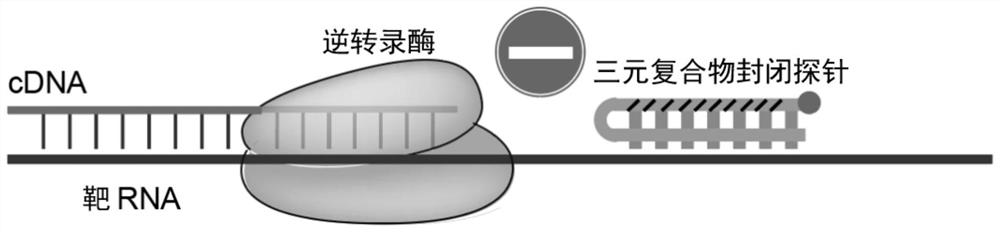
Probe for closing ribosome RNA or globulin RNA in RNA library building process and application thereof
A ribosome and globulin technology, applied in the field of probes for blocking ribosomal RNA or globulin RNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Inhibitory effect of different modification types of probes on target RNA reverse transcription
specific Embodiment approach
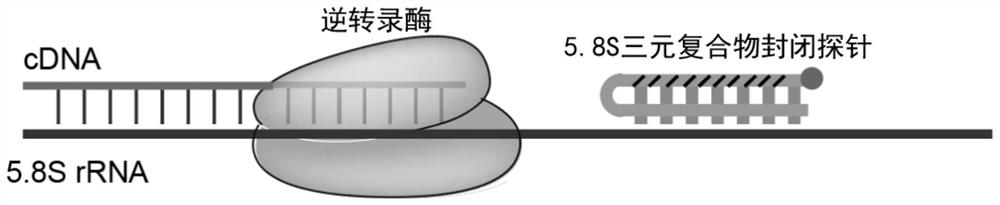
[0065] refer to figure 2 In this example, 5.8S-specific probes with different modifications (No. 1-5 in Table 1) were designed to inhibit the reverse transcription process of 5.8SrRNA. The specific implementation is as follows:
[0066] Table 2
[0067] components Dosage Total RNA 1μg 1 μM 5.8S modified probe (any one of No. 1-5 in Table 1) 1μL 1μM 5.8S rRNA RT primer 1μL 10mM dNTPs mix (with 0.5M KCl) 1μL Supplement DEPC H2O to 13μL
[0068] Mix and spin away. React at 75°C for 1 min and store at 4°C.
[0069] table 3
[0070]
[0071] Mix and spin away. React at 25°C for 10 minutes, at 42°C for 15 minutes, at 70°C for 5 minutes, and store at 4°C;
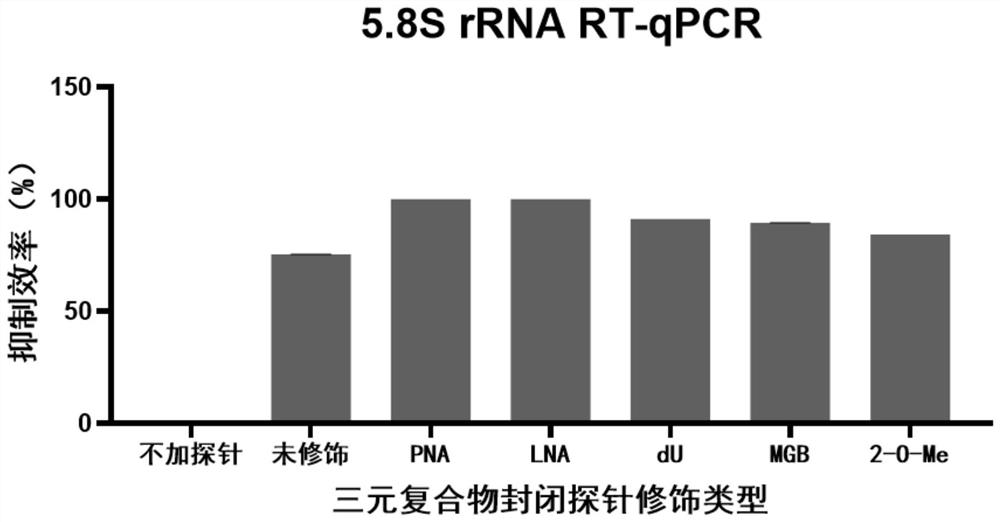
[0072] After diluting 1000 times, Hieff qPCR SYBR Green Master Mix (11200ES03) was used to quantify 5.8 ScDNA, and the quantitative primers are listed in Table 1. Quantitative results see image 3 .
[0073] From the quantitative results, it can be seen that peptide...
Embodiment 2
[0076] Example 2: The inhibitory effect of the distribution position of PNA in the probe on the reverse transcription of target RNA
[0077] In this example, 5.8S-specific probes (a mixture of sequences No. 6-8 in Table 1) were designed at different PNA modification positions to inhibit the reverse transcription process of 5.8S rRNA. The specific embodiment is with reference to embodiment 1, quantitative result sees Figure 4 .
[0078] It can be seen from the quantitative results that when the position of the PNA in the probe is close to the left end (ie, the 5' end) of the pairing region between the probe and the target RNA, the effect of hindering the reverse transcription of the target RNA is higher than that when the position of the PNA in the probe is close to the probe. The right end of the needle (i.e. the 3' end).
[0079] Table 5: Inhibition efficiency of target RNA reverse transcription by LNA distribution position in the probe
[0080] Distribution of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com