A Noise Reduction Method for Targeted Capture Enrichment of Sequencing Library Molecules with Small Panel Probes
A targeted capture and sequencing library technology, applied in the field of biomedicine, can solve the problems of difficult to meet the requirements of sequencing depth and waste of sequencing cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
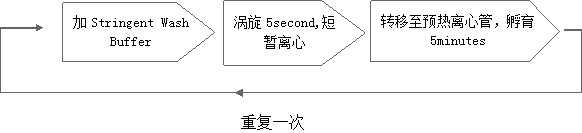
[0071] Aiming at the problems in the current library preparation, the present invention uses the probe to hybridize with the library molecules, and after the streptavidin magnetic beads capture the library molecules in the target region, the preheated Stringent Wash Buffer is added to the streptavidin during the heating and washing process. and magnetic beads, and vortex for about 5 seconds, then transfer the mixture to a pre-warmed clean low DNA binding centrifuge tube and incubate for 5 minutes. At the end of the incubation, vortex the magnetic beads for about 5 seconds, centrifuge briefly, place on the magnetic stand, aspirate the supernatant, add preheated Stringent Wash Buffer again, and vortex for about 5 seconds, then transfer the mixture to the preheated clean Incubate the centrifuge tube for 5 minutes. Subsequent steps are consistent with the existing process. The heating and washing process is an important link in the library capture process, which directly determin...
Embodiment 2
[0073] Use IDT’s kit to construct a DNA library, wash with heat, and make sure that 1× Wash Buffer 1 is always preheated in a 70°C water bath. After the 30 min incubation of the sample mixture containing the library, probes, and streptavidin magnetic beads, remove the sample from the thermostat. Transfer 80 μL of pre-warmed 1× Wash Buffer 1 to the sample, and mix by pipetting 10 times. Transfer all the above Mix (sample mixture) to new 1.5mL LoBind Tubes, vortex for 5sec, and let the sample stand still. Discard the supernatant after the solution is completely clear. Remove the sample from the magnetic stand, transfer 100μL of preheated 1×Stringent Wash Buffer to the sample, vortex for 8sec, avoid excessive air bubbles, and transfer to new 1.5mL LoBind Tubes. After incubating the new LoBind Tubes on the metal bath for 5 minutes, vortex for 3 seconds, put the sample on the magnetic stand, and discard the supernatant after the solution is completely clear. Repeat the steps of a...
Embodiment 3
[0077] To construct an RNA library, the steps are basically the same as in Example 2. During heating and washing, make sure that 1.5mL LoBindTubes and 1×Stringent Wash Buffer are always preheated in a 67°C metal bath.
[0078] 3.1 After the sample mixture containing library, probe and streptavidin magnetic beads has been incubated for 45 minutes, take the sample out of the PCR machine.
[0079] 3.2 Transfer 100 μL of preheated 1× wash buffer 1 (metal bath) to the sample, and mix by pipetting 10 times to avoid excessive air bubbles.
[0080] 3.3 Transfer all the above sample mixture (Mix) to new 1.5mL LoBind Tubes, vortex for 5sec, place the sample on the magnetic stand, and discard the supernatant after the solution is completely clear.
[0081]3.4 Remove the sample from the magnetic stand, transfer 150μL of preheated 1×Stringent Wash Buffer to the sample, vortex for 5sec to avoid excessive air bubbles, and transfer to new 1.5mL LoBind Tubes.
[0082] 3.5 After incubating on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com