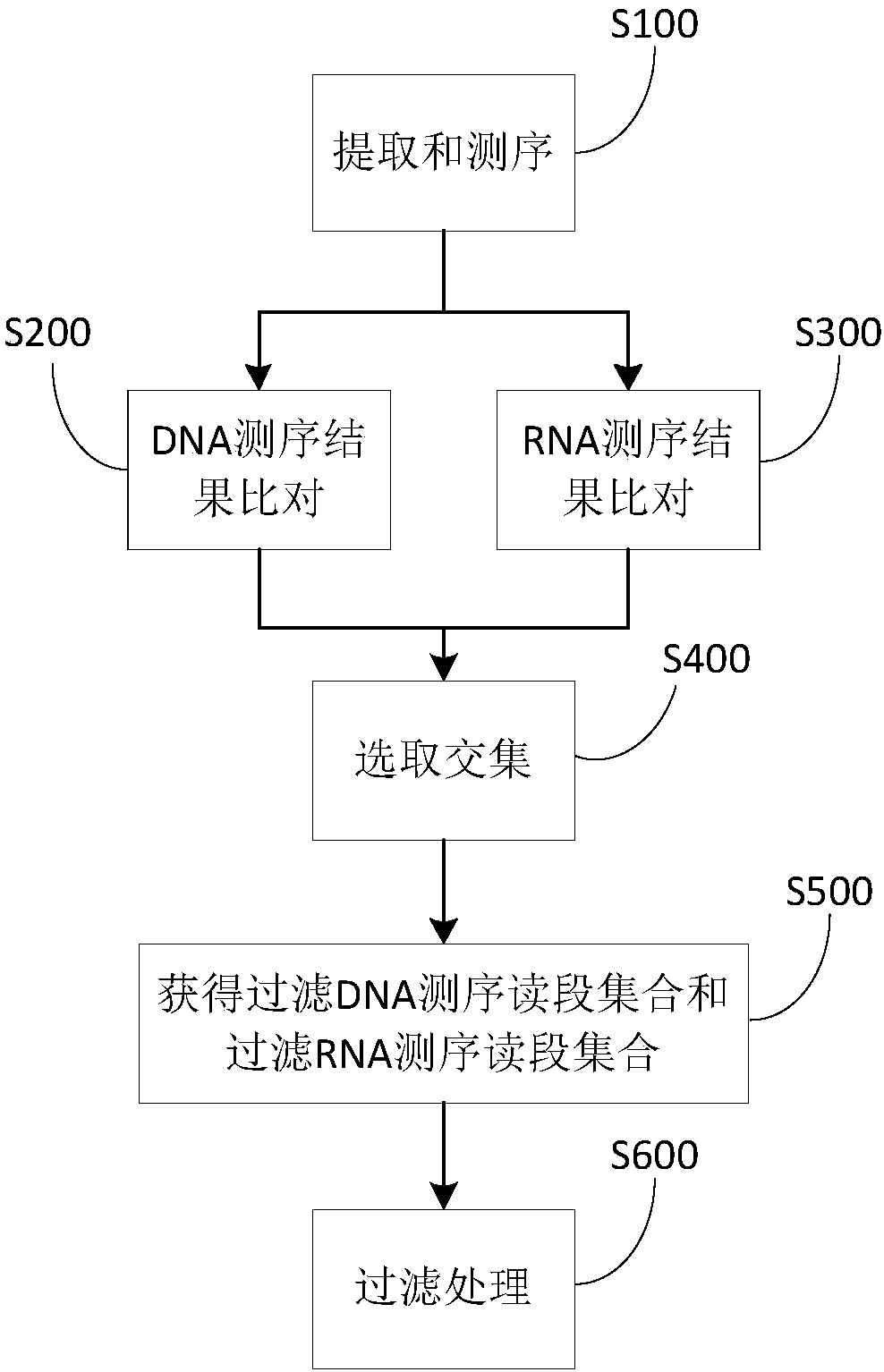
Method and device to identify microbial species from sample
A technology in microorganisms and samples, applied in enzymology/microbiology devices, biochemical cleaning devices, biochemical equipment and methods, etc., can solve the complex and changeable microorganisms, the interference of pathogenic microorganism infection treatment, and the method and device of microbial species Issues to be studied to achieve the effect of avoiding interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0110] In this step, the microorganisms in the sputum sample are identified as follows:
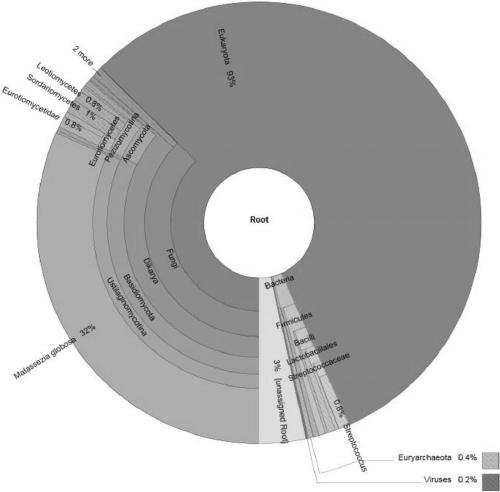
[0111] The amount of DNA sequencing data obtained by the above-mentioned general method of the sputum sample is about 47M, and the data amount of RNA sequencing is about 47M. After removing low-quality, ribosomal sequences, human sequences, and low-complexity sequences, the DNA sequencing results consisting of 9287 reads (such as image 3 ) and RNA sequencing results consisting of 12713 reads (such as Figure 4 ). There are 126 types of microorganisms (including bacteria, archaea, viruses, and eukaryotes) shared by DNA and RNA, and 43 types of microorganisms are filtered according to the number of reads (set to 1). Then divide the RNA and DNA data into sets of 1M reads for comparison. Filter the comparison results. The filtering standard for bacteria, archaea and viruses is that under the same amount of data, the ratio of the number of RNA and DNA reads of the same species is greater t...
Embodiment 2
[0113] In this step, microorganisms in CSF samples are identified as follows:
[0114] According to the general method above, the amount of DNA sequencing data of the cerebrospinal fluid sample is about 5M, and the data amount of RNA sequencing is about 4M. After removing low-quality, ribosomal sequences, human sequences, and low-complexity sequences, a DNA sequencing result consisting of 133,265 reads (such as Figure 6 ) and RNA sequencing results consisting of 974328 reads (such as Figure 7 ). There are 44 types of microorganisms (including bacteria, archaea, viruses, and eukaryotes) shared by DNA and RNA, and 11 types of microorganisms are filtered according to the number of reads (set to 1). Then divide the RNA and DNA data into sets of 1M reads for comparison. Filter the comparison results. The filtering standard for bacteria, archaea and viruses is that under the same amount of data, the ratio of the number of RNA and DNA reads of the same species is greater than or...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com