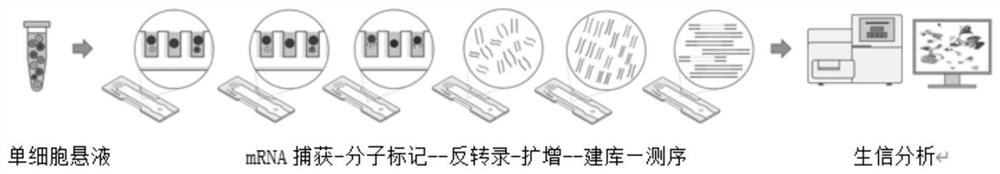
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
175 results about "Single cell sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
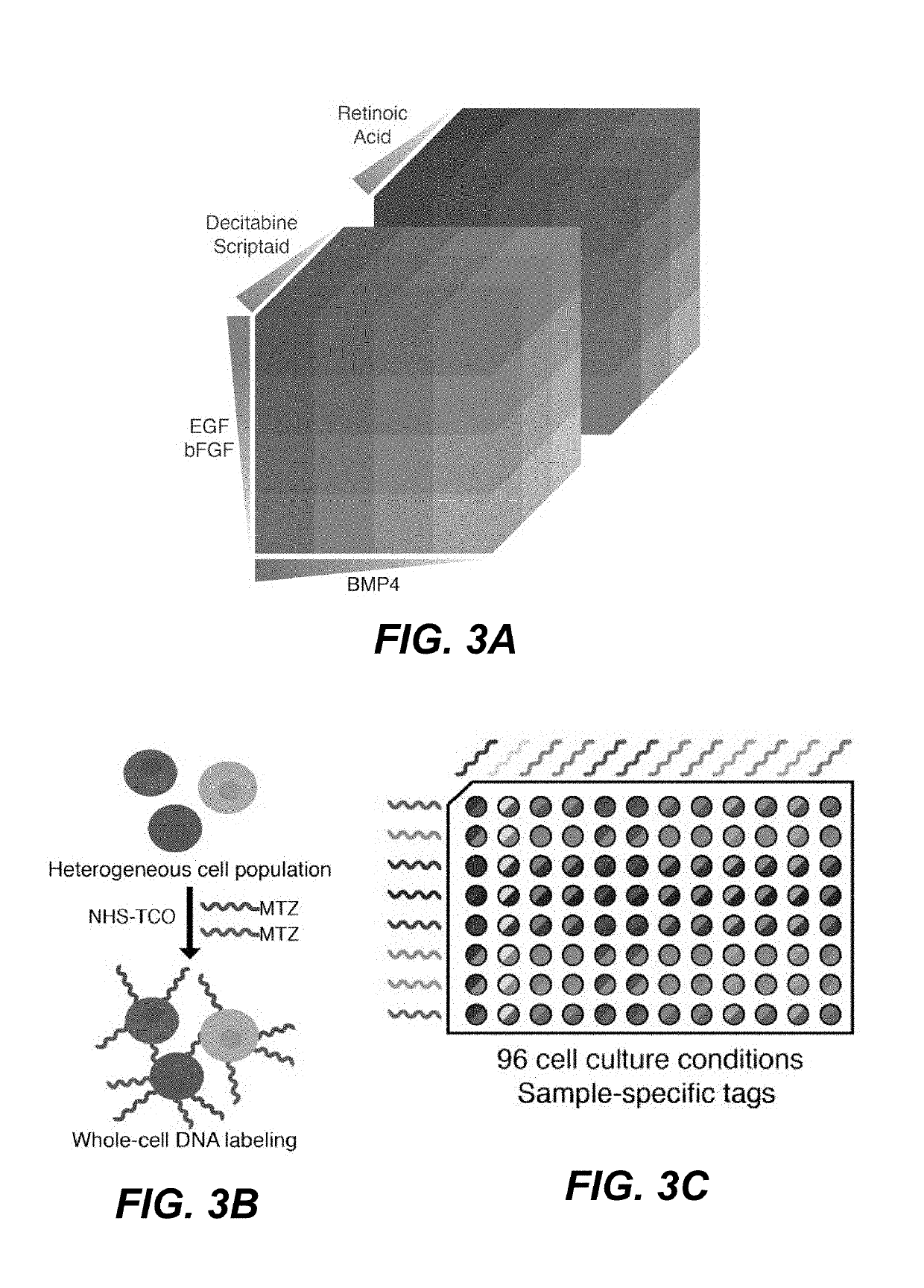
Single cell sequencing examines the sequence information from individual cells with optimized next generation sequencing (NGS) technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. Sequencing the DNA of individual cells can give information about mutations carried by small populations of cells, for example in cancer, while sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types, for example in development.
High-throughput single-cell analysis combining proteomic and genomic information
ActiveUS20160244828A1Predict susceptibilityMaterial analysis using sonic/ultrasonic/infrasonic wavesMicrobiological testing/measurementProteomicsPolynucleotide
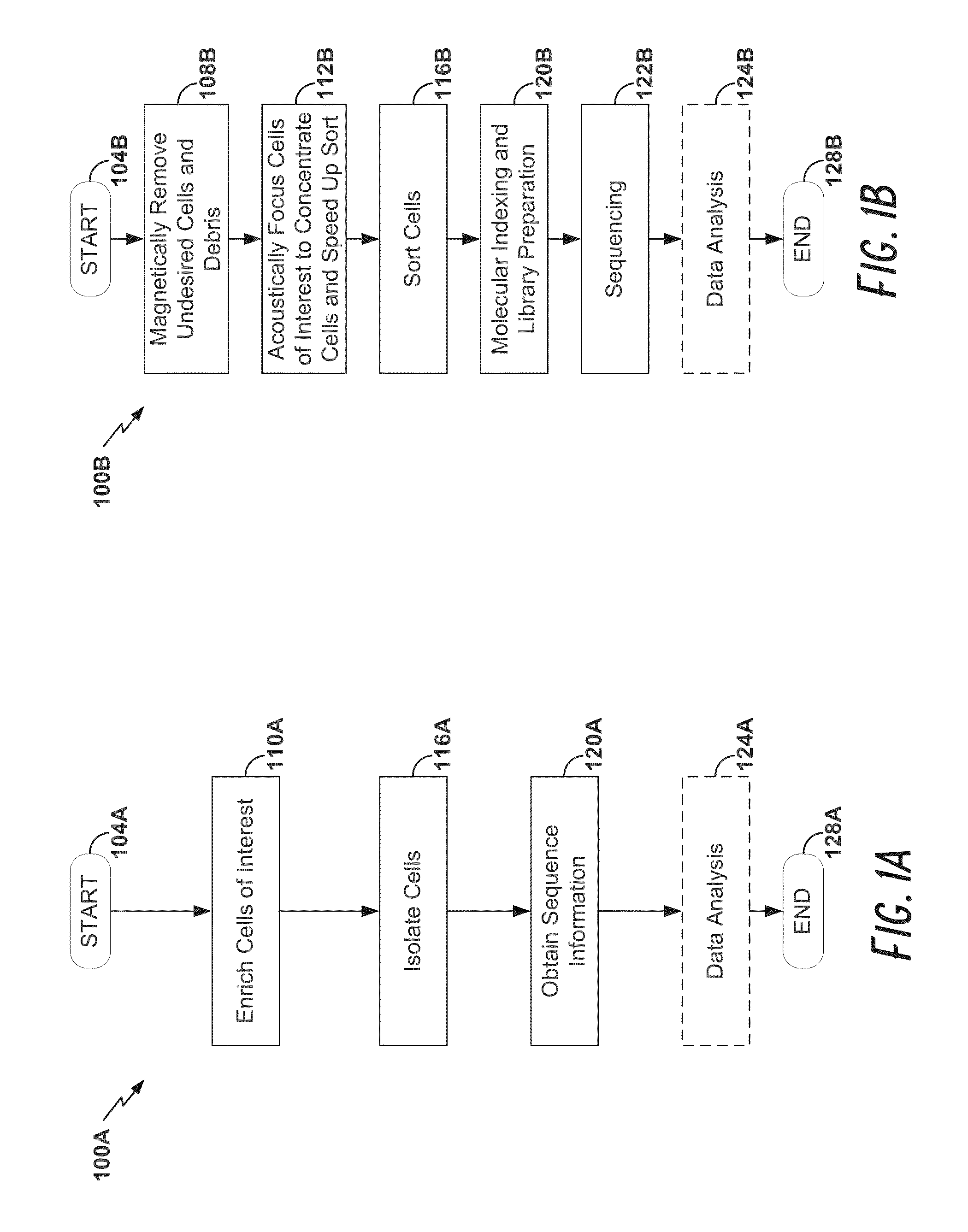
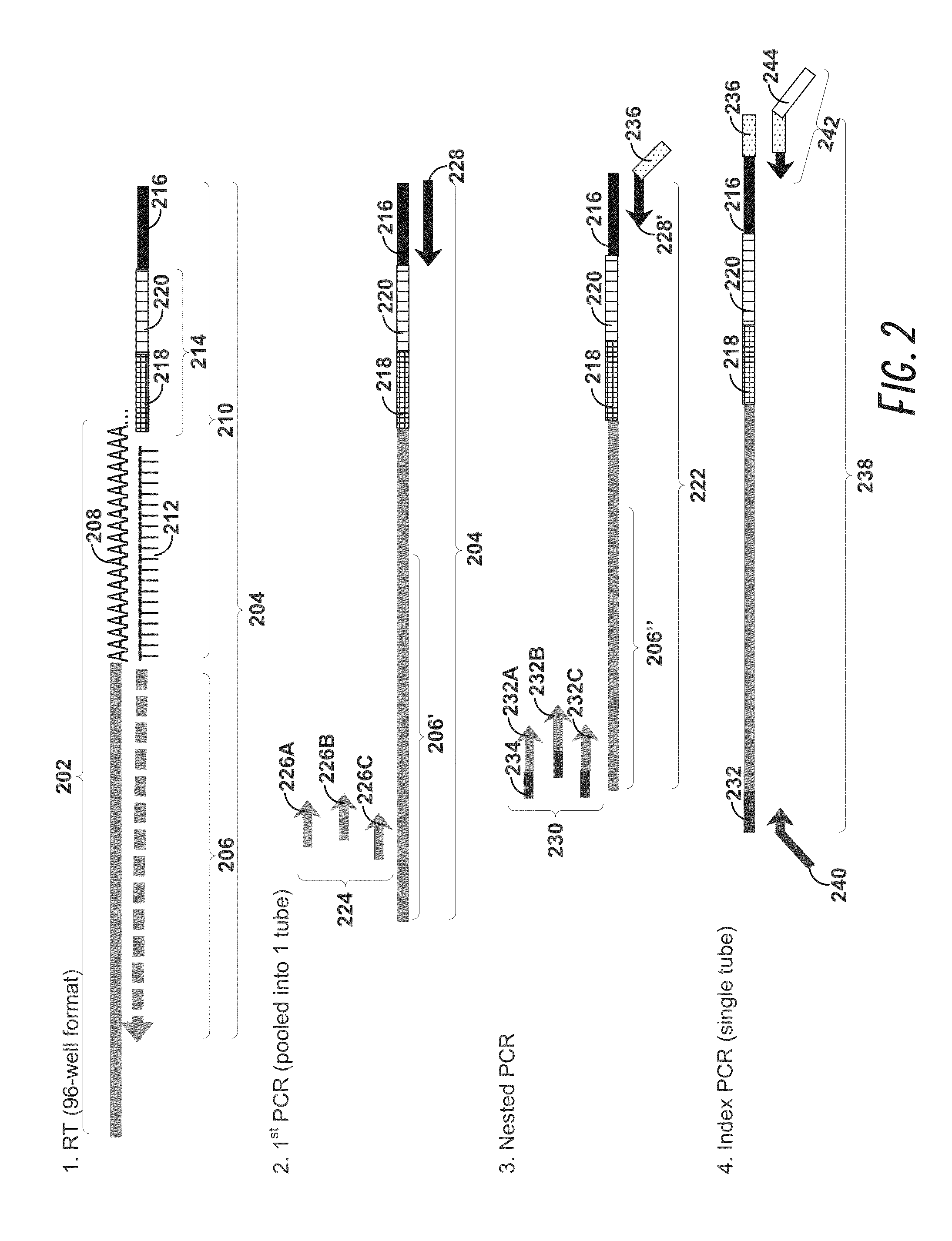
Disclosed herein are methods for single-cell sequencing. In some examples, the methods include enriching a sample comprising a plurality of cells for cells of interest to produce an enriched cell sample; isolating one or more cells of interest in the enriched cell sample; and obtaining sequence information of one or more polynucleotides from each of the one or more isolated cells. Obtaining sequence information may include generating a molecularly indexed polynucleotide library from the one or more isolated cells. Enriching the sample may include focusing cells of interest in the sample using acoustic focusing.
Owner:BECTON DICKINSON & CO
Tumor neoantigen screening method fused with single cell TCR sequencing data
ActiveCN113160887AOvercoming the high rate of misselection and omissionOvercoming problems such as insufficient immunogenicityMicrobiological testing/measurementProteomicsSingle cell transcriptomeCD8
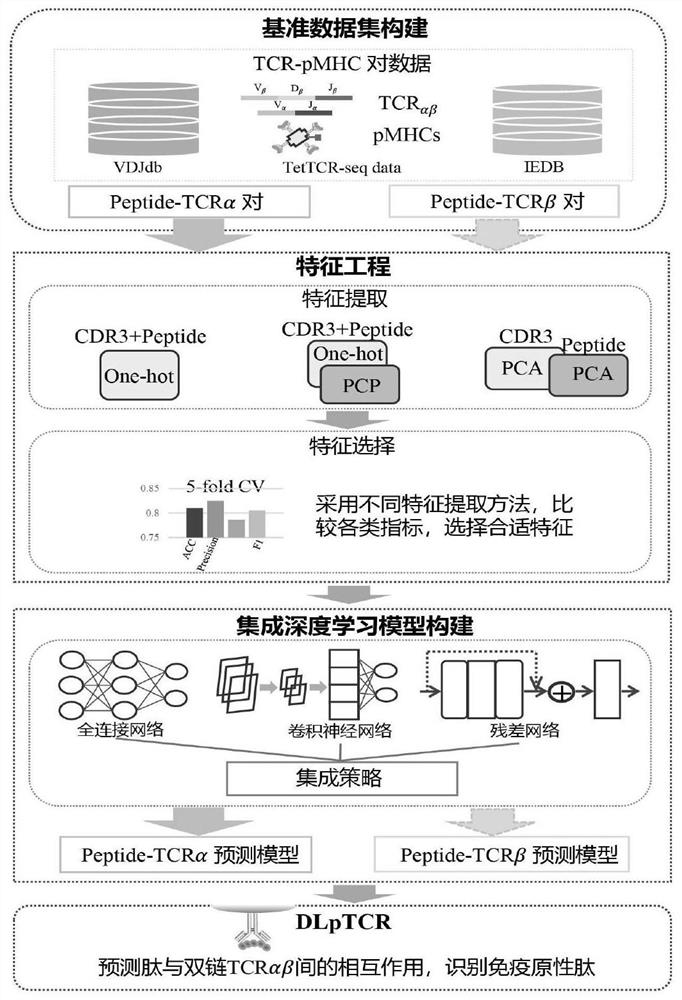
The invention discloses a tumor neoantigen screening method fused with single cell TCR sequencing data. The tumor neoantigen screening method comprises the following steps: based on whole exon sequencing data and transcriptome sequencing data, carrying out quality control, comparison and other steps through software to obtain a neomutant peptide library; predicting an HLA-I type typing by using HLA typing prediction software; in combination with single-cell TCR sequencing and single-cell transcriptome sequencing, finding cancer-specific CD8 + T cell receptors through cell type annotation and clone frequency analysis; meanwhile, based on integrated deep learning, the short peptide immunogenicity is identified through a peptide-TCR interaction prediction model, a tumor neoantigen screening method fused with single cell TCR sequencing data is provided, and the problems that a traditional tumor antigen screening method is high in neoantigen misselection and missed selection rate, insufficient in immunogenicity and the like are solved.
Owner:HARBIN INST OF TECH
Method for building high-flux single-cell full-length transcriptome sequencing library and application of method
ActiveCN109811045AGuaranteed accuracyIntegrity guaranteedMicrobiological testing/measurementLibrary creationTn5 transposaseLysis
The application discloses a method for building a high-flux single-cell full-length transcriptome sequencing library and application of the method. The method comprises the following steps: recognizing single cells by adopting a customized micro-hole chip and an ICELL8 platform, and performing cell lysis, mRNA reverse transcription, cDNA pre-amplification and Tn5 transposase library building on the single cells, wherein a product can be directly applied to sequencing; in a process of mRNA reverse transcription or Tn5 transposase library building, introducing a dual-terminal Barcode sequence with a 5' terminal and a 3' terminal in order to obtain a single cell full-length transcriptome. By adopting the method, the single cell recognition flux is high, and the product can be directly appliedto sequencing. When the method is applied to single-cell sequencing, only two days are required for efficiently obtaining thousands of single-cell full-length transcriptome libraries at one time, sothat the labor cost and time cost are lowered greatly; moreover, the reagent cost is lowered greatly through a trace reaction system, and a basis is laid for large-scale single-cell full-length transcript sequencing.
Owner:MGI TECH CO LTD
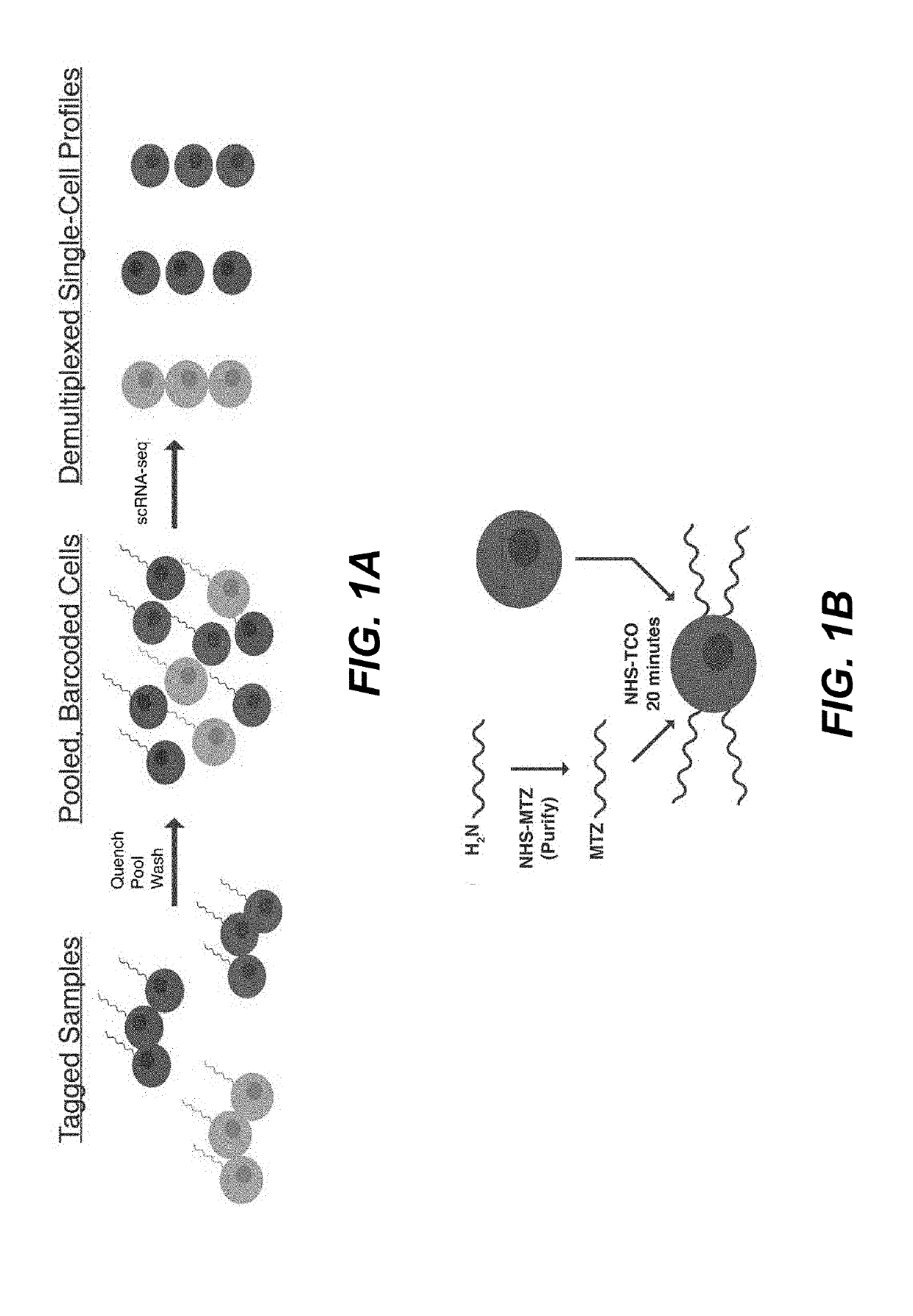
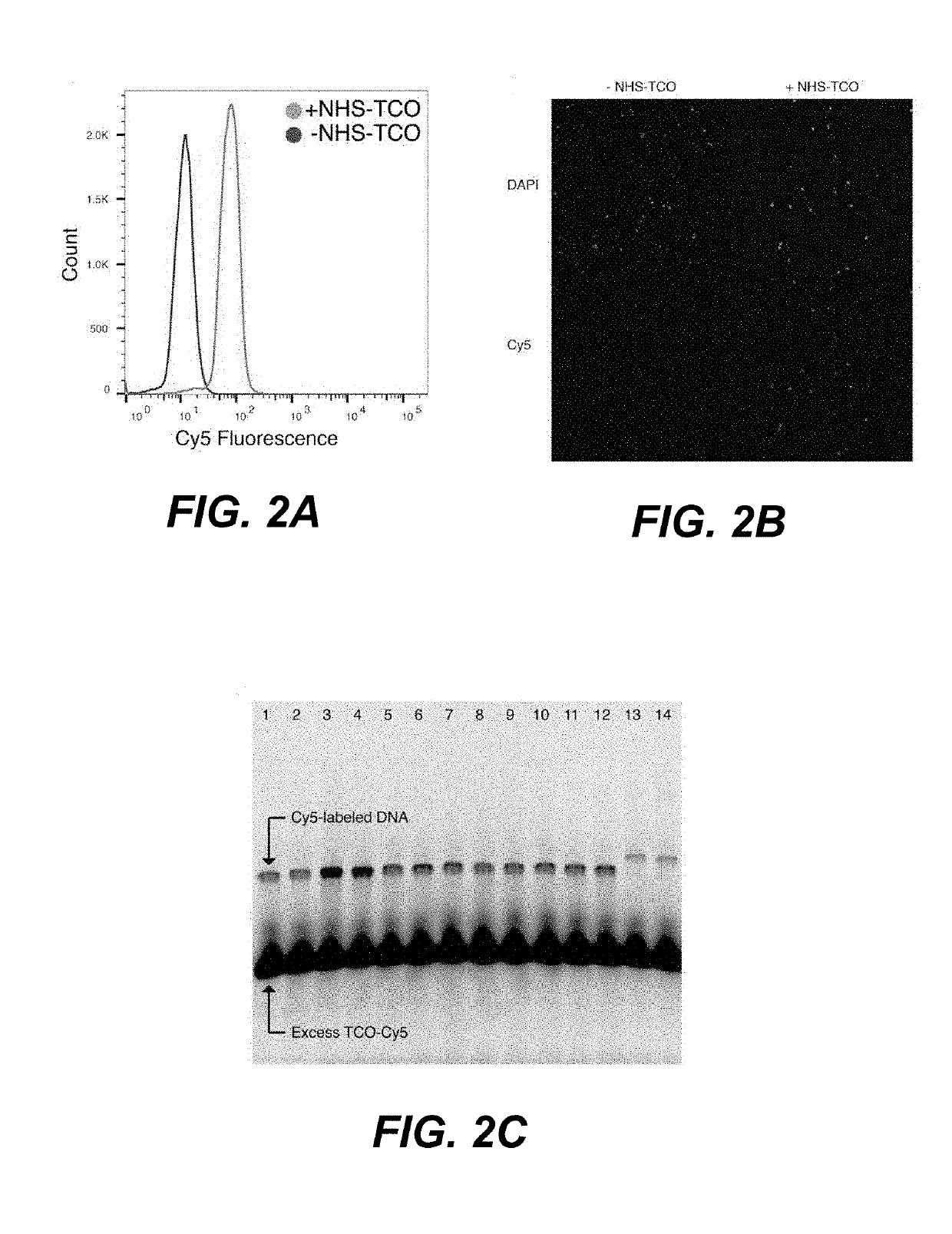
Sample multiplexing for single-cell RNA sequencing
ActiveUS20190276818A1Low costReduce the impactMicrobiological testing/measurementProtein nucleotide librariesNucleotideSingle cell sequencing
Disclosed herein include systems, methods, compositions, and kits for multiplexing single-cell RNA-sequencing (scRNA-seq) samples. In some embodiments, the methods comprise chemically tagging cells with identifying sample tags (e.g., barcoded DNA oligonucleotides).
Owner:CALIFORNIA INST OF TECH
DNA barcode compositions and methods of in situ identification in a microfluidic device
PendingUS20190345488A1Sequential/parallel process reactionsCell receptors/surface-antigens/surface-determinantsCell specificSequence analysis
Apparatuses, compositions and processes for DNA barcode deconvolution are described herein. A DNA barcode may be used to provide a bead specific identifier, which may be detected in situ using hybridization strategies. The DNA barcode provides identification by sequencing analysis. The dual mode of detection may be used in a wide variety of applications to link positional information with assay information including but not limited to genetic analysis. Methods are described for generation of barcoded single cell sequencing libraries. Isolation of nucleic acids from a single cell within a microfluidic environment can provide the foundation for cell specific sequencing library preparation.
Owner:BERKELEY LIGHTS
Magnetic microsphere with molecular tag sequence, and preparation method for magnetic microsphere
PendingCN112251504AImprove stabilityHigh precisionMicrobiological testing/measurementMicroballoon preparationMagnetic beadMicrosphere
The invention belongs to the field of the molecular biology, and particularly relates to a magnetic microsphere with a molecular tag sequence, and a preparation method for the magnetic microsphere. Amagnetic bead is taken as a molecular tag base, and a chemical bond and enzyme linking method is used for linking three-segment oligo to the magnetic bed in sequence so as to synthesize the magnetic microsphere with a unique oligo sequence. The magnetic microsphere prepared by the preparation method disclosed by the invention has high stability and accuracy. Meanwhile, the preparation technology of the magnetic microsphere is simple, time consumption is small, raw material sources are convenient, the cost is low, and the unicellular sequencing cost is greatly lowered. The oligo adopted by themagnetic microsphere is a known sequence, the three-segment oligo can be combined to form millions of molecular tags, batch differences are small, the stability and the accuracy of a unicellular sequencing result are obviously improved, and the magnetic microsphere can be widely applied to medical research, bioresearch and the like.
Owner:SINGLERON NANJING BIOTECHNOLOGIES LTD
Method of identifying cell types based on single-cell RNA sequencing data
ActiveCN110797089AEfficient clusteringImprove clustering effectBiostatisticsInstrumentsGraph regularizationCell type
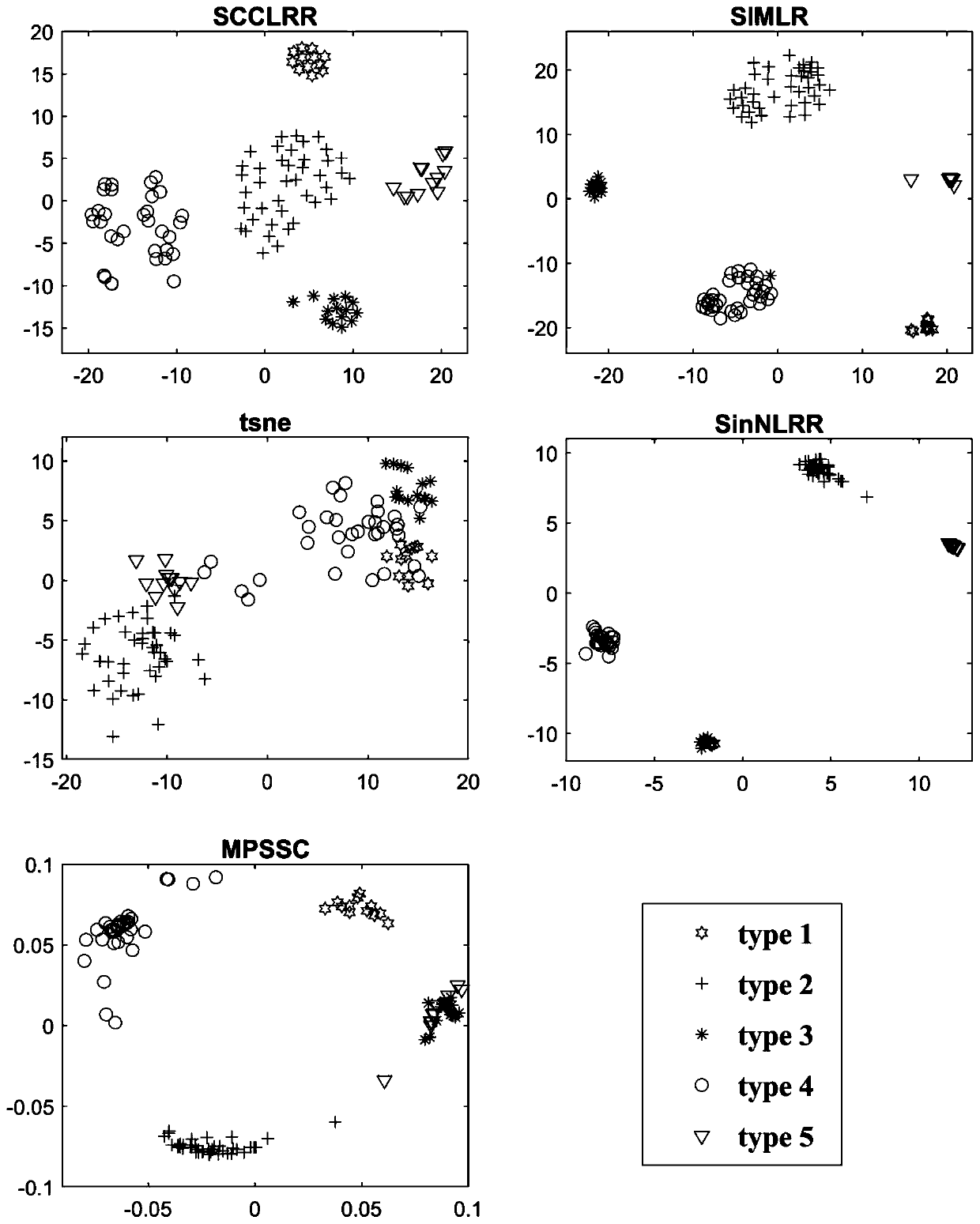
The invention provides a method of identifying cell types based on single-cell RNA sequencing data. According to the method, a low-rank representation model of a high-dimensional matrix is effectivelycombined with a graph regularization theory; an optimization model is constructed by considering the global structure and local structure characteristics of data, the model is solved by adopting an alternating direction multiplier method (ADMM) to obtain a reliable inter-cell similarity matrix, and then the similarity matrix is clustered by adopting a spectral clustering method, so that single cells are clustered, and the cell types are identified. According to the method, the clustering effect of the single-cell RNA sequencing data can be remarkably improved.
Owner:EAST CHINA JIAOTONG UNIVERSITY
Cell subset annotation method based on single cell transcriptome sequencing
ActiveCN112700820AComprehensive identificationFix the annotation problemBiostatisticsSequence analysisSingle cell transcriptomeCell subpopulations
The invention provides a cell subset annotation method based on single cell transcriptome sequencing. The cell subset annotation method comprises the following steps: 1) 10x barcode UMI identification; 2) genome comparison; 3) gene expression profile comparison; 4) low-quality cell filtering and data homogenization; 5) cell population clustering; 6) marker gene extraction; 7) cell subset annotation. The invention belongs to the technical field of biological information analysis, and provides a cell subset annotation method based on single-cell transcriptome sequencing, which solves the problem of single-cell subset annotation, so that single-cell sequencing data can support cell annotation according to a gene expression profile and / or a cell Marker gene after conventional analysis. Therefore, organic combination of different annotation methods is realized, and the distribution condition and related information of cell types are obtained.
Owner:广州华银医学检验中心有限公司 +1
Application of single cell sequencing in immune cell analysis
PendingCN110819706AActive autoimmune responseRaise the ratioBioreactor/fermenter combinationsBiological substance pretreatmentsDendritic cellT cell
The invention provides an application of single cell sequencing in immune cell analysis. The immune cells comprise any one or a combination of at least two of T cells, mononuclear cells, natural killer cells, B cells, macrophages, granulocytes, mast cells, megakaryocytes or dendritic cells. Wherein the immune cell analysis comprises analysis of any one or a combination of at least two of types, states or proportions of immune cells and / or immune cell subtypes. In the present invention, single cell sequencing is applied to the immune cell analysis, a method comprises the following steps: firstly, obtaining a sequencing result of a sample through single cell sequencing, then obtaining a distribution state, subtype composition and a cell track of immune cells by adopting a biological information analysis method, annotating the cell subtypes, and realizing analysis on the types, states and proportions of the immune cells and / or the immune cell subtypes by comparing different results.
Owner:SUZHOU SINGLERON BIOTECHNOLOGIES LTD
Single-cell RNA sequencing clustering method based on adversarial autoencoder
PendingCN111785329AImprove clustering effectBiostatisticsCharacter and pattern recognitionData setRNA Sequence
One or more embodiments of the present specification provide a single-cell RNA sequencing clustering method based on an adversarial autoencoder, integrating the advantages of specific bionoise modeling, variation inference, and deep clustering modeling. The model restrains a data structure, and clustering analysis is carried out through an AAE module. Experiments performed on three real scRNA-seqdata sets show that the clustering performance of the method is much better than that of the latest technology in clustering accuracy, standardized mutual information and Rand index adjustment.
Owner:NAT UNIV OF DEFENSE TECH
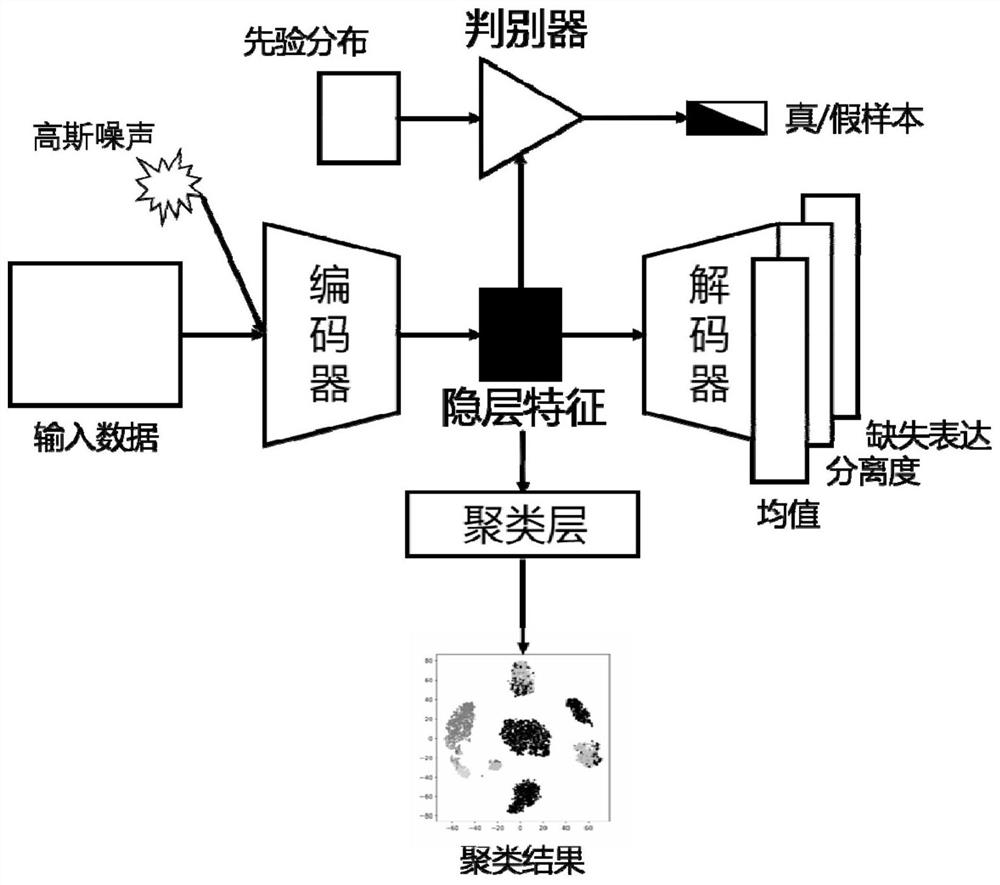
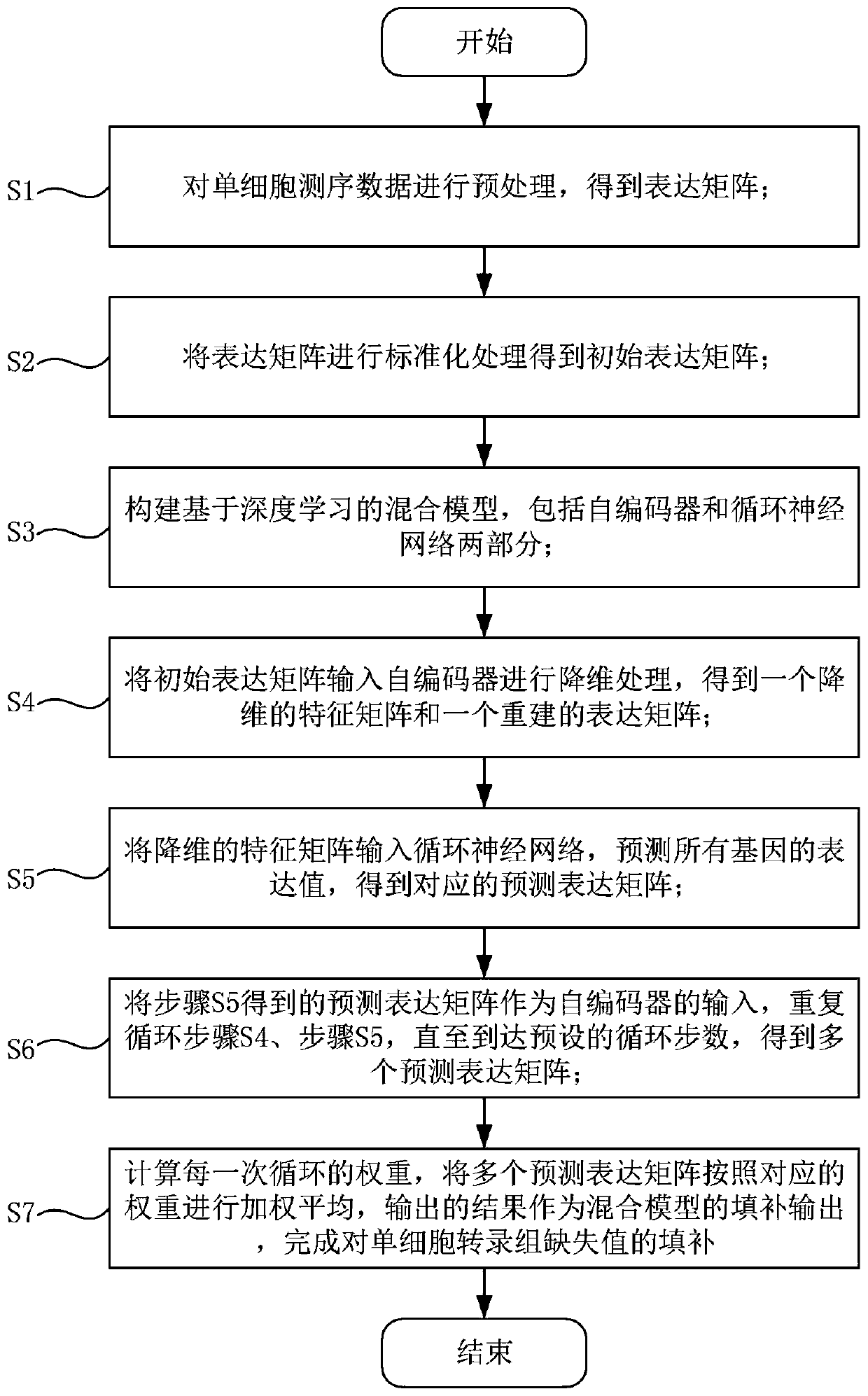
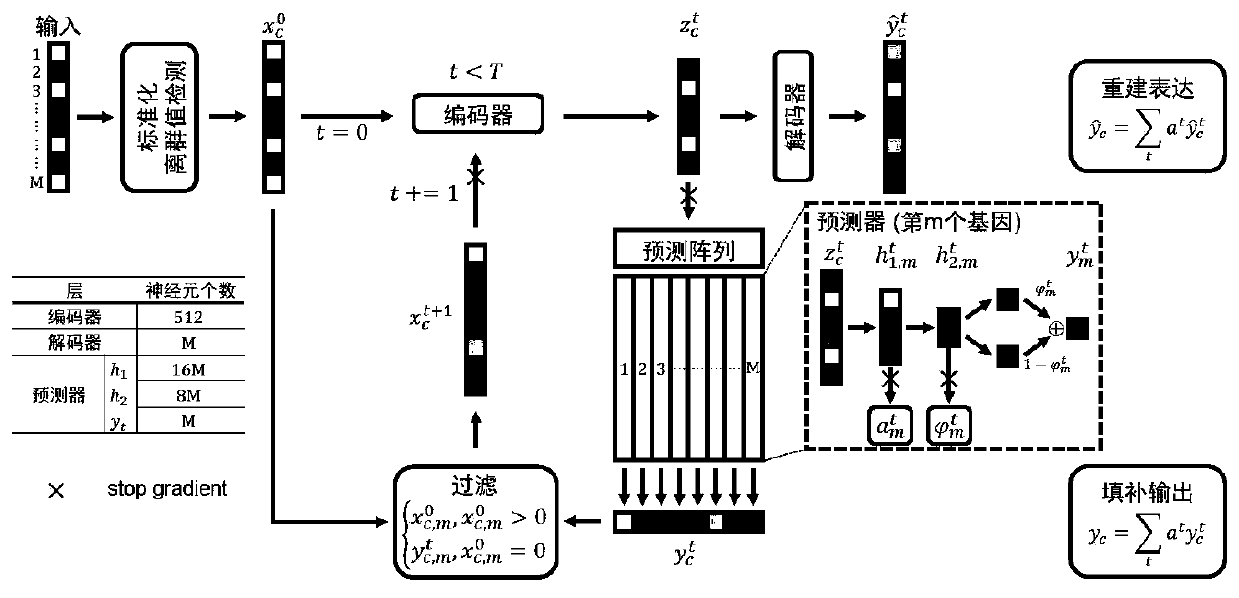
Single cell transcriptome missing value filling method based on deep hybrid network
ActiveCN110957009AImprove reliabilityGuaranteed versatilityBiostatisticsInstrumentsData setSingle cell transcriptome
The invention provides a single cell transcriptome missing value filling method based on a deep hybrid network. The method comprises the steps of: carrying out sequencing and preprocessing of a singlecell, obtaining an expression matrix, and carrying out standardization processing; constructing a hybrid model based on deep learning, and inputting the standardized expression matrix into the hybridmodel for cyclic calculation to obtain a plurality of prediction expression matrixes; calculating the weight of each cycle, performing weighted average on the multiple prediction expression matrixesaccording to the corresponding weights, wherein the obtained result is filling output of the hybrid model, and filling of missing values is completed. According to the filling method provided by the invention, the fitting capability of the deep neural network to a complex function is adapted to the expression distribution of the single cells, so that the universality of the filling method to various single cell transcriptome data is ensured; and moreover, the expansibility of deep learning on a data set with an ultra-large cell number is reserved, filling of the single cell transcriptome missing value is completed, and the reliability of single cell data interpretation is remarkably improved.
Owner:ZHONGSHAN OPHTHALMIC CENT SUN YAT SEN UNIV
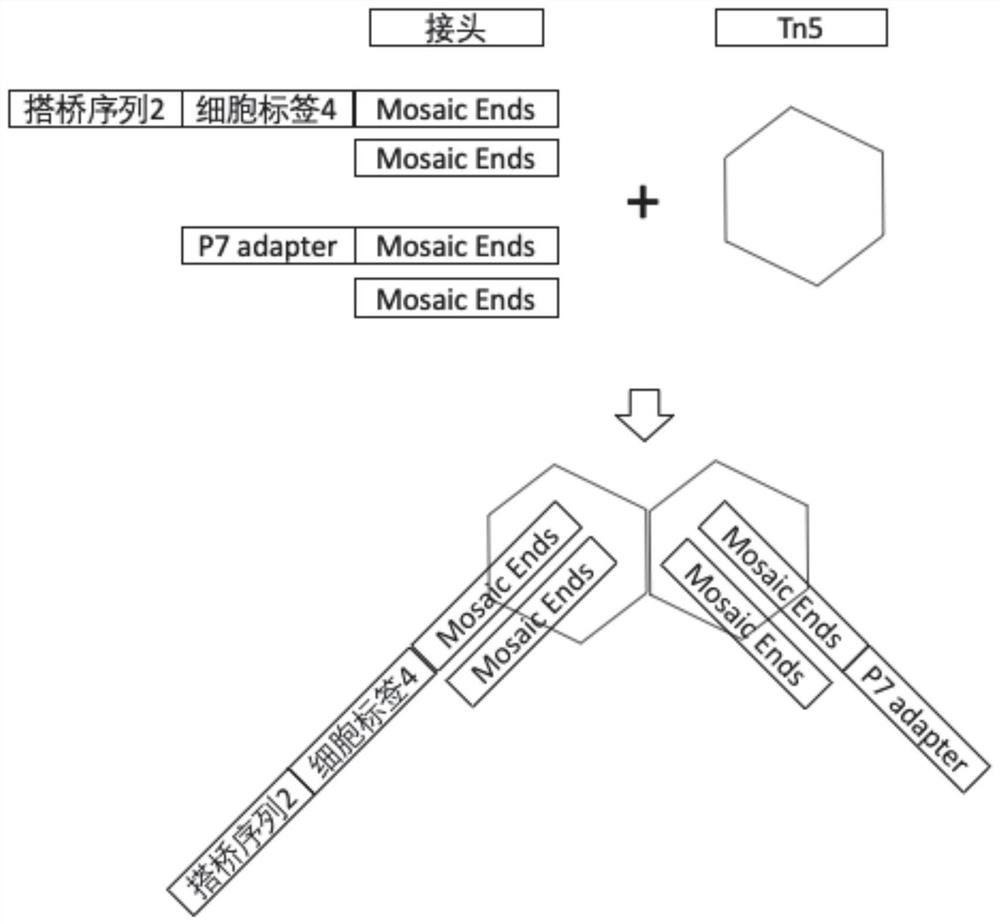
Ultrahigh-flux single-cell chromatin transposase accessibility sequencing method
The invention discloses an ultrahigh-flux single-cell chromatin transposase accessibility sequencing method. The ultrahigh-flux single-cell chromatin transposase accessibility sequencing method comprises the following steps that firstly, transposition of a chromatin transposase accessibility genome sequence in a cell nucleus is carried out by using a specific molecular tag transposase embedding compound; one molecular marker microbead and one or more cell nucleuses are located in a separated space through a microwell plate technology or a microfluidic technology, the cell nucleuses are split under the action of a splitting solution, and a specific molecular tag sequence is connected with a molecular marker sequence on the molecular marker microbead with the help of a bridging primer; a large number of sequences are obtained through PCR amplification, and a sequencing library is constructed and obtained; and then high-throughput sequencing is carried out, specific genome openness information of millions of single cells can be obtained through one-time sequencing, the single cell sequencing throughput is greatly improved, and the development of a single-cell high-flux chromatin transposase accessibility sequencing technology based on a microwell plate is actively promoted.
Owner:ZHEJIANG UNIV
High-throughput single-cell transcriptome sequencing method and kit
PendingCN110684829AReduce mutual contaminationIncrease the proportionMicrobiological testing/measurementGenomicsSingle cell transcriptome
The invention discloses a high-throughput single-cell transcriptome sequencing method and a kit. The high-throughput single-cell transcriptome sequencing method includes the steps that a droplet generation system is adopted to encapsulate a single cell and labeled microbeads in a droplet, and reverse transcription is performed in the droplet. According to the high-throughput single-cell transcriptome sequencing method, a throughput of 9,000 cells which is equivalent to 10 x genomics can be achieved, after the droplet is demulsified, the microbeads are less contaminated with each other, and theproportion of valid data is increased. According to the high-throughput single-cell transcriptome sequencing method, reverse transcription is performed in the droplet, fewer reagents are required, and the cost is low. In the preferred scheme of the high-throughput single-cell transcriptome sequencing method, a SMART template conversion technology is adopted in reverse transcription, and productscan be directly used for subsequent Tn5 library construction and BGISeq-500 platform sequencing without library conversion; and multiple amplification and introduction of deviations are avoided, a droplet-based microfluidics platform is used in conjunction with a BGISeq-500 sequencing platform, and large-scale single-cell sequencing is simplified and facilitated.
Owner:MGI TECH CO LTD
Ultrahigh-flux single cell sequencing method
ActiveCN113106150AImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationIntracellularMicrowell Plate
The invention discloses an ultrahigh-flux single-cell sequencing method, and the method comprises the following steps: by using molecular marker microspheres, a reverse transcription sequence and a bridging primer, carrying out primary intracellular reverse transcription in a cell by using the reverse transcription sequence; enabling one molecular marker microbead and one or more cells to be located in a separated space through a microwell plate technology or a micro-fluidic technology for the cells subjected to intracellular reverse transcription, splitting the cells under the action of a splitting solution, and connecting a sequence obtained after reverse transcription with a molecular marker sequence on the molecular marker microbead with the help of a bridging primer; obtaining a large number of sequences through PCR amplification, constructing and obtaining a cDNA sequencing library, and carrying out high-throughput sequencing, wherein specific transcriptome information of millions of single cells can be obtained through one-time sequencing. The single cell sequencing flux is greatly improved.
Owner:ZHEJIANG UNIV
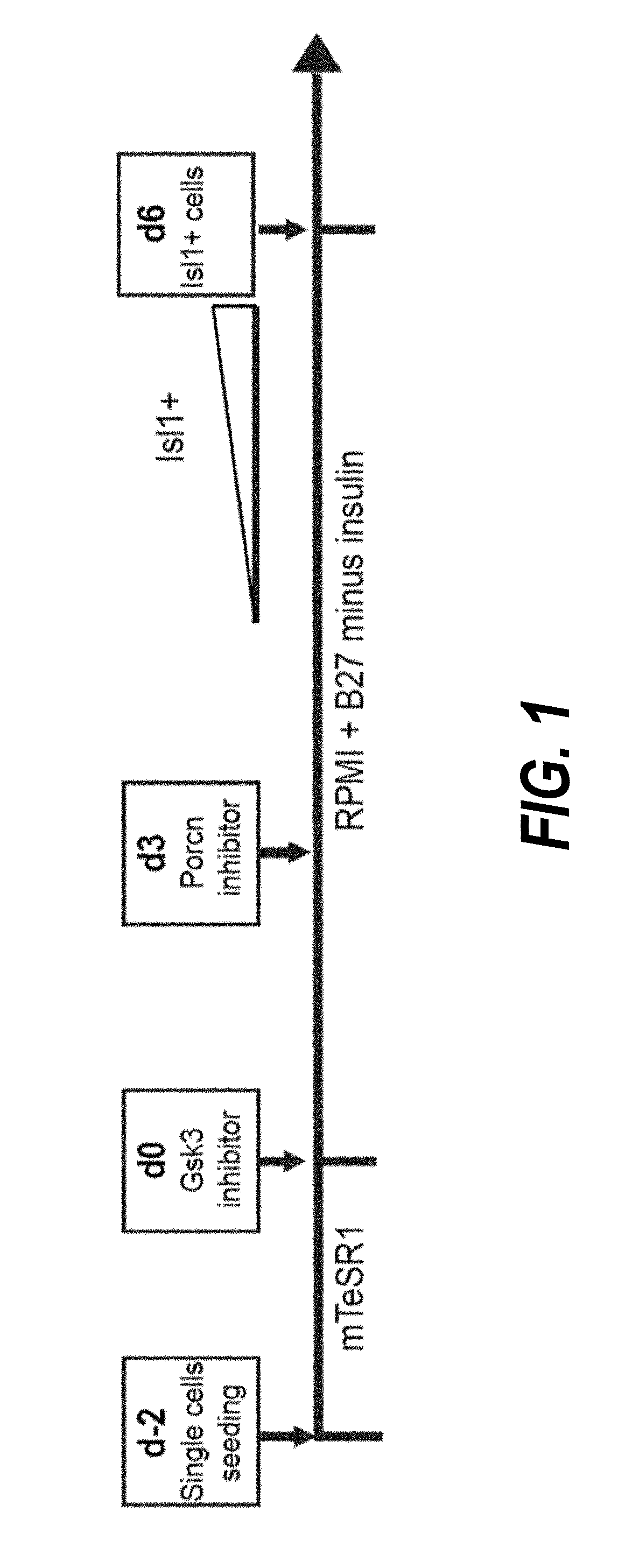
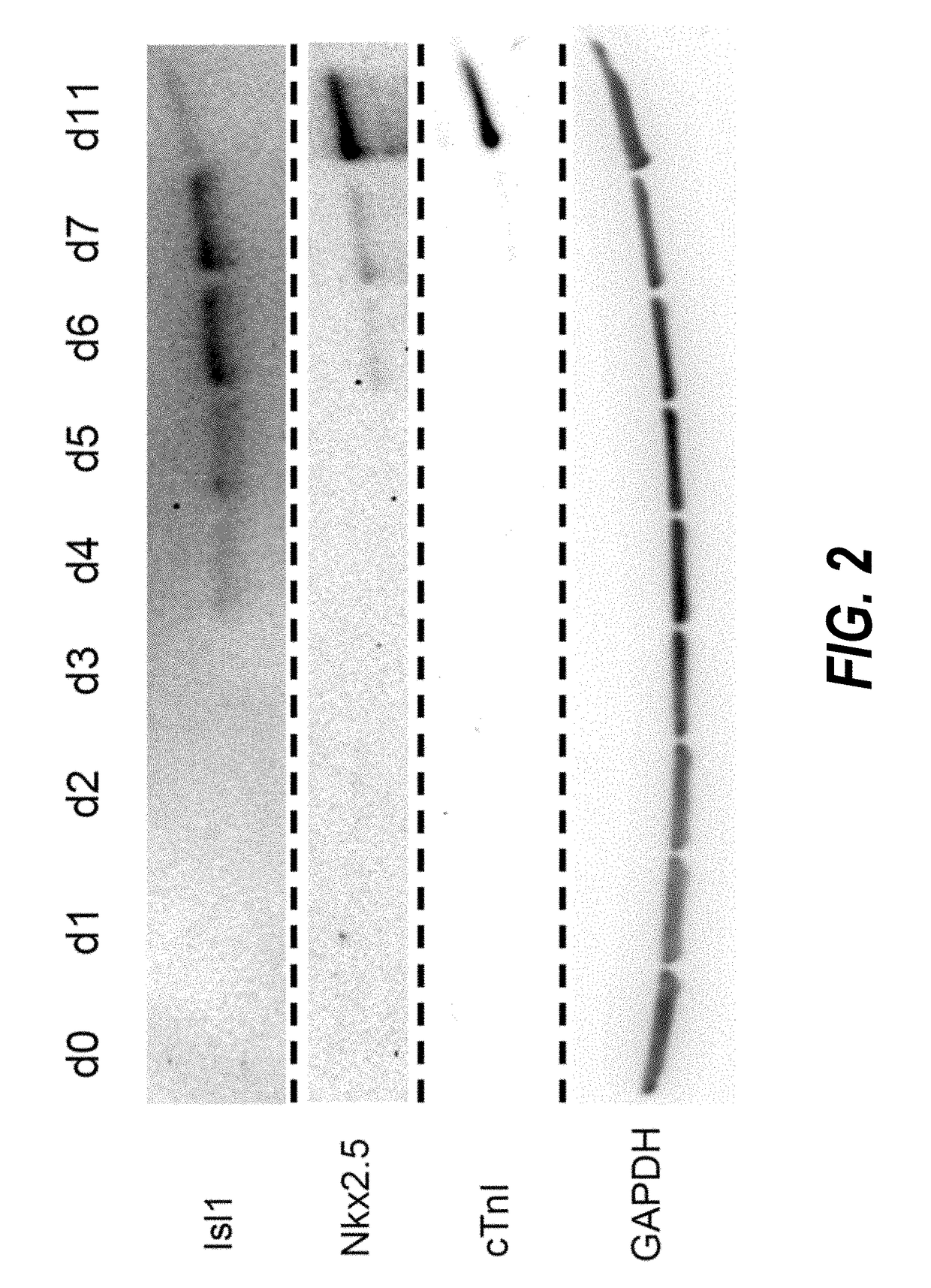
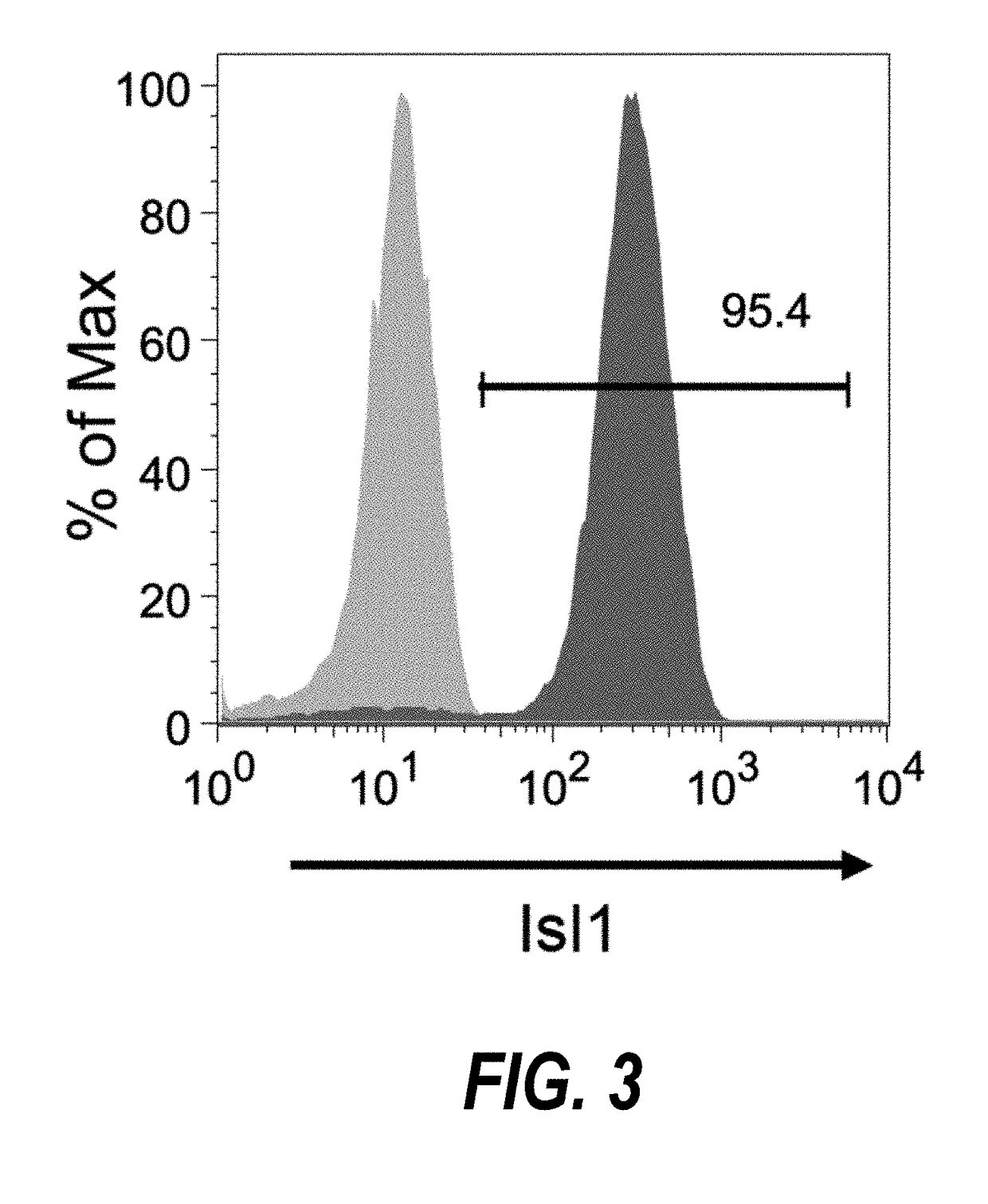
Use of neuropilin-1 (NRP1) as a cell surface marker for isolating human cardiac ventricular progenitor cells
ActiveUS20190062696A1Easy and rapid isolationFunction increaseCulture processCell culture supports/coatingProgenitorSurface marker
The present invention provides NRP1 as a cell surface marker for isolating human cardiomyogenic ventricular progenitor cells (HVPs), in particular progenitor cells that preferentially differentiate into cardiac ventricular muscle cells. Additional HVP cell surface markers identified by single cell sequencing are also provided. The invention provides in vitro methods of the separation of NRP1+ ventricular progenitor cells, and the large scale expansion and propagation thereof. Large clonal populations of isolated NRP1+ ventricular progenitor cells are also provided. Methods of in vivo use of NRP1+ ventricular progenitor cells for cardiac repair or to improve cardiac function are also provided. Methods of using the NRP1+ ventricular progenitor cells for cardiac toxicity screening of test compounds are also provided.
Owner:PROCELLA THERAPEUTICS AB
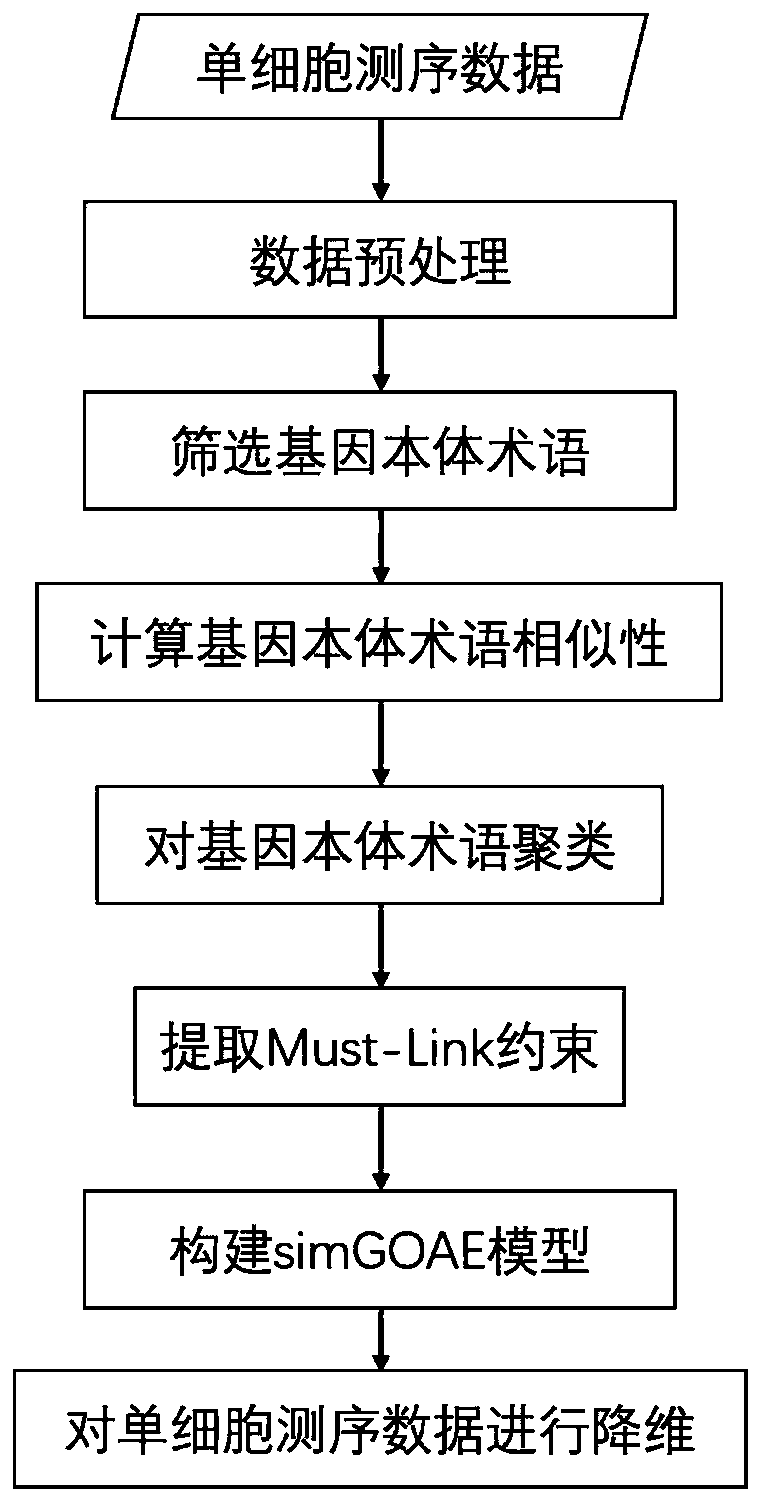
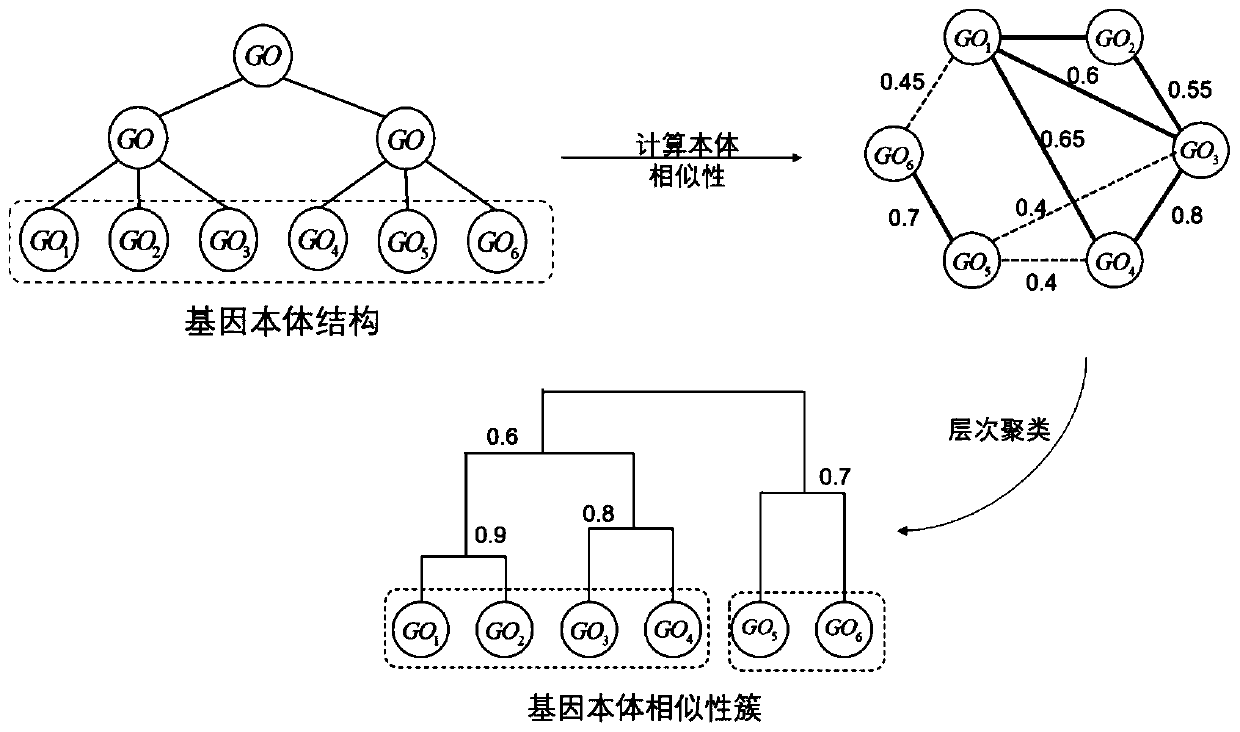
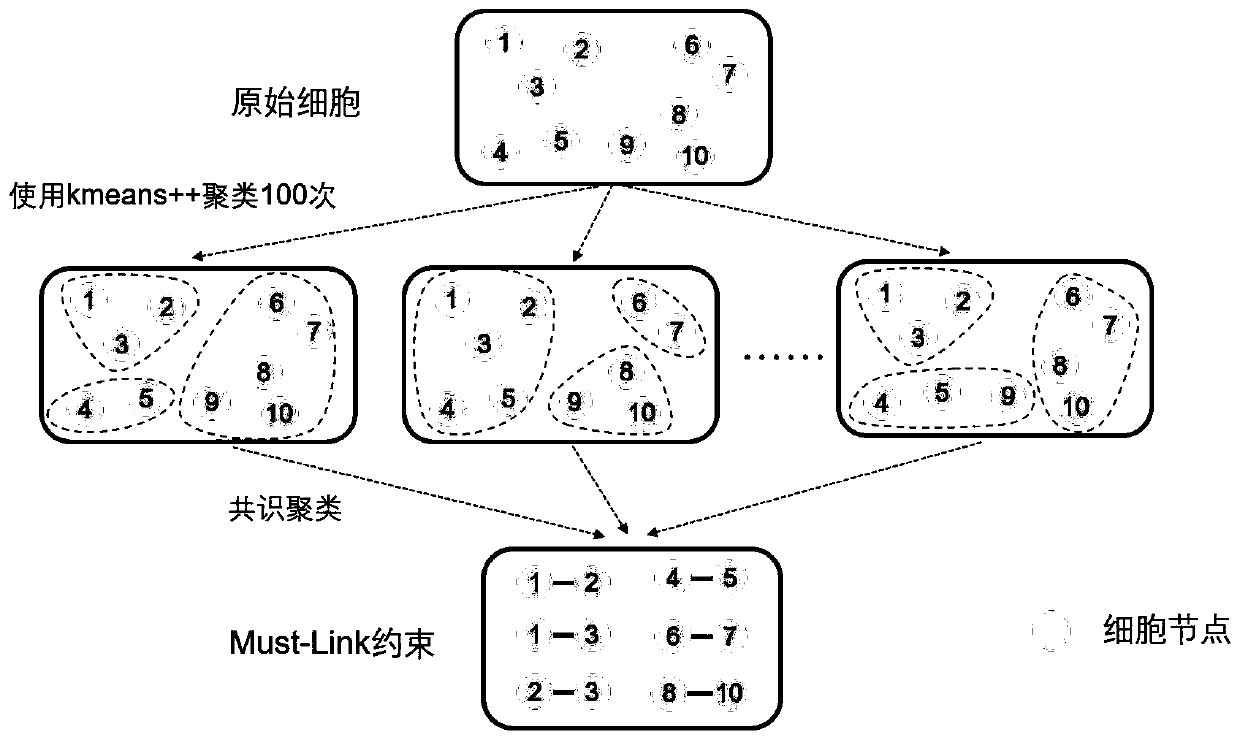
Single-cell sequencing data dimension reduction method fusing gene ontology and neural network
ActiveCN111564183AReduce training parametersAccelerated trainingBiostatisticsCharacter and pattern recognitionData setCell layer
The invention provides a single-cell sequencing data dimension reduction method fusing gene ontology and a neural network. The method comprises the following steps: firstly, extracting gene ontology terms as deep biological information priori knowledge; secondly, extracting Mut-Link constraints among cells as priori knowledge on a cell level; then, combining the two kinds of priori knowledge withan auto-encoder model, and proposing a simGOAE model; and finally, carrying out training dimension reduction on single-cell sequencing data by using the simGOAE model. The simGOAE model provided by the invention not only can adapt to the training of a large sample data set, but also can better mine the biological information of the cells and realize a better single-cell sequencing data dimension reduction effect.
Owner:NORTHWESTERN POLYTECHNICAL UNIV
Single cell sequencing chip automatic clamping device
ActiveCN110257235AReduce labor requirementsImprove clamping efficiencyBioreactor/fermenter combinationsBiological substance pretreatmentsSpare timeConveyor belt
The invention relates to a single cell sequencing chip automatic clamping device. The device is designed for a single cell sequencing chip for Shanghai NovelBio Technology Co., Ltd. All parts of the whole device are mounted and positioned through the support plates, and the chip and a chip clamping slot are respectively fed to the installation position through two conveyor belts in the horizontal direction and the longitudinal direction, a crank groove mechanism pushes a mounting slot and then completes the installation, when an experimenter uses the chip, the chip is only required to be taken in a guide groove at one side of the device, and the operation of the device during the spare time of single-cell sequencing and the condition whether the number of chips and slots is sufficient are observed. The single cell sequencing chip automatic clamping device improves the efficiency of the clamping while greatly reducing the labor demand.
Owner:XI'AN POLYTECHNIC UNIVERSITY
High-throughput single-cell sequencing with reduced amplification bias
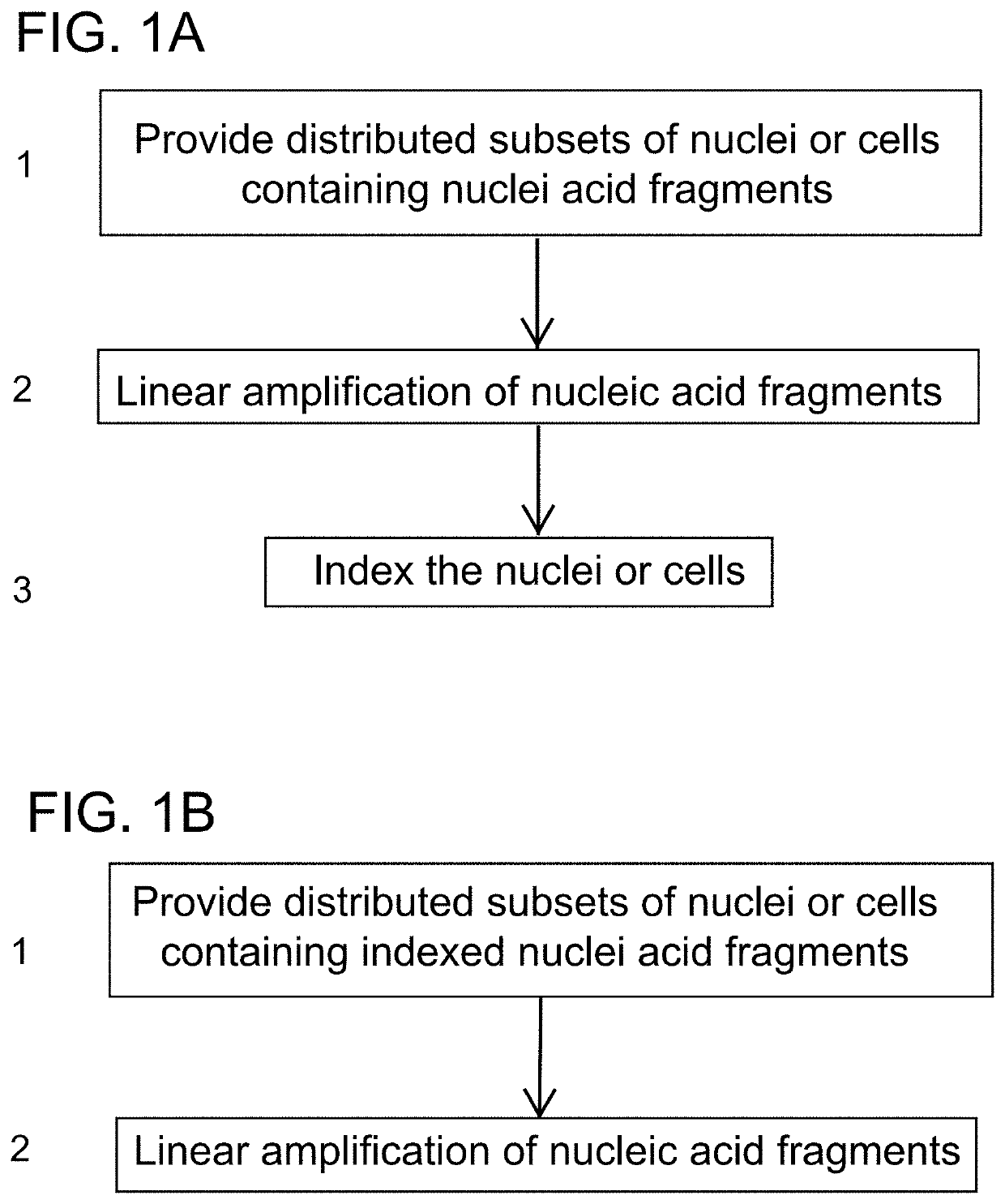
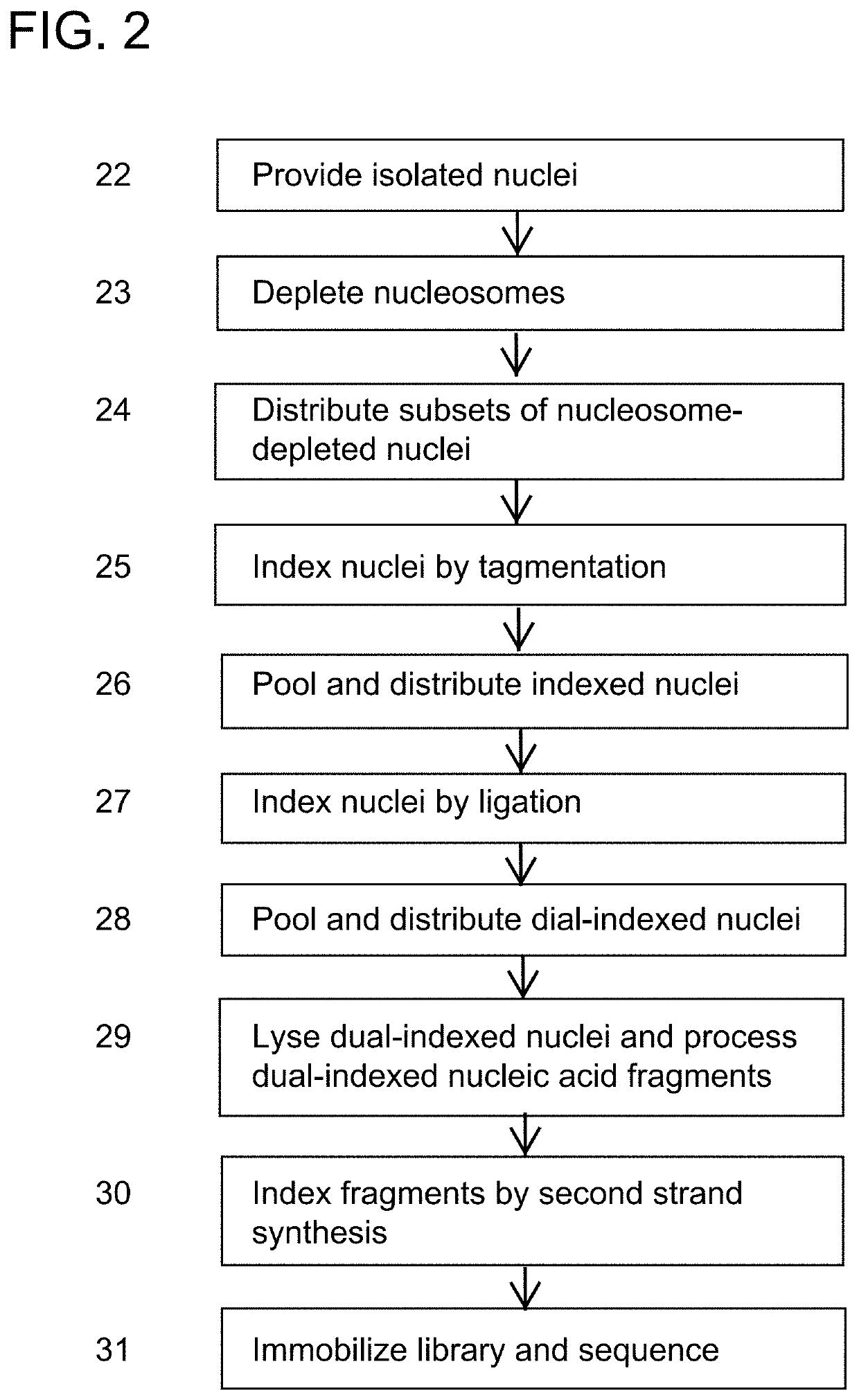
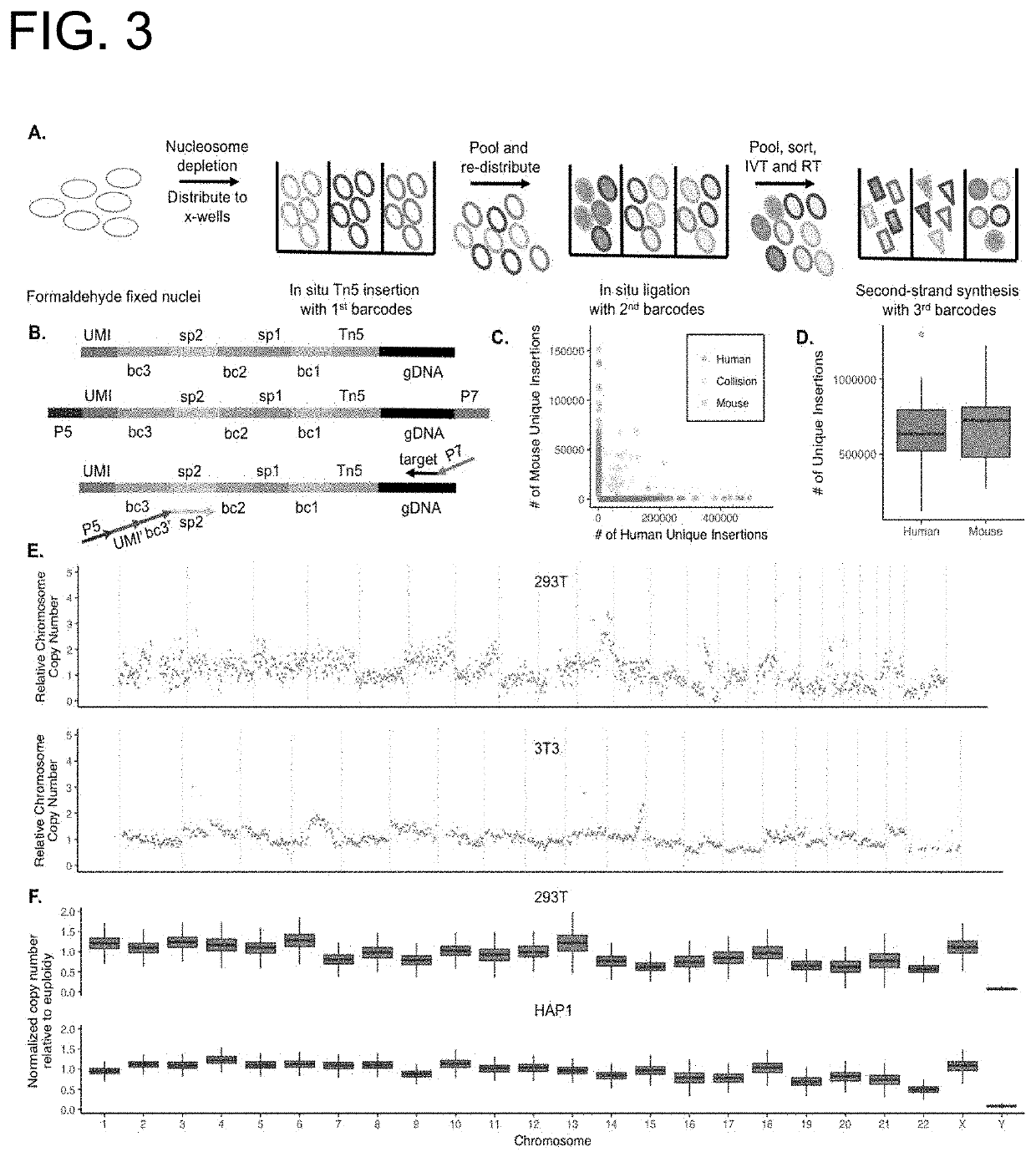
PendingUS20190382753A1Minimize amplification biasImprove throughputNucleotide librariesMicrobiological testing/measurementLinear amplificationAmplification bias
Provided herein are methods for preparing a sequencing library that includes nucleic acids from a plurality of single cells. In one embodiment, the methods include linear amplification of the nucleic acids. In one embodiment, the sequencing library includes whole genome nucleic acids from the plurality of single cells. In one embodiment, the nucleic acids include three index sequences. Also provided herein are compositions, such as compositions that include the nucleic acids having three index sequences
Owner:UNIV OF WASHINGTON
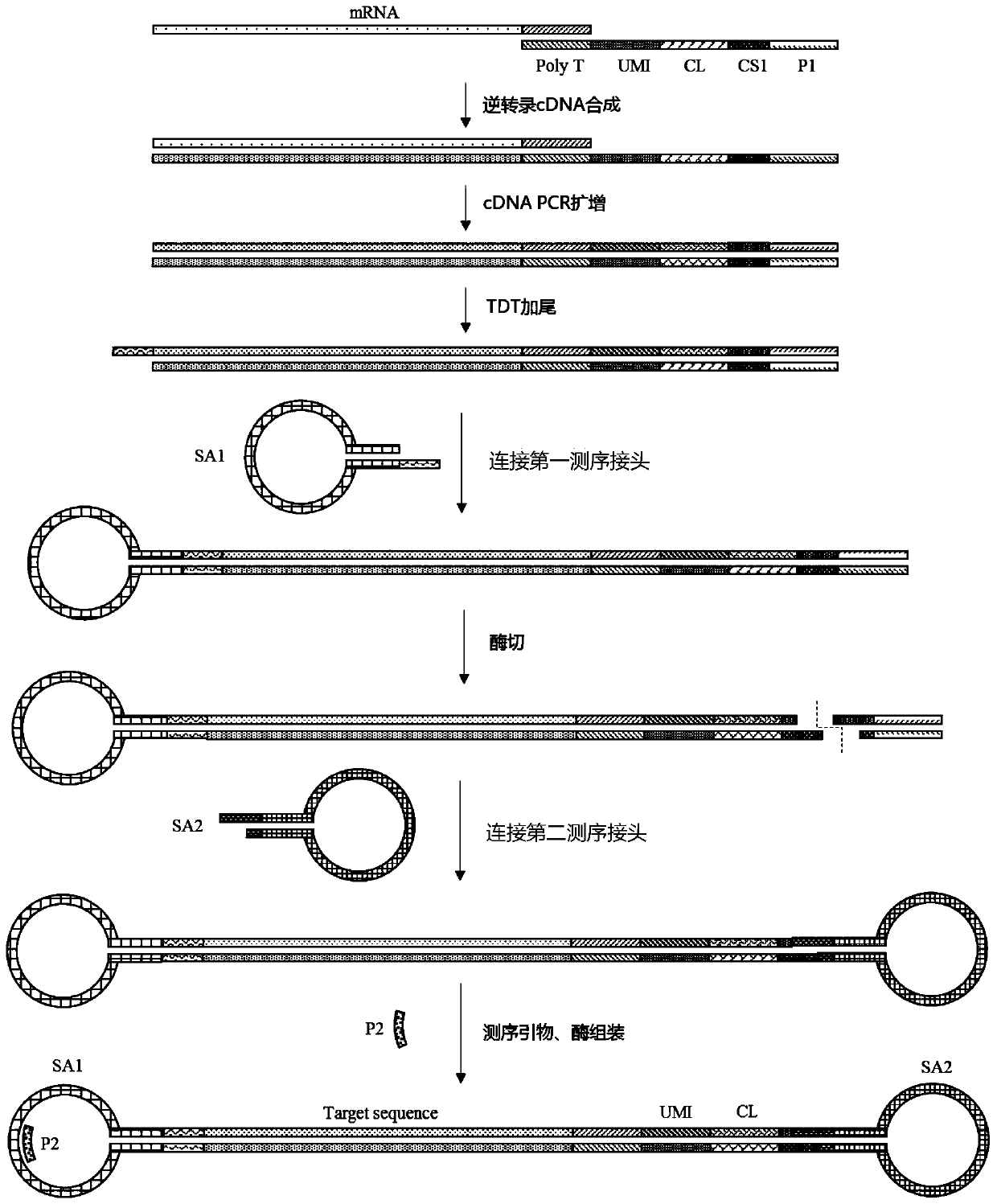
MRNA capture sequences, synthesis method of capture carrier and production method of high-throughput single-cell sequencing library
ActiveCN111088250AImprove capture efficiencySequencing guaranteeMicrobiological testing/measurementLibrary creationNucleotideCell trapping
The invention provides mRNA capture sequences. The mRNA capture sequences comprise universal primers, rare enzyme cutting sites, cell tags, random molecular tags and PolyT sequences, wherein by introducing rare enzyme cutting site sequences, sticky ends are provided for subsequent sequencing connector connection, so that two ends of a cDNA double strands are connected with two different sequencingconnectors. A synthesis method of a capture carrier used for mRNA capture and a production method of a high-throughput single-cell sequencing library are further provided. According to the provided capture carrier, by synthesizing the mRNA capture sequences of the rare enzyme cutting site sequences in situ and introducing the mRNA capture sequences of the rare enzyme cutting site sequences on a base material, the provided capture carrier is adopted for preparing the single-cell sequencing library, the single-cell capture efficiency and the labelling efficiency of oligonucleotide tags are improved, a process of constructing the library is simplified, and the two ends of the produced cDNA double strands are connected with the two different sequencing connectors, so that it is guaranteed that one dumbbell-shaped sequencing library is only assembled with a primer and DNA polymerase, and one-to-one correspondence of the dumbbell-shaped sequencing library, the primer and the DNA polymeraseis the premise of guaranteeing single-cell real-time sequencing.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
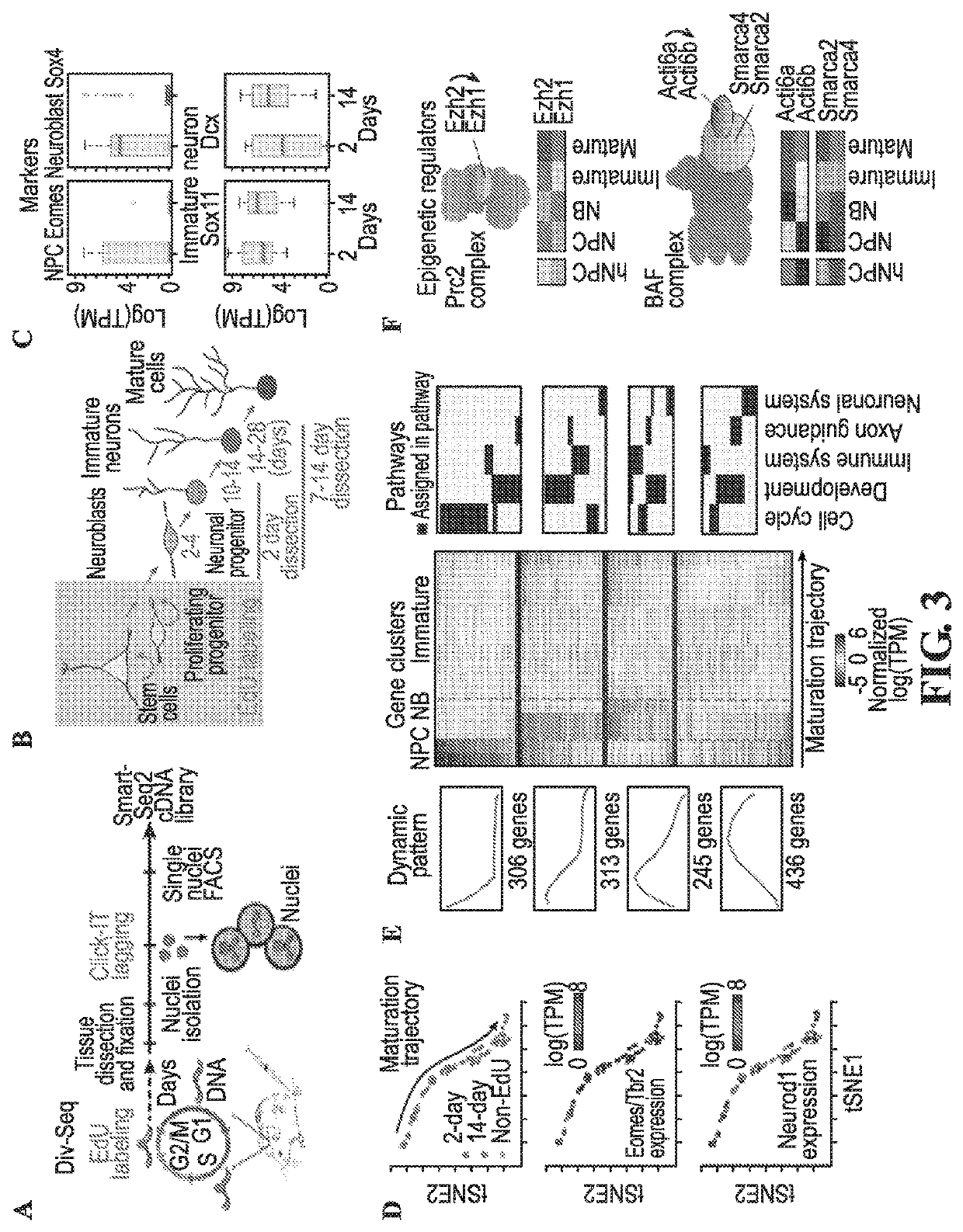
Methods for determining spatial and temporal gene expression dynamics during adult neurogenesis in single cells
PendingUS20200347449A1Microbiological testing/measurementPreparing sample for investigationCDNA libraryNuclear membrane
Provided herein are methods of recovering single nuclei from a tissue sample comprising chopping or dounce homogenizing the tissue sample in a nuclear extraction buffer at 4° C. to produce a tissue homogenate; centrifuging the tissue homogenate to produce a nuclear pellet; resuspending the nuclear pellet in a nuclear resuspension buffer comprising bovine serum albumin, RNase inhibitor, and salts to produce a resuspension; and filtering the resuspension through a strainer, wherein the single nuclei are present in a supernatant passed through the strainer. The invention also provides a method of single cell sequencing comprising extracting nuclei from a population of cells under conditions that preserve a portion of the outer nuclear envelope and rough endoplasmic reticulum; sorting single nuclei into separate reaction vessels; extracting RNA from the single nuclei; generating a cDNA library, whereby gene expression data from single cells are obtained. The tissue sample may be fresh or frozen.
Owner:HOWARD HUGHES MEDICAL INST +2
Method for separating cell nucleus from human frozen tumor tissue suitable for single cell sequencing
The invention discloses a method for separating a cell nucleus from a human frozen tumor tissue suitable for single cell sequencing and a method for extracting the cell nucleus. The method for extracting the cell nucleus comprises performing splitting treatment on a sample to be extracted in a first buffer solution; performing first centrifugation on a split product to obtain a cell nucleus precipitate; rinsing the cell nucleus and conducting resuspending treatment in a second buffer solution to obtain the cell nucleus; wherein the first buffer solution comprises: 116.8 mM of NaCl, 8 mM of Tris base at pH 7.8, 0.8 mM of CaCl2, 38 mM of MgCl2, 0.04% BSA, 0.16% Nonidet P-40, 1 mM of EDTA and 1 mg / mL of DAPI. The second buffer solution comprises 1*PBS, 1.0% BSA and 0.2U / [mu]L of RNase inhibitor. According to the method, only a simple reagent is required, no expensive separation equipment is required, and sufficient complete mononuclear can be obtained from the less frozen tumor tissue forsingle cell RNA sequencing.
Owner:杭州瑞普基因科技有限公司
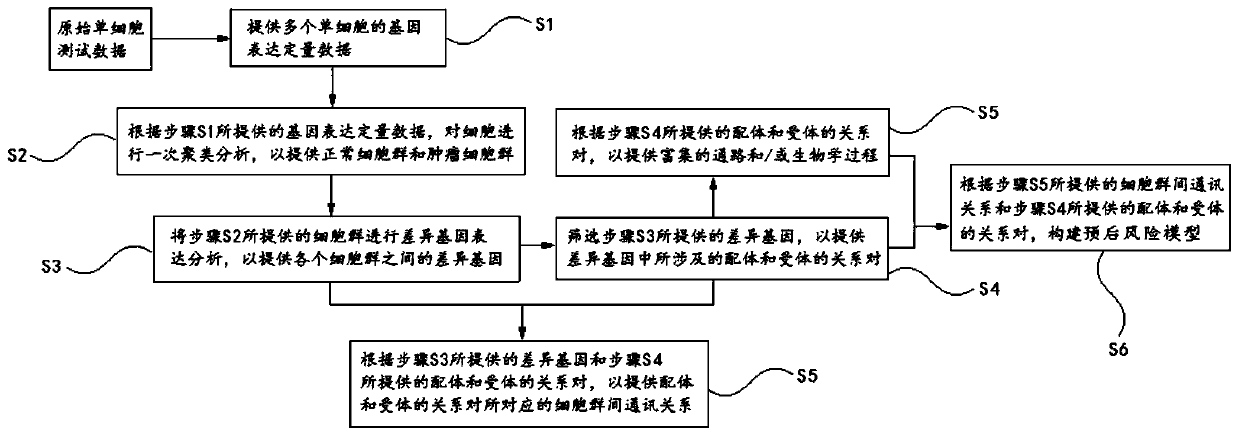
Receptor-ligand system analysis method influencing intercellular communication
The invention relates to bioinformatics, in particular to a receptor-ligand system analysis method influencing intercellular communication. The invention provides an analysis method which comprises the following steps: carrying out primary clustering analysis on cells according to provided gene expression quantitative data; carrying out differential gene expression analysis on the provided cell population; screening the provided differential genes; providing enriched pathways and / or biological processes according to the provided relationship pairs of ligands and receptors; and / or, according tothe provided differential genes and the provided relationship pairs of the ligands and the receptors, providing the inter-cell population communication relationship corresponding to the relationshippairs of the ligands and the receptors. The analysis method provided by the invention is based on single cell sequencing instead of traditional RNA-seq, can deeply understand transcriptome under single cell resolution, can deeply understand tumor heterogeneity, and can further construct a prognosis risk model in combination with an external common data set.
Owner:上海源兹生物科技有限公司
DNA barcode compositions and methods of in situ identification in a microfluidic device
ActiveCN110114520ASequential/parallel process reactionsCell receptors/surface-antigens/surface-determinantsSequence analysisLibrary preparation
Apparatuses, compositions and processes for DNA barcode de-convolution are described herein. A DNA barcode may be used to provide a bead specific identifier, which may be detected in situ using hybridization strategies. The DNA barcode provides identification by sequencing analysis. The dual mode of detection may be used in a wide variety of applications to link positional information with assay information including but not limited to genetic analysis. Methods are described for generation of bar coded single cell sequencing libraries. Isolation of nucleic acids from a single cell within a micro fluidic environment can provide the foundation for cell specific sequencing library preparation.
Owner:BERKELEY LIGHTS INC
Single-cell genome copy number variation detection method and kit
InactiveCN108410970ALower reaction costReduce the degree of reactionMicrobiological testing/measurementLibrary creationInformation analysisPollution
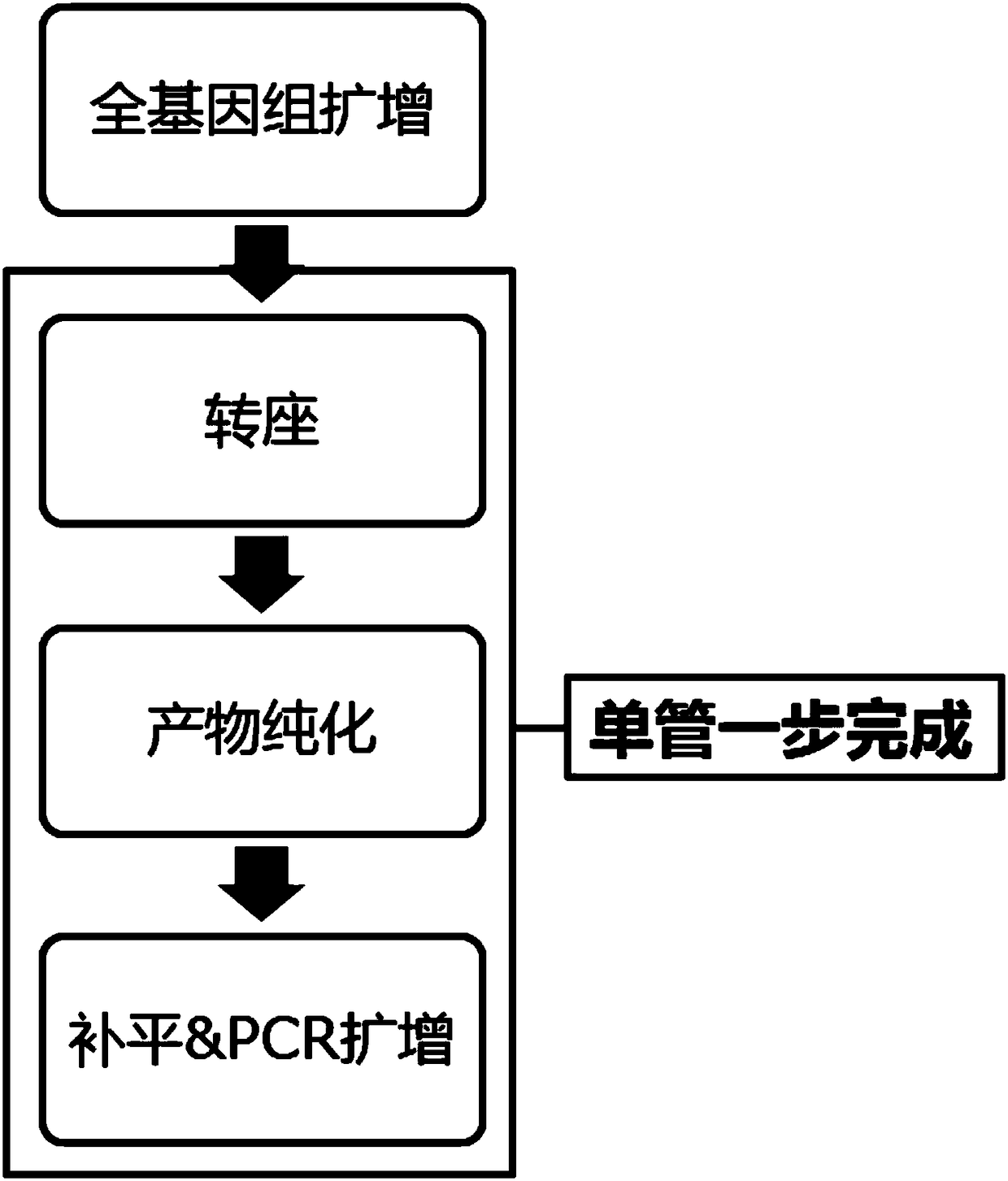
The invention relates to the technical field of biology, in particular to a combination, a kit and application of the kit. The application includes a solution of single-cell genome copy variation detection, comprising single cell amplification, library construction, high-throughput sequencing and information analysis; library construction based on transposase is applied to single cell sequencing;the use of single-tube one-step library construction process helps relieve operational complexity, thus avoiding pollution. In addition, the invention also provides optimal window length according tothe requirement on CNV (copy number variation) lowest detection resolution; run test is performed to count distribution differences, relative to coverage, of CNV area and normal area sequencing data;significance P value of each candidate CNV is output in final results; detection accuracy is effectively improved.
Owner:CAPITALBIO CORP
Method for manufacturing single cell sequencing reference material by utilizing oral epithelial cells
InactiveCN104404034AEasy to set upIncrease surface areaMicrobiological testing/measurementDNA preparationEnzyme digestionPhosphate
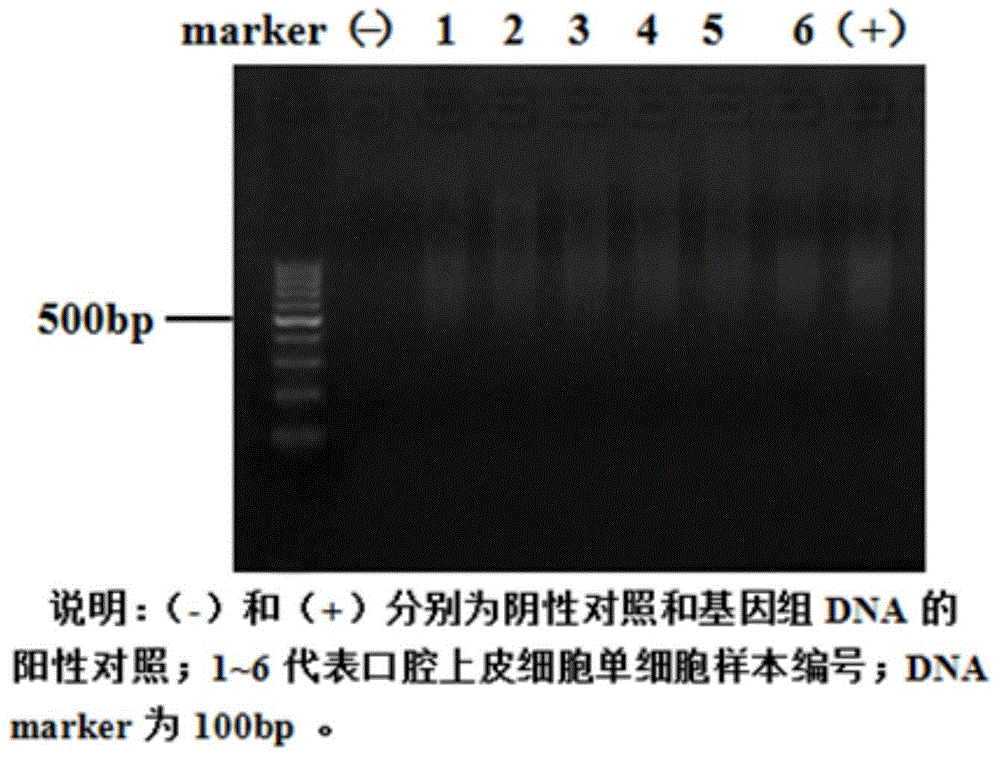
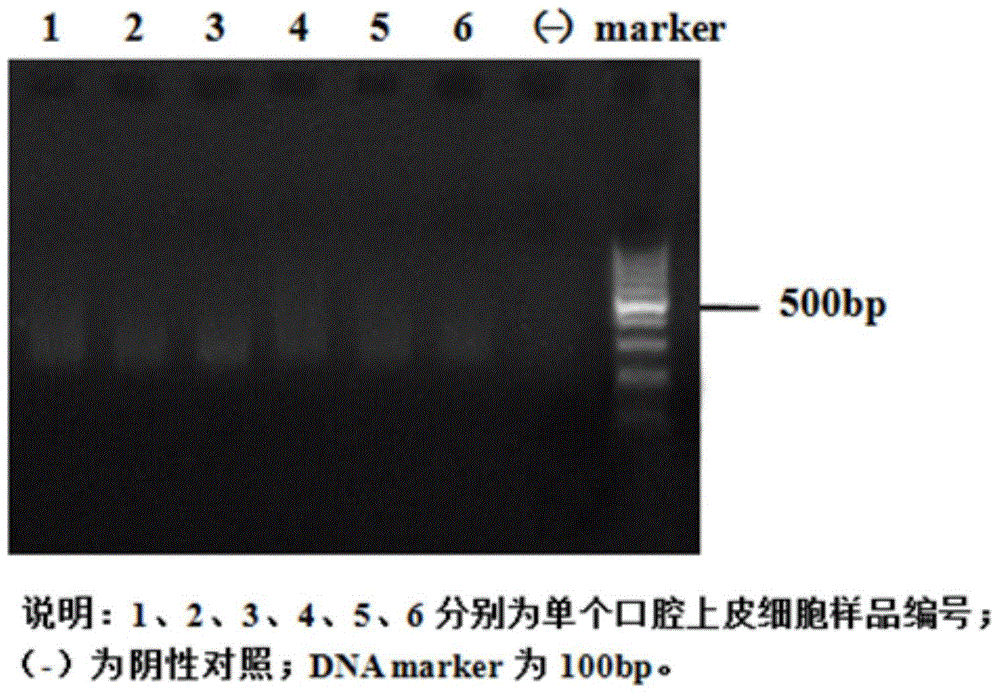
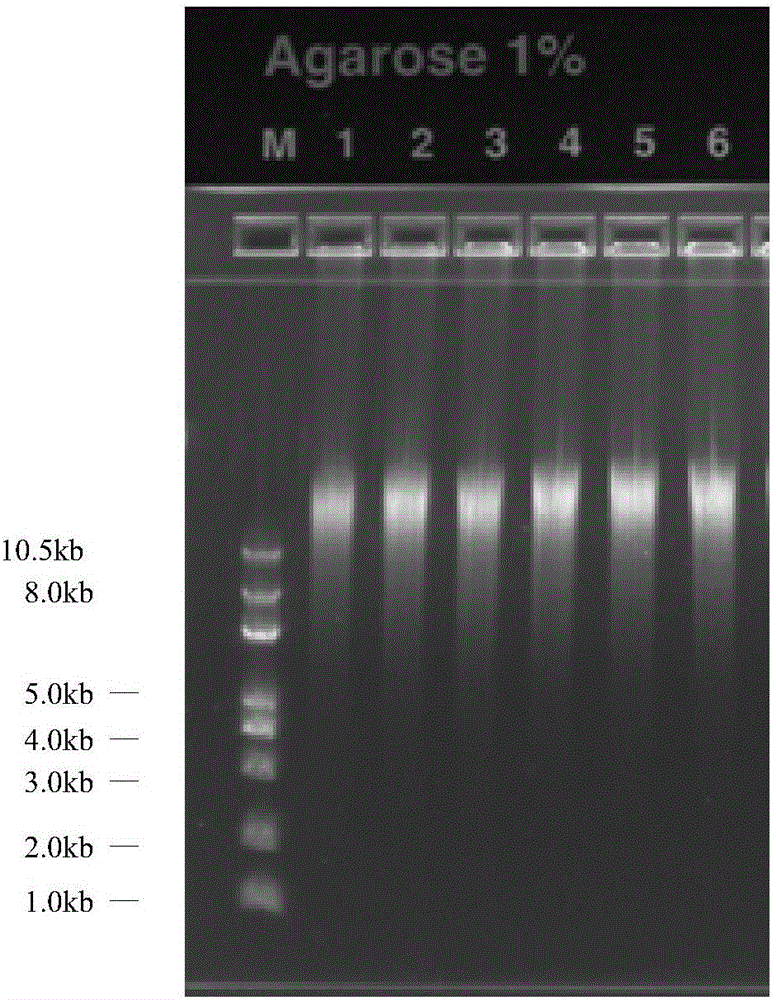
The invention relates to a method for manufacturing a single cell sequencing reference material by utilizing oral epithelial cells. The method comprises the following steps: (a) scrapping oral epithelial cells and saving in PBS (phosphate buffer solution); (b) sucking up a single oral epithelial cell by using capillary-opening suction pipe under a microscope; (c) amplifying a single-cell whole genome, and purifying the amplified single cell by using a paramagnetic particle method; (d) performing enzyme digestion on the amplified sample, stopping the experiment by using EDTA (ethylenediamine tetraacetic acid), and purifying by using the paramagnetic particle method; (e) constructing a Proton library; (f) performing computer sequencing on a Proton semiconductor high-throughput sequencing platform; (g) analyzing the sequencing data. The invention provides a new way of preparation of a standard product for single cell associated research; the method for manufacturing the single cell sequencing reference material by utilizing the oral epithelial cells has the characteristics that materials are simple to obtain, the operation is convenient and quick and the resolution is high, and is suitable for projects, such as PGS (preimplantation genetic screening) and tumor early screening.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Method for detecting trace fungus by using single-cell sequencing and kit
ActiveCN105925720AEfficient detectionMicroorganismsMicrobiological testing/measurementGenomic sequencingWater quality
The invention relates to the field of microorganism and molecular biology, and in particular relates to a method for detecting trace fungus by using single-cell sequencing and a fungus detection kit produced by using the method. The method comprises the steps of extraction of trace fungus cells, extraction of fungus protoplasm by fragmentation of fungus cell wall, extraction and amplification of trace fungus protoplasm gDNA, database creation of gDNA, genome sequencing, bioinformatic analysis comparison, and judgment on the type of the detected fungus. The method disclosed by the invention realizes effective detection on trace fungus, and can be directly applied to separation, detection and identification of trace difficult fungus samples or mixed samples and deep study of genetic information. The fungus detection method and the kit are applicable to the fields of industrial production, environmental monitoring, air detection, soil detection, water quality detection, food detection, drug detection, cosmetic detection, health care products detection and medical detection.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
Debonding protectant, method for preparing monocell suspension with high survival rate and application of debonding protectant
PendingCN111235090AGood effectShorten the timeCell dissociation methodsPancreatic cellsBiotechnologyAnimal science
Owner:CHENGDU DAOSHENG BIOTECH CO LTD
Method for constructing single cell sequencing library and use thereof
ActiveUS20190338279A1Reducing omics information lossEasy to analyzeMicrobiological testing/measurementLibrary creationTotal rnaTranscriptome Sequencing
Provided in the present invention is a method for constructing a single cell sequencing library, comprising the following steps: a) lysing a single cell to obtain a single cell lysate; b) separating the nucleus and the cytoplasm in the single cell lysate obtained in step a) to obtain a nuclear solution and a total RNA solution; and c) constructing a chromatin DNA library with the nuclear solution obtained in step b) to obtain a chromatin-accessibility sequencing library of the single cell; and constructing a transcriptome library with the total RNA solution obtained in step b) to obtain a transcriptome sequencing library of the single cell.
Owner:MGI TECH CO LTD
Preparation method and application of pulmonary artery tissue unicellular suspension
ActiveCN111621466AQuality assuranceEasy to operateCell dissociation methodsMicrobiological testing/measurementSingle cell suspensionPulmonary endarterectomy
The invention provides a preparation method of a pulmonary artery tissue unicellular suspension for unicellular sequencing. The unicellular suspension can be obtained by separating, cleaning, trimming, incubating and enzymatically digesting pulmonary endarterectomy tissues. According to the method disclosed by the invention, while the cell quality is ensured, a large number of separated cells derived from the pulmonary endarterectomy tissues can be obtained at a time; and a used reagent is simple and convenient to operate, and the method has good practical application value.
Owner:BEIJING CHAOYANG HOSPITAL CAPITAL MEDICAL UNIV
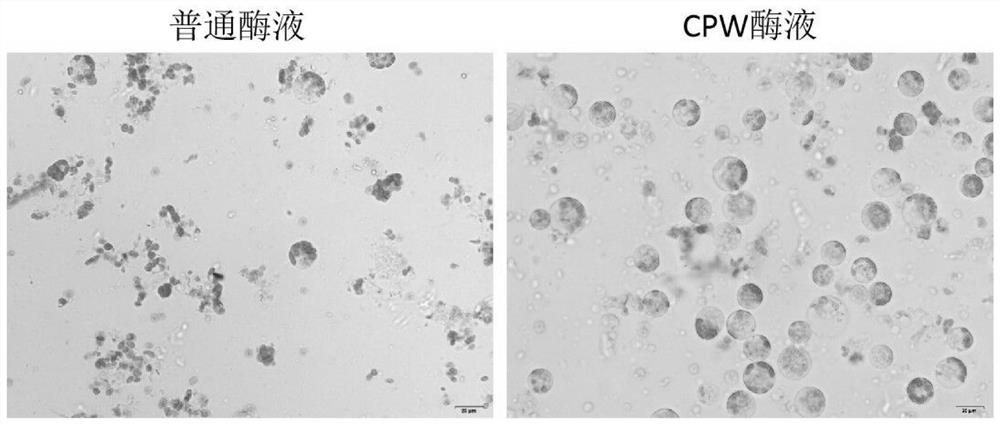
Rapid and efficient cymbopogon citratus protoplast preparation method
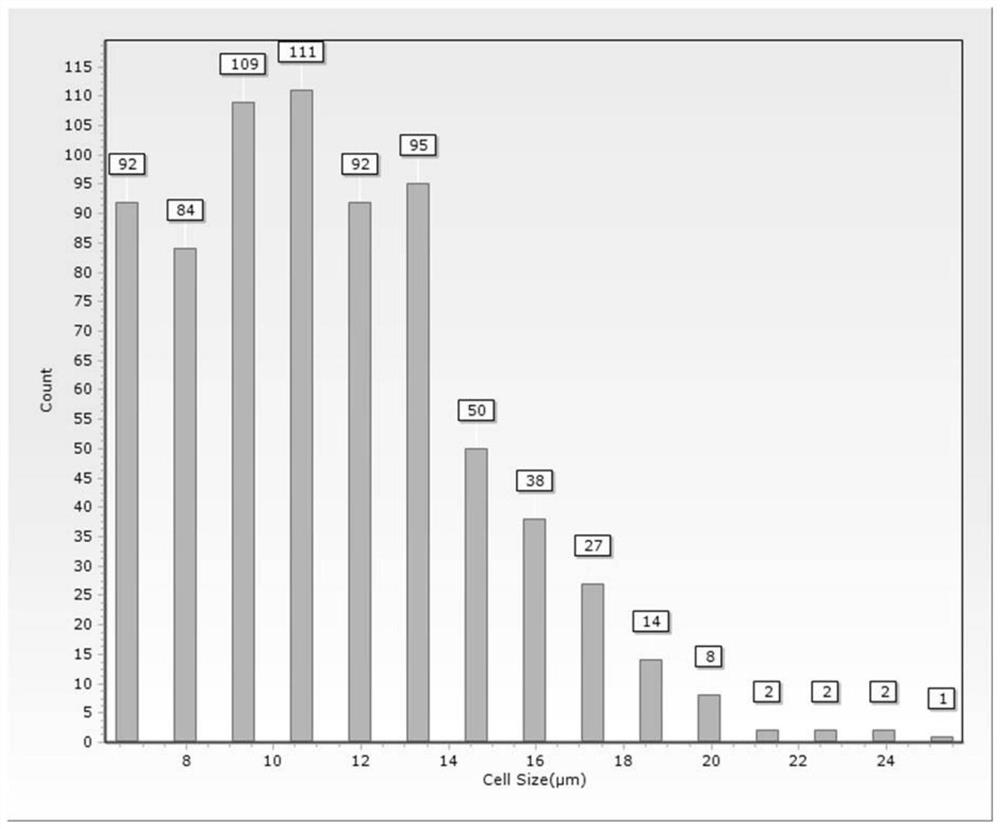
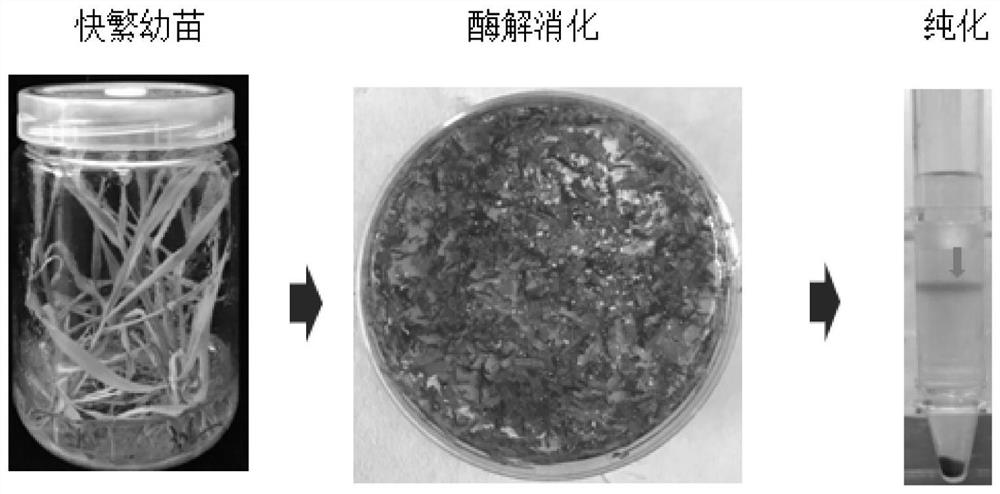
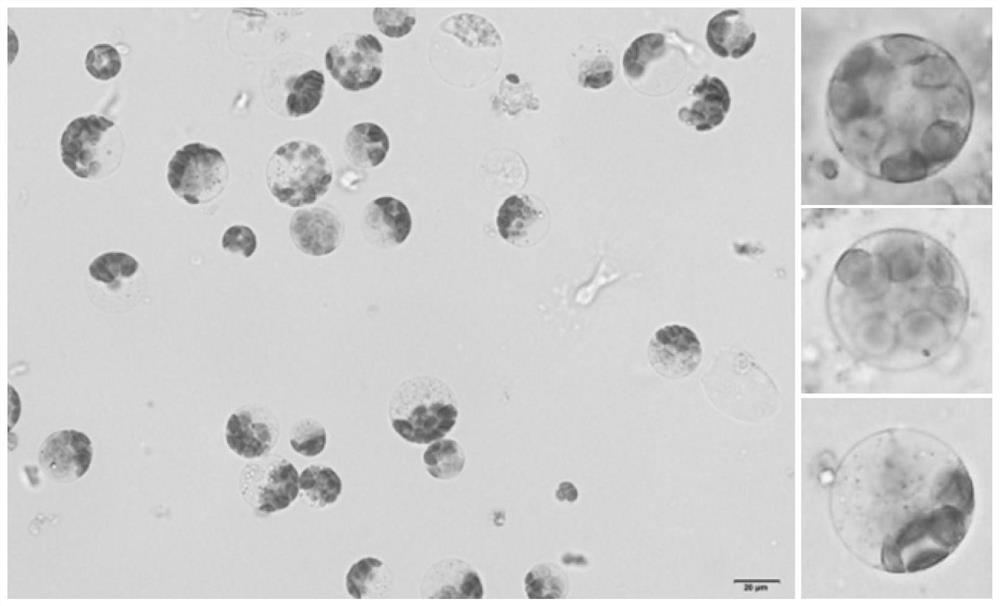
The invention discloses a rapid and efficient cymbopogon citratus protoplast preparation method, and belongs to the field of plant cell engineering. Healthy-growing cymbopogon citratus tissue culture seedlings are adopted as experimental materials, a CPW solution is used for preparing enzymatic hydrolysate, a low-speed oscillation enzymolysis method is adopted for carrying out enzymolysis digestion on cymbopogon citratus leaves, and the protoplast is efficiently prepared. And finally, purifying and collecting of the high-quality cymbopogon citratus protoplast is conducted by using a sucrose density gradient centrifugation method. The method is simple, convenient and easy to operate, and the separated protoplast is high in yield, complete in cell, spherical, rich in chloroplast and relatively high in activity. The invention provides an important basis for research and application in the aspects of gene function identification, subcellular localization, protein interaction, citronella cell fusion, single cell sequencing and the like by utilizing a cymbopogon citratus leaf instantaneous transformation system.
Owner:SHANGHAI CHENSHAN BOTANICAL GARDEN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com