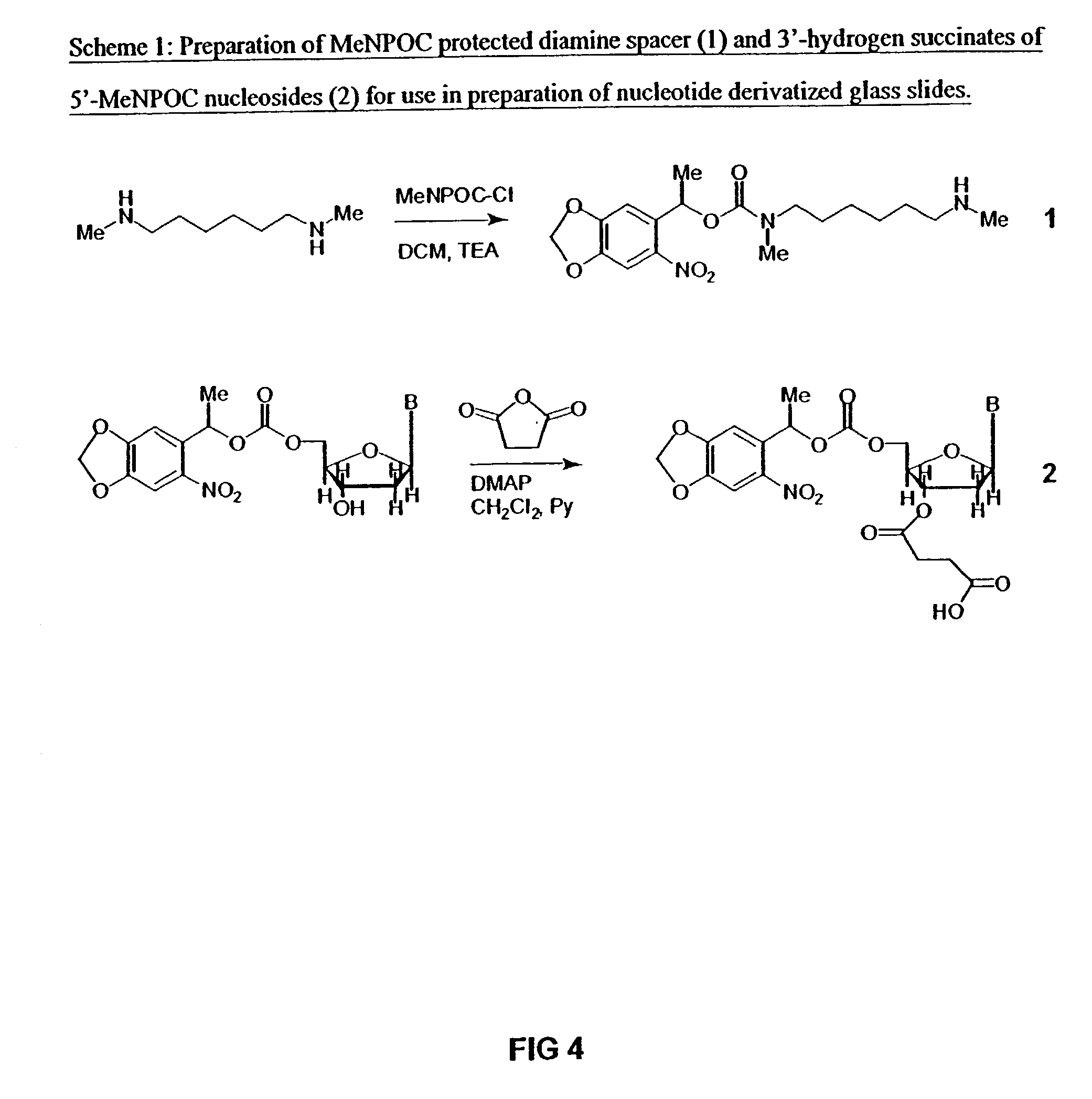
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
137 results about "DNA microarray" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
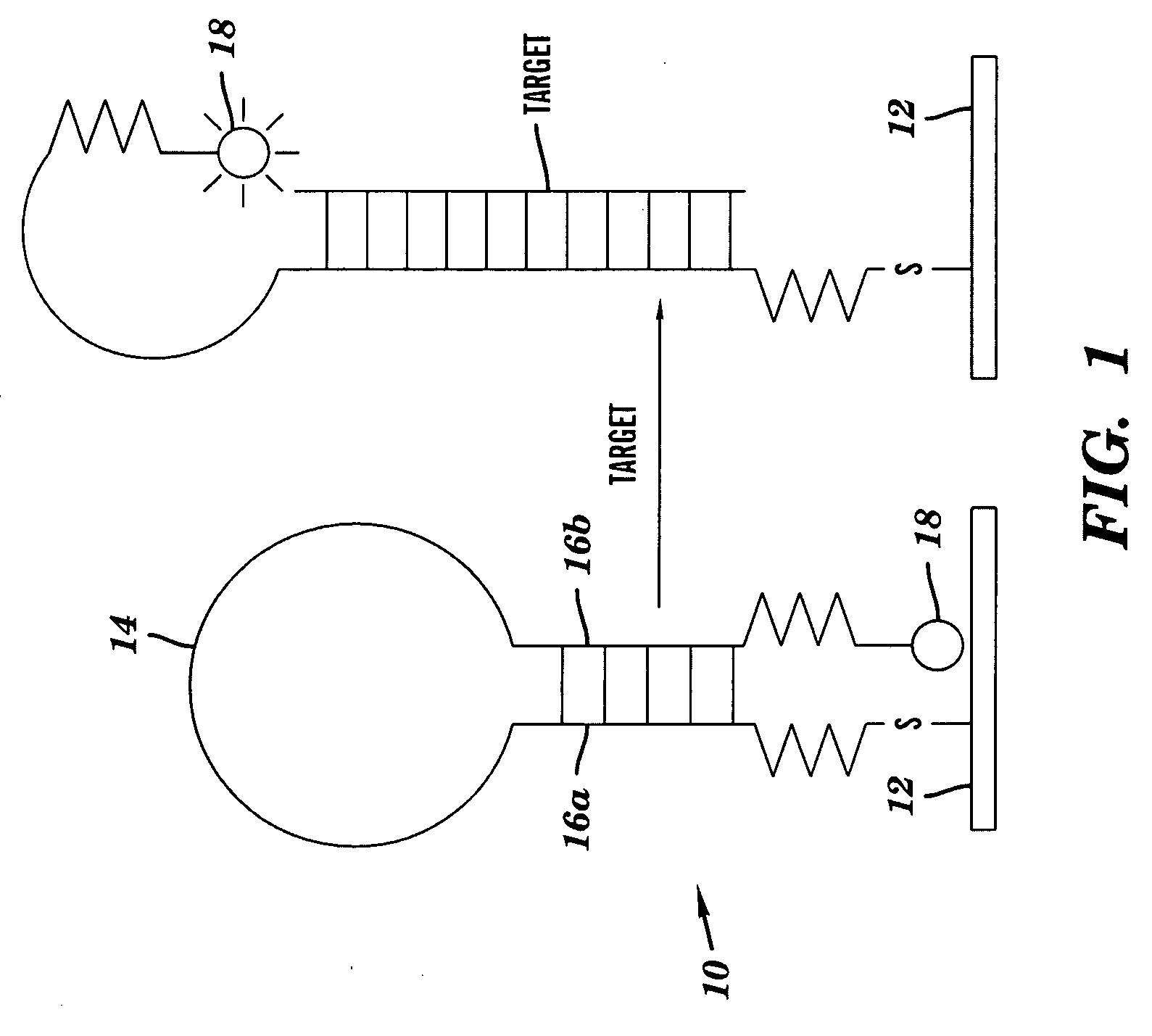
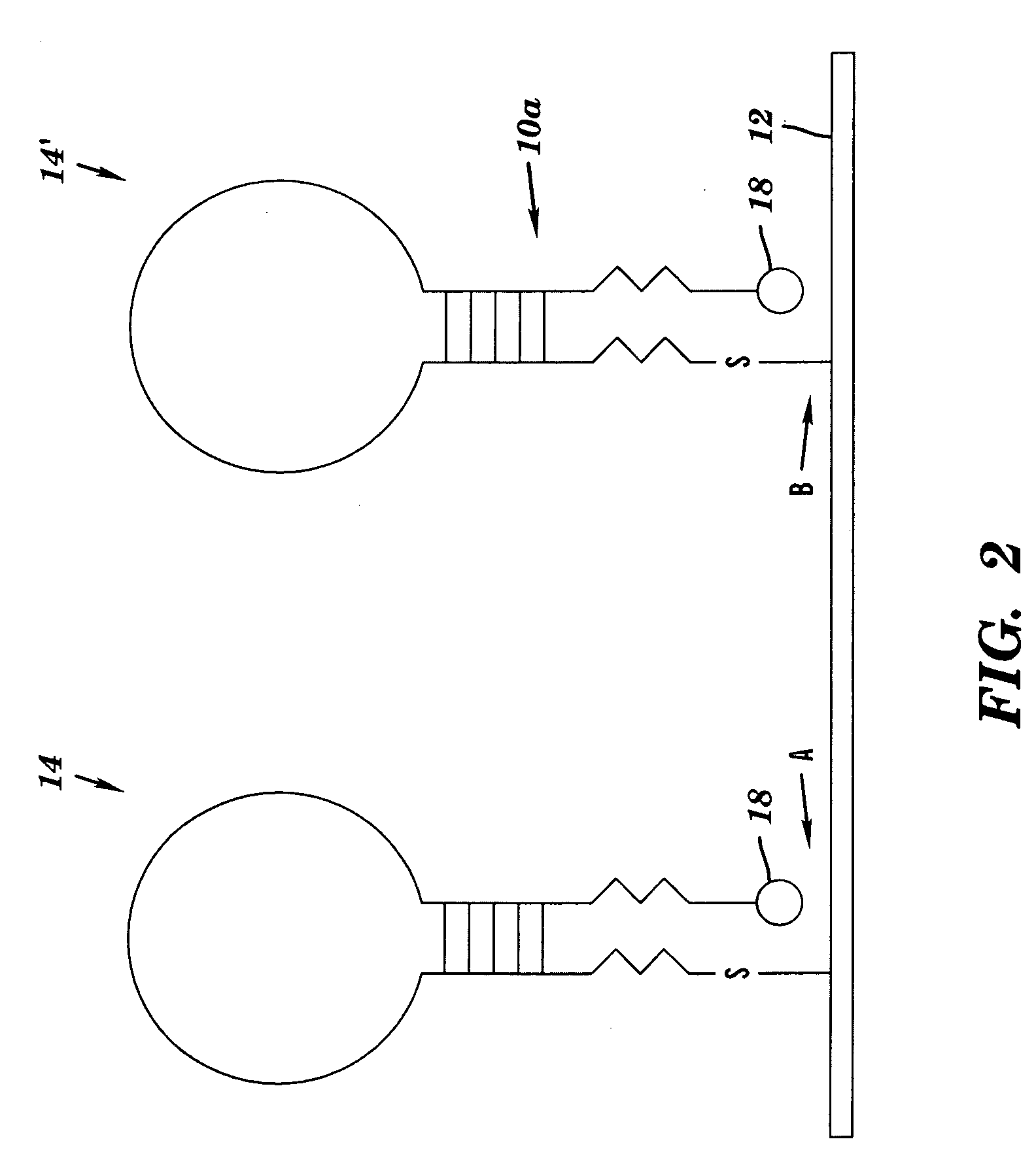
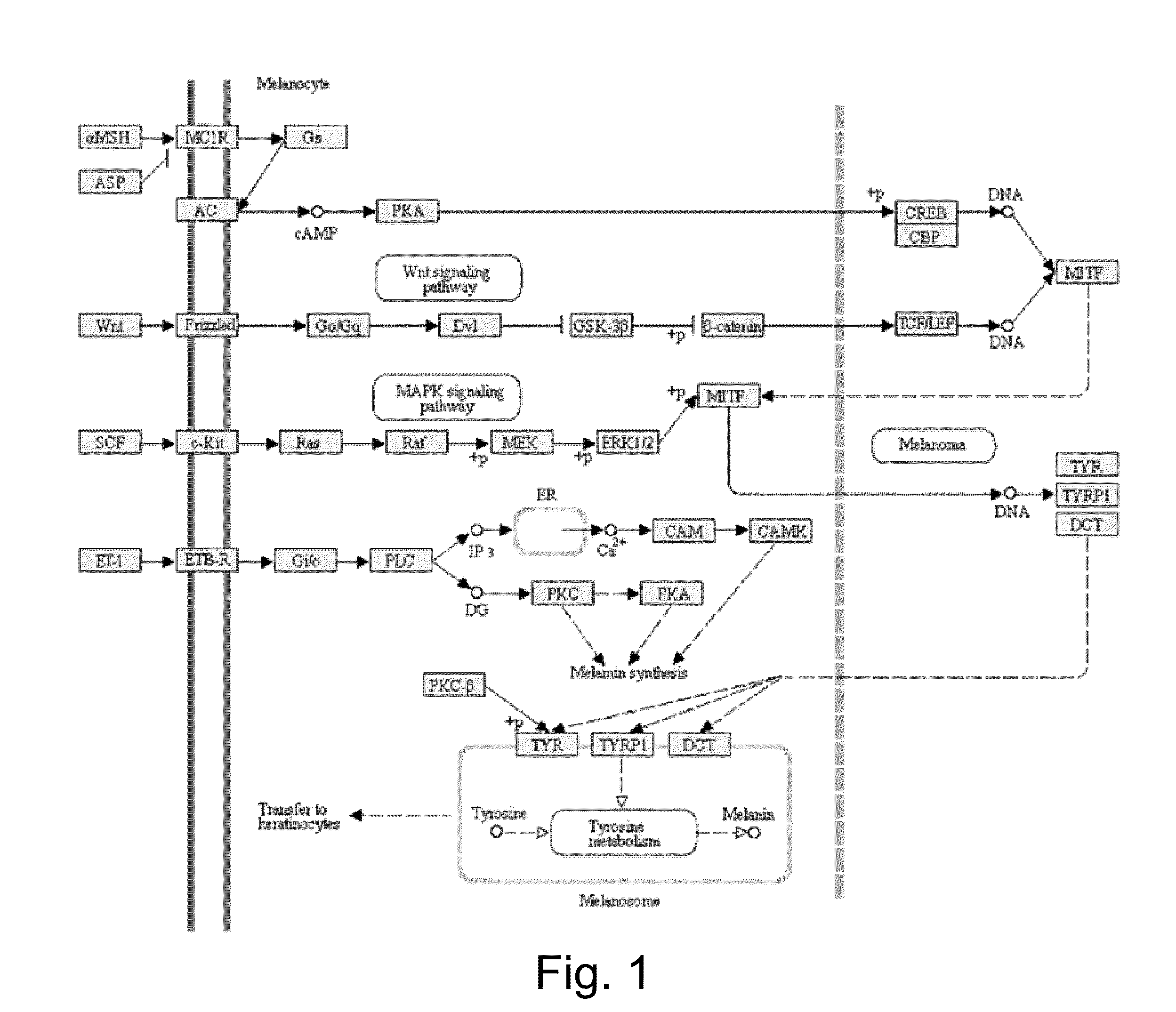
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10⁻¹² moles) of a specific DNA sequence, known as probes (or reporters or oligos). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called target) under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown.
Individualized cancer treatments
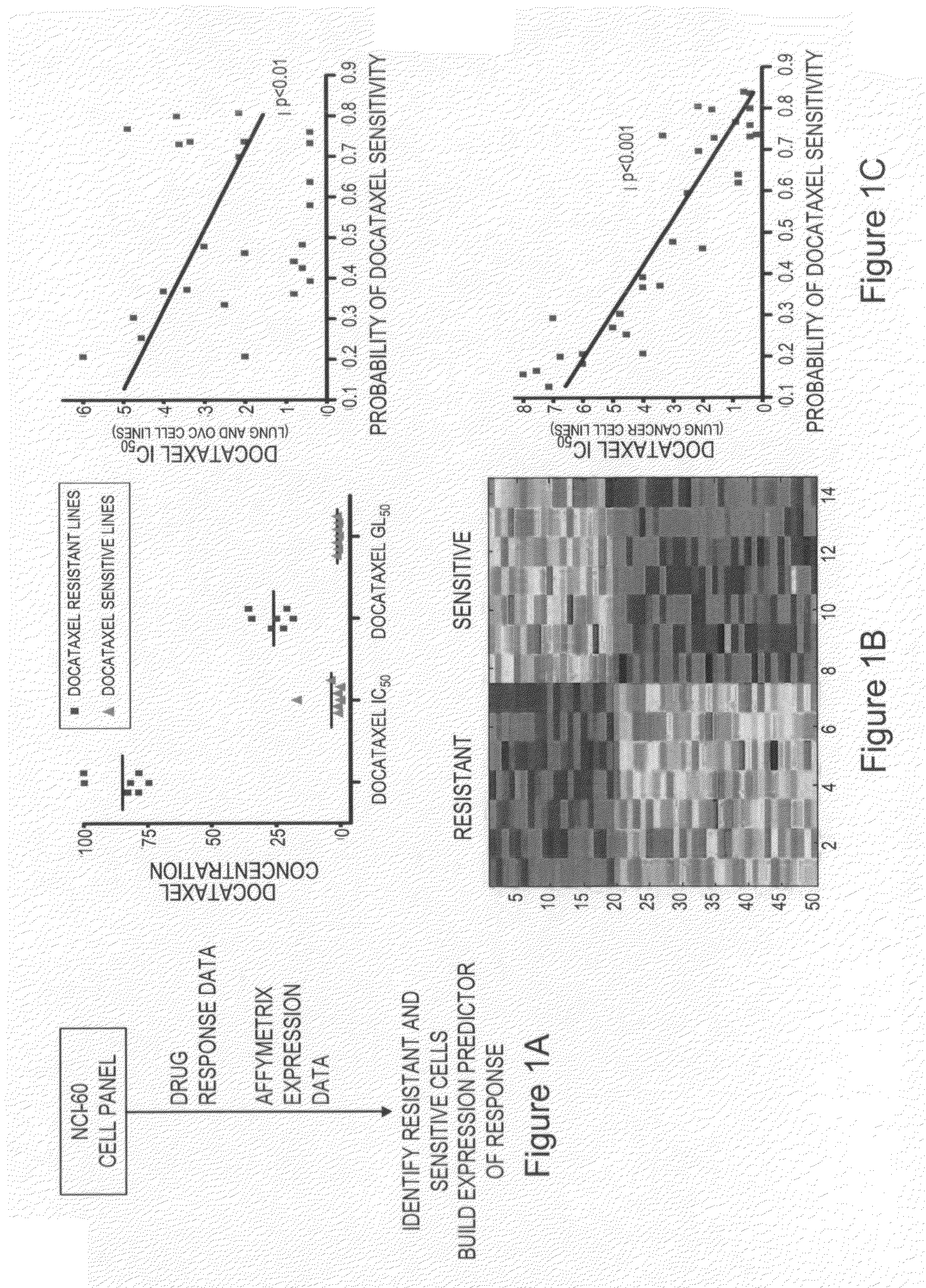
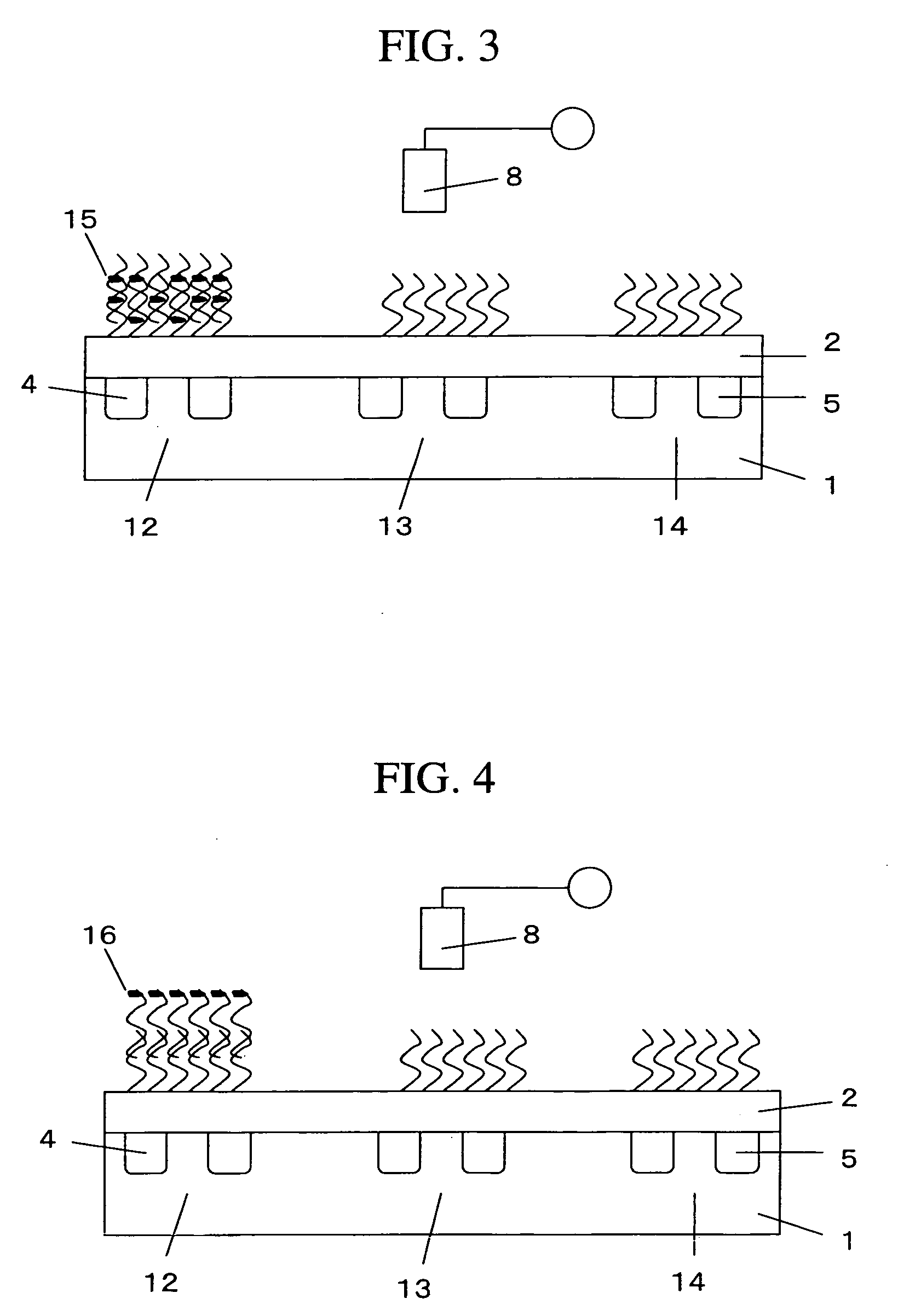
InactiveUS20070172844A1Increase probabilityLow toxicityBioreactor/fermenter combinationsBiological substance pretreatmentsDNA microarrayPlatinum
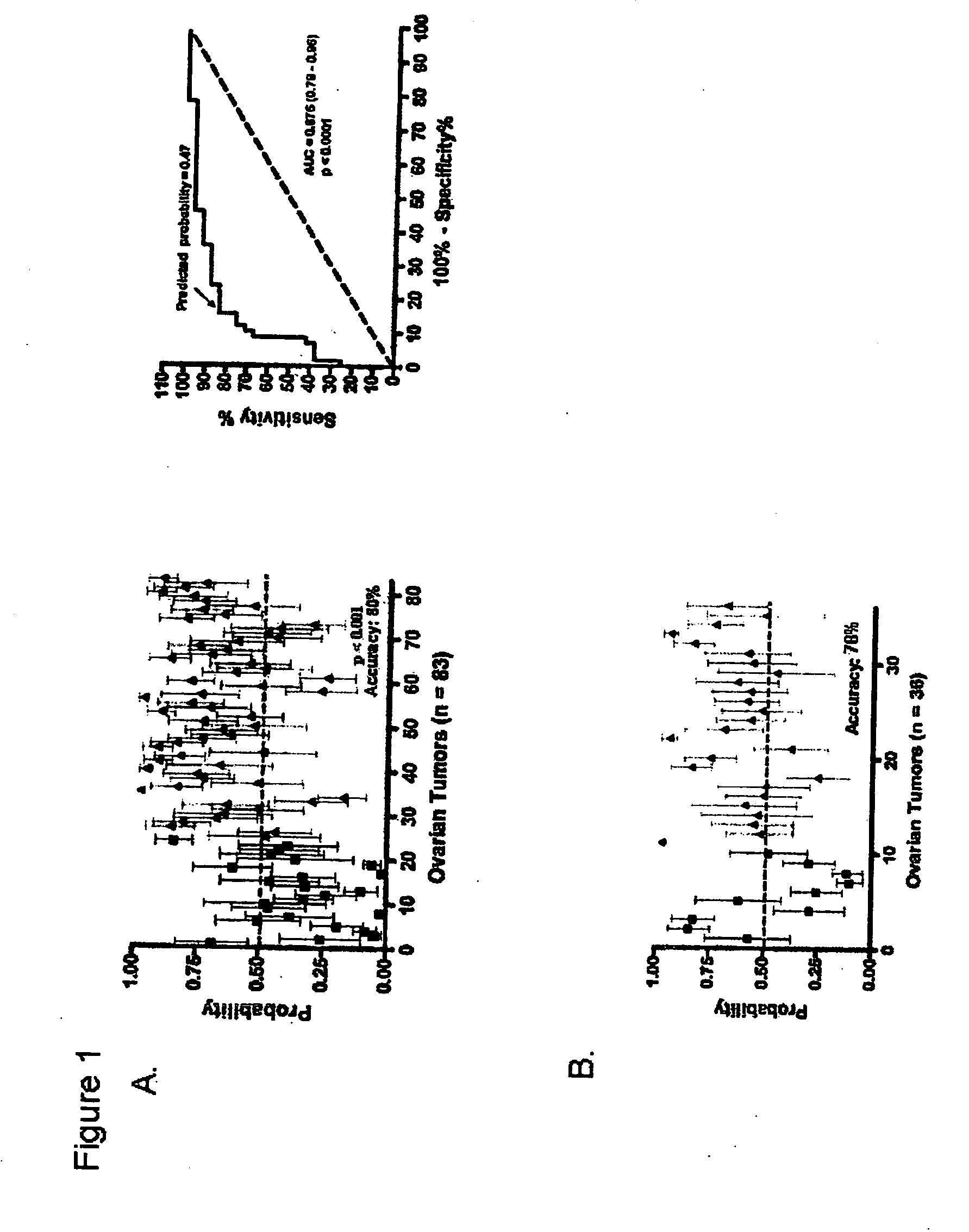
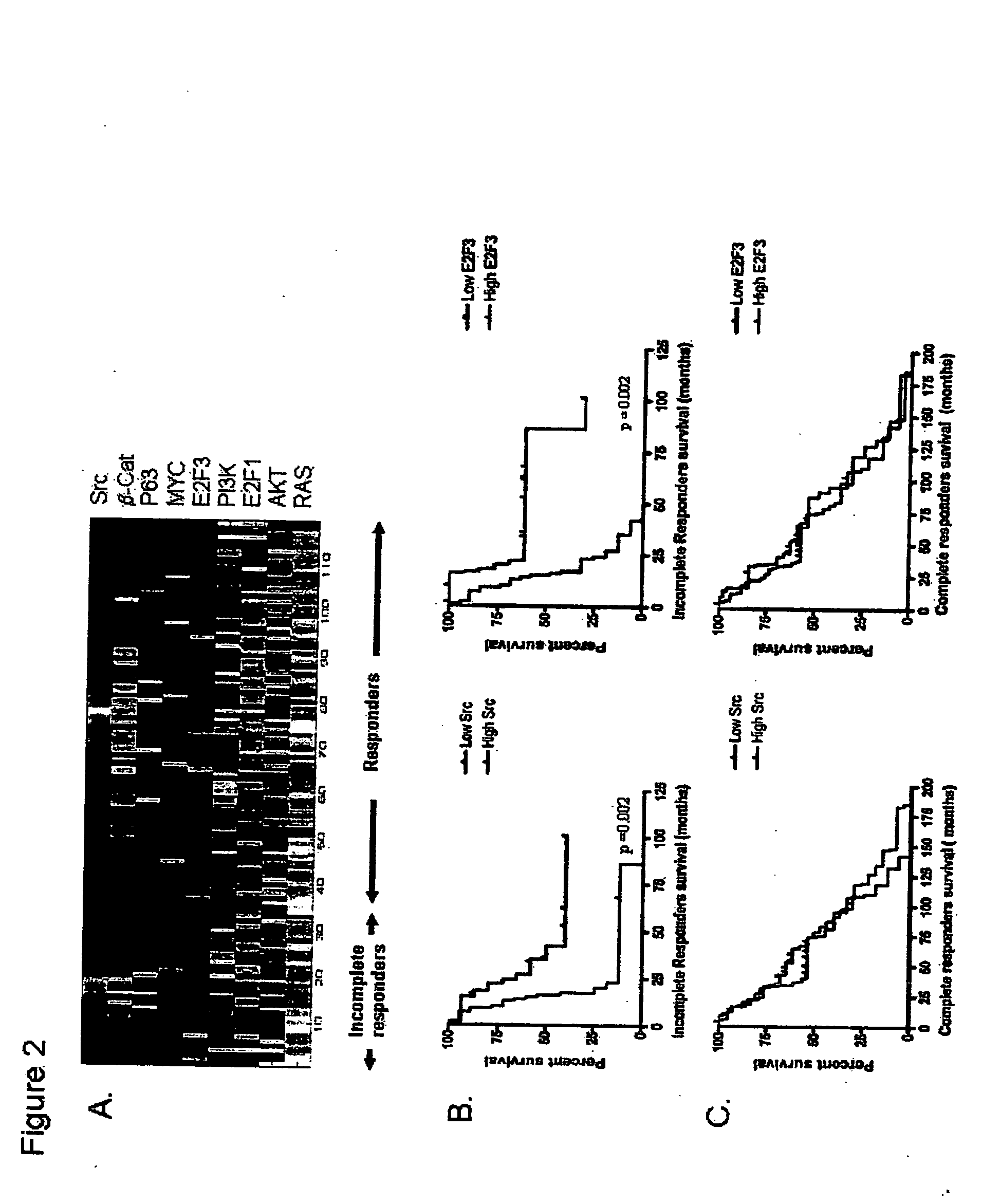
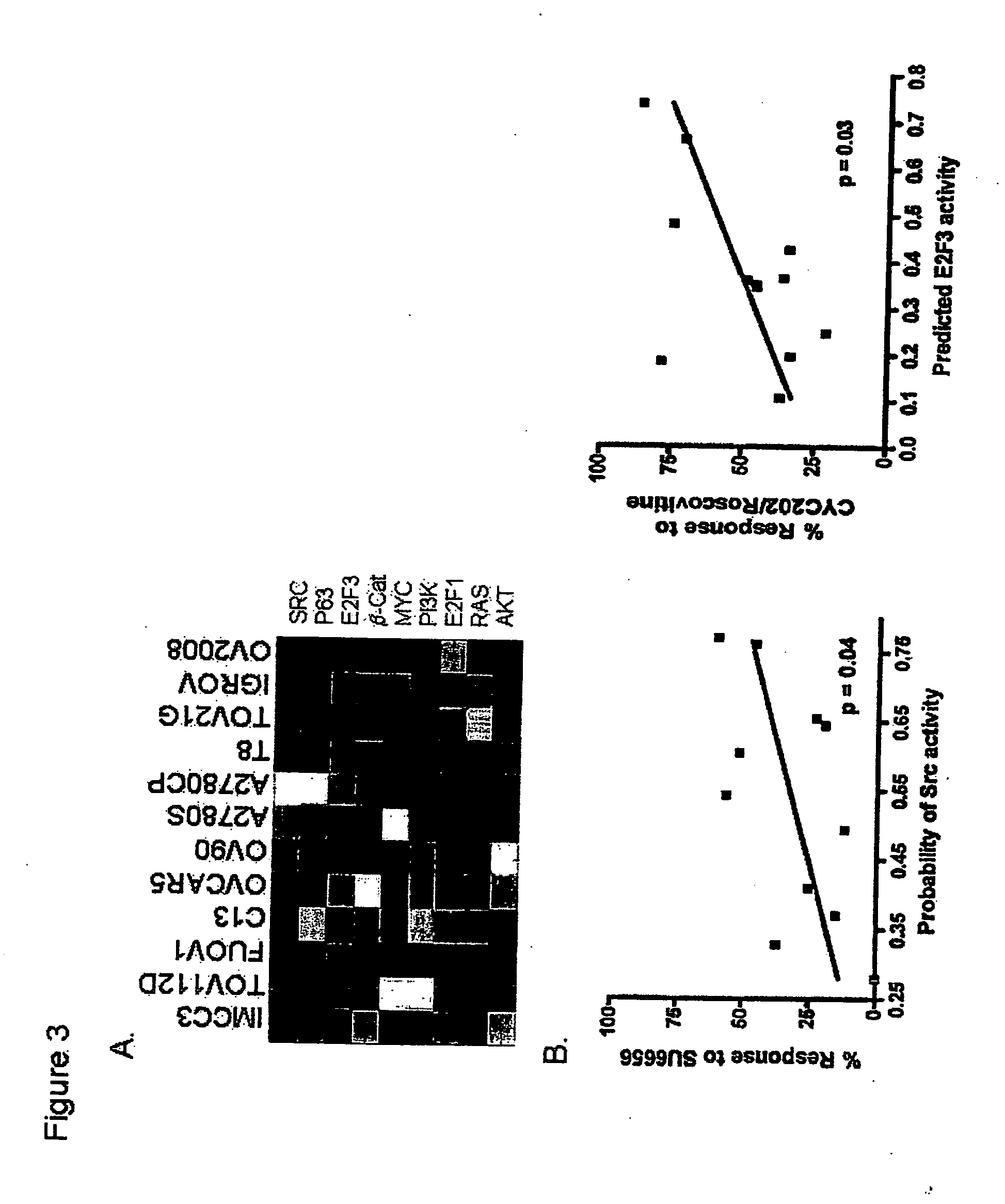
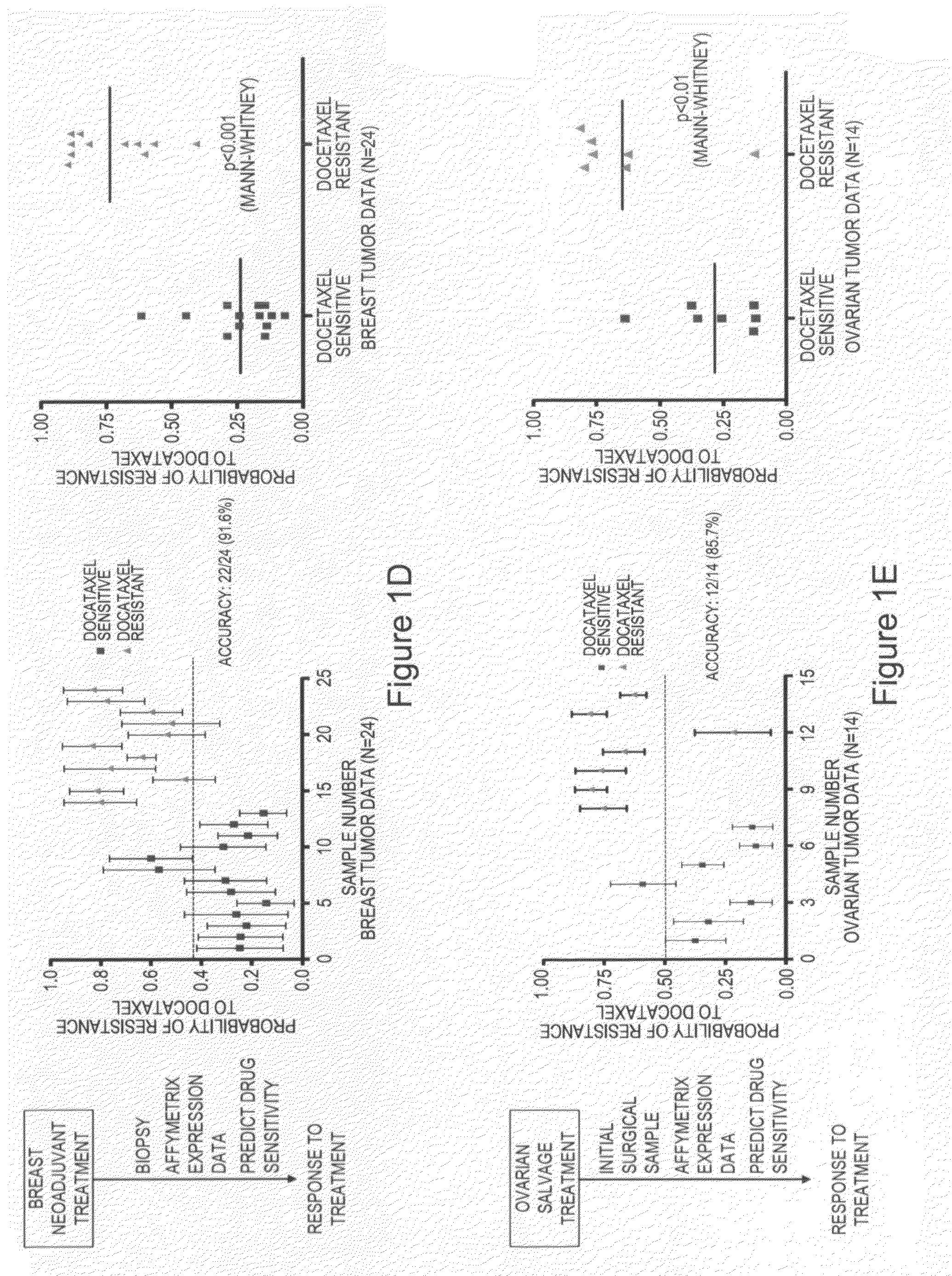
The invention provides for compositions and methods for predicting an individual's responsitivity to cancer treatments and methods of treating cancer. In certain embodiments, the invention provides compositions and methods for predicting an individual's responsitivity to chemotherapeutics, including platinum-based chemotherapeutics, to treat cancers such as ovarian cancer. Furthermore, the invention provides for compositions and methods for predicting an individual's responsivity to salvage therapeutic agents. By predicting if an individual will or will not respond to platinum-based chemotherapeutics, a physician can reduce side effects and toxicity by administering a particular additional salvage therapeutic agent. This type of personalized medical treatment for ovarian cancer allows for more efficient treatment of individuals suffering from ovarian cancer. The invention also provides reagents, such as DNA microarrays, software and computer systems useful for personalizing cancer treatments, and provides methods of conducting a diagnostic business for personalizing cancer treatments.
Owner:UNIV OF SOUTH FLORIDA +1
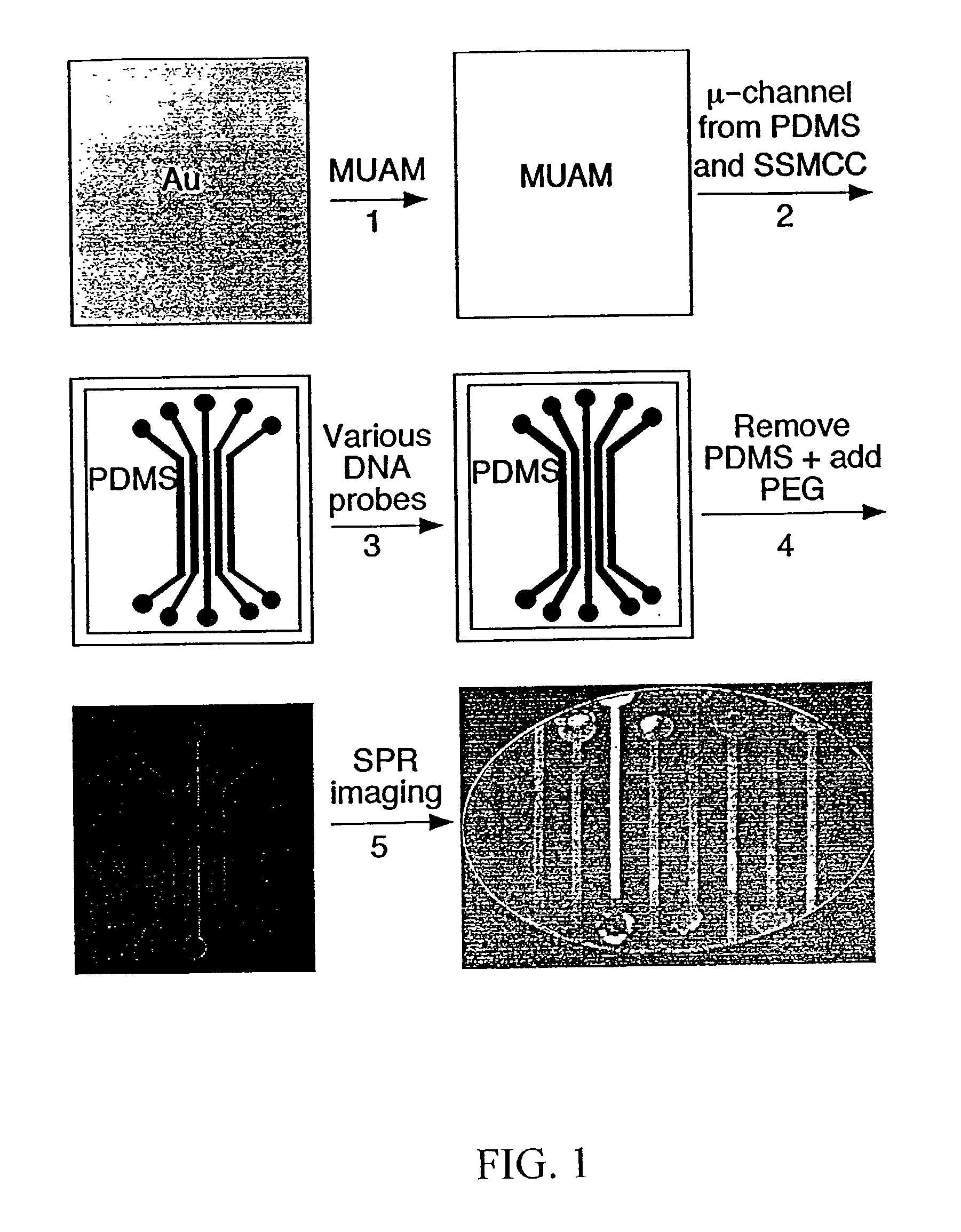
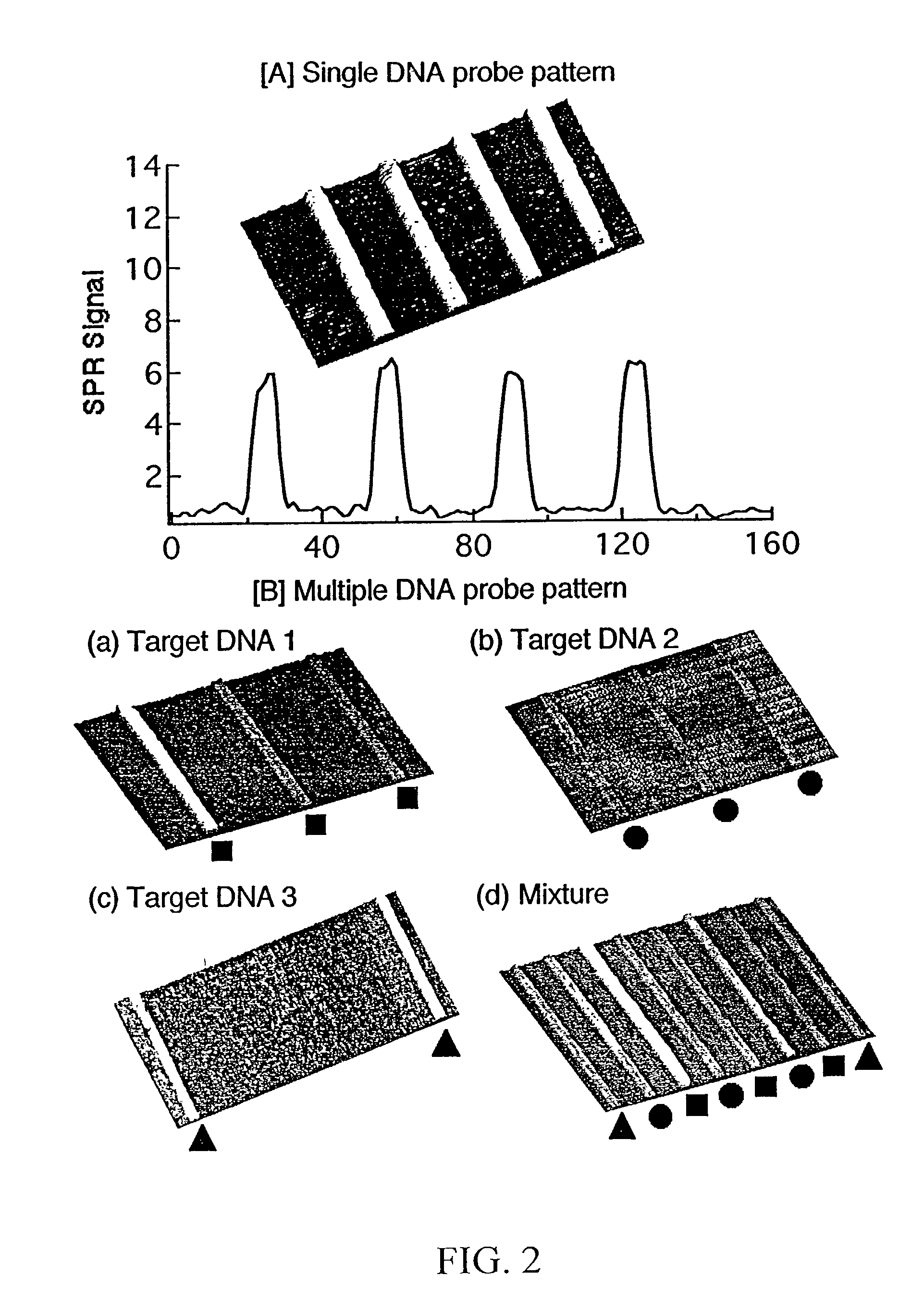
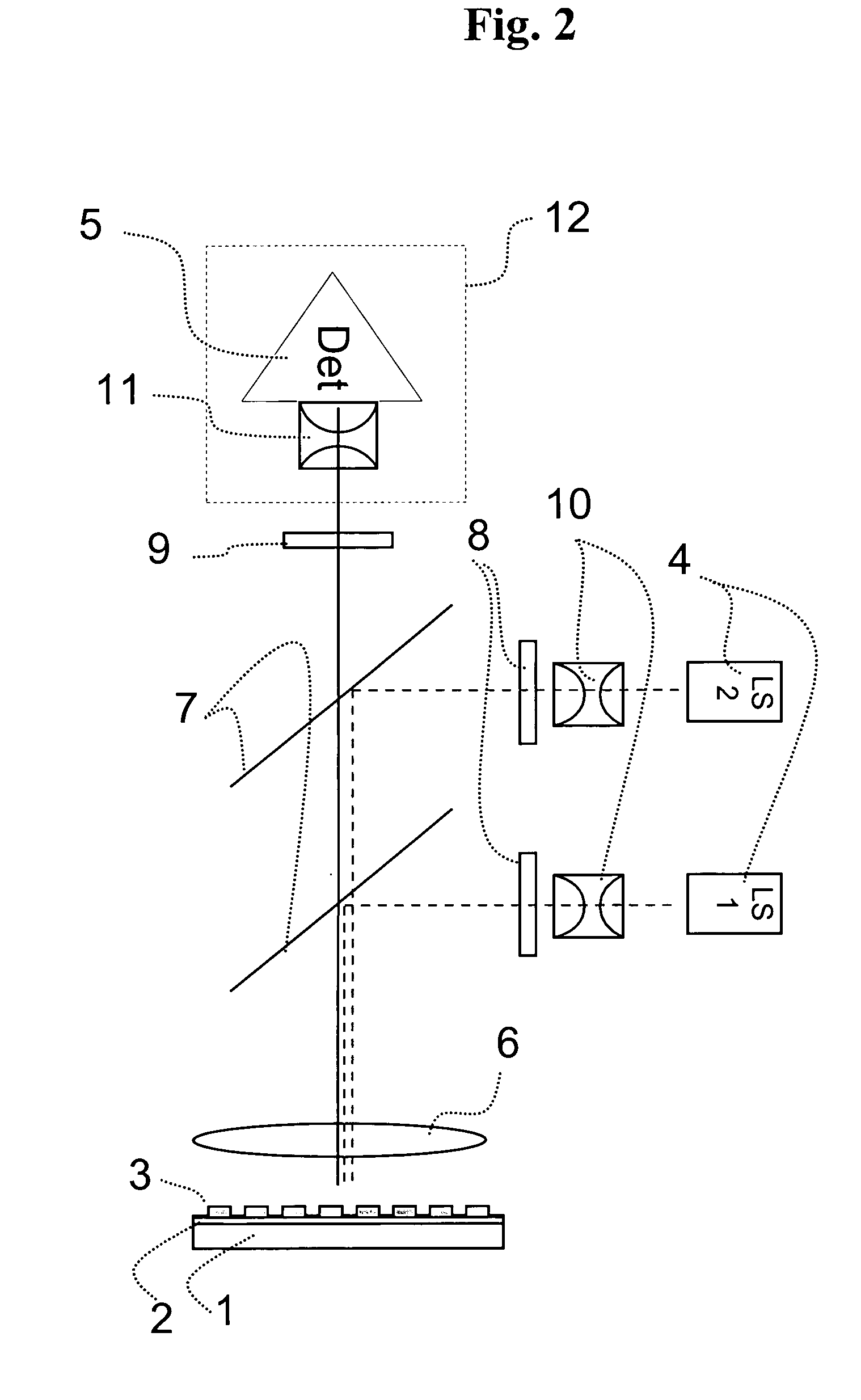
Surface plasmon resonance imaging of micro-arrays
InactiveUS20030017579A1Material nanotechnologyBioreactor/fermenter combinationsSpr imagingMicrofluidic channel
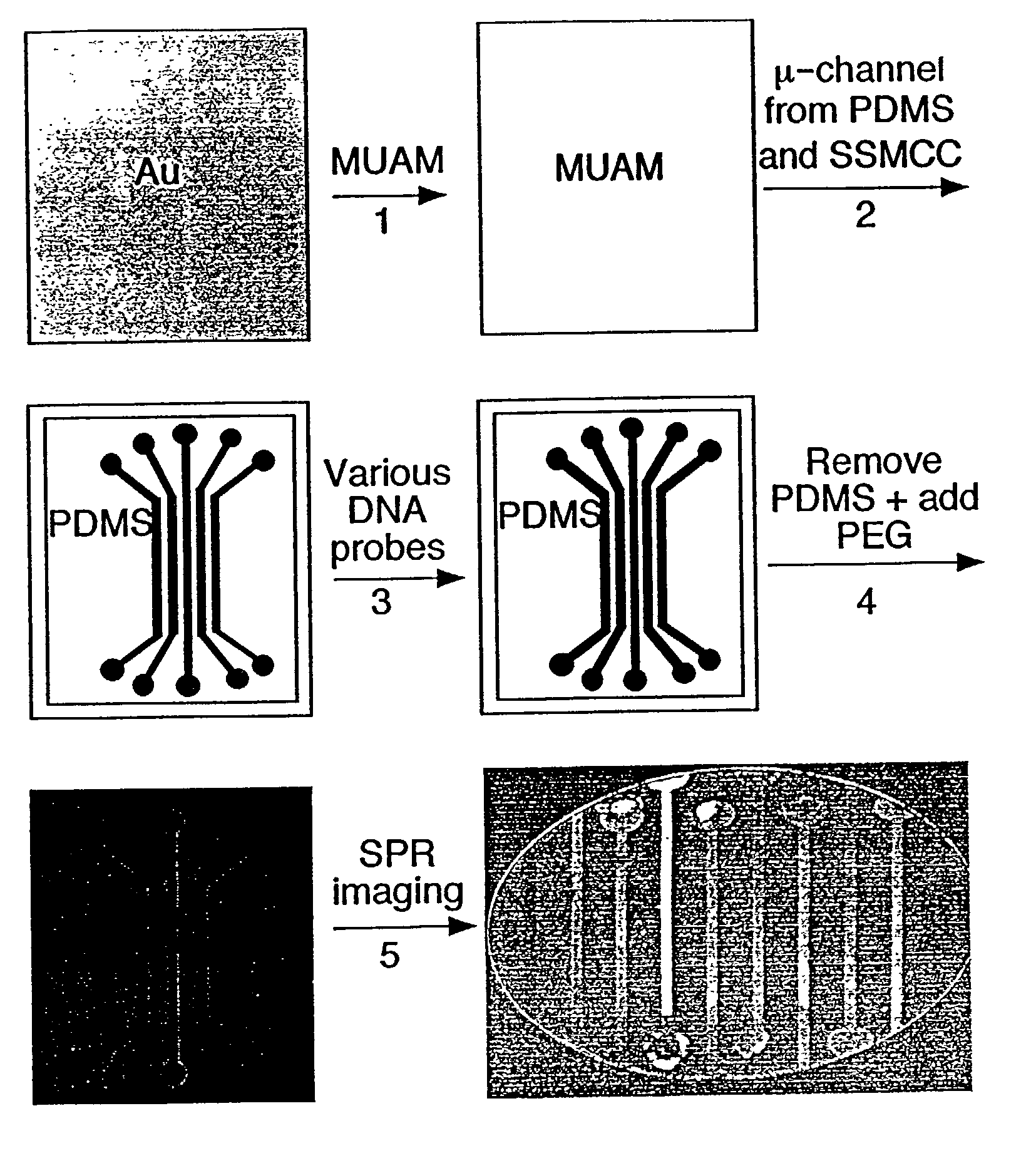
Disclosed is a method for fabricating 1-dimensional micro-arrays using parallel micro-fluidic channels on chemically-modified metal, carbon, silicon, and / or germanium surfaces; a muL detection volume method that uses 2-dimensional nucleic acid micro-arrays formed by employing the 1-dimensional DNA micro-arrays in conjunction with a second set of parallel micro-fluidic channels for solution delivery, and the 1-dimensional and 2-dimensional arrays used in the methods. The methodology allows the rapid creation of 1- and 2-dimensional arrays for SPR imaging and fluorescence imaging of DNA-DNA, DNA-RNA, DNA-protein, and protein-protein binding events. The invention enables very small volumes necessary for a variety of bioassay applications to be analyzed by SPR. Target solution volumes as small as 200 pL can be analyzed.
Owner:WISCONSIN ALUMNI RES FOUND
Method for the synthesis of DNA sequences
InactiveUS7183406B2Material nanotechnologyGroup 5/15 element organic compoundsDNA microarrayDNA construct
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will spontaneously hybridize together to form the desired DNA construct. This method makes possible the remote assembly of DNA sequence, through a process analogous to facsimile transmission of documents, since the information on DNA to be made can be transmitted remotely to an instrument which can then synthesize any needed DNA sequence from the information.
Owner:WISCONSIN ALUMNI RES FOUND
Potentionmetric DNA microarray, Process for producing same and method of analyzig nucleuic acid
InactiveCN1582334ABioreactor/fermenter combinationsSequential/parallel process reactionsDNA microarrayNucleic Acid Probes
A DNA microarray system whereby measurement can be performed at a low running cost, a low price and yet a high accuracy. A nucleic a cid probe 3is immobilized on the surface of a gate insulator of an electric field effect transistor and then hybridized with a target gene on the surface of the insulator. A change in the surface electric charge density thus arising is detected by using the electric effect.
Owner:HITACHI HIGH-TECH CORP
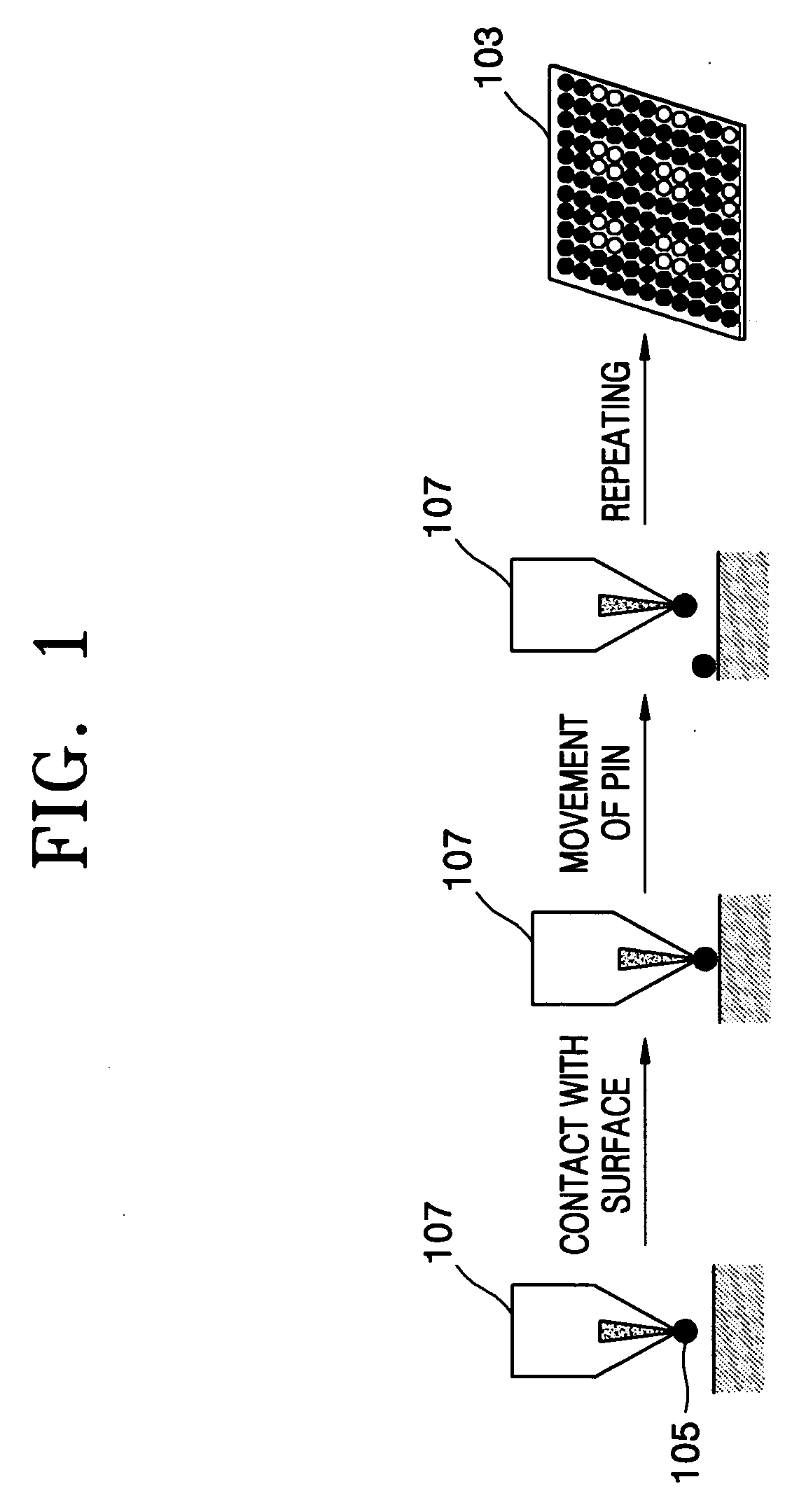
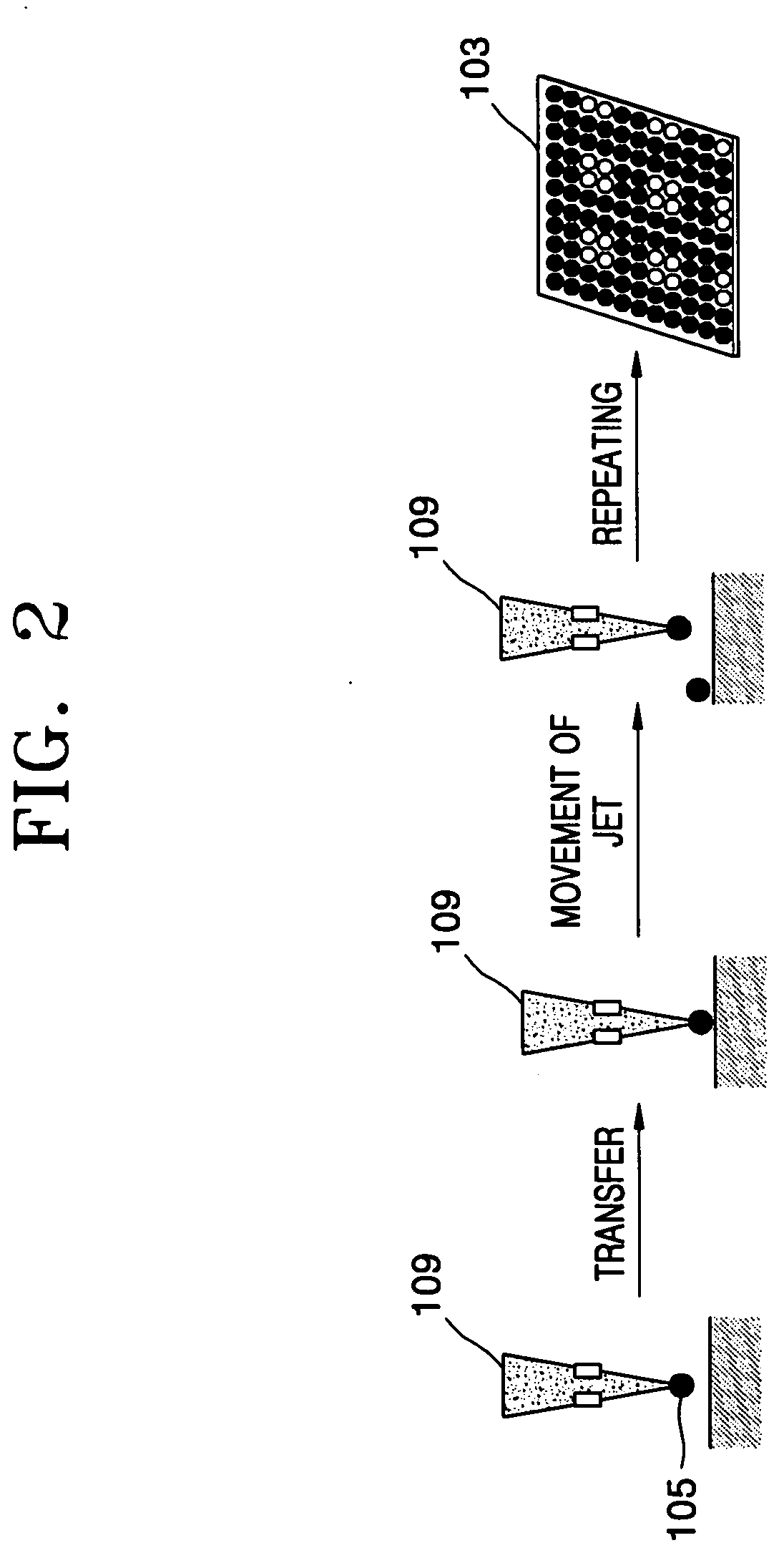
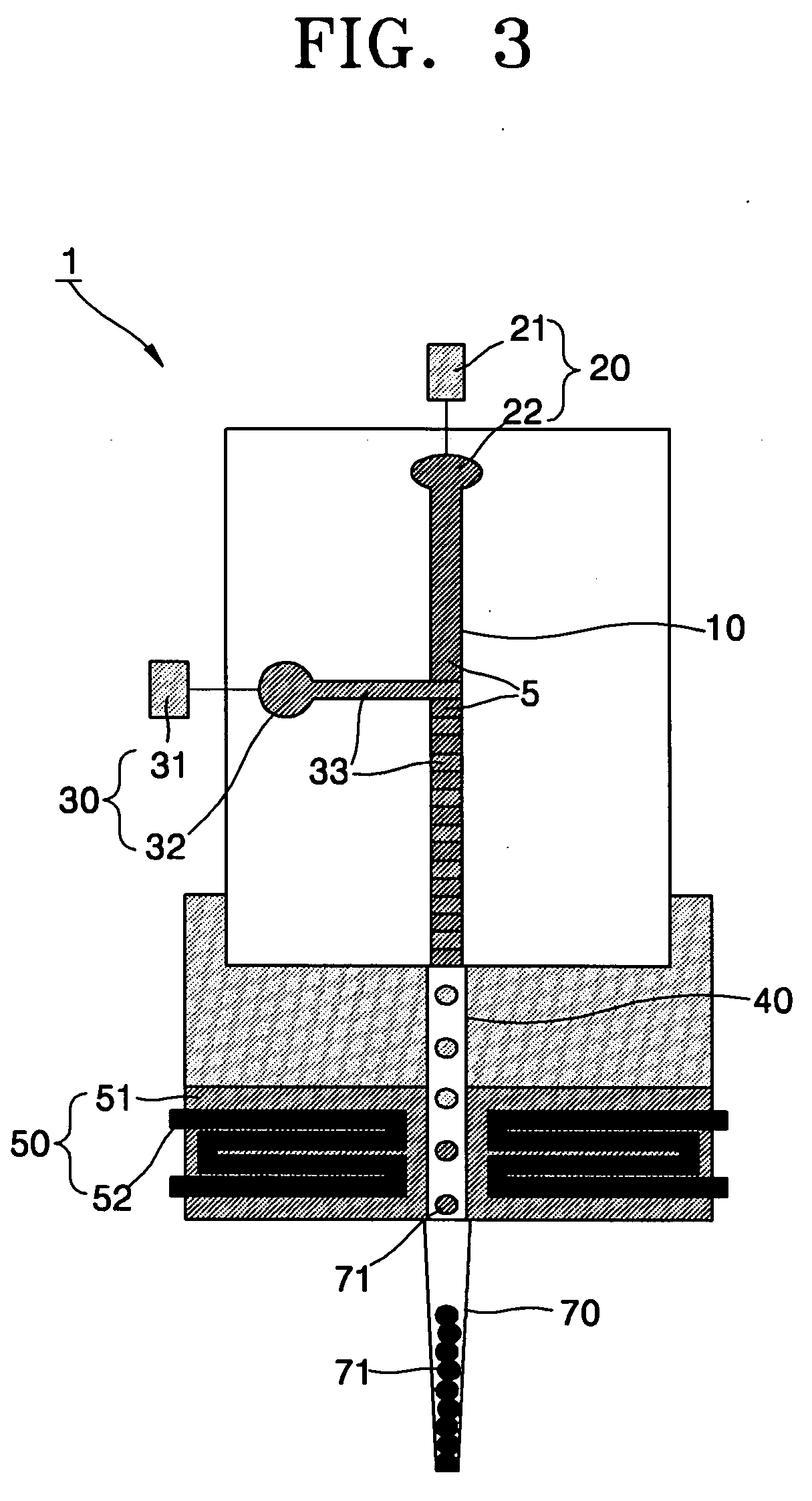
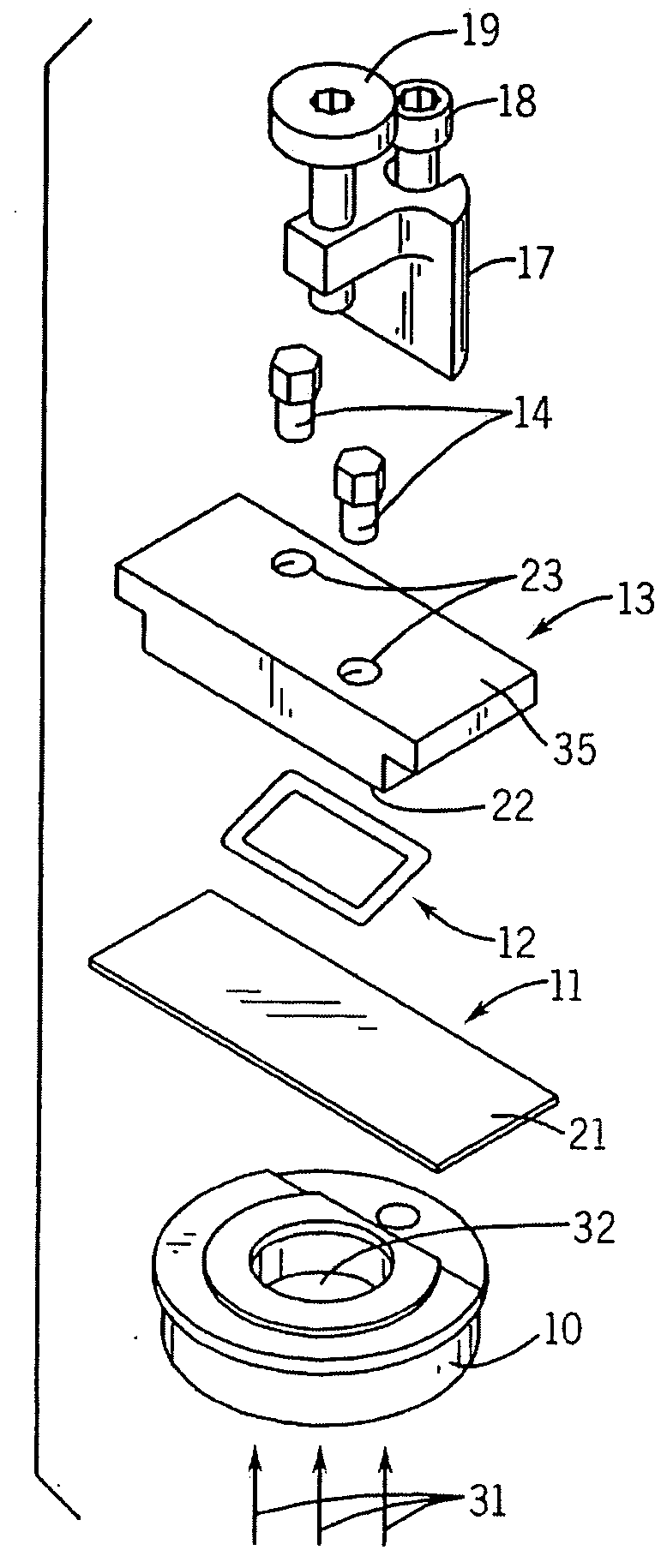
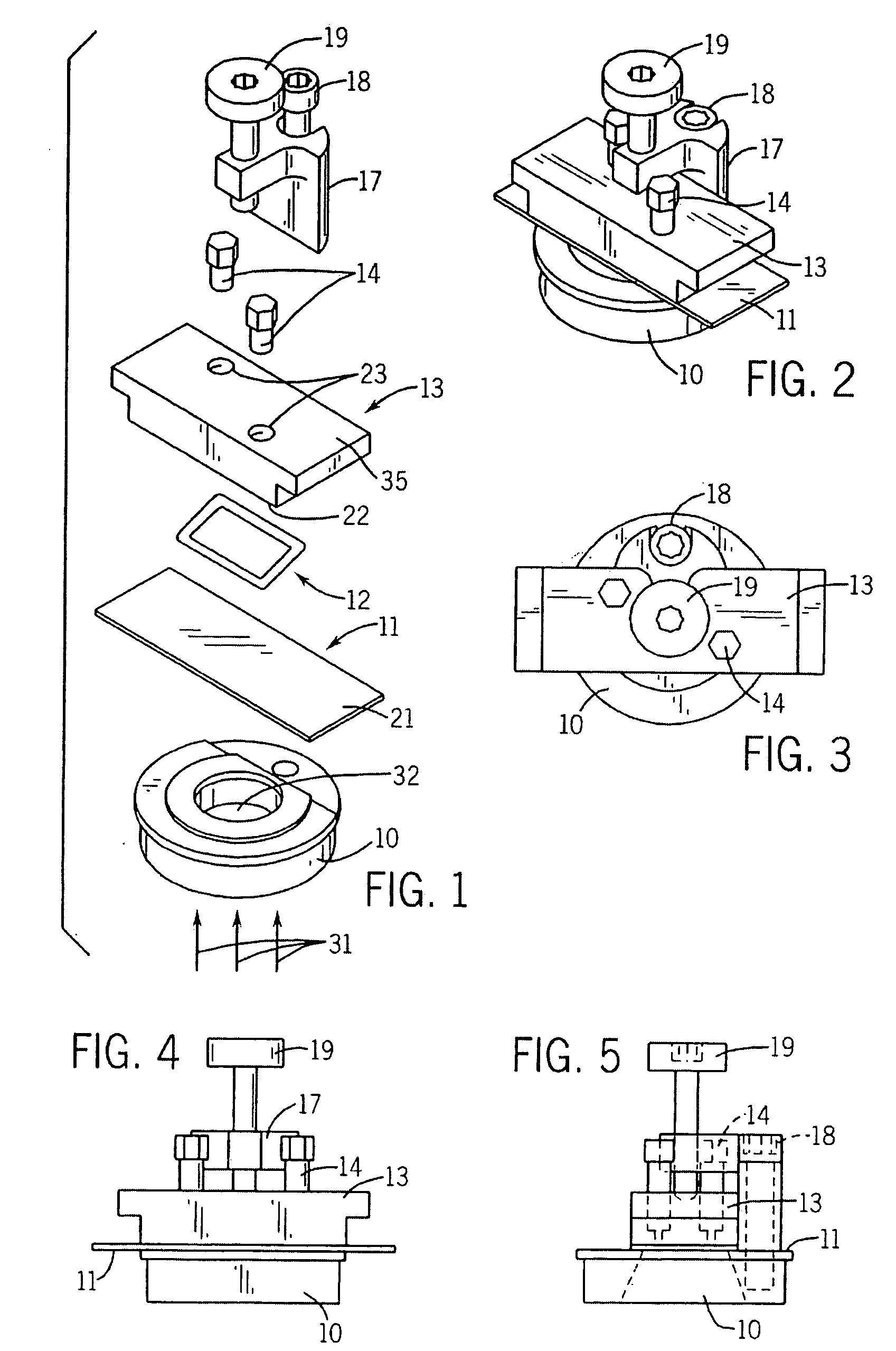
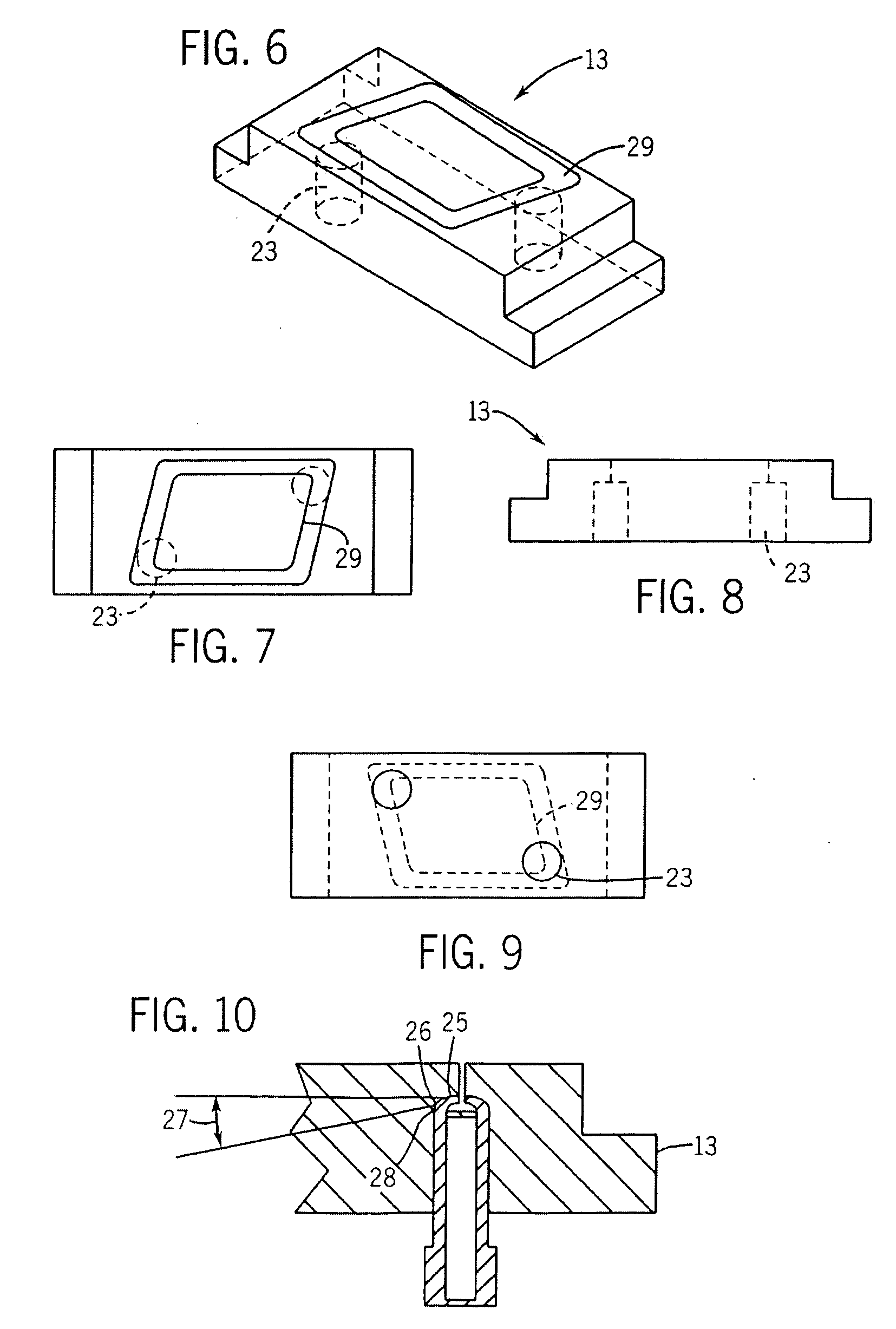
Spotting device for manufacturing DNA microarray and spotting method using the same
InactiveUS20050158755A1Shorten production timeUniform shapeBioreactor/fermenter combinationsMaterial nanotechnologyCross-linkDNA microarray
A spotting device for manufacturing a DNA microarray and a spotting method using the same are provided. The spotting device for dropping and immobilizing a solution of biomolecules, for example nucleic acids such as probe DNA, mRNA, and peptide nucleic acid (PNA), and proteins on a DNA microarray surface to manufacture a DNA microarray, includes a first microchannel with a tube shape; a supplying unit supplying the solution of biomolecules to the first microchannel; a biomolecule solution droplet forming unit cross-linked to the first microchannel and forming biomolecule solution droplets with a predetermined size by periodically jetting a gas toward the biomolecule solution flowing in the first microchannel; a second microchannel linked to the first microchannel and having a greater diameter than the first microchannel; a cooling unit surrounding at least a part of the second microchannel to freeze the biomolecule solution droplets which pass through the second microchannel; and a spotting unit thawing the frozen biomolecule solution droplets and dropping the thawed biomolecule solution droplets on a surface of the DNA microarray. The spotting device can form spots with uniform shape, minimize an effect of temperature on biomolecules, and easily manipulate biomolecules when manufacturing a DNA microarray.
Owner:SAMSUNG ELECTRONICS CO LTD
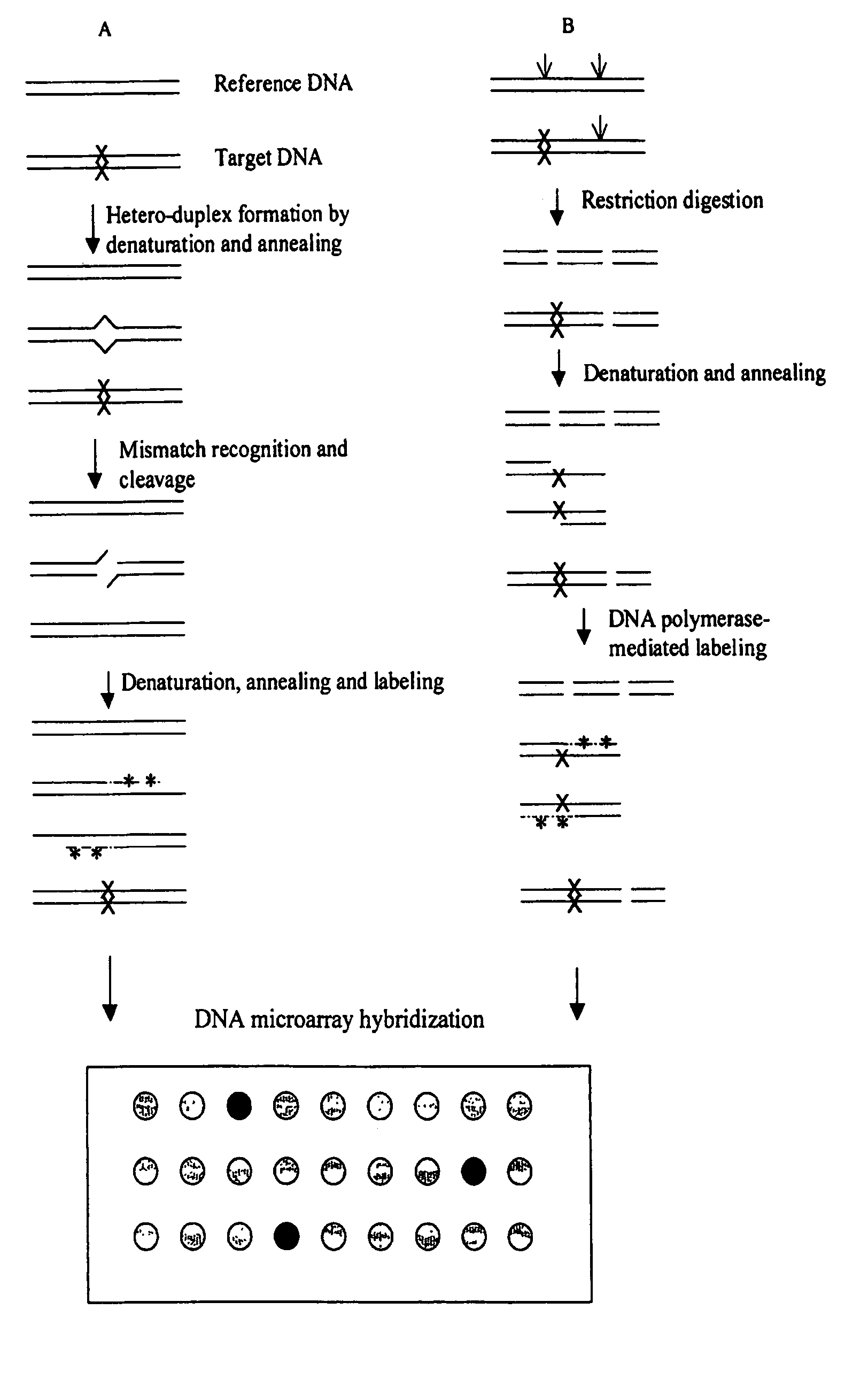
Methods for detecting and localizing DNA mutations by microarray
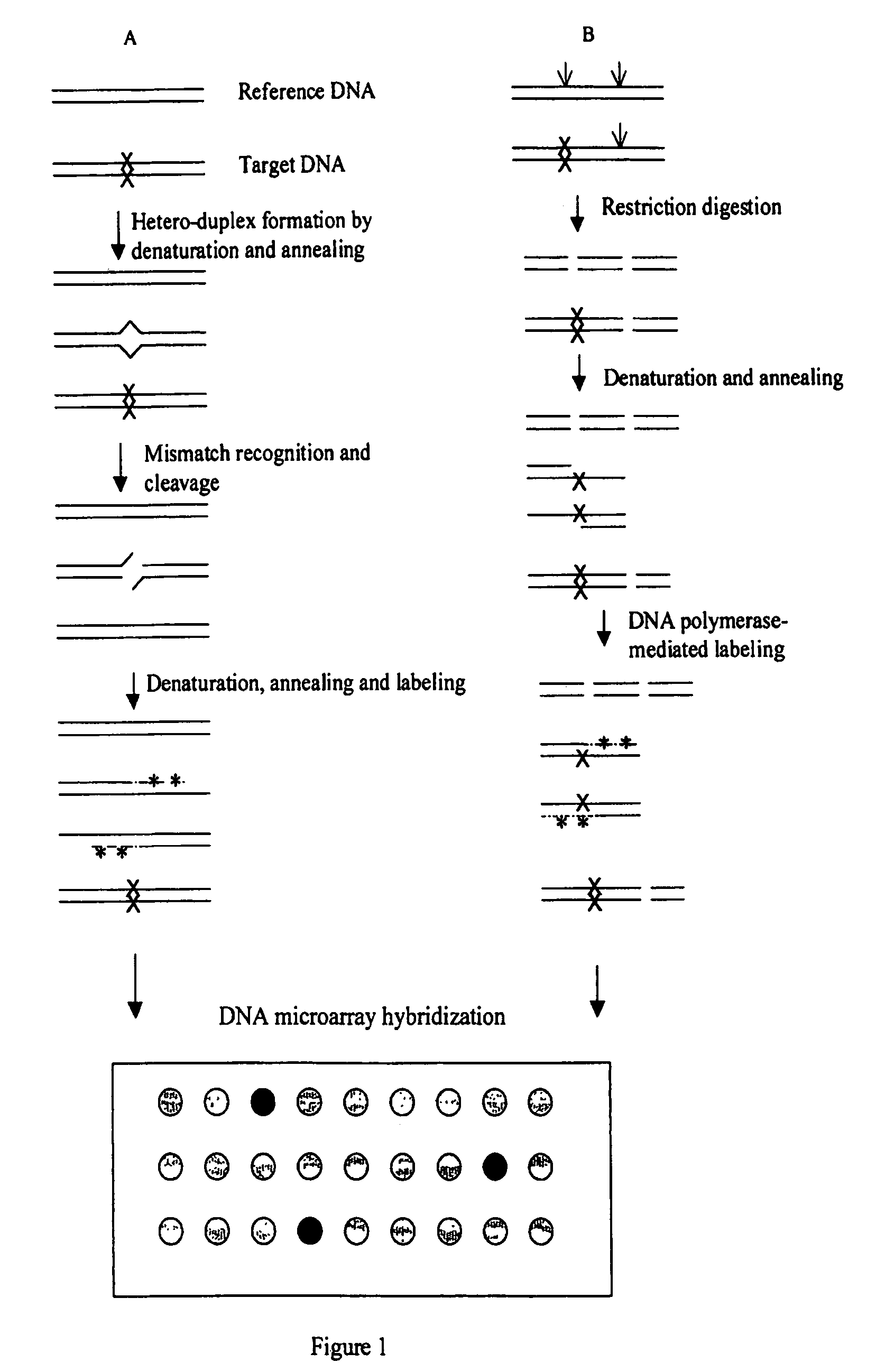
InactiveUS7378245B2Reduce impactMicrobiological testing/measurementFermentationDNA microarrayHeteroduplex
Owner:UNIVERSITY OF OREGON
Defining biological states and related genes, proteins and patterns
InactiveUS20020169562A1Reduce the valueSugar derivativesMicrobiological testing/measurementDiseaseProtein insertion
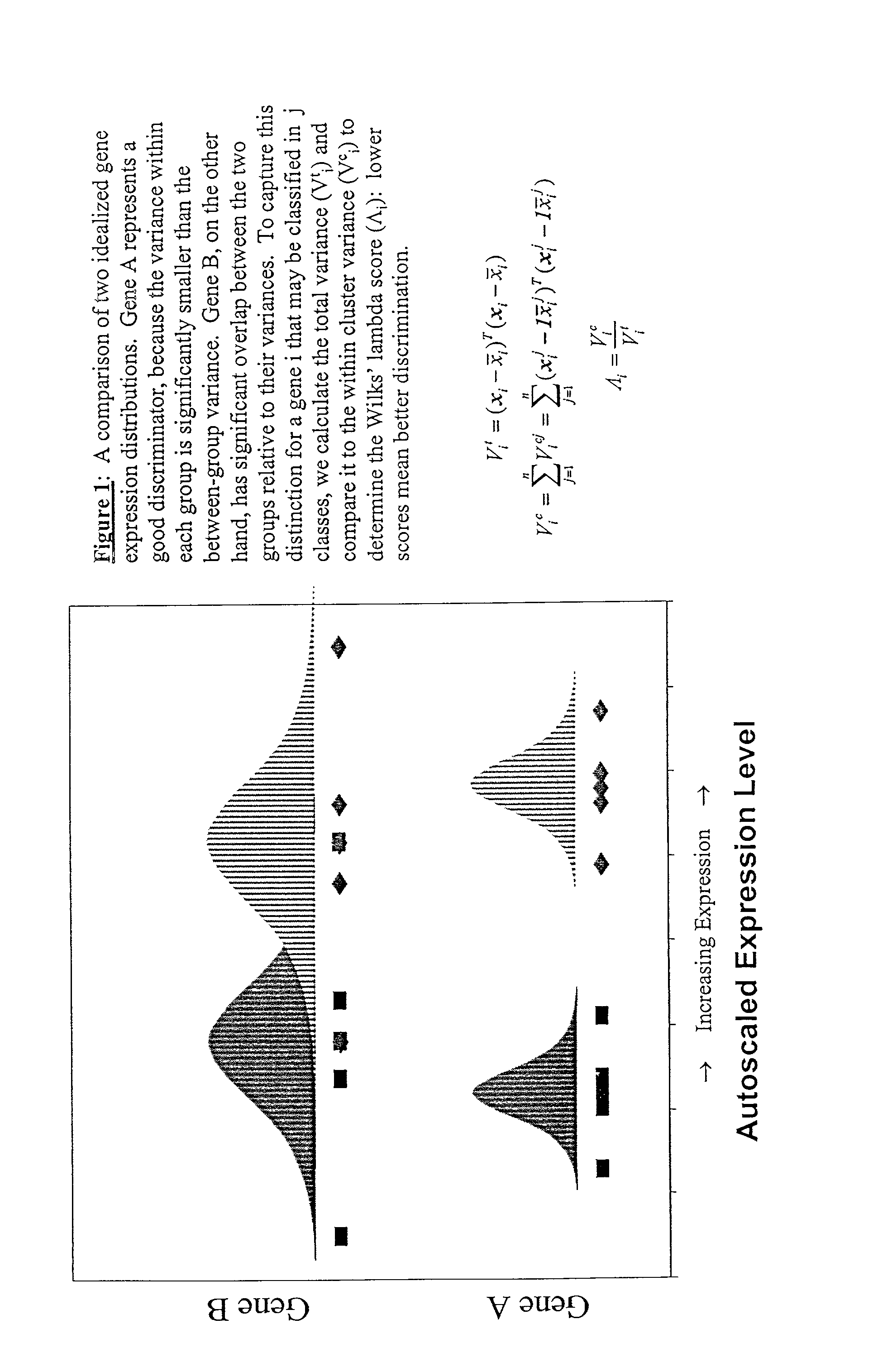
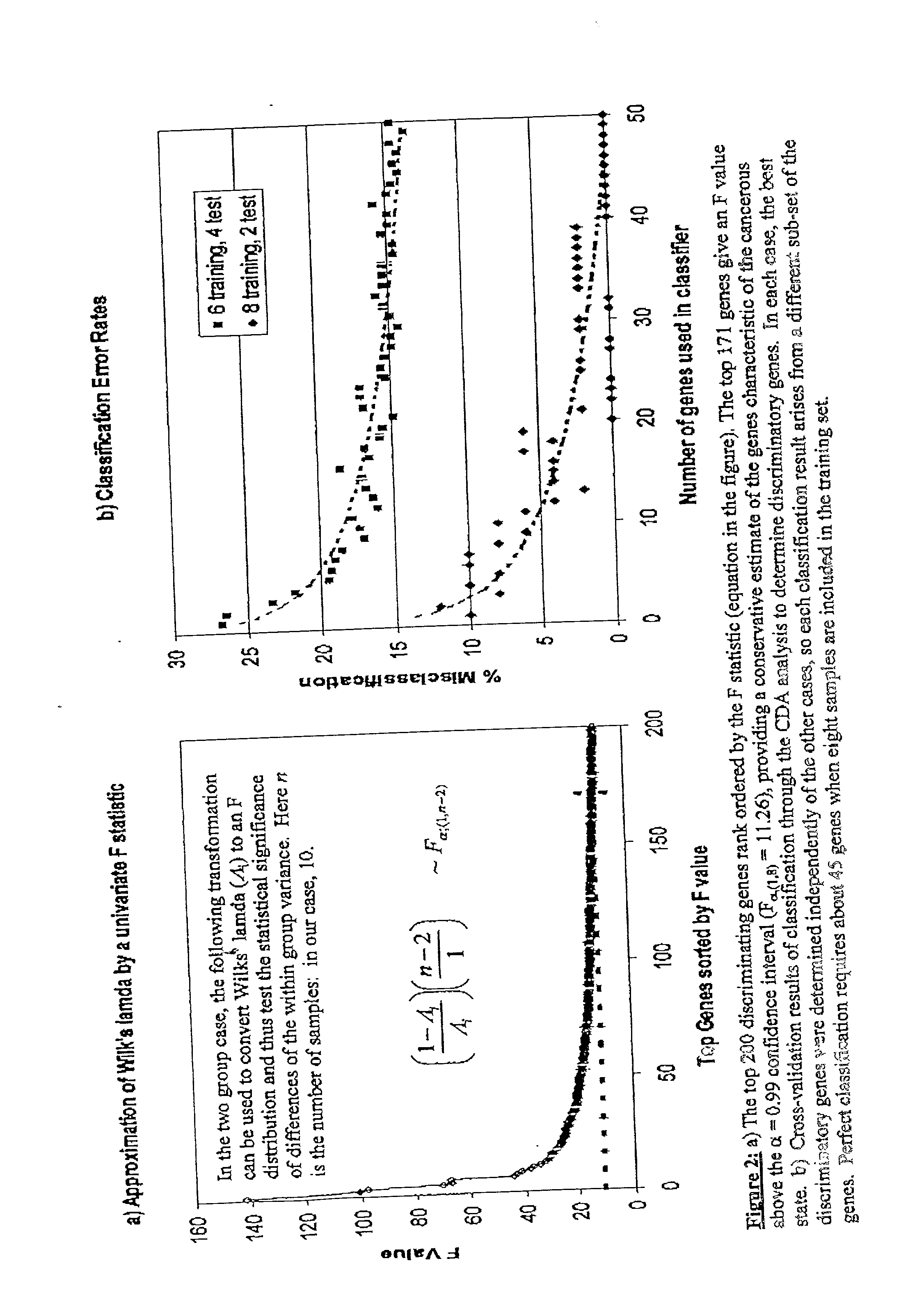
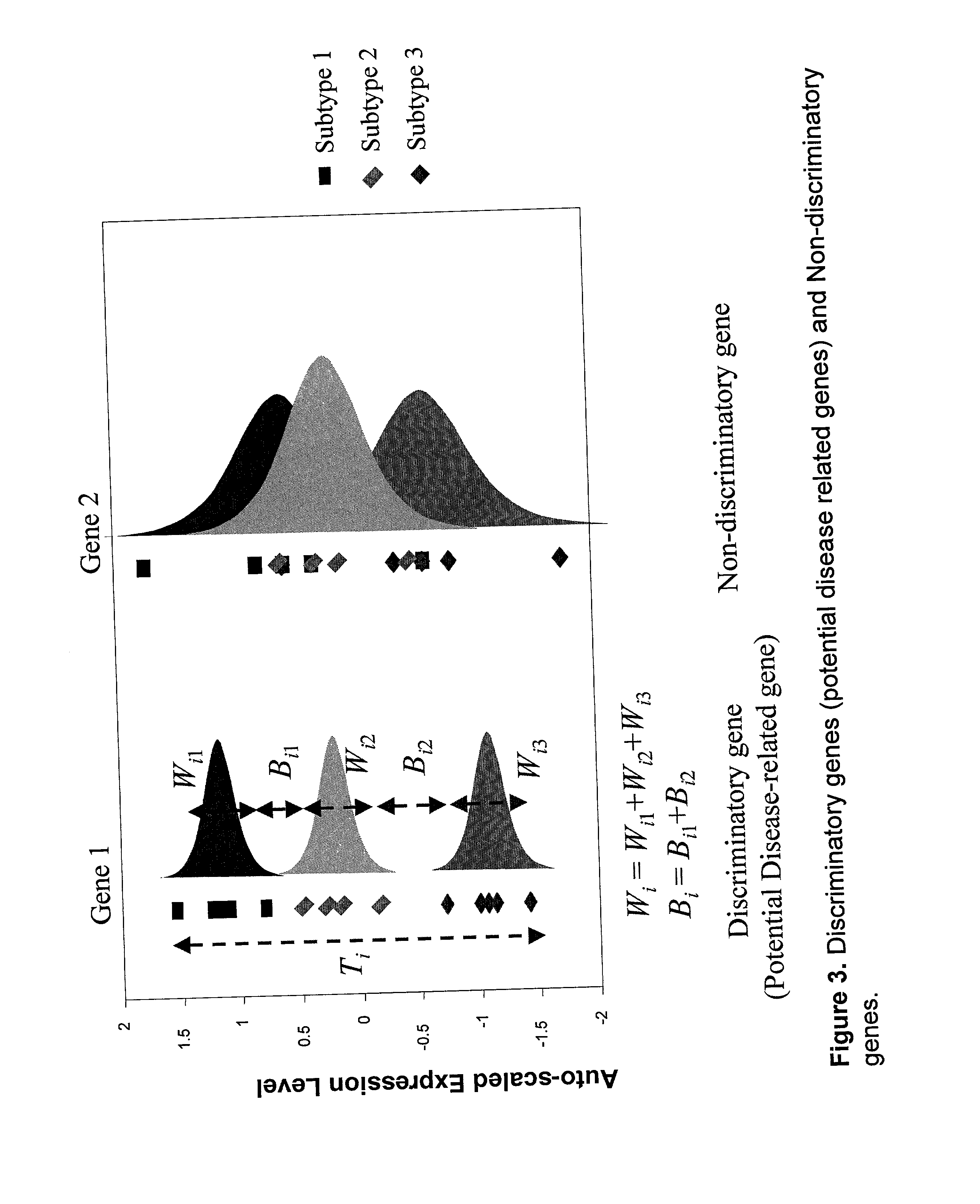
Disclosed are a variety of methods and computer systems for use in the analysis of gene and protein expression data. Also disclosed are methods for the definition of the cellular state of cells and tissues from multidimensional physiological data such as those obtained from gene expression measurements with DNA microarrays. A variety of classification methods can be applied to expression data to achieve this goal. Demonstrated is the application of several statistical tools including Wilks' lambda ratio of within-group to total variance, Fisher Discriminant Analysis, and the misclassification error rate to the identification of discriminating genes and the overall classification of expression data. Examples from several different cases demonstrate the ability of the method to produce well-separated groups in the projection space representing distinct physiological states. The method can be augmented and is useful in disease diagnosis, drug screening and bioprocessing applications.
Owner:MASSACHUSETTS INST OF TECH
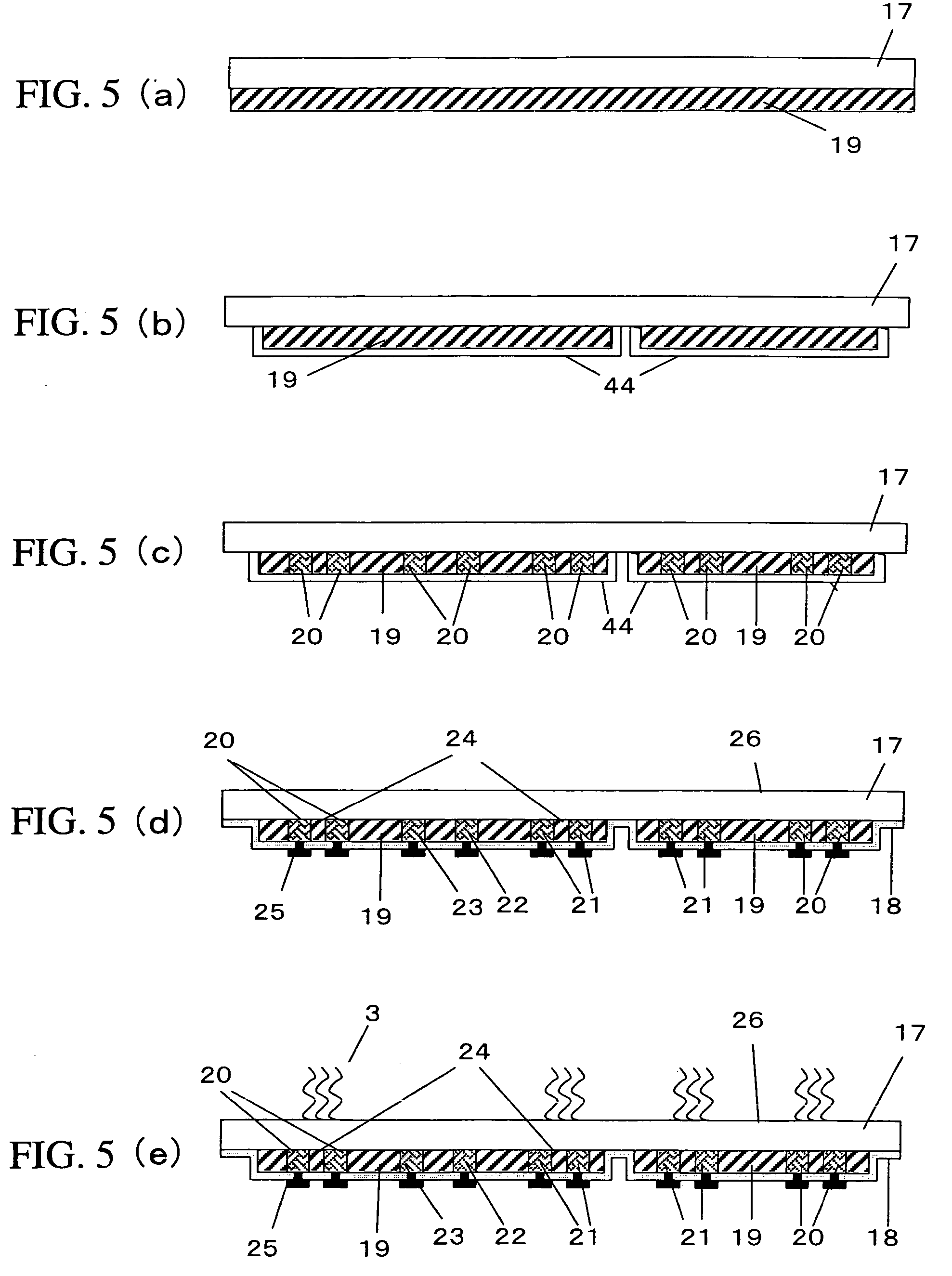
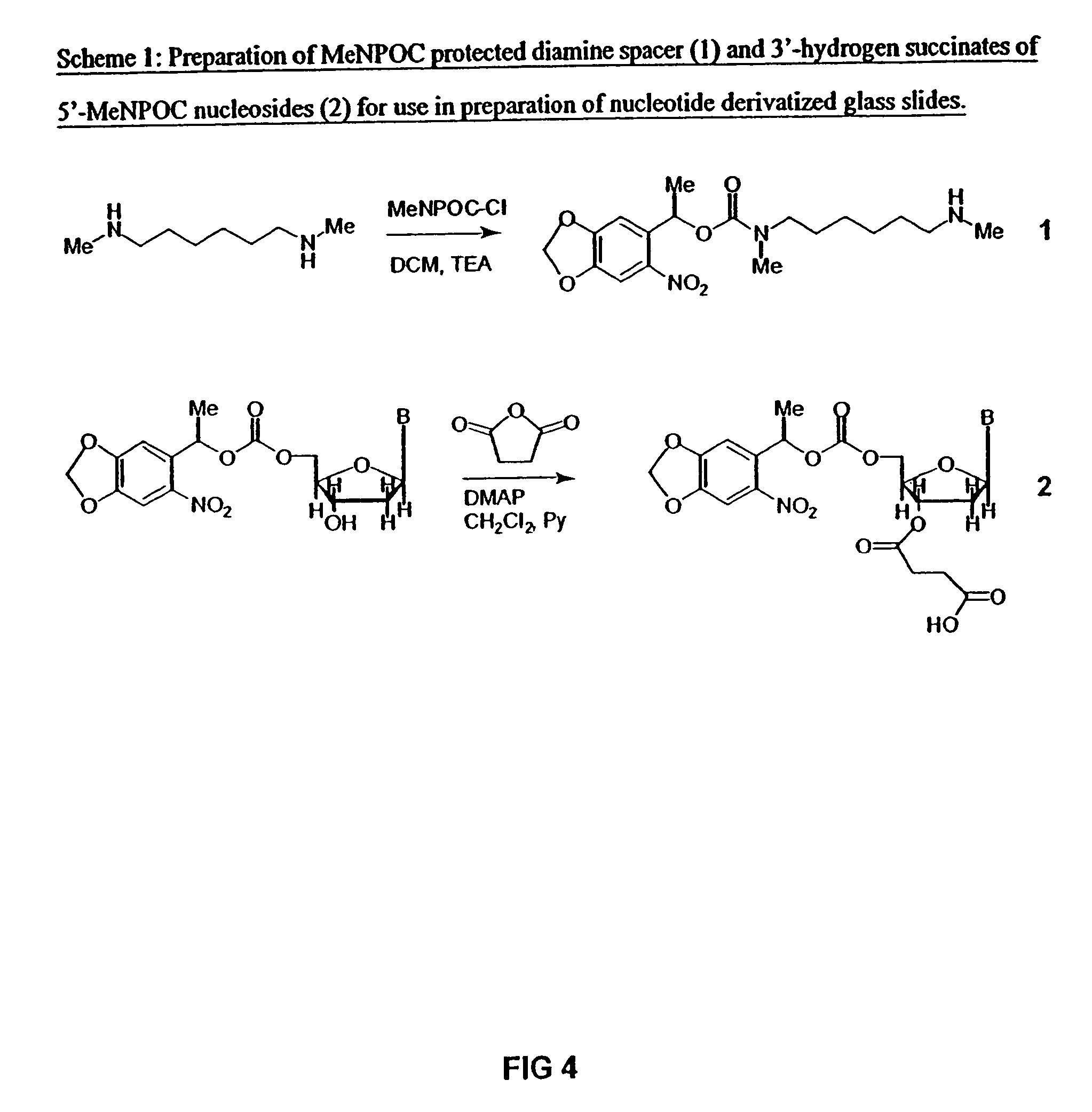
Nucleotide derivative and DNA microarray
ActiveUS7414117B2Easy to prepareSugar derivativesMicrobiological testing/measurementCytosineDNA microarray
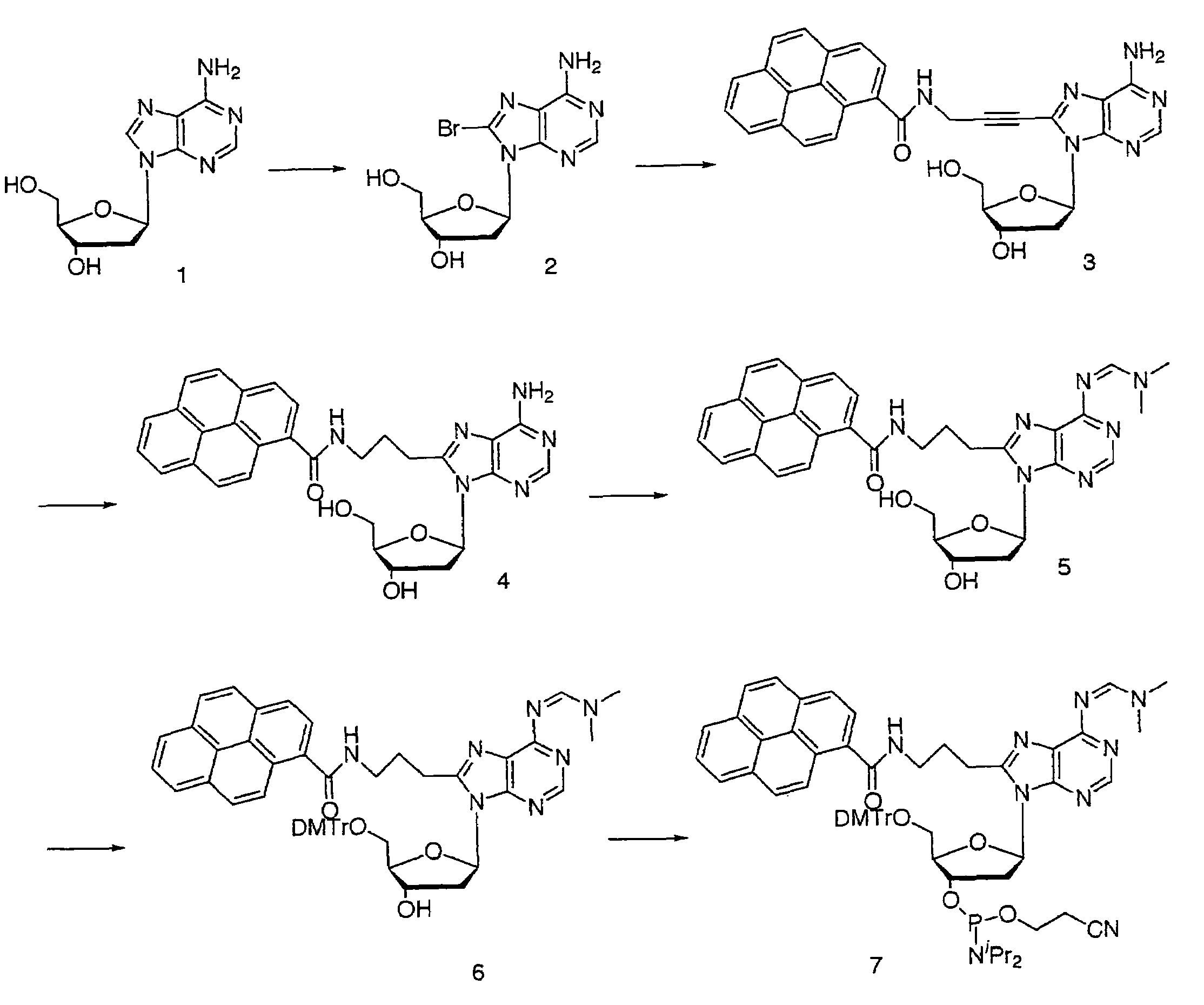
A novel nucleotide derivative, in case of existing as a member of a single-stranded sequence, undergoing a change in the fluorescent signal intendity depending on the corresponding base type in the partner strand with which the single-stranded sequence is hybridized, and which is a thymin / uracil derivative (1) emitting light most intensely when a confronting base in the partner strand with which the single-stranded nucleotide sequence is hybridized is adenine; a cytosine derivative (2) emitting light most intensely when the confronting base is guanine; an adenine derivative (3) emitting light most intensely when the confronting base is cytosine; a guanine derivative (4) emitting light most intensely when the confronting base is cytosine or thymine / uracil; and an adrnine derivative (5) emitting light most intensely when the confronting base is thymine / uracil / .
Owner:SAITO ISAO +1
Microarray synthesis instrument and method
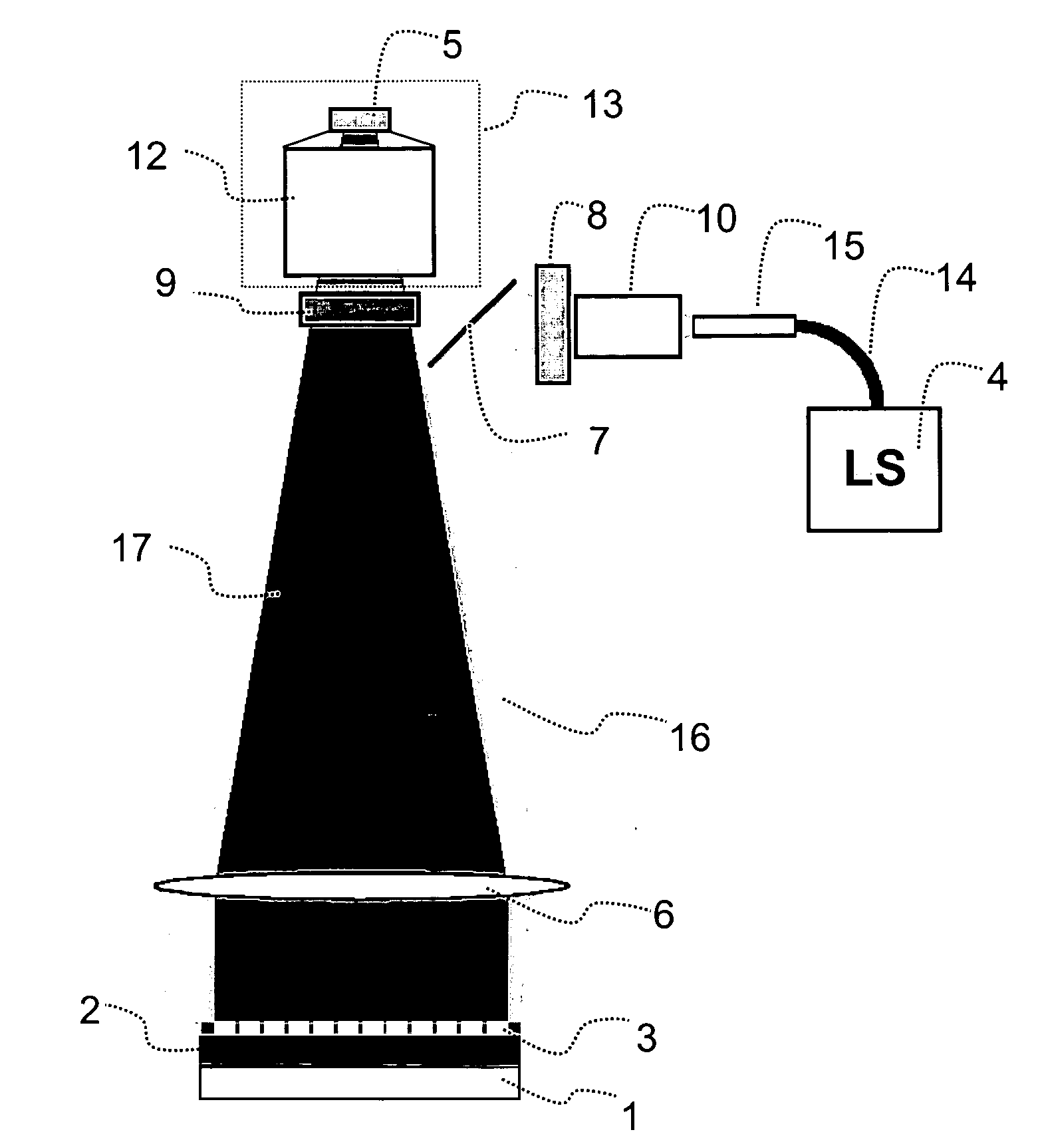
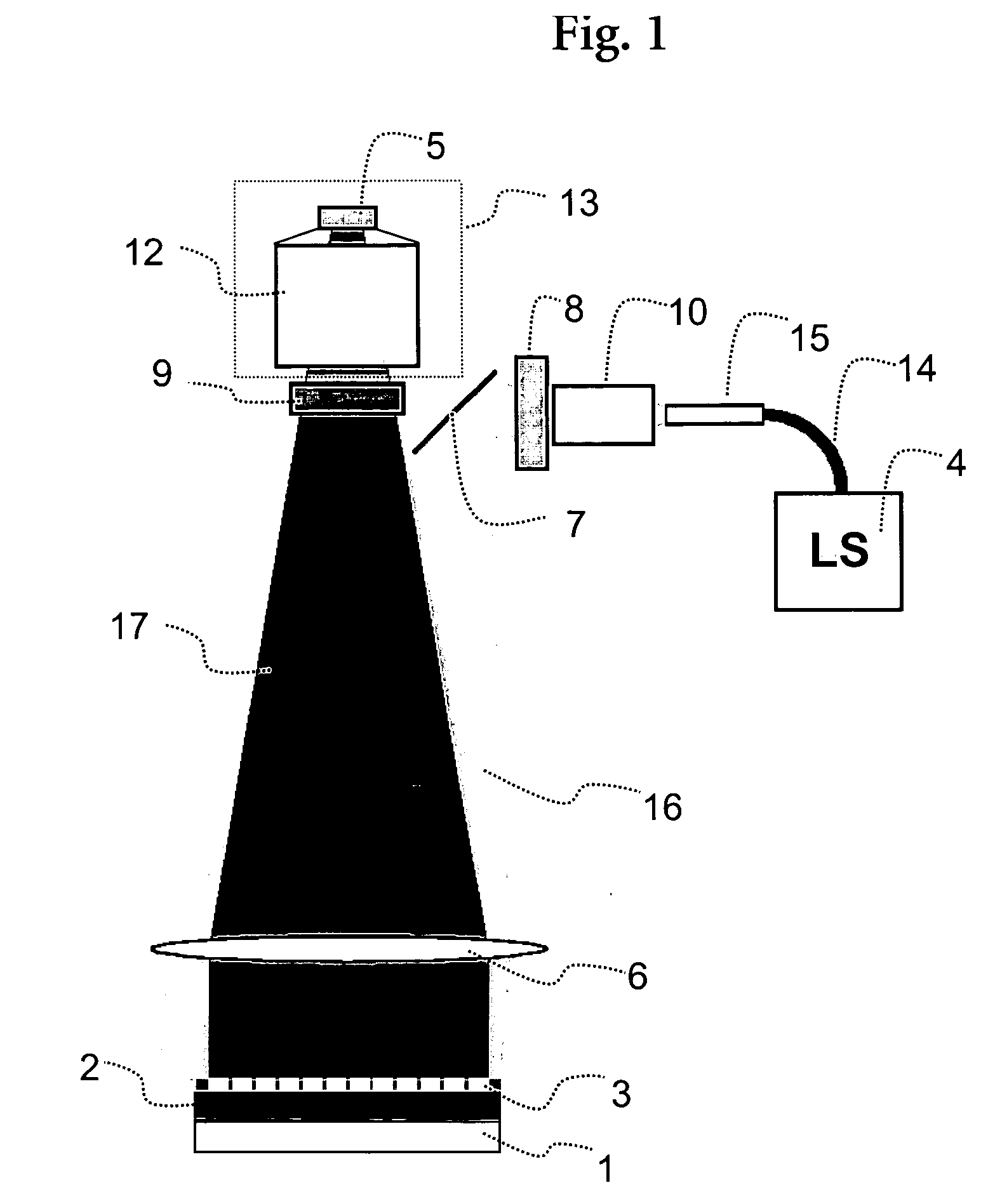
InactiveUS7083975B2Minimizes reflected lightUndesired reactionMaterial nanotechnologyBioreactor/fermenter combinationsDNA microarrayFlow cell
During the light illumination period of a monomer addition cycle in synthesizing an DNA microarray, undesirable reflections of illumination light from various interfaces that the illumination light passes through near the synthesis surface of the substrate may reduce the light-dark contrast, and negatively affect the precision and resolution of the microarray synthesis. The present invention provides an flow cell that reduces the undesired reflections by constructing certain flow cell structures with materials that have similar refractive indexes as that of the solution that is in the oligomer synthesis chamber during the illumination period and / or constructing certain flow cell structures or covering the structures with a layer of a material that has a high extinction coefficient.
Owner:ROCHE NIMBLEGEN
Predicting responsiveness to cancer therapeutics
InactiveUS20090105167A1Increase probabilityBiocideMicrobiological testing/measurementDNA microarrayComputerized system
The invention provides for compositions and methods for predicting an individual's responsitivity to cancer treatments and methods of treating cancer. In certain embodiments, the invention provides compositions and methods for predicting an individual's responsitivity to chemotherapeutics, including salvage agents, to treat cancers such as ovarian cancer. The invention also provides reagents, such as DNA microarrays, software and computer systems useful for personalizing cancer treatments, and provides methods of conducting a diagnostic business for personalizing cancer treatments.
Owner:UNIV OF SOUTH FLORIDA +1
Diagnostic tests for predicting prognosis, recurrence, resistance or sensitivity to therapy and metastatic status in cancer
InactiveUS20140342946A1Low costReduce morbidityMicrobiological testing/measurementProtein nucleotide librariesParanasal Sinus CarcinomaDiagnostic test
The present invention describes a method utilizing a set of genes or gene products whose altered expression in cancer tissue, particularly head and neck cancer and other carcinomas, or its adjacent normal tissues predicts (a) probability of recurrence in time after treatment (b) sensitivity or resistance to therapies or (c) probability of metastasis at the time of initial discovery of the tumor. Furthermore, the invention describes methods of determining the molecular signature in tumor tissues, tissues adjacent to the tumor, or in saliva by using DNA microarray techniques, quantitative real-time PCR, immunohistochemistry or other methods that are used for determining gene or gene product expression levels.
Owner:KURIAKOSE MONI ABRAHAM +1
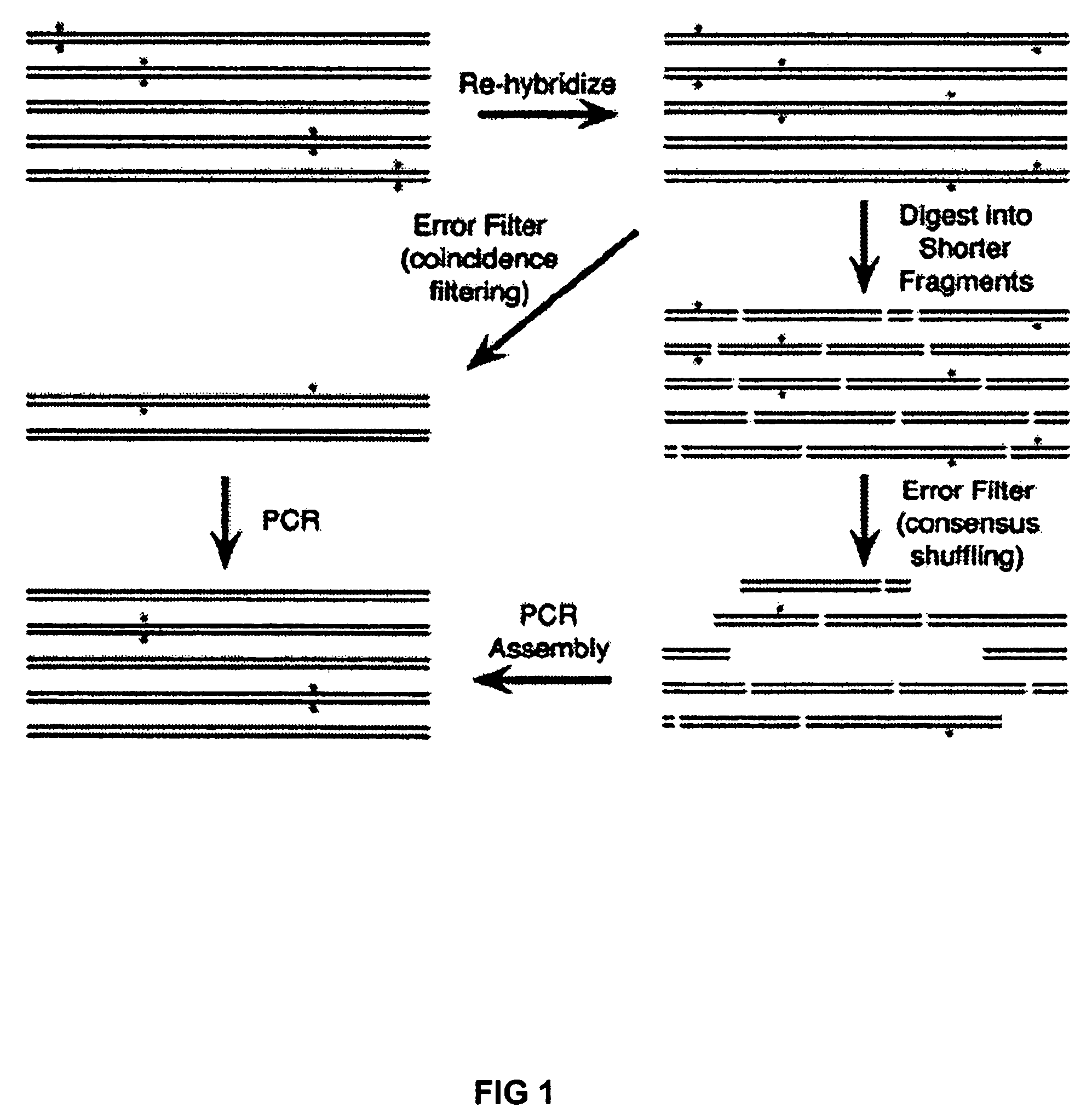
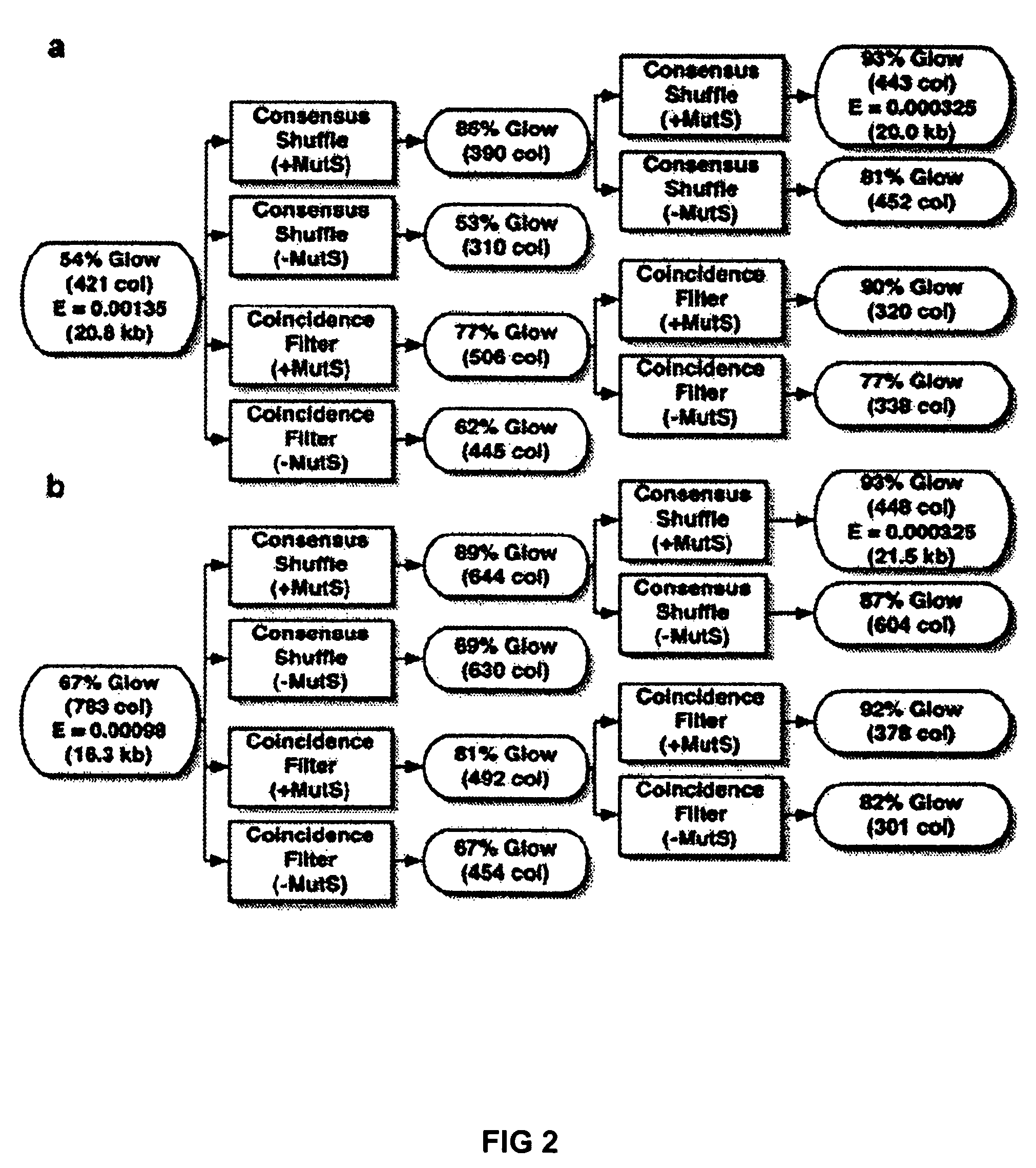
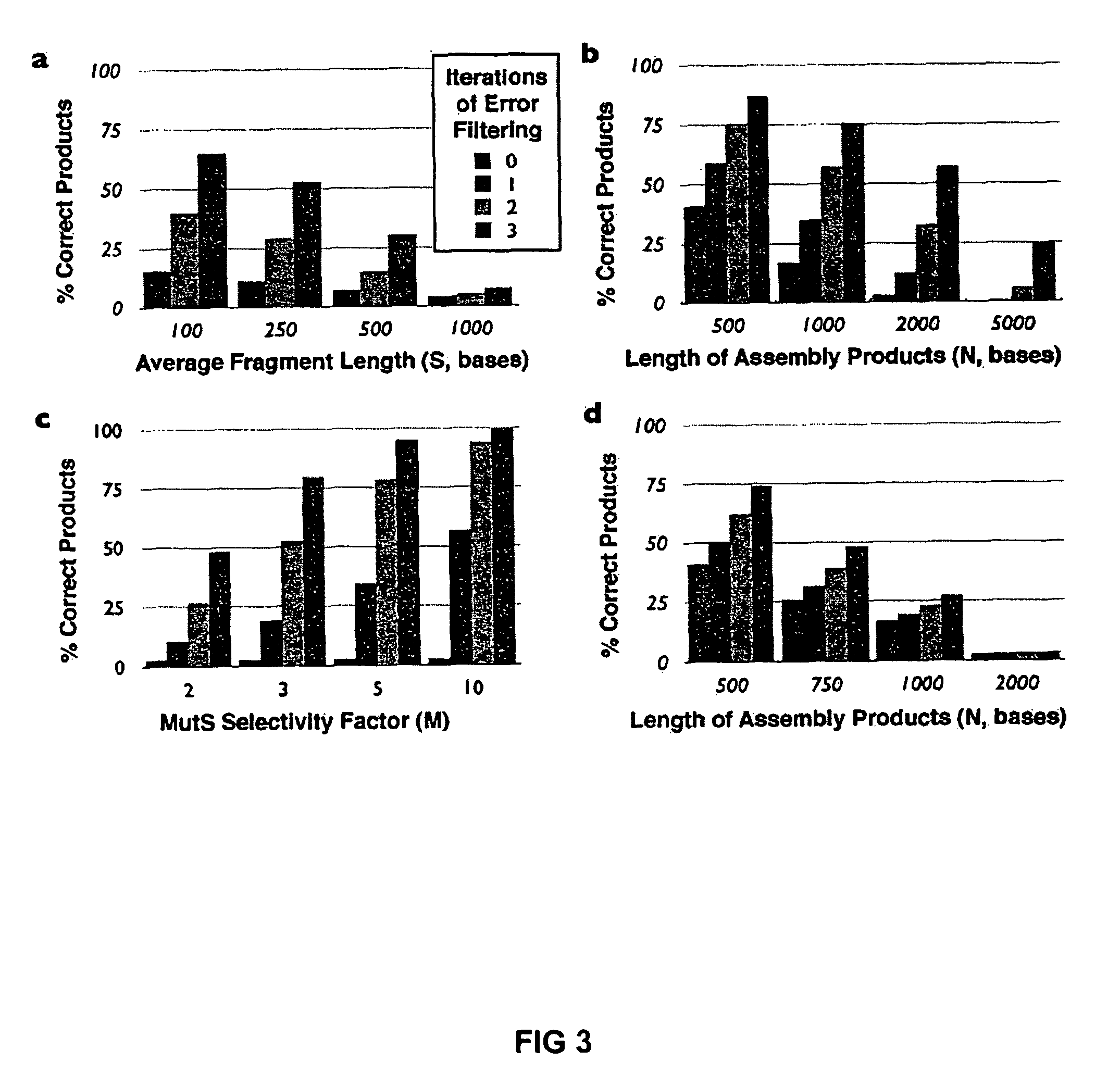
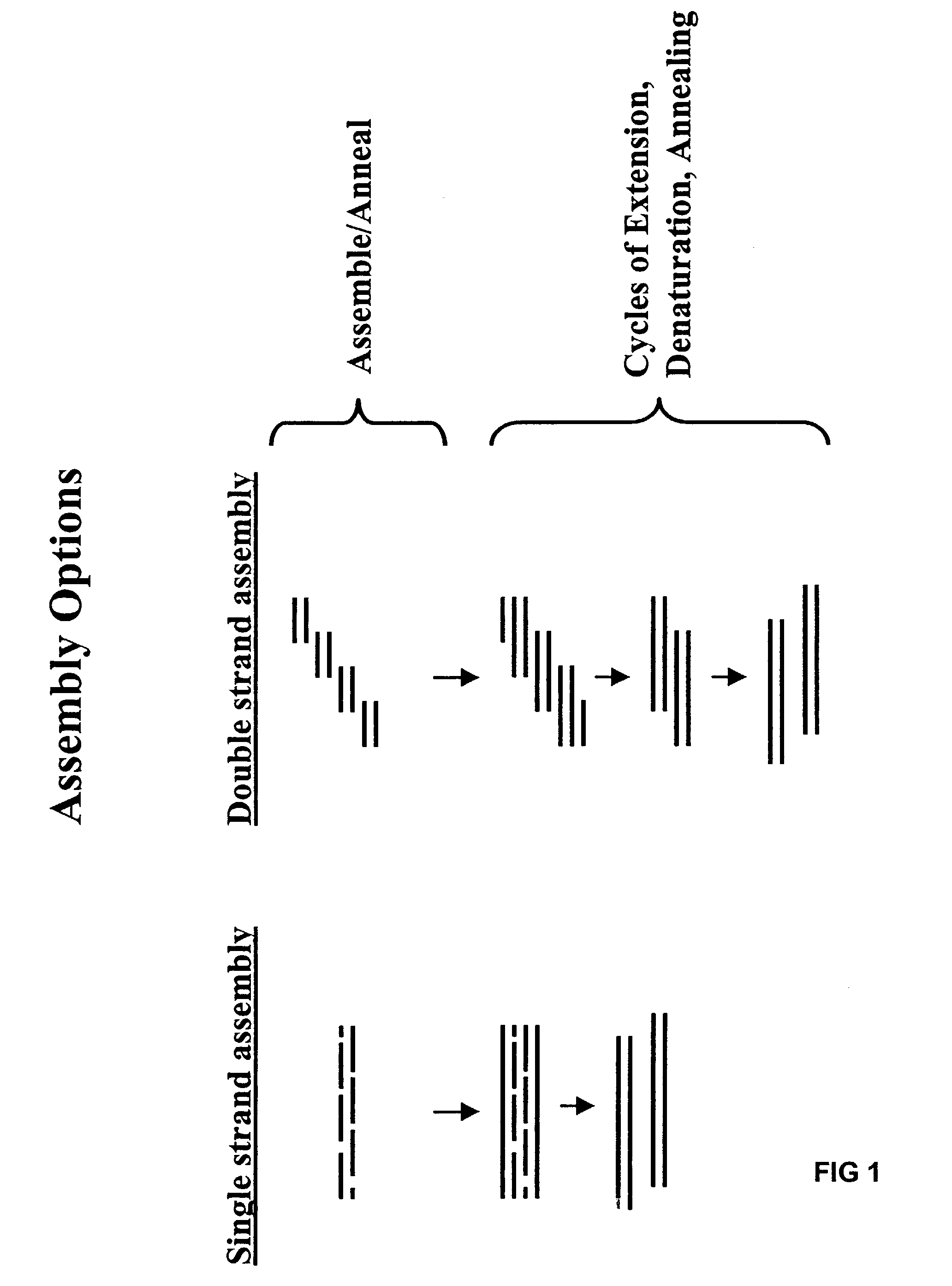
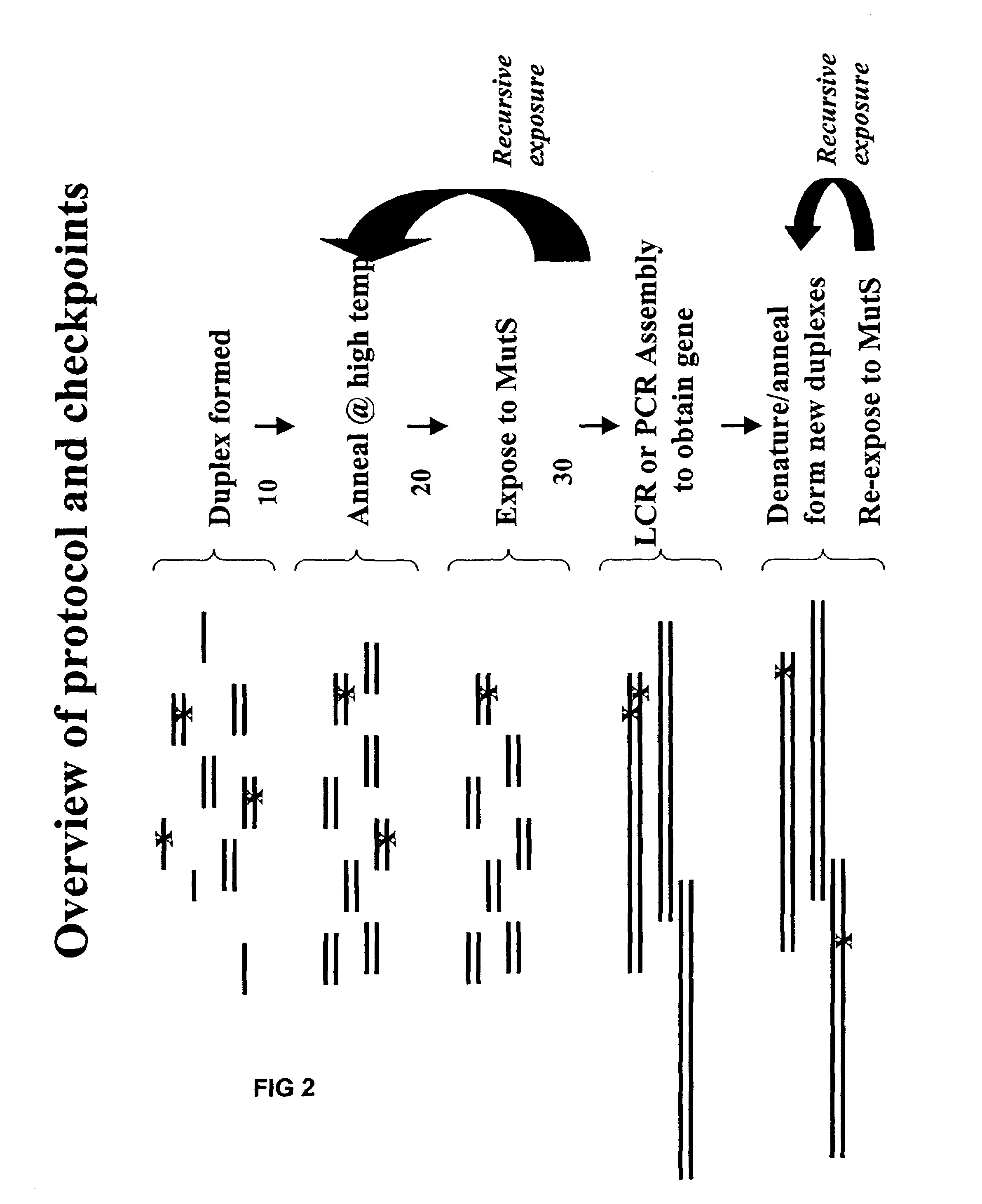
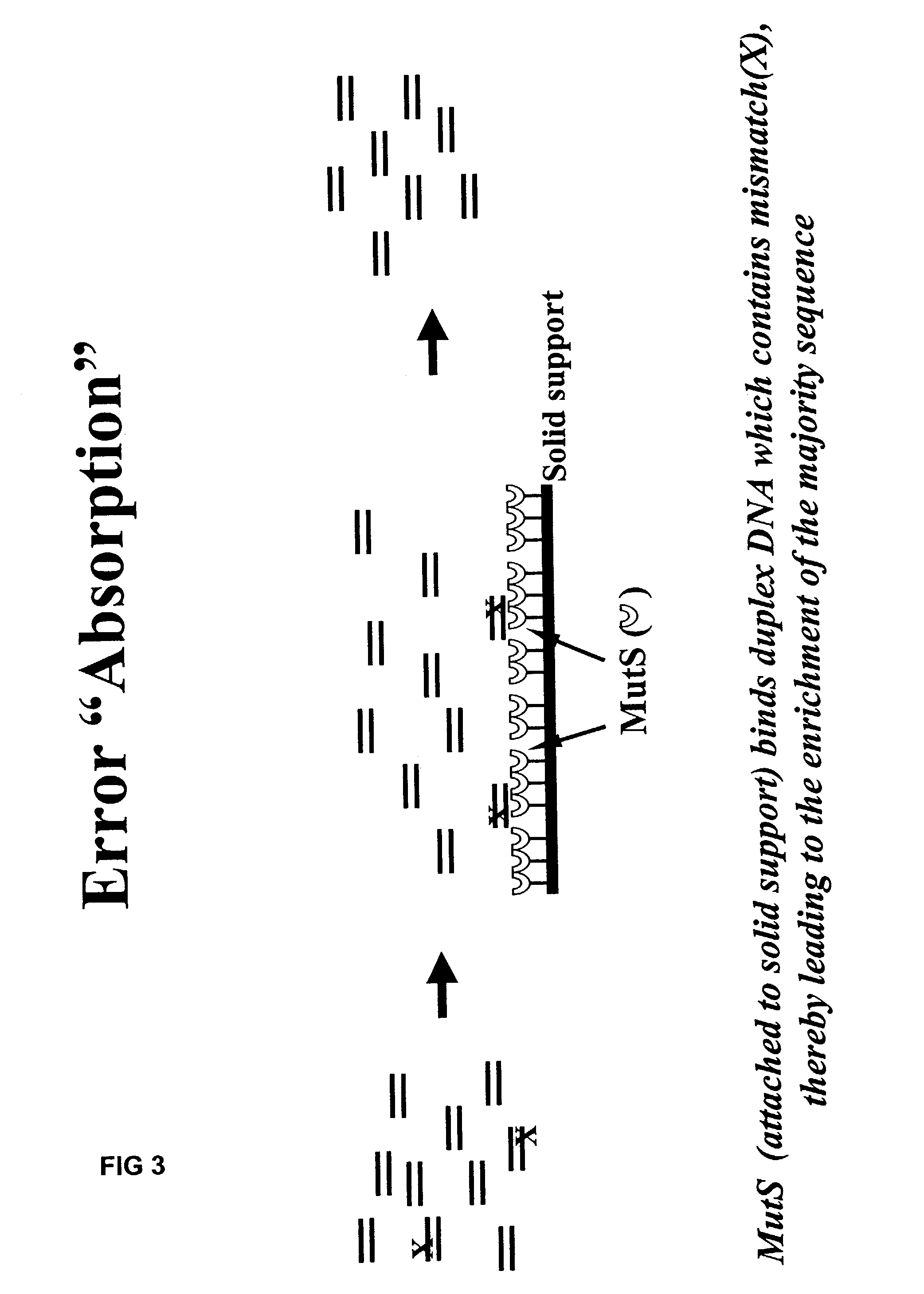
Method of error reduction in nucleic acid populations
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The complement of each segment is also made in the microarray. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will hybridize to form duplexes. The duplexes are then separated using a DNA binding agent which binds to improperly formed DNA helixes to remove errors from the set of DNA molecules. The segments can then be hybridized to each other to assemble the larger target DNA sequence.
Owner:WISCONSIN ALUMNI RES FOUND
Potentiometric dna microarray, process for producing the same and method of analyzing nucleic acid
InactiveUS20050170347A1Improve signal-to-noise ratioBioreactor/fermenter combinationsSequential/parallel process reactionsElectricityDNA microarray
A DNA microarray system whereby measurement can be performed at a low running cost, a low price and yet a high accuracy. A nucleic acid probe (3) is immobilized on the surface of a gate insulator of an electric field effect transistor and then hybridized with a target gene on the surface of the gate insulator. A change in the surface electric charge density thus arising is detected by using the electric effect.
Owner:HITACHI HIGH-TECH CORP
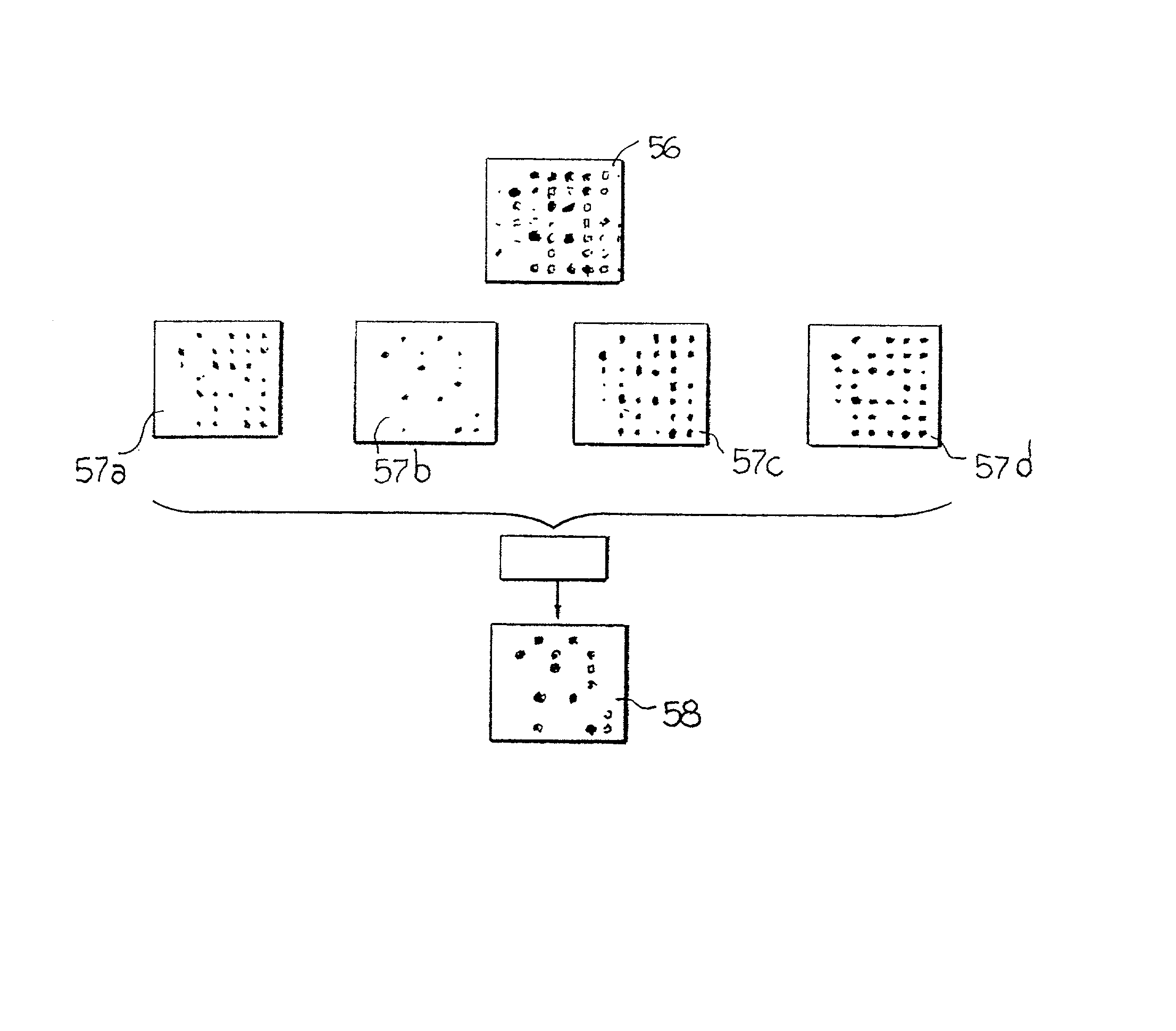
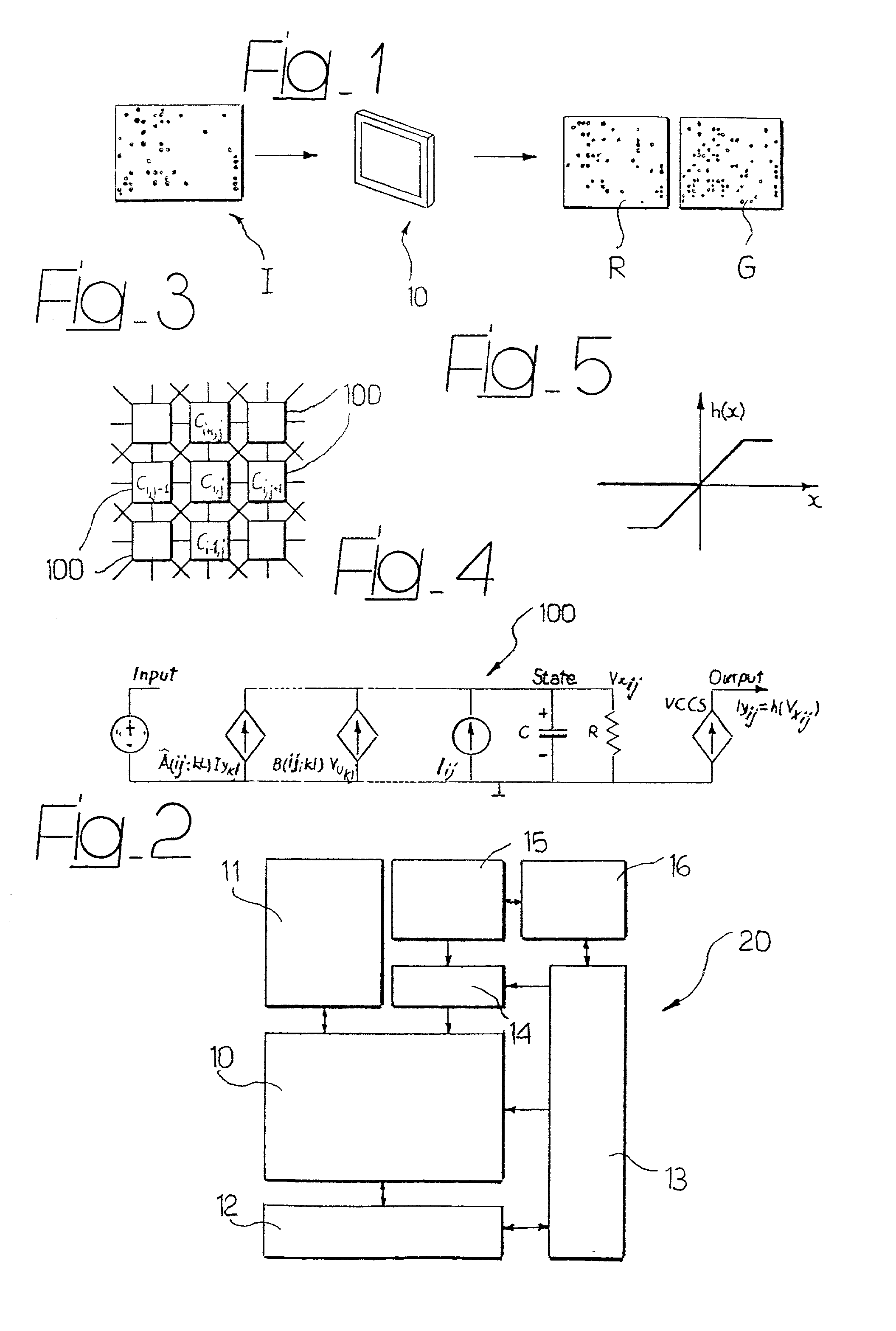
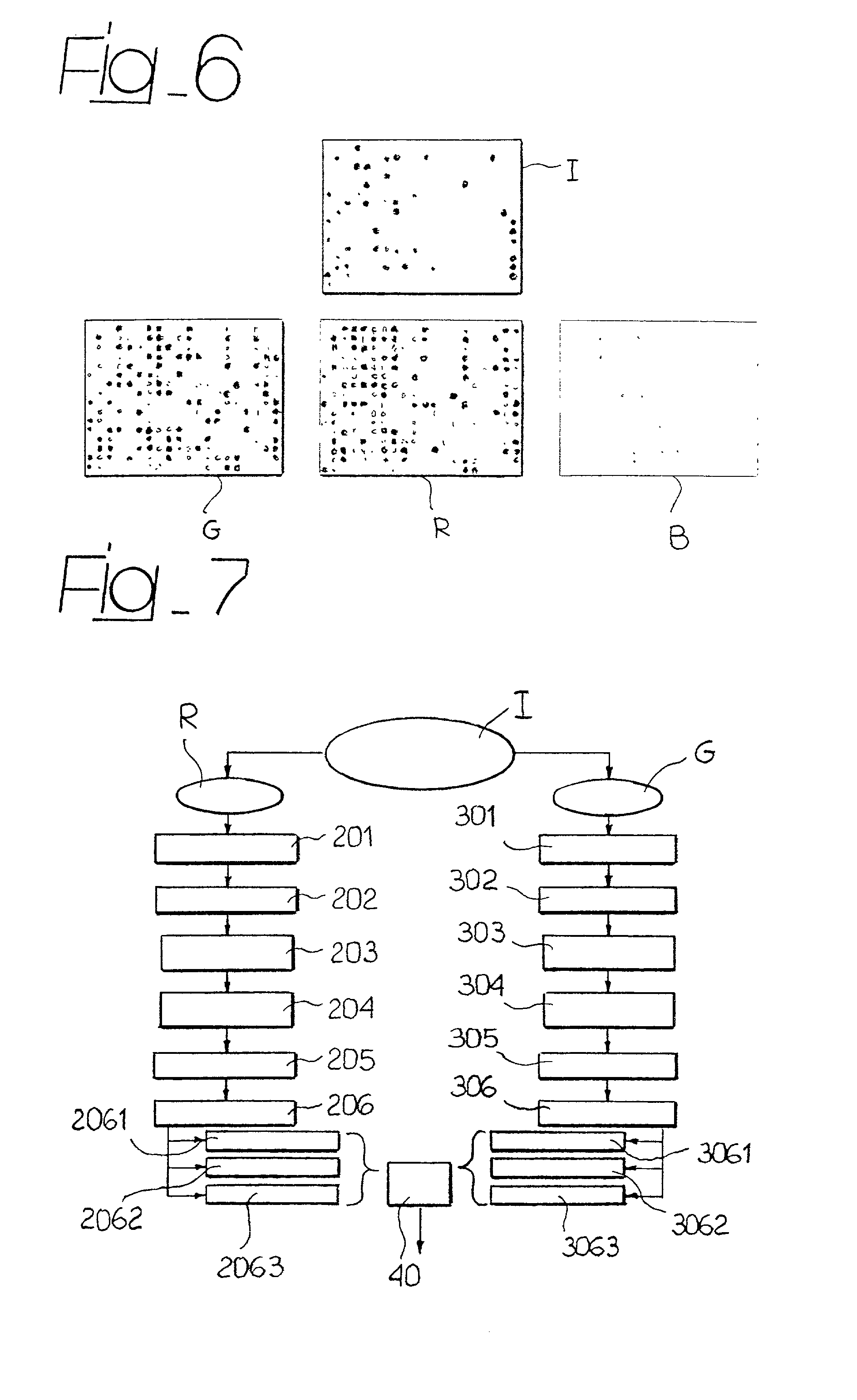
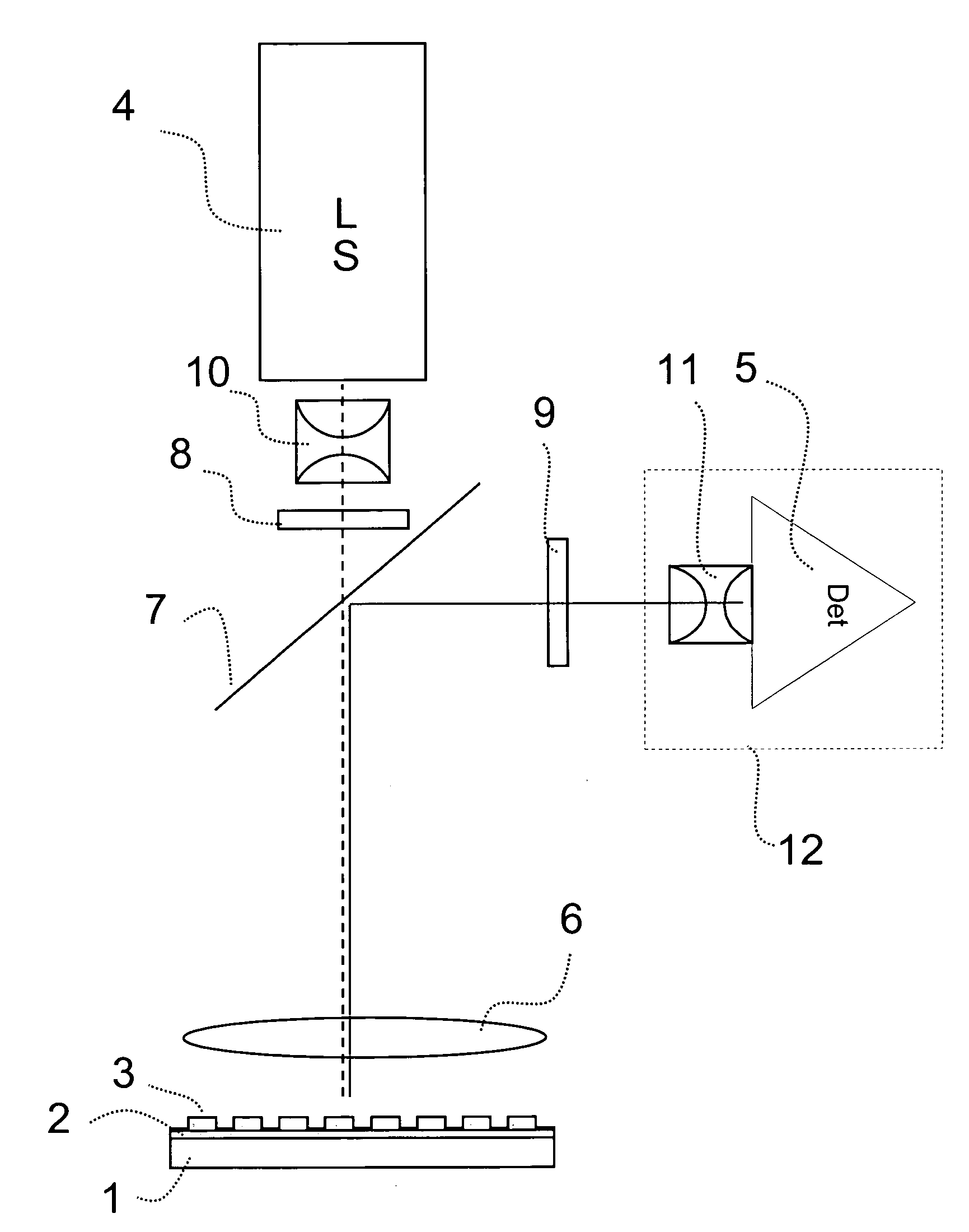
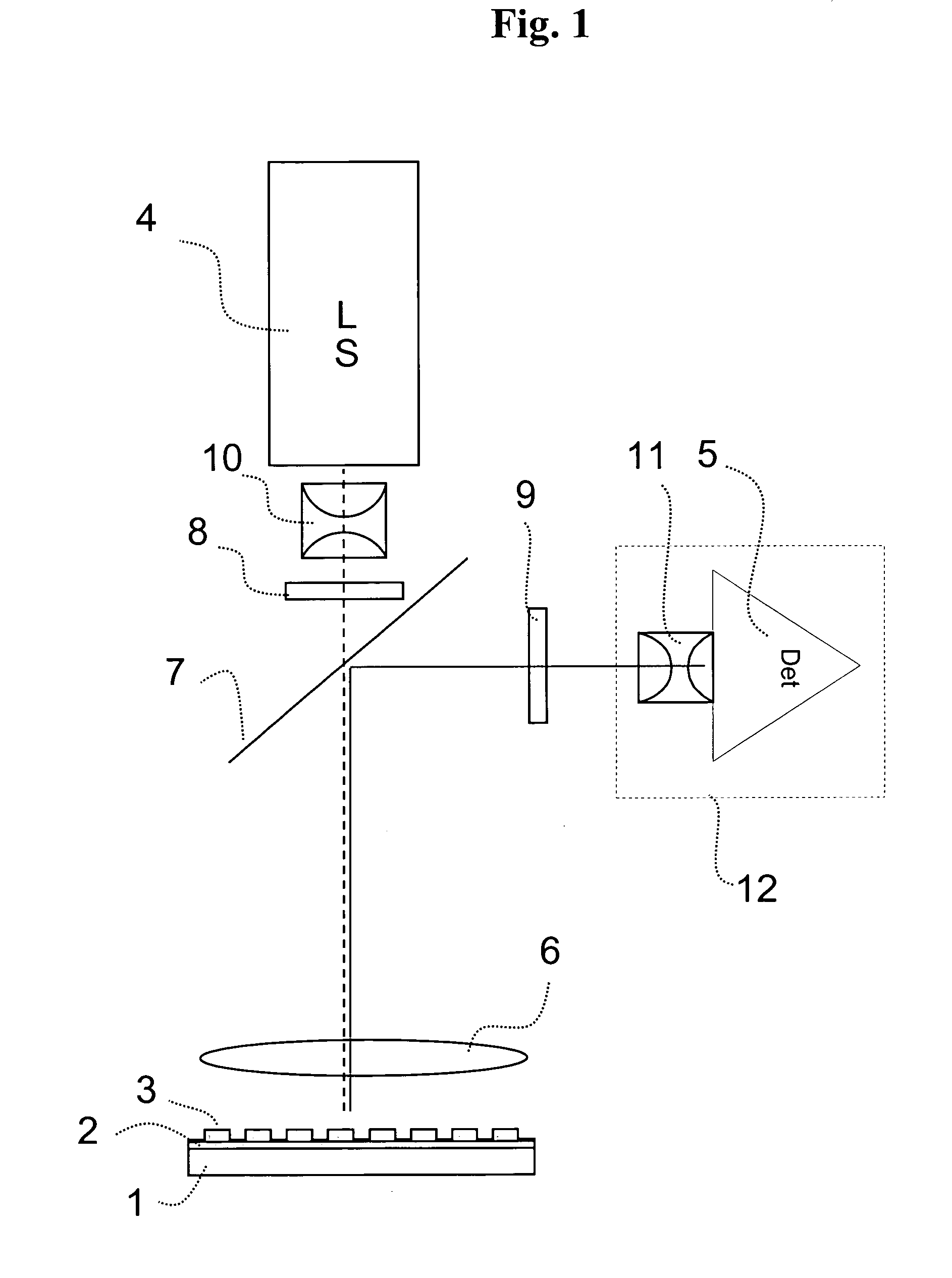
System for the automatic analysis of images such as DNA microarray images
InactiveUS20020097900A1Efficient and rapid automatic analysis of imageImage enhancementBioreactor/fermenter combinationsCMOSDNA microarray
The system can be used for the automatic analysis of images (I), comprising a matrix of spots, such as images of DNA microarrays after hybridisation. The system can be associated-and preferably integrated in a single monolithic component implementing VLSI CMOS technology-to a sensor (10) for acquiring said images (I). The system comprises a circuit (20) for processing the signals corresponding to the images (I), configured according to a cellular neural network (CNN) architecture for the parallel analogue processing of signals.
Owner:STMICROELECTRONICS SRL
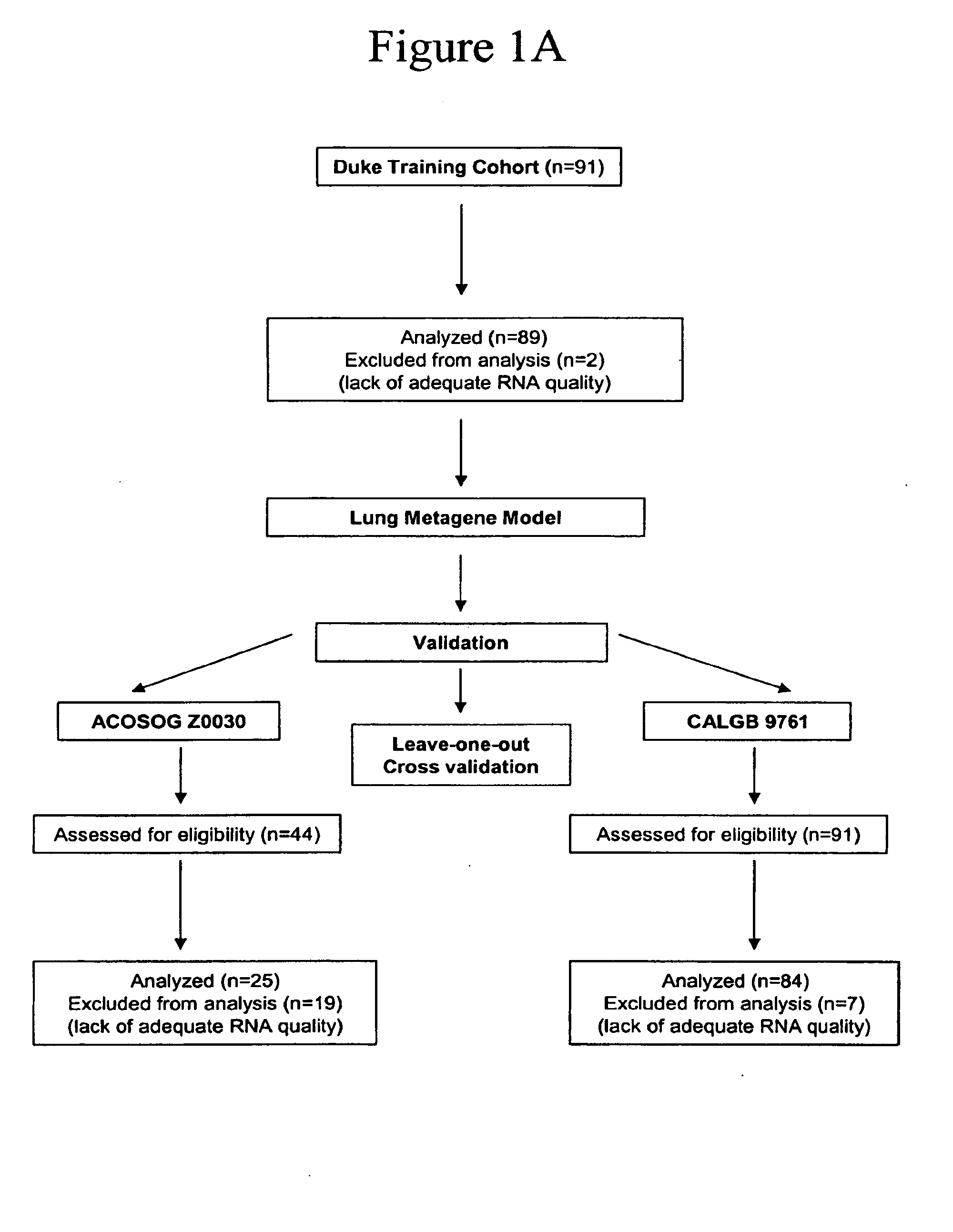
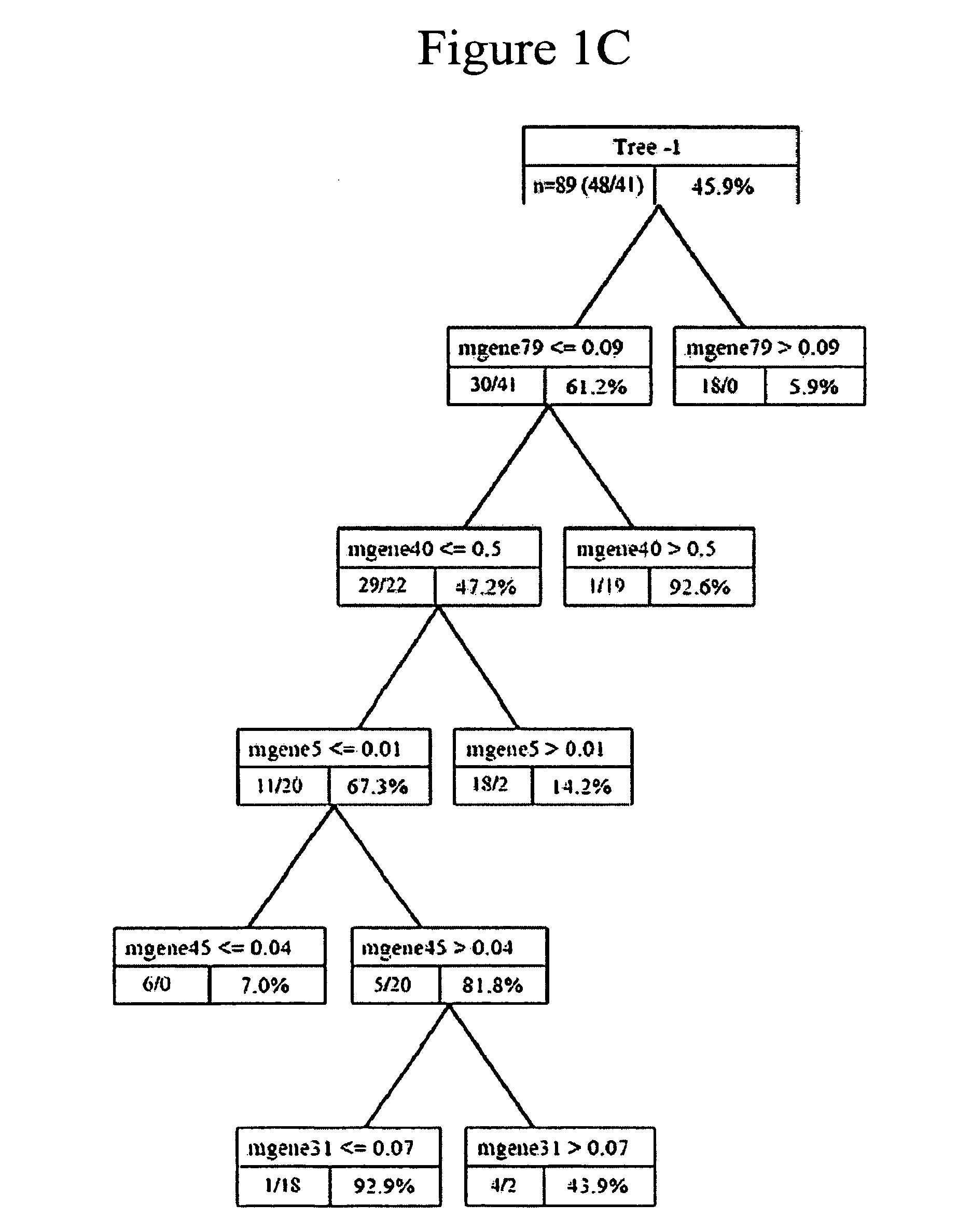
Prediction of lung cancer tumor recurrence
InactiveUS20100009357A1Accurate prognosisRisk of recurrenceMicrobiological testing/measurementDNA microarrayTumor recurrence
The invention provides methods of estimating the likelihood of lung cancer recurrence in a subject, including those afflicted with NSCLC. The methods of the invention are useful for developing a therapeutic treatment plan to prevent cancer recurrence for subjects deemed to be at high risk, and withholding treatments from those subjects deemed to be at low risk. The invention also provides methods of generating and using metagene-based prediction tree models for estimating the likelihood of lung cancer recurrence. The invention also provides reagents, such as DNA microarrays, software and computer systems useful for estimating cancer recurrence, and provides methods of conducting a diagnostic business for the prediction of cancer recurrence.
Owner:DUKE UNIV
Microarray synthesis instrument and method
InactiveUS20070037274A1Minimizes reflected lightEfficient use ofBioreactor/fermenter combinationsMaterial nanotechnologyDNA microarrayOligomer
During the light illumination period of a monomer addition cycle in synthesizing an DNA microarray, undesirable reflections of illumination light from various interfaces that the illumination light passes through near the synthesis surface of the substrate may reduce the light-dark contrast, and negatively affect the precision and resolution of the microarray synthesis. The present invention provides an flow cell that reduces the undesired reflections by constructing certain flow cell structures with materials that have similar refractive indexes as that of the solution that is in the oligomer synthesis chamber during the illumination period and / or constructing certain flow cell structures or covering the structures with a layer of a material that has a high extinction coefficient.
Owner:ROCHE NIMBLEGEN
DNA microarray having hairpin probes tethered to nanostructured metal surface
InactiveUS20090137418A1Avoid and minimize needFluorescence enhancementMaterial nanotechnologySequential/parallel process reactionsDNA microarraySurface plasmon
A sensor chip and detection device are disclosed. The sensor chip includes a substrate, at least a portion of which is covered by a metal nanoparticle film; a first nucleic acid molecule that is characterized by being able to (i) self-anneal into a hairpin conformation and (ii) hybridize specifically to a target nucleic acid molecule, the first nucleic acid molecule having first and second ends, which first end is tethered to the metal nanoparticle film; and a first fluorophore bound to the second end of the first nucleic molecule. When the first nucleic acid molecule is in the hairpin conformation, the metal nanoparticle film substantially quenches fluorescent emissions by the first fluorophore, and when the first nucleic acid molecule is in a non-hairpin conformation fluorescent emissions by the first fluorophore are surface plasmon-enhanced.
Owner:UNIVERSITY OF ROCHESTER
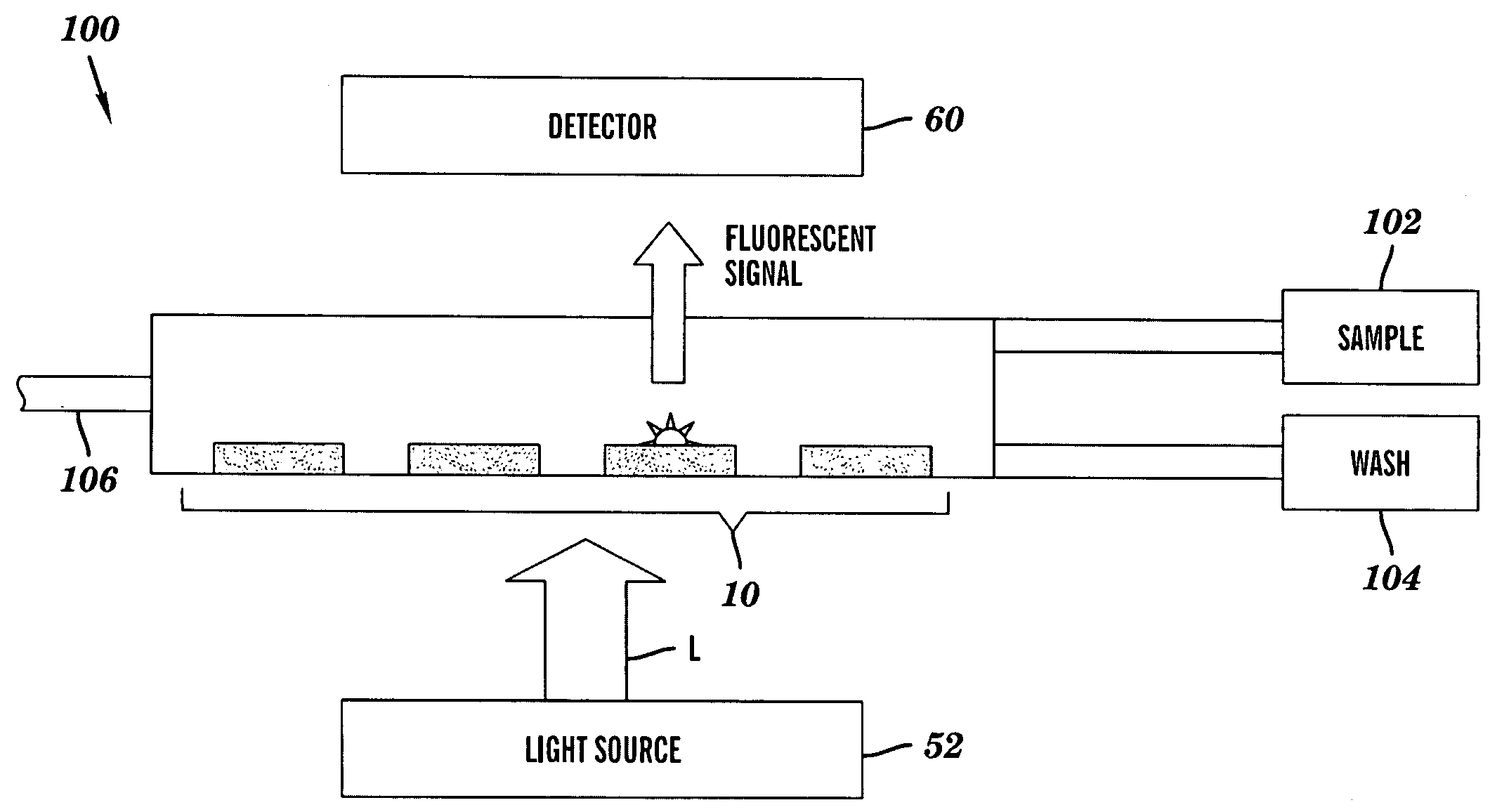
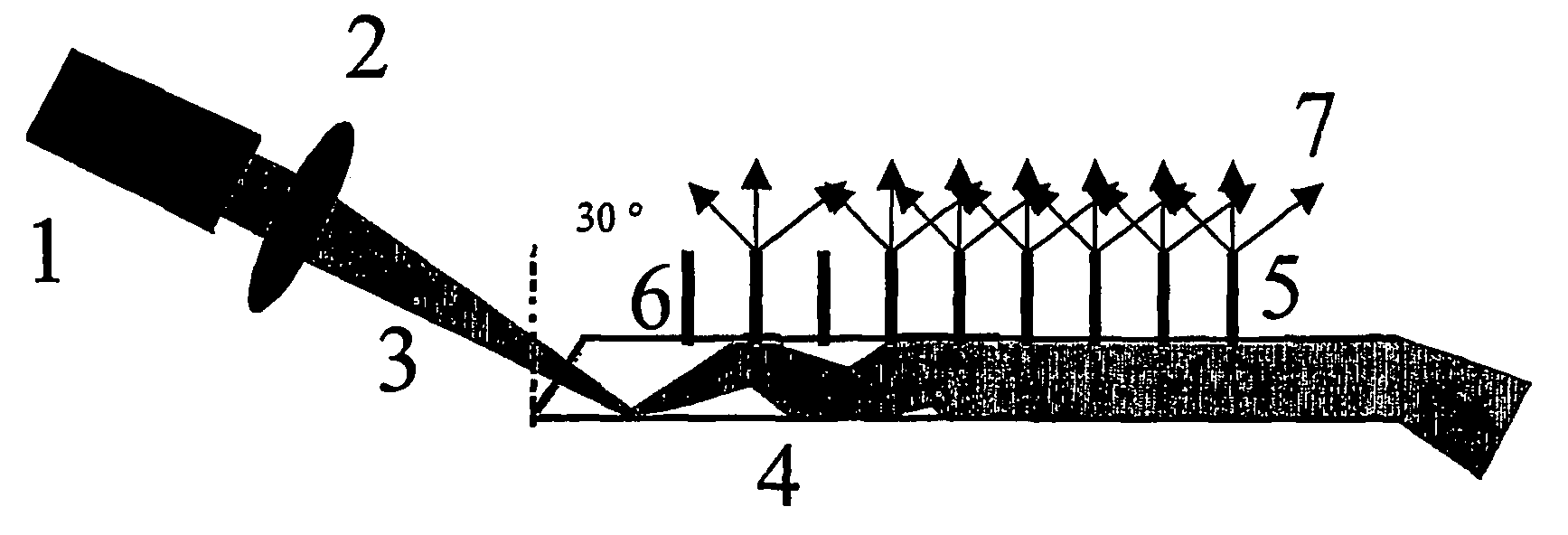
Imaging fluorescence signals using telecentric optics
ActiveUS7687260B2Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA microarrayParallel imaging
The present invention relates to the field of DNA analysis. In particular, the present invention is directed to a device for the parallel imaging of fluorescence intensities at a plurality of sites as a measure for DNA hybridization. More particular, the present invention is directed to a device to image multiplex real time PCR or to read out DNA microarrays.
Owner:ROCHE DIAGNOSTICS OPERATIONS INC
Method of error reduction in nucleic acid populations
InactiveUS7303872B2Minimum errorSugar derivativesMicrobiological testing/measurementDNA microarrayError reduction
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The complement of each segment is also made in the microarray. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will hybridize to form duplexes. The duplexes are then separated using a DNA binding agent which binds to improperly formed DNA helixes to remove errors from the set of DNA molecules. The segments can then be hybridized to each other to assemble the larger target DNA sequence.
Owner:WISCONSIN ALUMNI RES FOUND
Method for the synthesis of DNA sequences
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will spontaneously hybridize together to form the desired DNA construct. This method makes possible the remote assembly of DNA sequence, through a process analogous to facsimile transmission of documents, since the information on DNA to be made can be transmitted remotely to an instrument which can then synthesize any needed DNA sequence from the information.
Owner:WISCONSIN ALUMNI RES FOUND
Method For The Analysis Of Point Mutations
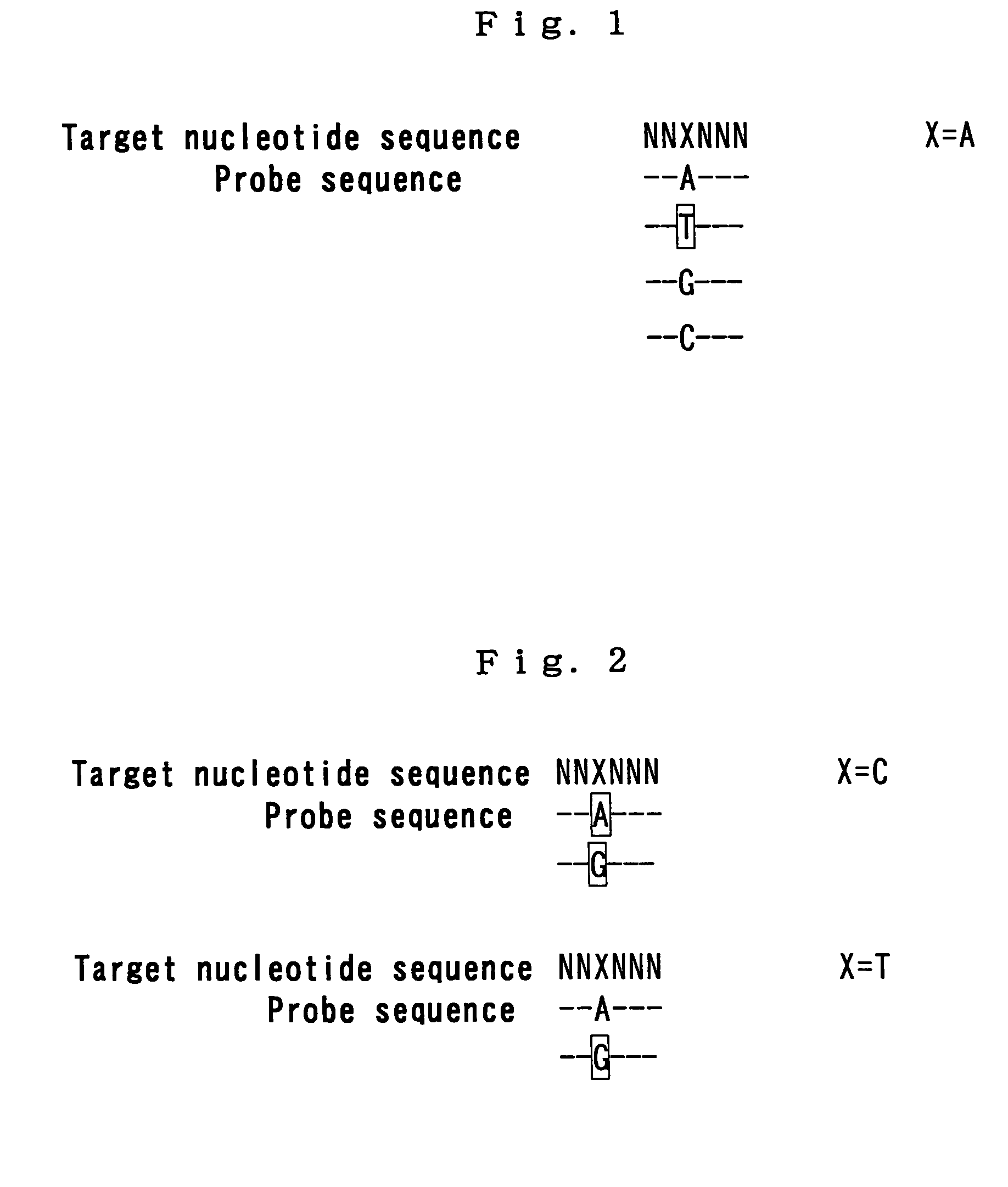
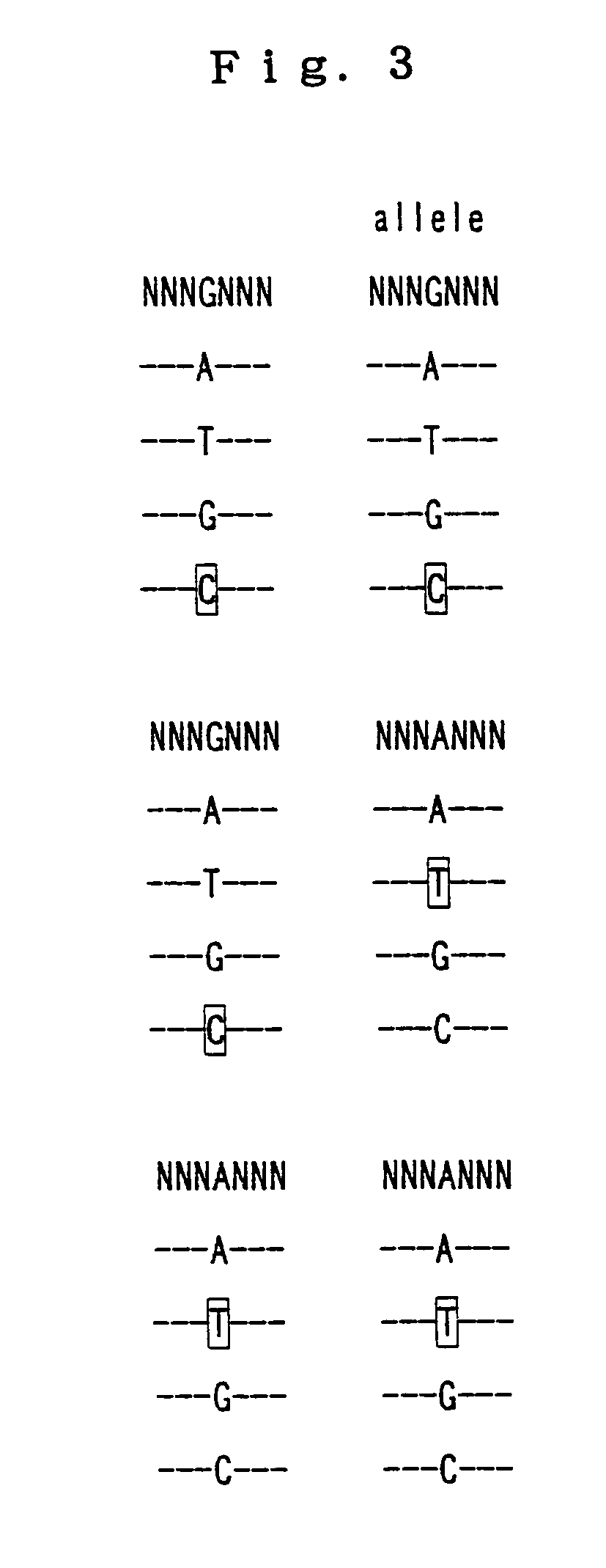
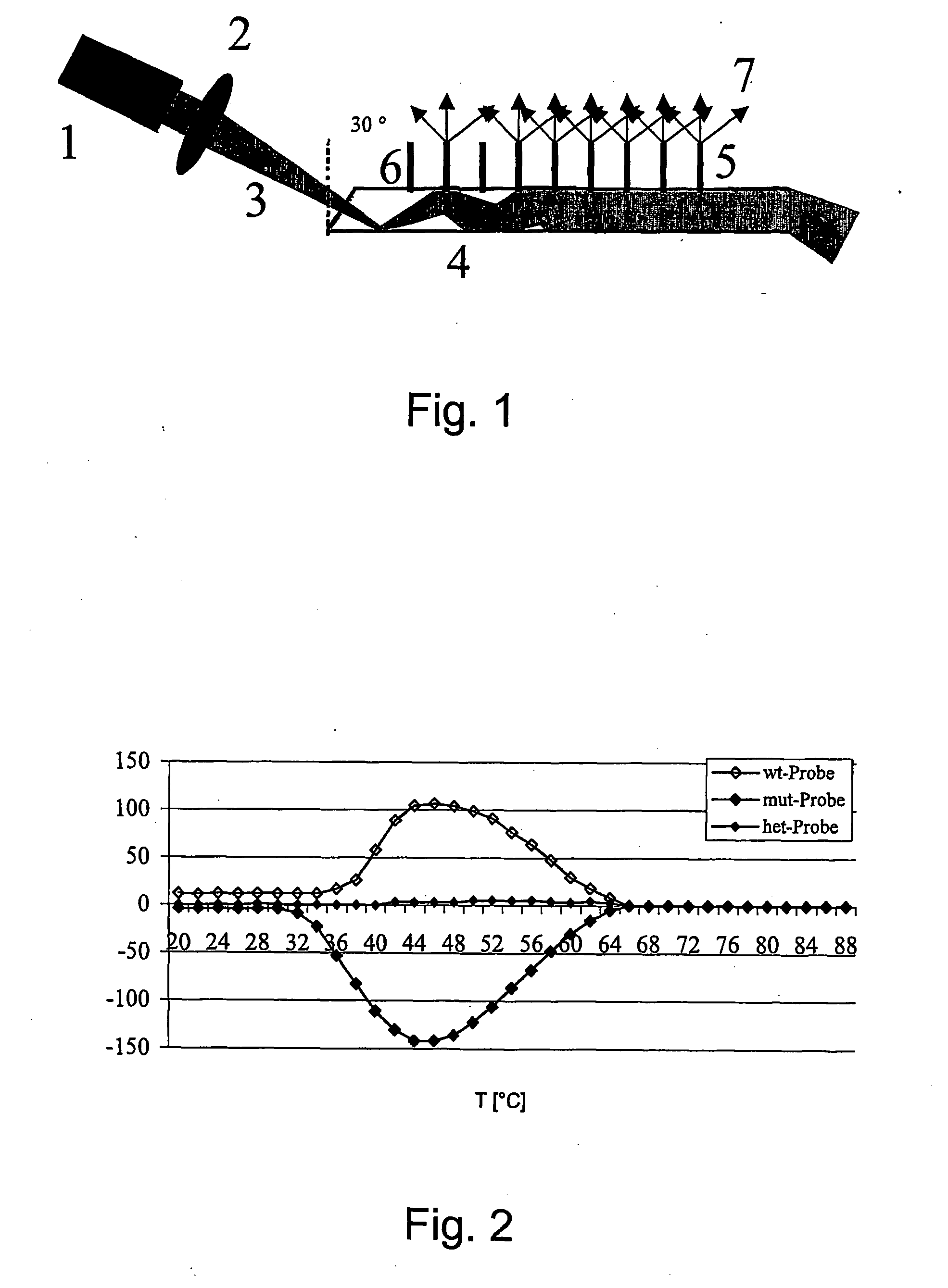
The invention relates to a method for determining point mutations by means of DNA microarrays and TIRF excitation. According to said method, bonding of nucleic acids to short DNA probes on a microarray is measured at different temperatures. Melting point curves are generated from the measured values and the difference in the melting point curves between the probe for the wild-type DNA and the probe for the corresponding mutated DNA is generated. The position of said curves makes it possible to unambiguously decide whether the point mutation is a homozygous DNA or a heterozygous DNA and what type of homozygosity it is.
Owner:KLAPPROTH HOLGER
Process for assay of nucleic acids by competitive hybridization using a dna microarray
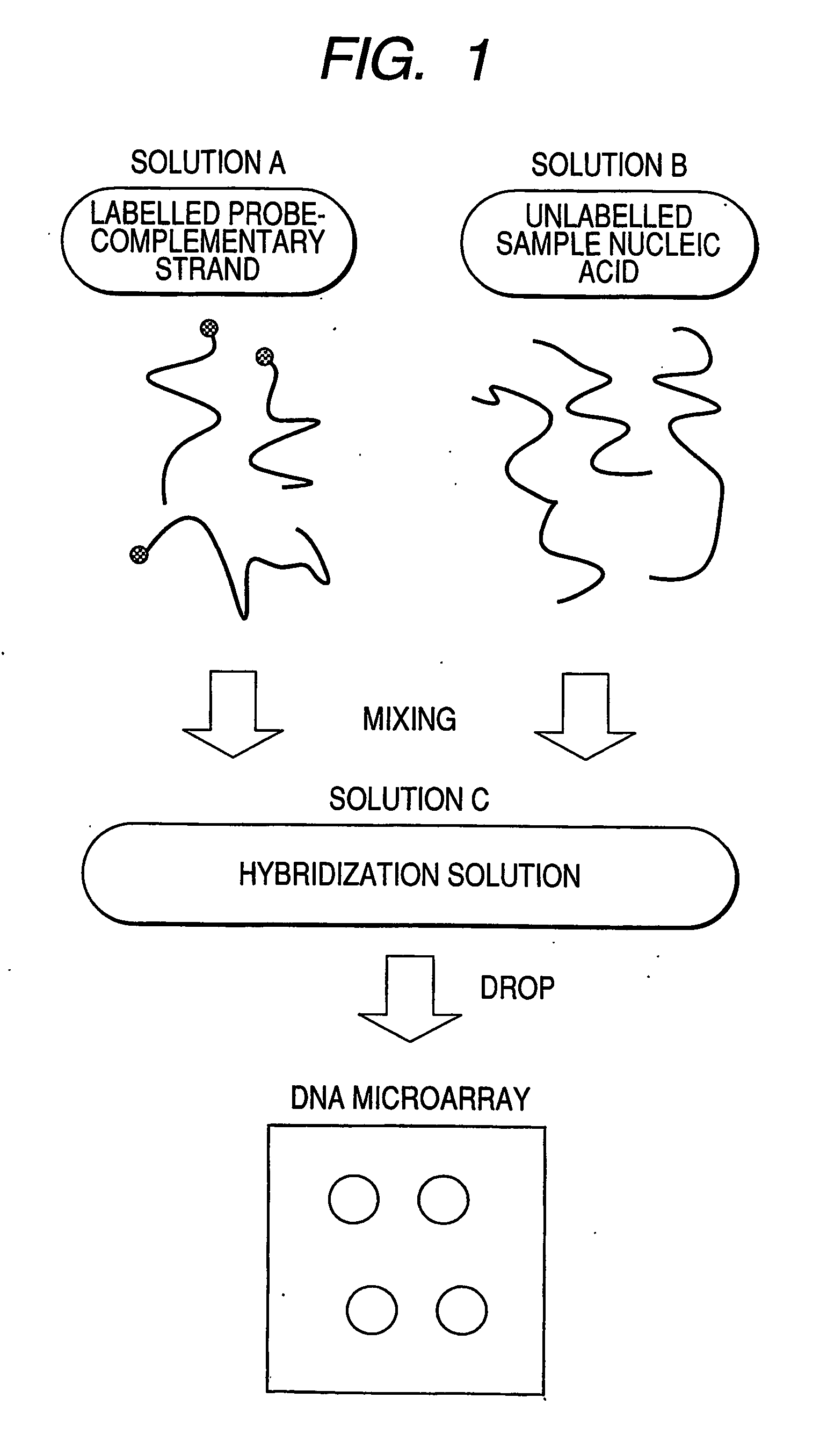
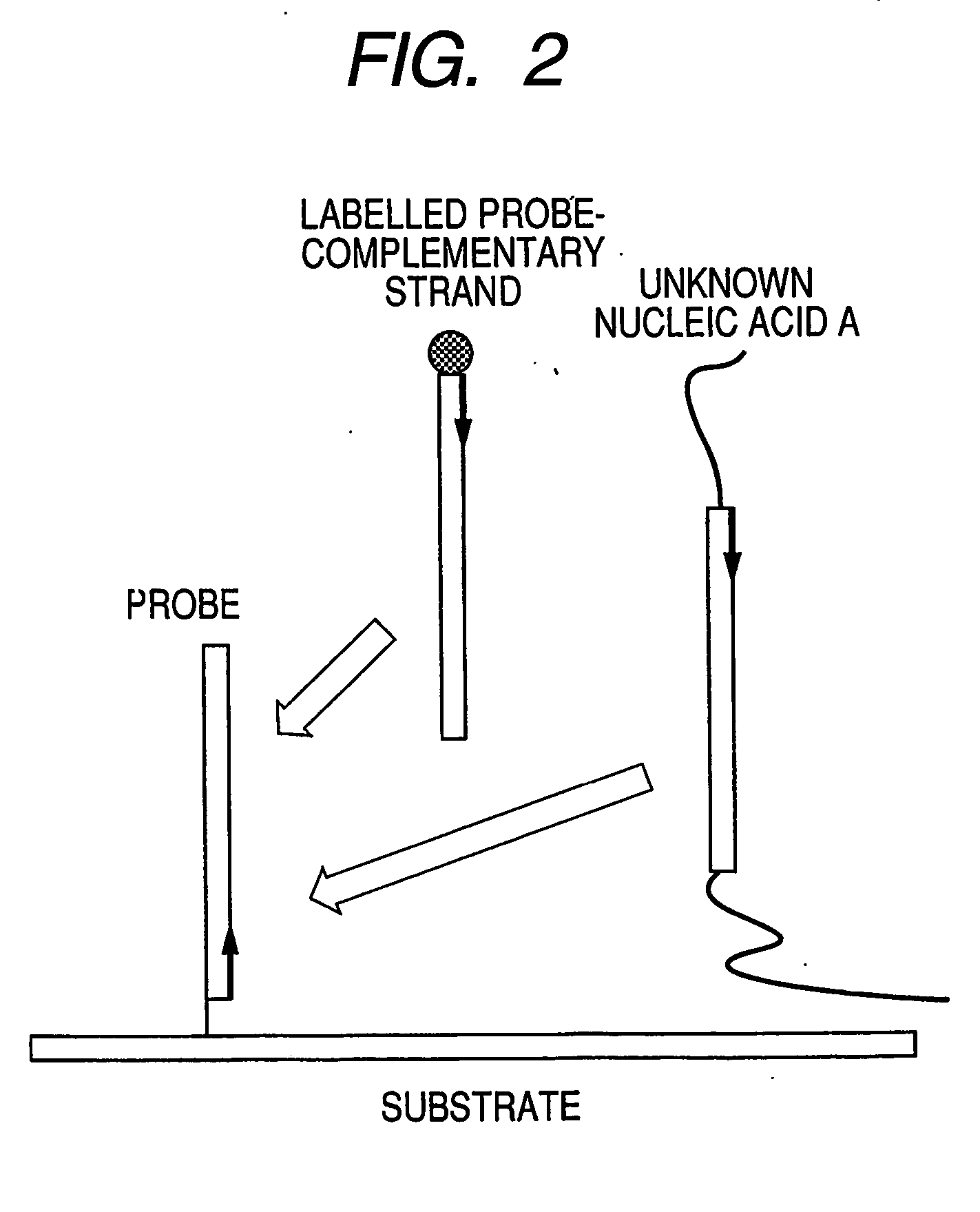
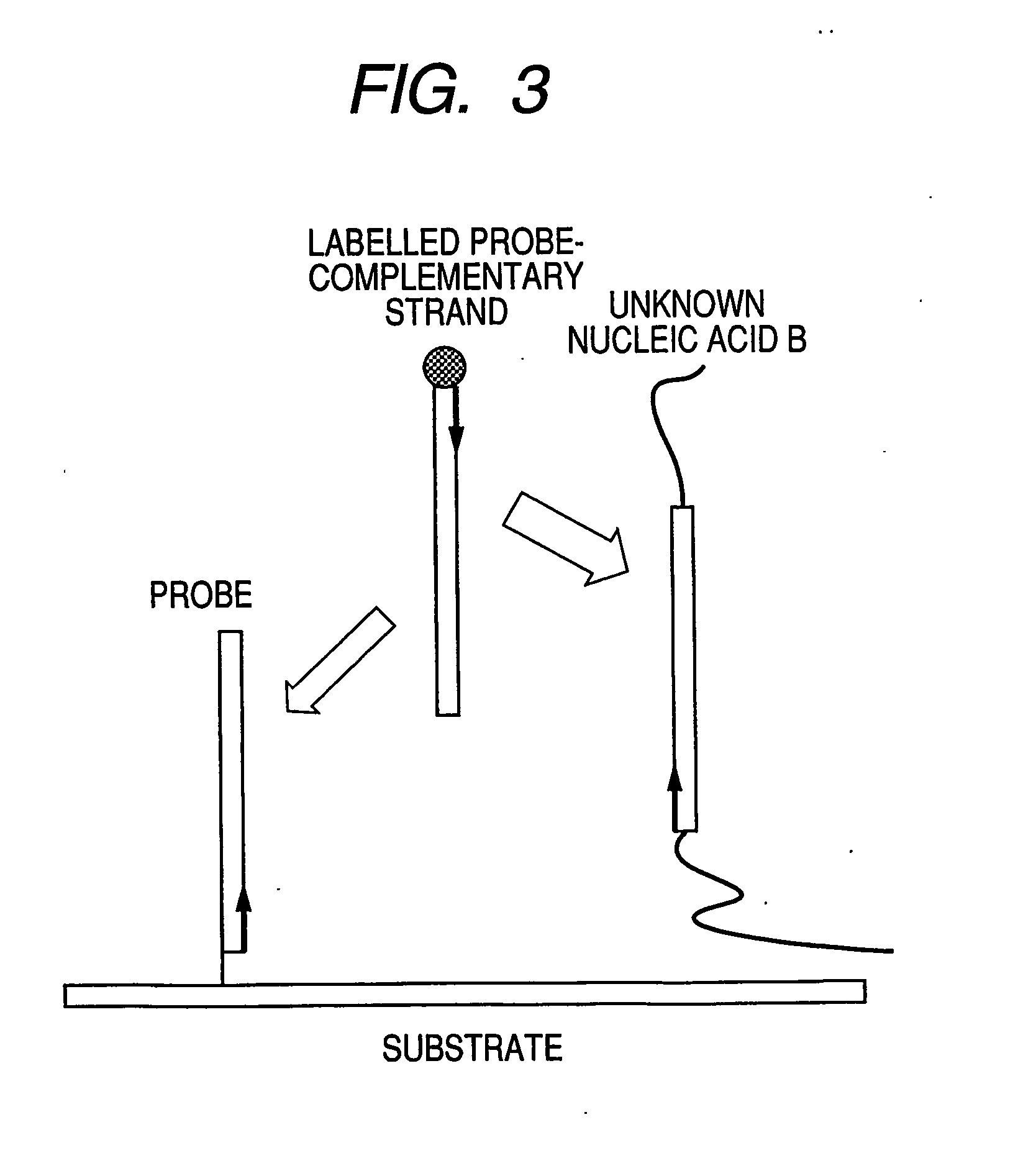
InactiveUS20070105100A1Shorten the timeLabor savingMicrobiological testing/measurementDNA microarrayA-DNA
A process for rapid and simple assay of nucleic acid on the basis of competitive hybridization using a DNA microarray that comprises a step of hybridizing a labeled nucleic acid other than the target nucleic acid with one of the immobilized probes on the substrate, where the labeled nucleic acid has a known sequence complementary to the probe and can bind thereto specifically.
Owner:CANON KK
Novel microarray techniques for nucleic acid expression analyses
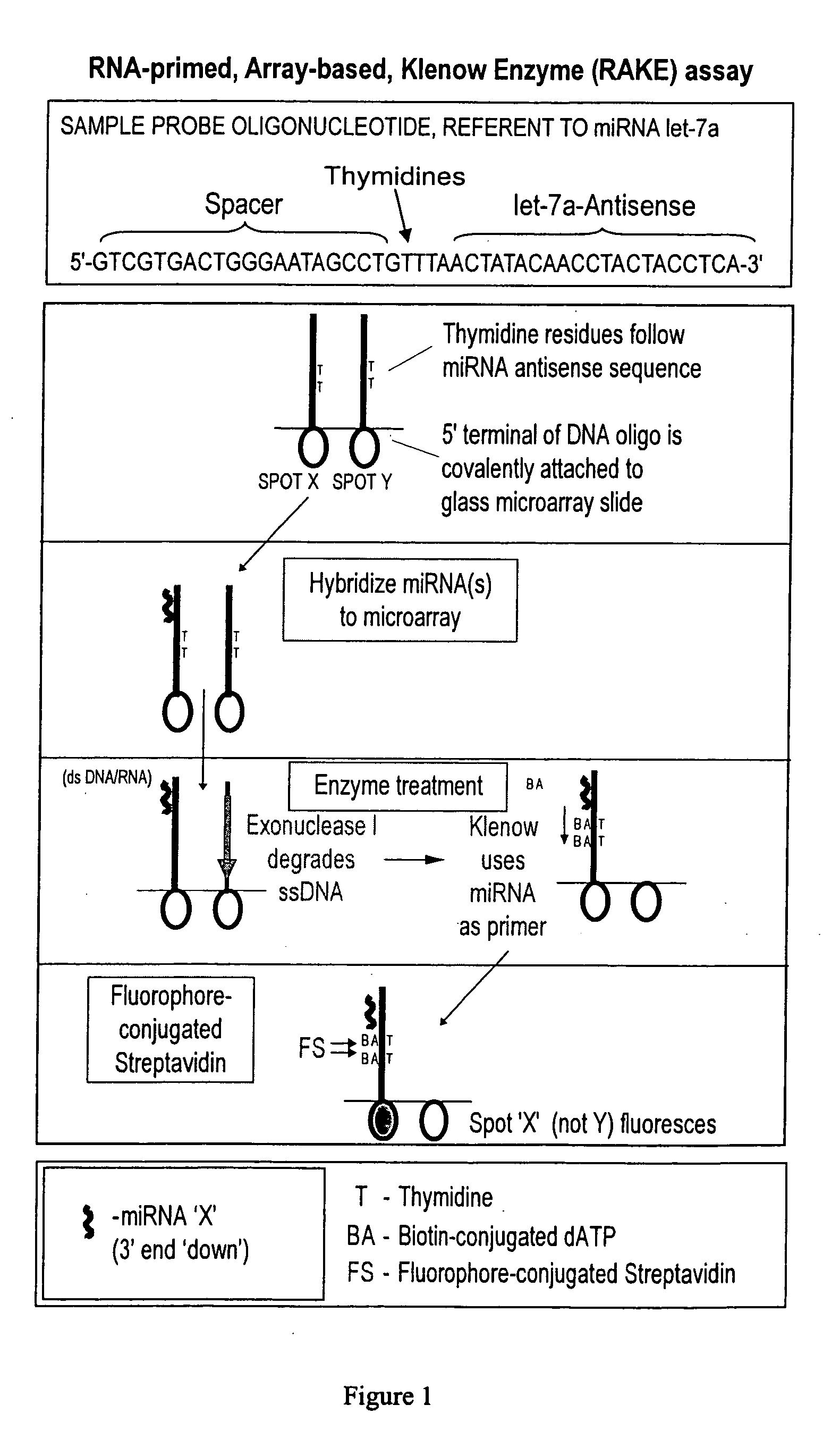
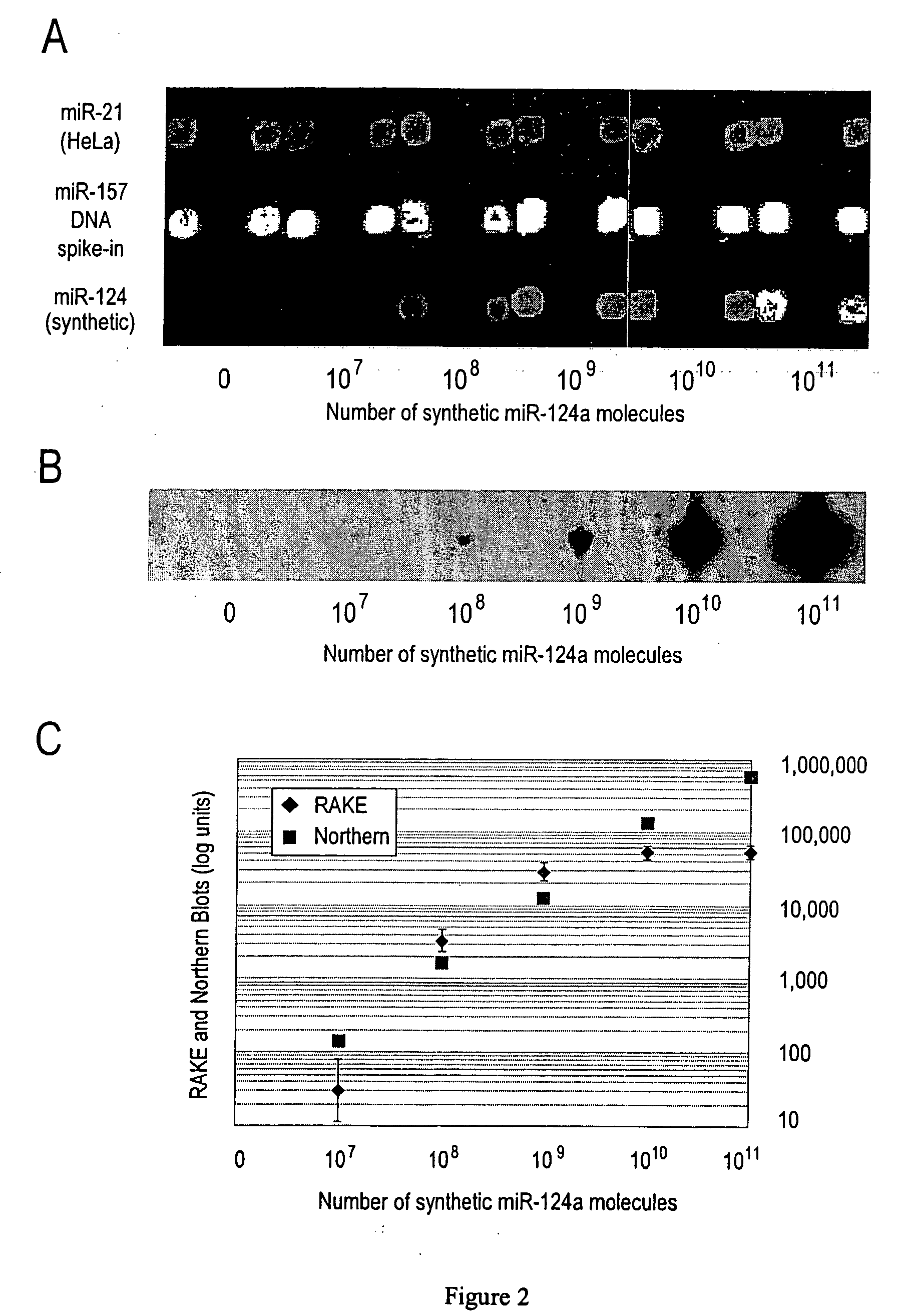
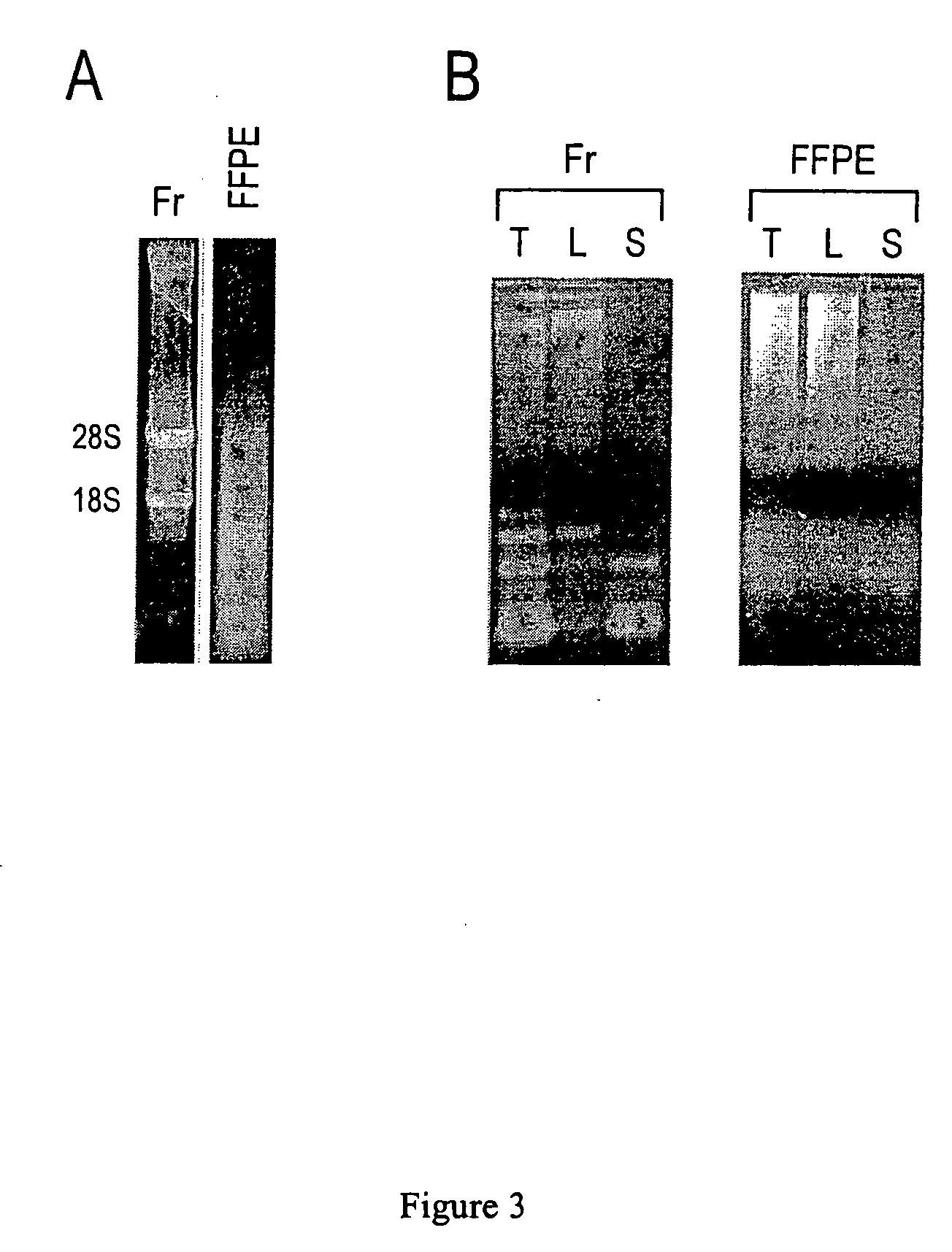
ActiveUS20060078925A1Eliminate biasConvenient experimentMicrobiological testing/measurementFermentationMiRNA GeneExonuclease I
Provided are DNA microarray techniques that allow hybridization without RNA amplification, without using cDNA, and without labeling the nucleic acid prior to hybridization. Referred to as the Double-stranded Exonuclease Protection (DEP) assay, the technique permits the sample RNA to be used directly for hybridization, without manipulation in any way. Further provided is a microarray technique for high-throughput miRNA gene expression analyses, termed the RNA-primed, Array-based, Klenow Enzyme (RAKE) assay. The RAKE assay is a sensitive and specific technique for assessing single-stranded DNA and RNA targets, and offers specific advantages over Northern blots.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Methods and mechanisms involving hyperpigmentation particularly for african american skin
Sets of genes are identified that show modulated activity in hyperpigmented sun-exposed (HE) and non-hyperpigmented sun-exposed (NHE) skin, when compared to non-hyperpigmented non-exposed (NHNE) skin. The modulated sets of genes reveal important information about the genetic changes that take place in skin as a result of environmental exposure and damage. The modulated sets of genes may be used to fabricate custom DNA microarrays for evaluating patients with skin diseases or disorders. The microarrays may also be used to screen new substances for treating skin diseases and disorders. The modulated gene sets, and substances that target them, may also be used to develop therapies for individuals who suffer from hypopigmentation, such as those with Fitzpatrick type I skin or vitiligo.
Owner:HAMPTON UNIVERSITY
Reusable substrate for DNA microarray production
InactiveUS7955798B2Sequential/parallel process reactionsElectrolysis componentsDNA microarrayBiopolymer
The invention relates to a method for producing biopolymer arrays comprising a porous membrane and bound biopolymers. In particular, said production of biopolymer arrays comprises electrochemical production cycles.
Owner:ROCHE DIAGNOSTICS OPERATIONS INC
Imaging fluorescence signals using telecentricity
ActiveUS20060194308A1Accurate imagingBioreactor/fermenter combinationsRadiation pyrometryDNA microarrayParallel imaging
The present invention relates to the field of DNA analysis. In particular, the present invention is directed to a device for the parallel imaging of fluorescence intensities at a plurality of sites as a measure for DNA hybridization. More particular, the present invention is directed to a device to image multiplex real time PCR or to read out DNA microarrays.
Owner:ROCHE DIAGNOSTICS OPERATIONS INC
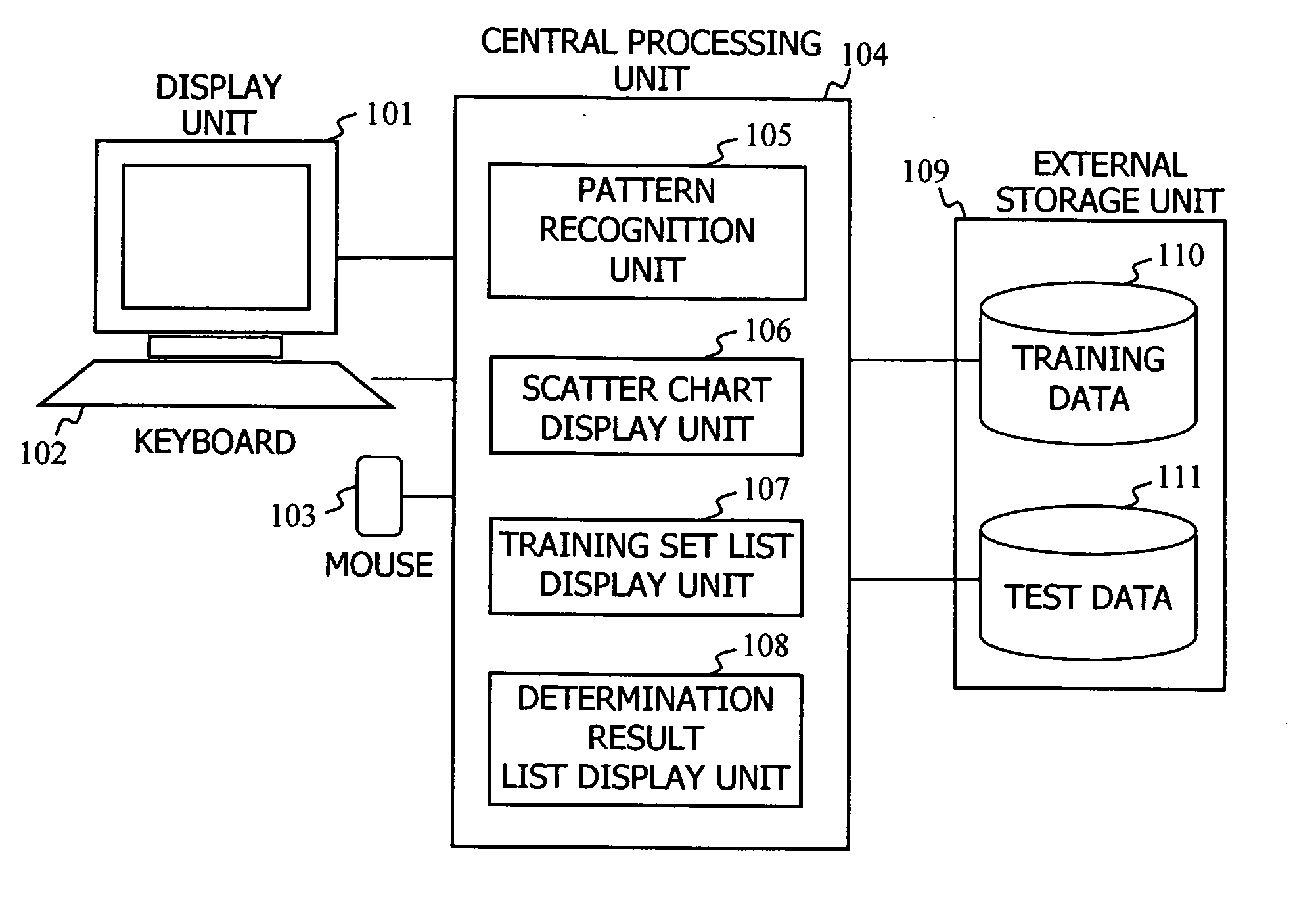
Pattern recognition system utilizing an expression profile
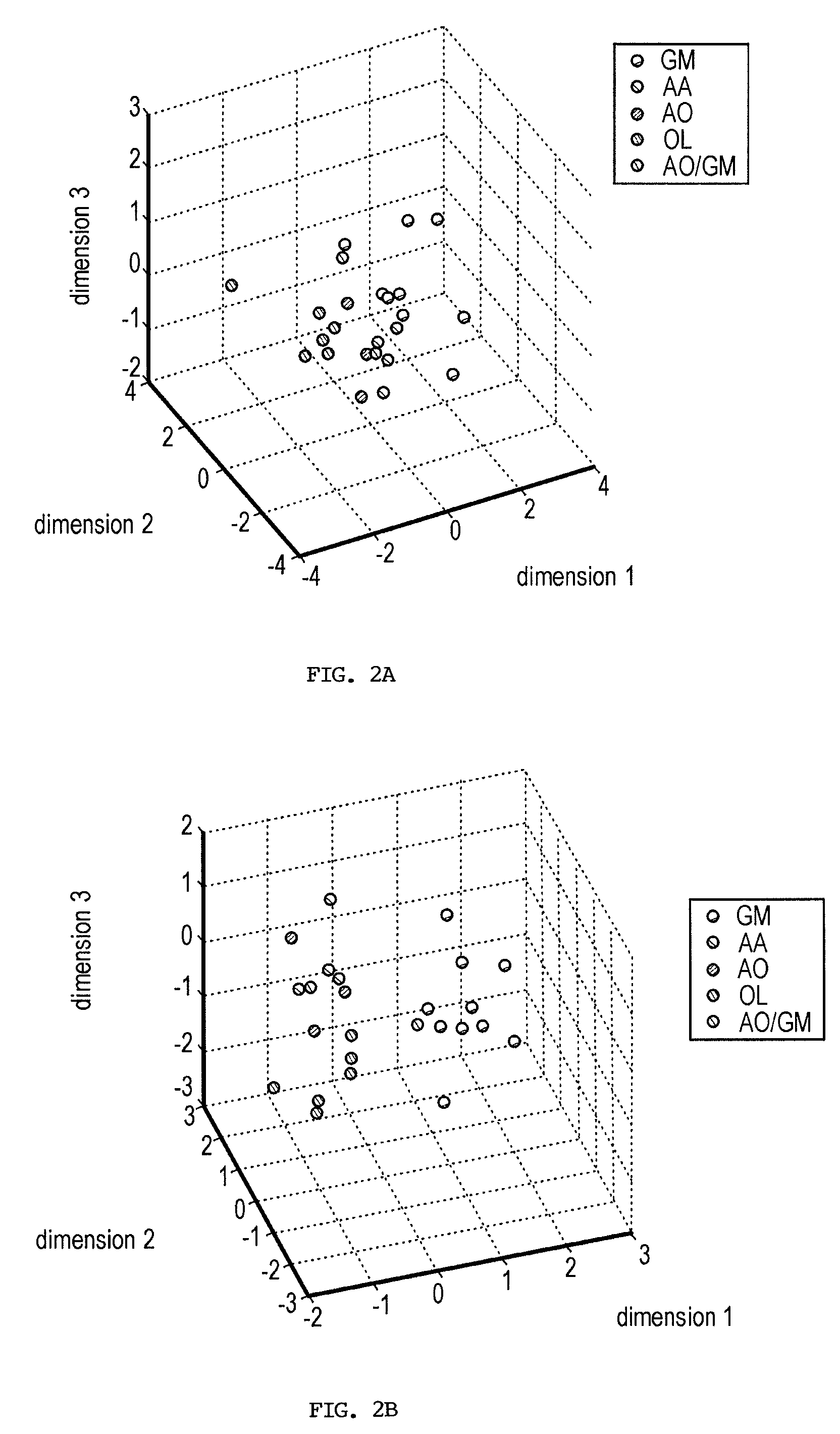
InactiveUS20050276485A1Data visualisationCharacter and pattern recognitionDNA microarrayMultidimensional data
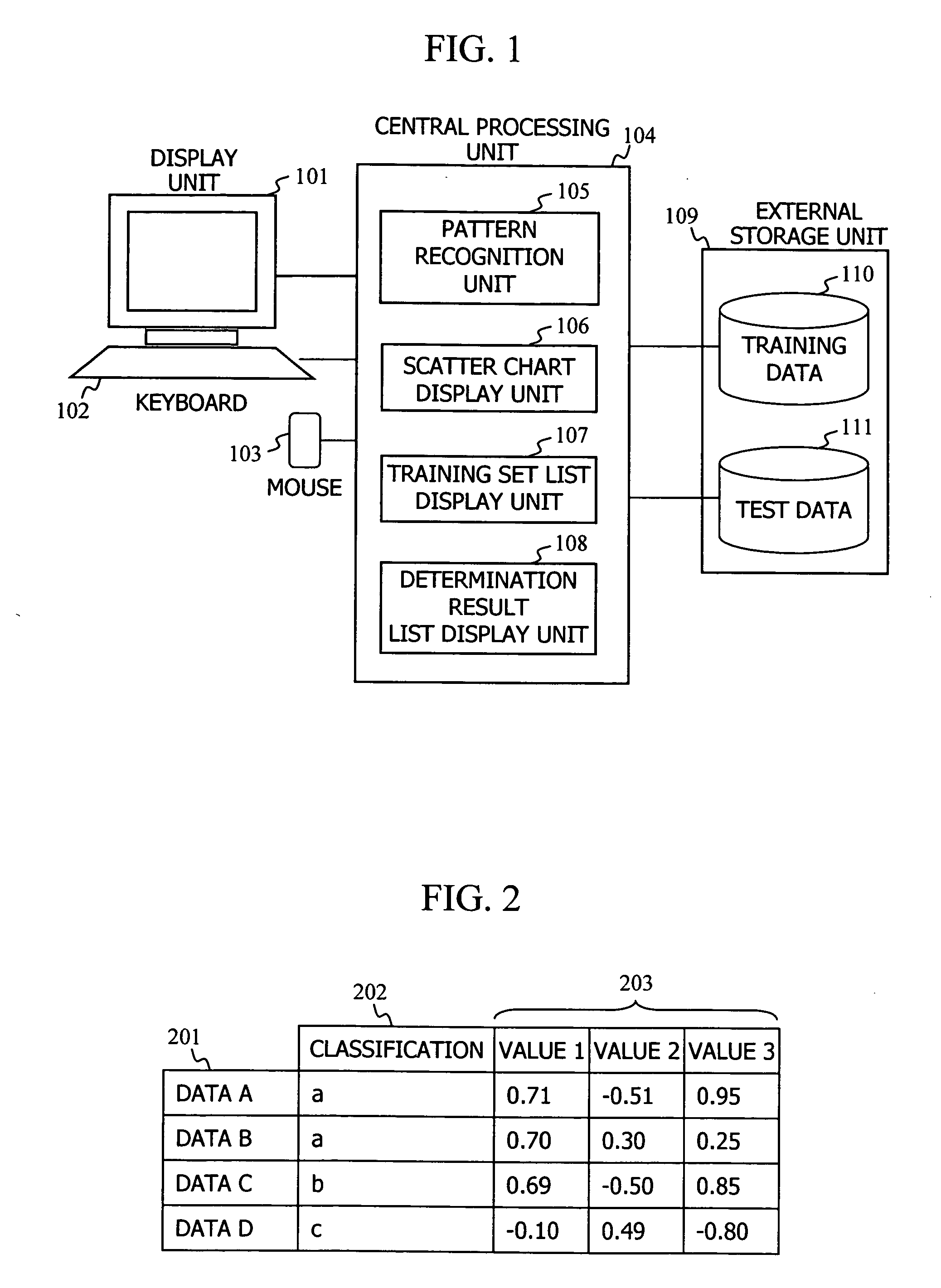
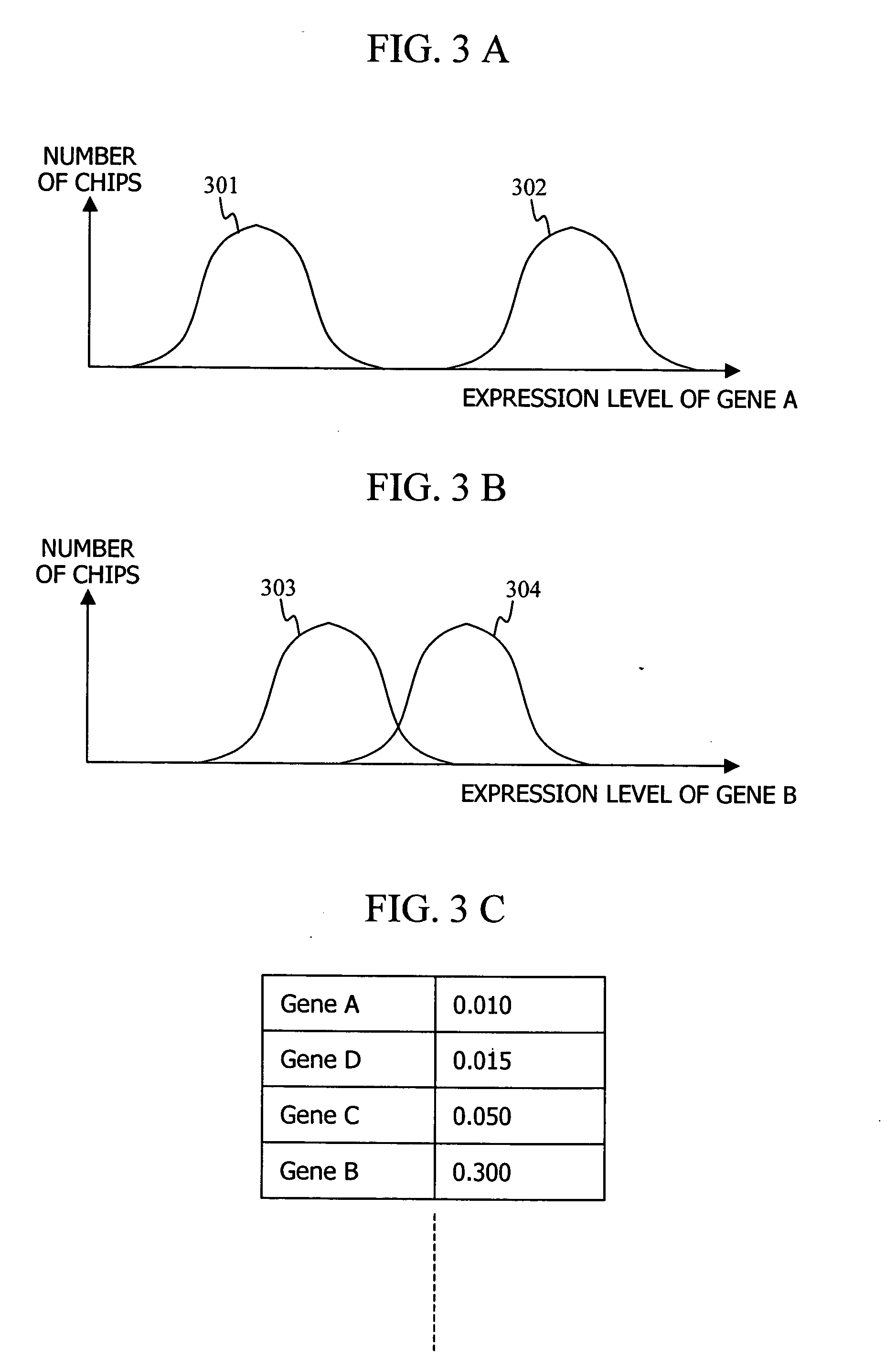
When making a clinical diagnosis using gene expression profiles or the like obtained from a DNA microarray, multidimensional data is visualized on a scatter chart so that outliers can be identified and the state of classifications can be recognized. A method comprises calculating said separating hyperplane by applying said pattern recognition algorithm to said training set that is entered; displaying the labels of two axes of said scatter chart in two or three dimensions; applying data of which the group it belongs to is unknown to said pattern recognition algorithm as a test set in order to determine the group the data belongs to; displaying a plot representing the data in said training set and a plot representing the data in said test set on a two- or three-dimensional scatter chart, in different manners for individual groups; and displaying said separating hyperplane by mapping it to said scatter chart.
Owner:HITACHI SOFTWARE ENG
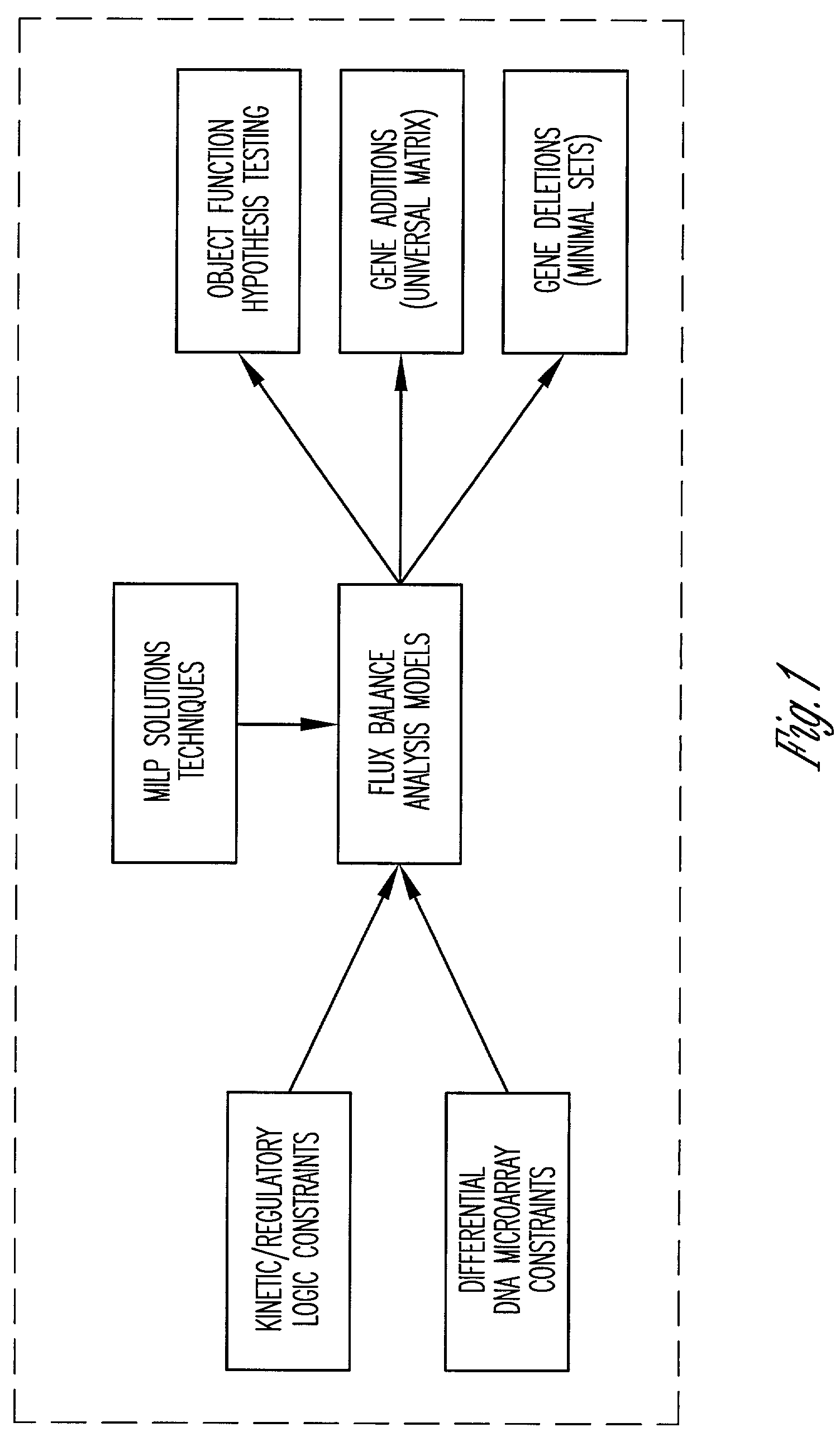
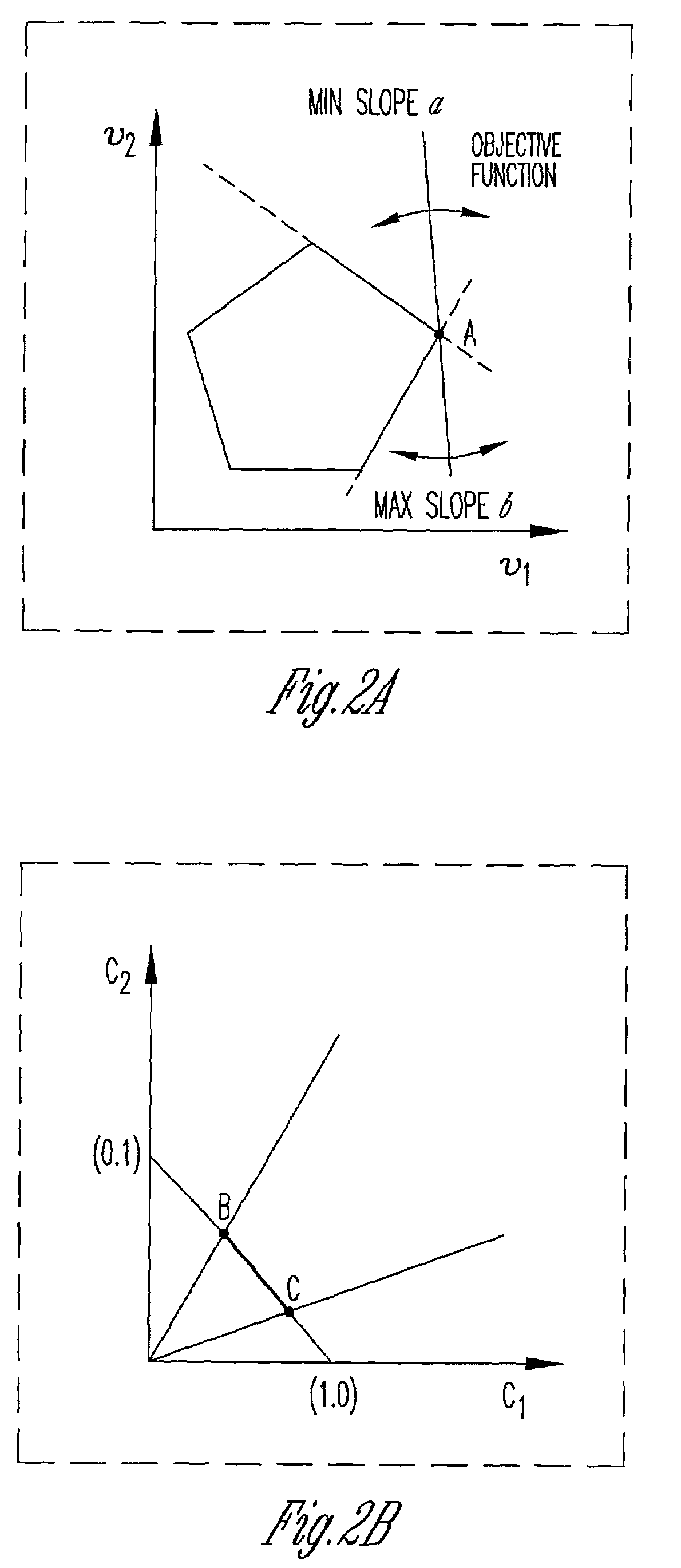
Method and system for modeling cellular metabolism
This invention relates to methods and systems for in silico or bioinformatic modeling of cellular metabolism. The invention includes methods and systems for modeling cellular metabolism of an organism, comprising constructing a flux balance analysis model, and applying constraints to the flux balance analysis model, the constraints selected from the set consisting of: qualitative kinetic information constraints, qualitative regulatory information constraints, and differential DNA microarray experimental data constraints. In addition, the present invention provides for computational procedures for solving metabolic problems.
Owner:PENN STATE RES FOUND
Gene sets for glioma classification
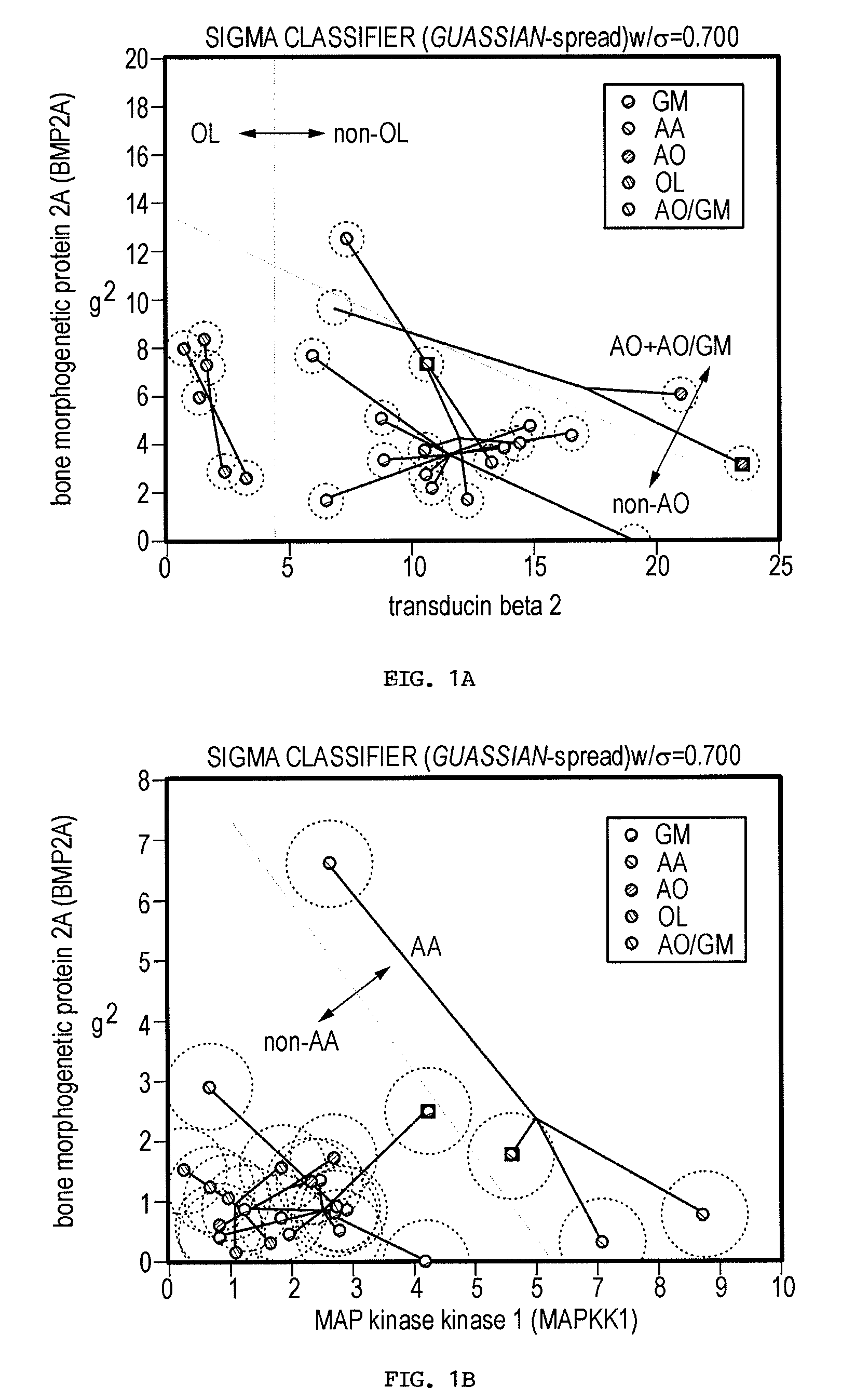
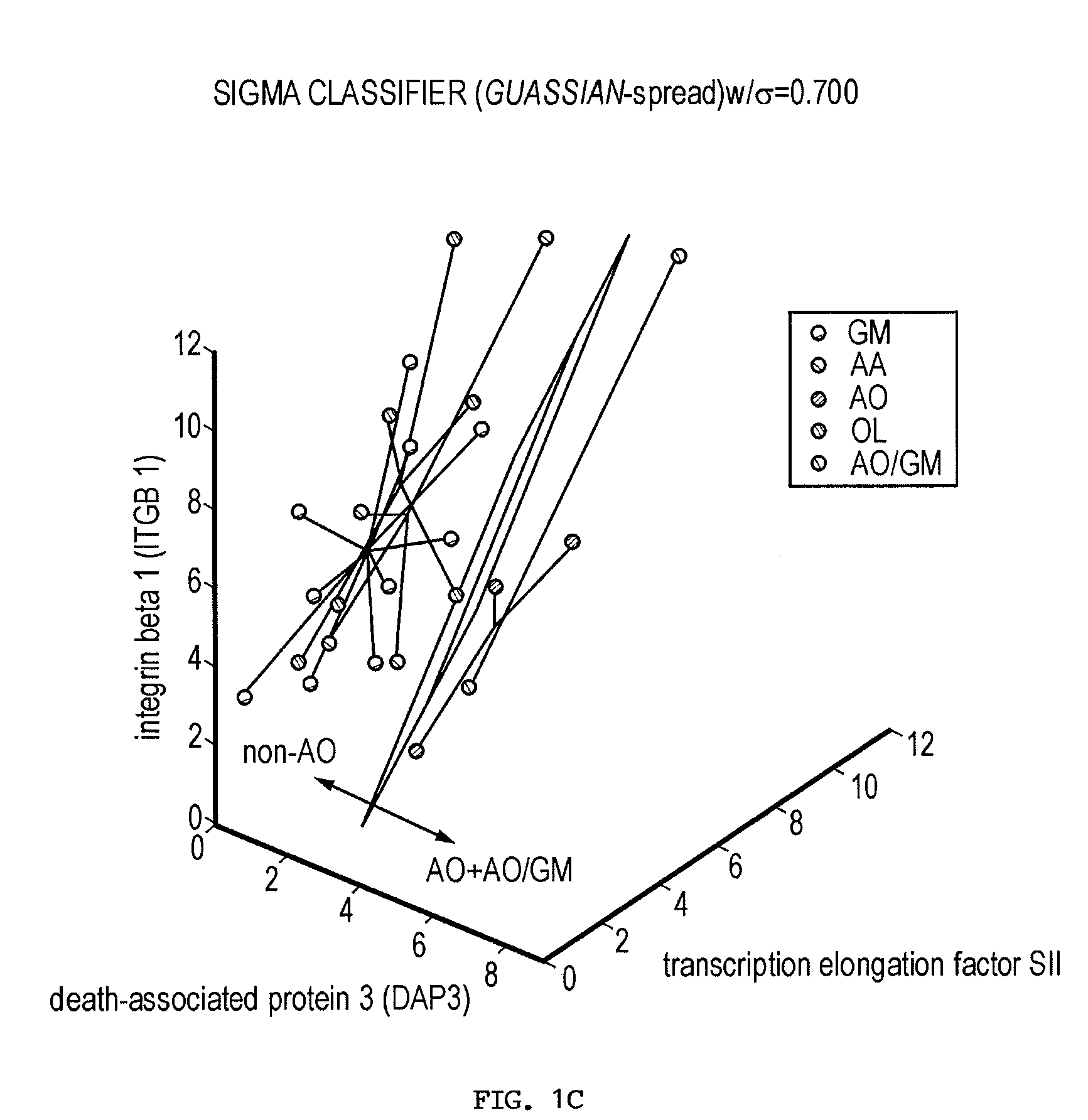
The present invention provides a number of gene markers whose expression is altered in various gliomas. In particular, by examining the expression these markers, one can accurately classify a glioma as glioblastoma multiforme (GM), anaplastic astrocytoma (AA), anaplastic oligodendroglioma (AO) or oligodendroglioma (OL). The diagnosis may be performed on nucleic acids, for example, using a DNA microarray, or on protein, for example, using immunologic means. Also disclosed are methods of therapy.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
DNA profiling and SNP detection utilizing microarrays
InactiveUS20060008823A1High sensitivityImprove accuracyMicrobiological testing/measurementDNA microarrayEnzymatic digestion
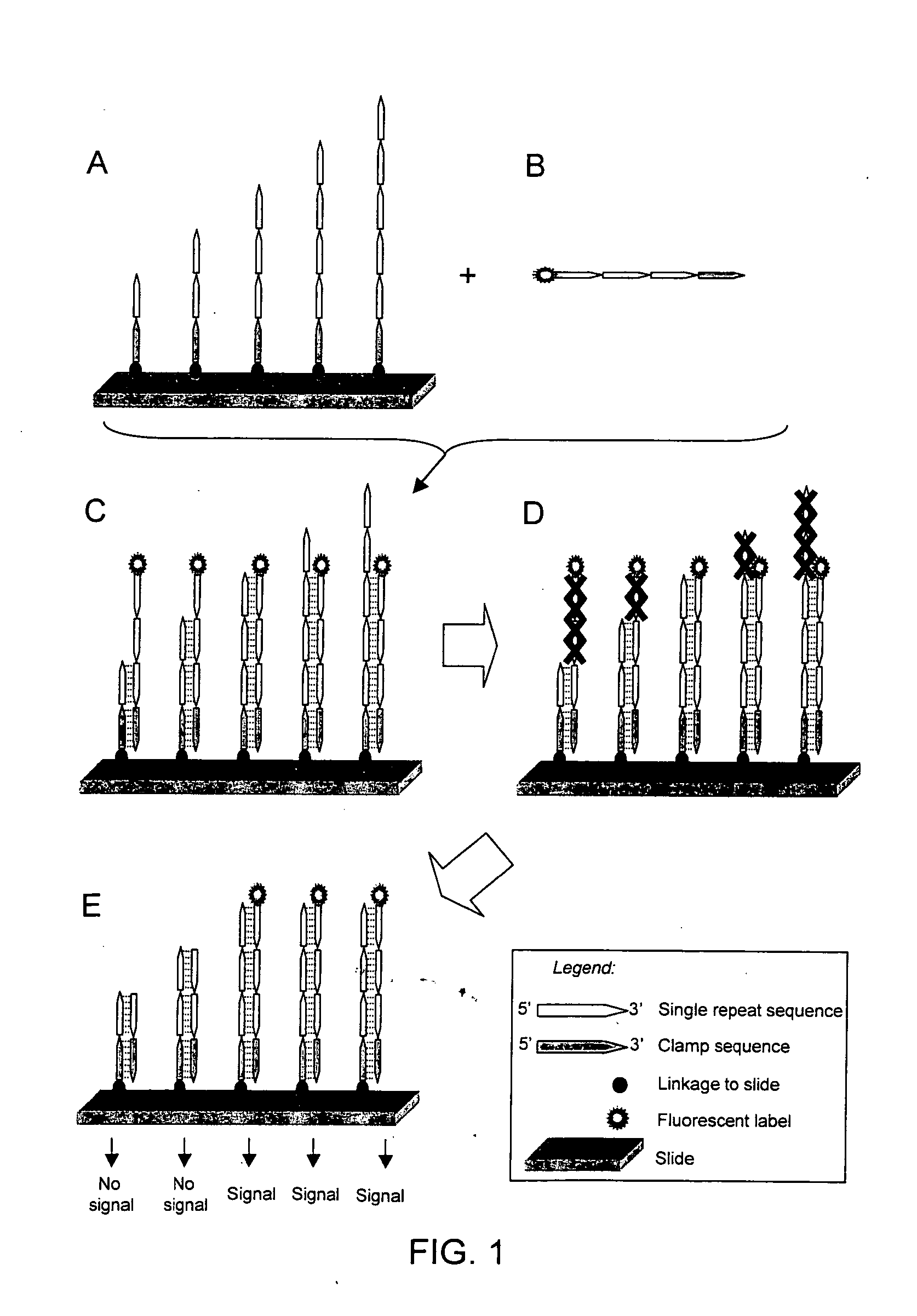
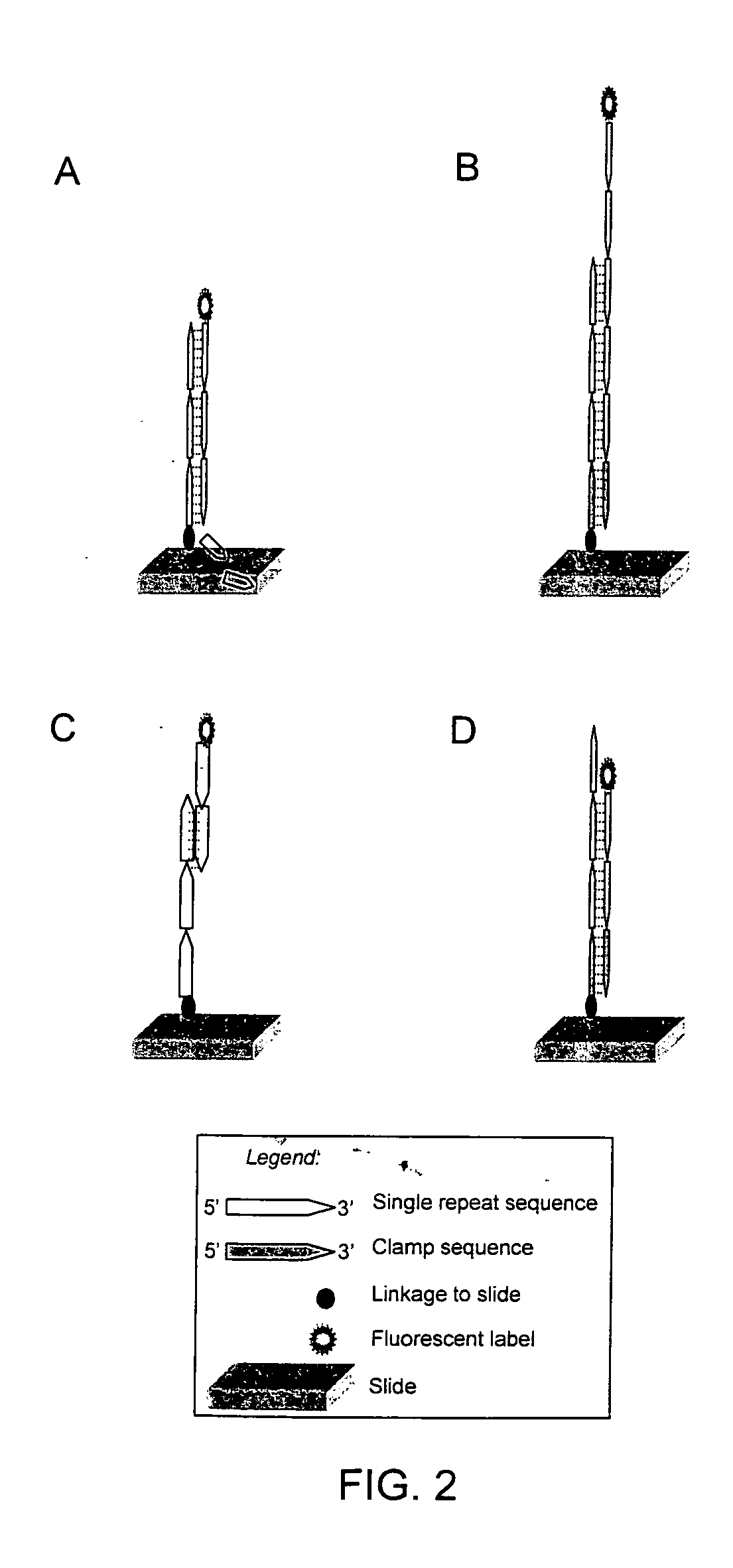
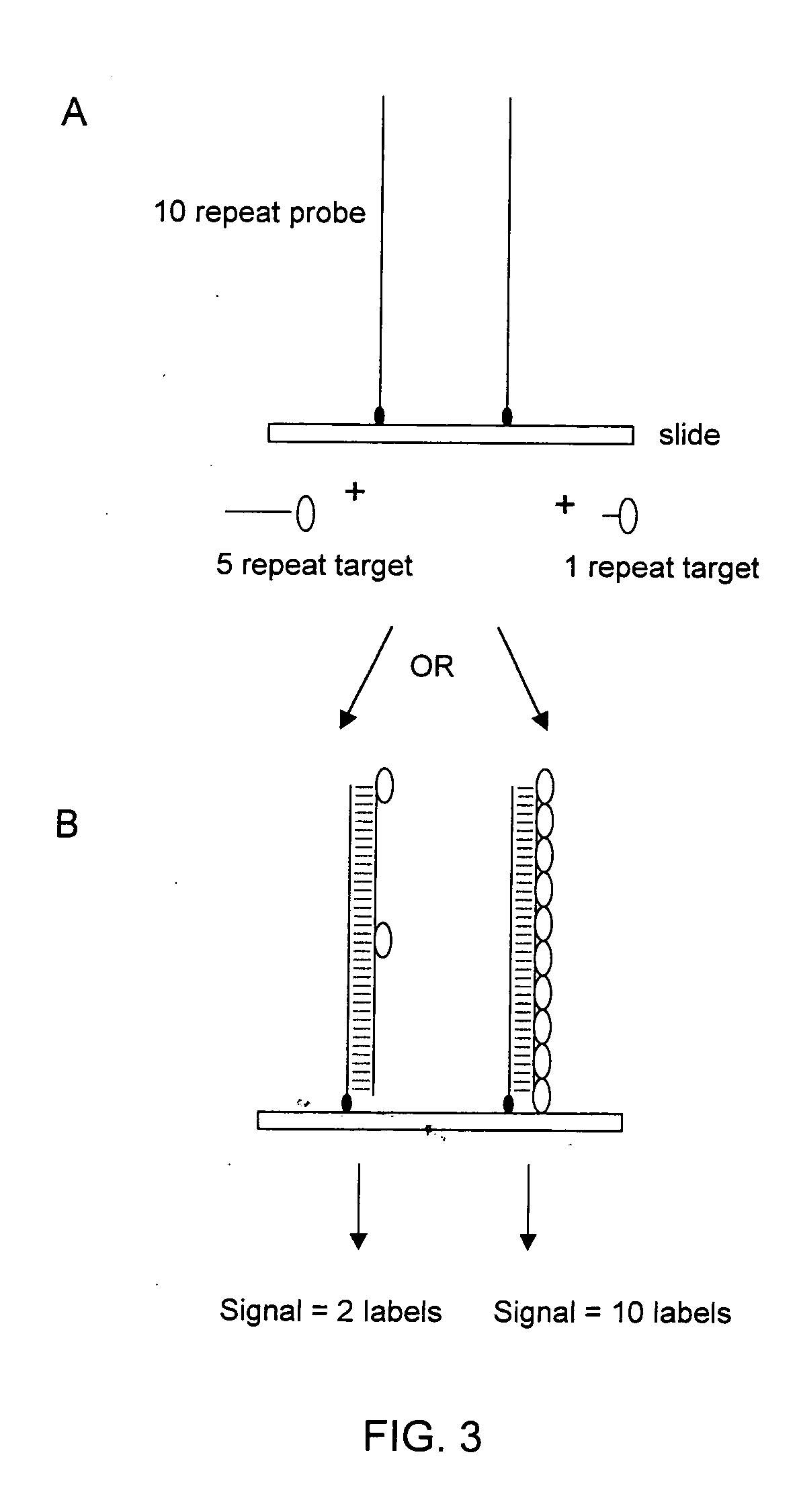
The present invention provides methods for rapidly identifying and distinguishing between different DNA sequences utilizing short tandem repeat (STR) analysis and DNA microarrays. Specifically, these methods facilitate the deduction of a target molecule's identity, length, and number of STRs. In an embodiment, a labeled STR target sequence is hybridized to a DNA microarray carrying complementary probes. These probes vary in length to cover the range of possible STRs. The labeled single-stranded regions of the DNA hybrids are selectively removed from the microarray surface utilizing a post-hybridization enzymatic digestion. The number of repeats in the unknown target is deduced based on the pattern of target DNA that remains hybridized to the microarray. The DNA profiling techniques described herein are useful for performing forensic analysis to uniquely identify individual humans or other species.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com