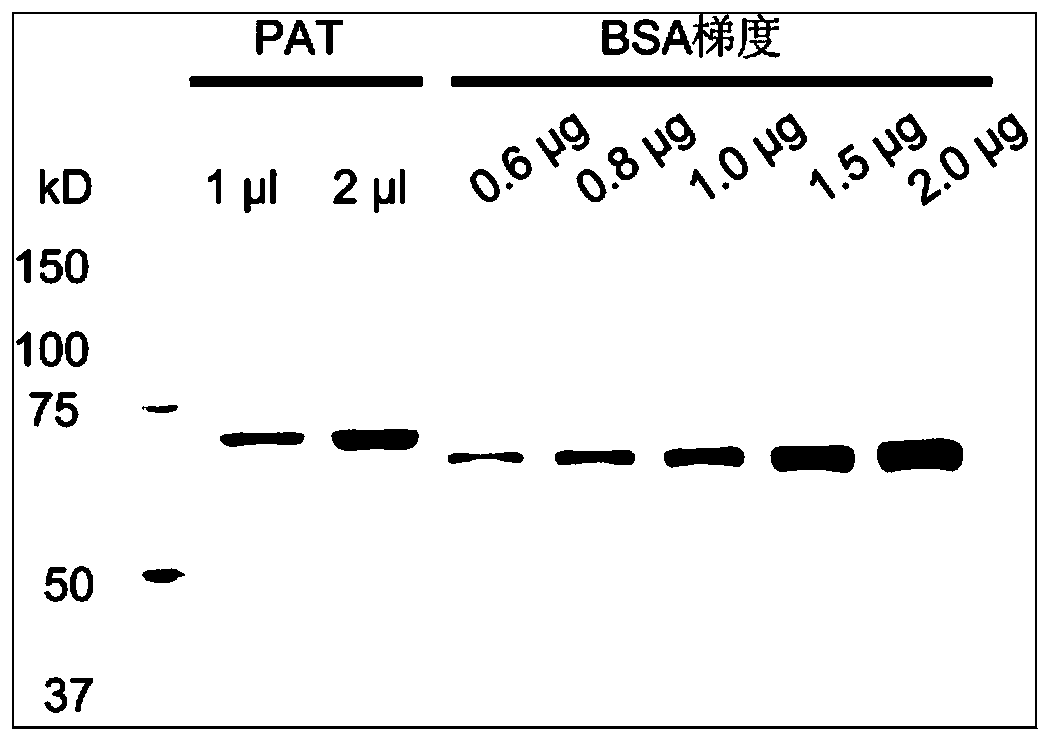
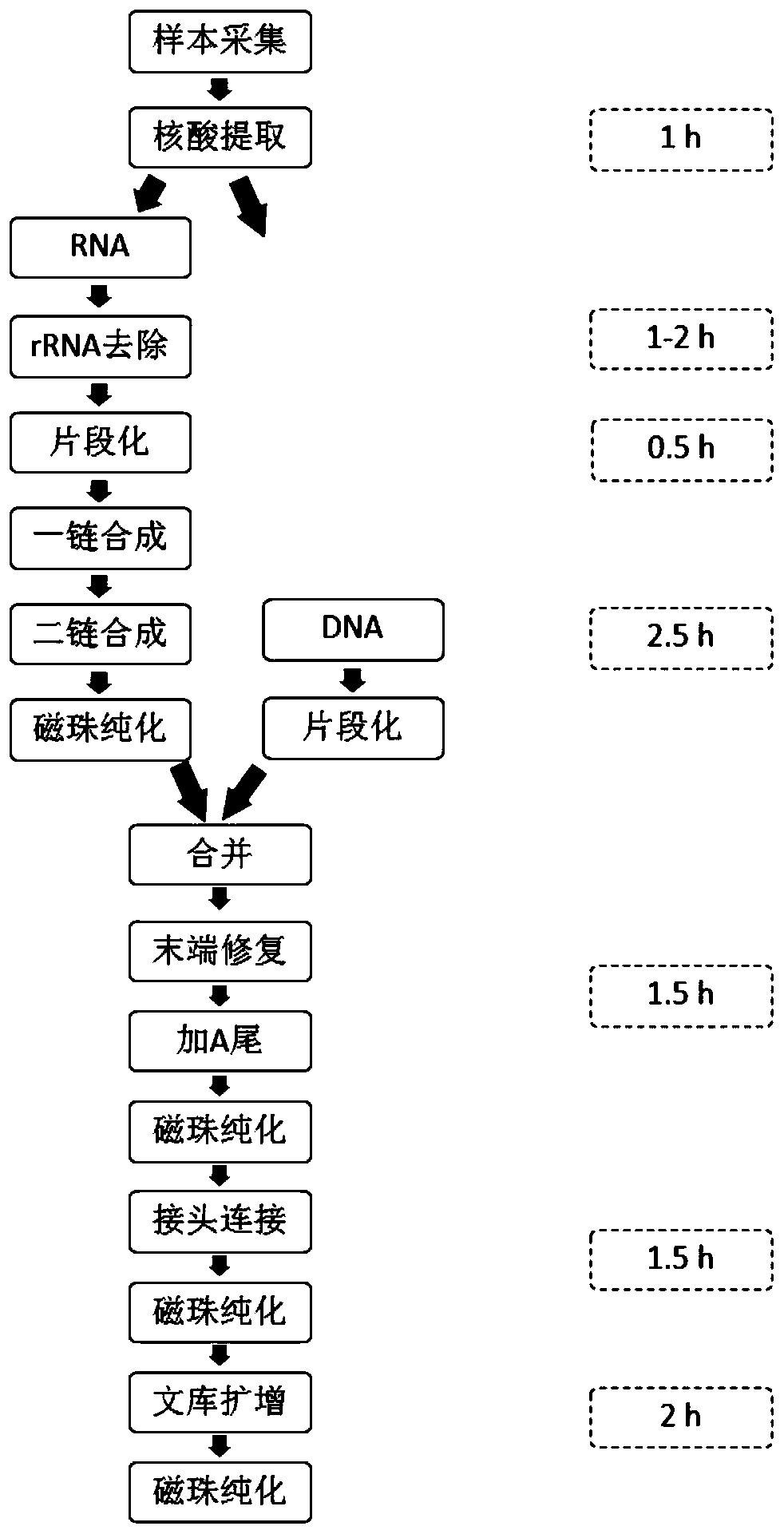
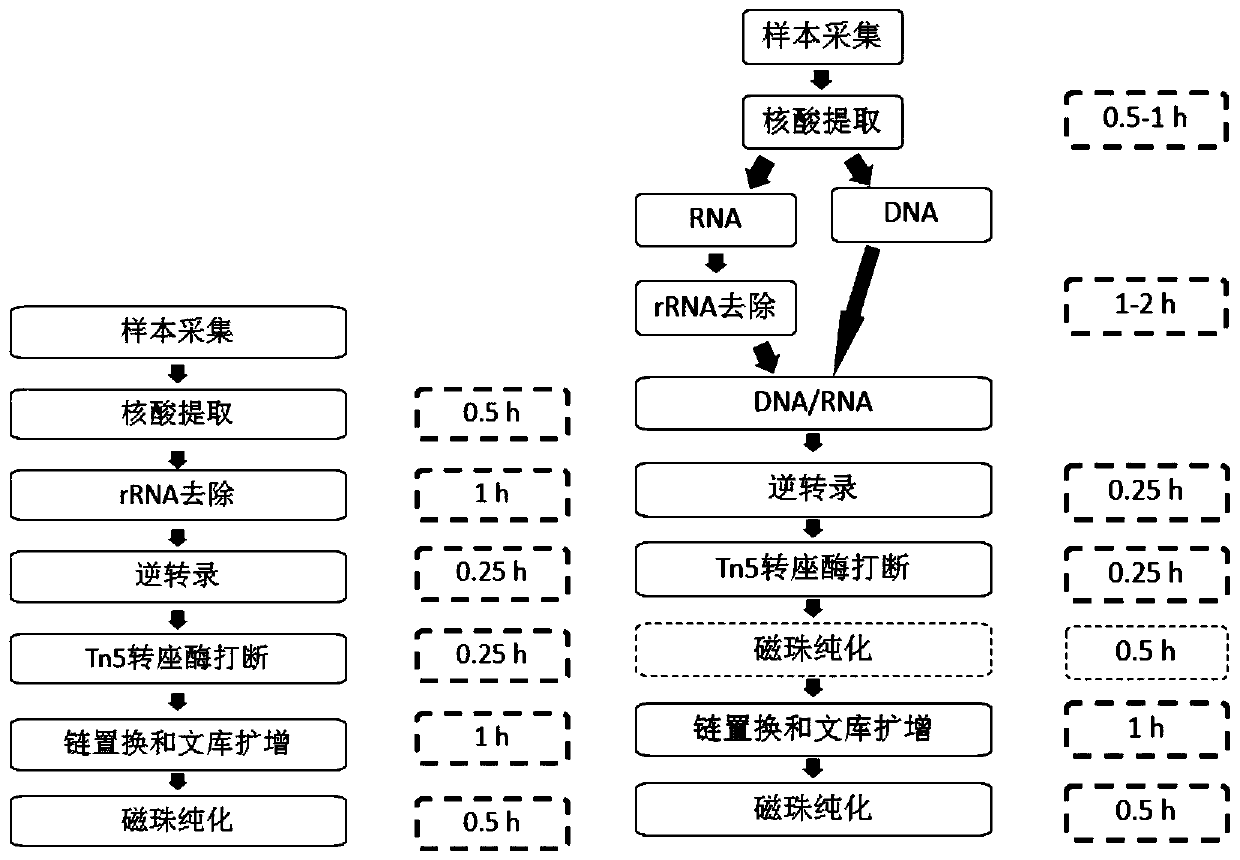
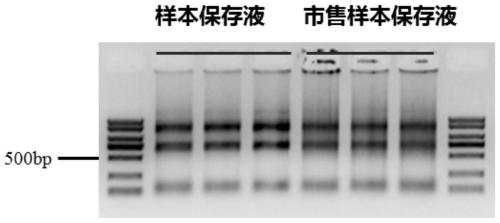
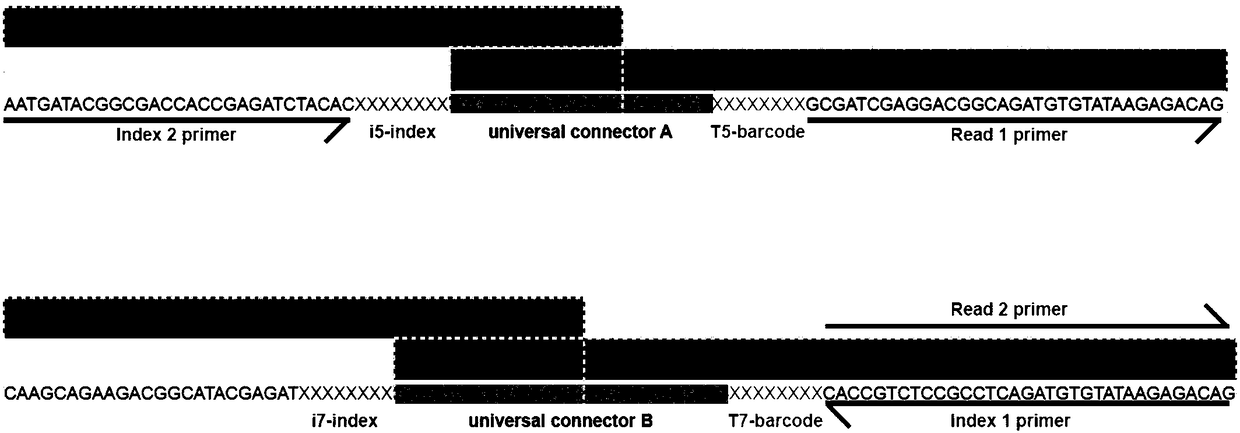
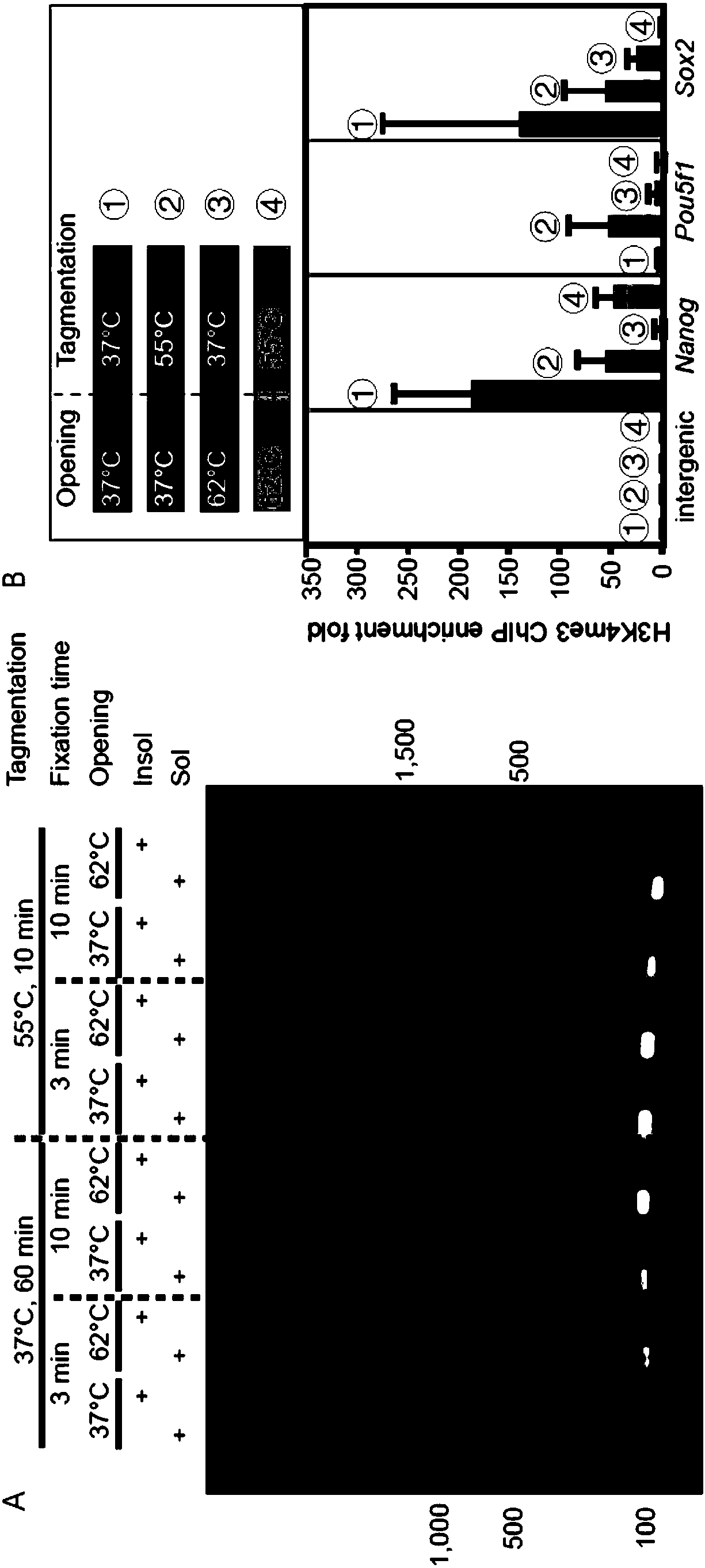
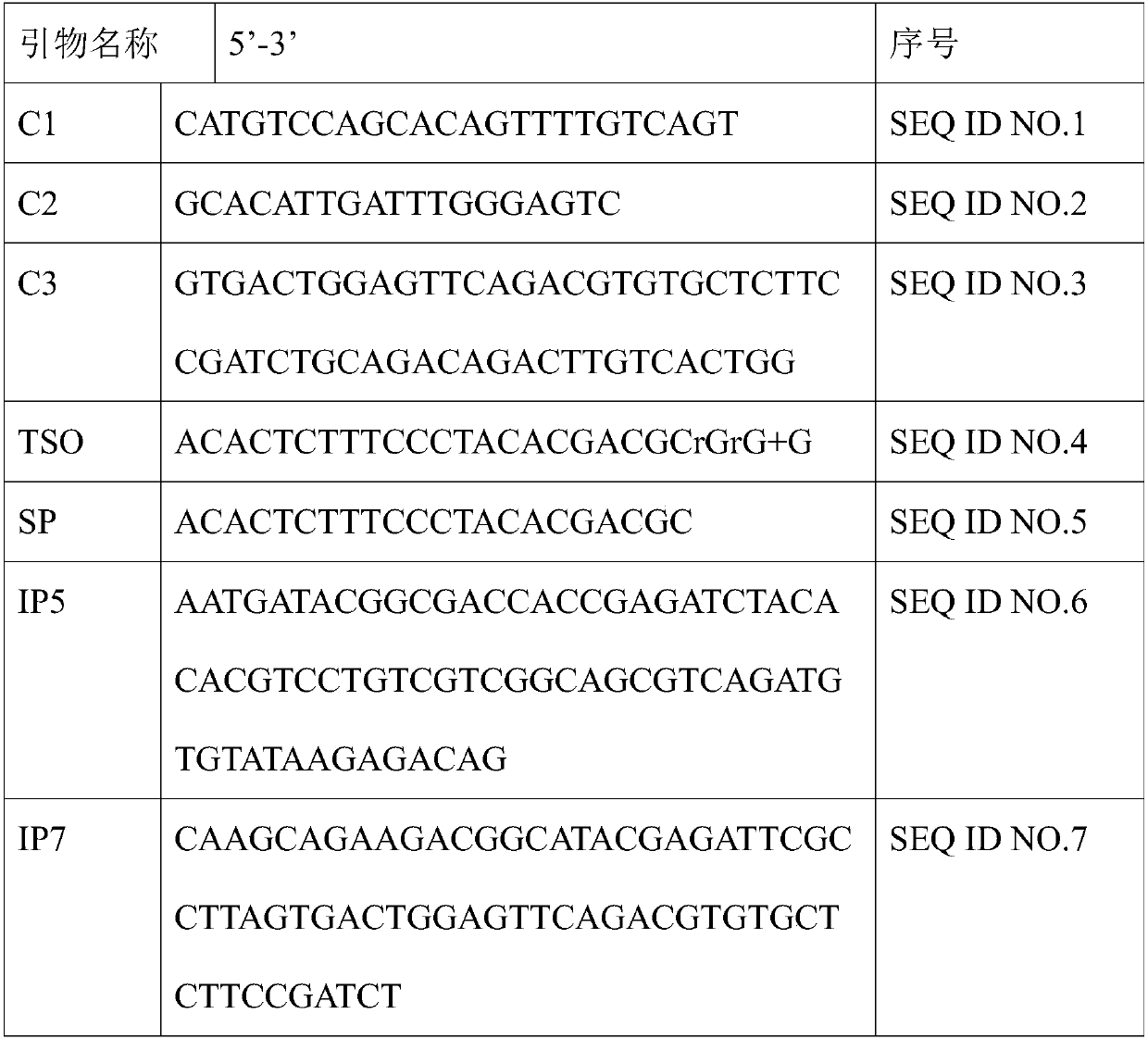
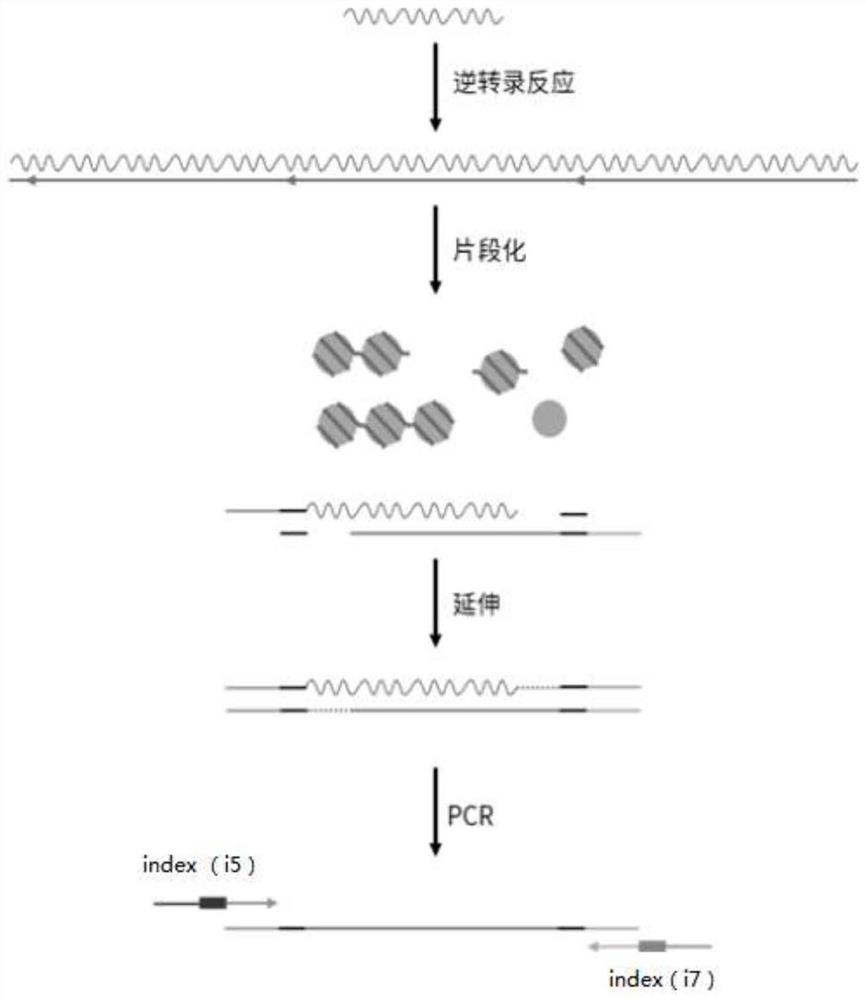
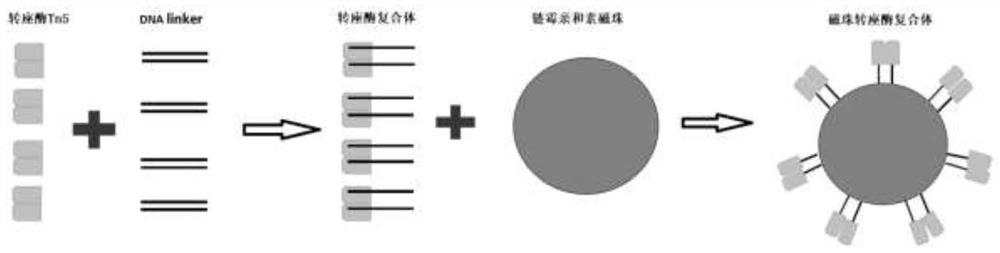
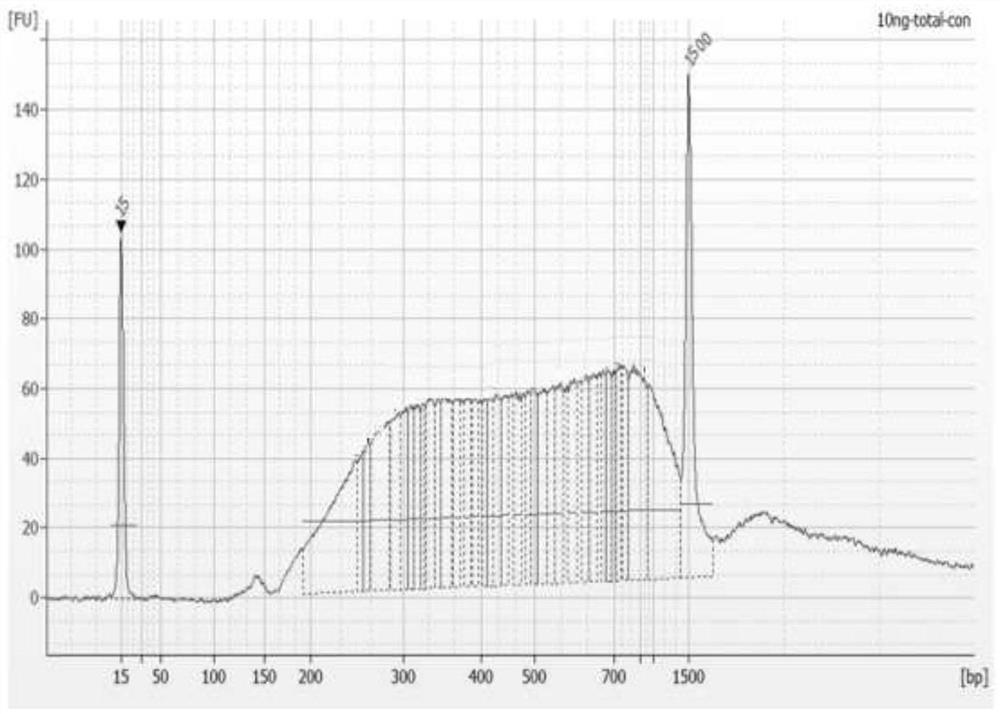
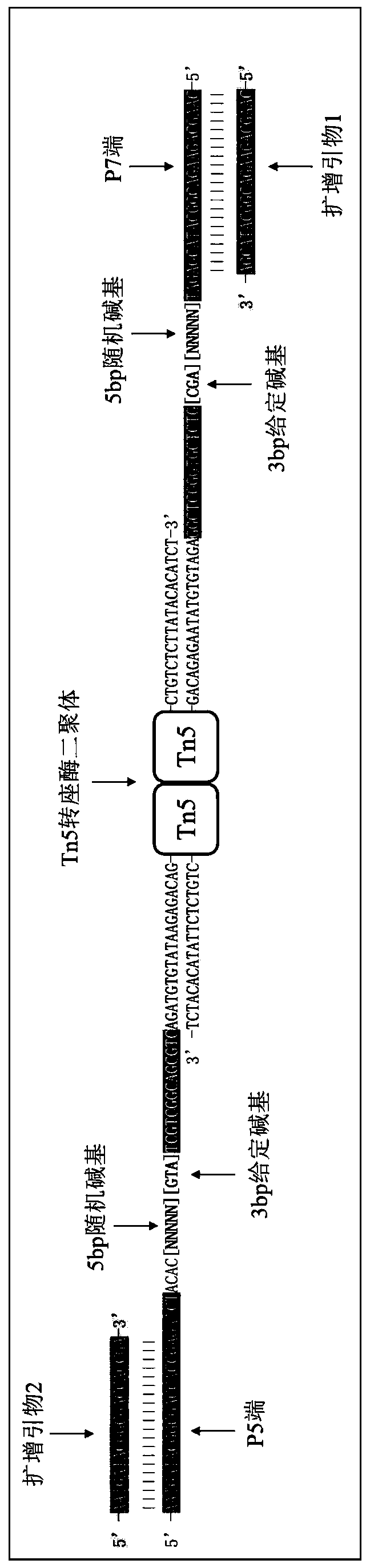
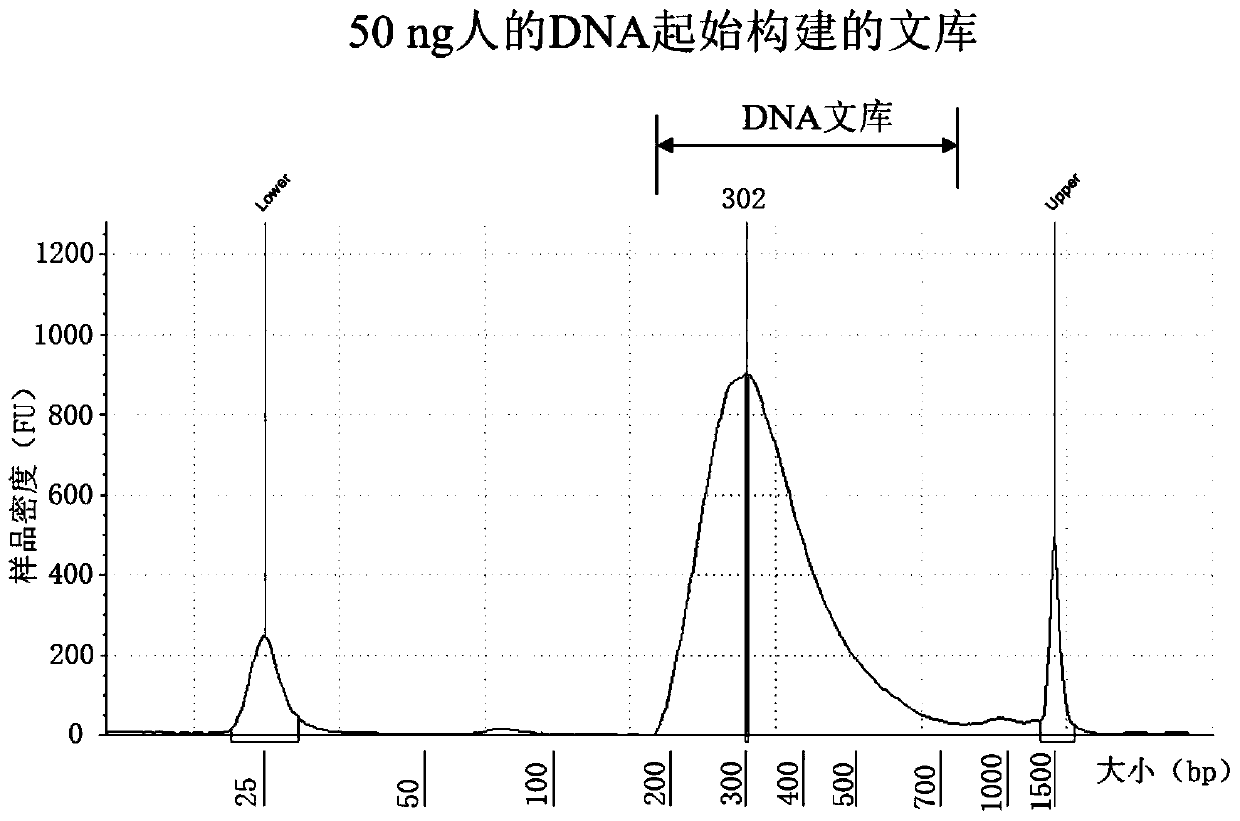
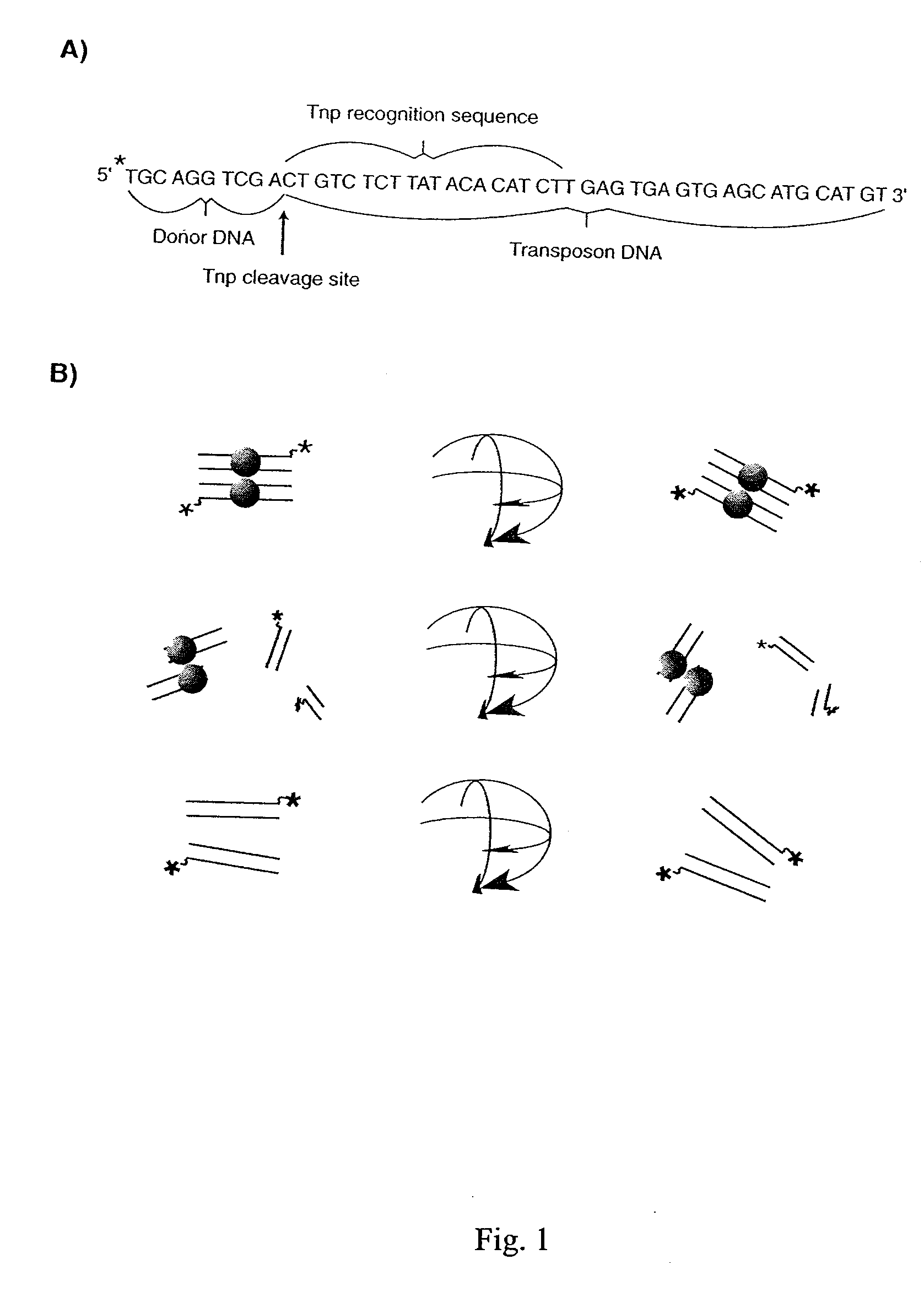Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
76 results about "Tn5 transposase" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Mutated Tn5 transposase proteins and the use thereof
Transposase proteins that are modified relative to and have higher transposase activities than the wild-type Tn5 transposase are disclosed. A transposase protein of the present invention differs from the wild-type Tn5 transposase at amino acid position 41, 42, 450, or 454 and has greater avidity than the wild-type Tn5 transposase for at least one of a Tn5 outside end sequence as defined by SEQ ID NO:3, a Tn5 inside end sequence as defined by SEQ ID NO:4, and a modified Tn5 outside end sequence as defined by SEQ ID NO:5. Also disclosed are various systems and methods of using the transposase proteins of the present invention for in vitro or in vivo transposition.
Owner:WISCONSIN ALUMNI RES FOUND
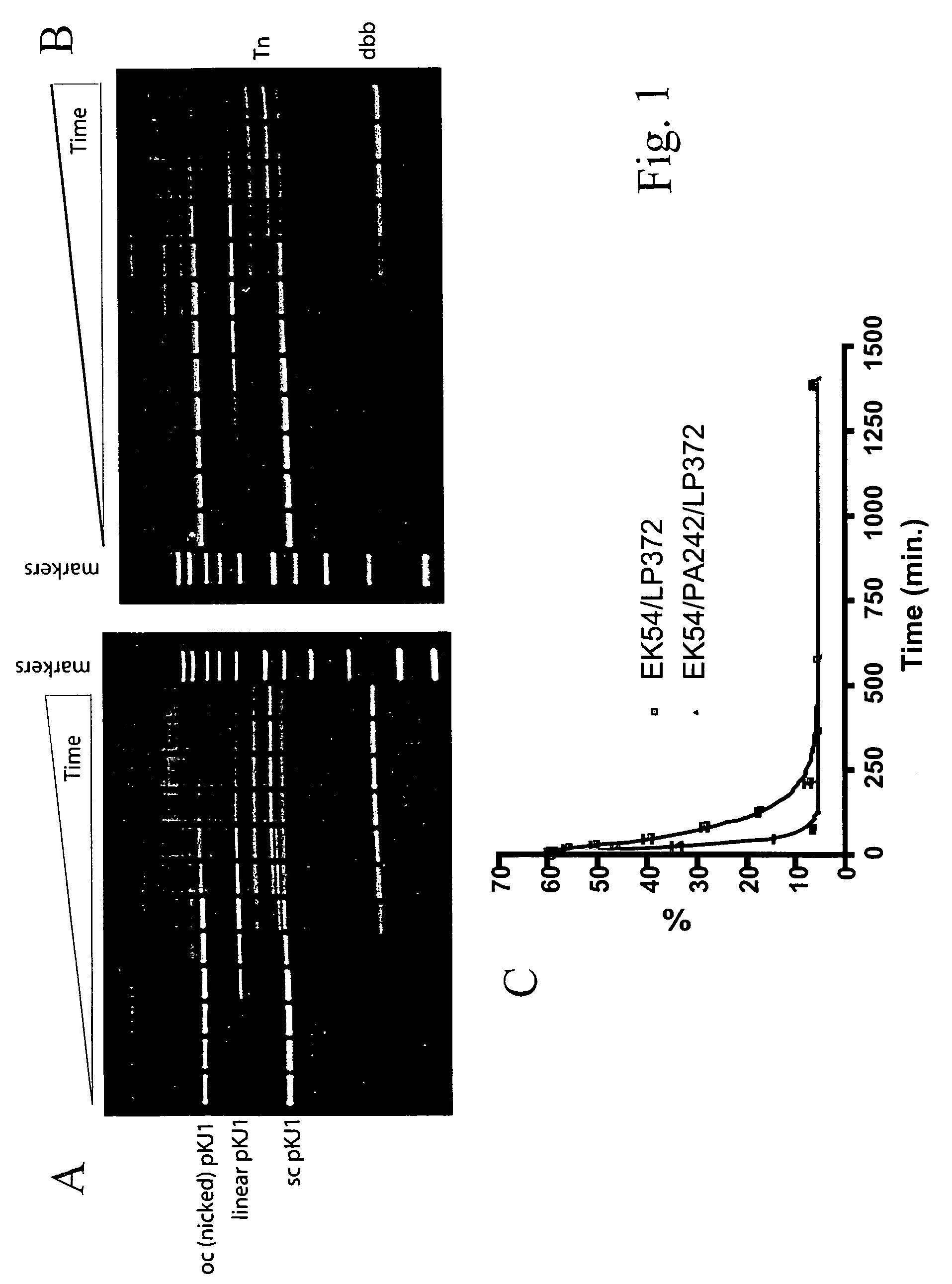
Tn5 transposase mutants and the use thereof
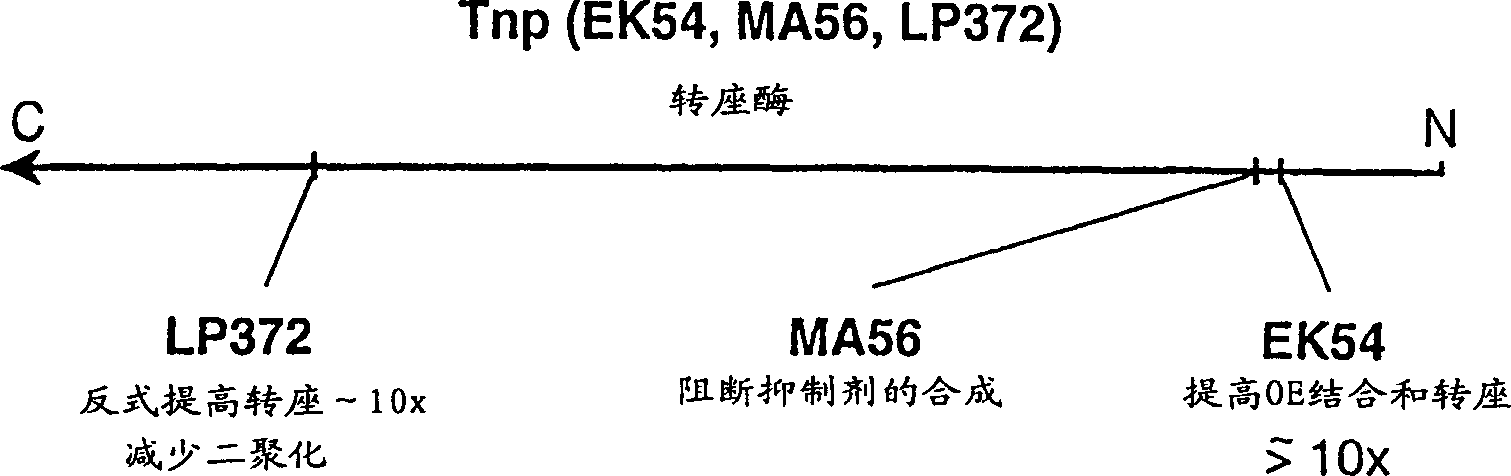
Tn5 transposase (Tnp) mutants that have higher transposase activities than the wild-type Tnp are disclosed. The Tn5 Tnp mutants differ from the wild-type Tnp at amino acid positions 54, 242, and 372 and have greater avidity than the wild-type Tnp for at least one of a wild-type Tn5 outside end sequence as defined by SEQ ID NO:3 and a modified Tn5 outside end sequence as defined by SEQ ID NO:5. Also disclosed are various systems and methods of using the Tnp mutants for in vitro or in vivo transposition.
Owner:WISCONSIN ALUMNI RES FOUND
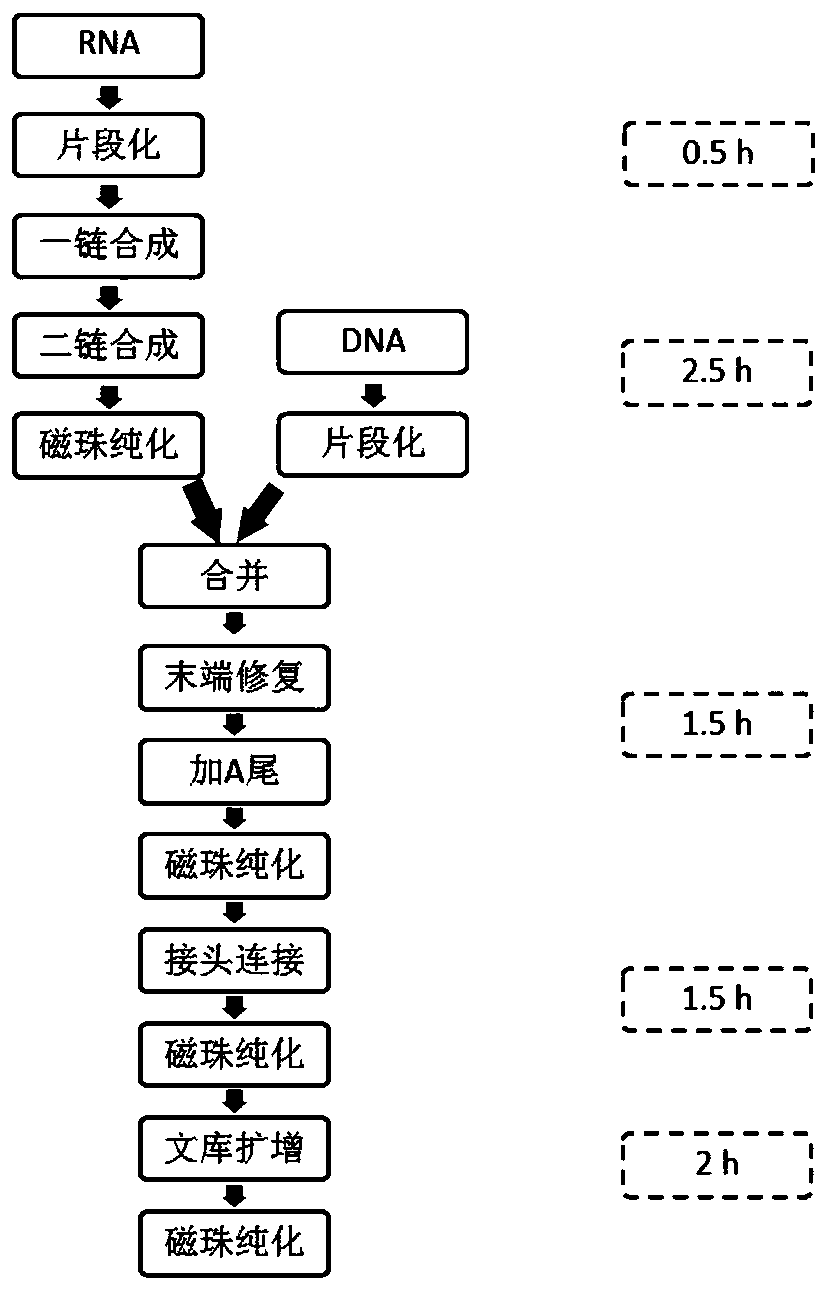
Method for building high-flux single-cell full-length transcriptome sequencing library and application of method
ActiveCN109811045AGuaranteed accuracyIntegrity guaranteedMicrobiological testing/measurementLibrary creationTn5 transposaseLysis
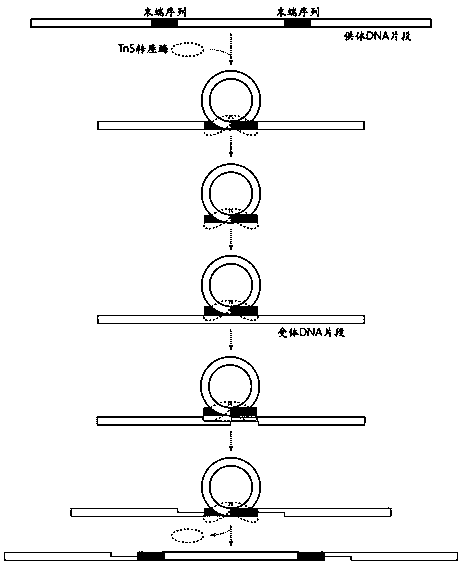
The application discloses a method for building a high-flux single-cell full-length transcriptome sequencing library and application of the method. The method comprises the following steps: recognizing single cells by adopting a customized micro-hole chip and an ICELL8 platform, and performing cell lysis, mRNA reverse transcription, cDNA pre-amplification and Tn5 transposase library building on the single cells, wherein a product can be directly applied to sequencing; in a process of mRNA reverse transcription or Tn5 transposase library building, introducing a dual-terminal Barcode sequence with a 5' terminal and a 3' terminal in order to obtain a single cell full-length transcriptome. By adopting the method, the single cell recognition flux is high, and the product can be directly appliedto sequencing. When the method is applied to single-cell sequencing, only two days are required for efficiently obtaining thousands of single-cell full-length transcriptome libraries at one time, sothat the labor cost and time cost are lowered greatly; moreover, the reagent cost is lowered greatly through a trace reaction system, and a basis is laid for large-scale single-cell full-length transcript sequencing.
Owner:MGI TECH CO LTD
Modulators of enzymatic nucleic acid elements mobilization
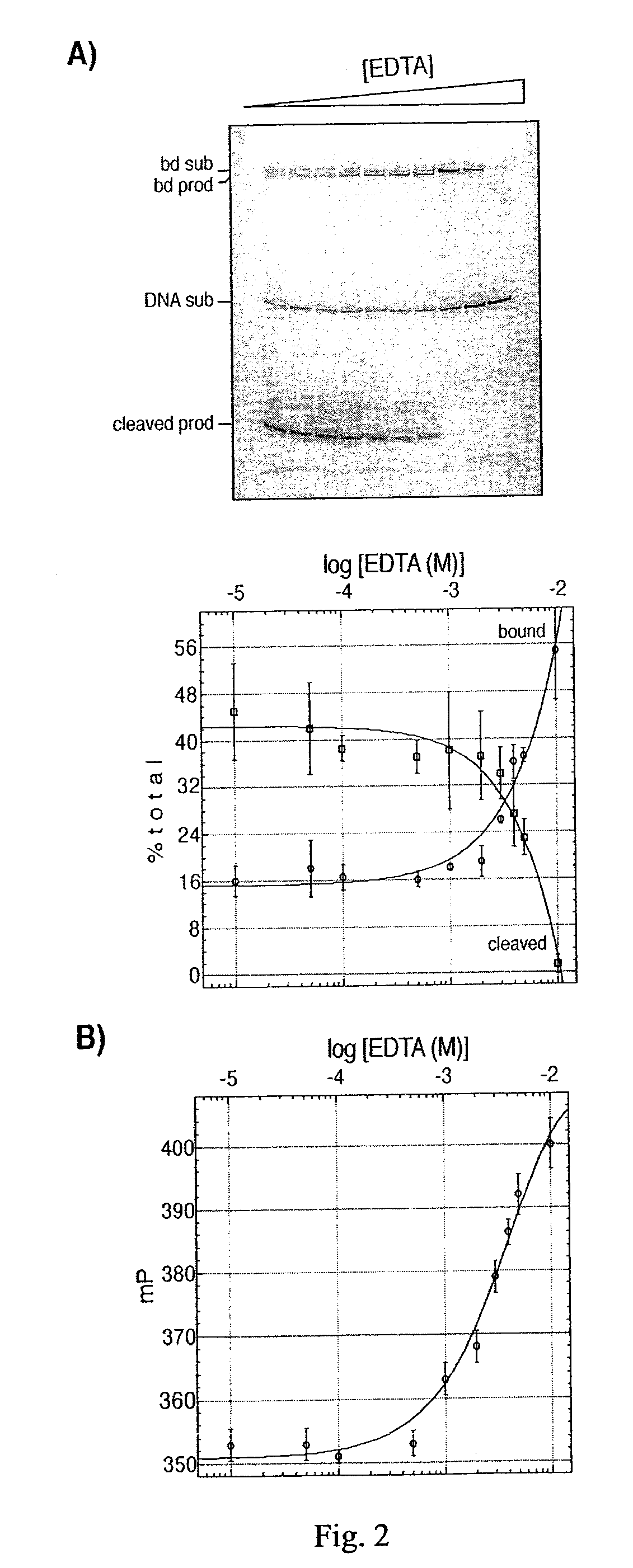
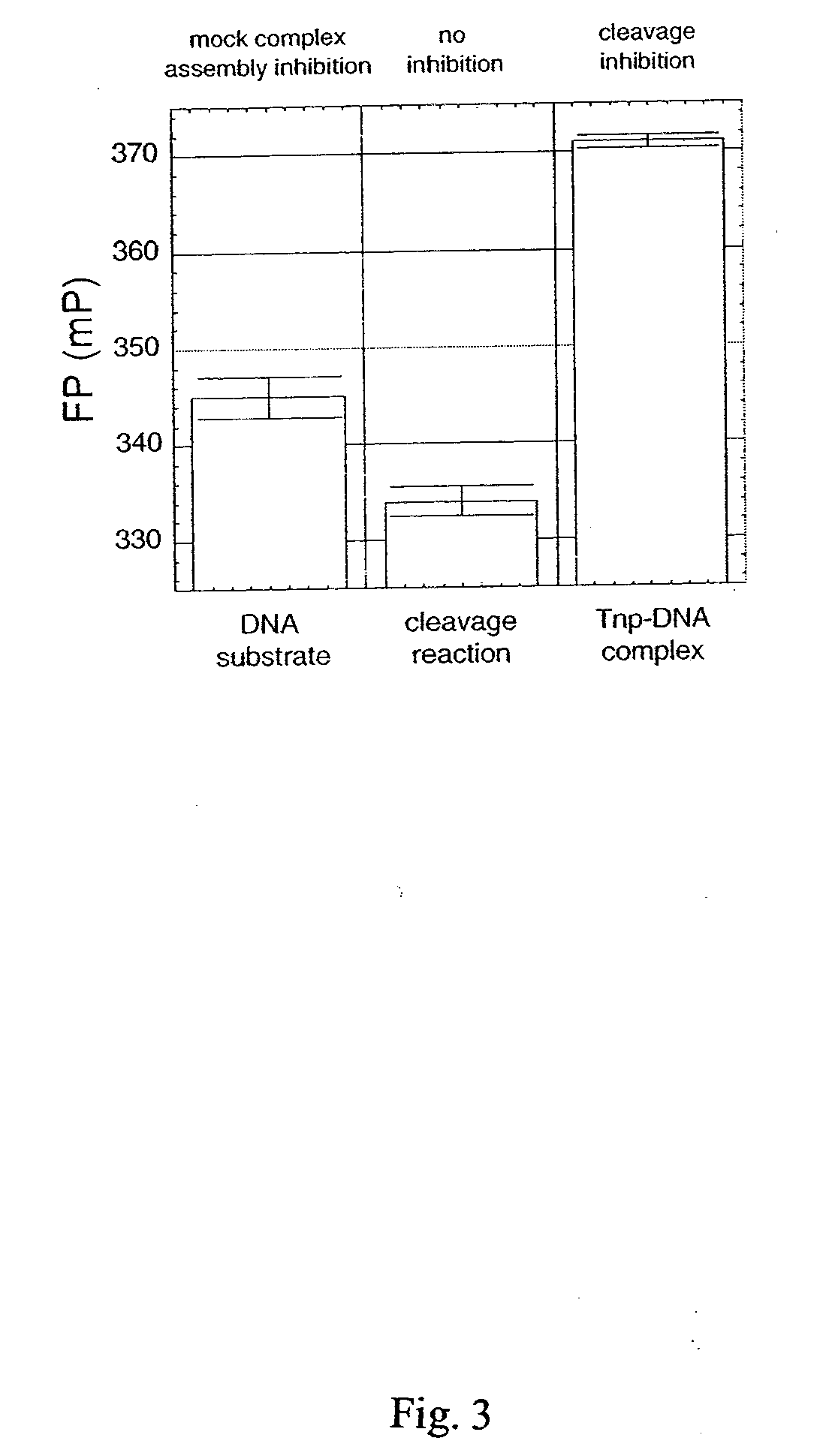
The present invention discloses a nucleic acid cleavage assay for members of the transposase / integrase superfamily. A method of using the assay to screen for modulators of the nucleic acid cleavage activity is also disclosed. The present invention further provides a method for screening for modulators of binding of a transposase / integrase to its corresponding recognition sequence. In addition, the present invention provides a method of identifying a modulator for a particular transposase / integrase such as HIV integrase based on modulators of other members of the transposase / integrase superfamily. Also disclosed are Tn5 transposase inhibitors and HIV integration inhibitors.
Owner:WISCONSIN ALUMNI RES FOUND
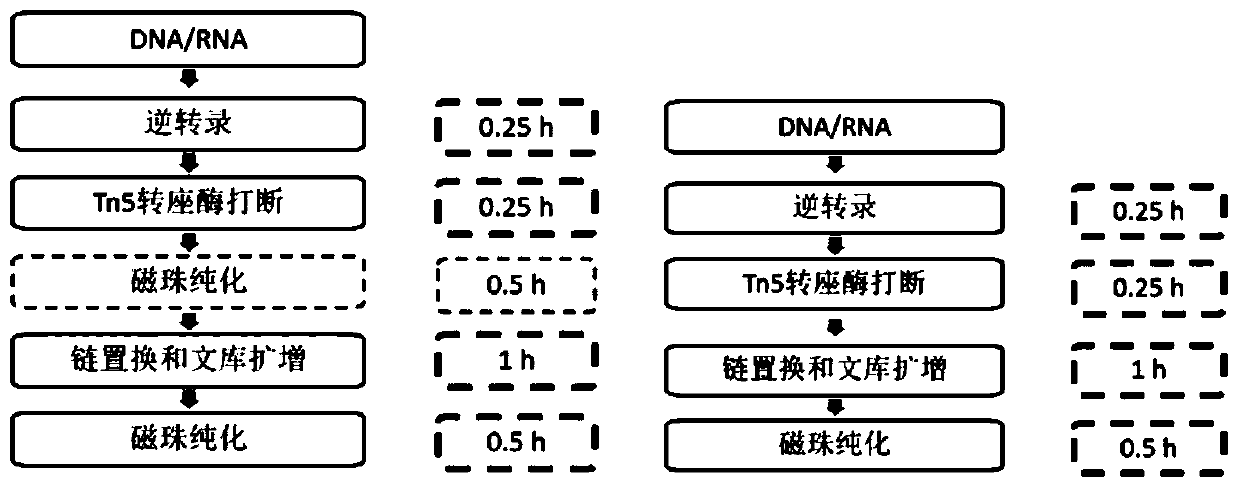
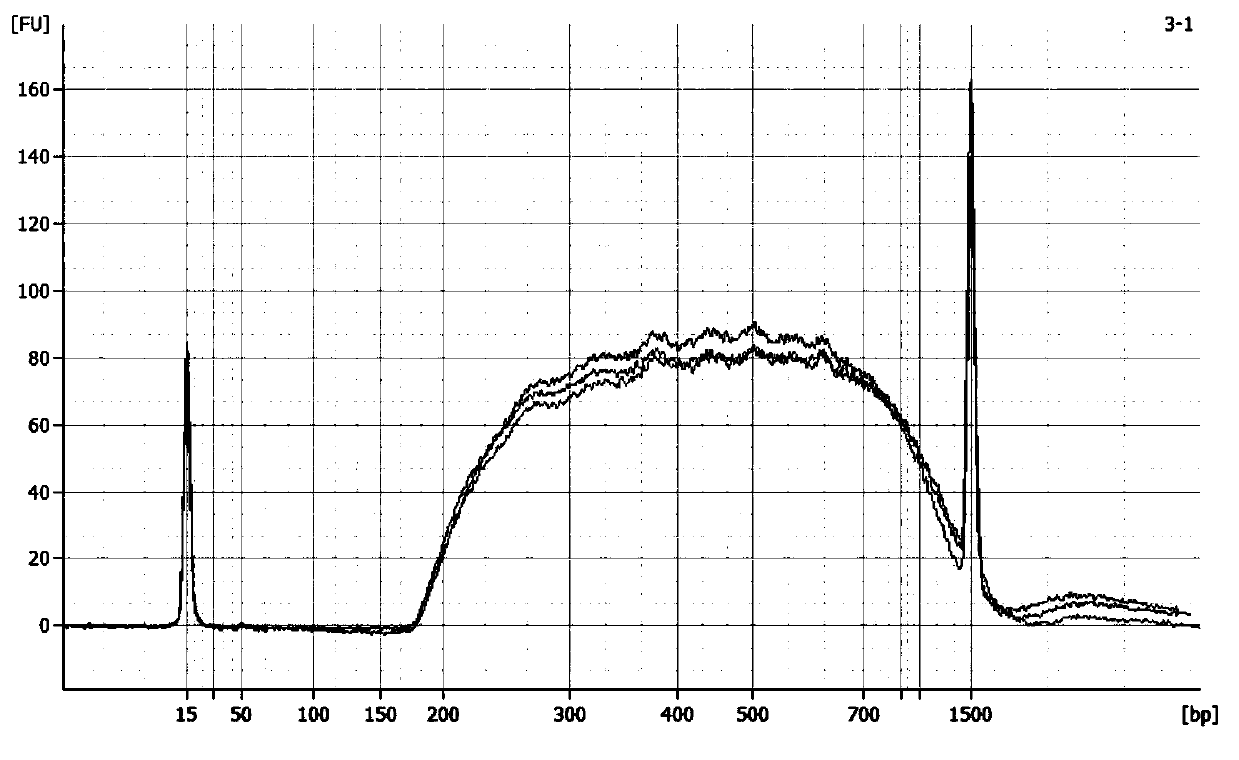
Method and kit for simultaneously constructing sequencing library by DNA and RNA
ActiveCN111139532AAvoid stickingHigh interrupt efficiencyTransferasesLibrary creationTn5 transposaseMagnetic bead
The invention discloses a method and a kit for simultaneously constructing a sequencing library by DNA and RNA. The method comprises the following steps: (1) carrying out reverse transcription on RNAin a reaction substrate to form RNA / cDNA composite double strands; (2) directly adding Tn5 transposase and a fragmentation buffer solution into a mixture of a reverse transcription product cDNA / RNA composite double strand and a DNA double strand for fragmentation, and adding a part of linker sequence to the 5' end of the double strands during fragmentation; (3) strand displacement and library amplification: adding an amplification buffer solution and a strand displacement and amplification enzyme mixture into the obtained product, and carrying out PCR amplification; and (4) carrying out magnetic bead purification to obtain a sequencing library of the original DNA / RNA. According to the method, the library building steps are greatly reduced, the library building time is shortened, the library output and offline data quality is improved, and more real information is helped to be obtained.
Owner:VAZYME BIOTECH NANJING
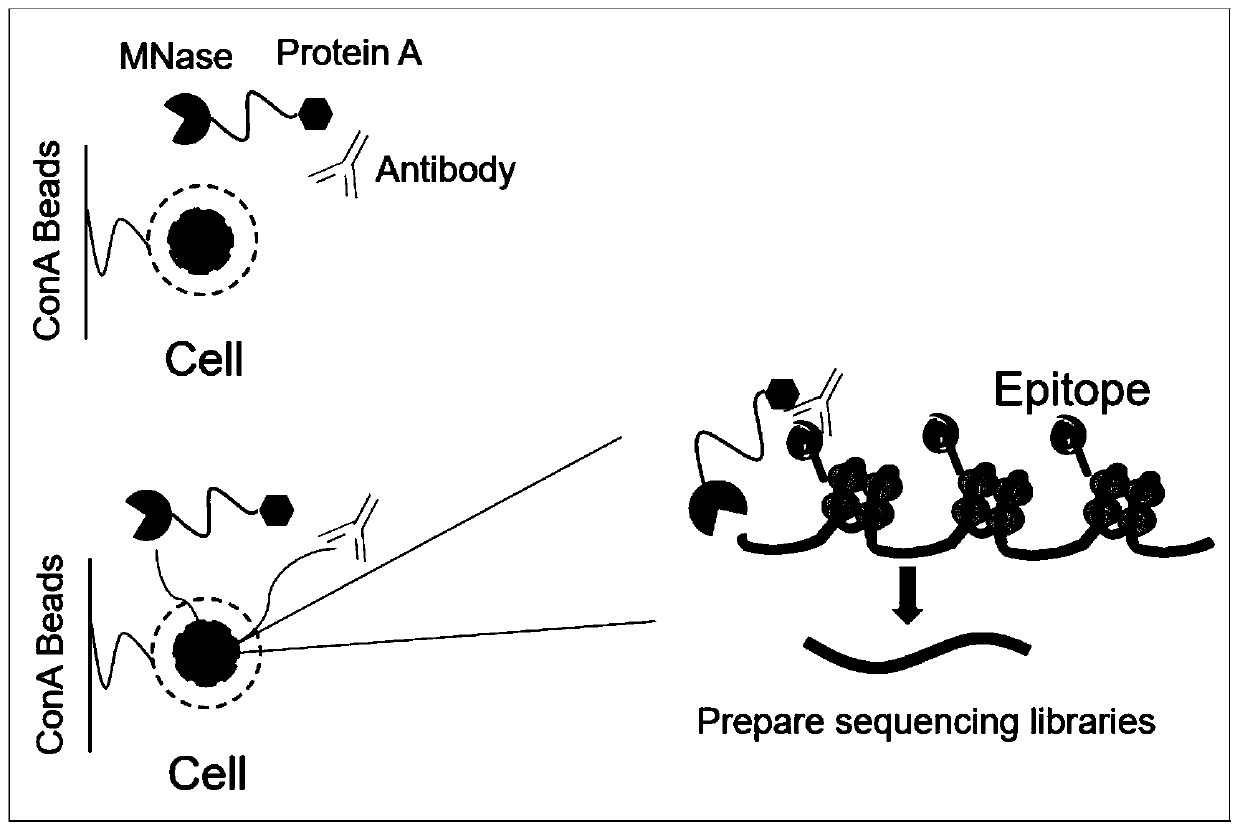
Fusion protein used for single-cell ChIP-seq library preparation and application thereof
ActiveCN110372799AImprove accuracySimplify the experimental processPolypeptide with affinity tagMicrobiological testing/measurementTn5 transposaseFc binding
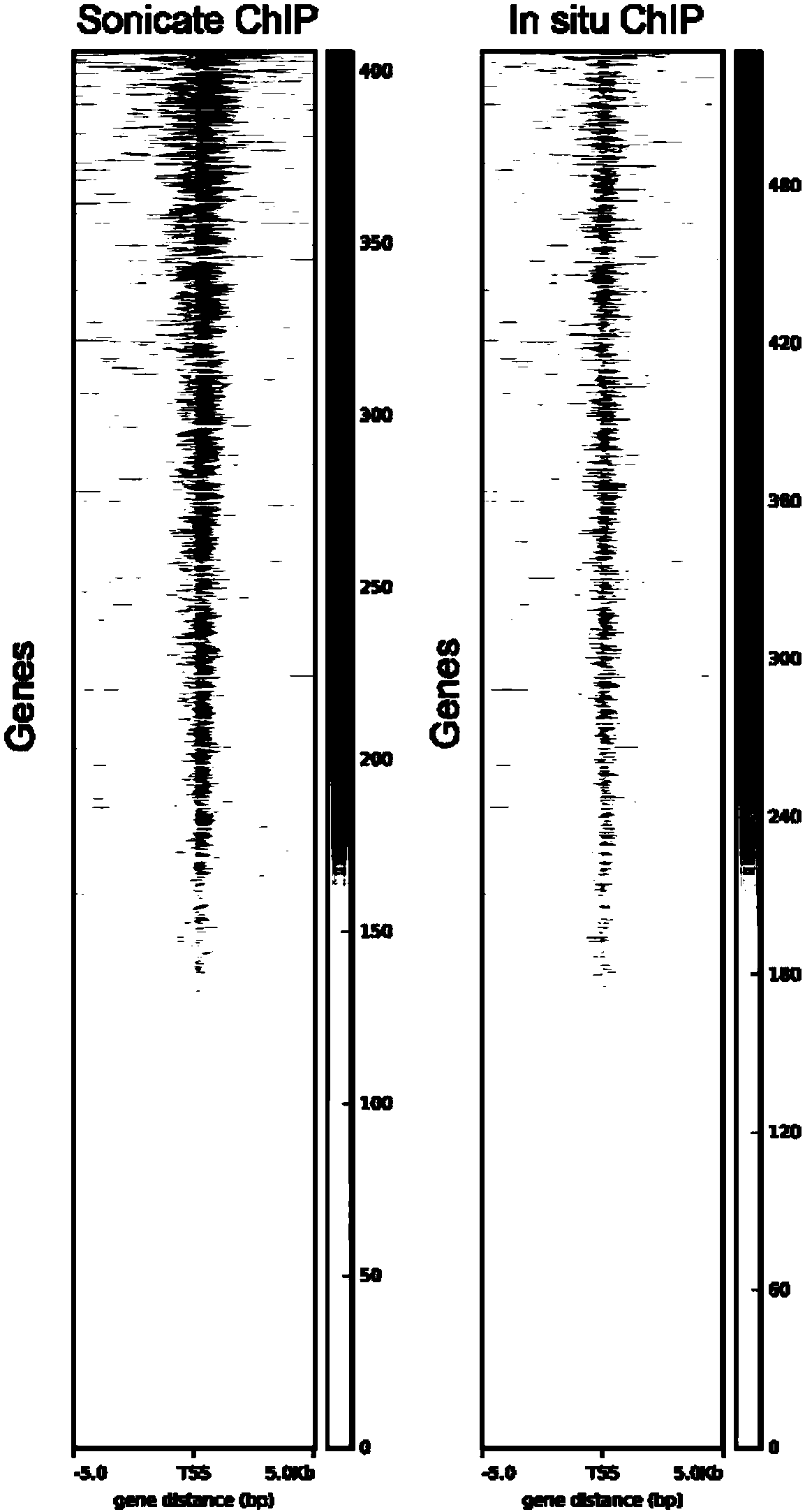
The invention relates to a fusion protein used for single-cell ChIP-seq library preparation and application thereof. The fusion protein comprises Tn5 transposase and an Fc binding protein; a kit comprises the fusion protein and other auxiliary detection reagents; a method uses the fusion protein or the kit for ChIP-seq detection. The fusion protein, the kit and the method can improve the library building efficiency and lower the library background in the ChIP-seq detection process, so that the accuracy of the ChIP-seq detection method can be improved, and the experimental flow of ChIP-seq canbe simplified.
Owner:PEKING UNIV
Sequencing library construction method and kit for pathogenic microorganism detection
ActiveCN111188094AHigh outputQuality improvementMicroorganism preservationLibrary creationReverse transcriptaseMagnetic bead
The invention discloses a sequencing library construction method and kit for pathogenic microorganism detection. The method comprises: (1) putting an extracted sample into a sample preservation solution, carrying out DNA / RNA co-extraction, and removing host rRNA; (2) adding reverse transcriptase to enable RNA in the sample to be subjected to reverse transcription to form RNA / cDNA composite doublestrands; (3) directly adding mutant Tn5 transposase and a breaking buffer solution into a mixture of the reverse transcription product cDNA / RNA composite double strand and a DNA double strand for fragmentation, and after the fragmentation is finished, carrying out a termination reaction by using a termination reaction solution 5*TS-plus; (3) adding an amplification buffer solution and a strand displacement and amplification enzyme mixture, and carrying out PCR amplification; and (4) carrying out magnetic bead purification to obtain a sequencing library of the original DNA / RNA. According to themethod, library building steps are greatly reduced, the library building time is shortened, the requirement on the initial quantity of samples is reduced, and the library output and offline data quality are improved.
Owner:VAZYME BIOTECH NANJING
Mutant type Tn5 transposase and preparation method and application thereof
ActiveCN106754811AChange combinationChange dependenciesMicrobiological testing/measurementTransferasesTn5 transposaseSequence Insertions
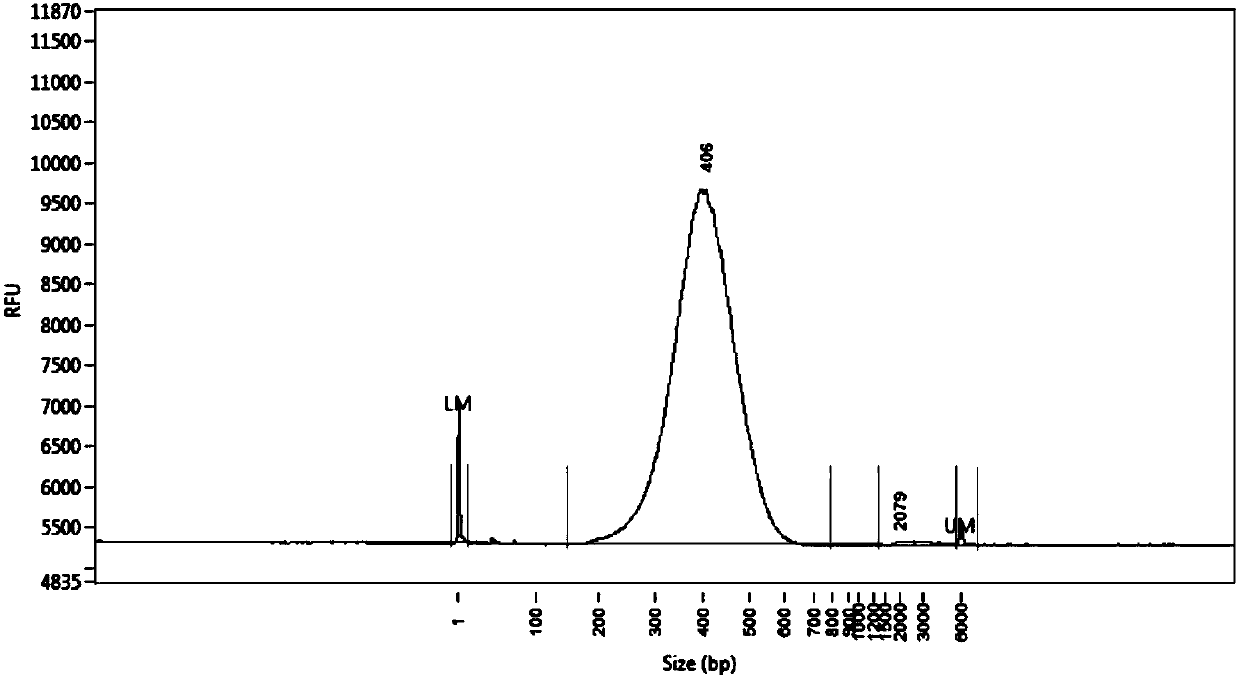
Disclosed are a mutant type Tn5 transposase and a preparation method and application thereof. The mutant type Tn5 transposase has the advantages that an existing EK / LP Tn5 transposase is subjected to site-directed mutagenesis for lowing DNA preference, site-directed mutagenesis refers to mutagenesis in at least two of D97E, D188E and E326D and improves a protein sequence and a conformation of the Tn5 transposase, and a reaction system is subjected to fine adjustment, so that preference of the Tn5 transposase to DNA target sequence insertion is improved, DNA library construction uniformity is improved remarkably, library construction results have better coverage degree, and requirements of high-throughput sequencing library construction are met.
Owner:VAZYME BIOTECH NANJING
TN5 library building primer group for Ion Proton sequencing platform, TN5 library building kit for Ion Proton sequencing platform and library building method
ActiveCN105524920AEfficient constructionQuick buildMicrobiological testing/measurementLibrary creationTn5 transposaseProton
The invention discloses a TN5 library building primer group for an Ion Proton sequencing platform, a TN5 library building kit for the Ion Proton sequencing platform and a library building method, wherein the primer group comprises sequences which are used for joint embedding and are shown by SEQ ID NO:1-3 and sequences which are used for PCR (Polymerase Chain Reaction) amplification and are shown by SEQ ID NO:4-5. The primer group provided by the invention has the advantages that TN5 transposase can be used for efficiently and fast building a trace amount of library applicable to Ion Proton; and the built library can be directly used for computer sequencing. The original complicated library building procedures are omitted; the operation is simple; and the sequencing result is stable.
Owner:SHENZHEN HUADA GENE INST
Mutant TN5 transposase enzymes and method for their use
InactiveCN1367840ASimple methodSolve problems of interestHydrolasesTransferasesTn5 transposaseIn vivo
Disclosed herein are modified Tn5 transposase proteins that prefer the inner end of the transposon Tn5 over the outer end and that can be combined with the outer end in an end-specifically directed transposition method in vivo or in vitro. The combination of transposases that prefer the inner end to the inner end.
Owner:WISCONSIN ALUMNI RES FOUND
System for in vitro transportation using modified TN5 transposase
InactiveCN1251135AShowed no measurable in vitro activityHydrolasesStable introduction of DNATn5 transposaseWild type
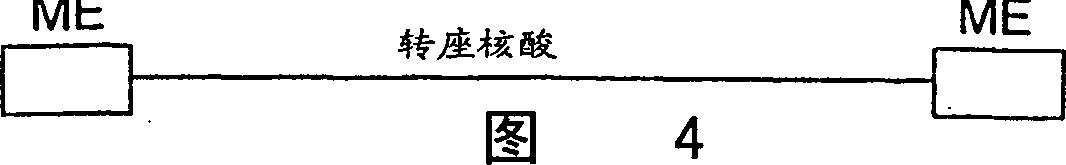
A system for in vitro transposition includes a donor DNA that includes a transposable element flanked by a pair of bacterial transposon Tn5 outside end repeat sequences, a target DNA into which the transposable element can transpose, and a modified Tn5 transposase having higher binding avidity to the outside end repeat sequences and being less likely to assume an inactive multimer form than wild type Tn5 transposase.
Owner:WISCONSIN ALUMNI RES FOUND
Fusion protein, kit and CHIP-seq detection method
ActiveCN108285494AImprove binding efficiencyImprove cutting efficiencyHydrolasesAntibody mimetics/scaffoldsTn5 transposaseFc binding
The invention relates to fusion protein, a kit and a CHIP-seq detection method. The fusion protein comprises Tn5 transposase and Fc binding protein, the kit comprises the fusion protein and other auxiliary detection reagents, and the method is the CHIP-seq detection method by using the fusion protein or the kit. According to the fusion protein, the kit and the method, the library establishment efficiency can be improved in the CHIP-seq detection process, the library background is reduced, the accuracy of the CHIP-seq detection method is improved accordingly, and the experimental process of CHIP-seq is simplified.
Owner:PEKING UNIV
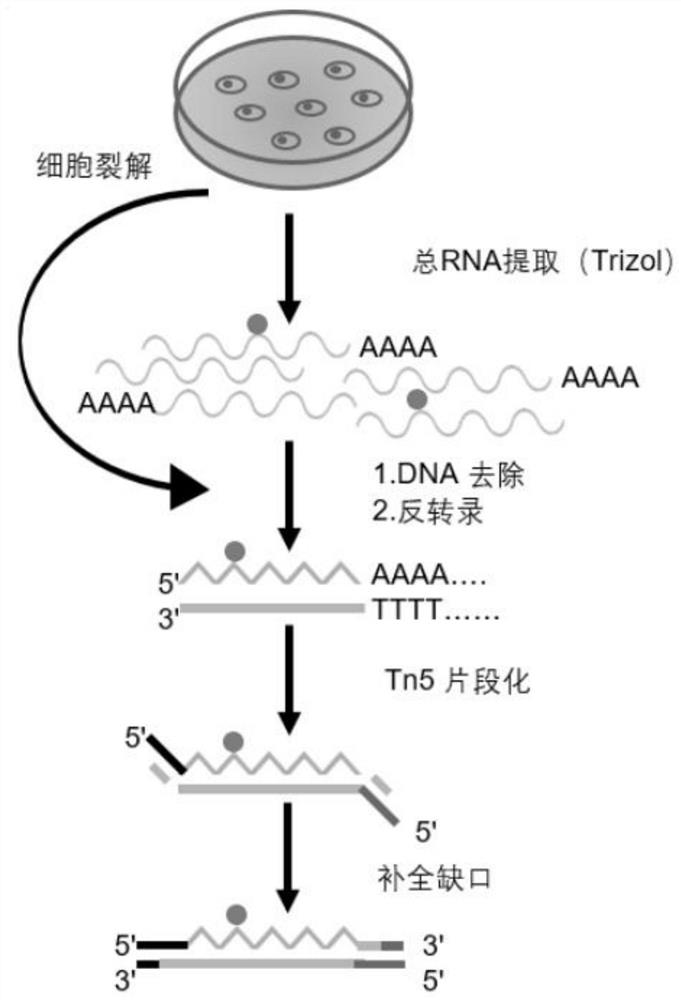
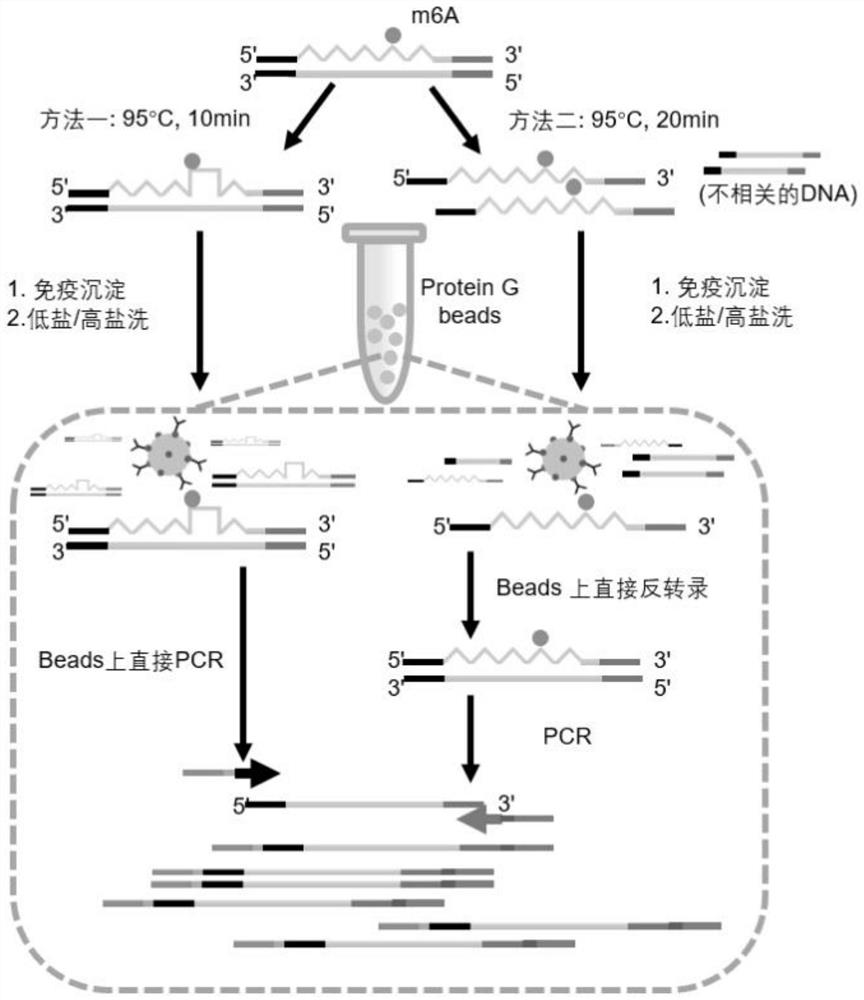
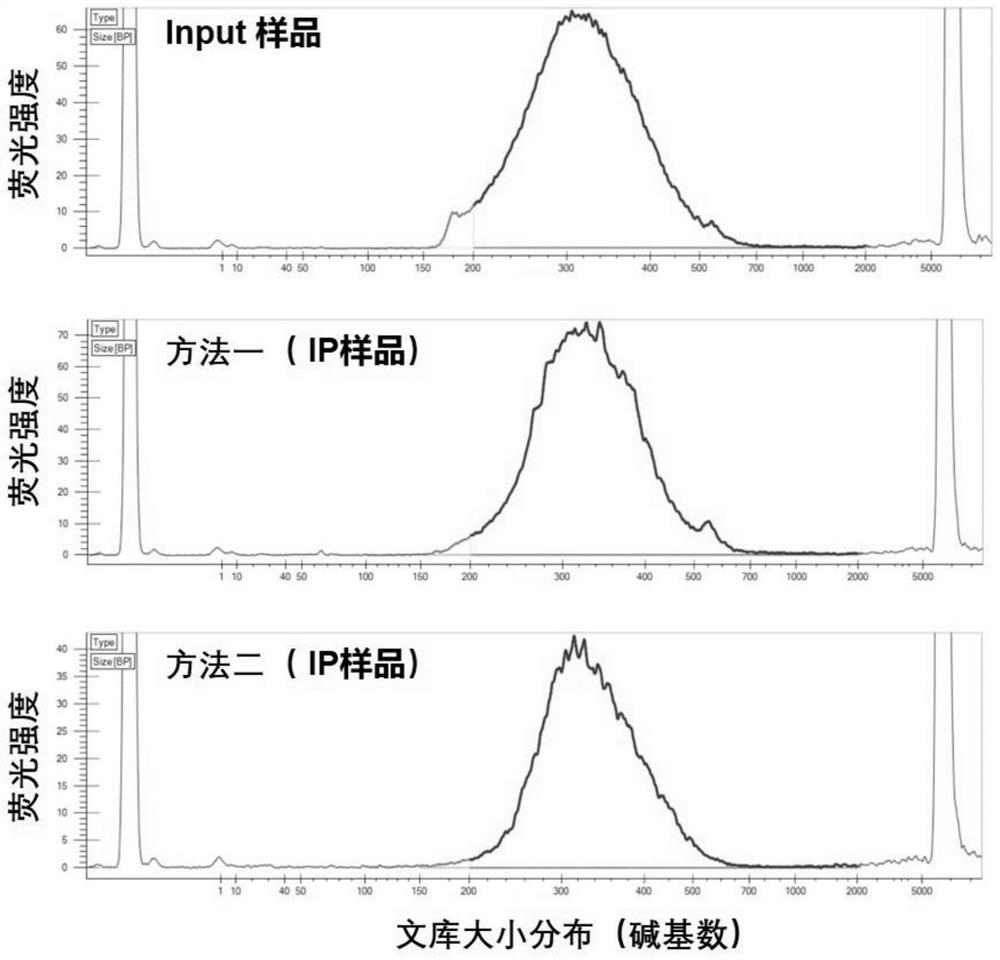
Method for constructing microscale sample m6A modification detection library under assistance of Tn5 transposase and application thereof
ActiveCN113061648AReduce initial investmentSave steps to removeMicrobiological testing/measurementLibrary creationTn5 transposaseRNA modification
The invention belongs to the technical field of RNA modification site detection, and particularly relates to a method for constructing a microscale sample m6A modification detection library under the assistance of Tn5 transposase and an application thereof. The m6A modified detection library constructed by the method can greatly reduce the initial input amount of a high-throughput sequencing sample, and the method disclosed by the invention can be used for detecting a trace sample which cannot be applied by the existing MeRIP-seq method, so that the initial total RNA input amount of the high-throughput sequencing sample is reduced to 0.06 microgram.
Owner:SUN YAT SEN UNIV
Modulators of enzymatic nucleic acid elements mobilization
The present invention discloses a nucleic acid cleavage assay for members of the transposase / integrase superfamily. A method of using the assay to screen for modulators of the nucleic acid cleavage activity is also disclosed. The present invention further provides a method for screening for modulators of binding of a transposase / integrase to its corresponding recognition sequence. In addition, the present invention provides a method of identifying a modulator for a particular transposase / integrase such as HIV integrase based on modulators of other members of the transposase / integrase superfamily. Also disclosed are Tn5 transposase inhibitors and HIV integration inhibitors.
Owner:ASON BRANDON L +3
Database establishing method suitable for plant transposase accessible chromatin analysis
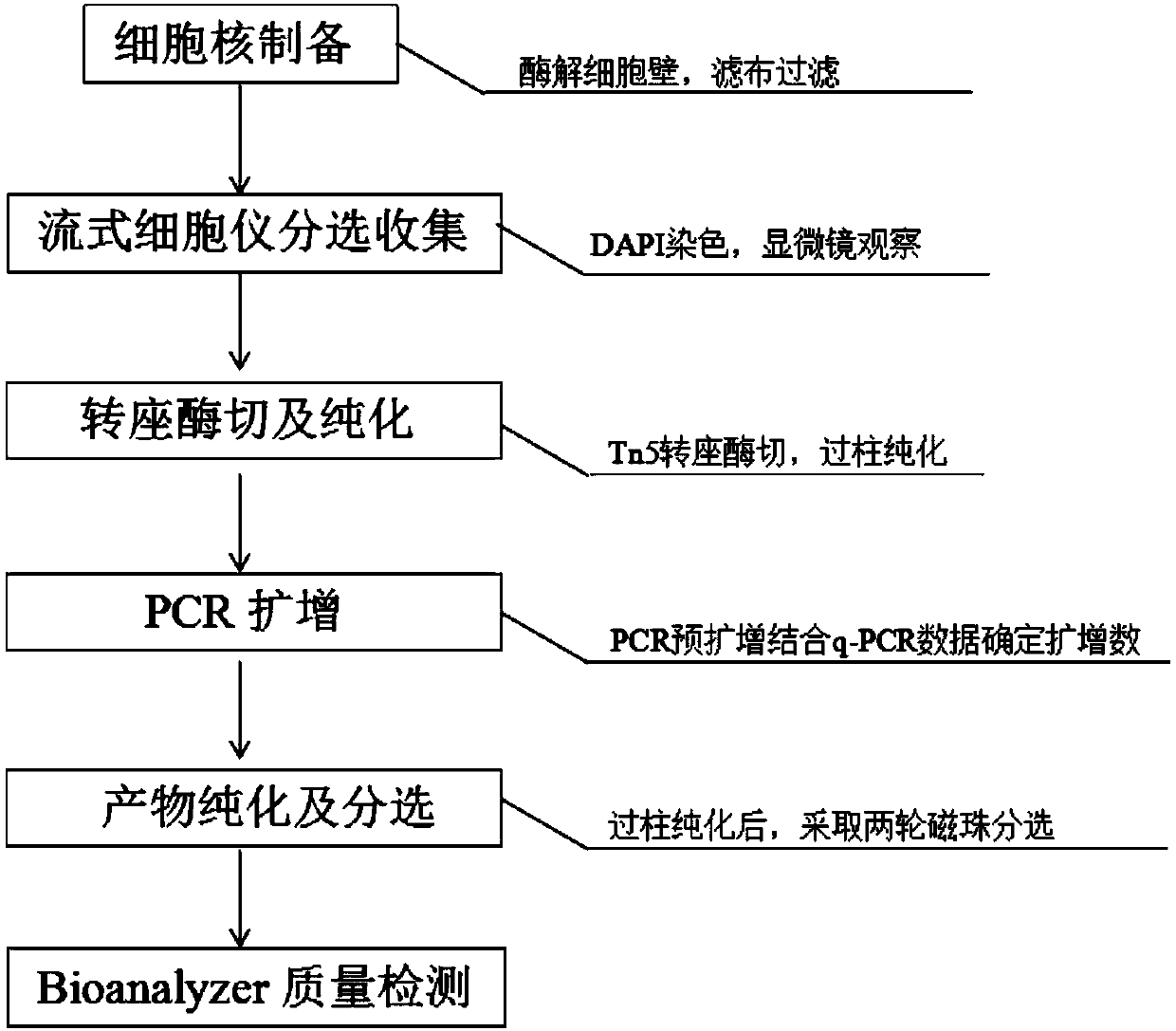
InactiveCN108130323AReduce distractionsQuality improvementLibrary creationDNA preparationCell wallDigestion
The invention discloses a database establishing method suitable for plant transposase accessible chromatin analysis. The database establishing method comprises following steps: firstly, plant tender tissue is subjected to pre-treatment, enzymatic hydrolysis is cell walls is carried out, filtering is carried out, DAPI dyeing is adopted to sort and collect nucleuses on a flow cytometer, the obtainednucleuses are filtered so as to reduce interference of cell walls and subcellular organelles on experiment data; Tn5 transposase digestion reaction and purification are carried out, qPCR is adopted to determine circulating number needed by database establishing; at last obtained single library is subjected to equimolar mixing. According to the database establishing method, the high quality on computer library is obtained via optimized two circles of magnetic cell sorting at a certain ratio, so that important experiment method reference is provided for study of scientific research personnel onplant tender tissue accessible transposase nucleus chromatin high flux sequencing.
Owner:奥明(杭州)基因科技有限公司
Trace cell ChIP (Chromatin Immunoprecipitation) method
ActiveCN108315387AHigh catchImprove the efficiency of database constructionMicrobiological testing/measurementProtein targetEnzyme digestion
The invention relates to a trace cell ChIP (Chromatin Immunoprecipitation) method. The method comprises the following steps: 1) dividing a cell sample to be detected into n groups, wherein n is a natural number which is not equal to 0; 2) carrying out crosslinking fixation on the nth group sample; 3) treating the nth group sample by utilizing a cell membrane perforating agent; 4) carrying out enzyme digestion by utilizing Tn5 transposase and breaking a chromatin segment of the nth group sample, and connecting a barcode sequence and a primer sequence to two ends of a production segment DNA (Deoxyribonucleic Acid), wherein when the n is greater than or equal to 2, the barcode sequence and / or the primer sequence used in each group of step 4) are / is different; 5) when the n is greater than orequal to 2, combining samples of all groups; 6) enriching a DNA segment combined with target protein and taking a primer as an index for creating a database; sequencing and analyzing. When the methodis used for sequencing, the database creating efficiency is high; the capturing of transcription factor sites of an extremely trace amount of cells can be realized; relatively good efficiency is ensured and the stability and the accuracy are relatively good.
Owner:PEKING UNIV
Reagent for transposase fragmentation and application of reagent
InactiveCN110564705AEasy to useLow costMicrobiological testing/measurementTransferasesForward primerPolymerase L
The invention discloses a reagent for transposase fragmentation. The reagent comprises the following assemblies independently packaged: Tn5transposase, a first connector, a second connector and a transposition reaction buffer solution. Further, the reagent also comprises a forward primer set, a reverse primer set, DNA polymerase, a DNA polymerase reaction buffer solution, as well as assemblies ofa fragmentation reaction buffer solution, a stop solution and the like, wherein the forward primer set, the reverse primer set, the DNA polymerase and the DNA polymerase reaction buffer solution are independently packaged. The invention also provides a reagent kit including the reagent. The reagent and the reagent kit provided by the invention are convenient to use, low in cost, good in use flexibility and suitable for various actual requirements, and the defects that a conventional commercialization transposition system is poor in stability, single in purposes, high in cost and the like can be avoided; and particularly when the reagent and the reagent kit are used for constructing a fragmentation nucleic acid library, the process of DNA fragmentation, terminal repair and connector connection can be completed only within 10min, the library construction time can be notably shortened, and excellent sequencing quality can be obtained.
Owner:苏州璞瑞卓越生物科技有限公司
Method for making insertional mutations
InactiveCN1319135AIncreased transpositionEasy to manufactureHydrolasesMicrobiological testing/measurementTn5 transposaseNucleotide
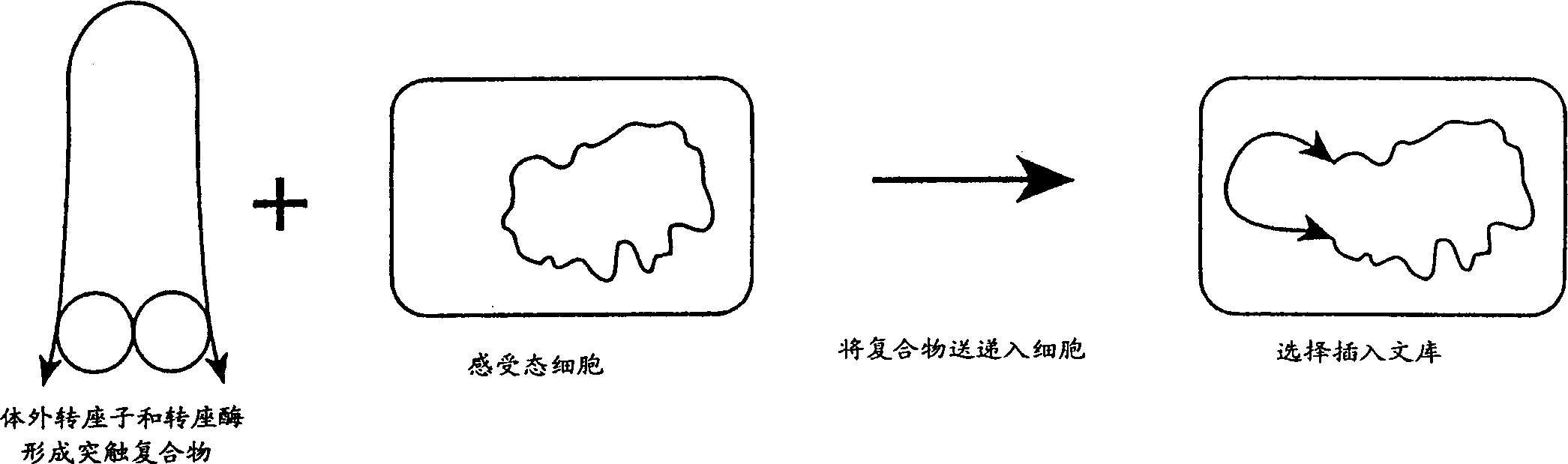
A method for making insertional mutations at random or quasi-random locations in the chromosomal or extra-chromosomal nucleic acid of a target cell includes the step of combining, in the target cell, cellular nucleic acid with a synaptic complex that comprises (a) a Tn5 transposase protein and (b) a polynucleotide that comprises a pair of nucleotide sequences adapted for operably interacting with Tn5 transposase and a transposable nucleotide sequence therebetween, under conditions that mediate transpositions into the cellular DNA. In the method, the synaptic complex is formed in vitro under conditions that disfavor or prevent the synaptic complexes from undergoing productive transposition.
Owner:WISCONSIN ALUMNI RES FOUND
SNP sites related to number of pig nipples and detection method and application thereof
ActiveCN110714082AQuality improvementEarly predictionMicrobiological testing/measurementProteomicsBiotechnologyTn5 transposase
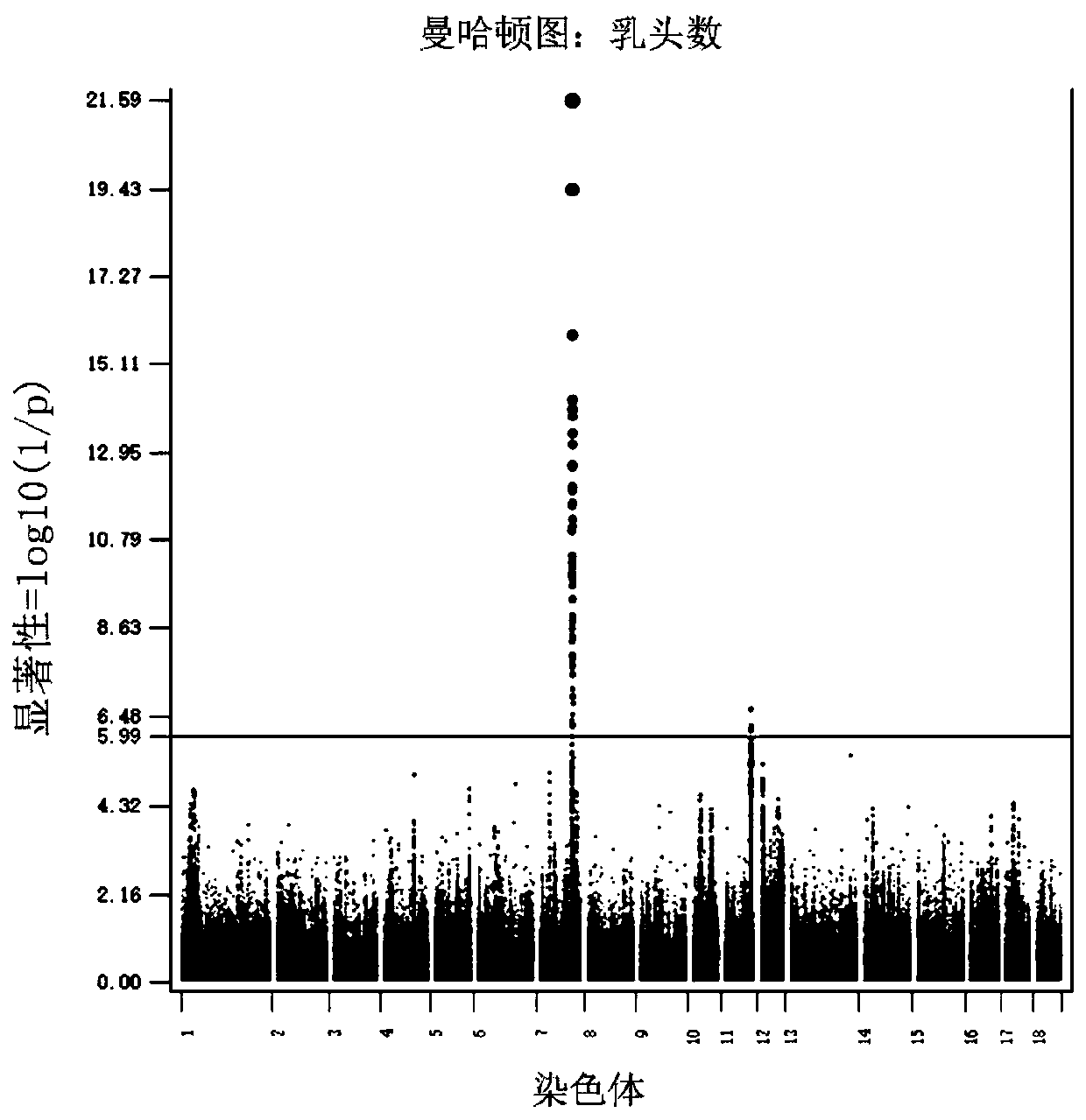
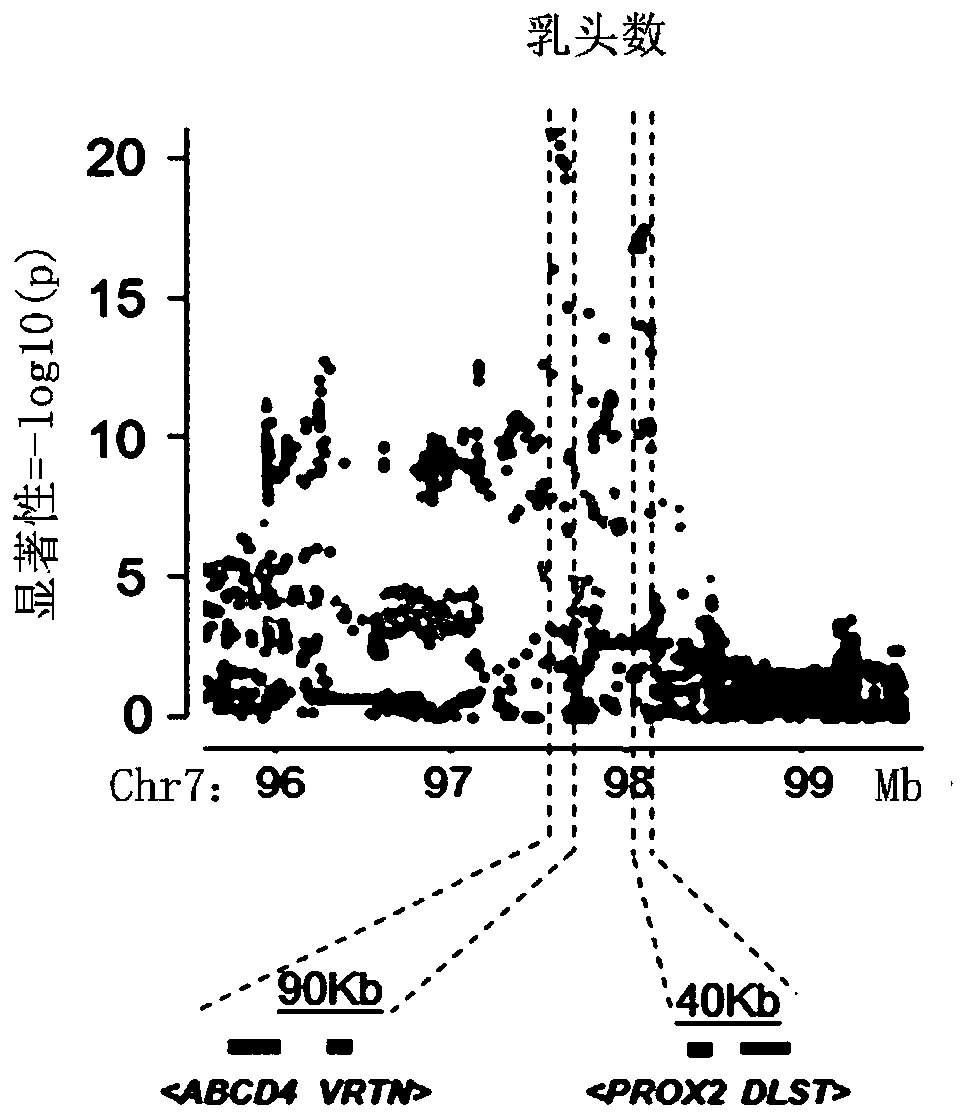
The invention relates to SNP sites related to the number of pig nipples and a detection method and application thereof. The SNP sites are located at a chr7:97570528 of a genomic version Ensembl Sscrofa 11.1, and alleles of the sites are A and G. According to the SNP sites related to the number of the pig nipples and the detection method and application thereof, Tn5 transposase is used for establishing a library, low-depth sequencing is carried out on a core population of 3,616 Duroc pigs with important economic performance records, and high-quality whole-genome resequencing data are obtained through padding. Through GWAS analysis of the characteristics of the number of the pig nipples, seven SNP sites which significantly affect the number of the pig nipples are detected, and a newly discovered SNP site SSC7: 97570528 with the most significant effect is included. The SNP sites can be used as molecular markers for the breeding of dominant populations of the number of the pig nipples.
Owner:CHINA AGRI UNIV
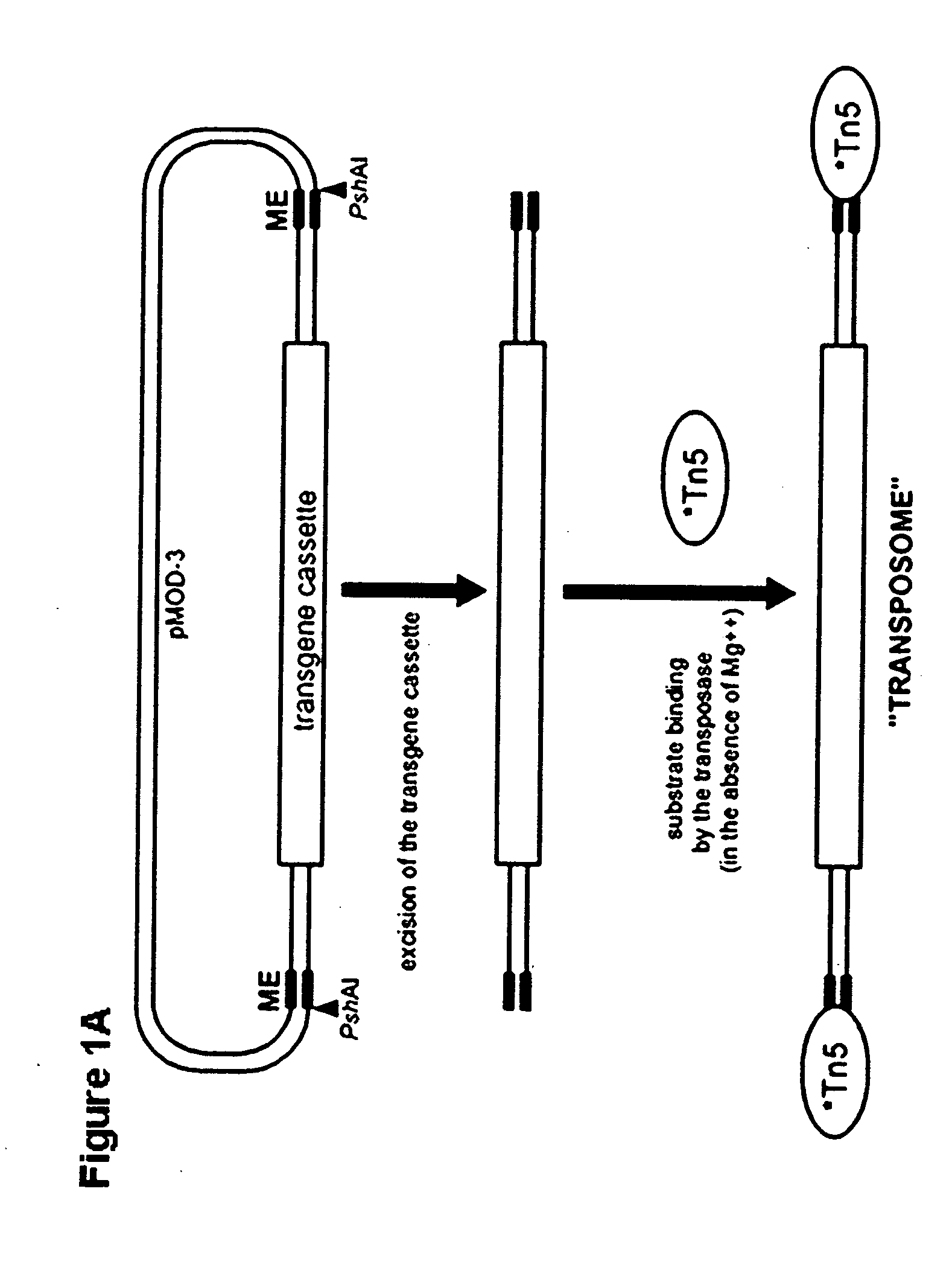
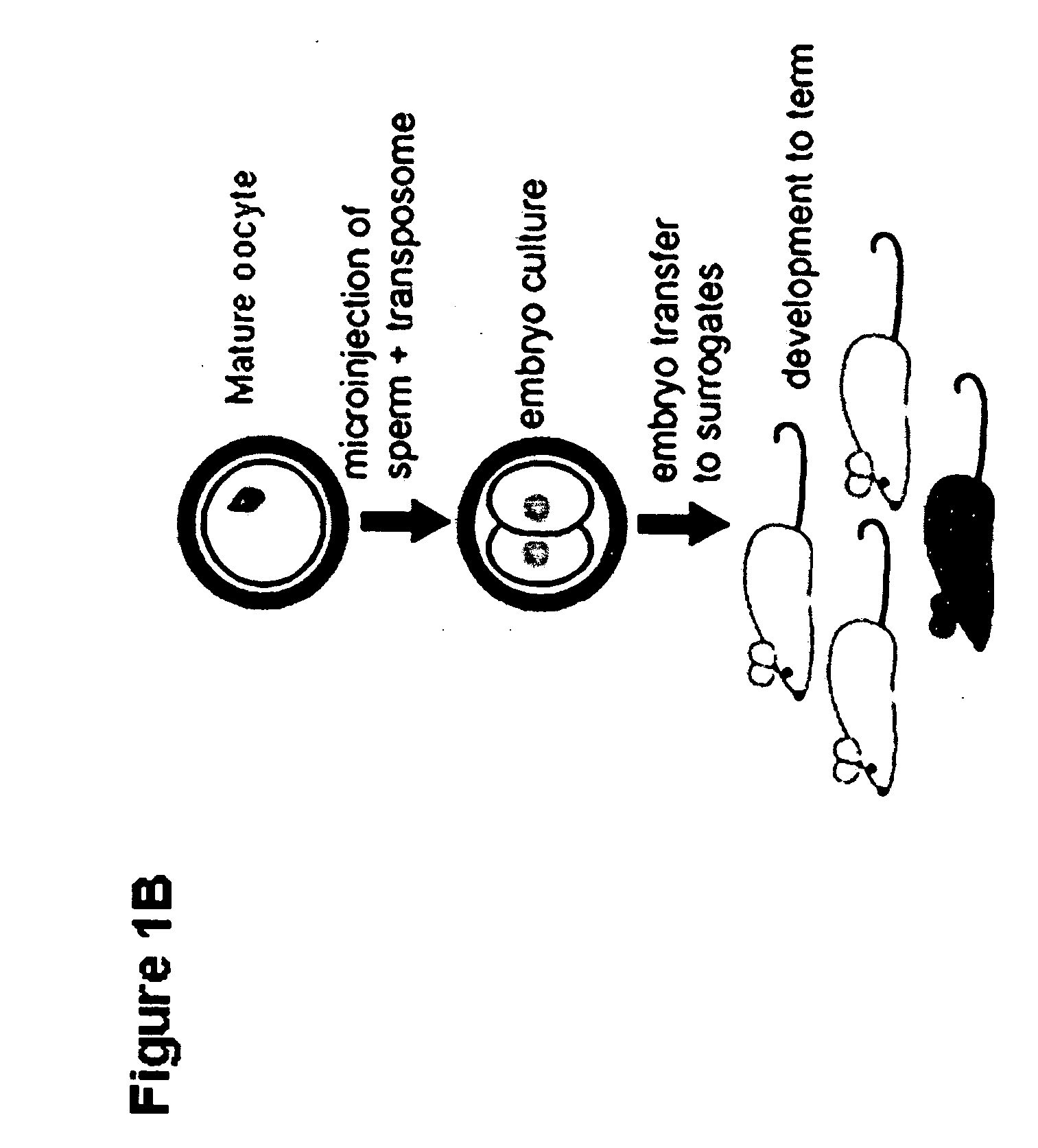
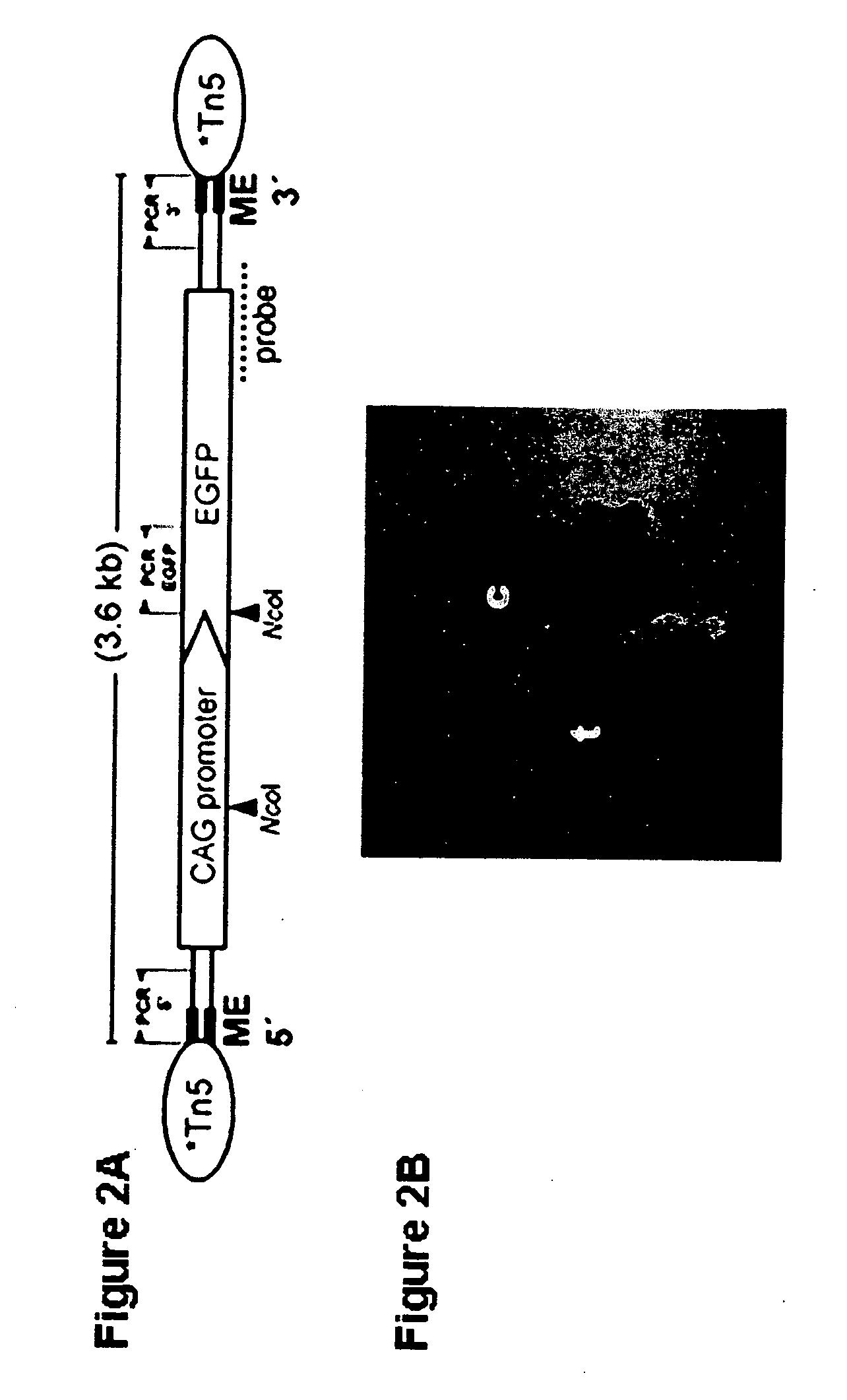
Tn5 transposase-mediated transgenesis
Owner:MOISYADI ISTEFO +2
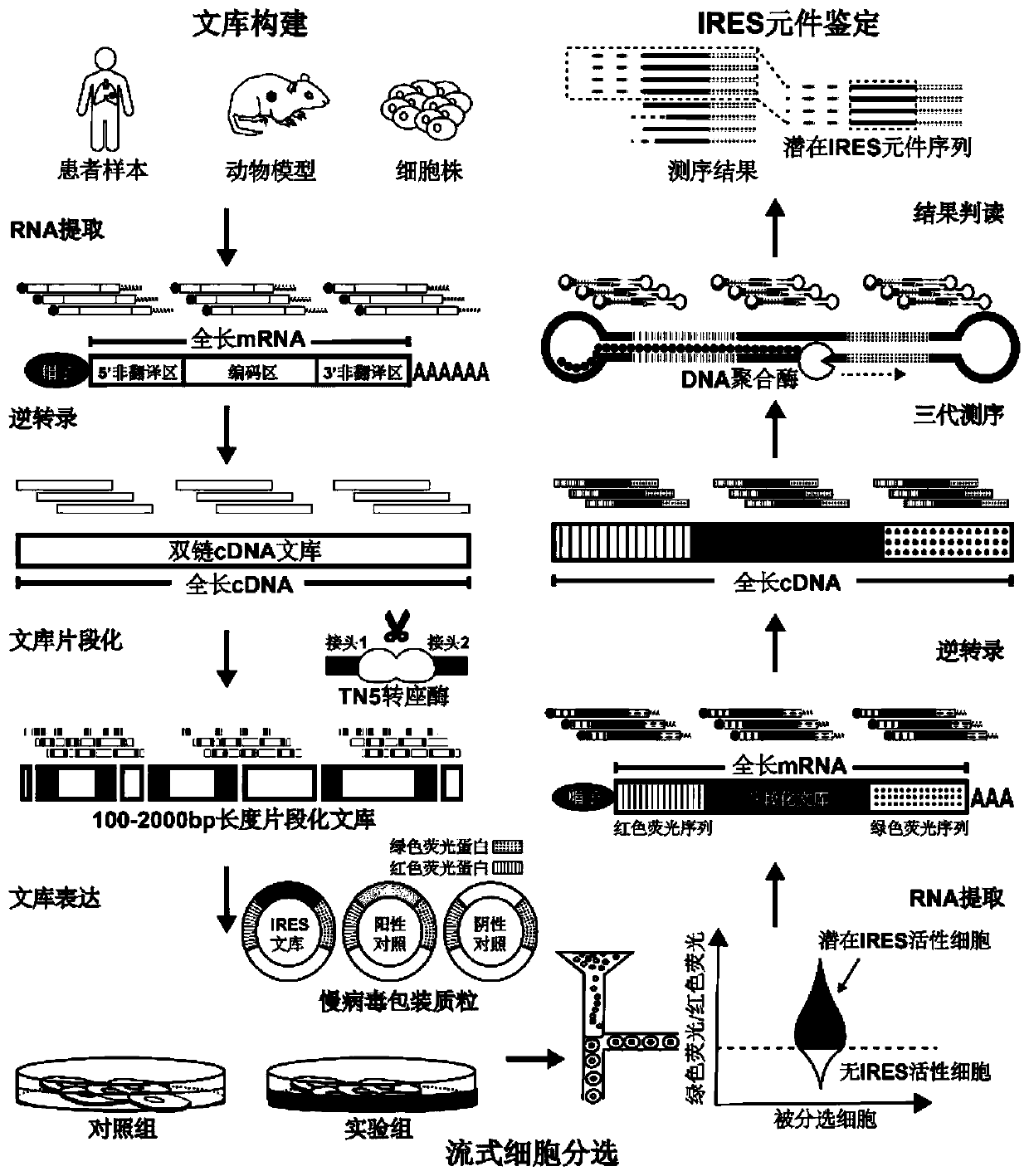
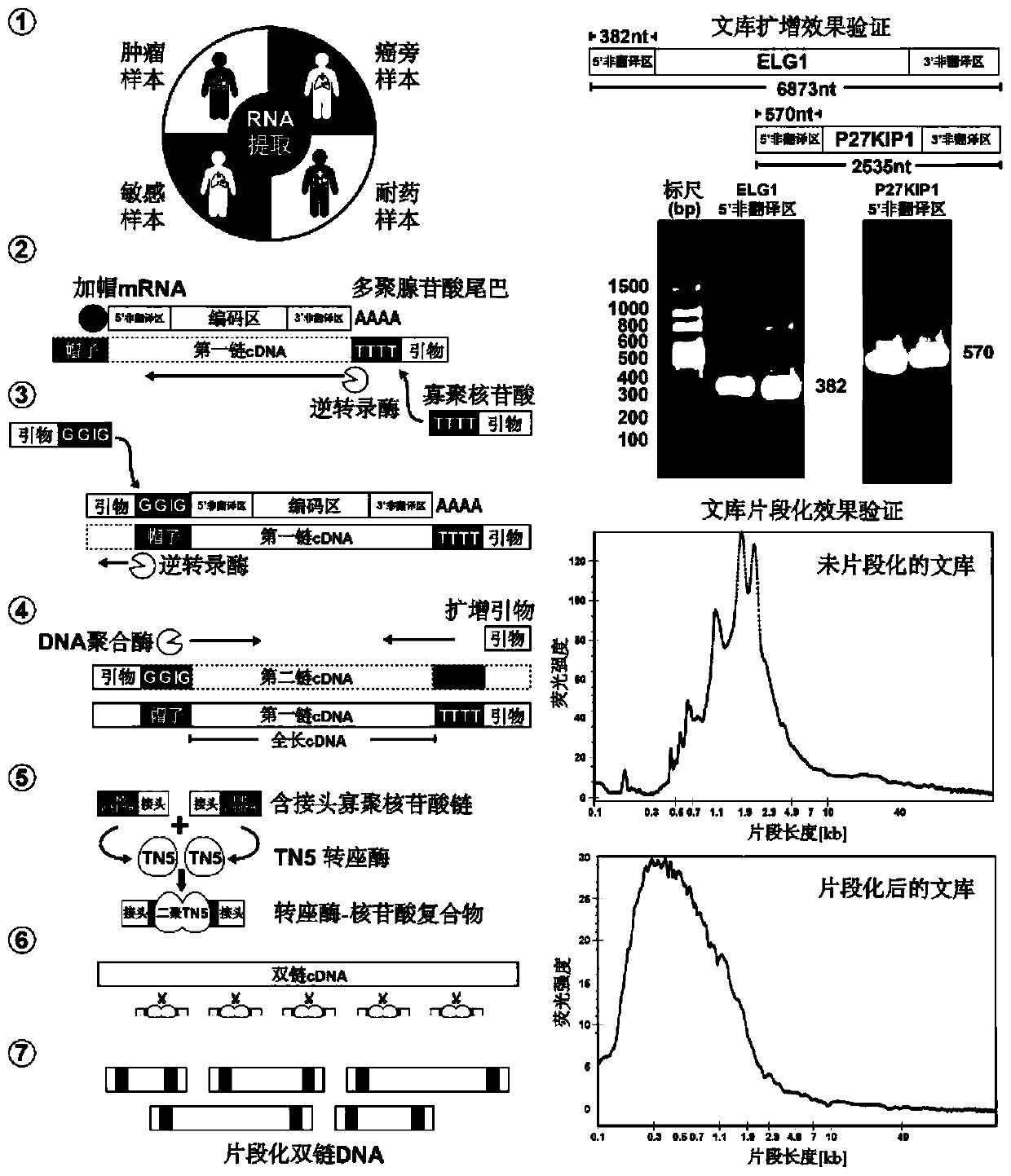
High-throughput identification method of internal ribosome entry site (IRES) elements in various source cell samples
ActiveCN110819707AImprove accuracyAvoid Consistency EffectsMicrobiological testing/measurementNucleic acid vectorCDNA libraryBinding site
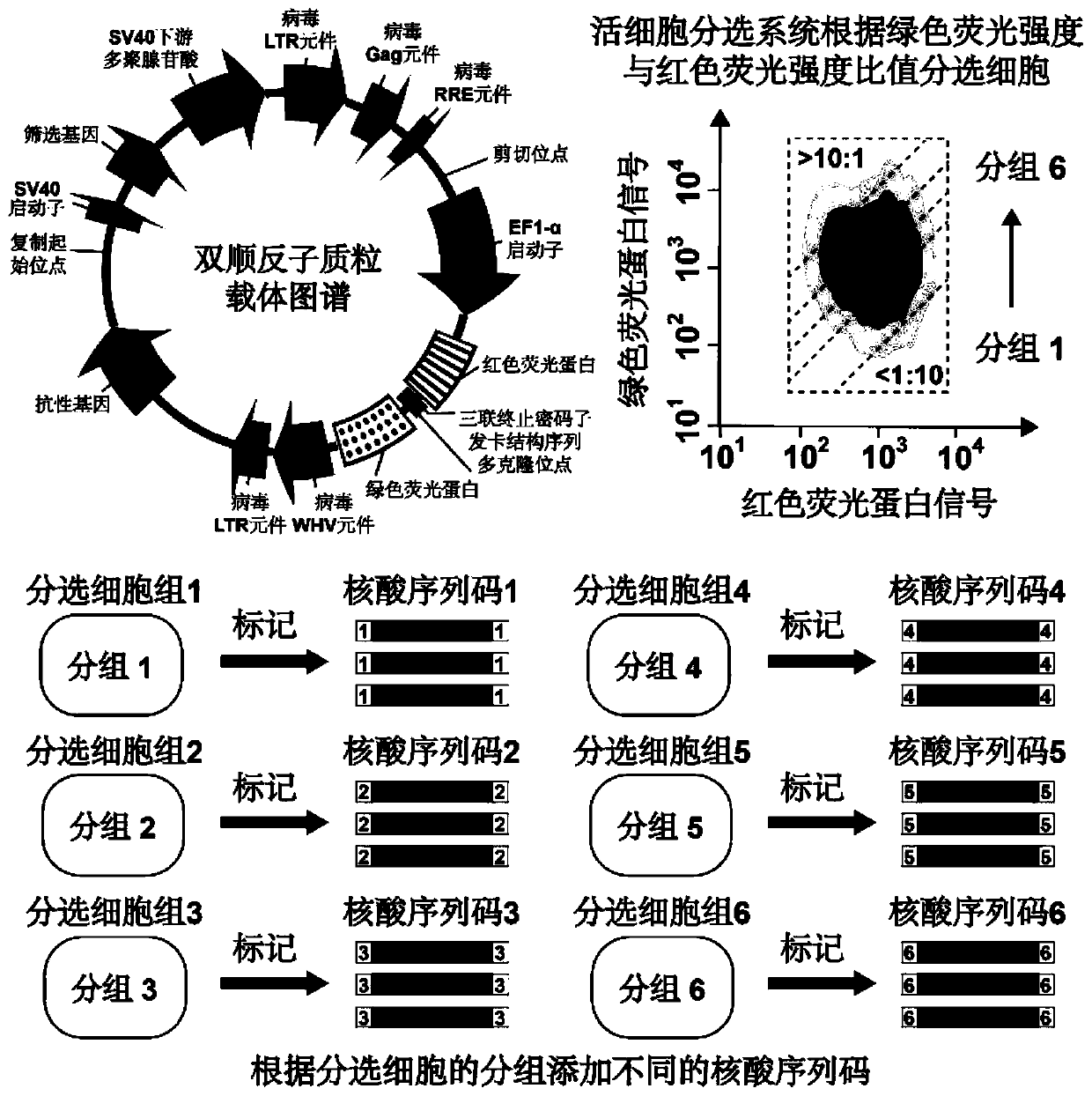
The invention discloses a high-throughput identification method of internal ribosome entry site (IRES) elements in various source cell samples. The method comprises the following steps: a full-lengthcDNA library is obtained from a sample by utilizing oligonucleotides designed for an mRNA chain 5'- terminal cap structure and a 3'-terminal polyadenylic acid tail through reverse transcription; the cDNA library is fragmented by utilizing TN5 transposase and a designed nucleic acid fragment containing a special linker sequence; active IRES elements are screened by utilizing a lentivirus reporter gene plasmid vector containing red fluorescent proteins, green fluorescent proteins and special elements, and false positive results caused in a translation process are eliminated; and the active IRESelements are identified by utilizing a long reading and long sequencing technology and false positive results caused by a promoter and variable shearing in the transcription process are eliminated. The method pushes high-throughput identification of IRES elements to clinical application, so that a role played by the IRES elements in a process of disease occurrence and development can be deeply studied.
Owner:XIANGYA HOSPITAL CENT SOUTH UNIV
Method for quickly and stably measuring enzyme activity of Tn5 transposase
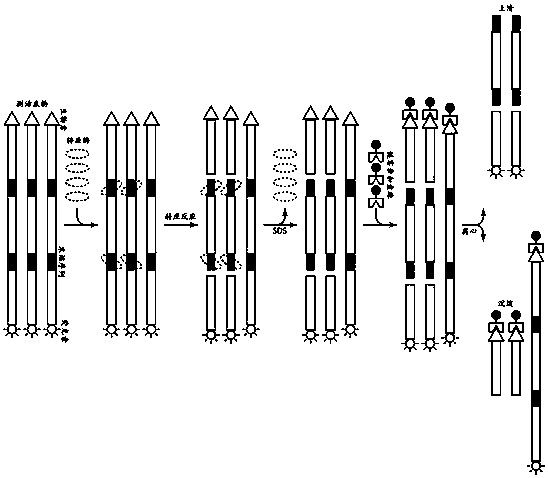
InactiveCN108265101AEasy to operateNo training requiredMicrobiological testing/measurementBiological material analysisSulfonateTn5 transposase
The invention discloses a method for quickly and stably measuring the enzyme activity of Tn5 transposase. The method includes steps of (1), mixing activity measuring substrates, the transposase and reaction buffer solution with one another; (2), carrying out transposition reaction; (3), adding denaturants [such as SDS (sodium dodecyl sulfonate) and guanidine hydrochloride] into reaction products to carry out denaturation on the transposase, and separating the transposase from DNA (deoxyribonucleic acid) fragments; (4), adding media capable of identifying labels of substrates and binding the media with the unreacted substrates; (5), removing the media bound with the substrates by selective means (such as centrifuging, filtering and concentrating); (6), detecting the content of released DNAwith labels, and comparing the content to standard numerical values to obtain the activity of the Tn5 transposase. The method for measuring the enzyme activity has the advantages that the method is easy and convenient to operate and quite suitable for detecting the enzyme activity of the Tn5 transposase during mass production, and is speedy, and results are good in repeatability.
Owner:天津强微特生物科技有限公司 +1
Mutated Tn5 transposase proteins and the use thereof
ActiveUS20060040355A1High transposaseGreat aviditySugar derivativesHydrolasesTn5 transposaseWild type
Transposase proteins that are modified relative to and have higher transposase activities than the wild-type Tn5 transposase are disclosed. A transposase protein of the present invention differs from the wild-type Tn5 transposase at amino acid position 41, 42, 450, or 454 and has greater avidity than the wild-type Tn5 transposase for at least one of a Tn5 outside end sequence as defined by SEQ ID NO:3, a Tn5 inside end sequence as defined by SEQ ID NO:4, and a modified Tn5 outside end sequence as defined by SEQ ID NO:5. Also disclosed are various systems and methods of using the transposase proteins of the present invention for in vitro or in vivo transposition.
Owner:WISCONSIN ALUMNI RES FOUND
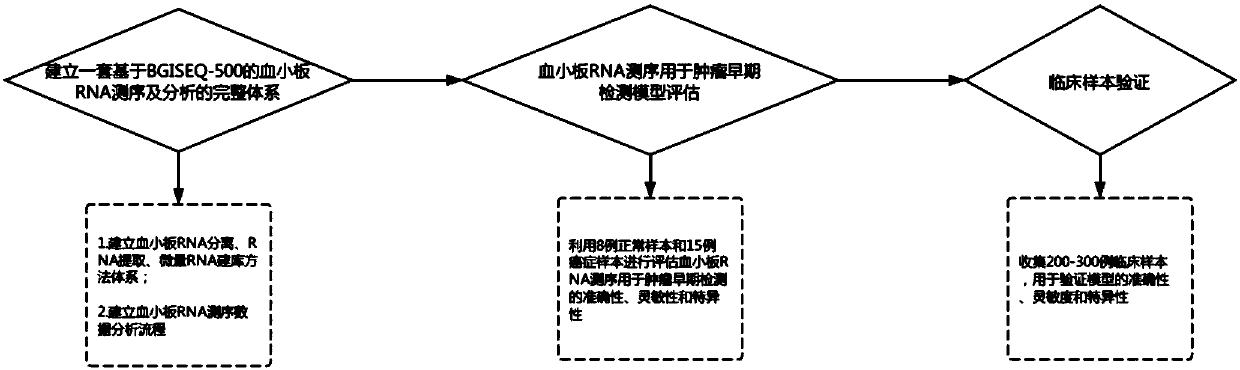
Micro-platelet RNA sequencing detection kit based on BGISEQ-500
The invention discloses a micro-platelet RNA sequencing detection kit based on BGISEQ-500. The invention also provides a method for constructing a platelet RNA sequencing library based on BGISEQ-500.The method comprises the following steps: 1) extracting platelet RNA from a sample to be detected, and performing reverse transcription to obtain dscDNA; 2) fragmenting the dscDNA by a linker-embeddedTn5 transposase to obtain a fragmented product; 3) carrying out PCR amplification of the fragmented product to obtain an amplification product; and 4) cyclizing the amplification product to obtain the platelet RNA sequencing library. Experiments prove that below 1 ml of whole blood is adopted to construct the micro-platelet mRNA library, and an autonomously established platelet analysis procedureis used to carry out highly-sensitive and highly-specific early screening of tumors.
Owner:SHENZHEN HUADA GENE INST
Preparation and application of Tn5 mutant enzyme
ActiveCN113186174AHigh sequencing saturationNo apparent preferenceMicrobiological testing/measurementTransferasesTn5 transposaseMutant enzyme
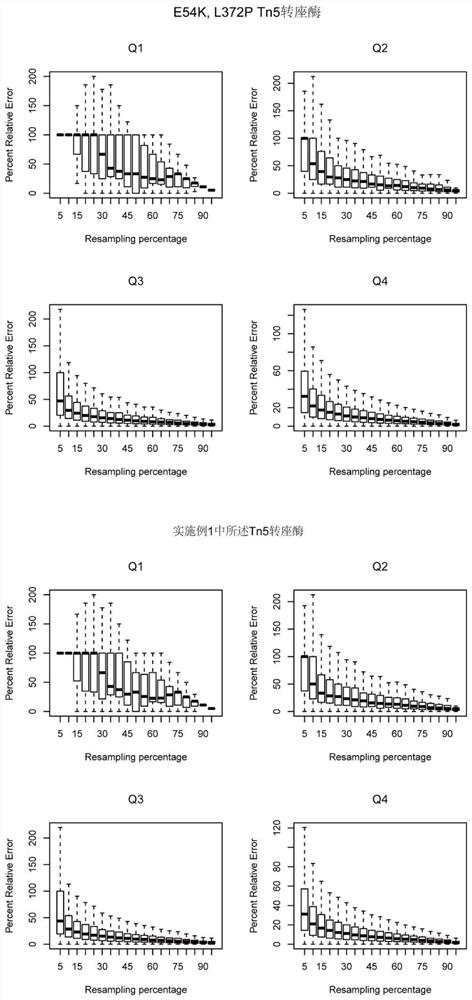
The invention discloses preparation and application of a Tn5 mutant enzyme, and belongs to the technical field of biology. Existing E54K and L372P mutated Tn5 transposase is mutated, and the efficiency of fragmentation reaction is remarkably improved. The mutation changes the protein sequence of the Tn5 transposase, so the redundant amino acid sequence of the Tn5 transposase is deleted, and the problems of low efficiency and preference of the original Tn5 tagging reaction are solved. According to the mutant Tn5 transposase, the reaction efficiency can be more effectively improved, and the stability of the transposase is remarkably improved.
Owner:JIANGNAN UNIV
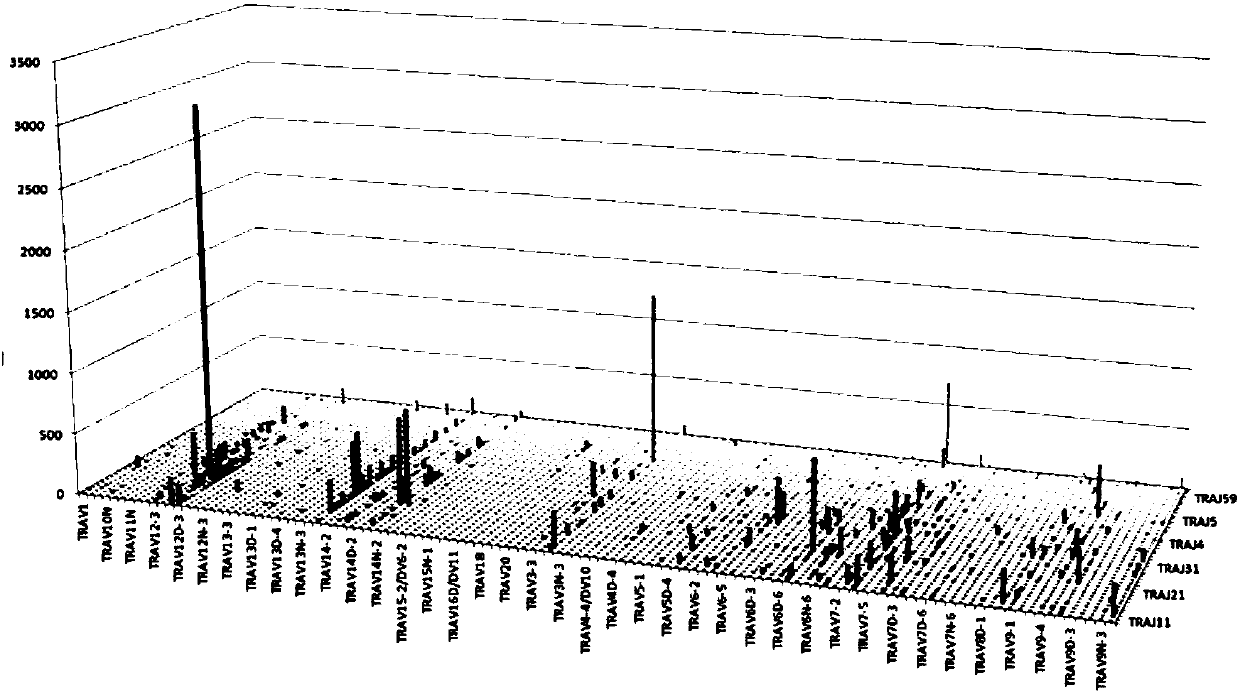
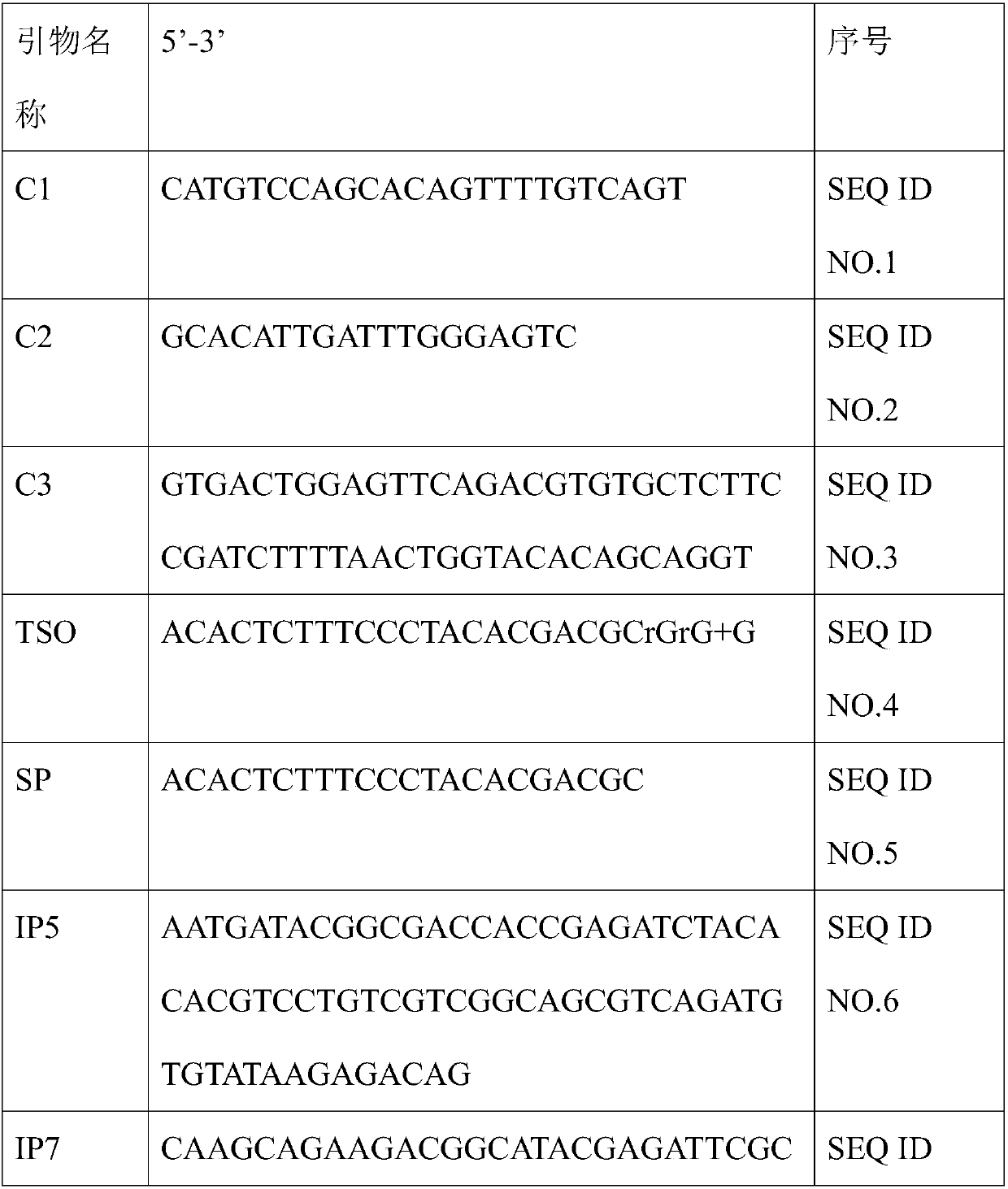
Method for constructing mouse TCR alpha CDR3 region library
PendingCN107829145AAccurate detectionMicrobiological testing/measurementLibrary creationTotal rnaMouse tissue
The invention relates to a method for constructing a mouse TCR alpha CDR3 region sequencing library. The method mainly comprises the following steps: (1) extracting total RNA from mouse tissues or whole blood; (2) reversely transcribing the RNA into cDNA, ligating a linker sequence to the end of the long-stranded cDNA, and amplifying TCR alpha containing a CDR3 region through a universal C-terminal primer and a linker sequence primer; (3) fragmenting the TCR alpha through Tn5 transposase, and enriching a CDR3 region sequence; and (4) carrying out sequencing by an illumina high-throughput sequencing platform. The method is suitable for library construction of tissues containing various types of T cells, and body fluids, can highly-efficiently amplify the mouse TCR alpha through a 5'RACE technology in an agonic manner, and solves the problems of difficulty in control of the template copy number and the offset of the product, caused by multiplex PCR amplification; and the Tn5 enzyme's ability to simultaneously complete DNA fragmentation and linker ligation and a specific primer are used to fast and accurately enrich and construct a library of the hypervariable region (CDR3 region) ofthe TCR alpha, so pertinent sequencing, easy data analysis and sequencing cost saving are achieved.
Owner:重庆天科雅生物科技有限公司
Method for constructing human TCR alpha CDR3 region library
The invention relates to a method for constructing a human TCR alpha CDR3 region sequencing library. The method mainly comprises the following steps: (1) extracting total RNA from human tissues or whole blood; (2) reversely transcribing the RNA into cDNA, ligating a linker sequence to the end of the long-stranded cDNA, and amplifying TCR alpha containing a CDR3 region through a universal C-terminal primer and a linker sequence primer; (3) fragmenting the TCR alpha through Tn5 transposase, and enriching a CDR3 region sequence; and (4) carrying out sequencing by an illumina high-throughput sequencing platform. The method is suitable for library construction of tissues and body fluids, containing various types of T cells, can highly-efficiently amplify the human TCR alpha through a 5'RACE technology in an agonic manner, and solves the problems of difficulty in control of the template copy number and the offset of the product, caused by multiplex PCR amplification; and the Tn5 enzyme's ability to simultaneously complete DNA fragmentation and linker ligation and a specific primer are used to fast and accurately enrich and construct a library of the hypervariable region (CDR3 region) ofthe TCR alpha, so pertinent sequencing, easy data analysis and sequencing cost saving are achieved.
Owner:重庆天科雅生物科技有限公司
RNA library building method
PendingCN112176421AReduce dependenceInput flexibleMicrobiological testing/measurementLibrary creationTn5 transposaseRNA library
The invention discloses an RNA library building method and a kit, and relates to the field of gene sequencing. The method comprises the following steps of: 1) synthesizing First cDNA to form a DNA / RNAhybrid chain; 2) breaking the First cDNA hybrid chain by using streptavidin magnetic beads immobilized with a Tn5 transposase complex, and connecting a joint; and 3) supplementing gaps of the fragmented hybrid chains and enriching a complete library through PCR amplification. According to the method provided by the invention, the library building process is simple to operate, and the RNA librarybuilding process can be completed within two hours at the soonest; moreover, the input amount of the initial RNA is relatively flexible, the constraint that the traditional Tn5 transposase amount onlyaims at the input amount of specific RNA to establish a library is broken through, the dependence of the transposase on template input is reduced, and 10ng-1[mu]g of RNA with different amounts can beinput according to own requirements.
Owner:上海英基生物科技有限公司
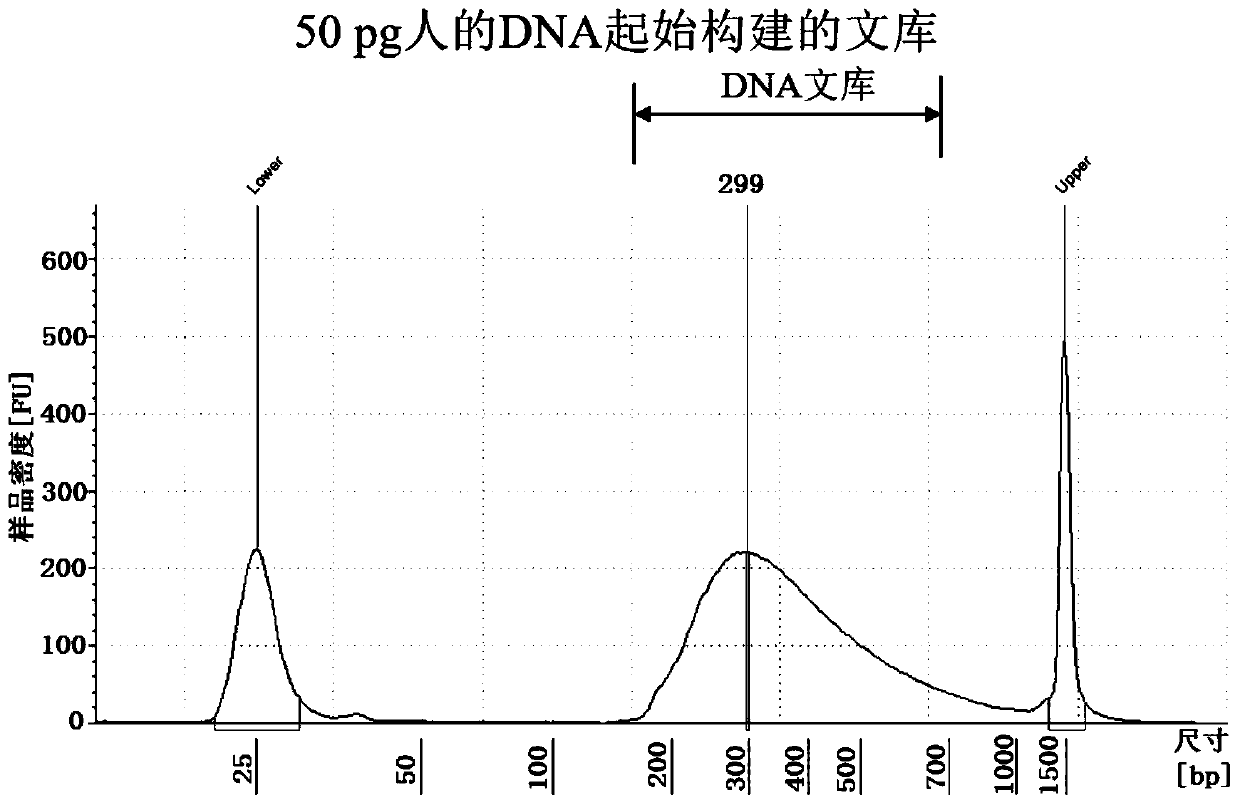
Method for introducing molecular tag in library construction suitable for Illumina next-generation sequencing platform, linker sequence and application thereof
PendingCN110628889AAvoid errorsAccurate quantitative analysisMicrobiological testing/measurementTn5 transposaseRecognition sequence
The invention provides a method for introducing a molecular tag in library construction suitable for an Illumina next-generation sequencing platform, a linker sequence and a design method, a method for constructing a library by using the linker sequence, and a kit containing the linker sequence. Wherein the linker sequence comprises 5'-P-E-F-Q-3', P is a sequencing platform pairing sequence, E isa random base sequence composed of the same base, F is a given base sequence, and Q is a Tn5 transposase terminal recognition sequence. The linker sequence and the library construction method providedby the invention can successfully construct the library for DNA samples as low as 50 pg, and are suitable for preparing DNA library construction kits and disease diagnosis kits.
Owner:远辰生物科技(苏州)有限公司
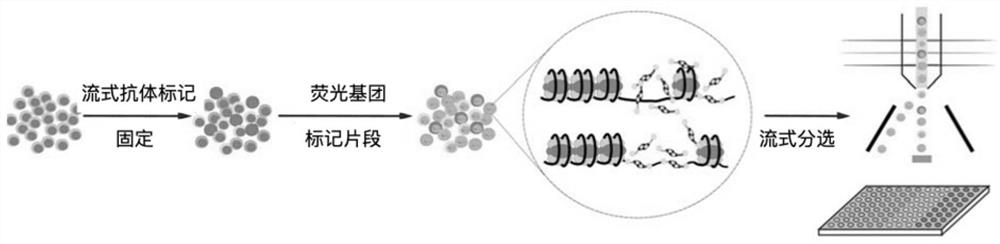
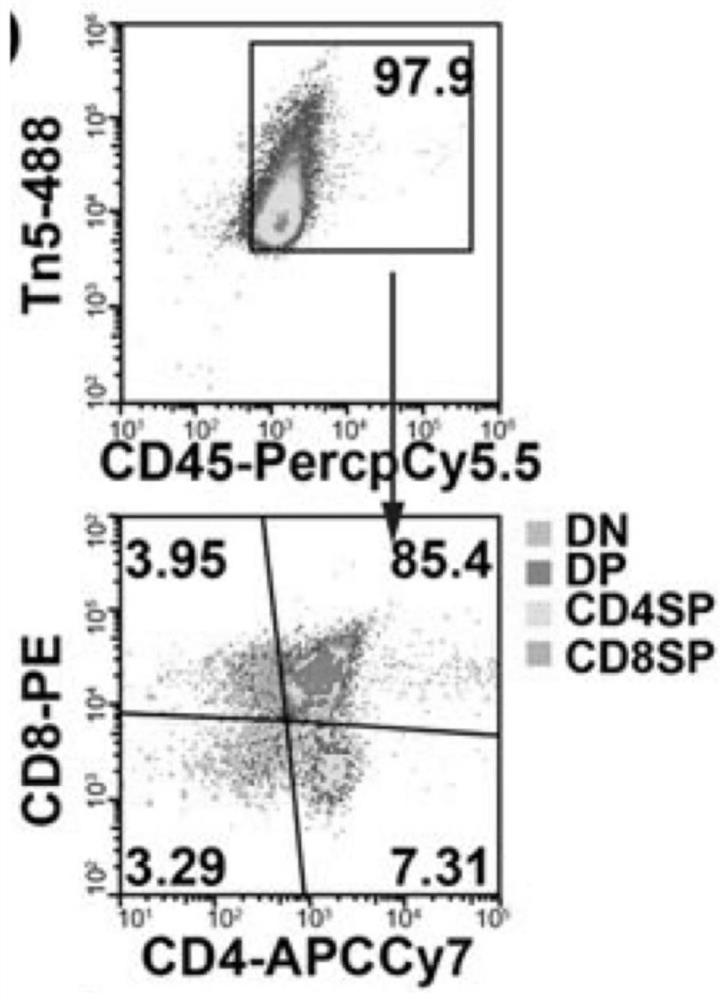
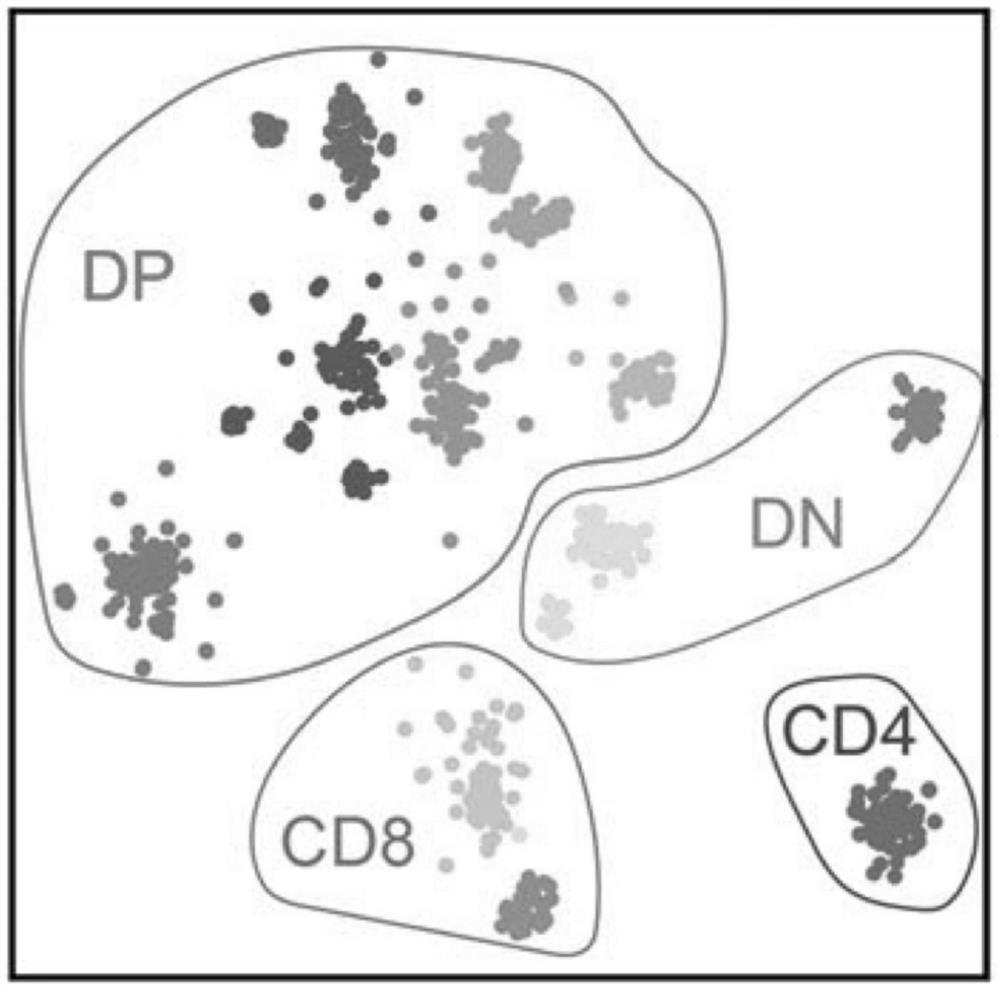
Library building method for accessibility of unicellular chromatin
The invention relates to a library building method for accessibility of unicellular chromatin. The method comprises the following steps of: designing a pair of oligonucleotide chains capable of beingcoupled fluorescently, preparing a fluorescent transposon by combining with Tn5 transposase, dissociating single-cell suspension, adding a fluorescent antibody aiming at specific protein of a target population for incubation, carrying out fluorescence labeling, sorting and enriching, carrying out PCR library building, and carrying out library sequencing. According to the library building method for the accessibility of unicellular chromatin provided by the invention, a target cell population can be screened under the condition of ensuring the activity in a cell body; a set of oligonucleotide chains capable of realizing fluorescence coupling is designed and is combined with Tn5 transposase to prepare the fluorescence transposon, quantitative operation is carried out, the applicability is wide, and the method is especially suitable for constructing tissues with few species or tissues and poorer preservation states; and the method is suitable for cell populations with extremely small proportions.
Owner:HANGZHOU HANGENE BIOTECH CO LTD
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com