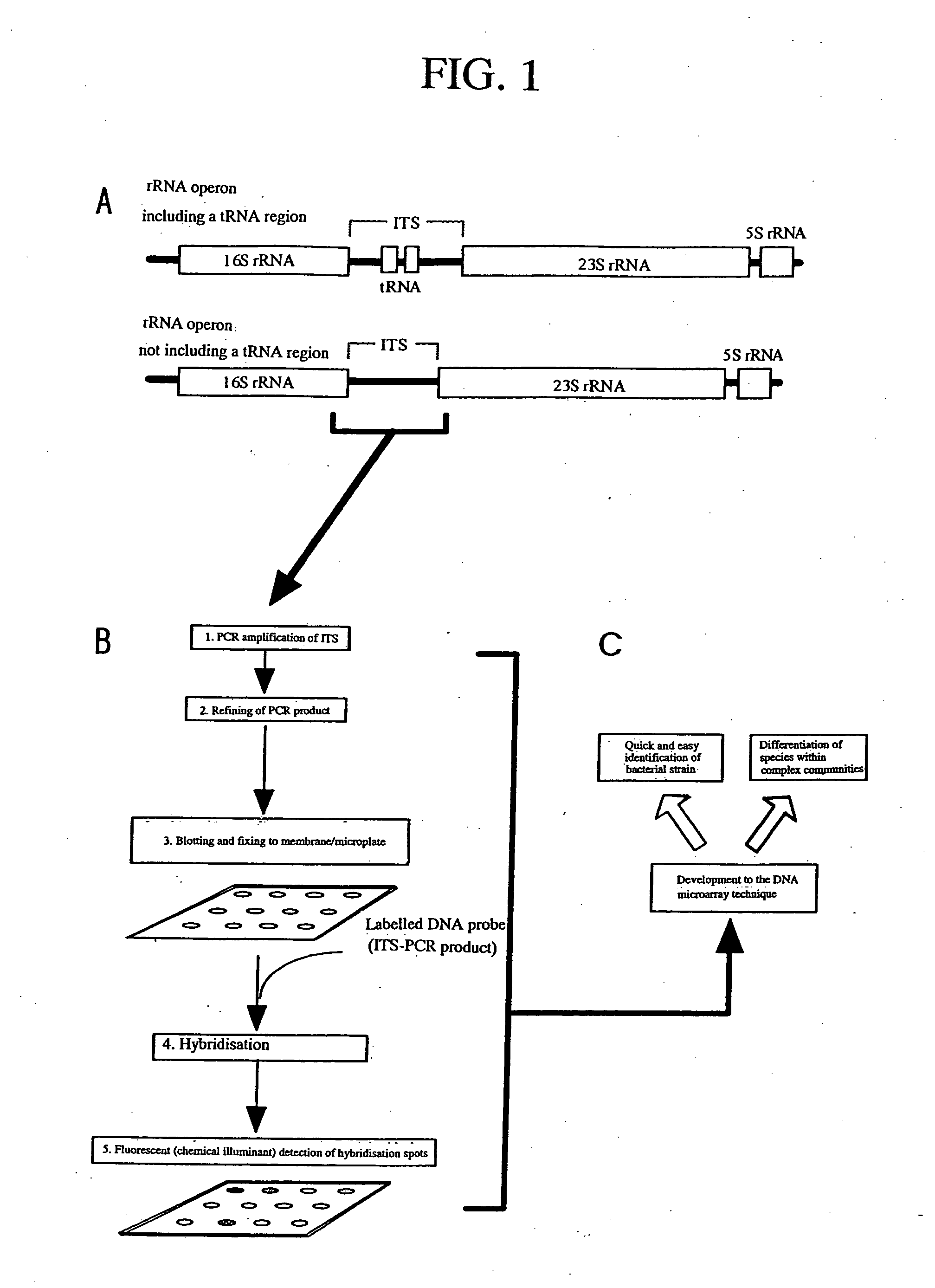
Molecular biological identification techniques for microorganisms
InactiveUS20050048524A1Easily and accuratelyHigh correlationMicrobiological testing/measurementRecombinant DNA-technologyDNA microarrayMicroorganism
By using an ITS-DNA hybridization technique, microorganisms being quickly differentiated and identified at the bacterial species level. By applying the principle to DNA microarray, environmental microorganisms are detected and identified. By using ITS-DNA hybridization and by applying the ITS-DNA hybridization technique to the DNA microarray, differentiation at the bacterial species level or the relative species level becomes possible.
Owner:HIRAISHI AKIRA +1
Gene chip for specific detection of bacillus thuringiensis
InactiveCN106367427ARealize classification and identificationReliable resultsMicrobiological testing/measurementTransferasesAureobasidium sp.Bacillus thuringiensis
The invention discloses a gene chip for specific detection of bacillus thuringiensis, and belongs to a soil microorganism gene detection technology. An rpoB unique gene fragment and a 16S unique gene fragment of the bacillus thuringiensis are selected as target sequences for detection according to a reported 16S DNA sequence and rpoB gene sequence of the bacillus thuringiensis, and corresponding multiple PCR amplification primer pairs, probes and detection chips are designed. Accordingly, qualitative and quantitative detection can be conducted on the bacillus thuringiensis in a short time through the gene detection chip, and classification and determination on the bacillus thuringiensis on the species level are achieved; a powerful tool is provided for tracking colonization and reproduction of the bacillus thuringiensis in fertilizer or soil.
Owner:SHANXI UNIV
Degraded wetland waterfowl diversity recovery method and system
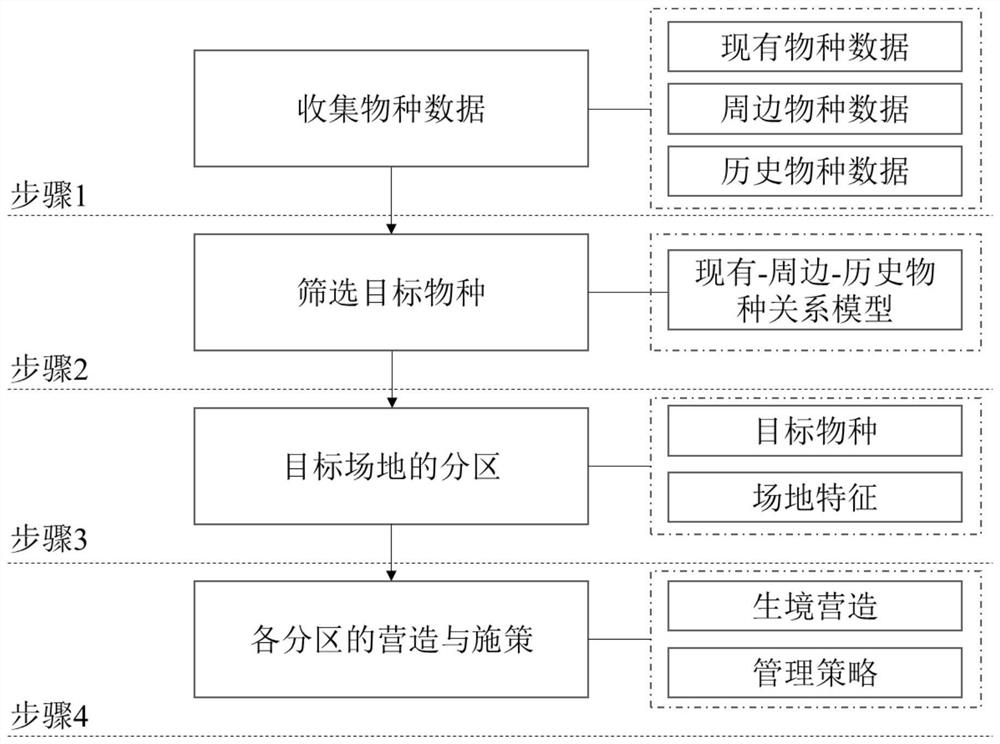
PendingCN111861831ADecision scienceSimple and fast operationData processing applicationsClimate change adaptationEngineeringEnvironmental engineering
The invention discloses a degraded wetland waterfowl diversity recovery method and system. The method comprises the following steps: step 1, collecting existing, peripheral and historical waterfowl species data of a degraded wetland; step 2, according to the three types of waterfowl data, establishing a relationship model, and screening out target species; step 4, according to the target species and the site characteristics, performing partitioning; and step 5, designing and creating each subarea, and applying a strategy in a centralized manner. According to the method, the waterfowl in degraded wetland is innovatively recovered to the original species level before degradation, and degradation of the wetland habitat is restrained, so that waterfowl diversity recovery engineering of the degraded wetland is carried out according to site characteristics on the basis of establishing a historical-surrounding-target species relationship model. The method is clear in target, scientific, simple and convenient, and plays an important role in improving decision-making and engineering efficiency in degraded wetland habitat restoration and waterfowl diversity restoration work.
Owner:INST OF ZOOLOGY GUANGDONG ACAD OF SCI +1
Application of madder polysaccharide in preparation of health-protection food with effects of preventing and treating neurodegenerative diseases
ActiveCN103564288AInhibit aggregationInhibition of aggregation toxicityFood ingredient functionsFood preparationDiseaseReactive oxygen radicals
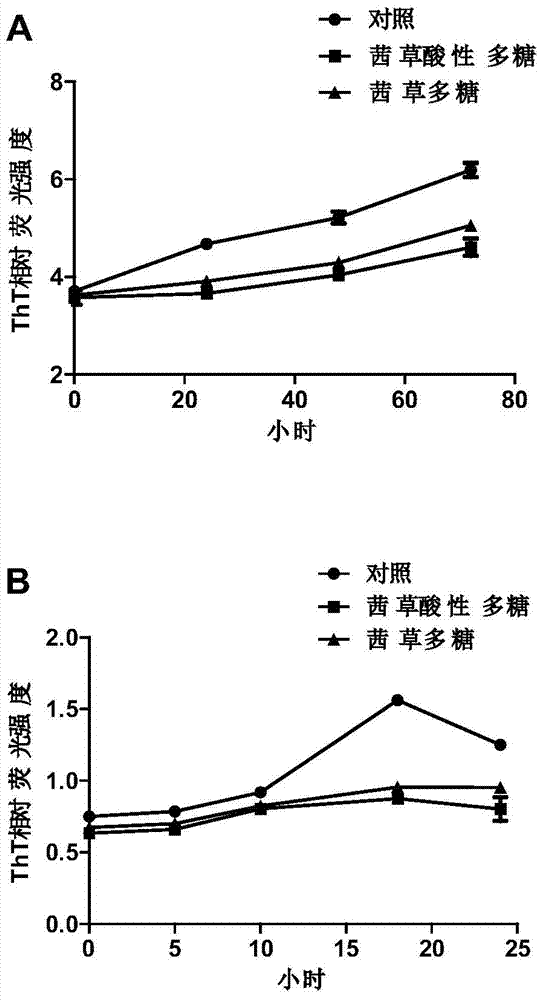
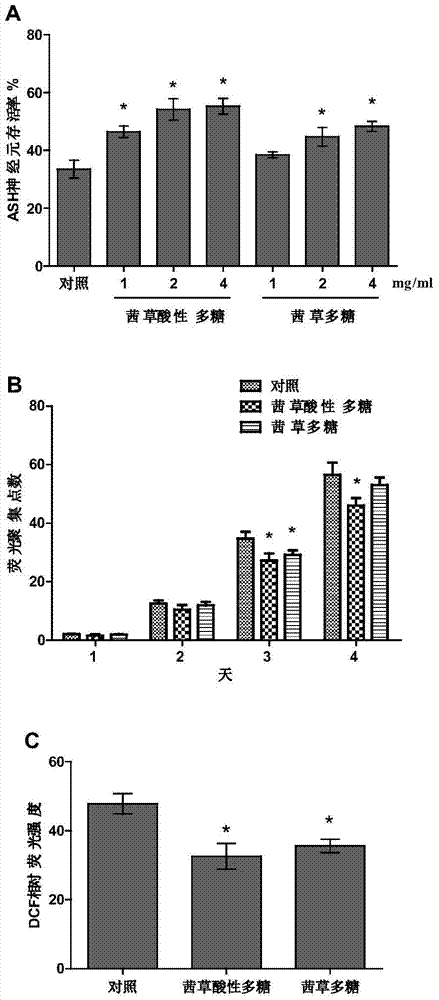
The invention discloses application of a madder polysaccharide in the preparation of a health-protection food with effects of preventing and treating neurodegenerative diseases. The madder polysaccharide can be a commercially available product or a product prepared by an applicant and is preferably a madder acidic polysaccharide. The invention also discloses a preparation method of the madder acidic polysaccharide. The madder acidic polysaccharide prepared by using the method disclosed by the invention has the characteristics of high polysaccharides content, low impurity content and high uniformity. Furthermore, the preparation method is simple and high in efficiency and is suitable for industrial production. Experiments on an in-vitro aggregation model, mammalian cells and a caenorhabditis elegans disease model show that the madder polysaccharide can effectively suppress the abnormal aggregation of polyglutamine and beta-amyloid proteins and the neurotoxicity of the polyglutamine and the beta-amyloid proteins and also obviously reduce the reactive oxide species level of the disease model, so that the madder polysaccharide has a good preventing and treating effect on the neurodegenerative diseases.
Owner:INFINITUS (CHINA) CO LTD
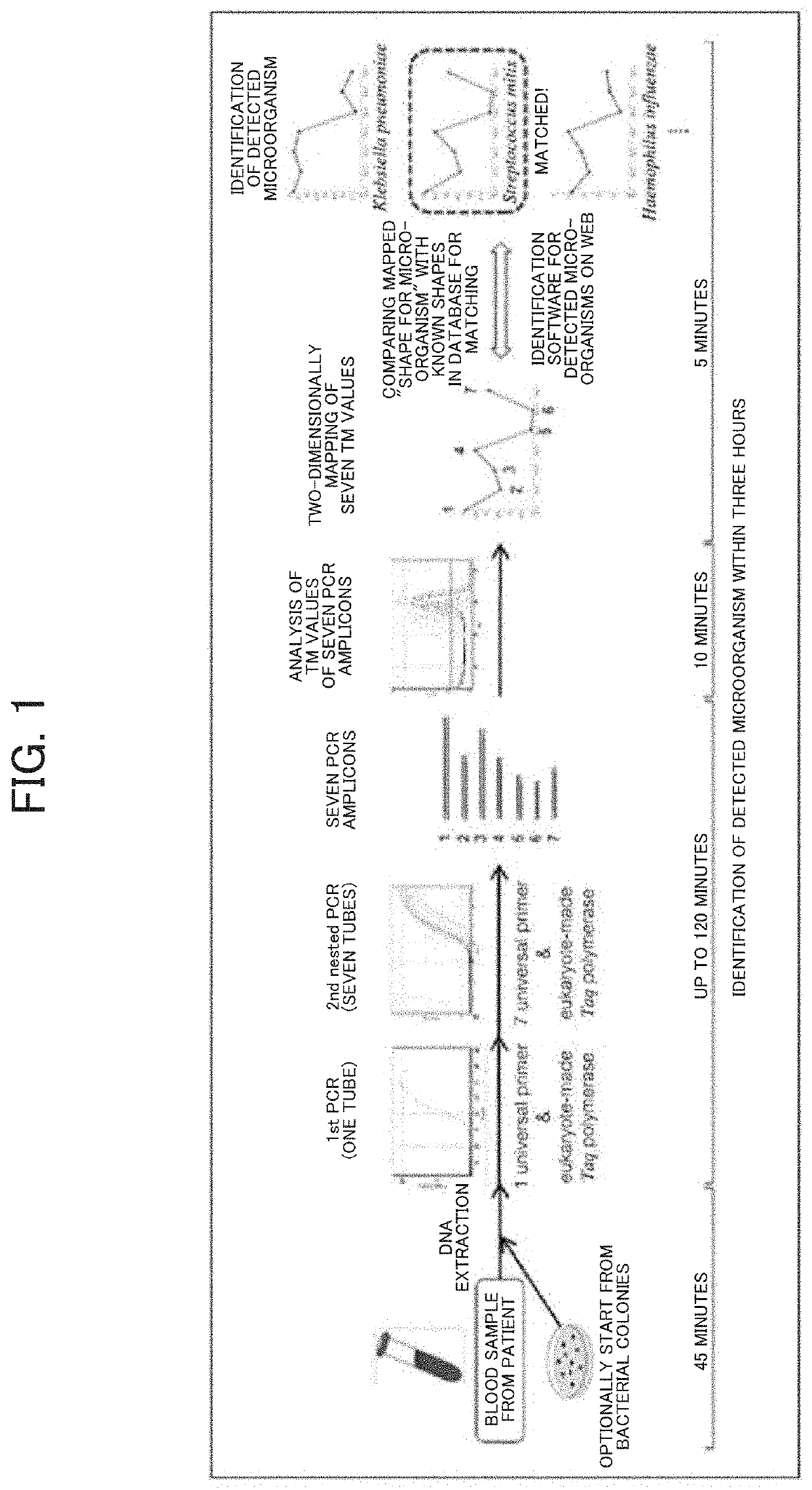
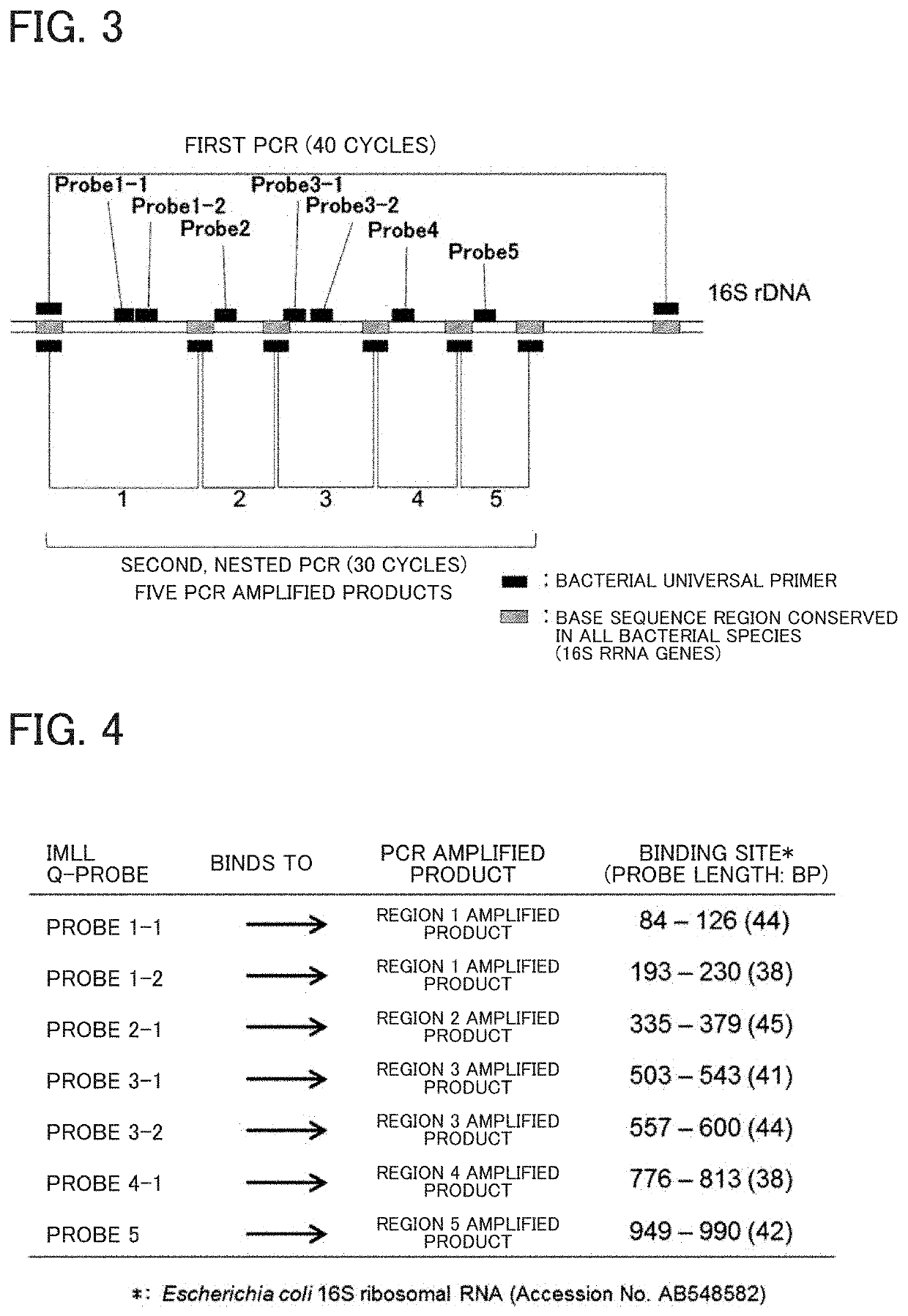
Tm mapping method
PendingUS20210032686A1Microbiological testing/measurementRecombinant DNA-technologyPathogenic microorganismMedicine
The improved Tm mapping method using imperfect-match linear long quenching probes can accurately distinguish among and identify microorganisms at least at the genus level and often at the species level even in a real-time PCR instrument having measurement errors of Tm values between PCR tubes within ±0.5° C. Therefore, the Tm mapping method can be performed in almost all real-time PCR instruments and can identify unspecified infection-causing pathogenic microorganisms in about 4 hours after sample collection.
Owner:UNIVERSITY OF TOYAMA
Water level detection and installation method of a cordless electric kettle
ActiveCN103968914BImprove securitySimple structureLevel indicators by physical variable measurementElectrical resistance and conductanceSpecies level
A water level detection and installation method for a cordless electric kettle, which is characterized in that the water level electrode wrapped by silica gel or other insulator materials is installed on the bottom or side of the kettle body, the NTC circuit and the water level electrode wire share a connection port of the connector, and the power supply is grounded The wire and the PCB water level detection incoming wire share a connection port of the connector, and the water level electrode wire connection port of the upper connector and the ground wire connection port are connected by resistance. An anti-dry device is added to the cordless electric kettle to provide a technical premise for automatic water addition.
Owner:陈宗仁
Interpretation method based on pathogen metagene next-generation sequencing
PendingCN110335656AEasy to operateLow heterogeneityMedical automated diagnosisSequence analysisSpecies levelColonization
The invention discloses an interpretation method based on pathogen metagene next-generation sequencing. A large number of detected sequences accord with at least one of the following sequences: coverage: the seed level being greater than 10 times of that of any other seed; the abundance being greater than 60%; sequence number: under the condition that the value of the total sequence number is normal, SMRN or SDMRN being greater than 10,000. A medium number of detected sequences accord with at least one of the following sequences: coverage: the species level being 4-10 times of that of any other species; abundance: 40%-60%; sequence number: under the condition that the value of the total sequence number is normal, the SMRN or SDMRN: 5,000-10,000. A small number or an extremely small numberof detected sequences accord with at least one of the following sequences: colonization and pathopoiesis cannot be distinguished; the bacteria are not common background bacteria of the specimens or donot accord with general characteristics of the background bacteria. The conclusion is more objective, the interpretation can be conducted at any time according to the standard, the method has the clinical practicability and objectivity, and the reported conclusion can be directly used for clinical practice.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV
Quick identification method of magnoliaceae plant seed
InactiveCN1231757CSeed and root treatmentMaterial analysis by optical meansMagnoliaceaeAgricultural science
The invention belongs to a method for identifying seeds, and mainly relates to a method for identifying seeds of magnolia plants. Magnoliaceae plants are mostly tall evergreen trees with beautiful tree shapes, which are excellent urban street trees and garden greening tree species, but the identification of Magnoliaceae seeds is a major difficulty encountered by garden workers when selecting species. The endocarp has a special shape in the chalazal area, which can be used to accurately and quickly determine whether it is a Magnoliaceae seed or not. And it can be identified to the level of genus or species, which has strong practicability in garden seed selection and seedling cultivation, and is also of great help to the collection and identification of field specimens. There is no relevant technology and method for the identification of Magnoliaceae seeds at home and abroad. The invention provides a rapid identification method for Magnoliaceae plant seeds, the genus or species can be identified by the method, and the method has great significance in practice.
Owner:SOUTH CHINA PLANT INST CHINESE ACAD OF SCI
A construction method and database of unknown plant species identification database based on matk gene
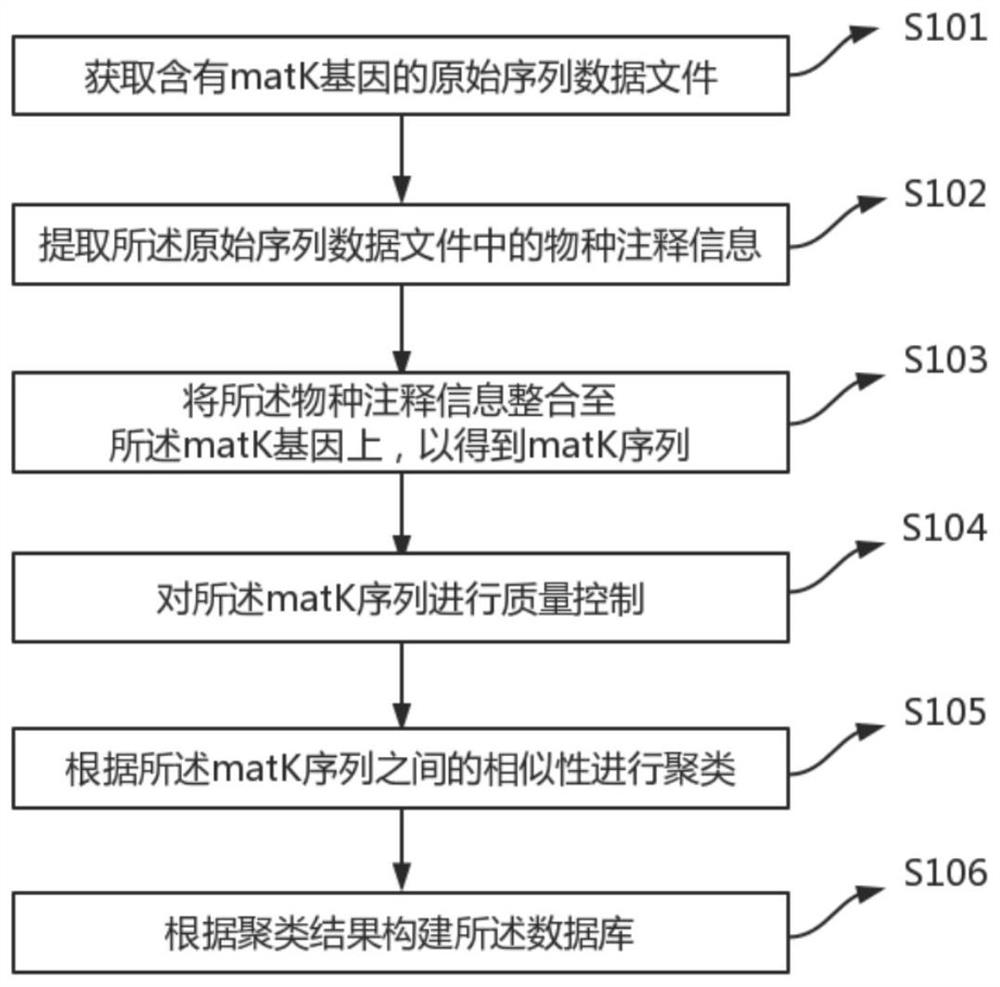
ActiveCN111681704BWide coverageImprove data qualityCharacter and pattern recognitionProteomicsData fileSpecies level
A construction method and database of an unknown plant species identification database based on the matk gene, the method comprising the steps of: obtaining an original sequence data file containing the matk gene; extracting species annotation information in the original sequence data file; The annotation information is integrated into the matK gene to obtain the matK sequence; quality control is performed on the matk sequence; clustering is performed according to the similarity between the matk sequences; and the database is constructed according to the clustering results. The construction method and database advantages of a kind of unknown plant species identification database based on matk gene provided by the application are: (1) high detection throughput; (2) high sensitivity and selectivity; (3) database coverage constructed by the method (4) Bioinformatics methods can be used to identify unknown species at the species level.
Owner:EZHOU INST OF IND TECH HUAZHONG UNIV OF SCI & TECH +1
A kind of m65 resistance welding sleeve and its manufacturing method
ActiveCN106319359BGood welding performanceImprove impact toughnessElectric resistance weldingContinuous rolling
The invention discloses an M65-grade electric resistance welding casing pipe and a manufacturing method thereof. The M65-grade electric resistance welding casing pipe comprises, by weight percentage, 0.14-0.18% of C, 0.15-0.30% of Si, 1.10-1.40% of Mn, less than or equal to 0.020% of P, less than or equal to 0.008% of S, 0.06-0.12% of Ti, 0.02-0.05% of Als, less than or equal to 0.008% of N, less than or equal to 0.40% of carbon equivalents and the balance Fe and unavoidable elements. A continuous casting sheet billet is heated to 1150-1200 DEG C through a heating furnace and subjected to hot continuous rolling. The finishing rolling temperature is 780-840 DEG C. After being rolled, a steel belt is cooled at the speed of 8-12 DEG C / s. The steel belt is coiled at the temperature of 560-620 DEG C. The steel belt is subjected to high frequency / medium frequency electric resistance welding and made into a steel pipe through an ERW unit. The whole pipe is subjected to the normalizing heat treatment process, wherein the whole pipe is heated to 880-910 DEG C, the temperature is preserved for 30-50 minutes, and air cooling is conducted. The steel belt is good in weldability and impact toughness; normalizing treatment is adopted, so that banded structures and residual stress are eliminated; the heat treatment process is simplified; and performance consistency of a pipe body and weld joints is guaranteed.
Owner:ANGANG STEEL CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com