Preparation method of target sequence random sgRNA full-coverage group
A full-coverage, target-sequence technology, applied in DNA/RNA fragments, recombinant DNA technology, combinatorial chemistry, etc., to achieve the effect of less bias, simple production, and uniform coverage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Design of sgRNA backbone and flow of RPTS
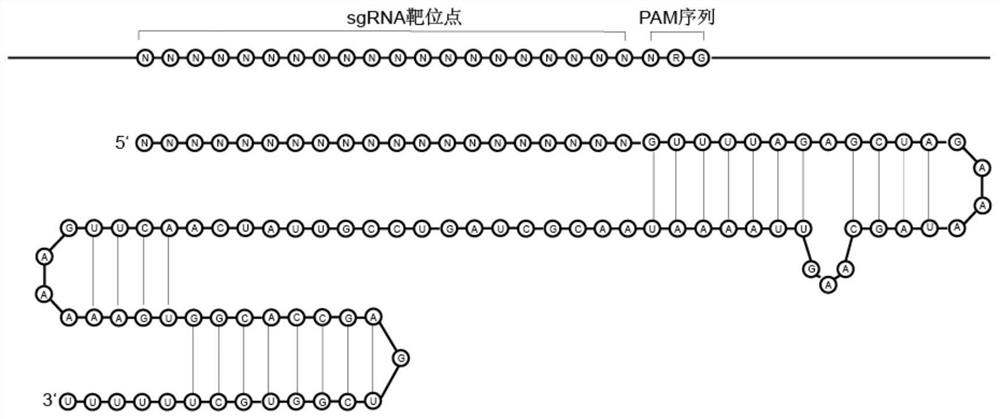
[0055] In this example, the backbone of the traditional sgRNA is modified, and the enzyme cutting site MmeI or EcoP15I is incorporated, but the structure of the sgRNA and the binding of Cas9 are not changed. See the modified sgRNA sequence and structure Figure 1-Figure 3 shown.
[0056] The process of testing RPTS, the process is as follows Figure 4 Shown:
[0057] (1) Restriction enzyme cleavage that recognizes the PAM sequence:
[0058] Table 2 restriction enzyme digestion system
[0059] components Dosage PUC19 plasmamid DNA 1μg 10×rCutSmart buffer 8μL ScrFI (NEB) 5U MspI (NEB) 5U HpaII (NEB) 5U BstNI (NEB) 5U BfaI (NEB) 5U DdeI (NEB) 5U Hydrate to 80 μL
[0060] React overnight at 37°C. After the reaction, 160 μL of Ampure DNA beads (Beckman) was added to recover the fragmented DNA. 45 μL water for elution.
[0061] Table 3 end cu...
Embodiment 2
[0092] Example 2: RPTS preparation of 18S rRNA or 28S rRNA.
specific Embodiment approach
[0093] In this example, we used RPTS to prepare a random sgRNA library covering 18S rRNA or 28S rRNA. The specific implementation is as follows:
[0094] (1) Acquisition of DNA fragments
[0095] Table 10 reverse transcription system
[0096] components Dosage 293 cellular RNA 1μg 10μM reverse transcription primer 18S-R or 28S-R 1μL 10mM dNTPs 1 Reaction at 75°C for 5 minutes 5×FS Buffer 4μL 0.1M DTT 1μL SuperScript IV (Thermo) 2μL total capacity 20 μL
[0097] 15 minutes at 42°C, 15 minutes at 50°C, 15 minutes at 55°C, 15 minutes at 50°C, 15 minutes at 55°C, and 15 minutes at 70°C.
[0098] Table 11 PCR amplification system
[0099] components Dosage The above reverse transcription product 1μL 10μM 18S-F / R or 28S-F / R 5μL Phusion High-Fidelity PCR Master Mix 25 μL Add water to 50μL
[0100] After denaturation at 98°C for 3 min, library cycles were amplified by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com