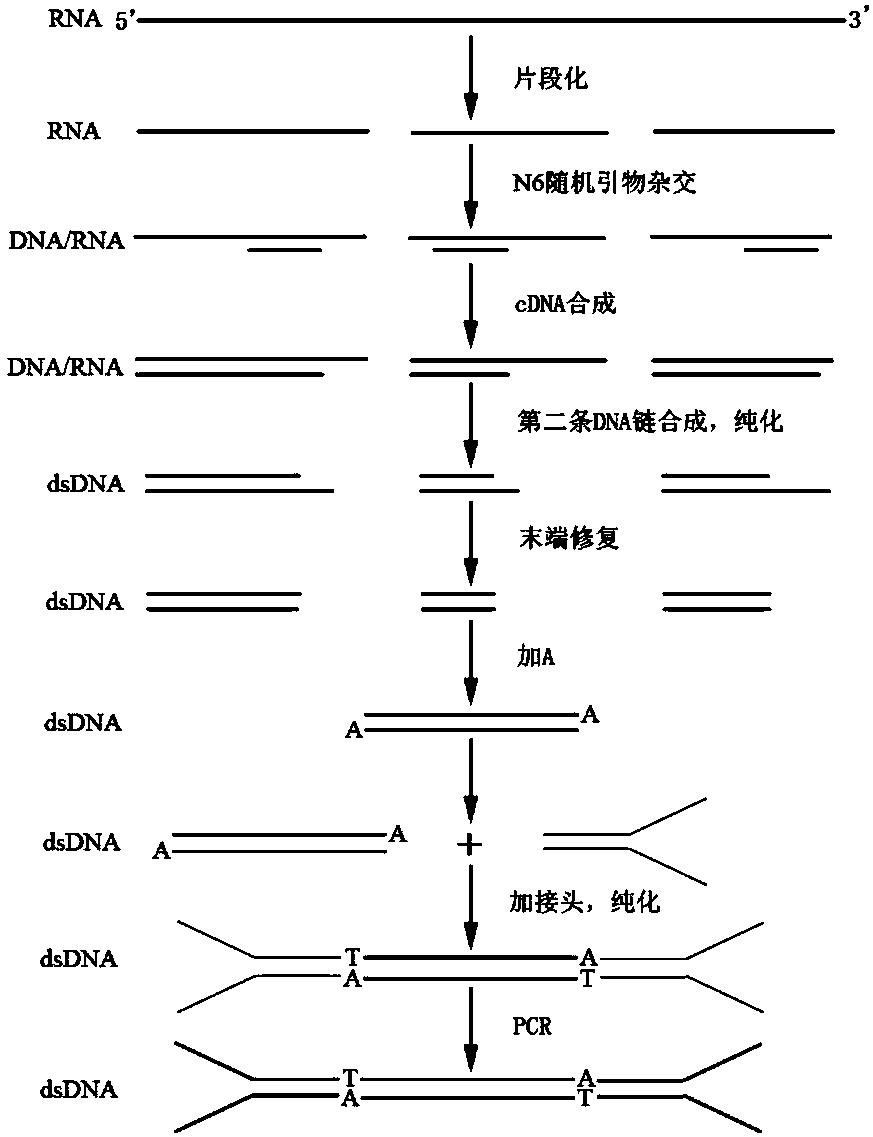
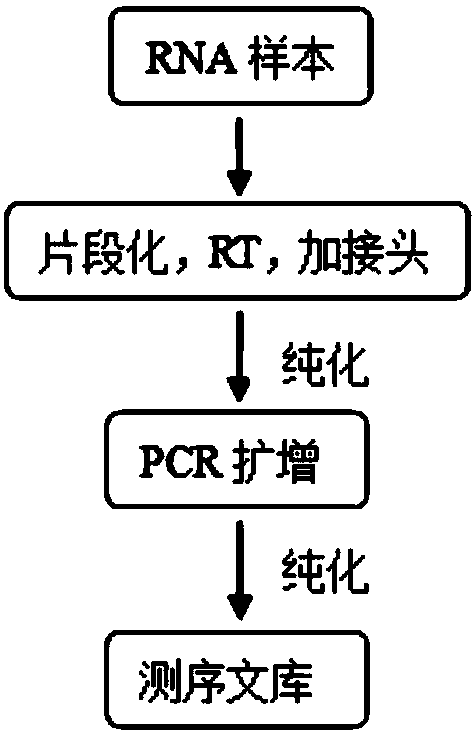
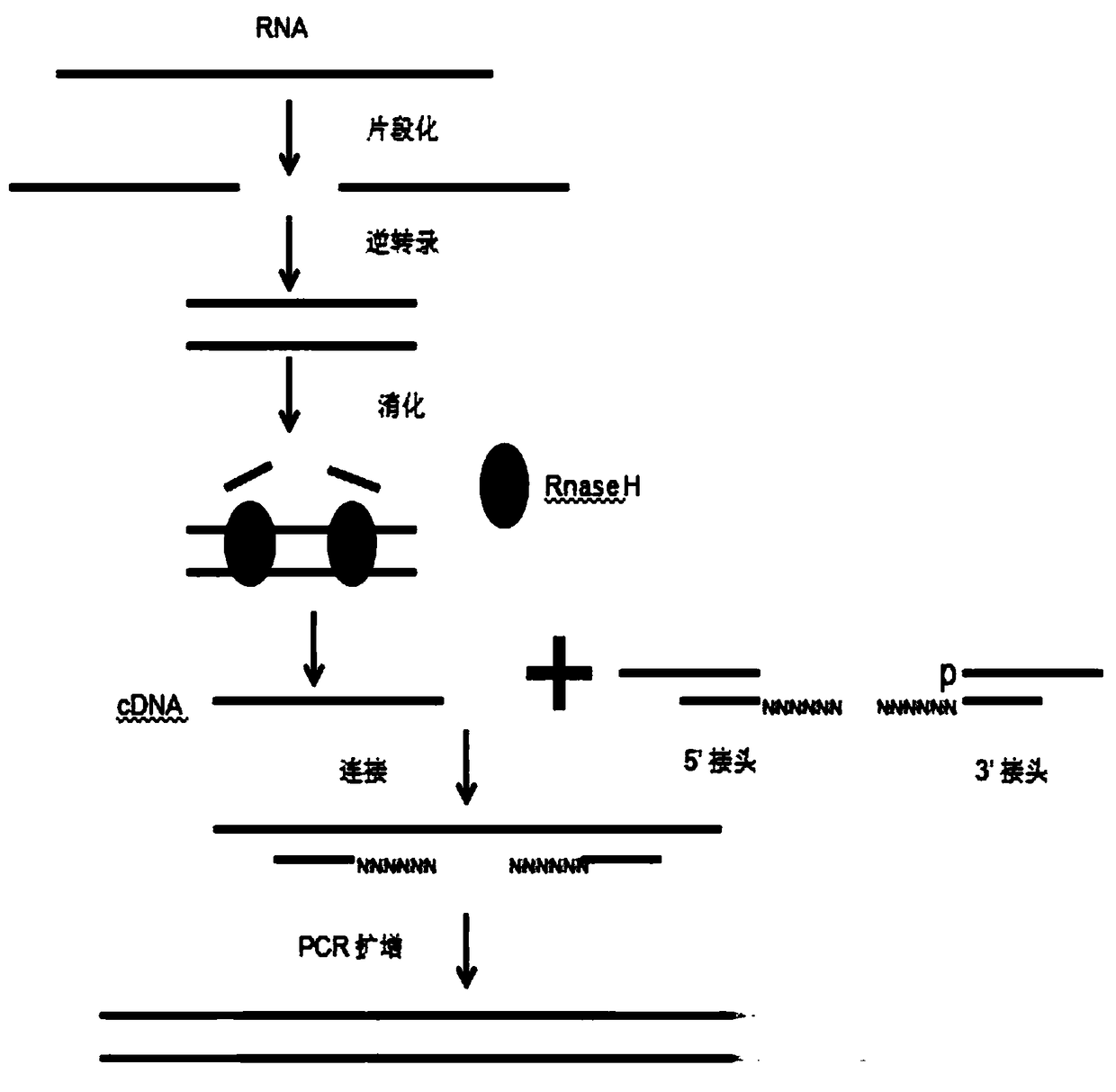
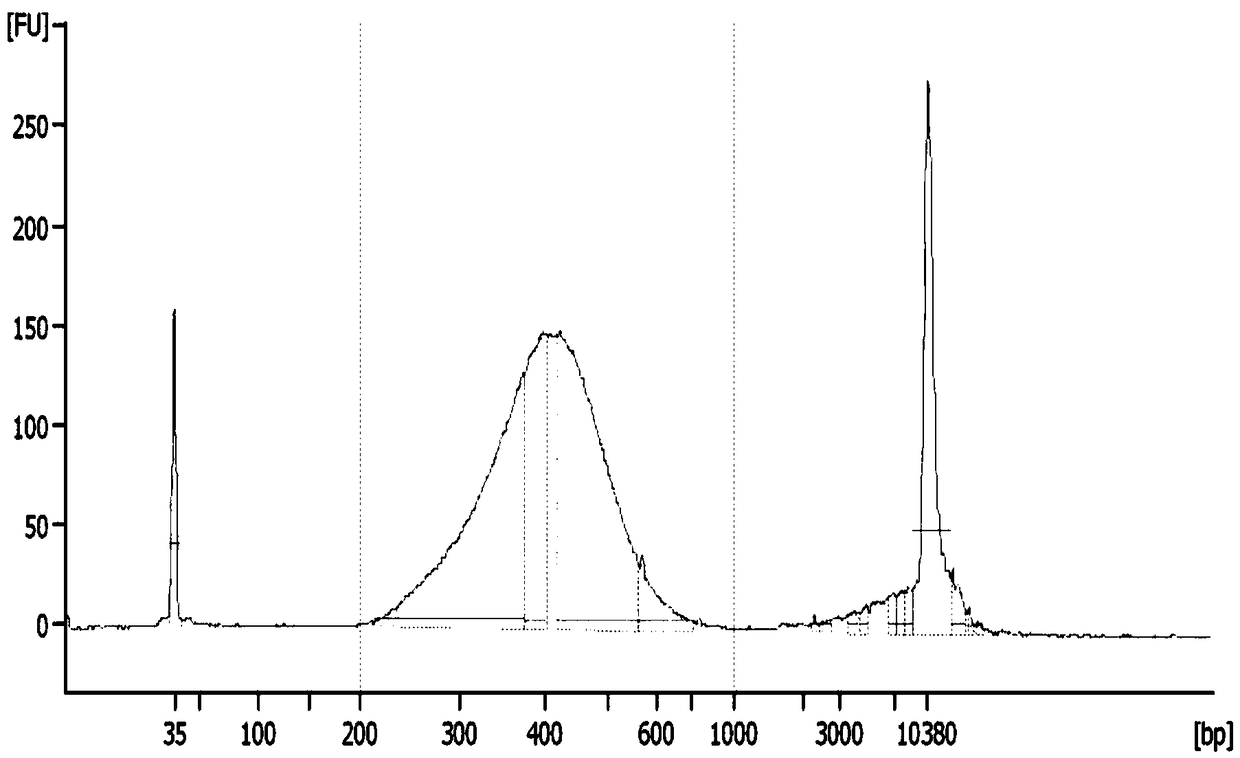
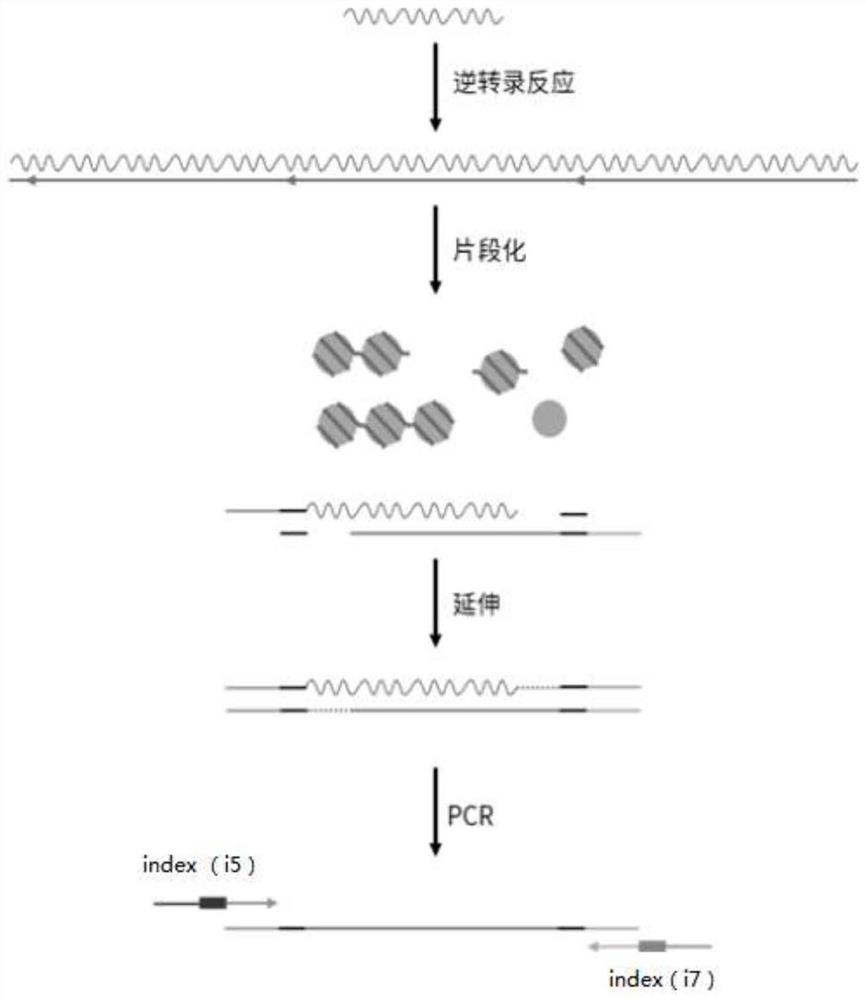
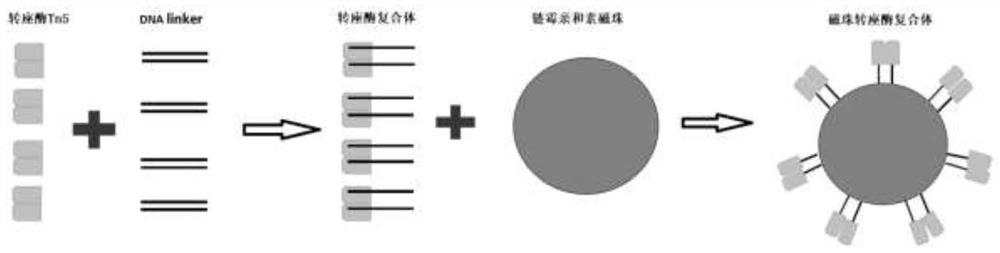
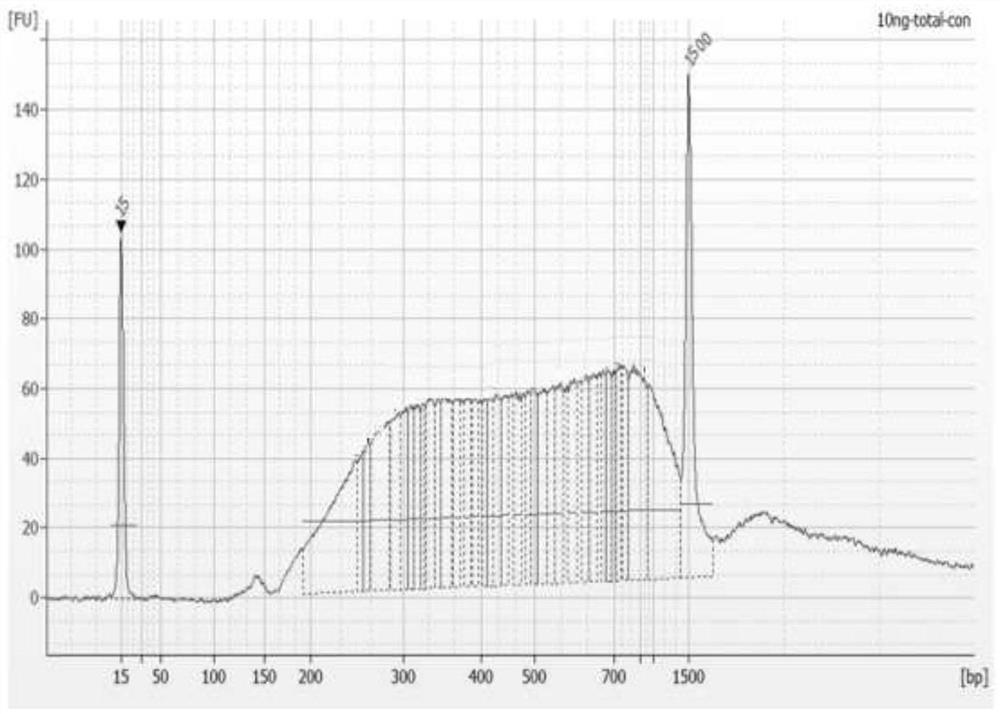
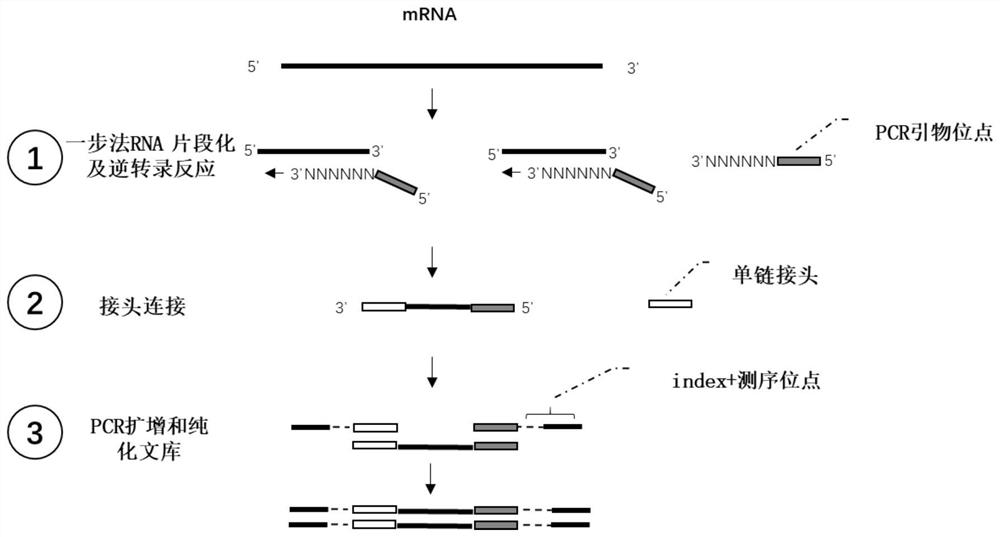
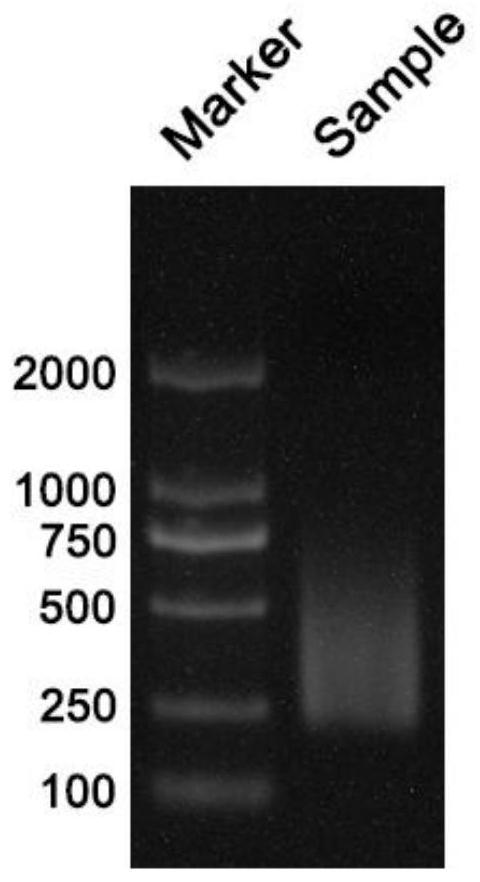
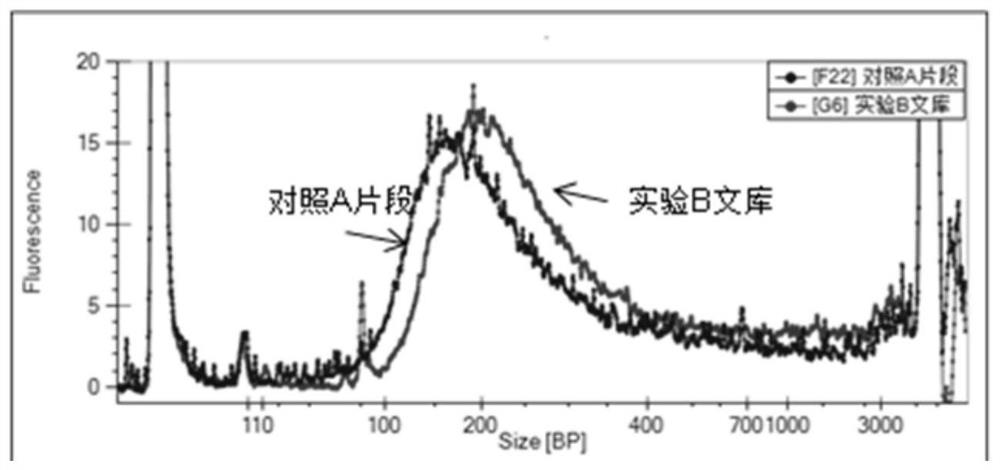
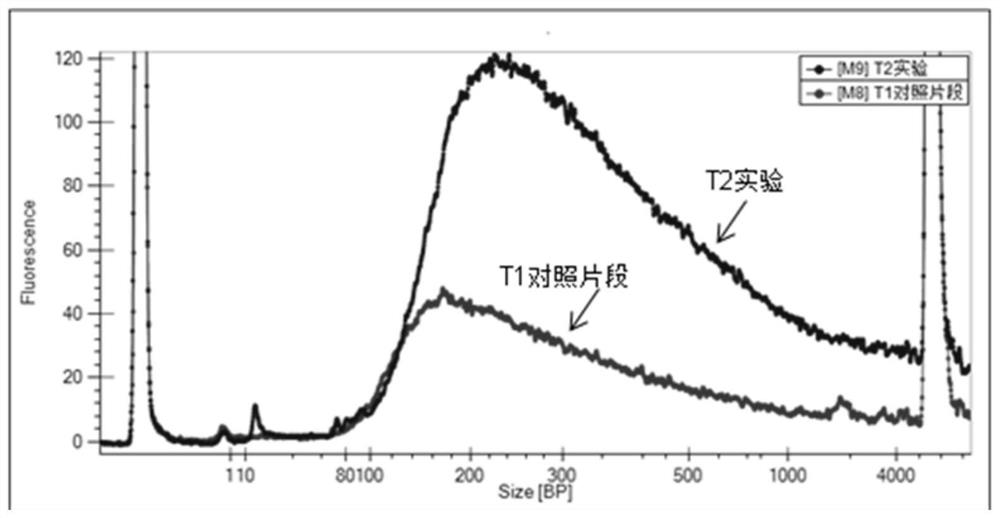
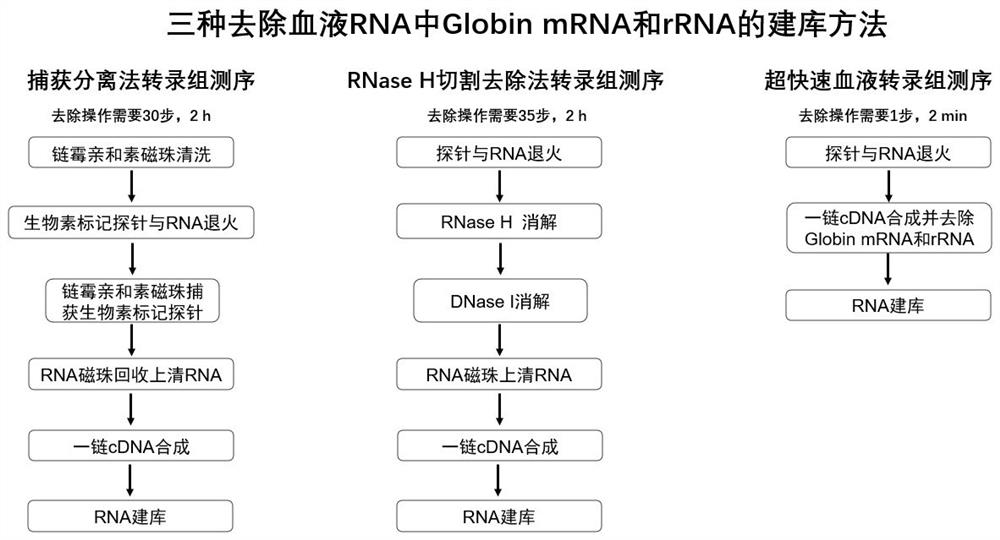
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
59 results about "RNA library" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
RNA adaptor capable of restraining bacillus typhosus from invading to cells of human body, and preparation method
InactiveCN1563401APrevent intrusionPrevention and treatment of enteric feverAntibacterial agentsGenetic material ingredientsBacillus typhosusHuman body
This invention discloses a RNA adaptor preventing typhoid typhi from intruding into human body cells. This invention discloses a method for preparing the RNA adaptors by preparing and purifying TVB type cilium structure fusion protein-agarose 4B, constructing single strand DNA library randomly, comining double-strand DNA library, amplifying PCR, preparing RNA libraries, screen SELEX, selecting RCR to get RNA adaptors, which can be the agonist of typhi.
Owner:WUHAN UNIV
Reverse transcription obstruction probe for rapidly removing target RNA in RNA library construction and application thereof
ActiveCN112626176AKeep intactImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideComplementary pair
The invention provides a reverse transcription obstruction probe for rapidly removing target RNA in RNA library construction and application thereof. The probe has a length of 20-40 nt, is strictly complementarily paired with a target RNA sequence, and has no complementary pairing continuously exceeding 15 nt with other non-target RNAs, the Tm value of the probe is not lower than 80 DEG C, the self complementary value is less than 5, and 3'-OH of the probe is sealed by modified deoxyribonucleotide. According to the probe, a reverse transcription inhibition probe method is utilized to realize high-efficiency and high-specificity inhibition on reverse transcription of the target RNA during synthesis of one strand of cDNA, so that the target RNA is prevented from participating in a downstream library construction process. The probe has the advantages of simplicity in operation (one-step operation), extremely short consumed time (2 minutes), completeness in retention of the non-target RNAs, high specificity, high efficiency, low cost, high gene detection number and the like, and is very suitable for the fields of industrial automatic library building and rapid disease diagnosis.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Construction method of RNA library for enriching original transcript information and application thereof
ActiveCN105506747AHigh gene coverageEfficiently buildLibrary creationProtein nucleotide librariesRNA libraryGlobulin
The invention provides a construction method of a high-quality RNA library for enriching original transcript information by using RNase H and selectively removing lots of rRNA and blood globulin and an application thereof. The RNA library constructed by the method has very low rRNA residual volume (which is only 1 / 500 of residual volume in a conventional oligo(dT) method), and has very high gene coverage.
Owner:BGI SHENZHEN CO LTD
Optimized method for constructing RNA high-throughput sequencing library and application of optimized method
InactiveCN107385018AGood extensibilityImprove stabilityLibrary creationReverse transcriptaseRNA library
The invention belongs to the field of molecular biological techniques and application, and particularly relates to an optimized method for constructing an RNA high-throughput sequencing library and application of the optimized method. The optimized method for constructing the RNA high-throughput sequencing library is characterized in that the method adopts RNase III to break to-be-sequenced RNA, adopts RNA ligase to connect linkers and adopts reverse transcriptase and a high-fidelity DNA polymerase system to conduct one-step reverse transcription and labeling PCR to construct the high-throughput sequencing library. The method can be applied to construction of the RNA high-throughput sequencing library. According to the optimized method for constructing the RNA high-throughput sequencing library, RNA is directly adopted to add the linkers onto single-strands, the step of random primer amplification is omitted, the influences of random primers can be eliminated, and more comprehensive RNA information is obtained; and an one-step method is adopted to conduct reverse transcription and labeling PCR, intermediate steps are reduced, and the preparation efficiency of the RNA library and the sequencing quality are enhanced effectively.
Owner:哈尔滨博泰生物科技有限公司
High-throughput method based on DNA (desoxyribonucleic acid) and RNA (ribonucleic acid) gene mutation detection
ActiveCN110079594ASimple designIncrease expression abundanceMicrobiological testing/measurementRNA libraryA-DNA
The invention discloses a high-throughput method based on DNA (desoxyribonucleic acid) and RNA (ribonucleic acid) gene mutation detection. The method comprises the steps that (1) desoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are extracted from a biological sample collected from a subject through utilizing a reagent, and a DNA library is constructed based on desoxyribonucleic acid; ribonucleic acid is reversely transcribed into double-stranded cDNA firstly, and then is constructed into an RNA library; (2) a first probe set is mixed with the DNA library, and meanwhile, a second probe set is mixed with the RNA library; (3) a complex of probes and nucleic acid is separated and obtained from mixtures obtained in step (2) by utilizing markers in the first probe set and second probe set; (4) sequencing is performed on target nucleic acid in the complex by utilizing a high-throughput technique. The method is based on NGS methodology and optimizes the NGS methodology, so that the specificity and sensitivity of detection are further improved. In addition, a DNA detection result can also be verified by detecting RNA.
Owner:GENEIS TECH BEIJING CO LTD
Absolute quantitative transcriptome library construction method based on specific recognition sequence
ActiveCN110396516AReduce the cost of building a libraryImprove the speed of database constructionMicrobiological testing/measurementLibrary creationReverse transcriptaseRNA library
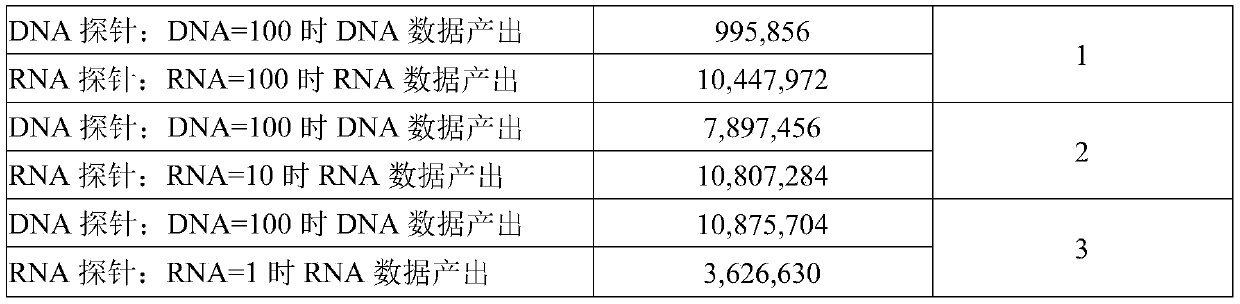
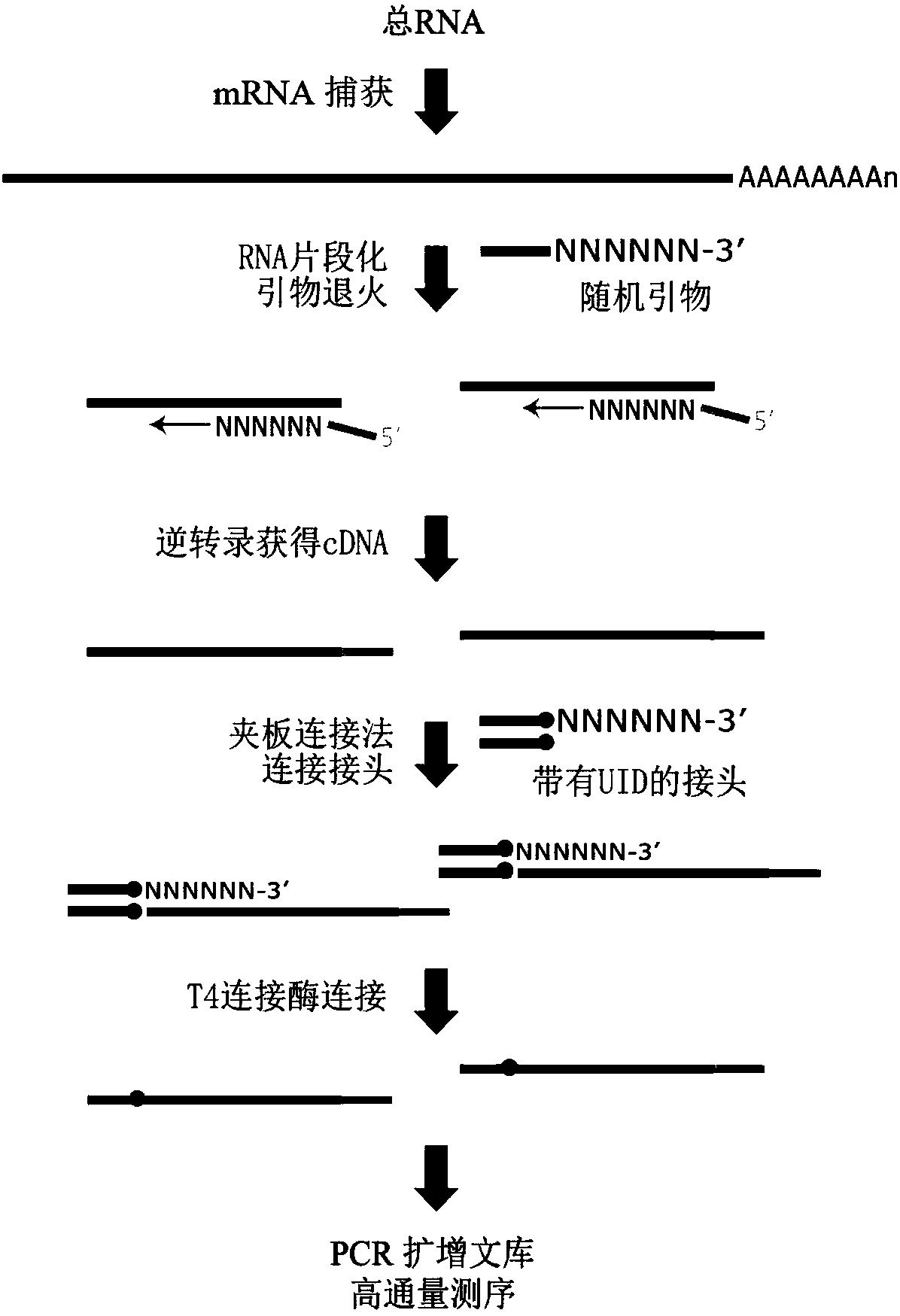
The invention discloses an absolute quantitative transcriptome library construction method based on a specific recognition sequence. Fragmented mRNA is used as a template, a first cDNA chain is synthesized under the action of reverse transcriptase by a primer pool with a general joint sequence, a library construction joint with a special recognition UID sequence is added to cDNA 3' end through anenzymatic reaction, so that each cDNA carries a unique sequence label; and finally, PCR amplification is performed by the general library construction joint to obtain an RNA library. The method constructs the RNA library on the basis of single-stranded cDNA by a splint connection method for the first time, at the same time, a UID sequence is used to precisely reduce the cDNA composition before PCRamplification and can realize precise quantification of transcripts. Library construction is performed by the single-stranded cDNA as a raw material, the step of second strand synthesis is omitted, the template loss rate is reduced, the cost and time are saved, and the defect that the prior art can relatively quantify transcripts is thoroughly solved.
Owner:武汉康测科技有限公司
Library building method based on high-throughput sequencing
ActiveCN111321208AFirmly connectedGuaranteed specificityMicrobiological testing/measurementLibrary creationMultiplexNucleotide
The invention discloses a library building method based on high-throughput sequencing. The library building method based on the high-throughput sequencing is applicable to fragmented nucleotide samples, such as fragmented genomic DNA / FFPE RNA / cfDNActDNA and the like. Furthermore, the fragmented nucleotide samples are subjected to modification, denaturation and joint connection, followed by enrichment of target regions by two specific PCR reactions, so as to obtain a sequencing library; and the sequencing library is subjected to quality control and quantitative analysis so as to carry out high-throughput sequencing. The invention further provides a simulated double-chain connection joint. The library building method based on the high-throughput sequencing is simple and convenient, and retains advantages of multiplex PCR library building methods based on amplicons; and moreover, the library building method based on the high-throughput sequencing can also be used for detecting unknownfusion, and is especially applicable for detecting nucleotide samples (e.g. cfDNA / ctDNA sample) with relatively high degree of fragmentation. The library building method based on the high-throughput sequencing is also feasible for building libraries of DNA molecules with damages. The library building method based on the high-throughput sequencing disclosed by the invention is capable of retaining all original nucleotide information to the maximum extent; and moreover, no two-chain synthesis is required when the method is adopted for carrying out RNA library building.
Owner:上海厦维医学检验实验室有限公司
Method for rapidly constructing strand-specific RNA high-throughput sequencing library
InactiveCN108251503AQuality improvementEasy to operateMicrobiological testing/measurementLibrary creationRNA libraryGenetic engineering
The invention belongs to the technical field of genetic engineering, and particularly relates to a method for rapidly constructing a strand-specific RNA high-throughput sequencing library. The methodprovided by the invention completes construction of an RNA library in only four steps as follows: fragmentation, RT and PCR enrichment and library purification. Compared with a traditional RNA libraryconstruction method, the method provided by the invention simplifies operation flows, shortens time, has high efficiency and can produce a high-quality strand-specific RNA-seq library by only two enzymes.
Owner:HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
Method for constructing chain specific RNA library and application
ActiveCN109234813AOvercoming complexityOvercoming cumbersomenessMicrobiological testing/measurementLibrary creationRNA libraryDigestion
The invention relates to a new method for constructing a chain specific RNA library and especially relates to a method for constructing the chain specific RNA library and an application. The method comprises the following steps: performing fragmentation treatment on RNA, thereby acquiring a fragmented RNA; performing reverse transcription on the fragmented RNA and then adopting RNase H for digesting a RNA / DNA heterozygote; connecting a digestion product with a joint, thereby acquiring a connected product; performing PCR amplification on the connected product, thereby acquiring an amplificationproduct, namely, the chain specific RNA library. According to the invention, RNase H is adopted for digesting the RNA / DNA heterozygote and then double-end joint connection is directly performed, so as to acquire the chain specific RNA library; due to an optimized reaction system, the process from sample RNA to joint connection can be finished by only adding reaction reagents in turn in a tube; compared with the conventional method for constructing the chain specific RNA library, the method provided by the invention is capable of overcoming complexity and tediousness of process and saving a large amount of manpower, time and reagent cost.
Owner:广东安科华南生物科技有限公司
Multi-nucleic acid co-labeling support, preparation method therefor, and application thereof
PendingCN114096678ANucleotide librariesMicrobiological testing/measurementMultiplexChemical reaction
Provided are a variety of nucleic acid co-labeling supports, a preparation method therefor, and an application thereof. The supports comprise a support body, and a variety of nucleic acid labels located on the surface and / or inside of the support body. The nucleic acid labeled on a single support at least comprises: one or more first nucleic acid labels having a function that at least comprises of trapping a specific compound in a reaction system onto the surface of the support; and one or more second nucleic acid labels having a function that at least comprises participating in a specified biochemical reaction process of the specific compound trapped onto the surface of the support. The foregoing variety of nucleic acid co-labeled supports can be used for 5'-terminus single-cell RNA expression profile analysis, construction of a 5'-terminus single-cell VDJ library for a microwell array platform, construction of a 3'-terminus single-cell RNA library, construction of a single-cell transcriptome library, single-cell multi-omics research, multiplex PCR and / or construction of a multiplex PCR sequencing library, etc.
Owner:BEIJING SEEKGENE BIOSCIENCES CO LTD
RNA library building method
PendingCN112176421AReduce dependenceInput flexibleMicrobiological testing/measurementLibrary creationTn5 transposaseRNA library
The invention discloses an RNA library building method and a kit, and relates to the field of gene sequencing. The method comprises the following steps of: 1) synthesizing First cDNA to form a DNA / RNAhybrid chain; 2) breaking the First cDNA hybrid chain by using streptavidin magnetic beads immobilized with a Tn5 transposase complex, and connecting a joint; and 3) supplementing gaps of the fragmented hybrid chains and enriching a complete library through PCR amplification. According to the method provided by the invention, the library building process is simple to operate, and the RNA librarybuilding process can be completed within two hours at the soonest; moreover, the input amount of the initial RNA is relatively flexible, the constraint that the traditional Tn5 transposase amount onlyaims at the input amount of specific RNA to establish a library is broken through, the dependence of the transposase on template input is reduced, and 10ng-1[mu]g of RNA with different amounts can beinput according to own requirements.
Owner:上海英基生物科技有限公司
Library preparation method, kit and sequencing method
PendingCN113337576AMathematical modelsMicrobiological testing/measurementLibrary preparationRNA library
The invention provides a library preparation method, a kit and a sequencing method. The library preparation method comprises the following steps: obtaining a target nucleic acid molecule, namely performing reverse transcription on RNA (Ribonucleic Acid) from a nucleic acid sample to be detected to obtain a reverse transcription product which comprises RNA: DNA (Deoxyribose Nucleic Acid) hybrid double strands and RNA: DNA hybrid double strands enriched in the reverse transcription product; providing a transposal complex, wherein the transposal complex comprises a linker and a transposase, the linker is a double-stranded nucleic acid molecule with a known sequence; contacting the target nucleic acid molecule and the transposition complex under conditions suitable for a transposition reaction to obtain a transposition product, wherein the transposition product comprises a double-stranded nucleic acid molecule with a joint at the tail end and a gap; providing a first primer configured to hybridize with at least a portion of the linker; and hybridizing the transposon with the first primer and placing under conditions suitable for strand displacement reaction to obtain the library. The method is particularly suitable for rapid construction of RNA libraries with low initial quantities.
Owner:GENEMIND BIOSCIENCES CO LTD
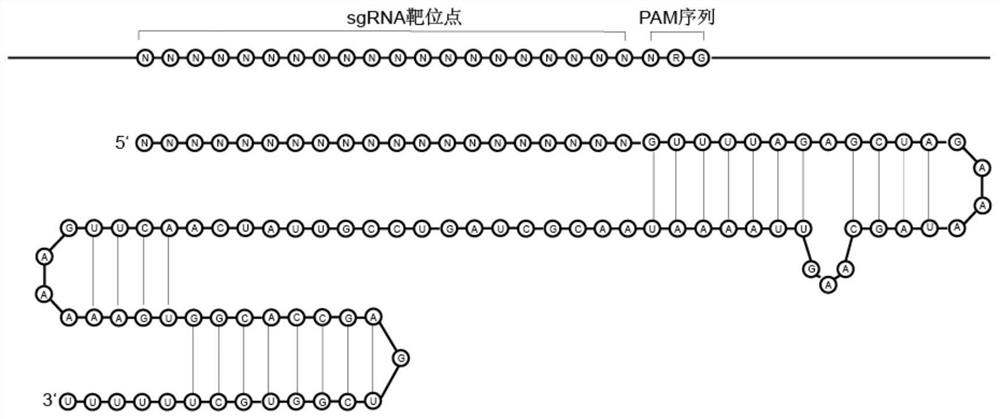
Preparation method of target sequence random sgRNA full-coverage group
The invention provides a preparation method of a target sequence random sgRNA full-coverage group. The preparation method comprises the following steps: cutting a PAM region of a target sequence by using restriction enzyme; connecting an upper sgRNA skeleton; obtaining a sequence of a targeting original spacer region through a side-cut activity restriction enzyme site on the sgRNA skeleton; connecting a T7 promoter; performing amplification to obtain an sgRNA library with a T7 promoter; and carrying out in-vitro transcription to obtain a target sequence sgRNA library. The invention also discloses a preparation method of the sgRNA library of the ribosomal RNA, a method for removing the ribosomal RNA in the RNA library, and a method for removing a human whole genome from a host genome. The method disclosed by the invention has the advantages of low cost, simplicity in manufacturing, uniform coverage, small preference, no limitation by the length of a target sequence, no need of designing sgRNA on a large scale and the like.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Method for obtaining male and female differential expression genes of early embryos or larvae of prawns
ActiveCN110964797AMeet the sequencing concentration requirementsMicrobiological testing/measurementSequence analysisObstetricsPhysiology
According to the invention, prawns have obvious sex bimorphism; sex determination and differentiation of the prawns often occur in an early development stage; thus, sex determination and sex differentiation related genes need to be screened in the early development stage of male and female individuals. Since prawn individuals at the early development stage are extremely small, and cannot be distinguished in sex, so a screening method for male and female differential expression genes of early embryos or larvae of the prawns is not established yet. According to the invention, co-extracting of DNA and RNA of a single individual of a prawn zoae is realized; a sex probe is used for sex identification on a DNA level; RNAs of a same gender are mixed to respectively construct a male RNA library and a female RNA library in a zoae stage; a micro transcriptome sequencing technology is utilized to obtain the female and male larva differential expression genes; thus, the screening method of the male and female differential expression genes of the early embryos or larvae of the prawns is established. The screening method provided by the invention promotes the research on sex determination and sex differentiation of the prawns and provides a basis for analyzing a sex determination and differentiation mechanism.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
RNA library construction method
ActiveCN112176422AEasy to operateSave man hoursLibrary creationProtein nucleotide librariesCyclaseGenetics
The invention provides a RNA library construction method. The construction method comprises the following steps: carrying out reverse transcription on fragmented RNA by adopting a reverse transcription primer to obtain a first strand cDNA, wherein the reverse transcription primer sequentially comprises a known sequence and a random sequence from 5' end to 3' end, carrying out single-chain linker connection on the first chain cDNA by adopting cyclase to obtain a connection product, carrying out PCR amplification on the connection product to obtain an RNA library, introducing a first sequencingprimer sequence while RNA is subjected to reverse transcription to synthesize first-chain cDNA, then directly carrying out single-chain linker connection through cyclase to introduce a second sequencing primer sequence, and finally, designing a proper amplification primer according to the first sequencing primer sequence and the second sequencing primer sequence to carry out PCR amplification on alinker connection product so that RNA libraries can be obtained. According to the method, synthesis of second chain cDNA is not needed, chain specificity is directly achieved without other digestionprocesses, operation is easy, and working hours are saved.
Owner:天津诺禾致源生物信息科技有限公司
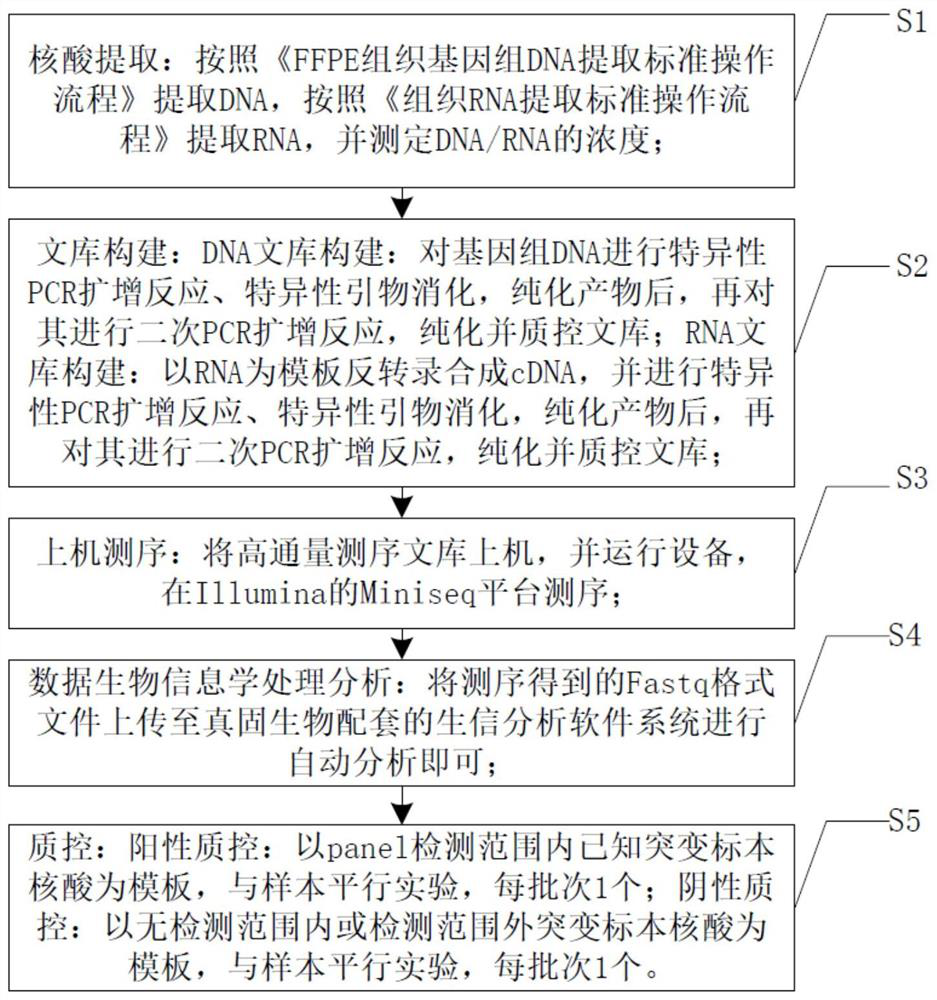
Method and system for NGS calibration of lung cancer targeted drug and chemotherapy drug genomes by DNA and RNA double library construction
PendingCN112226496AReduce outputGuarantee the quality of inspectionMicrobiological testing/measurementLibrary creationA-DNAChemo therapy
The invention discloses a method and system for NGS calibration of lung cancer targeted drug and chemotherapy drug genomes by DNA and RNA double library construction. The method comprises the following steps: directly amplifying genes with known mutation sites, then constructing a library, performing sequencing detection on NGS, and determining and verifying the existence of mutant genes in combination with bioinformatics analysis. The method comprises the following steps: S1, extracting nucleic acid, and determining the concentration of DNA / RNA; S2, constructing a DNA or RNA library; S3, performing on-machine sequencing: loading a high-throughput sequencing library on a machine, operating equipment, and performing NGS sequencing; S4, performing data bioinformatics processing and analysis:uploading a Fastq format file obtained by sequencing to a bioinformatics analysis software system matched with a true solid organism for automatic analysis; and S5, performing quality control. According to the invention, the problem that the existing lung cancer drug gene detection system can only aim at a targeted drug or a chemotherapeutic drug is solved, lung cancer chemotherapy and targeted drug gene detection are realized in one project, the detection cost is low, and the detection efficiency is high.
Owner:合肥金域医学检验实验室有限公司
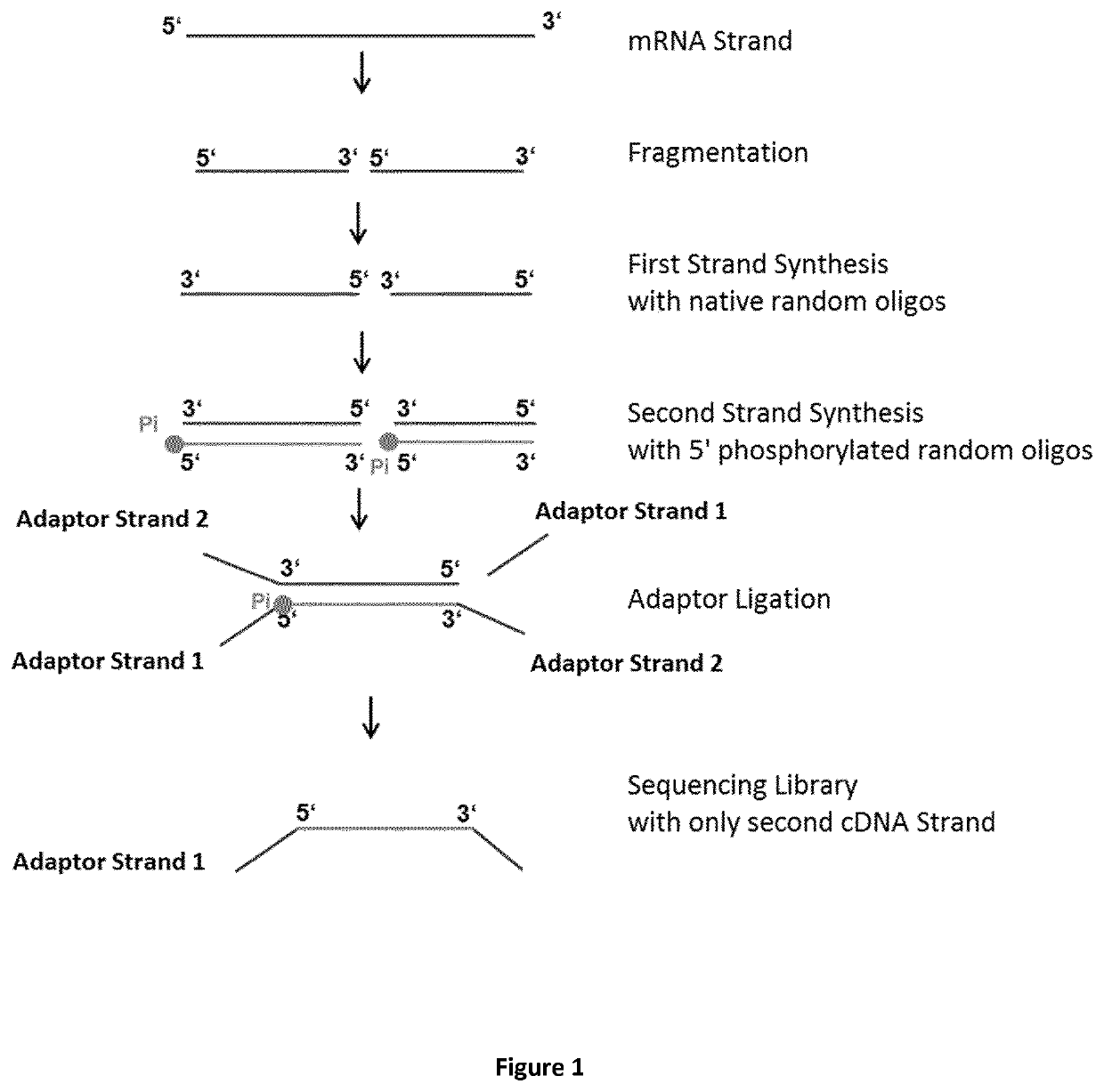
Method for generating a stranded RNA library
ActiveUS20190119671A1Microbiological testing/measurementLibrary creationReverse transcriptaseNucleotide
The invention relates to a method for preparing a strand-specific library from an nucleic acid or preferably RNA sample, for RNA comprising the steps of: (i) optionally fragmenting said RNA sample, (ii) generating a plurality of first cDNA strands by subjecting said fragmented RNA to reverse transcription by using a reverse transcriptase and first oligonucleotide primers, (iii) generating a plurality of second cDNA strands by using a DNA polymerase, second oligonucleotide primers, and the plurality of first cDNA strands, and (iv) ligating adapters to the 3′ and 5′ termini of the of double-stranded cDNA, (v) wherein the first cDNA strand allows no adapter ligation at its 5′ terminus and said second cDNA strand allows adapter ligation at its 5′ terminus, or vice versa, and, (v) optionally cloning, sequencing or otherwise using the strand-specific library. The invention also relates to a kit for preparing a strand-specific library from an RNA sample, wherein said kit comprises, (i) random oligonucleotide primers comprising a 5′ terminus nucleotide which allows no adaptor ligation, (ii) random oligonucleotide primers comprising a 5′ terminus nucleotide which allows adaptor ligation, (iii) optionally a reverse transcriptase for synthesizing a first cDNA strand complementary to the template RNA, (iv) optionally dNTPs, and (v) optionally a DNA polymerase
Owner:QIAGEN GMBH
An Absolute Quantitative Transcriptome Library Construction Method Based on Unique Recognition Sequences
ActiveCN110396516BReduce the cost of building a libraryImprove the speed of database constructionMicrobiological testing/measurementLibrary creationReverse transcriptaseRNA library
The invention discloses a method for constructing an absolute quantitative transcriptome library based on a unique recognition sequence. Using the fragmented mRNA as a template, the first cDNA strand is synthesized under the action of reverse transcriptase using a primer pool with a universal linker sequence, and a unique recognition UID sequence is added to the 3' end of the synthesized cDNA by an enzymatic reaction. Build a library linker so that each cDNA has a unique sequence tag; finally, use a universal library linker to perform PCR amplification to obtain an RNA library. For the first time, the present invention uses the splint ligation method to construct an RNA library based on single-stranded cDNA. At the same time, the UID sequence is used to accurately restore the cDNA composition before PCR amplification, which can realize accurate quantification of transcripts; the present invention uses single-stranded cDNA as a raw material for library construction. The step of second-strand synthesis is omitted, the template loss rate is reduced, cost and time are saved, and the existing technology can only relatively quantify transcripts.
Owner:武汉康测科技有限公司
RNA library preparation method, sequencing method and kit
PendingCN112501249AImprove genome coverageImprove effective utilizationMicrobiological testing/measurementLibrary creationRNA libraryRNA Sequence
The invention relates to the technical field of biology, in particular to an RNA library preparation method, a sequencing method and a kit. The RNA library preparation method comprises the following steps of performing reverse transcription by taking RNA as a template to generate first chain cDNA, thereby obtaining an RNA / cDNA heterozygote; and adding a first preset sequence to at least one tail end of the RNA / cDNA heterozygote to form an RNA / DNA heterozygote, wherein the RNA / DNA heterozygote is an RNA library, and the first preset sequence is located at at least one tail end of the cDNA chain. The RNA library prepared with the method is simple and rapid to operate, the constructed RNA library can be directly used for sequencing, and the method has no double-stranded cDNA synthesis process, namely no cDNA amplification and enrichment process, so that bias and errors caused by PCR amplification and enrichment can be avoided. Meanwhile, the invention discloses the RNA sequencing method which is used for directly measuring cDNA sequence information to obtain RNA sequence information. The invention also discloses the kit for RAN testing, and the kit can be used for constructing the RNAlibrary and sequencing the RNA library.
Owner:GENEMIND BIOSCIENCES CO LTD
Simple RNA library building method
PendingCN114410741ASimple processSimplify time consumingMicrobiological testing/measurementLibrary creationRNA libraryMutant
The invention provides a simple RNA library building method which is characterized by comprising the following steps: (1) extracting RNA in a sample, adding a reverse transcription primer and an rRNA reverse transcription hindering probe, and fragmenting; (2) carrying out RNA reverse transcription to obtain DNA / RNA hybrid double strands; (3) linker connection: connecting a blunt-end double-chain DNA linker with adenylation modification to the 3'end of the DNA / RNA hybrid chain cDNA by using a T4 DNA ligase mutant K159L; and (4) library amplification and recovery. According to the simple RNA library building method, the principle that DNA ligase mutants K159L can be efficiently connected with DNA / RNA hybrid chains is utilized, only five steps of RNA fragmentation, reverse transcription, linker connection, library amplification and magnetic bead recovery are needed, the RNA library building process is greatly simplified, and consumed time is greatly shortened. In combination with a technology for rapidly removing rRNA by using an rRNA reverse transcription hindering probe, rRNA removal and RNA library establishment can be completed in one tube. The whole process only needs 2 hours, the operation is simple, the RNA loss is small, the cost is low, and the method is very suitable for automatic RNA library building and low-abundance RNA library building.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Probe composition for hindering Globin mRNA reverse transcription, and application of probe composition
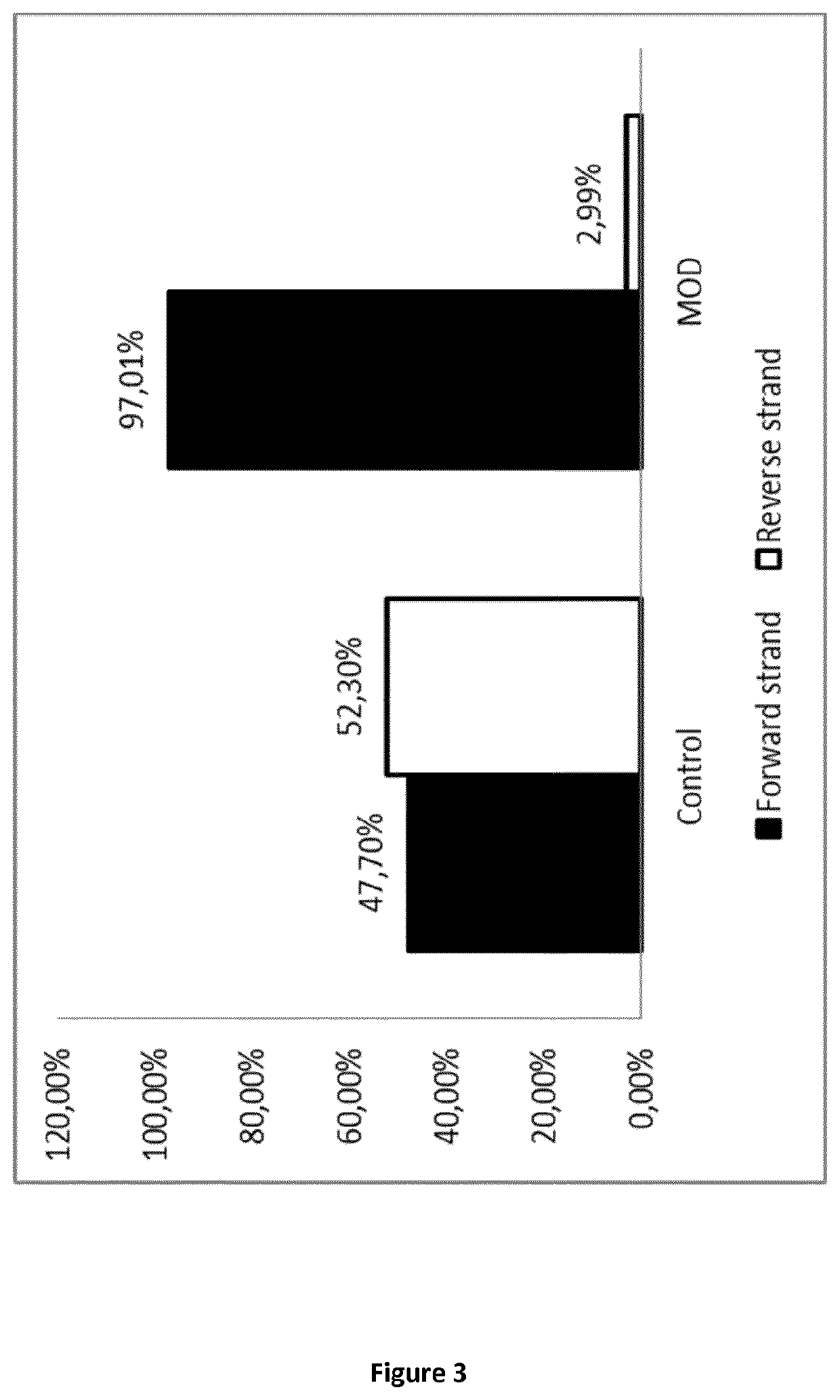
PendingCN113373201AEfficient removalEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationAlpha globulinGenetics
The invention provides a probe composition, which comprises 20 probes, for hindering Globin mRNA reverse transcription, and discloses application of the probe composition. According to the invention, reverse transcription of a target RNA is inhibited with high efficiency and high specificity during synthesis of one-chain cDNA by using a reverse transcription hindering probe method, so that the target RNA is hindered from participating in a downstream library building process. The probe can specifically recognize and bind to the human globulin RNA and hinder the reverse transcription of the globulin RNA so as to realize the purpose of removing the globulin RNA in the RNA library building process, 99.5% or above of Globin mRNA sequences in the blood sample RNA can be effectively removed in the reverse transcription process, and the probe has the advantages of being easy in operation (one step) and short in time consumption (2 min), and is very suitable for large-scale industrial automation.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
DNA display of folded RNA libraries enabling RNA-selex without reverse transcription
PendingUS20200002699A1Promote RNA displayMicrobiological testing/measurementTransferasesRNA libraryMolecular binding
The present invention is directed to the method for selecting an RNA molecule that binds to a target molecule and a kit for carrying out the method. This method includes: providing a pool of oligonucleotide complexes that each comprise a ds-DNA molecule and an RNA molecule, the ds-DNA molecule comprising a first DNA strand at least partially annealed to a first region of the RNA molecule, whereby a second region of the RNA molecule is free to adopt a secondary structure; exposing the pool to a target molecule and allowing the second region of the RNA to bind the target molecule; and selecting from the pool one or more oligonucleotide complexes comprising an RNA molecule having the second region bound to the target molecule.
Owner:BRANDEIS UNIV
Digital microfluidic library construction system
A chip is constructed through a full-flow full-automatic digital micro-fluidic library, full-flow library construction of a sample is included on a single chip, and the chip comprises a low-temperature area used for keeping effectiveness of a reagent in the waiting process; the system comprises: a temperature change zone which is used for providing temperature conditions required by each reaction in library construction; and liquid storage and distribution areas 2 which can be used for respectively distributing washing liquid, magnetic bead solution and eluent drops. A user only needs to load a sample to the chip to realize full-process DNA or RNA library construction.
Owner:张研
Probe for closing ribosome RNA or globulin RNA in RNA library building process and application thereof
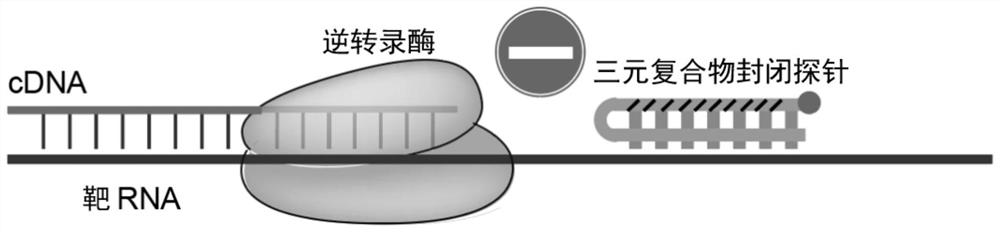
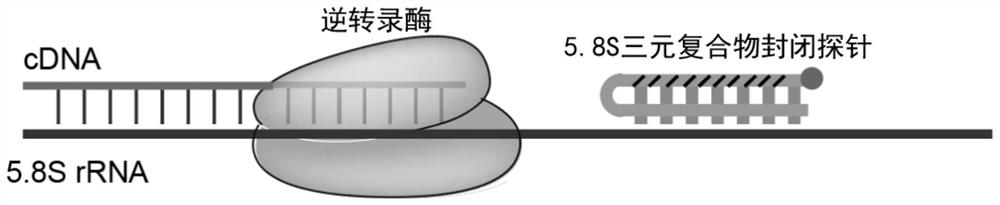
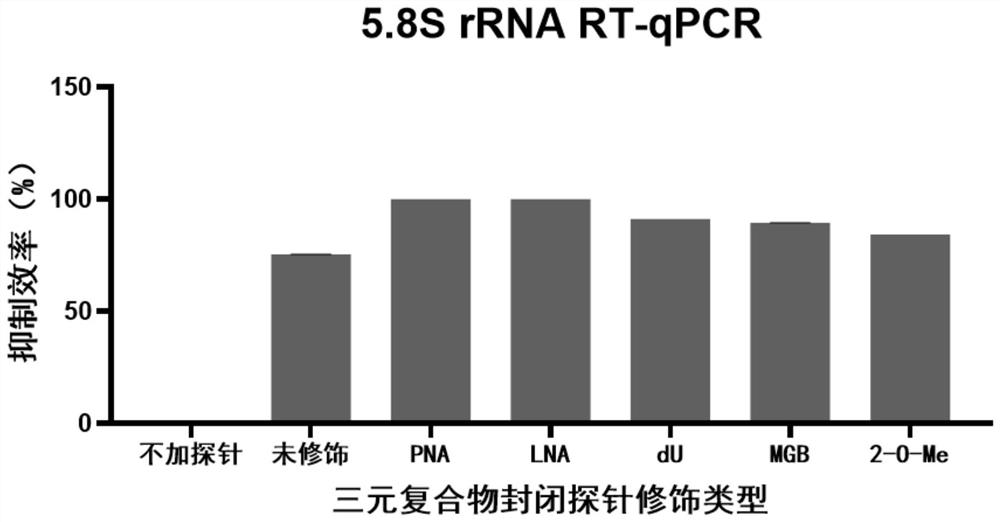
PendingCN113862334AImprove stabilityCombined with high efficiency and stabilityMicrobiological testing/measurementLibrary creationBase JTernary complex
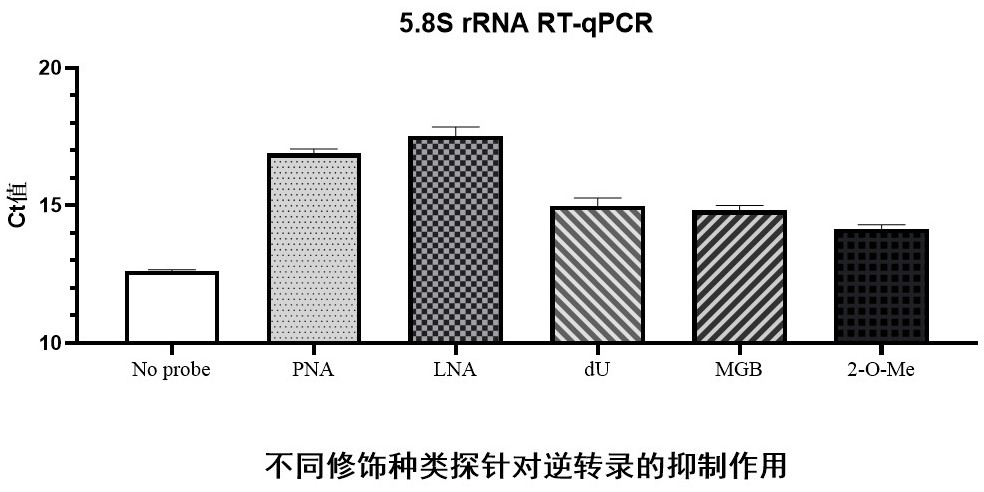
The invention provides a probe for blocking ribosome RNA or globulin RNA in the RNA library building process, the length of the probe is 35-60 nt, and the probe and the ribosome RNA or globulin RNA form a ternary blocking compound through complementary pairing. The rear half region of the probe is in strict complementary pairing with the target RNA, and 5-9 bases at the 5' end of the probe pairing region are replaced by modified bases; and the front half region of the probe is a pyrimidine base which is used for being embedded into a large groove of a DNA / RNA double strand to form a ternary complex. The 3'-OH of the probe is sealed by MGB for preventing extension of the probe as a primer; and meanwhile, the MGB can be embedded into a small groove of DNA / RNA double strands, so that the stability of the ternary complex is improved. The invention also discloses an application of the gene in rapid removal of ribosome RNA or globulin RNA in RNA library construction. The probe has the advantages of simplicity in operation, extremely short time consumption, high specificity, wide applicability, high removal efficiency, high gene detection number and the like, and is suitable for the fields of industrial automatic library building and rapid disease diagnosis.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Application of tag protein covalently bonded to substrate in CLIP
ActiveCN107828876ASimple and fast operationShorten the timeMicrobiological testing/measurementBiological testingCross-linkMagnetic bead
The invention discloses an application of a tag protein covalently bonded to a substrate in CLIP (cross-linking immunoprecipitation), and provides an application of a tag fusion protein covalently bonded to the substrate in a CLIP technology in a denaturant purifying manner. An RNA library used for sequencing is obtained by using the tag fusion protein covalently bonded to the substrate in a protein denaturant washing manner, and a result proves that an experiment result published in previous literatures can be well repeated. Steps of membrane transfer and gel extraction for recovering fragments in traditional CLIP processes are omitted. Labels covalently bonded to the substrate and a specific binder immobilized on a medium form covalent bonds, so strong washing of magnetic beads, which cannot be directly used previously, is added to the experiment operating steps, thereby the operation is simple, the operating time is saved, the CLIP process is optimized, and the loss possibly causedin the operating process is reduced.
Owner:INSITUTE OF BIOPHYSICS CHINESE ACADEMY OF SCIENCES
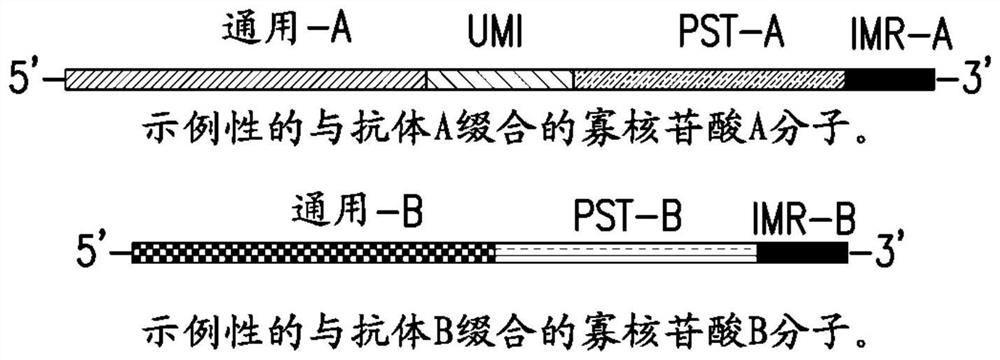
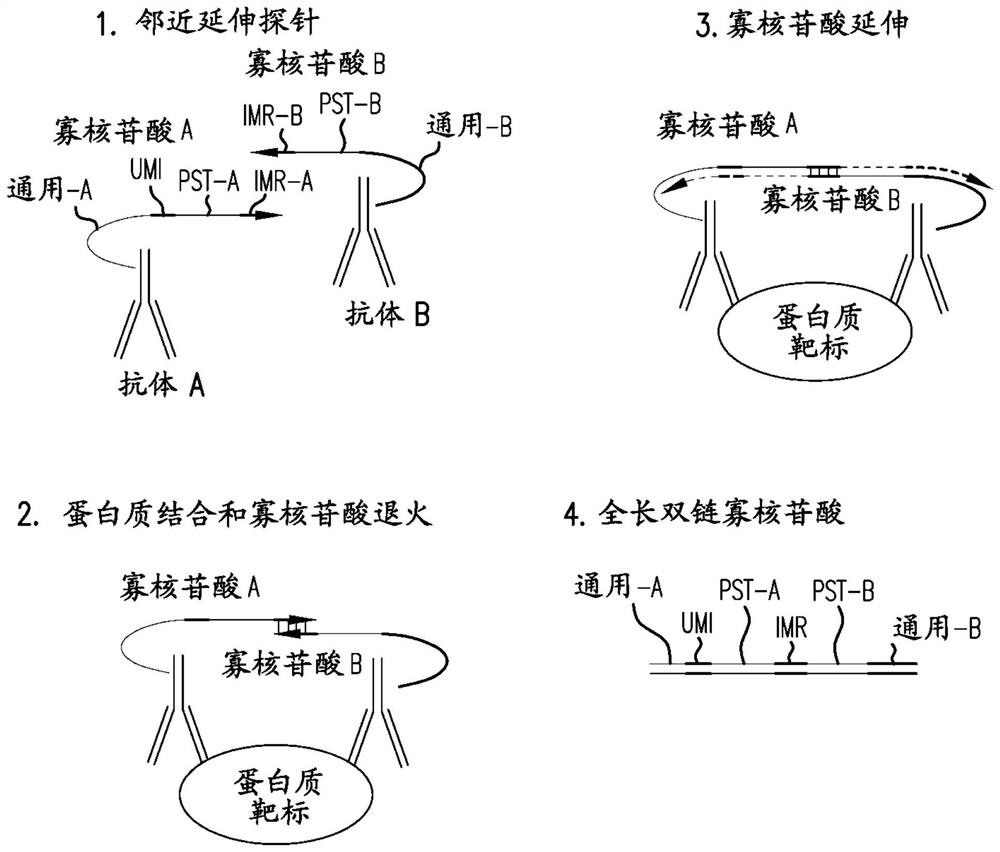
Methods of detecting analytes and compositions thereof
The invention relates to methods of detecting analytes in samples by generating analyte-based DNA libraries amenable for sequencing. The methods include the use of proximity probe pairs, each probe comprising an analyte binding domain and oligonucleotide domain. The methods further provide for integrated DNA and RNA library preparations and methods of making and uses thereof. The invention also provides compositions useful in the methods.
Owner:凯杰科学有限责任公司
Method for generating a stranded RNA library
ActiveUS11015192B2Microbiological testing/measurementLibrary creationReverse transcriptaseNucleotide
The invention relates to a method for preparing a strand-specific library from an nucleic acid or preferably RNA sample, for RNA comprising the steps of: (i) optionally fragmenting said RNA sample, (ii) generating a plurality of first cDNA strands by subjecting said fragmented RNA to reverse transcription by using a reverse transcriptase and first oligonucleotide primers, (iii) generating a plurality of second cDNA strands by using a DNA polymerase, second oligonucleotide primers, and the plurality of first cDNA strands, and (iv) ligating adapters to the 3′ and 5′ termini of the of double-stranded cDNA, (v) wherein the first cDNA strand allows no adapter ligation at its 5′ terminus and said second cDNA strand allows adapter ligation at its 5′ terminus, or vice versa, and, (v) optionally cloning, sequencing or otherwise using the strand-specific library. The invention also relates to a kit for preparing a strand-specific library from an RNA sample, wherein said kit comprises, (i) random oligonucleotide primers comprising a 5′ terminus nucleotide which allows no adaptor ligation, (ii) random oligonucleotide primers comprising a 5′ terminus nucleotide which allows adaptor ligation, (iii) optionally a reverse transcriptase for synthesizing a first cDNA strand complementary to the template RNA, (iv) optionally dNTPs, and (v) optionally a DNA polymerase.
Owner:QIAGEN GMBH
Ribosome print sequencing library construction method
PendingCN112899345ALow costSimplify the experimental steps of library constructionMicrobiological testing/measurementLibrary creationRNA libraryEngineering
The invention provides a ribosome print sequencing library construction method. The library construction method provided by the invention comprises the following steps: (1) extracting RFP-RNA from a sample; (2) repairing structures of the two ends of the RFP-RNA; and (3) subjecting normal-structure RNA to library construction by employing a small RNA library construction kit. According to the method, the RFP-RNA can restore to a normal RNA structure through repairing the two ends of the RFP-RNA, the normal RNA structure can be in seamless engagement with the small RNA kit on the market for direct library construction, rRNA is not required to be removed, a complicated RFP-RNA library construction method is not required to be employed, experimental steps for library construction are simplified, the material cost is reduced, and the experimental stability is improved; and the experimental time is shortened by 4-5 days, and the time cost is reduced by 50%. The library construction method provided by the invention is applicable to all classes of species; and obtained sequencing data are high in quality and are consistent with a theoretical level.
Owner:JINAN UNIVERSITY
A Noise Reduction Method for Targeted Capture Enrichment of Sequencing Library Molecules with Small Panel Probes
The invention belongs to the field of biomedicine, and relates to a noise reduction method for targeted capture and enrichment of sequencing library molecules with small panel probes. The noise reduction method includes: acquiring library molecules, adding washing buffer to the mixture containing library, probe and streptavidin magnetic beads, removing non-specific hybridization by physical shaking, and removing the hybridization after decontamination. The product is moved to a new container and the rinsed library molecules are obtained. The noise reduction method for the probe of the present invention to target capture and enrich the sequencing library molecules, optimize the capture efficiency, reduce non-specific hybridization, improve the target rate, and can effectively reduce the rRNA ratio for the RNA library, and the present invention provides high-quality sequencing. library.
Owner:SHANGHAI ORIGIMED CO LTD +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com