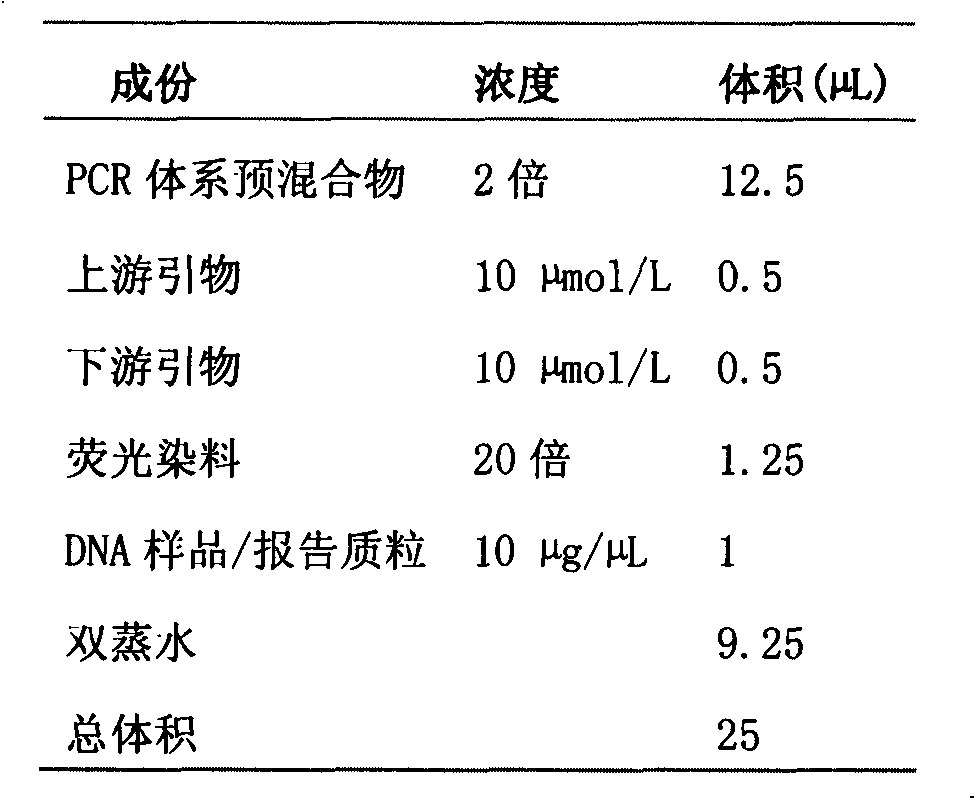
Fluorescent PCR (polymerase chain reaction) quick detection primer and kit for pear fire blight pathogenic bacteria
A technology for detection of Phytophthora amylovora and detection primers, which is applied in the directions of fluorescence/phosphorescence, measurement/inspection of microorganisms, biochemical equipment and methods, etc., and can solve the problem of inability to detect all strains of bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1: Effect detection of the present invention
[0035] The specific primers of the present invention detect the standard bacterial strain of E. amylovora by reporting fluorescent signals through fluorescent chimeric PCR. During the amplification process, a fluorescent PCR instrument is used to measure the real-time fluorescent light intensity, and the synthetic DNA fragment is measured after the PCR reaction. Melting temperature, transfer the data to the computer for analysis through the supporting software of the fluorescent PCR instrument, the fluorescence generated by the specific amplification of E. amylovora can be observed, and the specific fragments generated by the amplification of E. amylovora can be obtained. The held melting curve (main peak 84 ± 0.5). The results show that the primers and the kit of the invention can accurately detect the positive samples carrying E. amylovora without false negatives.
Embodiment 2
[0036] Example 2: Negative Control
[0037] With the primers and kit of the present invention, according to the detection steps of the present invention, the bacterial wilt of corn (Erwinia stewartii), soft rot of cabbage (Erwinia carotovora subsp.carotovora), bacterial wilt of cassava (Xanthomonas axonopodis pv.manihotis) , Clavibacter michiganensis subsp.michiganensis, and Pseudomonas syringae pv.pisi and other five negative control strains were tested for fluorescence, and a fluorescent PCR instrument was used for real-time fluorescence light intensity measurement during the amplification process , and measure the melting temperature of the synthesized DNA fragments after the PCR reaction finishes, the data is transmitted to the computer and analyzed by the supporting software of the fluorescent PCR instrument, and no similar melting curve (main peak 84 ± 0.5), and the fluorescent signal of the positive control reporter plasmid and the resulting melting curve can be observe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com