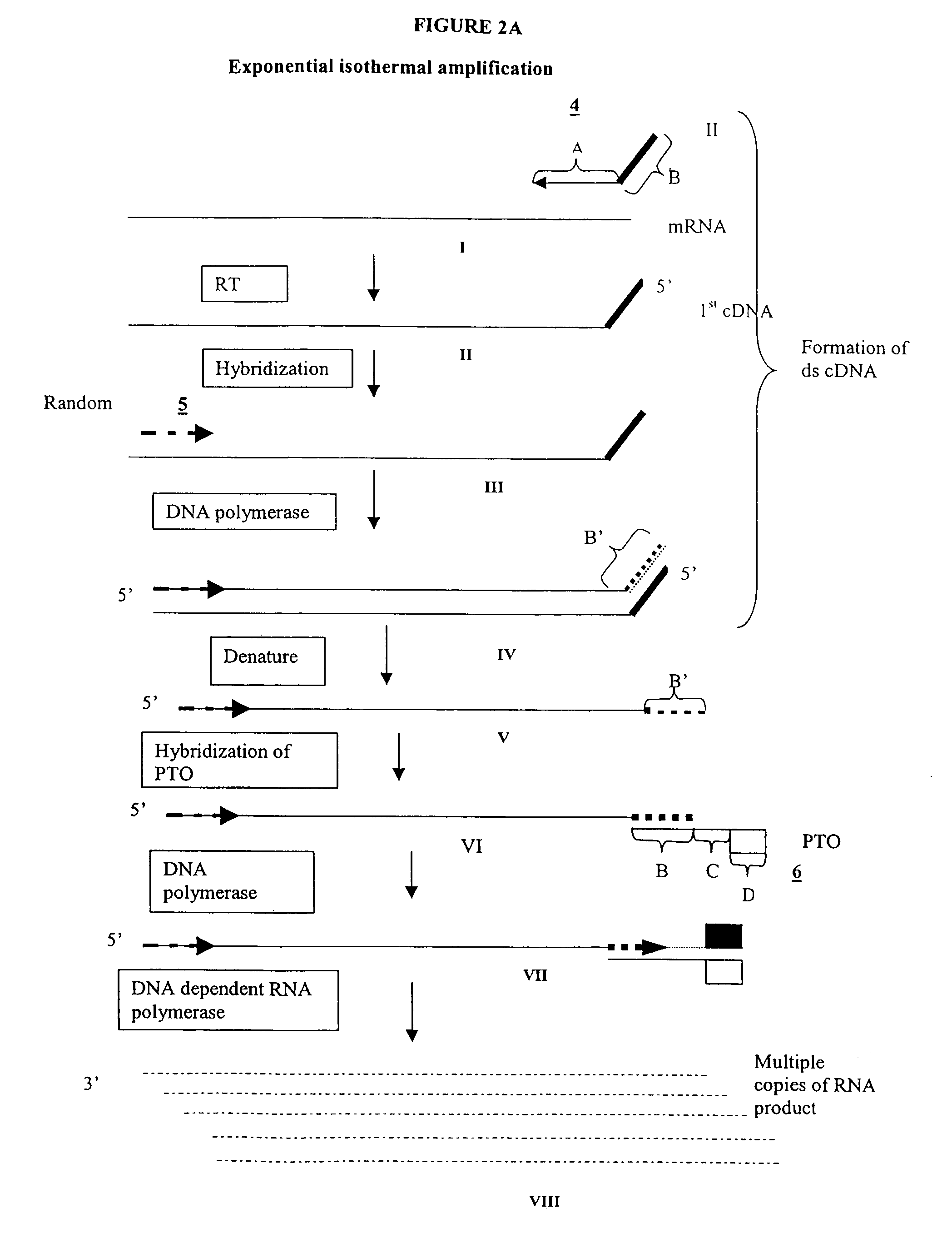
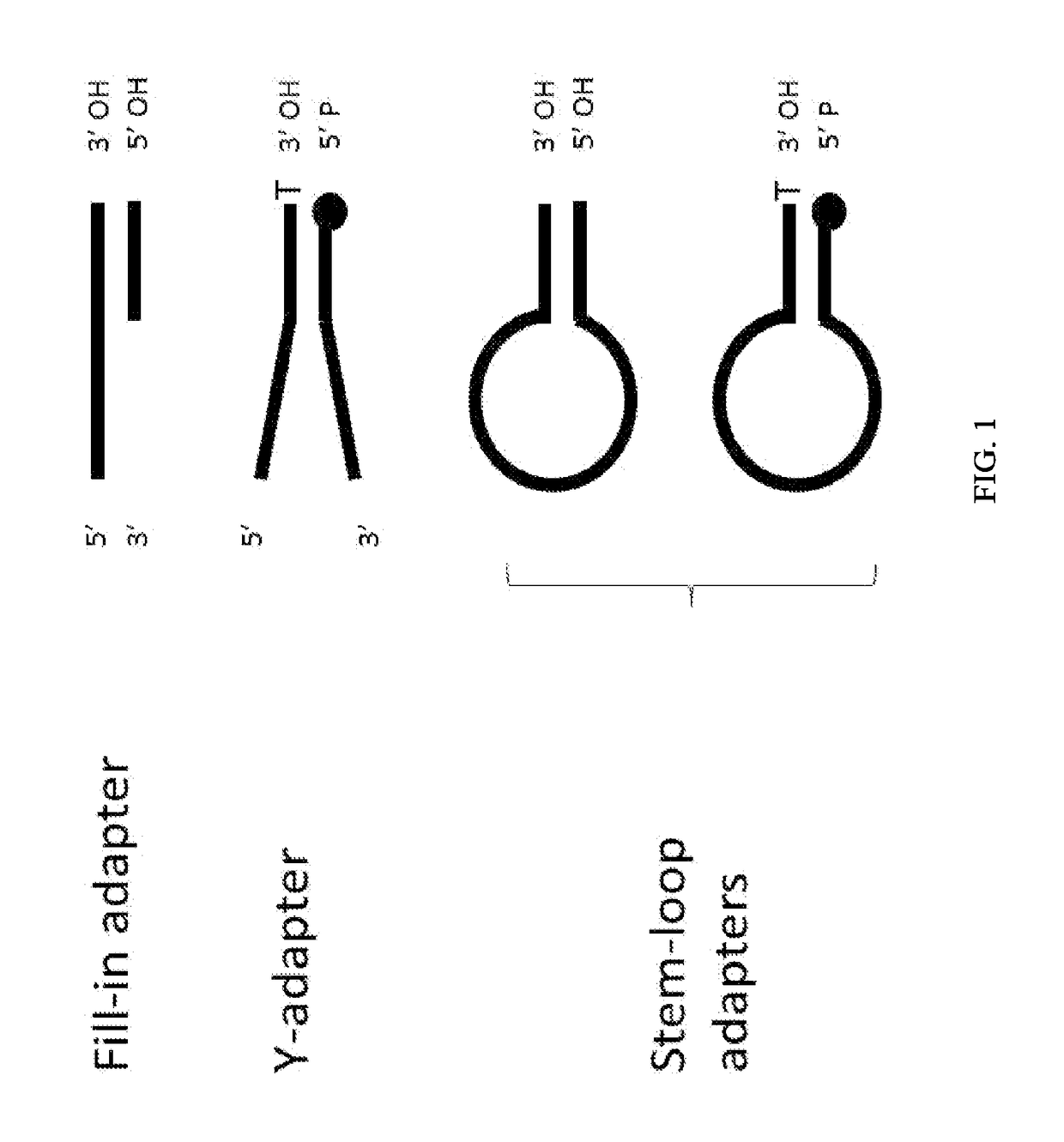
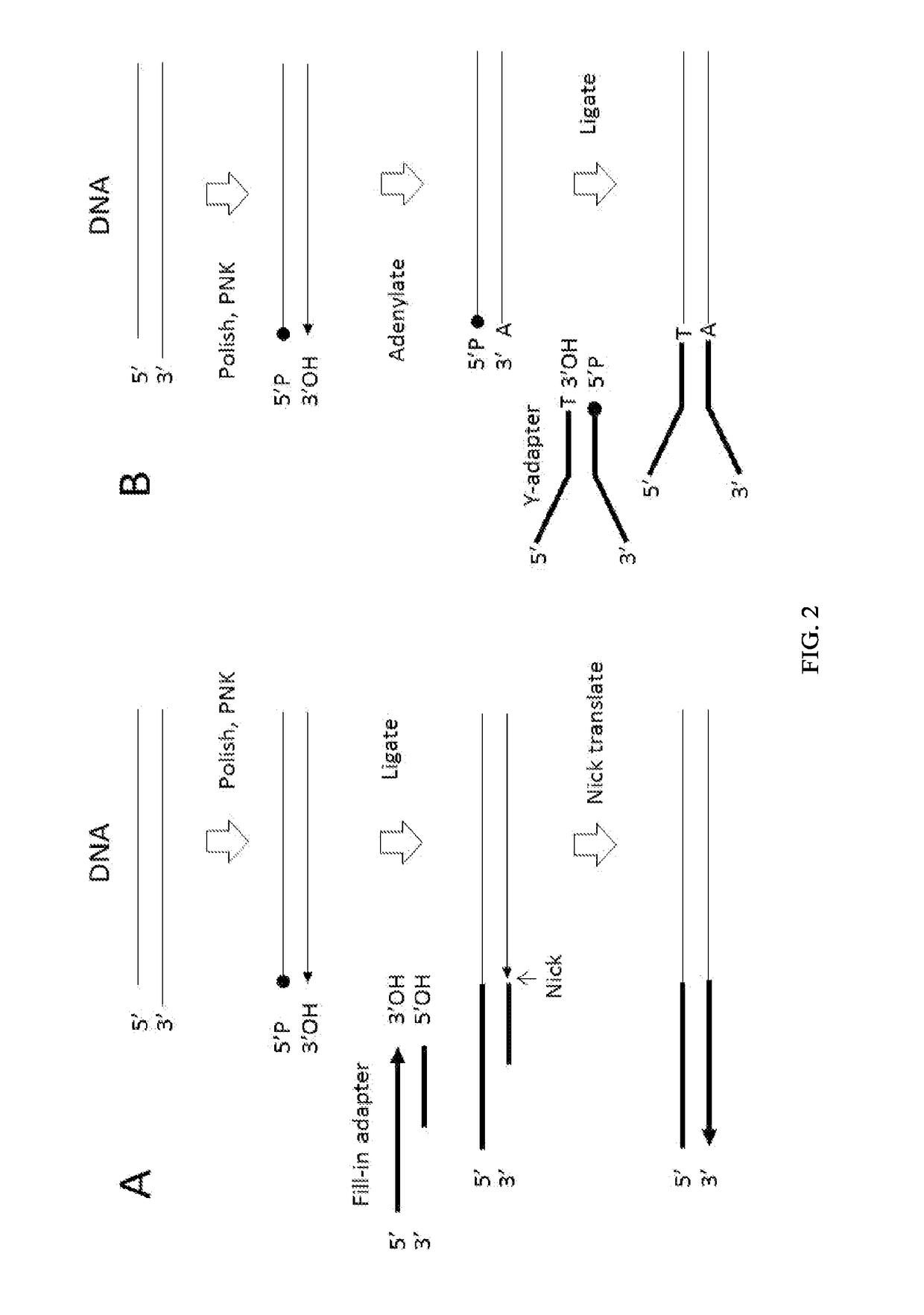
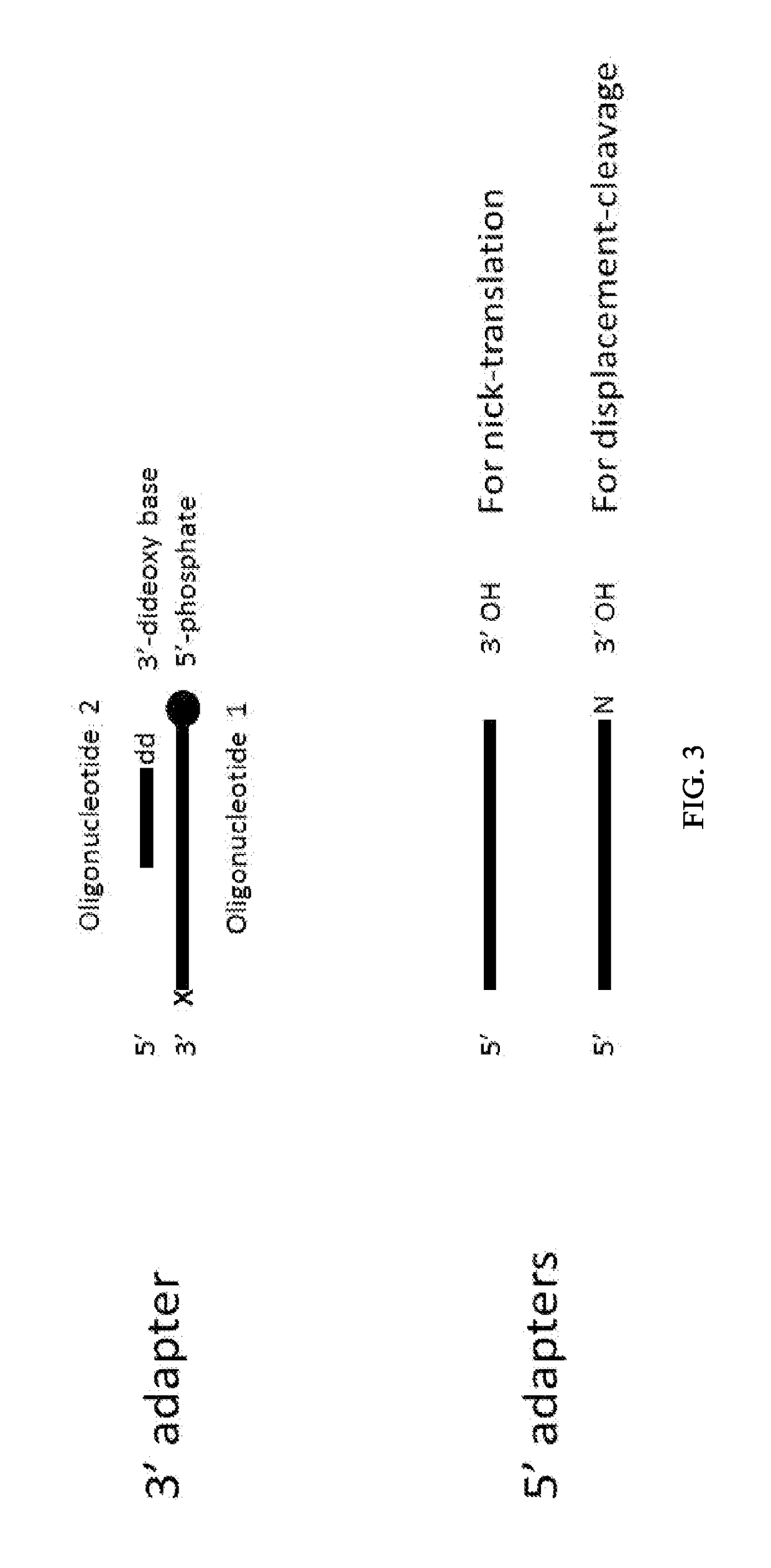
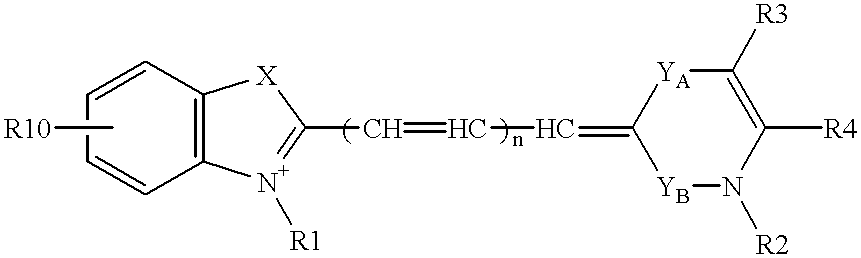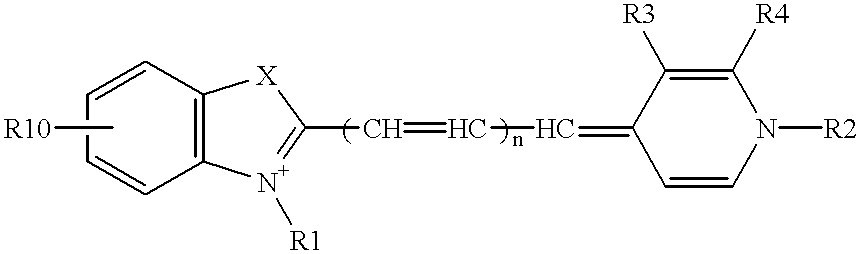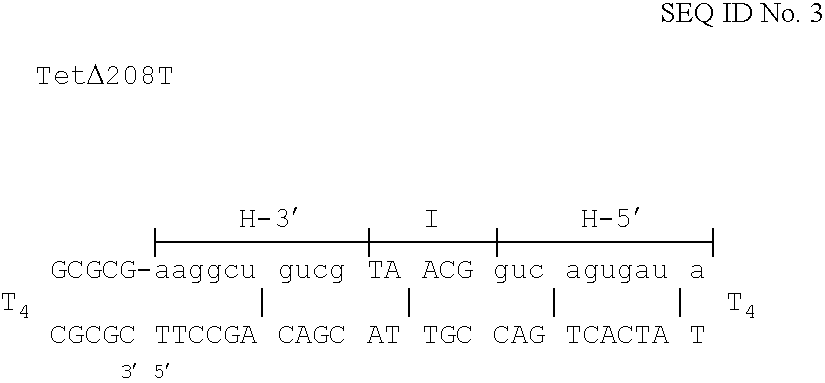Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
904 results about "Double strand dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
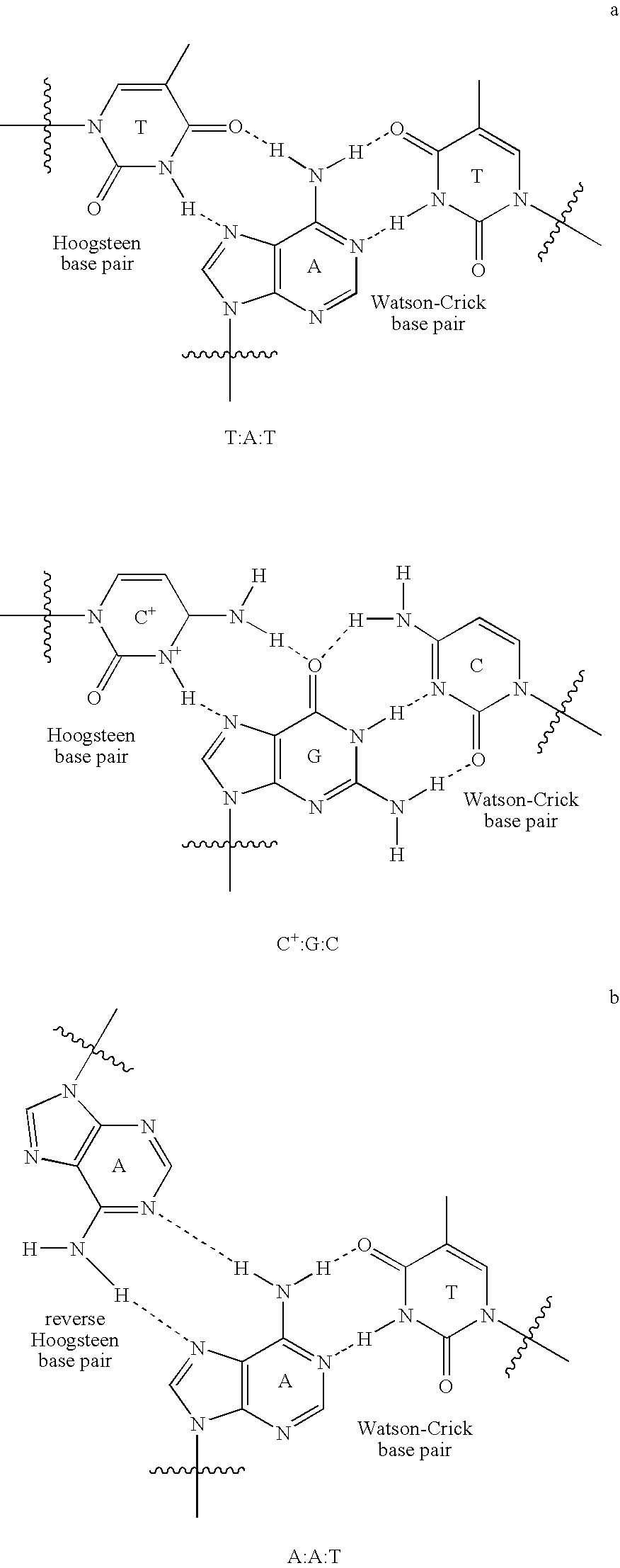
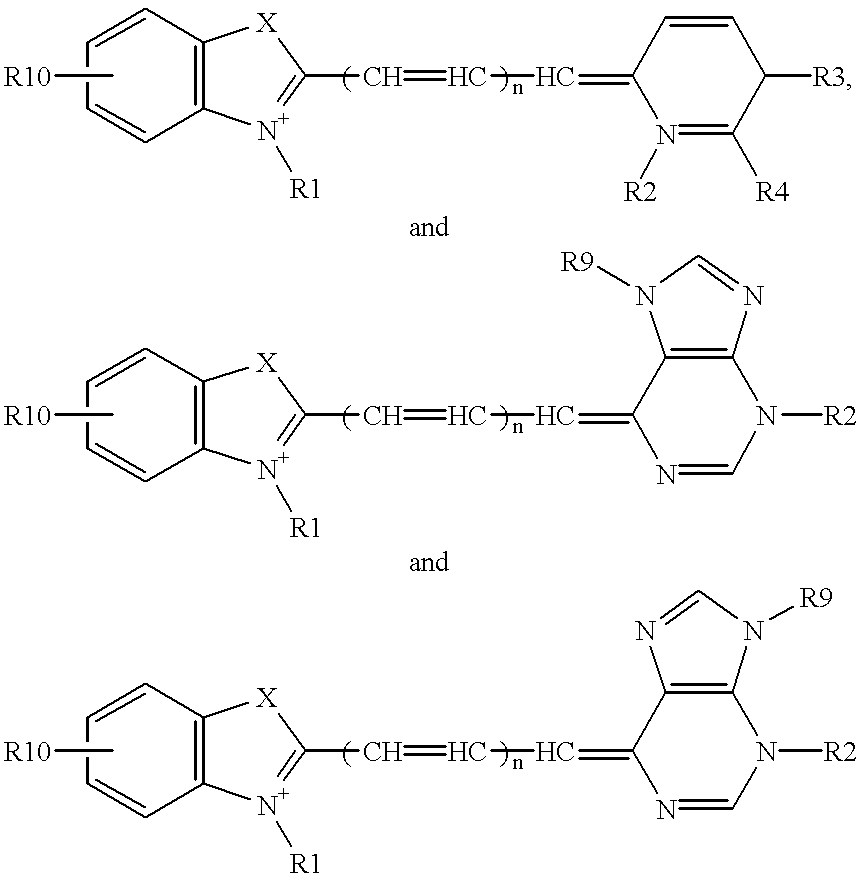
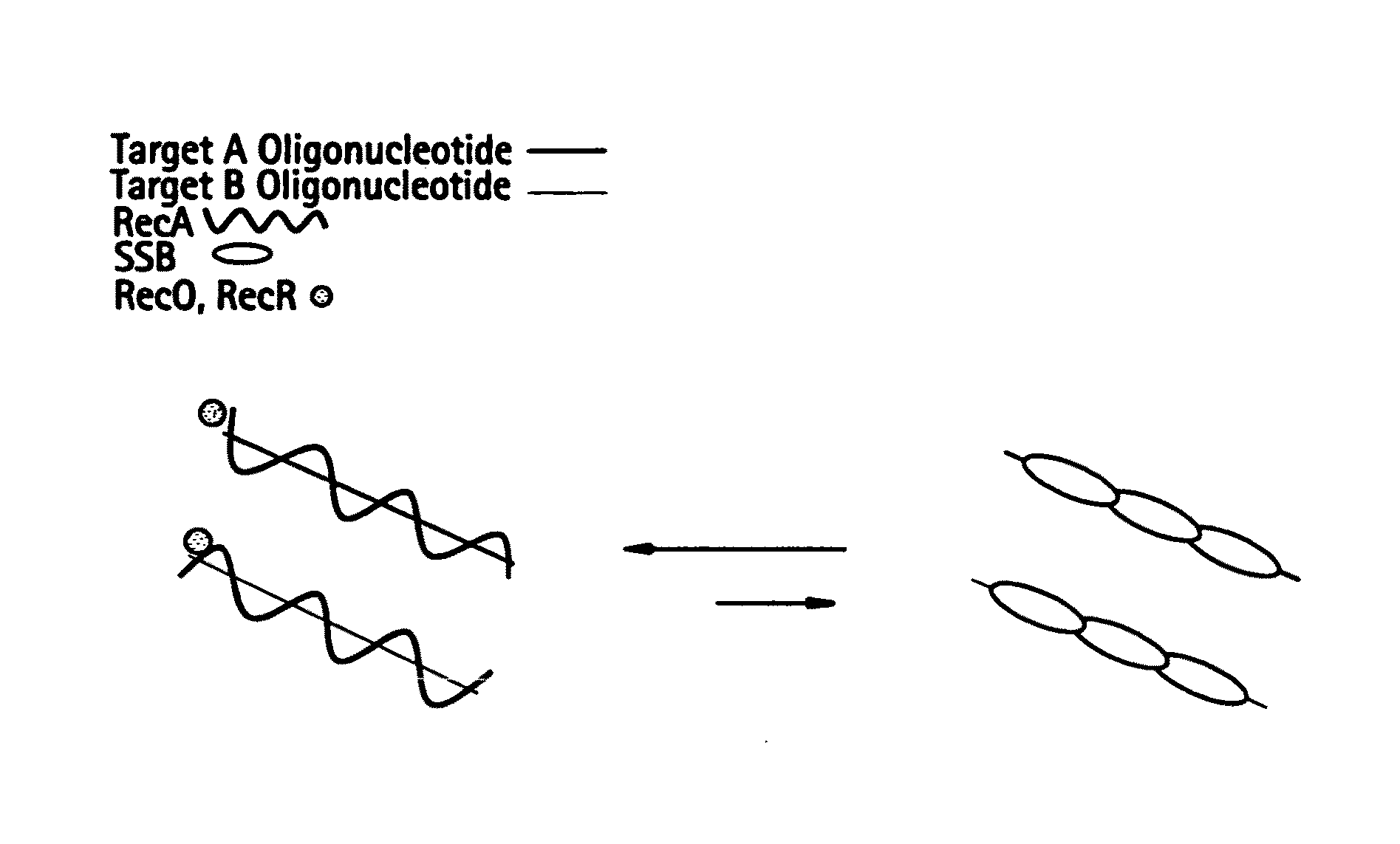
Double-stranded DNA (dsDNA), a molecule of DNA consisting of two parallel strands joined by hydrogen bonds between complementary purines and pyrimidines; a double helix, the form in which DNA occurs in chromosomes.
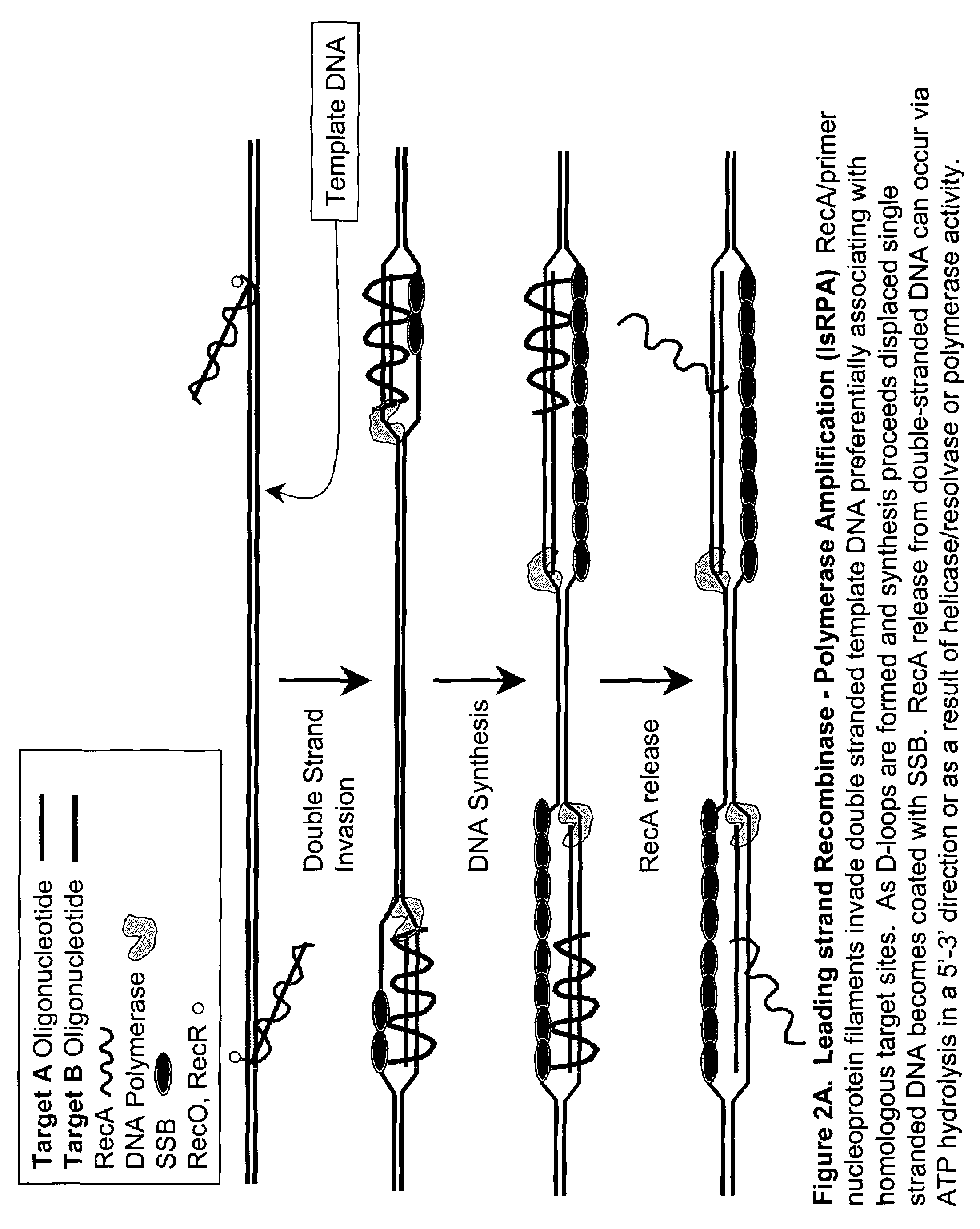
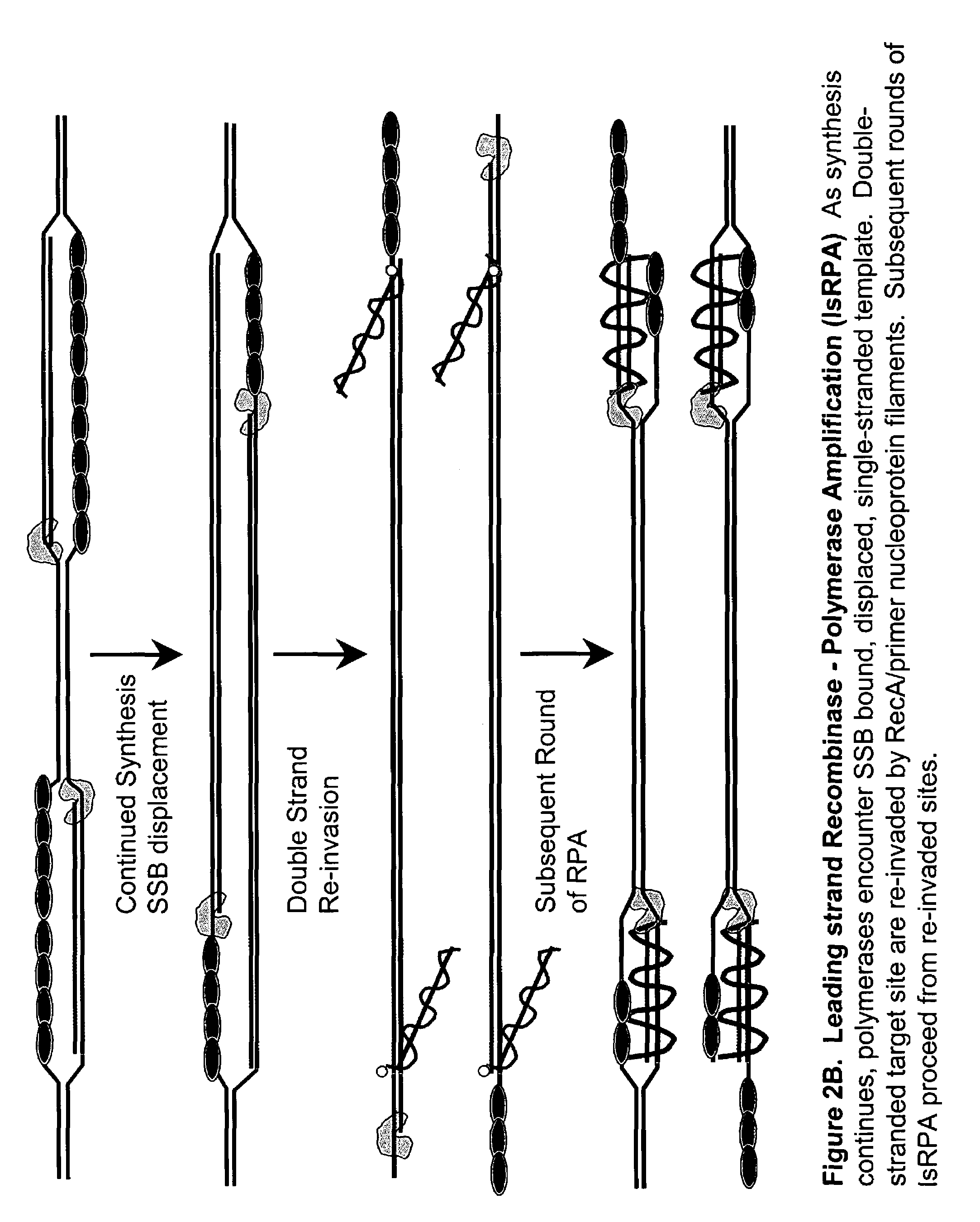
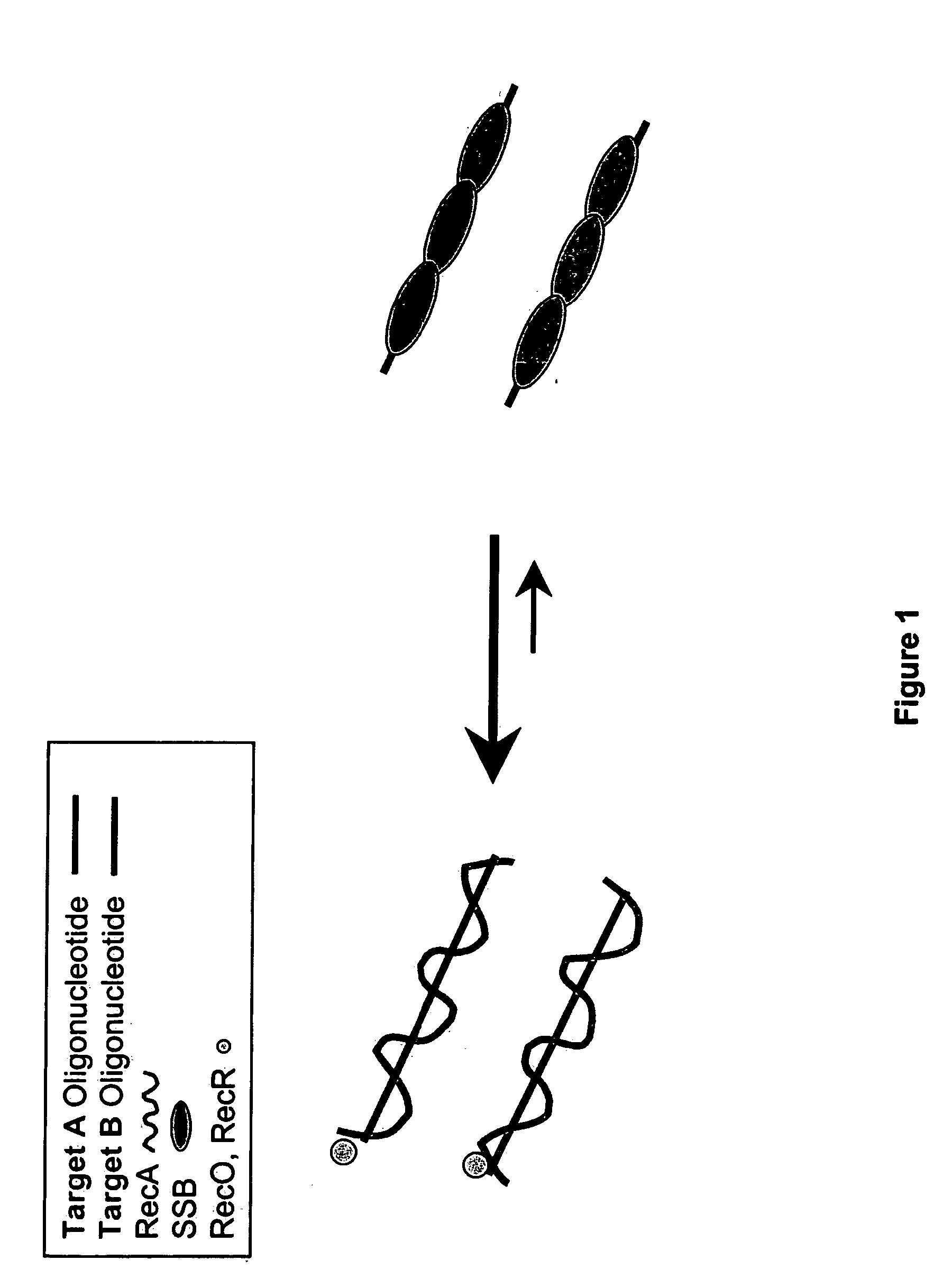
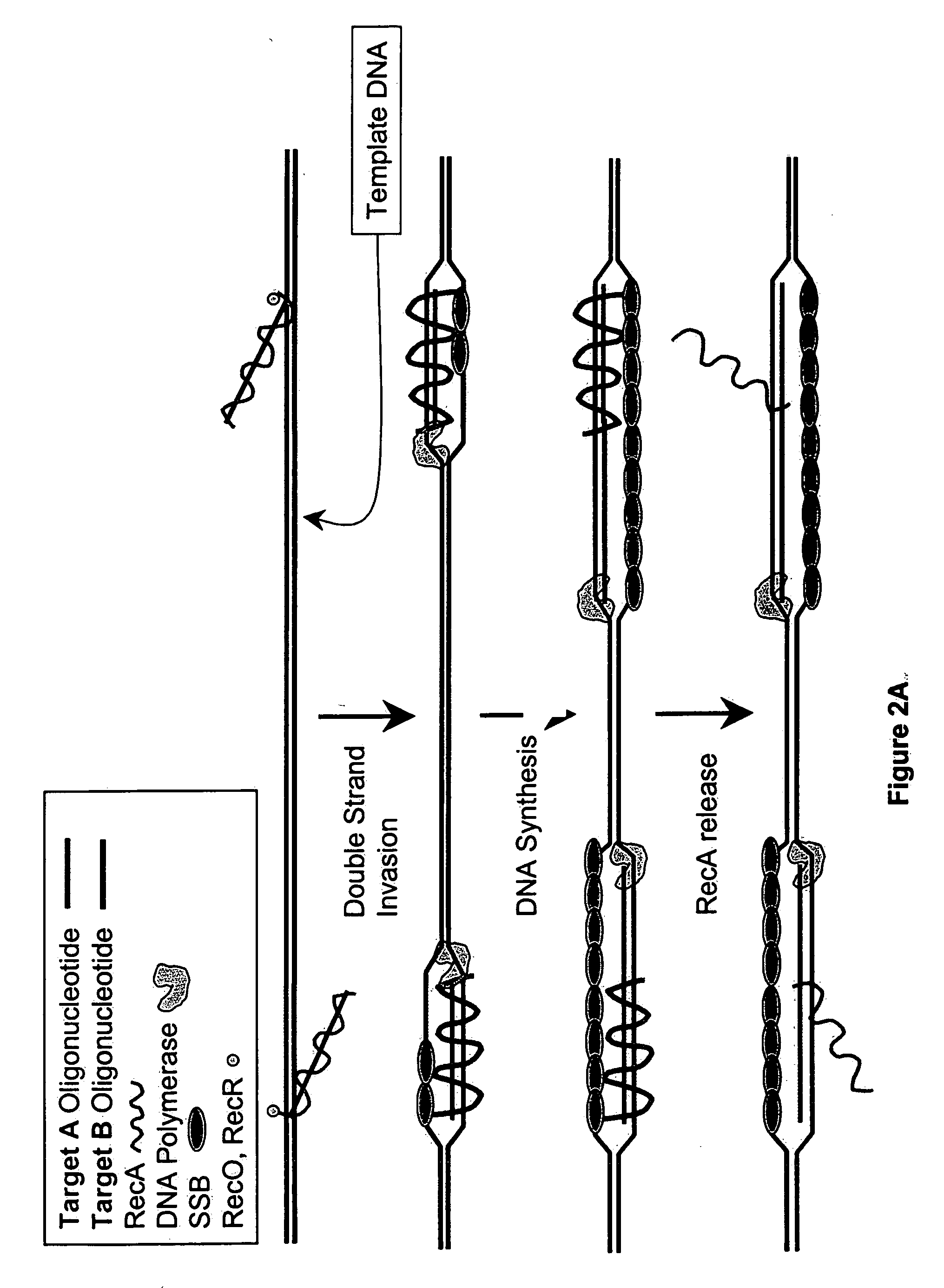
Recombinase polymerase amplification
ActiveUS7399590B2HydrolasesMicrobiological testing/measurementRecombinase Polymerase AmplificationSingle strand
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Recombinase polymerase amplification
ActiveUS7270981B2Web data indexingMicrobiological testing/measurementSingle strandRecombinase Polymerase Amplification
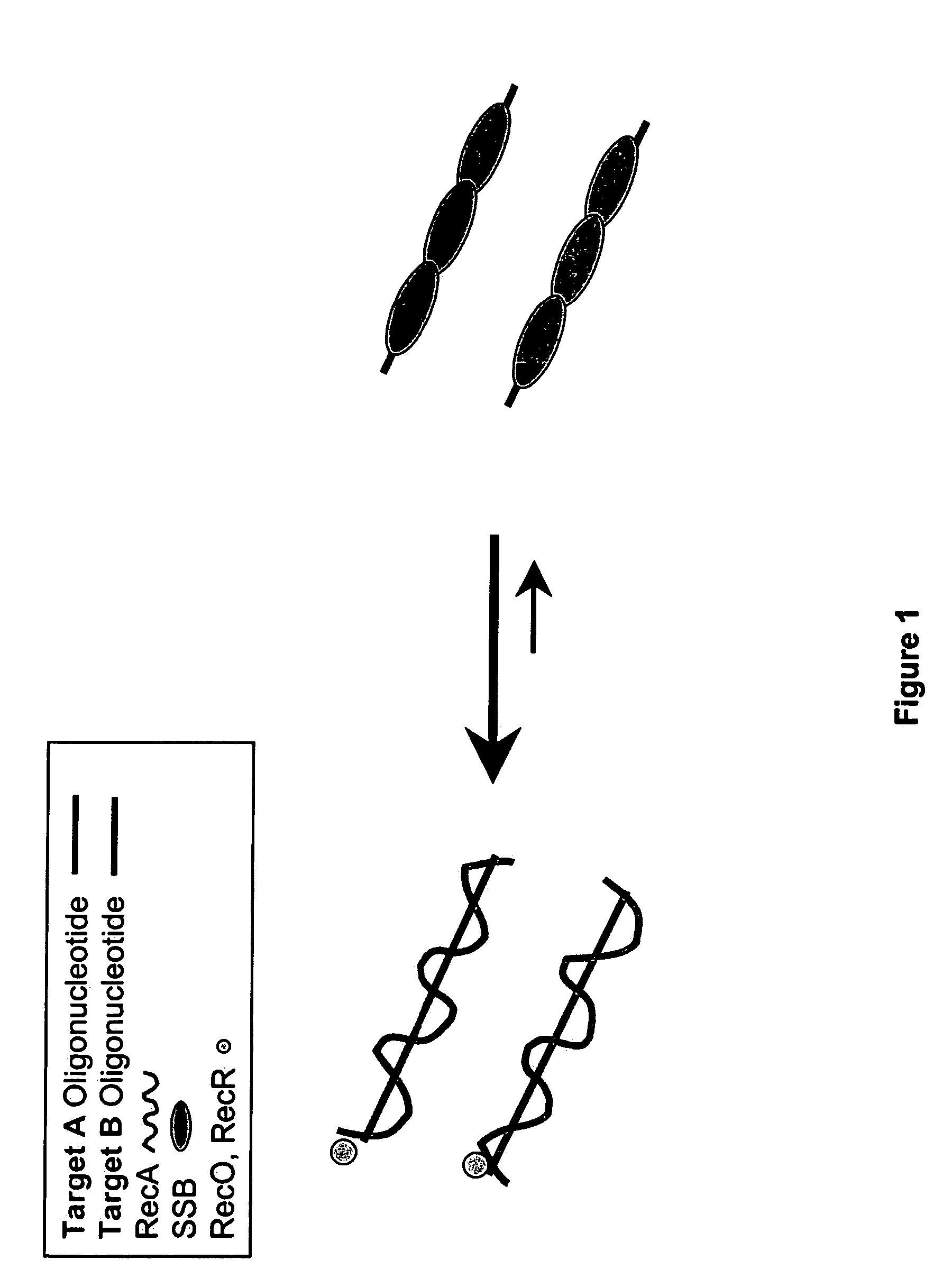
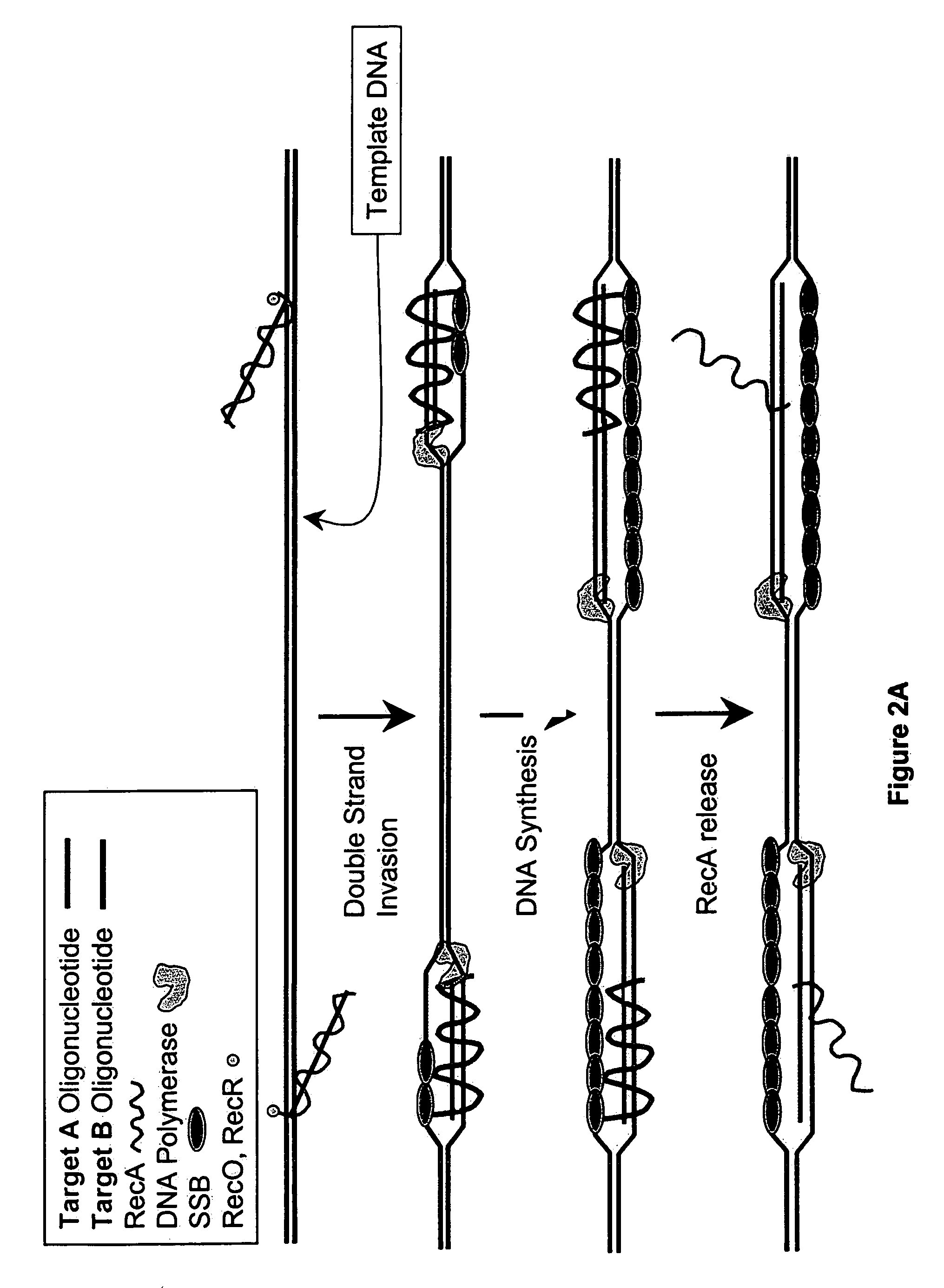
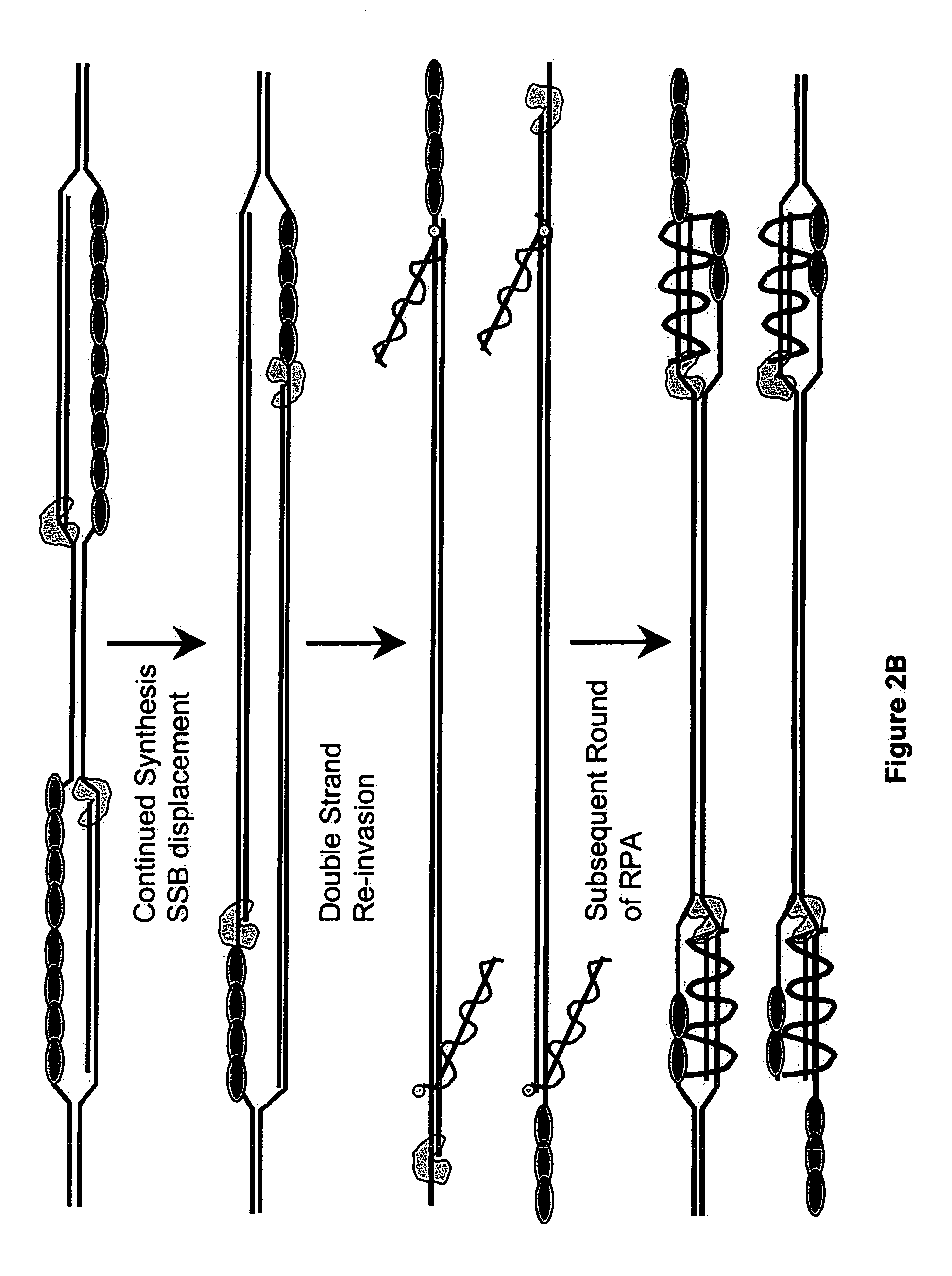
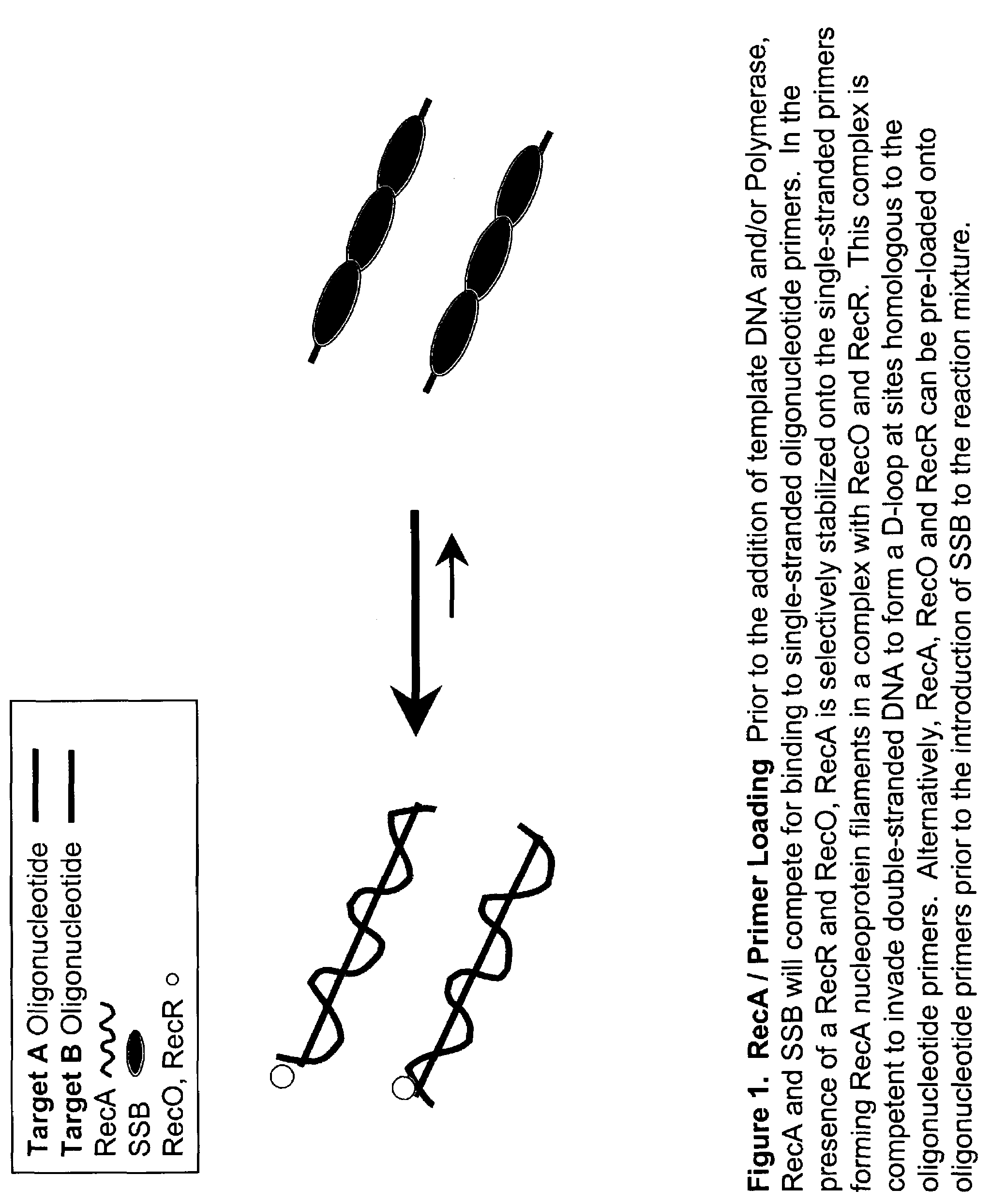
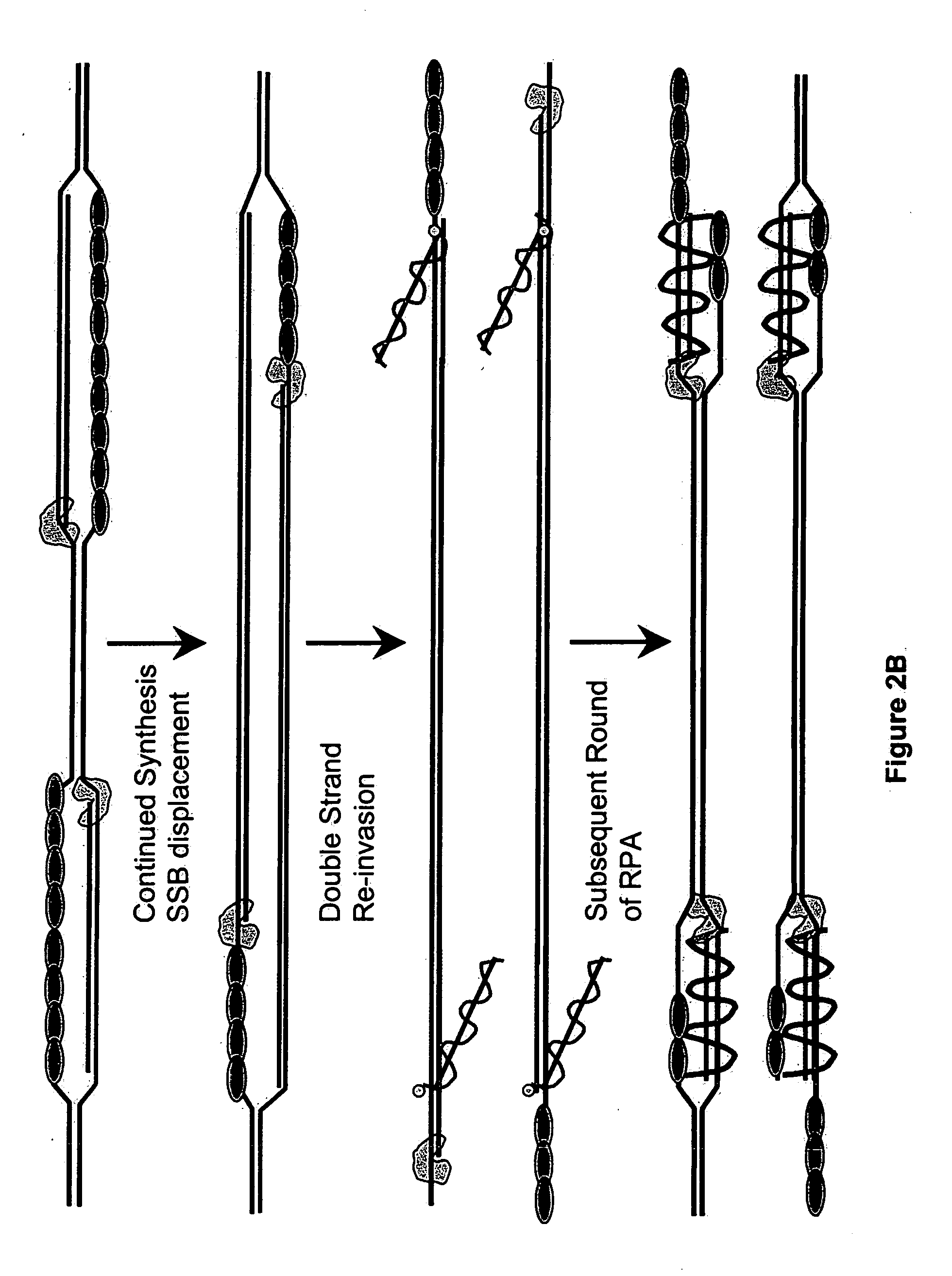
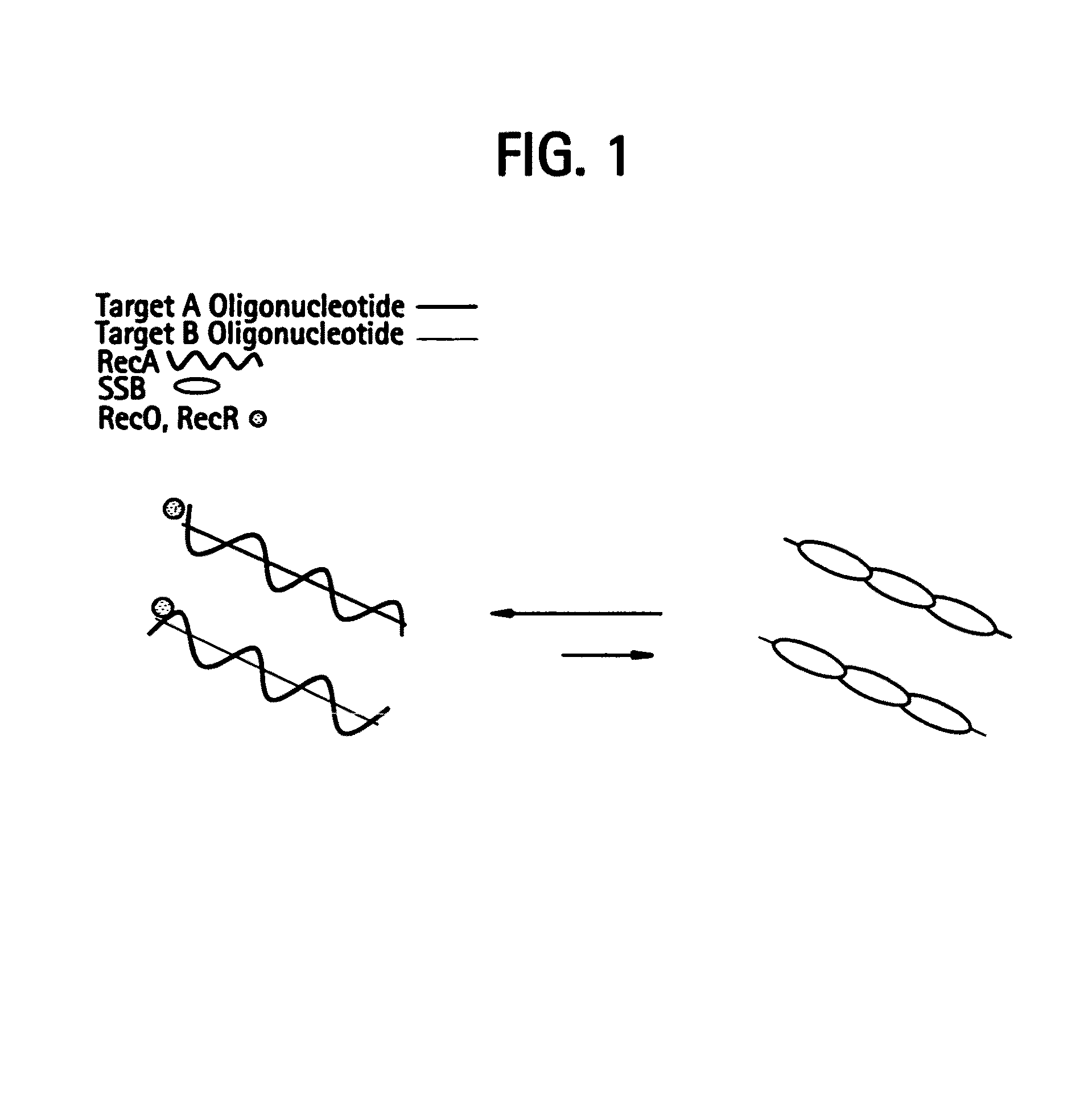
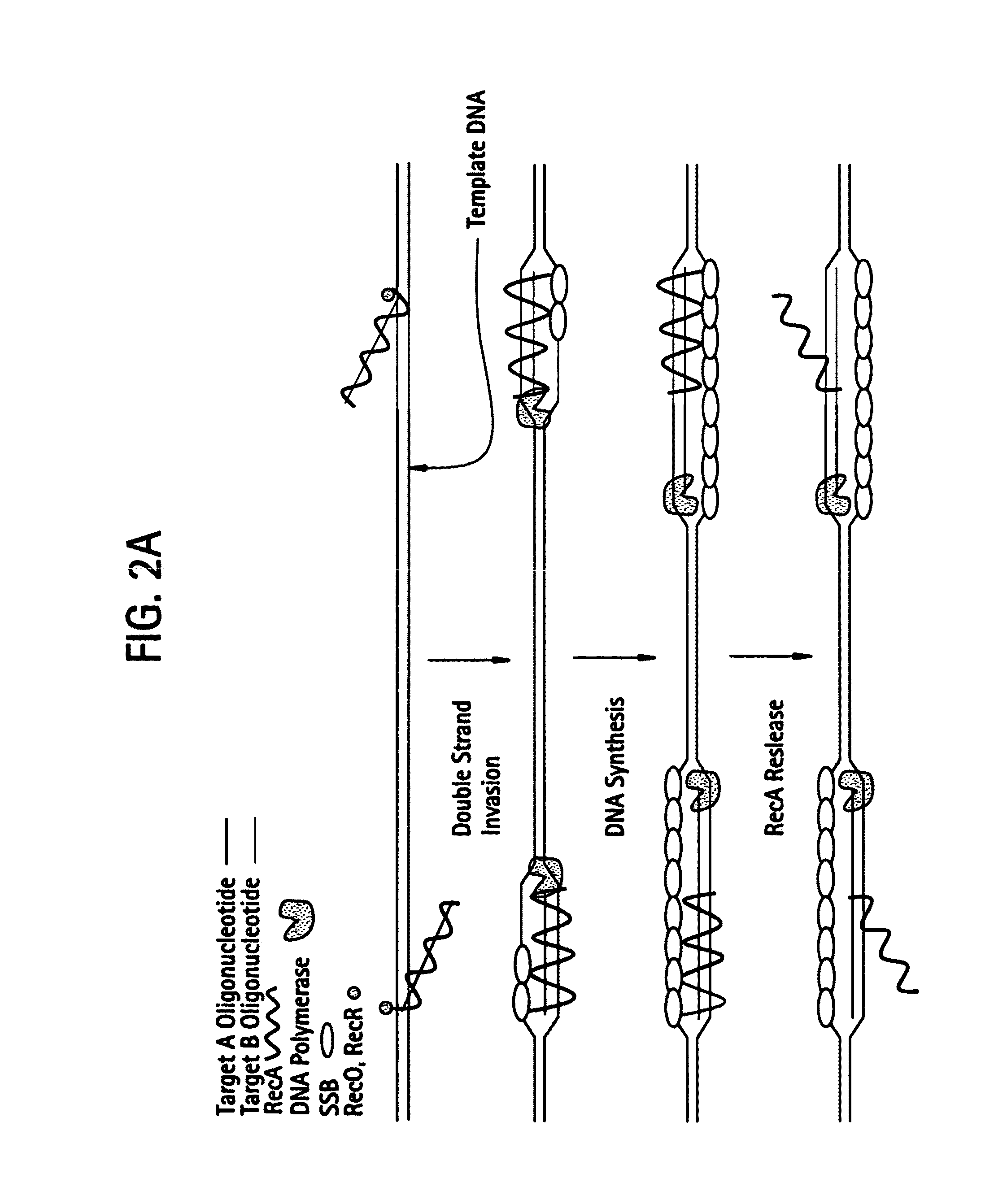
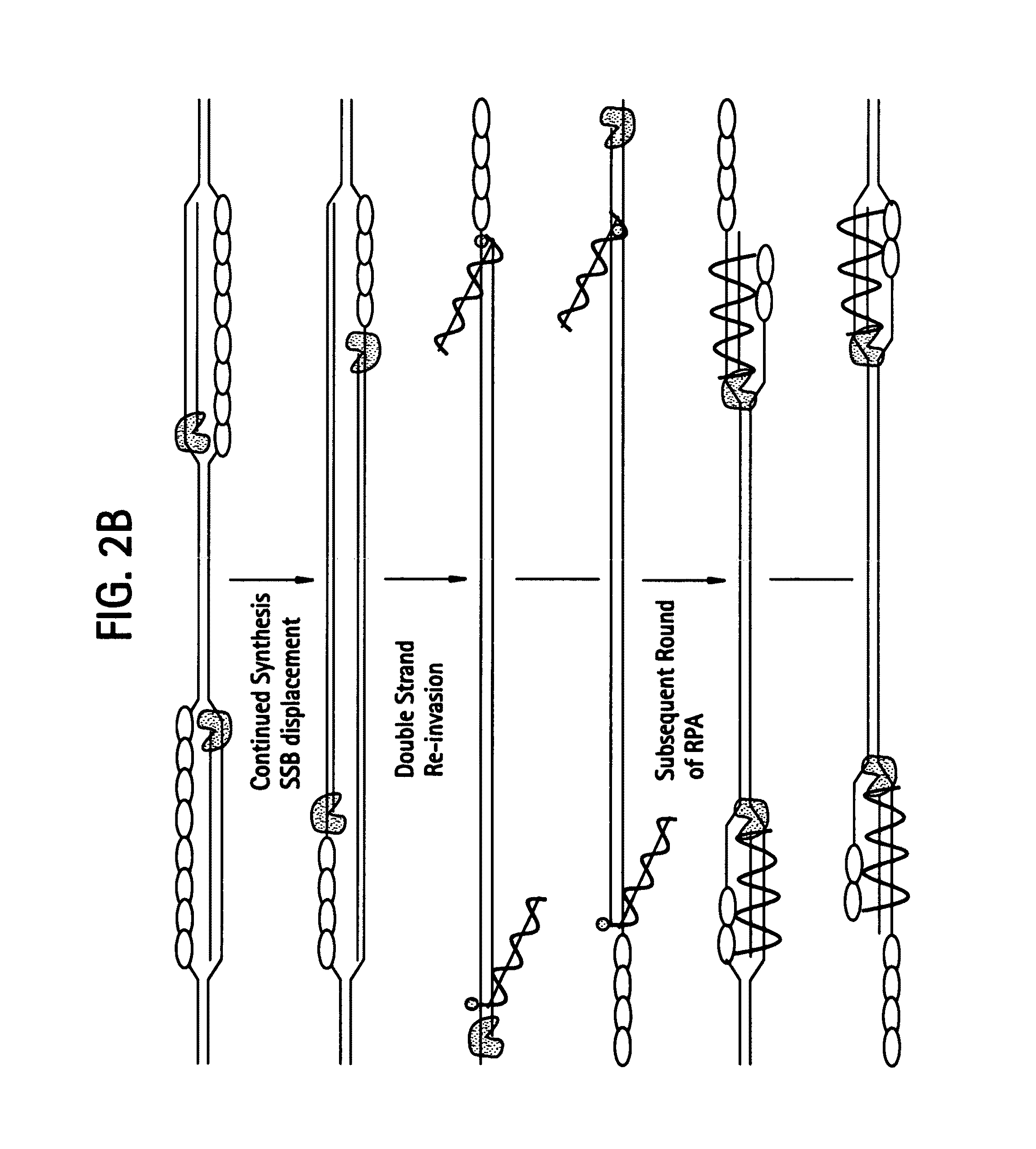
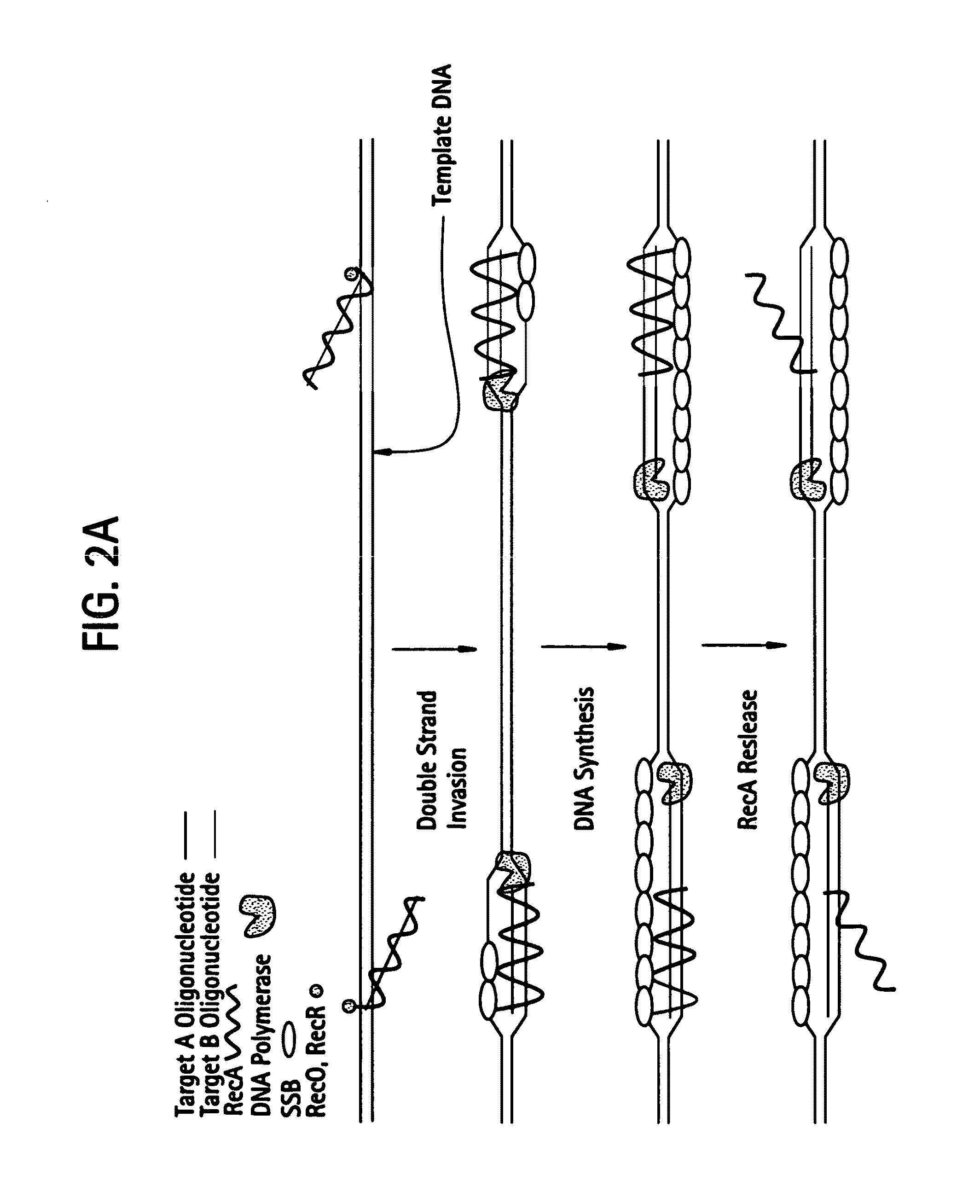
This disclosure describe three related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of the bacterial RecA and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods has the advantage of not requiring thermocycling or thermophilic enzymes. Further, the improved processivity of the disclosed methods allow amplification of DNA up to hundreds of megabases in length.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Recombinase polymerase amplification
ActiveUS20050112631A1HydrolasesMicrobiological testing/measurementRecombinase Polymerase AmplificationSingle strand
This disclosure describe three related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of recombinase and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods have the advantage of not requiring thermocycling or thermophilic enzymes. Further, the improved processivity of the disclosed methods may allow amplification of DNA up to hundreds of megabases in length.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Method of analyzing DNA sequence using field-effect device, and base sequence analyzer
ActiveUS7888013B2Bioreactor/fermenter combinationsHeating or cooling apparatusAnalysis dnaFluorescence
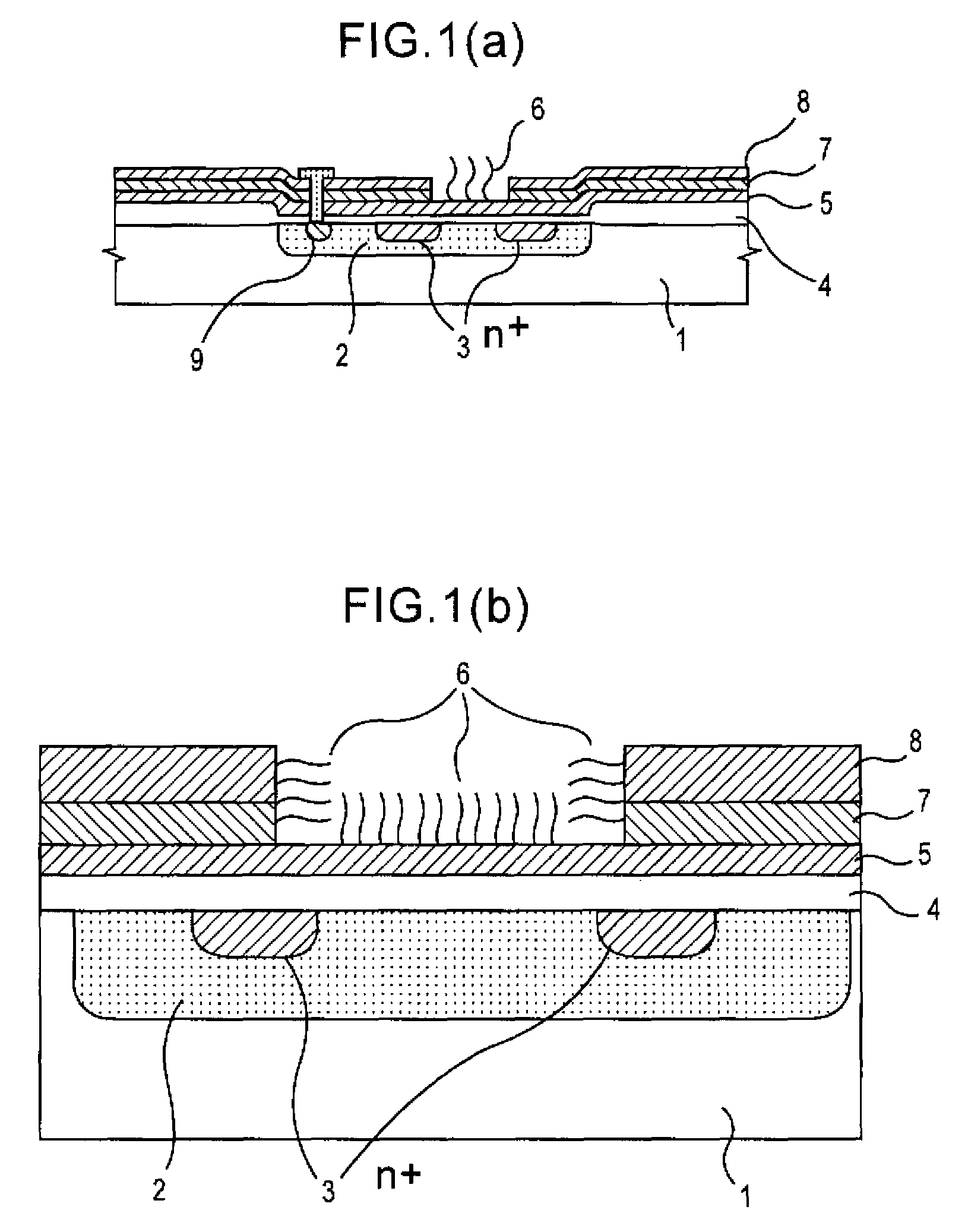
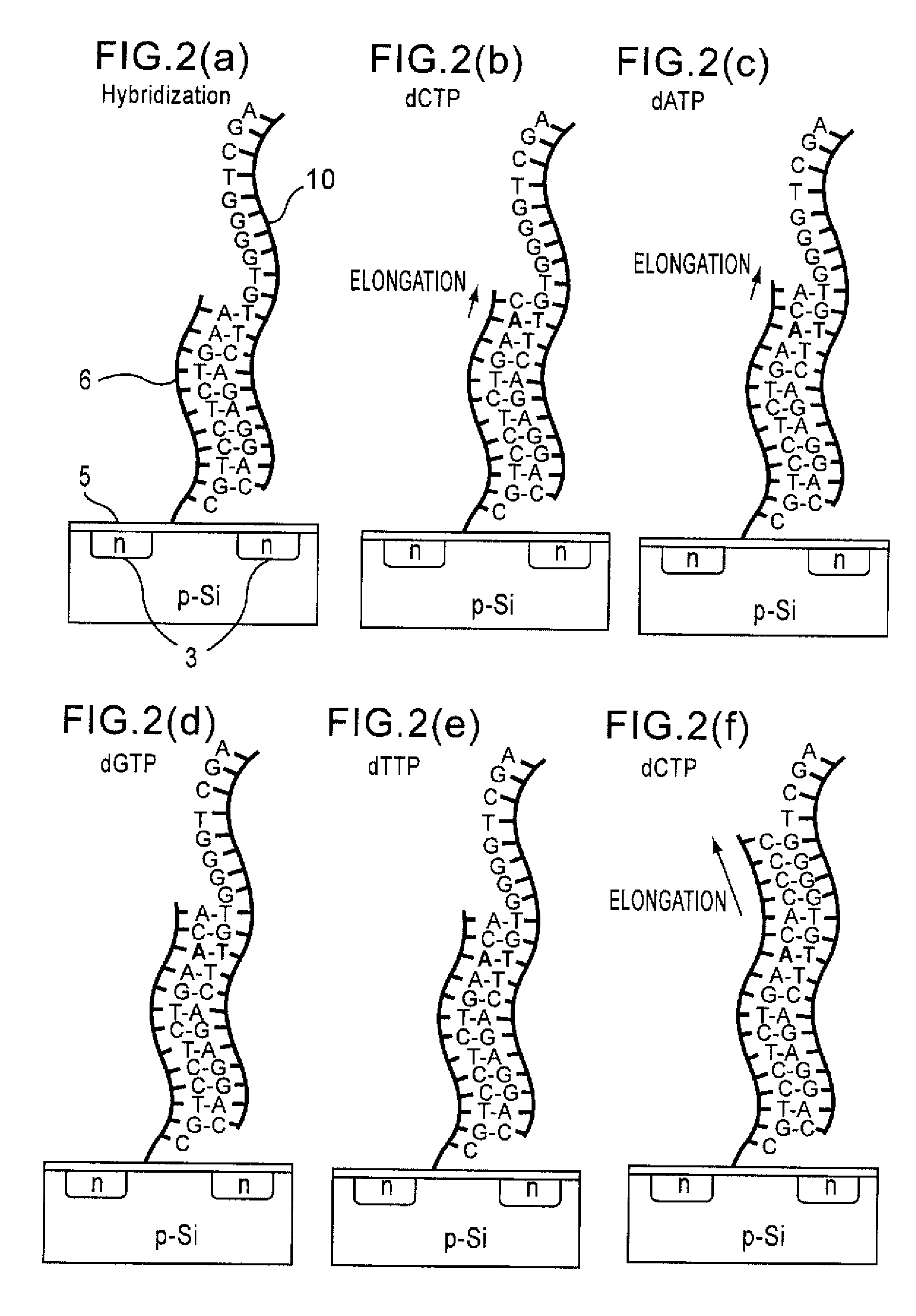
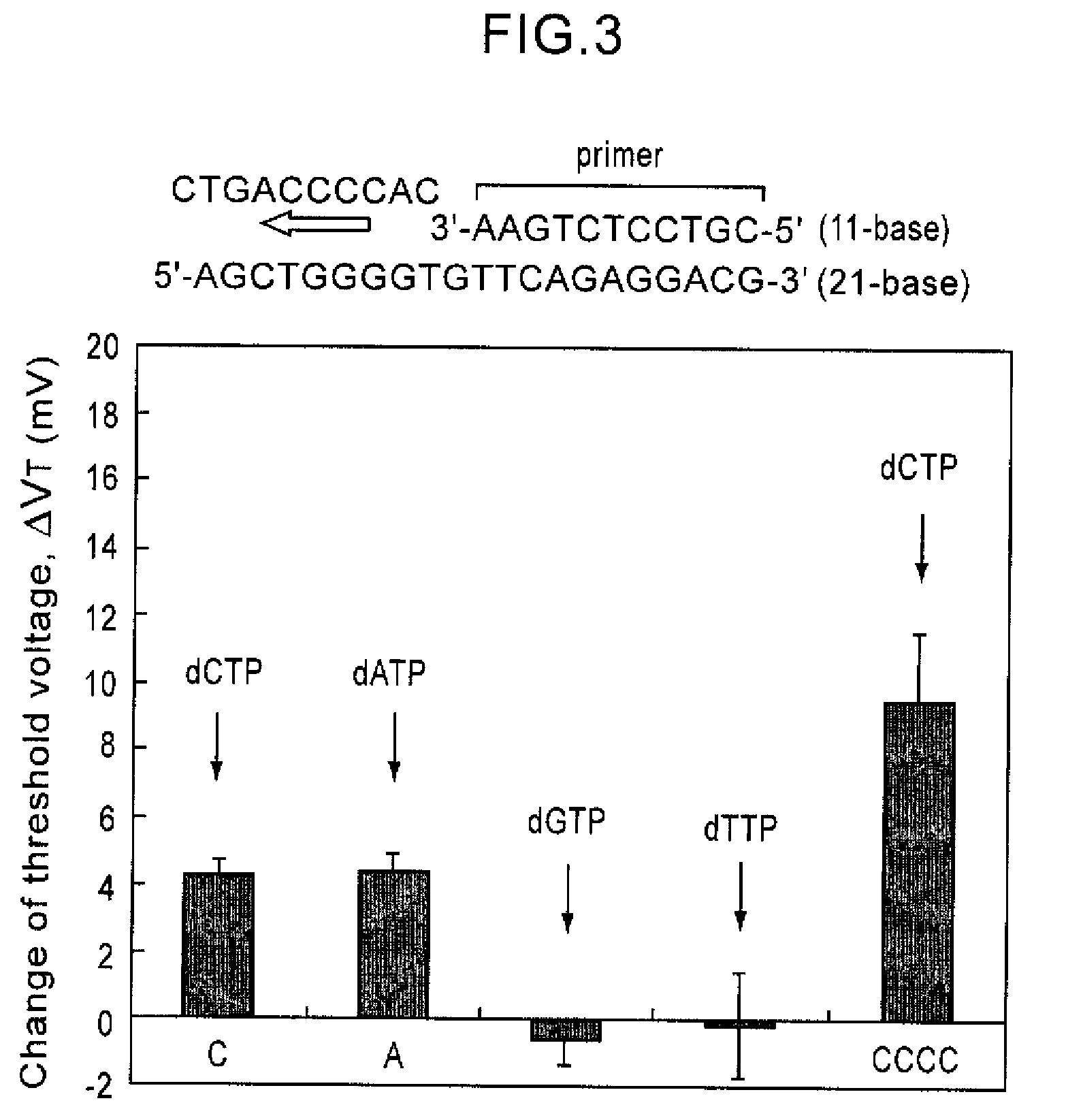
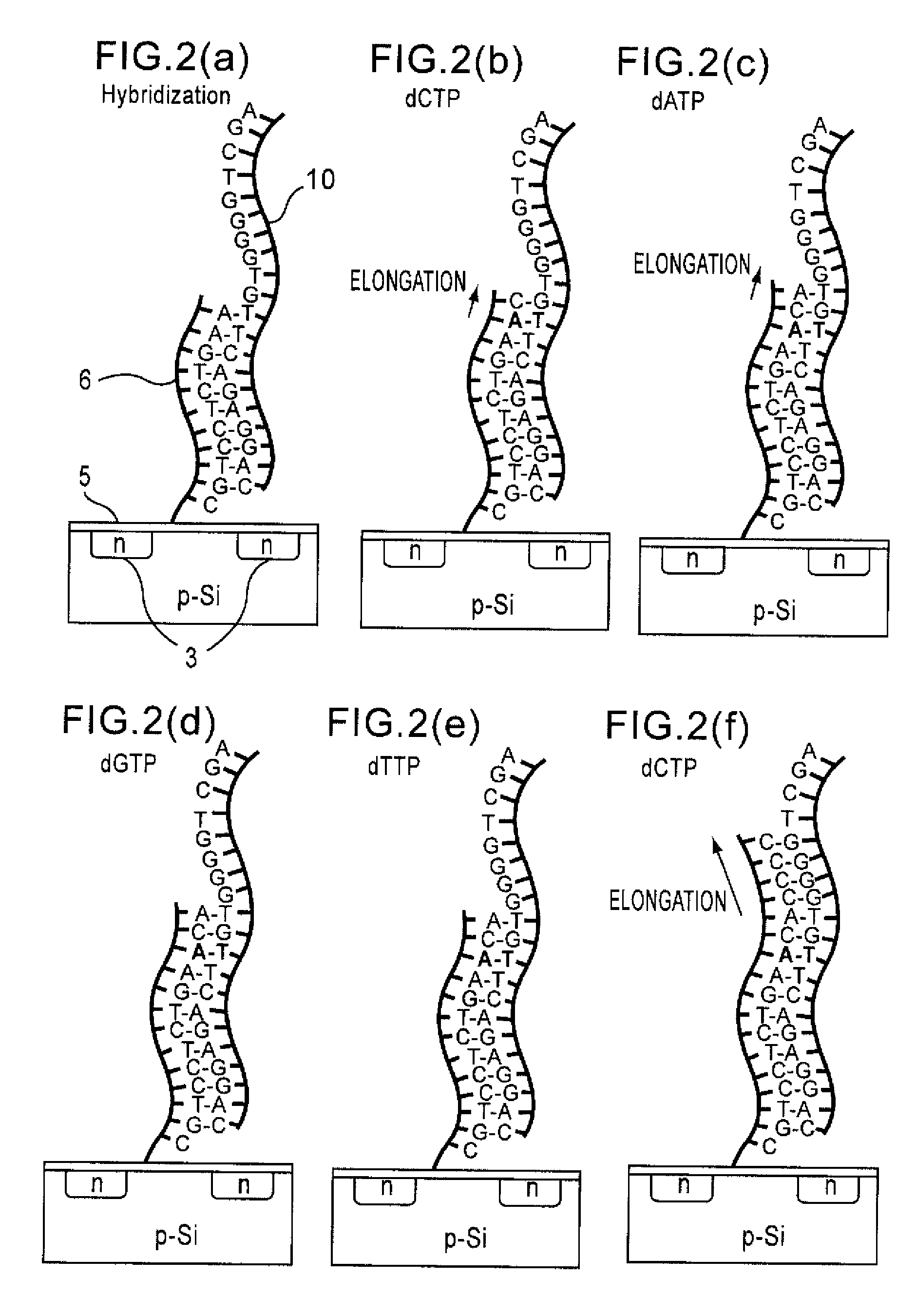
Since conventional DNA sequence analyzing technologies are based on the fundamental principle of fluorescent detection, expensive, complex optical systems and laser sources have been necessary.A field-effect device for gene detection of the present invention analyzes a base sequence by immobilizing a single-strand nucleic acid probe at a gate portion, inducing hybridization at the gate portion to form a double-stranded DNA, inducing elongation reaction by adding a DNA polymerase and one of the substrates, and measuring the electrical characteristic of the field-effect device caused by elongation reaction.Since the elongation reaction of one base induced at the gate portion can be directly converted to an electrical signal, expensive lasers or complex optical systems are not needed. Thus, a small gene polymorphism detection system that can conduct measurement at high precision can be provided.
Owner:NAT INST FOR MATERIALS SCI
Method for the generation of compact tale-nucleases and uses thereof
ActiveUS20130117869A1Simple processSimple and efficient vectorizationFusion with DNA-binding domainHydrolasesDNA-binding domainNuclease
The present invention relates to a method for the generation of compact Transcription Activator-Like Effector Nucleases (TALENs) that can efficiently target and process double-stranded DNA. More specifically, the present invention concerns a method for the creation of TALENs that consist of a single TALE DNA binding domain fused to at least one catalytic domain such that the active entity is composed of a single polypeptide chain for simple and efficient vectorization and does not require dimerization to target a specific single double-stranded DNA target sequence of interest and process DNA nearby said DNA target sequence. The present invention also relates to compact TALENs, vectors, compositions and kits used to implement the method.
Owner:CELLECTIS SA
Method of Analyzing Dna Sequence Using Field-Effect Device, and Base Sequence Analyzer
ActiveUS20080286767A1Bioreactor/fermenter combinationsHeating or cooling apparatusAnalysis dnaFluorescence
Since conventional DNA sequence analyzing technologies are based on the fundamental principle of fluorescent detection, expensive, complex optical systems and laser sources have been necessary.A field-effect device for gene detection of the present invention analyzes a base sequence by immobilizing a single-strand nucleic acid probe at a gate portion, inducing hybridization at the gate portion to form a double-stranded DNA, inducing elongation reaction by adding a DNA polymerase and one of the substrates, and measuring the electrical characteristic of the field-effect device caused by elongation reaction.Since the elongation reaction of one base induced at the gate portion can be directly converted to an electrical signal, expensive lasers or complex optical systems are not needed. Thus, a small gene polymorphism detection system that can conduct measurement at high precision can be provided.
Owner:NAT INST FOR MATERIALS SCI
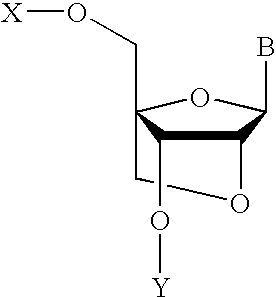
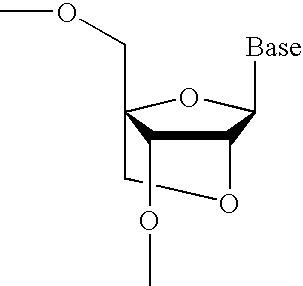
Nucleoside analogs and oligonucleotide derivatives containing these analogs
Nucleoside analogues expressed by the following general formula where B represents an aromatic base having carbonyl oxygen at the 2-position, or a 2-hydroxyphenyl group, or oligonucleotide derivatives containing one or more of the nucleoside analogues are provided.The oligonucleotide derivatives are triplex-forming oligonucleotide derivatives which bind specifically to target double-stranded DNA with high affinity in the antigene method to form triplexes, and can thereby control and inhibit the expression of relevant genes efficiently, and which show high resistance to nucleases.
Owner:IMANISHI TAKESHI
Recombinase polymerase amplification
This disclosure describes related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of recombinase and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods have the advantage of not requiring thermocycling or thermophilic enzymes, thus offering easy and affordable implementation and portability relative to other amplification methods. Further RPA reactions using light and otherwise, methods to determine the nature of amplified species without a need for gel electrophoresis, methods to improve and optimize signal to noise ratios in RPA reactions, methods to optimize oligonucleotide primer function, methods to control carry-over contamination, and methods to employ sequence-specific third ‘specificity’ probes. Further described are novel properties and approaches for use of probes monitored by light in dynamic recombination environments.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Probe for analysis of target nucleic acids
The invention is a probe for detecting nucleic acids having a particular sequence. It is composed of two joint units. One unit is chemically different from natural nucleic acids, but has the ability to recognize a particular sequence of bases or base pairs in single or double-stranded DNA of RNA. The other unit is a compound whose detectable properties are altered upon binding to nucleic acids.
Owner:LIGHTUP TECH
Detection of nucleic acids
ActiveUS9273349B2Enhanced hybridizationConfidenceSugar derivativesMicrobiological testing/measurementTissue sampleSingle strand
Owner:AFFYMETRIX INC
Methods, cells & organisms
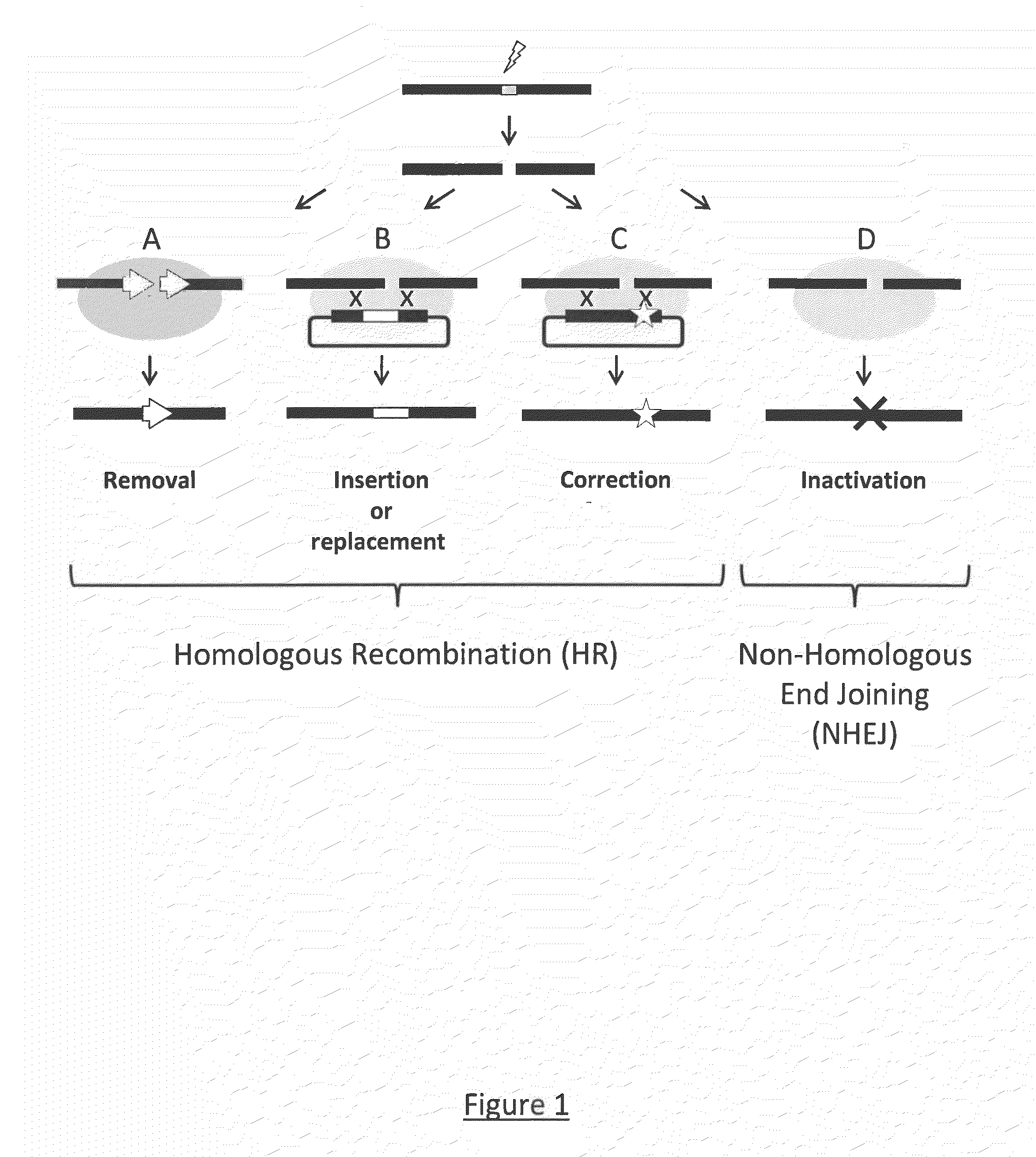
InactiveUS20150079680A1Reduce chanceGenetically modified cellsStable introduction of DNABiological bodyDNA fragmentation
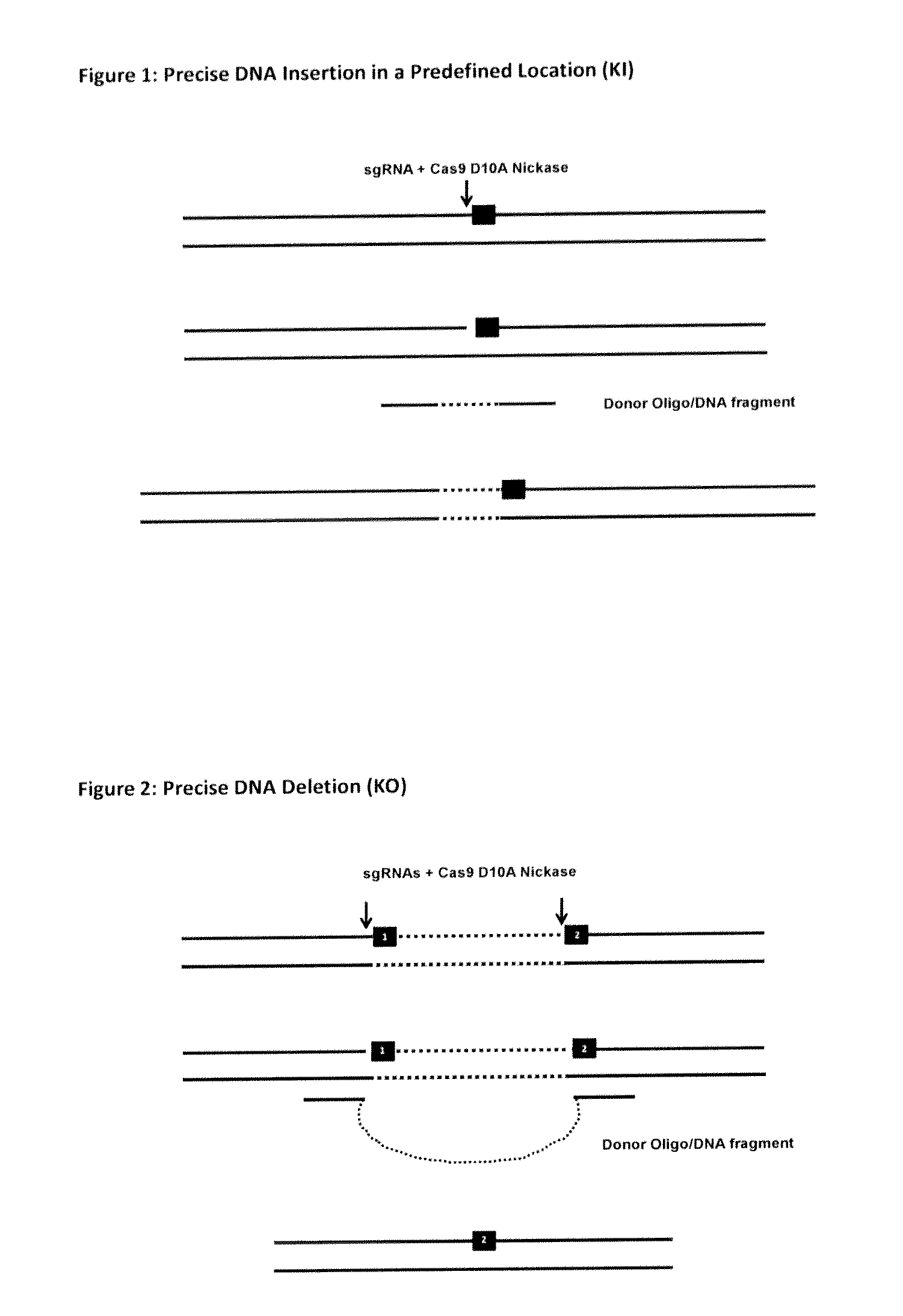
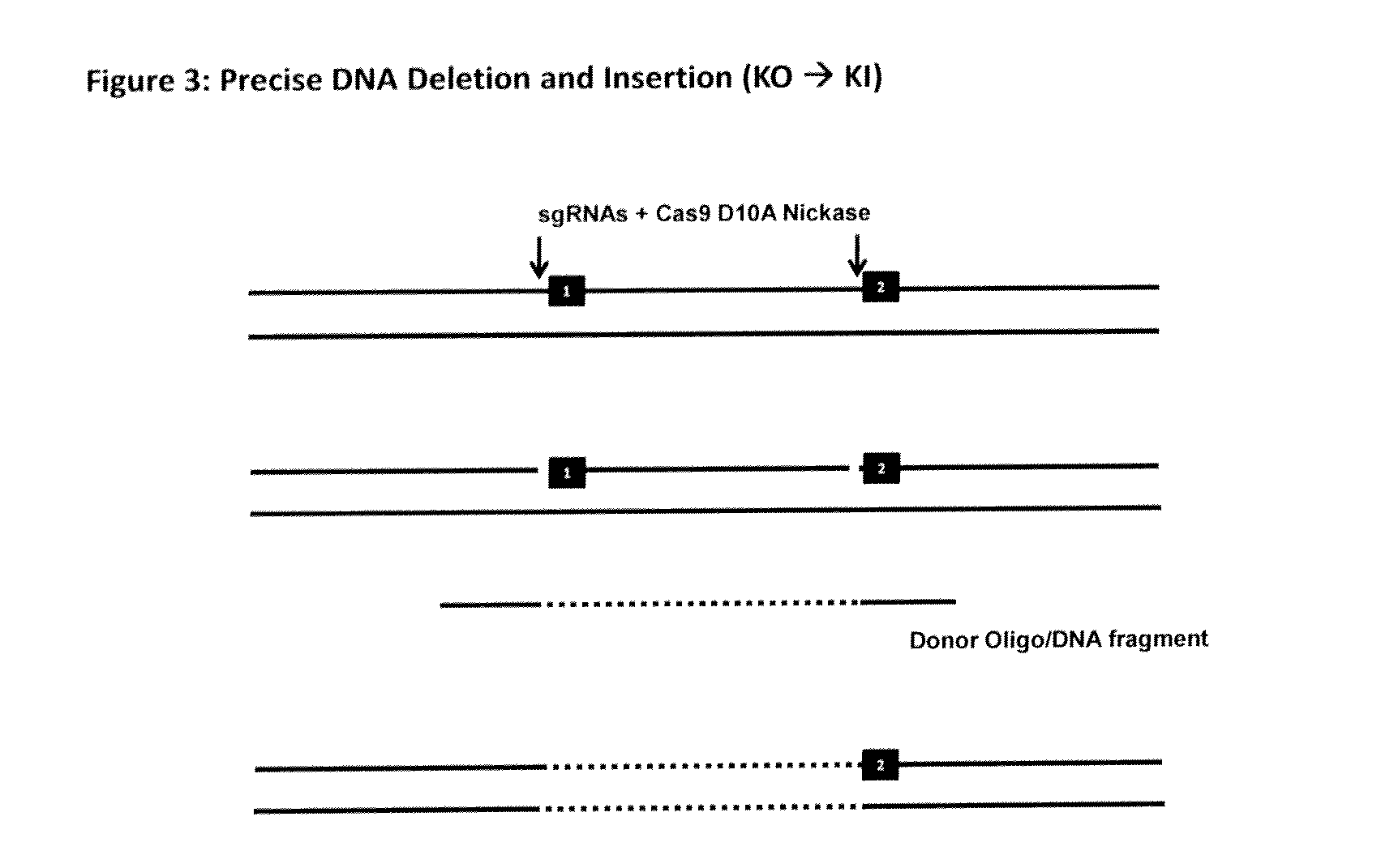
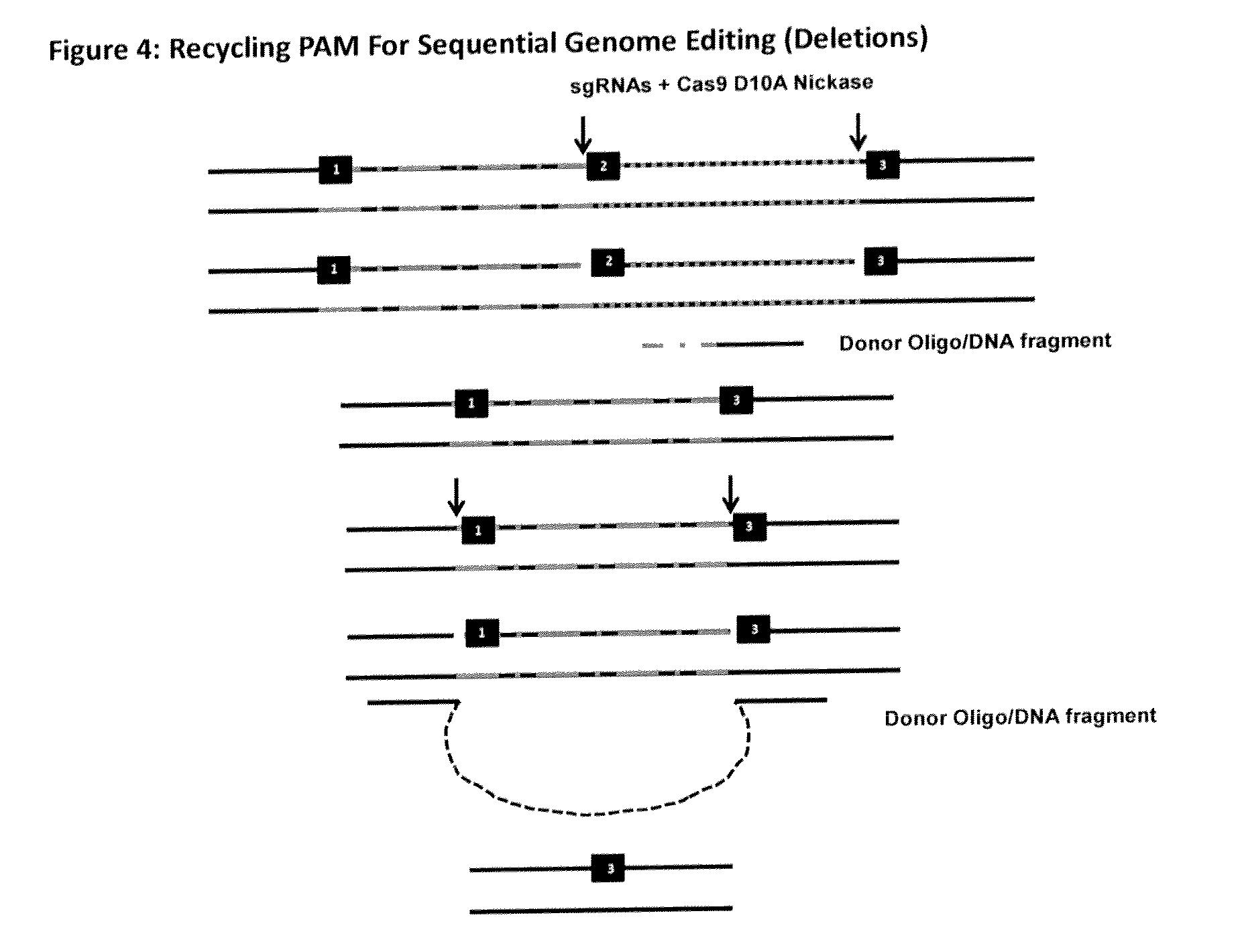
The invention relates to an approach for introducing one or more desired insertions and / or deletions of known sizes into one or more predefined locations in a nucleic acid (eg, in a cell or organism genome). They developed techniques to do this either in a sequential fashion or by inserting a discrete DNA fragment of defined size into the genome precisely in a predefined location or carrying out a discrete deletion of a defined size at a precise location. The technique is based on the observation that DNA single-stranded breaks are preferentially repaired through the HDR pathway, and this reduces the chances of indels (eg, produced by NHEJ) in the present invention and thus is more efficient than prior art techniques. The invention also provides sequential insertion and / or deletions using single- or double-stranded DNA cutting.
Owner:KIMAB LTD
Methods and devices for characterizing duplex nucleic acid molecules
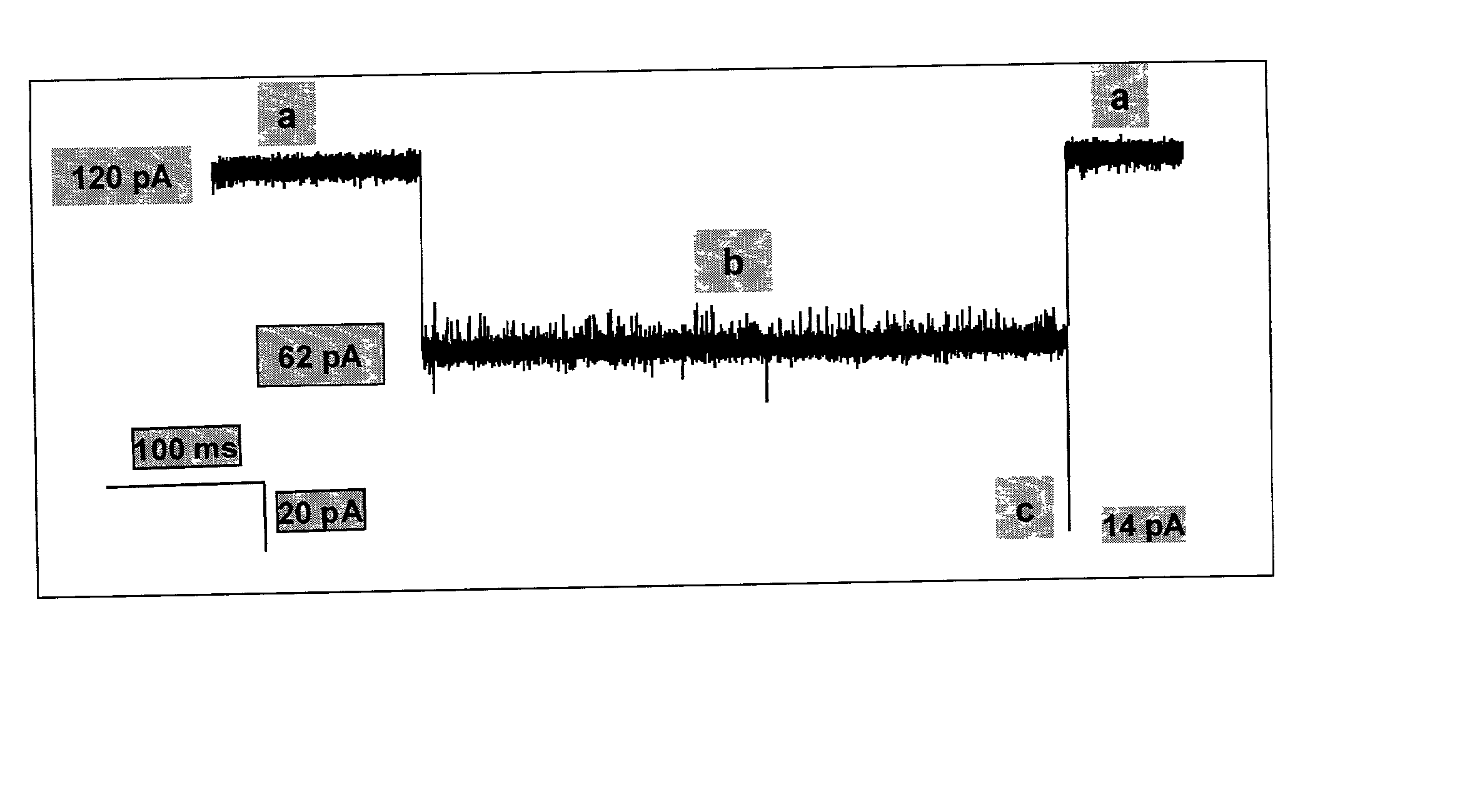
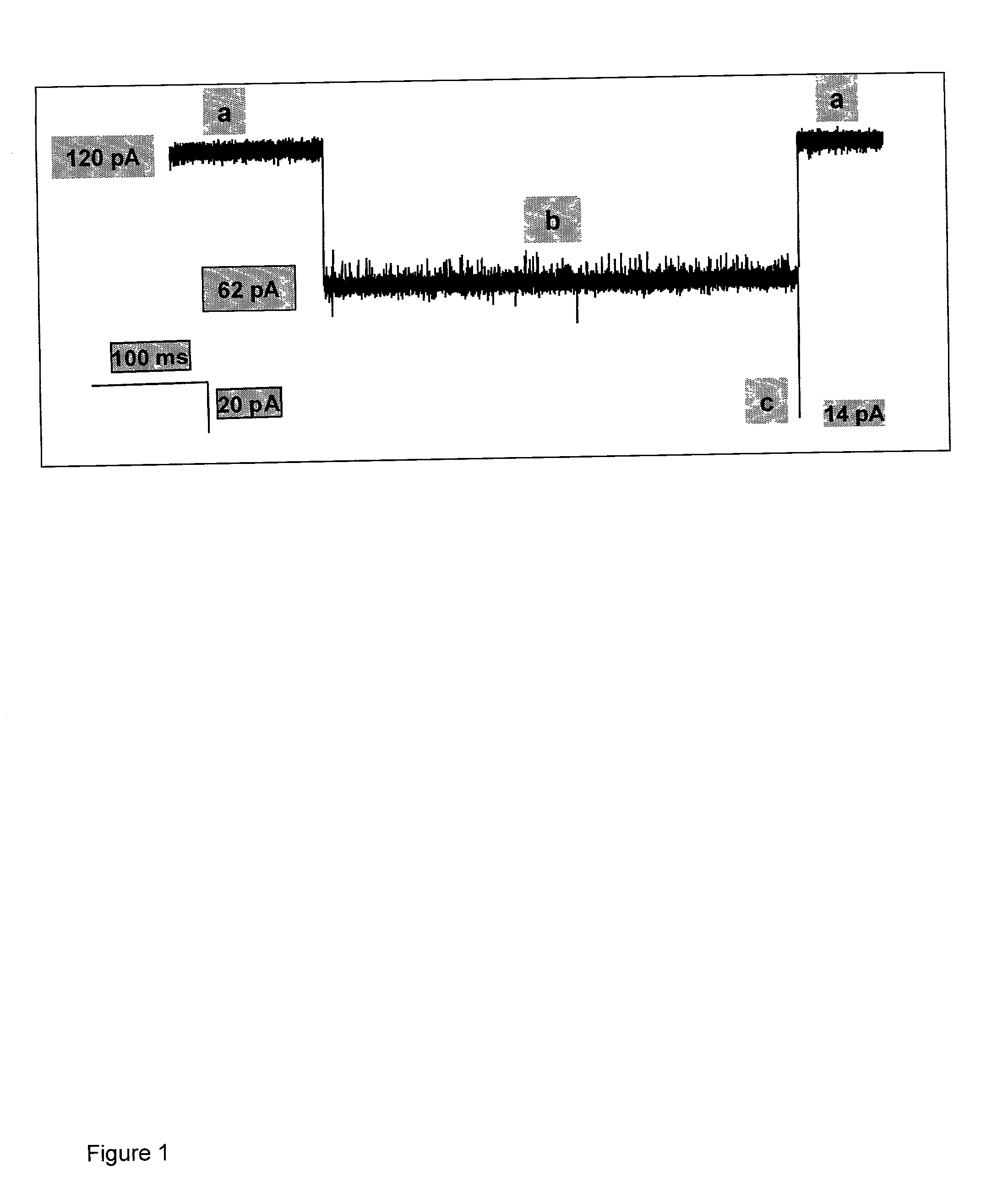
Methods and devices are provided for characterizing a duplex nucleic acid, e.g., a duplex DNA molecule. In the subject methods, a fluid conducting medium that includes a duplex nucleic acid molecule is contacted with a nanopore under the influence of an applied electric field and the resulting changes in current through the nanopore caused by the duplex nucleic acid molecule are monitored. The observed changes in current through the nanopore are then employed as a set of data values to characterize the duplex nucleic acid, where the set of data values may be employed in raw form or manipulated, e.g., into a current blockade profile. Also provided are nanopore devices for practicing the subject methods, where the subject nanopore devices are characterized by the presence of an algorithm which directs a processing means to employ monitored changes in current through a nanopore to characterize a duplex nucleic acid molecule responsible for the current changes. The subject methods and devices find use in a variety of applications, including, among other applications, the identification of an analyte duplex DNA molecule in a sample, the specific base sequence at a single nulceotide polymorphism (SNP), and the sequencing of duplex DNA molecules.
Owner:RGT UNIV OF CALIFORNIA
In vitro recombination method
ActiveUS20070037197A1HydrolasesMicrobiological testing/measurementSingle-strand DNA-binding proteinSequence identity
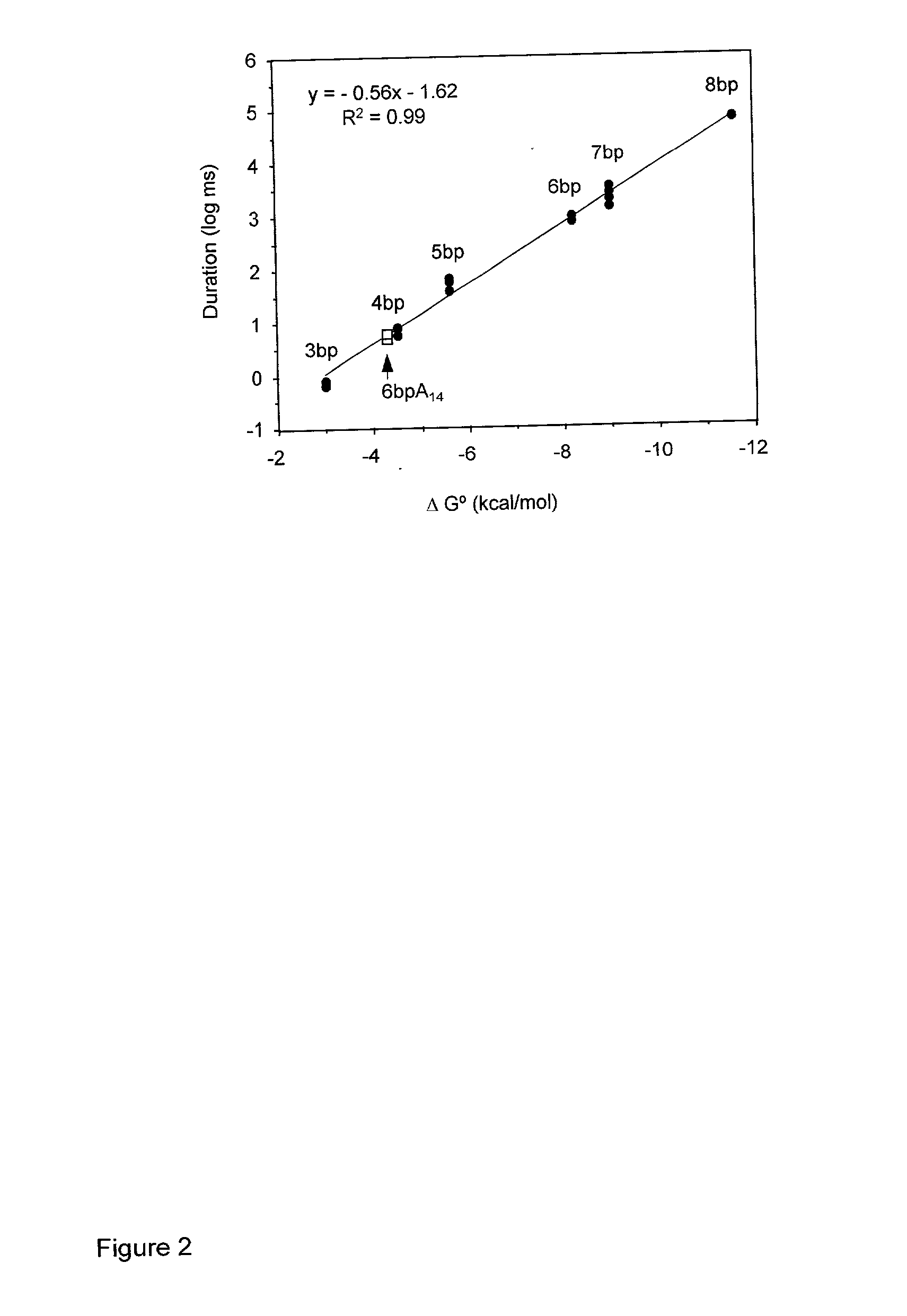
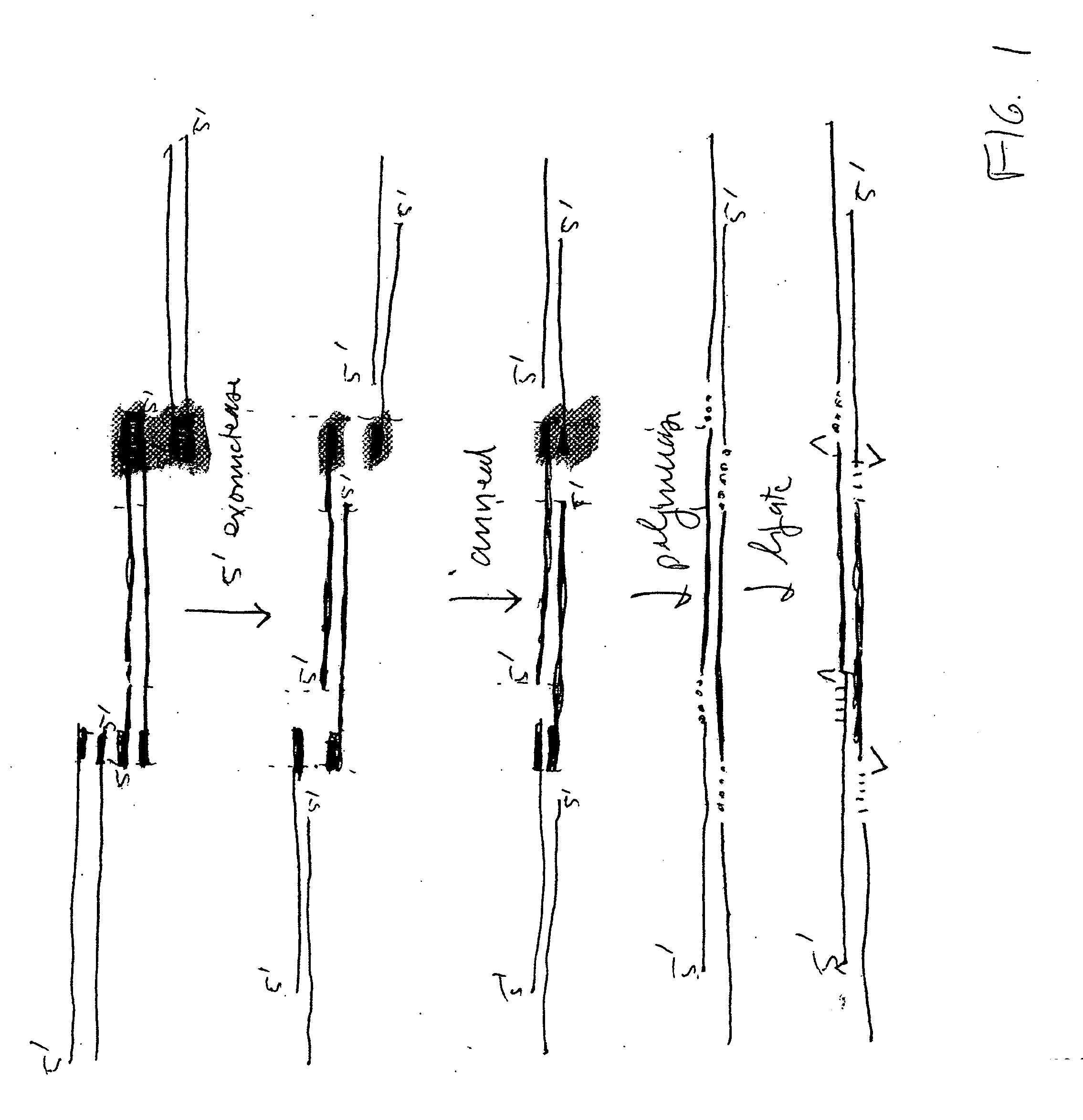
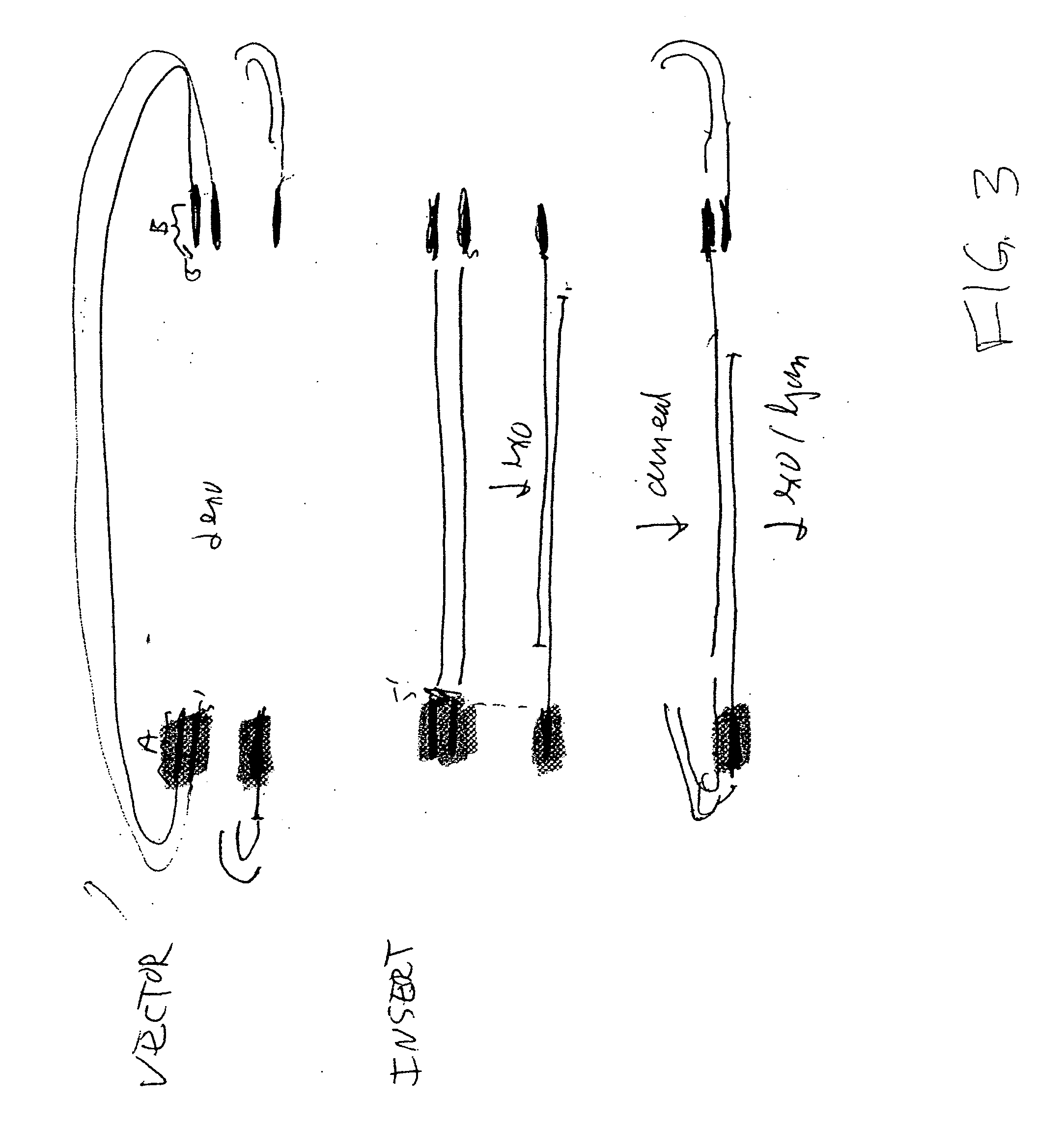
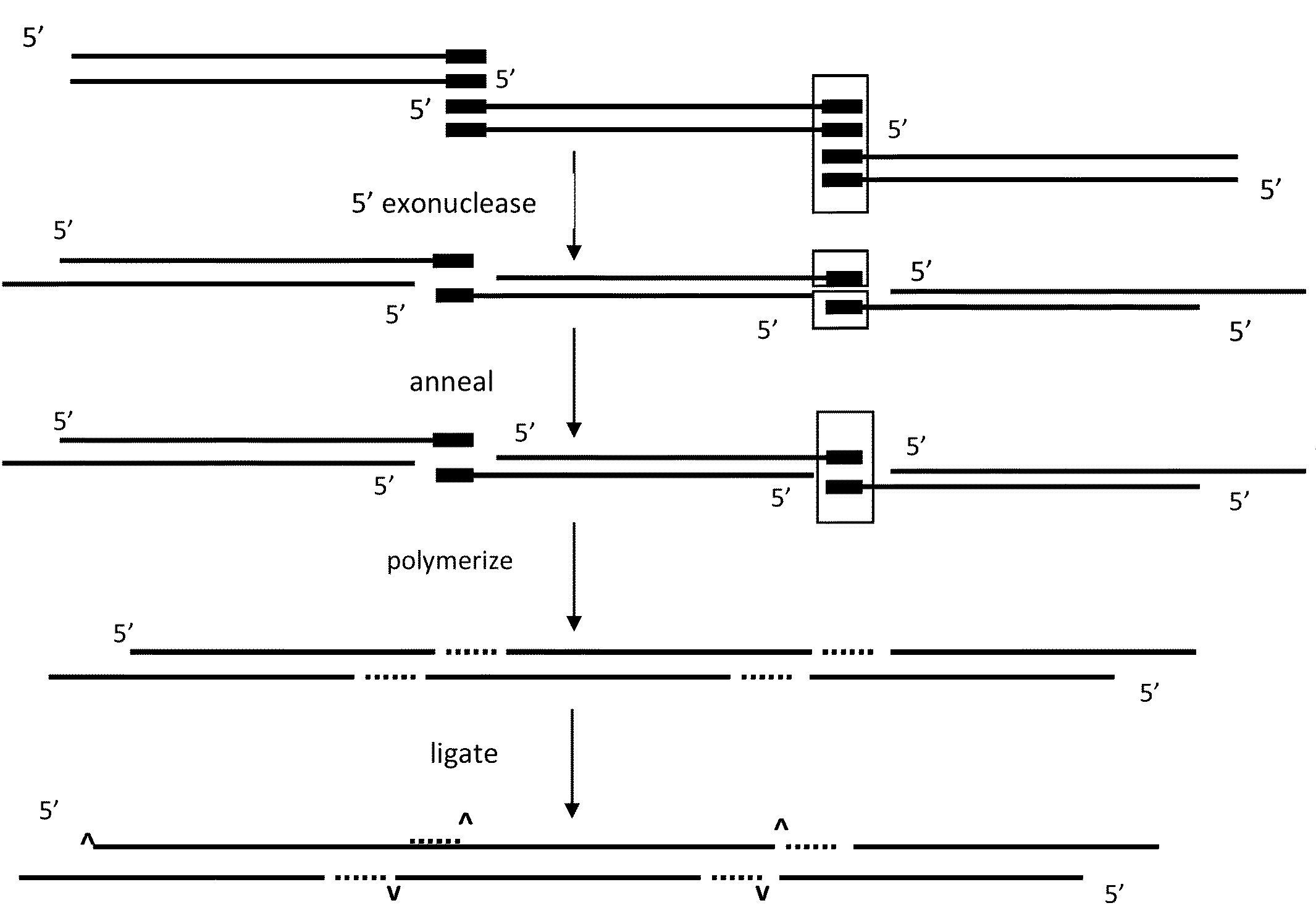
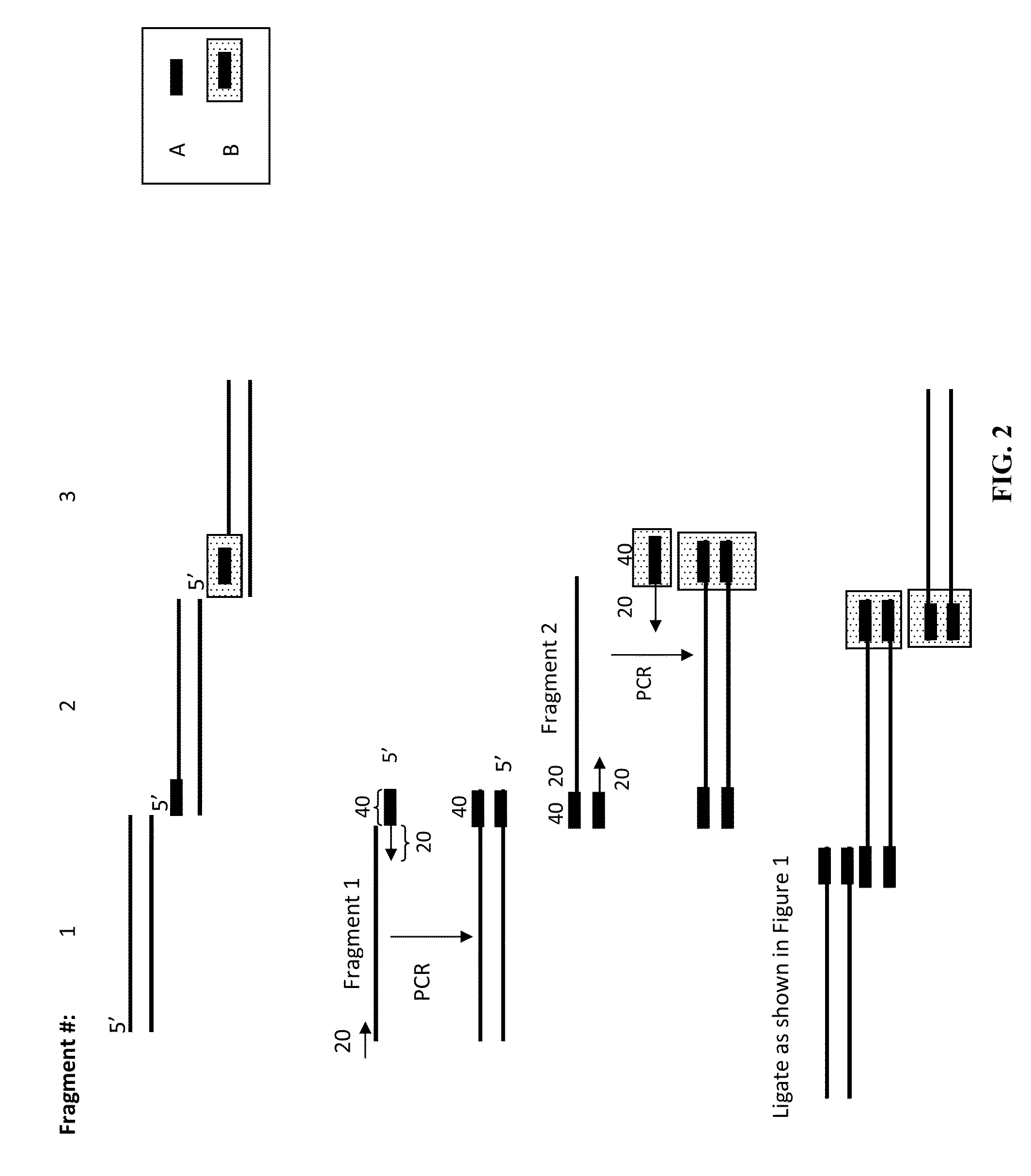
The present invention relates, e.g., to in vitro method, using isolated protein reagents, for joining two double stranded (ds) DNA molecules of interest, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule share a region of sequence identity, comprising contacting the two DNA molecules in a reaction mixture with (a) a non-processive 5′ exonculease; (b) a single stranded DNA binding protein (SSB) which accelerates nucleic acid annealing; (c) a non strand-displacing DNA polymerase; and (d) a ligase, under conditions effective to join the two DNA molecules to form an intact double stranded DNA molecule, in which a single copy of the region of sequence identity is retained. The method allows the joining of a number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes.
Owner:TELESIS BIO INC
Selected amplification of polynucleotides
InactiveUS8137936B2Reducing background and spurious amplificationSave materialMicrobiological testing/measurementFermentationPolynucleotideDouble stranded
The invention provides methods and compositions for selectively amplifying one or more target polynucleotides in a sample. In one aspect, a plurality of selection oligonucleotides are provided that are capable of simultaneously annealing to separate regions of a target polynucleotide to form a complex that is enzymatically converted into a closed double stranded DNA circle that incorporates the sequence region between the two separate regions. Sequences that fail to form such complexes may be removed by nuclease digestion and the sequences of the remaining DNA circles may be amplified by a variety of techniques, such as rolling circle replication after nicking, PCR amplification after linearization, or the like.
Owner:MACEVICZ STEPHEN C
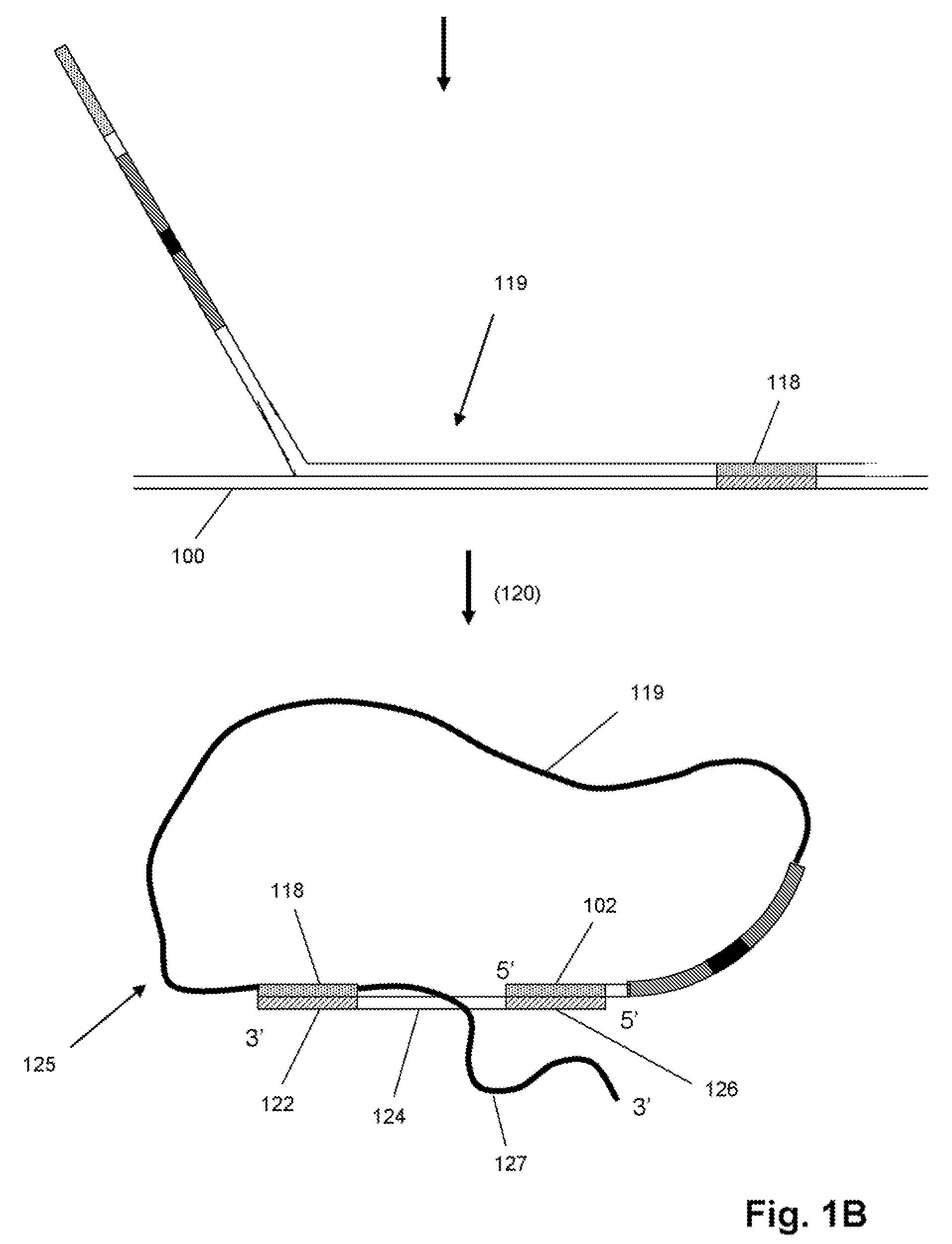
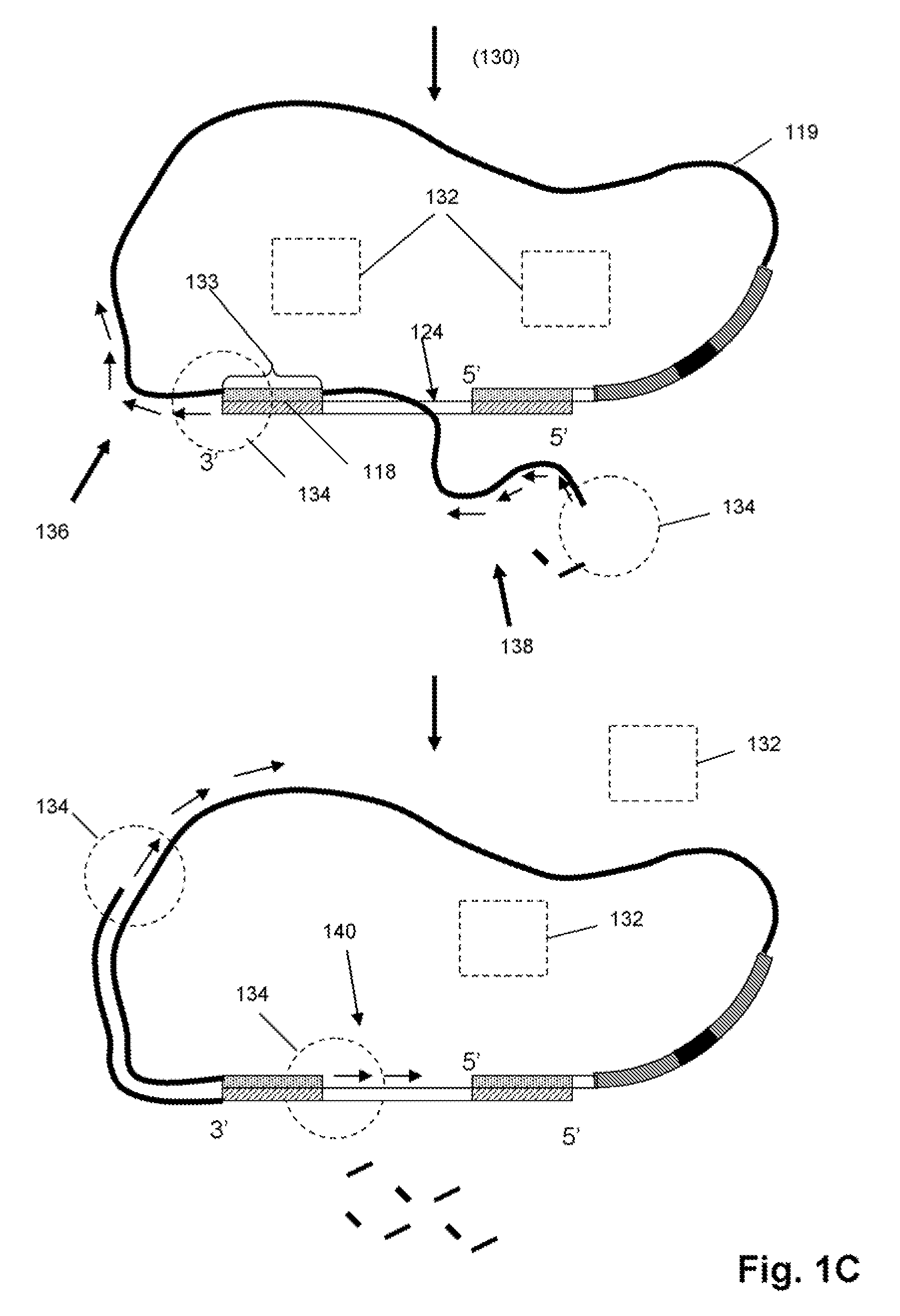
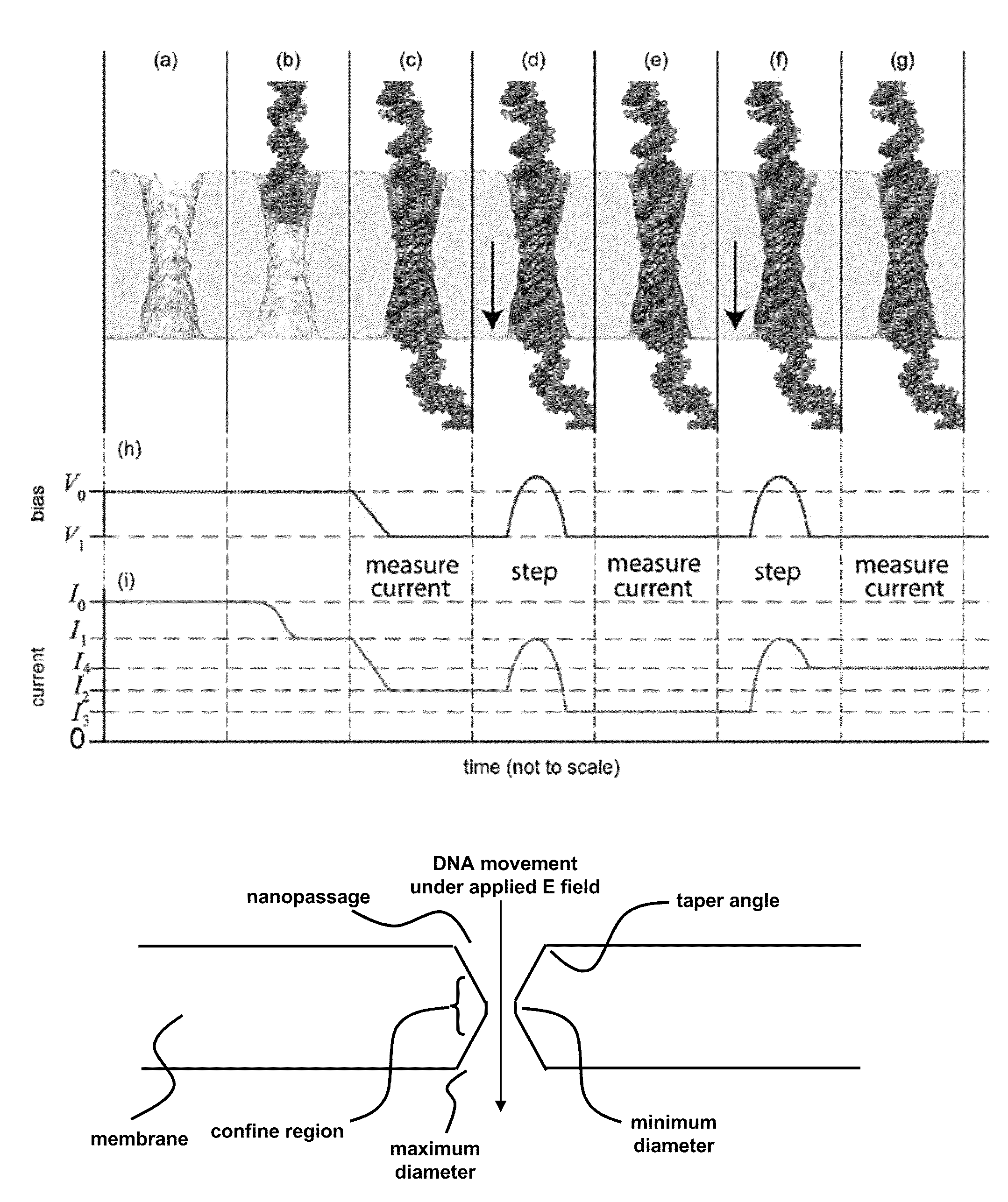
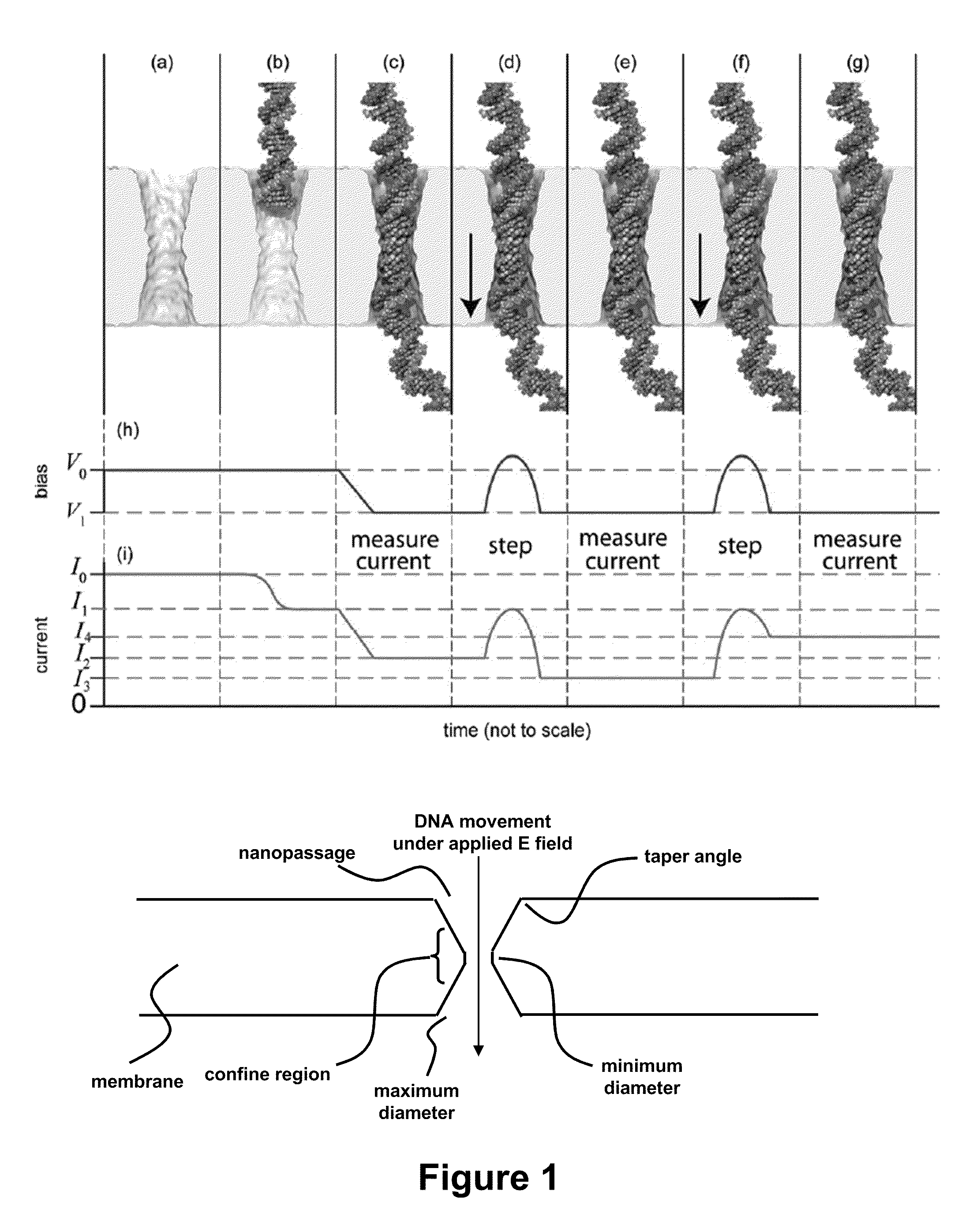
Characterizing Stretched Polynucleotides in a Synthetic Nanopassage
ActiveUS20110226623A1Longer read lengthIncrease speedElectrolysis componentsVolume/mass flow measurementFluid compartmentsTrapping
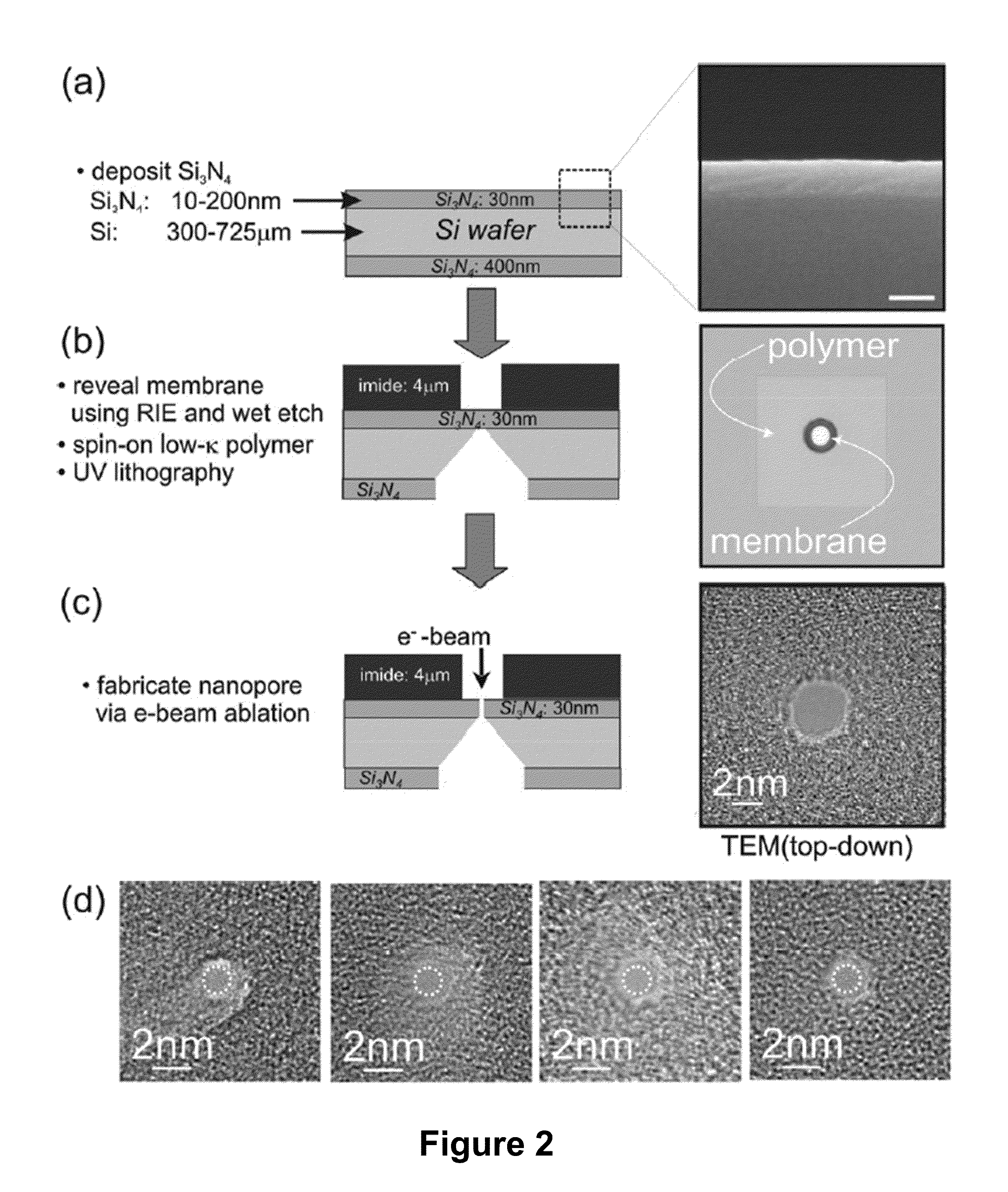
Methods of trapping a deformed portion of a double-stranded polynucleotide in a membrane nanopassage are provided. In an aspect, the membrane has a nanopassage that defines a confine region, wherein the membrane separates a first fluid compartment from a second fluid compartment, and the nanopassage is in fluid communication with the first and second compartments. A polynucleotide is provided to the first fluid compartment and optionally a threshold voltage for the membrane and the polynucleotide is determined. A driving voltage across the membrane that is greater than the threshold voltage is applied to force a portion of the polynucleotide sequence into the nanopassage confine region, and decreased to a holding voltage bias to trap the polynucleotide portion in the nanopassage confine region. In particular, at least one nucleotide base-pair is fixably positioned in the nanopassage confine volume. In further embodiments, any of the trapping methods are used to characterize or sequence double stranded DNA.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
Method for the synthesis of DNA sequences
InactiveUS7183406B2Material nanotechnologyGroup 5/15 element organic compoundsDNA microarrayDNA construct
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will spontaneously hybridize together to form the desired DNA construct. This method makes possible the remote assembly of DNA sequence, through a process analogous to facsimile transmission of documents, since the information on DNA to be made can be transmitted remotely to an instrument which can then synthesize any needed DNA sequence from the information.
Owner:WISCONSIN ALUMNI RES FOUND
Therapeutic methods for benzodiazepine derivatives
This invention provides an antibody with high affinity for single-stranded DNA, low or no affinity for double-stranded DNA, and capable of specifically binding a DNA hairpin and the hybridoma cell lines which produces these monoclonal antibodies. A chimeric mouse comprising these hybridoma cell lines and a histocompatible mouse is further provided. A method for screening for an agent which will inhibit anti-DNA antibody.DNA binding. One such agent is a benzodiazepine derivative. This invention therefore provides a method of inhibiting the binding of an anti-DNA antibody to its DNA ligand in a sample by contacting the sample with an effective amount of a benzodiazepine derivative.
Owner:RGT UNIV OF MICHIGAN
Methods, compositions, and kits for generating nucleic acid products substantially free of template nucleic acid
InactiveUS20110224105A1Simple methodHydrolasesMicrobiological testing/measurementDouble strandedComputational biology
Owner:NUGEN TECH
In vitro recombination method
The present invention relates, e.g., to in vitro method, using isolated protein reagents, for joining two double stranded (ds) DNA molecules of interest, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule share a region of sequence identity, comprising contacting the two DNA molecules in a reaction mixture with (a) a non-processive 5′ exonculease; (b) a single stranded DNA binding protein (SSB) which accelerates nucleic acid annealing; (c) a non strand-displacing DNA polymerase; and (d) a ligase, under conditions effective to join the two DNA molecules to form an intact double stranded DNA molecule, in which a single copy of the region of sequence identity is retained. The method allows the joining of a number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes.
Owner:TELESIS BIO INC
Recombinase polymerase amplification
InactiveUS20100311127A1High activitySimple designHeating or cooling apparatusHydrolasesOligonucleotide primersSingle strand
This disclosure describes related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of recombinase and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods have the advantage of not requiring thermocycling or thermophilic enzymes, thus offering easy and affordable implementation and portability relative to other amplification methods. Further RPA reactions using light and otherwise, methods to determine the nature of amplified species without a need for gel electrophoresis, methods to improve and optimize signal to noise ratios in RPA reactions, methods to optimize oligonucleotide primer function, methods to control carry-over contamination, and methods to employ sequence-specific third ‘specificity’ probes. Further described are novel properties and approaches for use of probes monitored by light in dynamic recombination environments.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Methods of producing carbon nanotubes using peptide or nucleic acid micropatterning
InactiveUS20050151126A1Facilitates a predominantly 5′-attachmentMaterial nanotechnologyNanoinformaticsMolecular wireDouble stranded
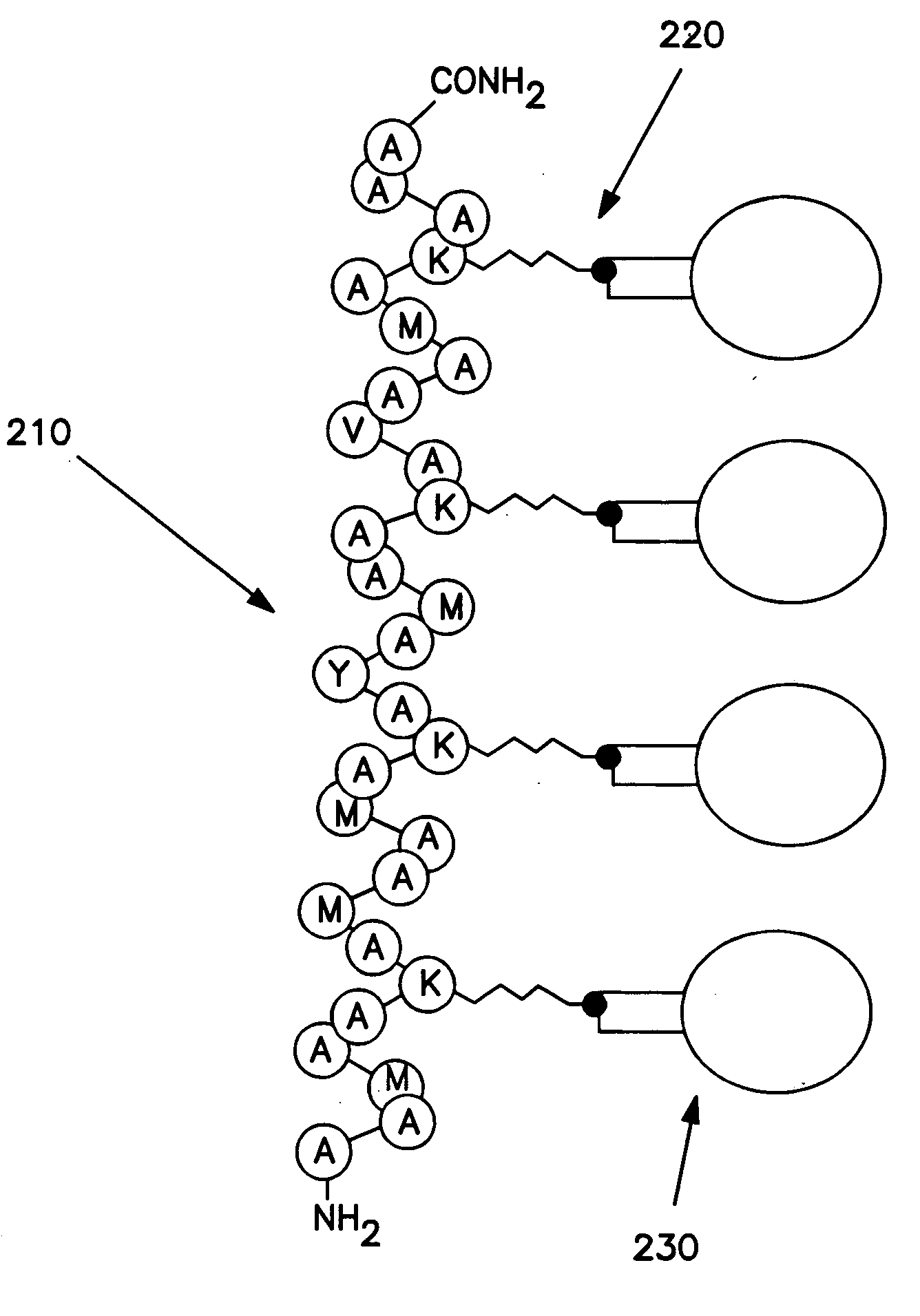
The methods, apparatus and systems disclosed herein concern ordered arrays of carbon nanotubes. In particular embodiments of the invention, the nanotube arrays are formed by a method comprising attaching catalyst nanoparticles 140, 230 to polymer 120, 210 molecules, attaching the polymer 120, 210 molecules to a substrate, removing the polymer 120, 210 molecules and producing carbon nanotubes on the catalyst nanoparticles 140, 230. The polymer 120, 210 molecules can be attached to the substrate in ordered patterns, using self-assembly or molecular alignment techniques. The nanotube arrays can be attached to selected areas 110, 310 of the substrate. Within the selected areas 110, 310, the nanotubes are distributed non-randomly. Other embodiments disclosed herein concern apparatus that include ordered arrays of nanotubes attached to a substrate and systems that include ordered arrays of carbon nanotubes attached to a substrate, produced by the claimed methods. In certain embodiments, provided herein are methods for aligning a molecular wire, by ligating the molecular wire to a double stranded DNA molecule.
Owner:INTEL CORP
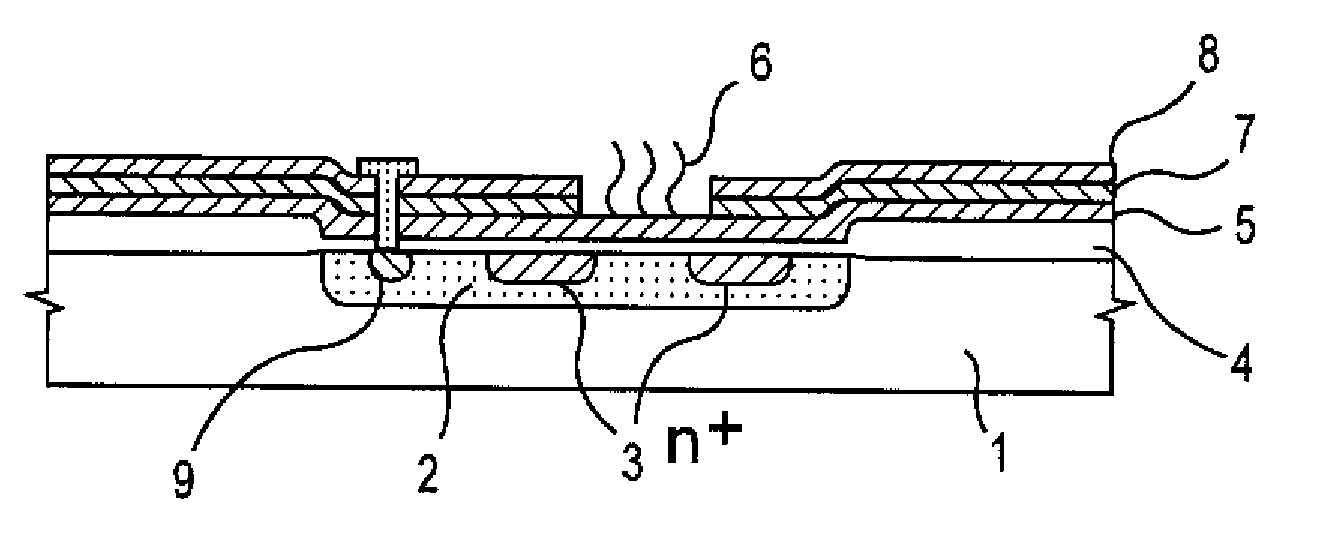
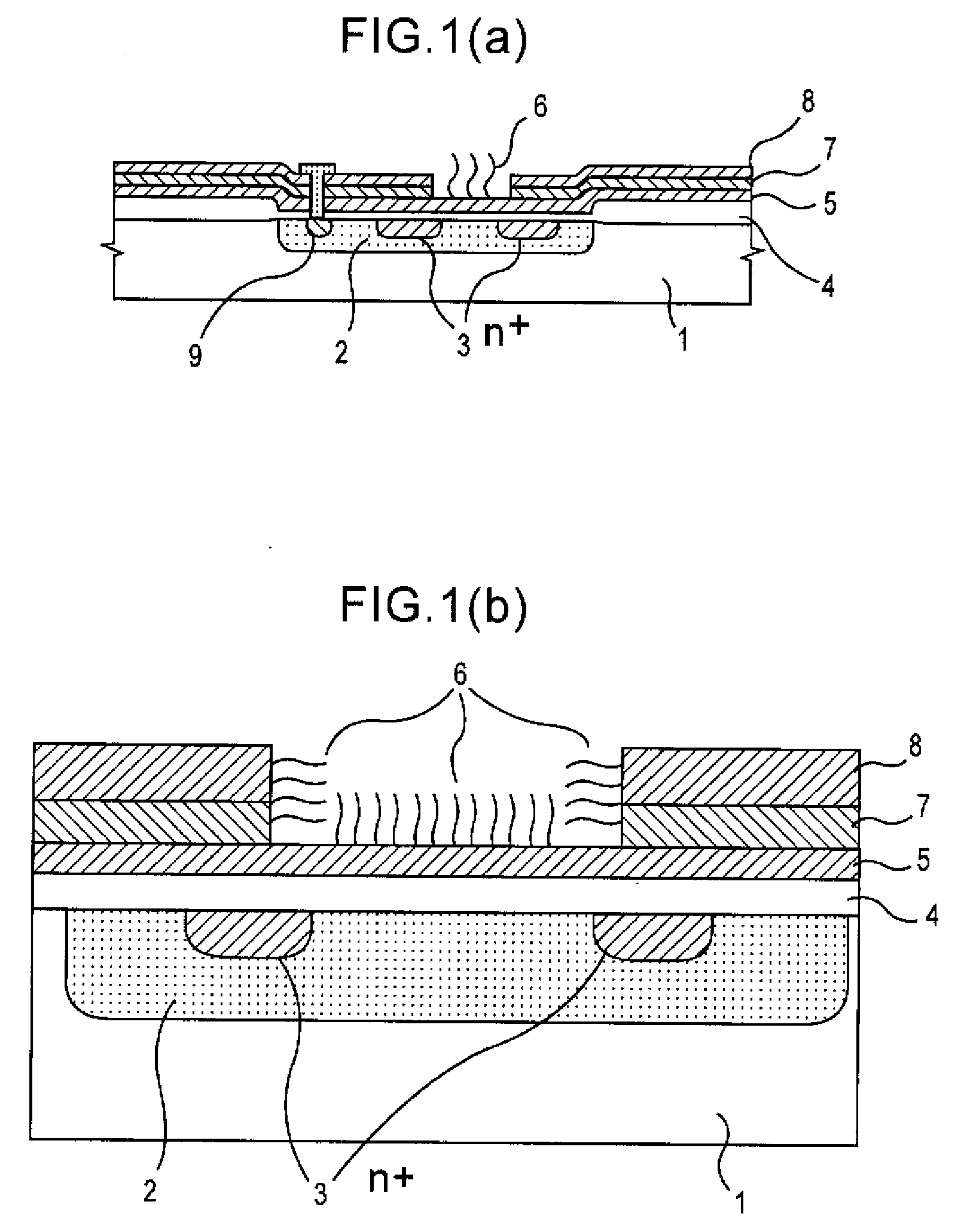
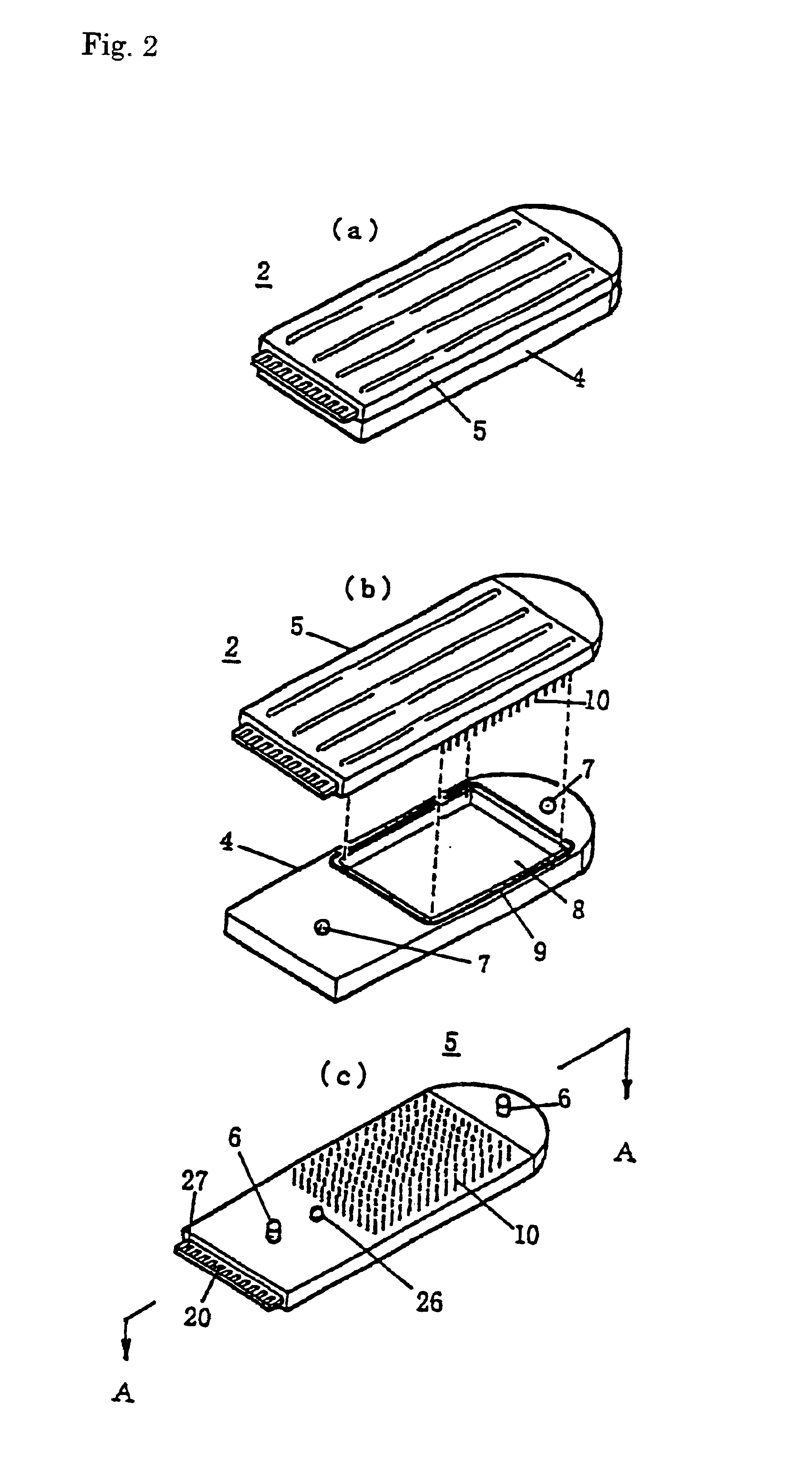
Gene detecting chip, detector, and detecting method
InactiveUS6916614B1Increase the areaImprove precisionImmobilised enzymesBioreactor/fermenter combinationsEngineeringOligonucleotide
A detecting chip (2) comprising a body (5) and a frame (4) both detachable from each other. A large number of projecting pin electrodes (10) are arranged in a matrix inside the body (5). Oligonucleotide consisting of different gene sequences is fixed to the pin electrode (10). A common electrode is so disposed in a recess (8) in a frame (4) as to be out of contact with the pin electrodes (10). A DNA sample is placed in the recess (8). By applying a voltage between the common electrode and the pin electrodes (10), a current is detected to detect a hybridized two-strand DNA, thus analyzing a gene DNA.
Owner:TOPPAN PRINTING CO LTD
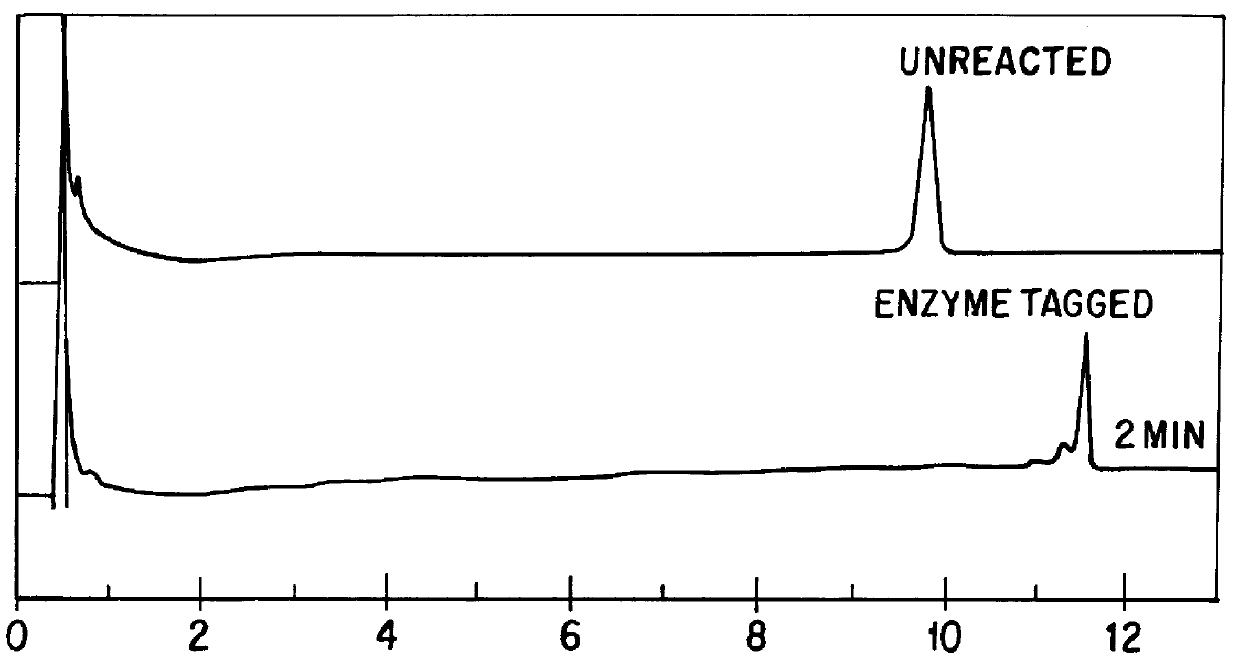
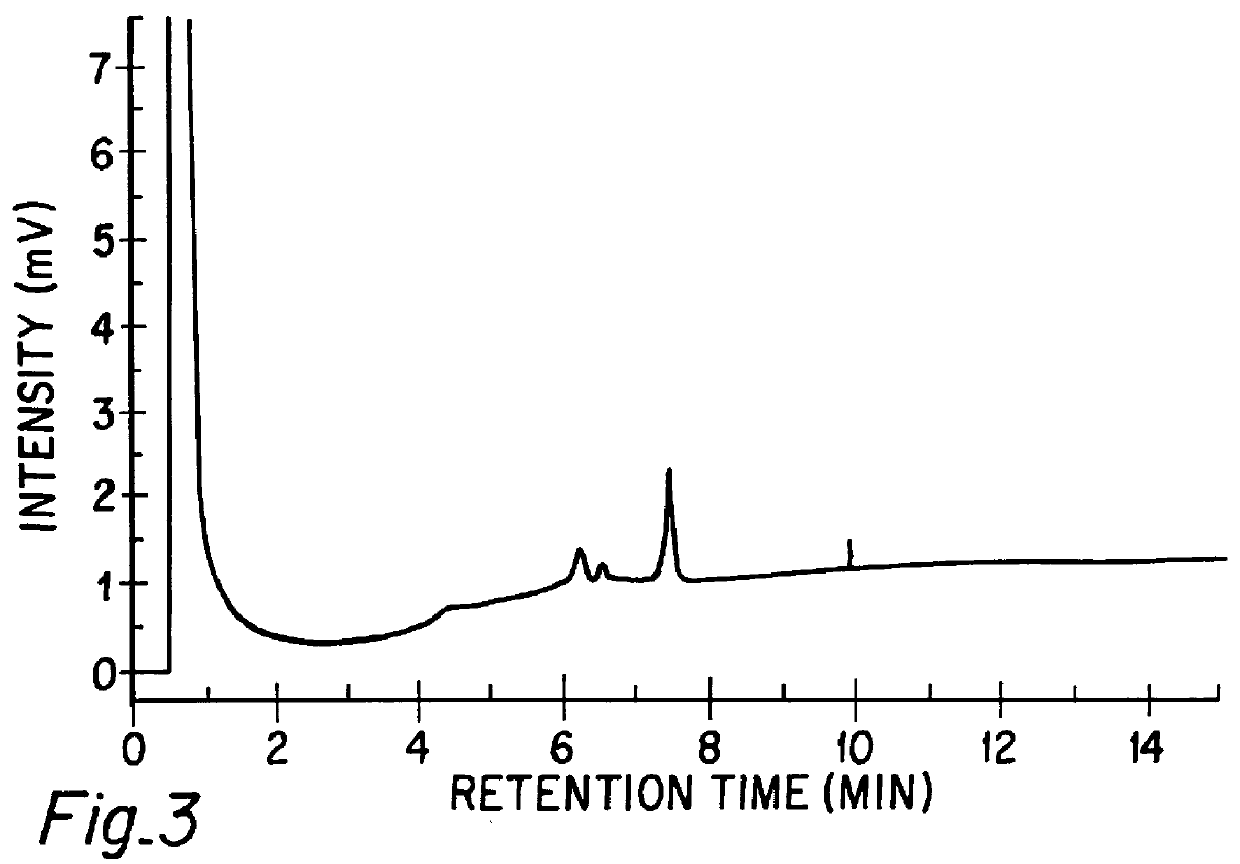
Chromatographic method for mutation detection using mutation site specifically acting enzymes and chemicals
InactiveUS6027898AHighly reproducibleEasy to implementSugar derivativesMicrobiological testing/measurementChromatographic separationRetention time
A method for analyzing a sample of double stranded DNA to determine the presence of a mutation therein comprises contacting the sample with a mutation site binding reagent, and chromatographically separating and detecting the product. The chromatographic separation can be performed using Matched Ion Polynucleotide Chromatography, size exclusion chromatography, ion exchange chromatography, or reverse phase chromatography. The mutation site binding reagent can be an enzyme or a non-proteinaceous chemical reagent. In one embodiment, a mutation site binding reagent binds to the site of mutation and alters the chromatographic retention time. In another embodiment, a mutation site binding reagent cleaves at the site of mutation, resulting in an increase in the number of fragments.
Owner:ADS BIOTEC INC
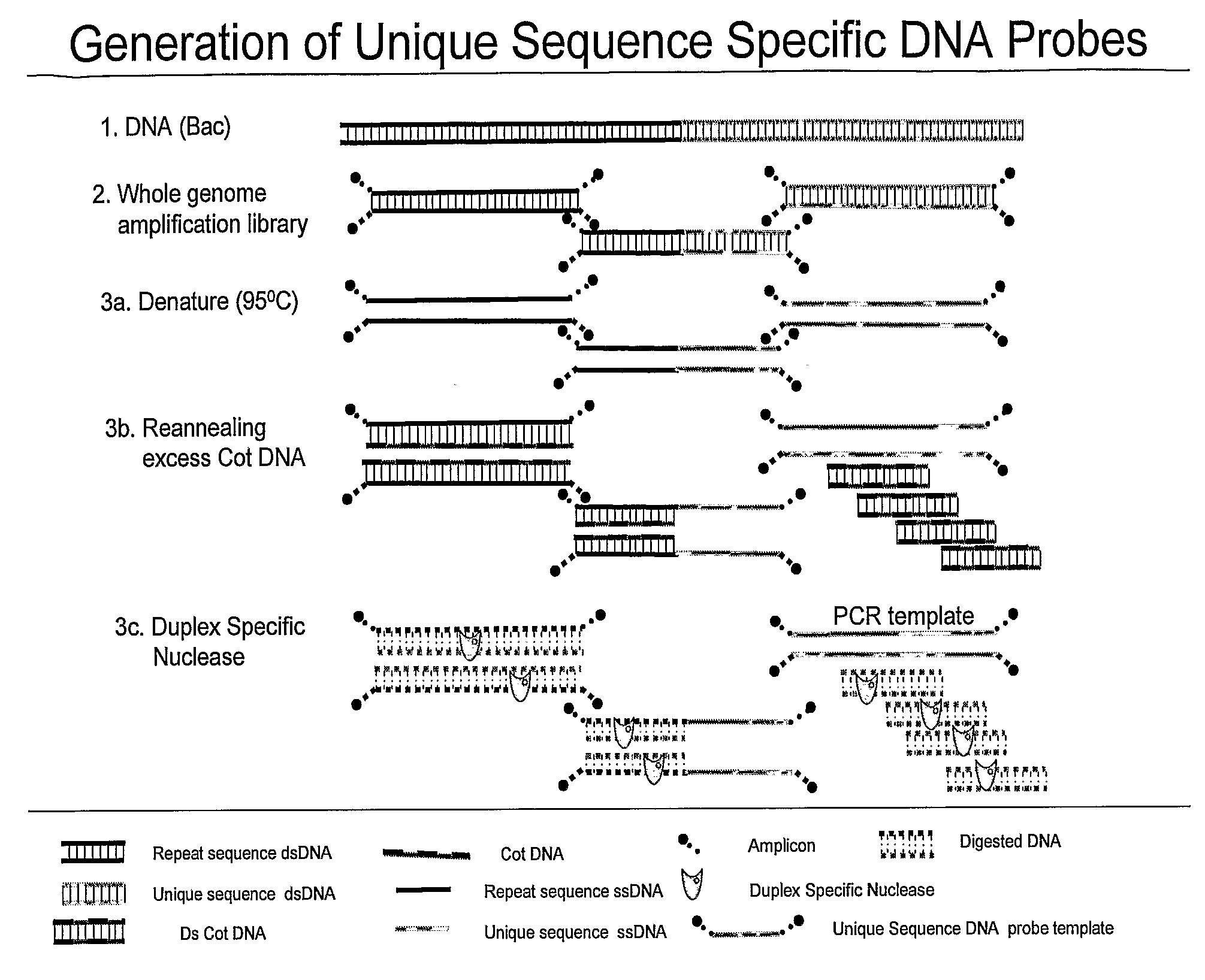
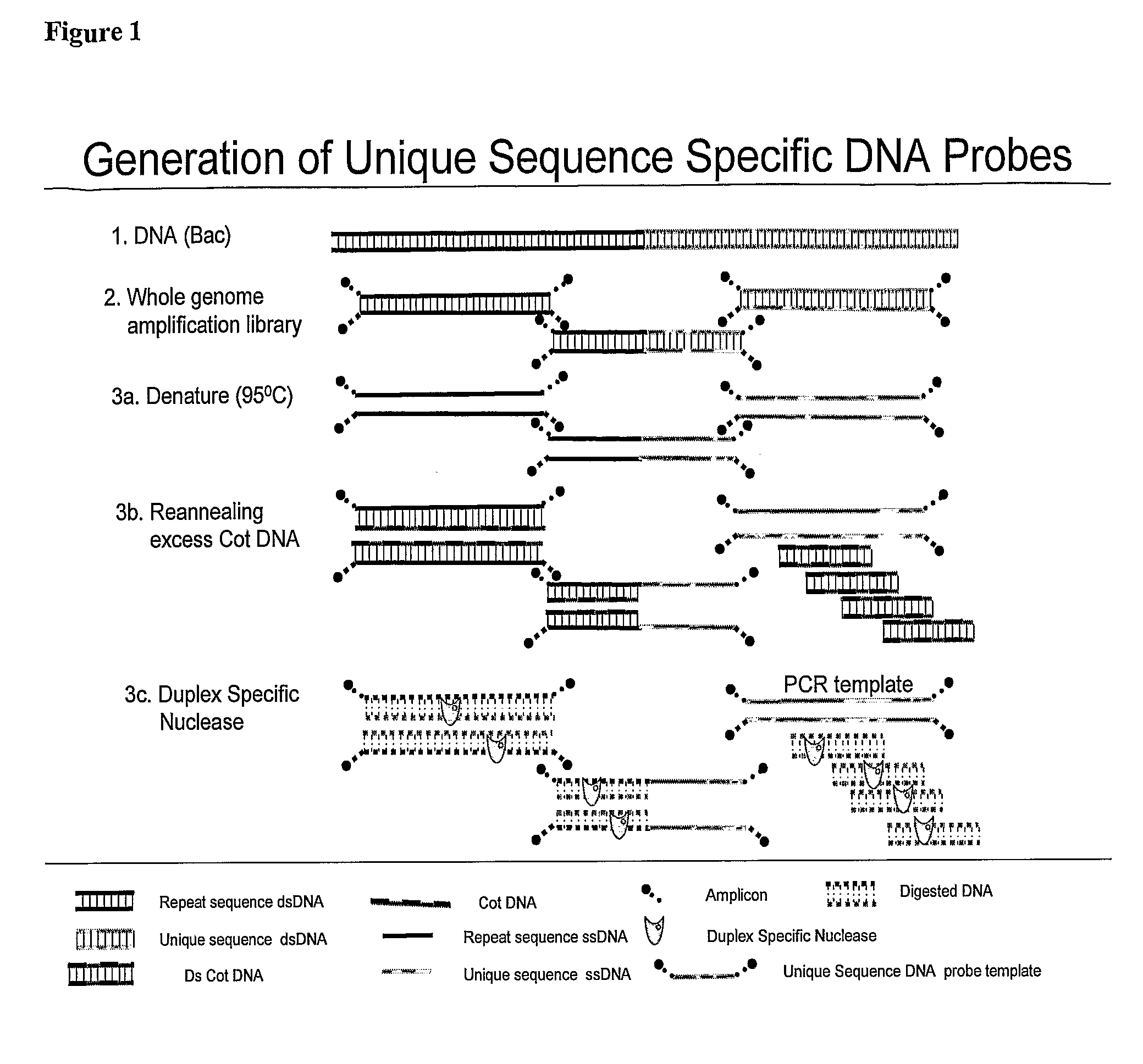
Methods and composition to generate unique sequence DNA probes labeling of DNA probes and the use of these probes
ActiveUS20090220955A1Strong specificityIncreased riskBioreactor/fermenter combinationsBiological substance pretreatmentsFluorescenceDouble stranded
The invention relates generally to the field of identification of DNA sequences, genes or chromosomes. Methods and composition to obtain Unique Sequence DNA probes are provided. Composition comprises of any double stranded DNA containing Unique Sequences from which the repetitive sequences are eliminated according to the method described in this invention. The invention also relates to the preservation of cells that have been identified after immunuomagnetic selection and fluorescent labeling in order to further interrogate the cells of interest. Furthermore the invention relates to genetic analysis of cells that have been identified after immunomagnetic selection and fluorescent labeling.
Owner:MENARINI SILICON BIOSYSTEMS SPA
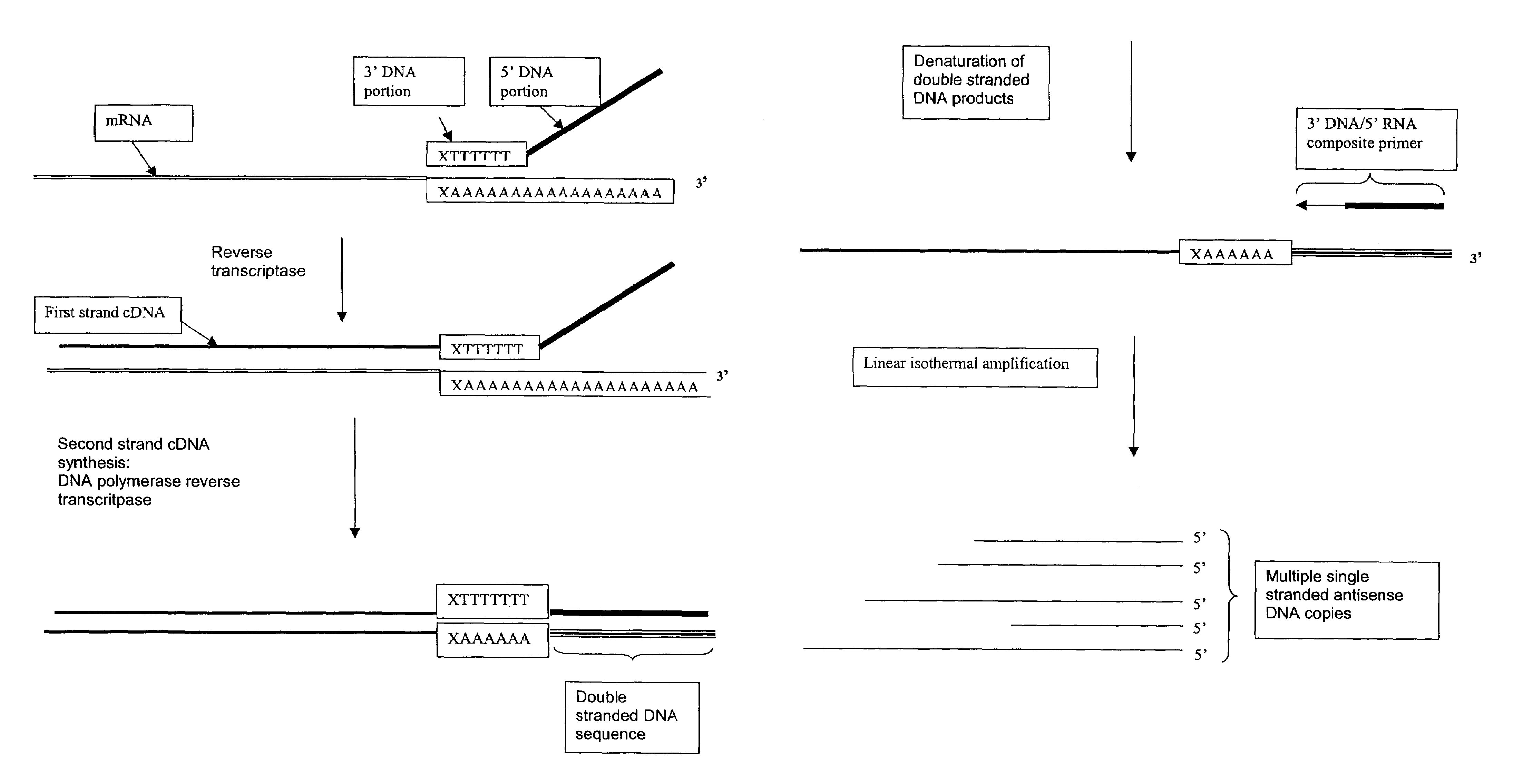
Amplification of RNA sequences using composite RNA-DNA primers and strand displacement
The invention provides methods for linear amplification of RNA that has an RNA sequence of interest. The methods are based on using a first DNA primer with a 3′ portion that is complementary to the RNA and a 5′ portion that is not complementary to the RNA in that region. The primer and an RNA-dependent DNA polymerase are used to make a DNA-RNA complex from the RNA. The RNA is cleaved from the complex with an enzyme, then a second primer and a DNA-dependent DNA polymerase are used to make double stranded DNA. The double stranded DNA is then denatured, and an RNA-DNA composite primer, a DNA-dependent DNA polymerase, and strand displacement are used to isothermally produce multiple copies of the complementary sequence to the RNA sequence of interest.
Owner:NUGEN TECH
Enhanced Adaptor Ligation
ActiveUS20170088887A1Simple methodAdapter ligation efficiency is restoredMicrobiological testing/measurementTransferasesDouble strandedDouble strand dna
Owner:INTEGRATED DNA TECHNOLOGIES
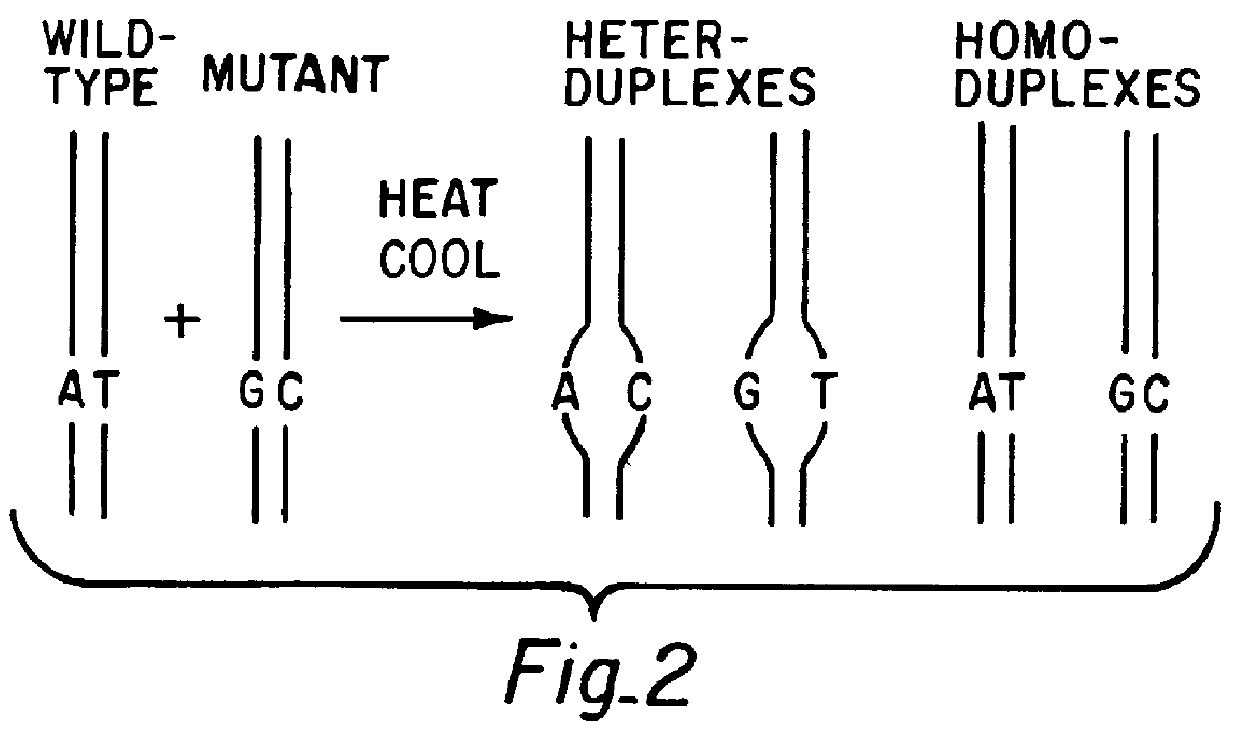
Eukaryotic use of improved chimeric mutational vectors
The invention is based on the reaction of recombinagenic oligonucleotides in a cell-free system containing a cytoplasmic cell extract and a test duplex DNA on a plasmid. The reaction specifically converts a mutant kanr gene to recover the resistant phenotype in transformed MutS, RecA deficient bacteria and allows for the rapid and quantitative comparison of recombinagenic oligonucleobases. Using this system a type of Duplex Mutational Vector termed a Heteroduplex Mutational Vector, was shown to be more active in than the types of mutational vectors heretofore tested. Further improvements in activity were obtained by replacement of a tetrathymidine linker by a nuclease resistant oligonucleotide, such as tetra-2'-O-methyl-uridine, to link the two strands of the Duplex Mutational Vector and removal of the DNA-containing intervening segment. The claims concern Duplex Mutational Vectors that contain the above improvements. In an alternative embodiment the claims concern a reaction mixture containing a recombinagenic oligonucleobase, a cell-free enzyme mixture and a duplex DNA containing a target sequence. In yet an alternative embodiment, the invention concerns the use of such mixture to test improvements in recombinagenic oligonucleobases, as well as to test the effects of compounds on the activity of the cell-free enzyme mixture and also to make specific changes in the target DNA sequence.
Owner:CIBUS
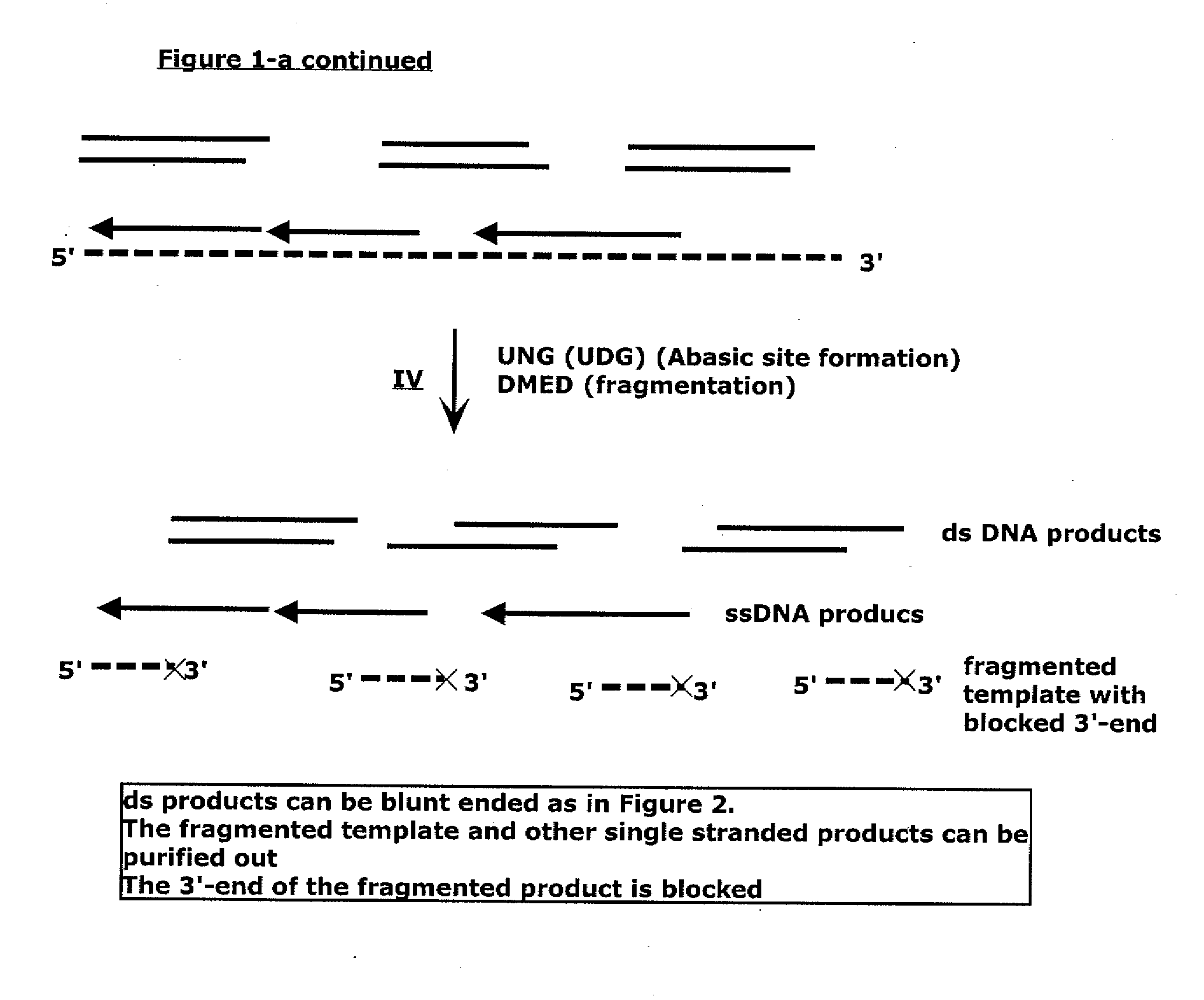
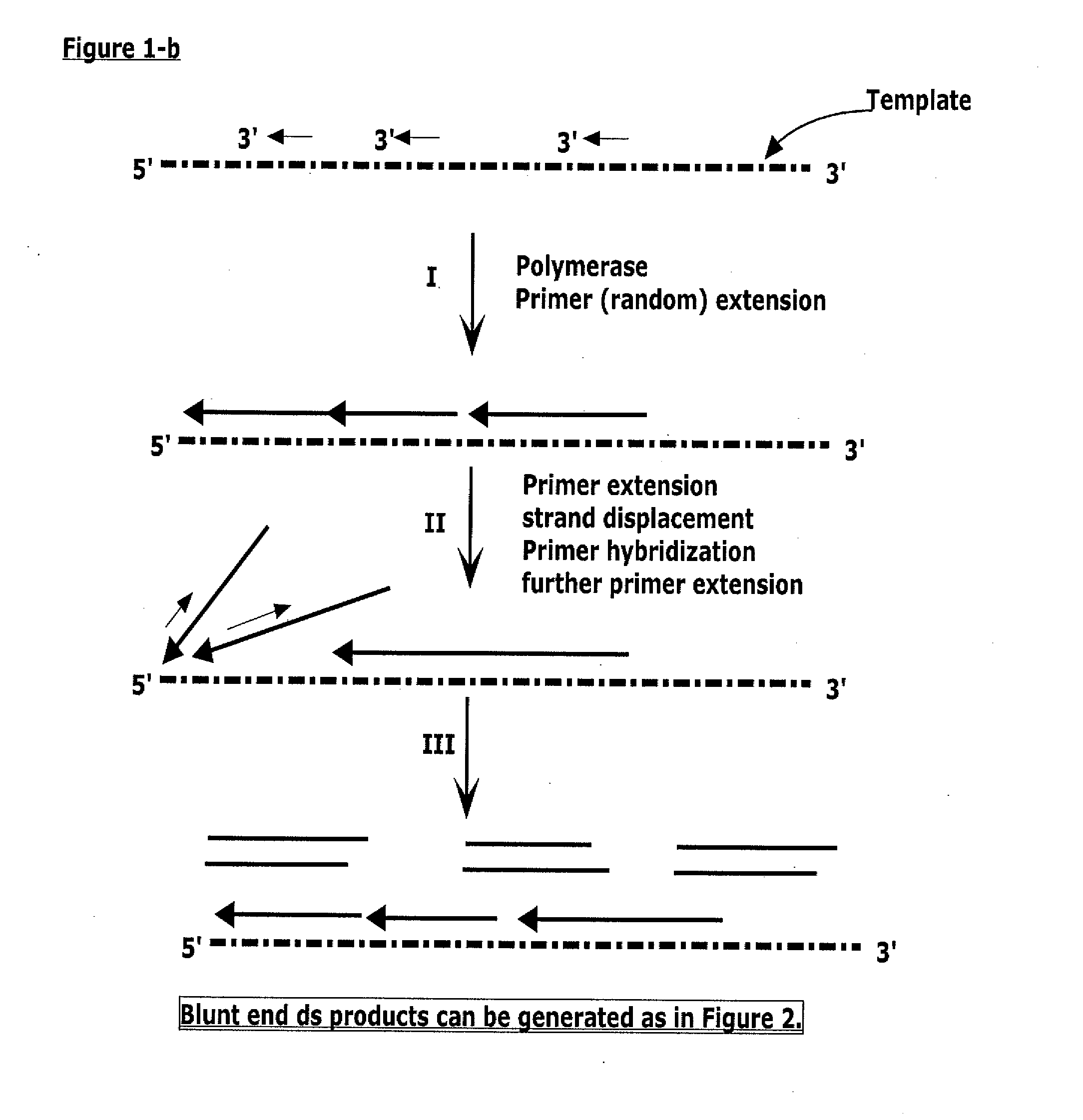
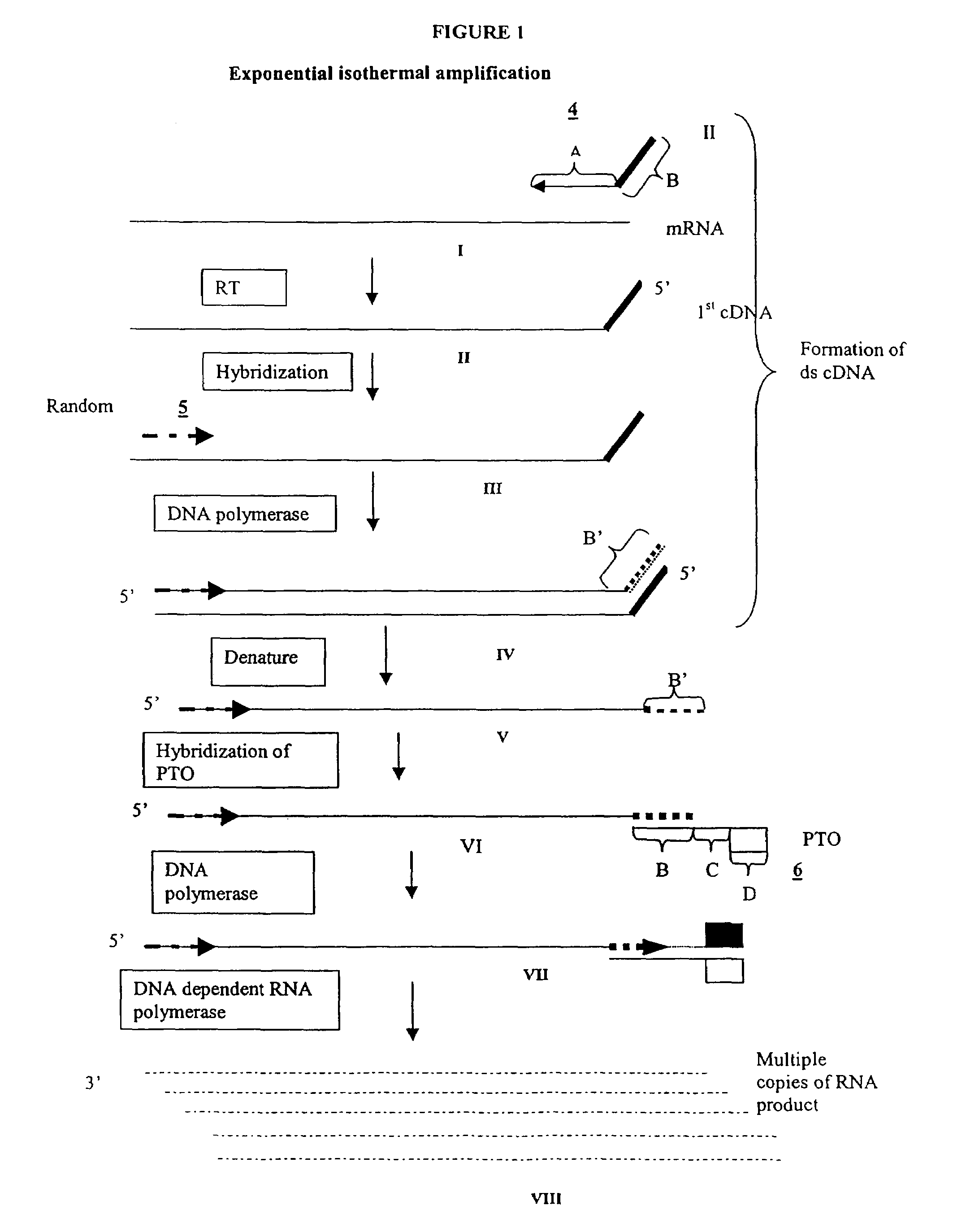
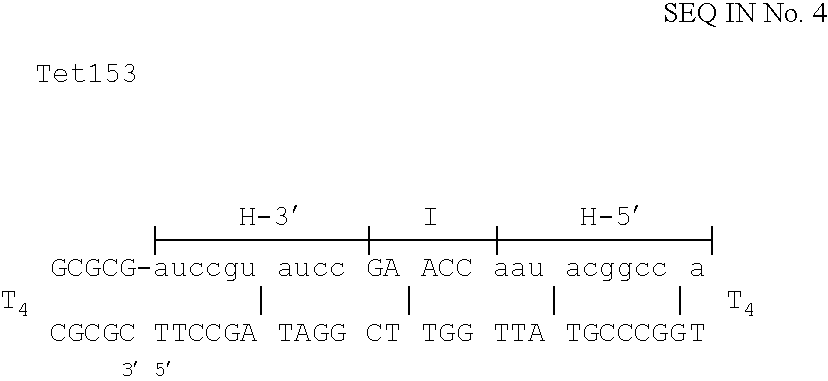
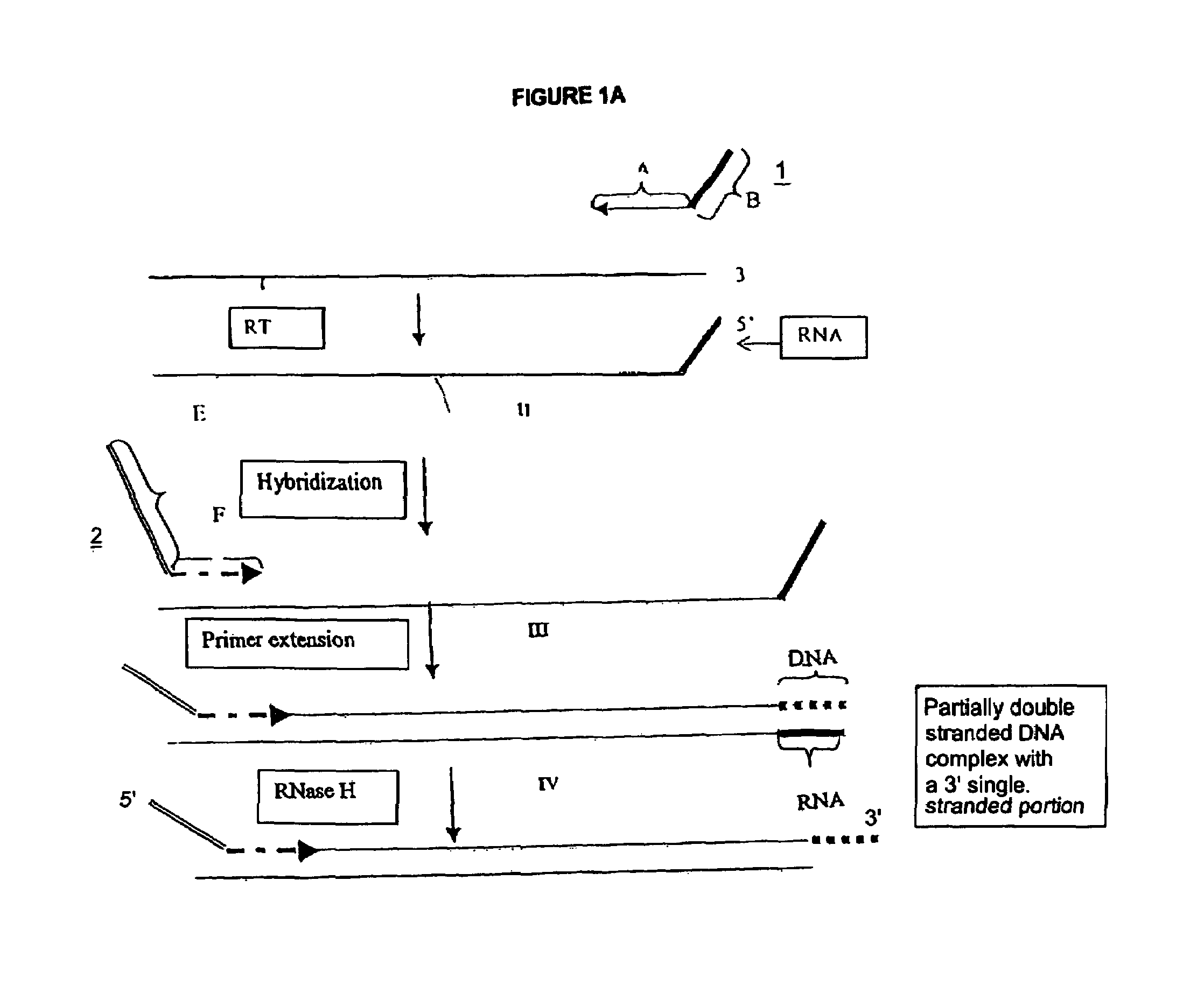
Methods for generating double stranded DNA comprising a 3' single stranded portion and uses of these complexes for recombination
Methods of recombining nucleic acids are provided. In particular, methods for the production of partially double stranded nucleic acids comprising a 3′ overhang from an RNA target and use in methods of recombining polynucleotides is described. These methods do not require thermocycling. The present invention also provides methods of recombining and selection which allow for identification of proteins comprising improved or desired characteristics.
Owner:NUGEN TECH
Methods for preparing nucleic acid samples useful for screening dna arrays
InactiveUS7432084B2High sensitivityStrong specificityMicrobiological testing/measurementLibrary member identificationEnzymatic synthesisImmobilized Nucleic Acids
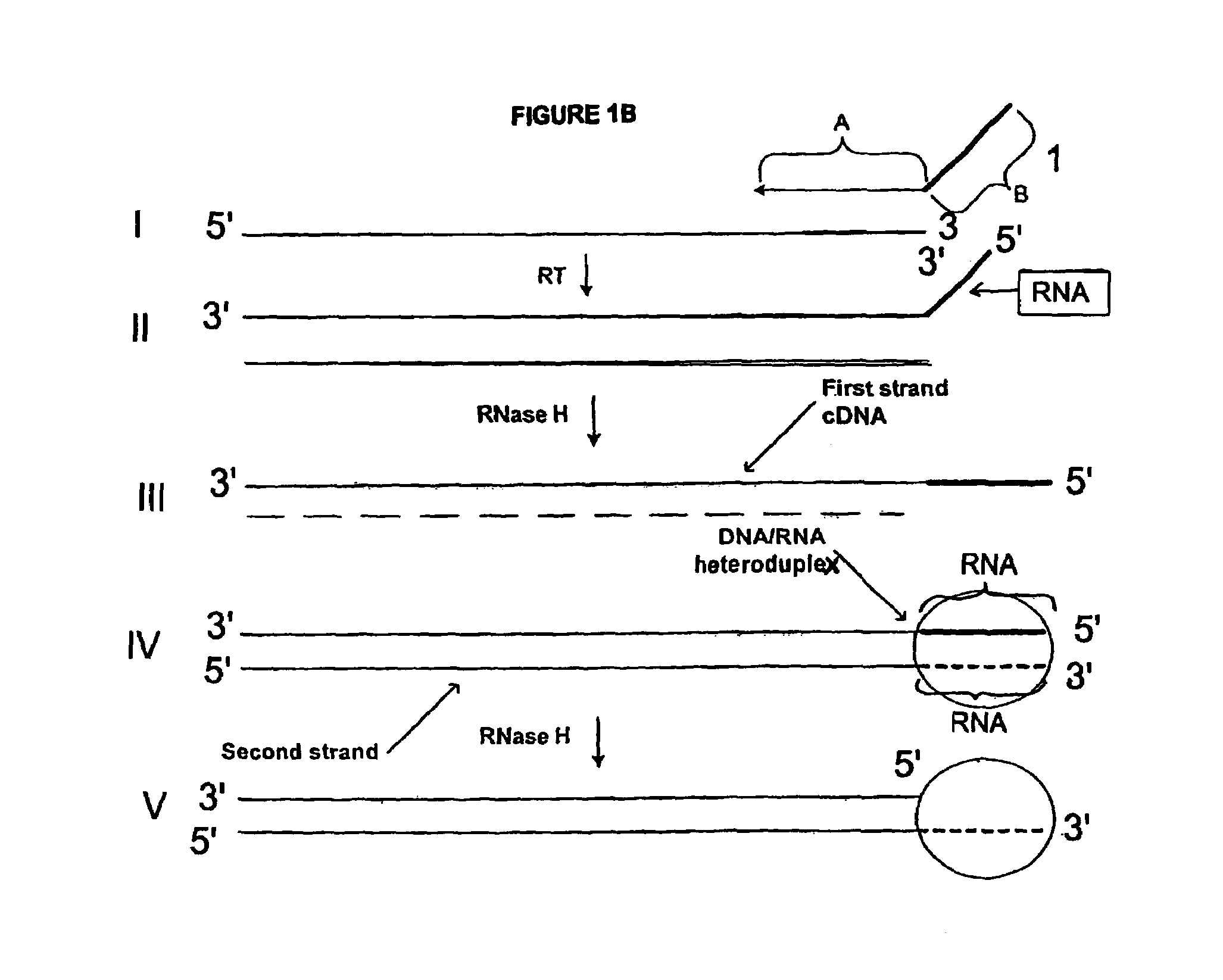
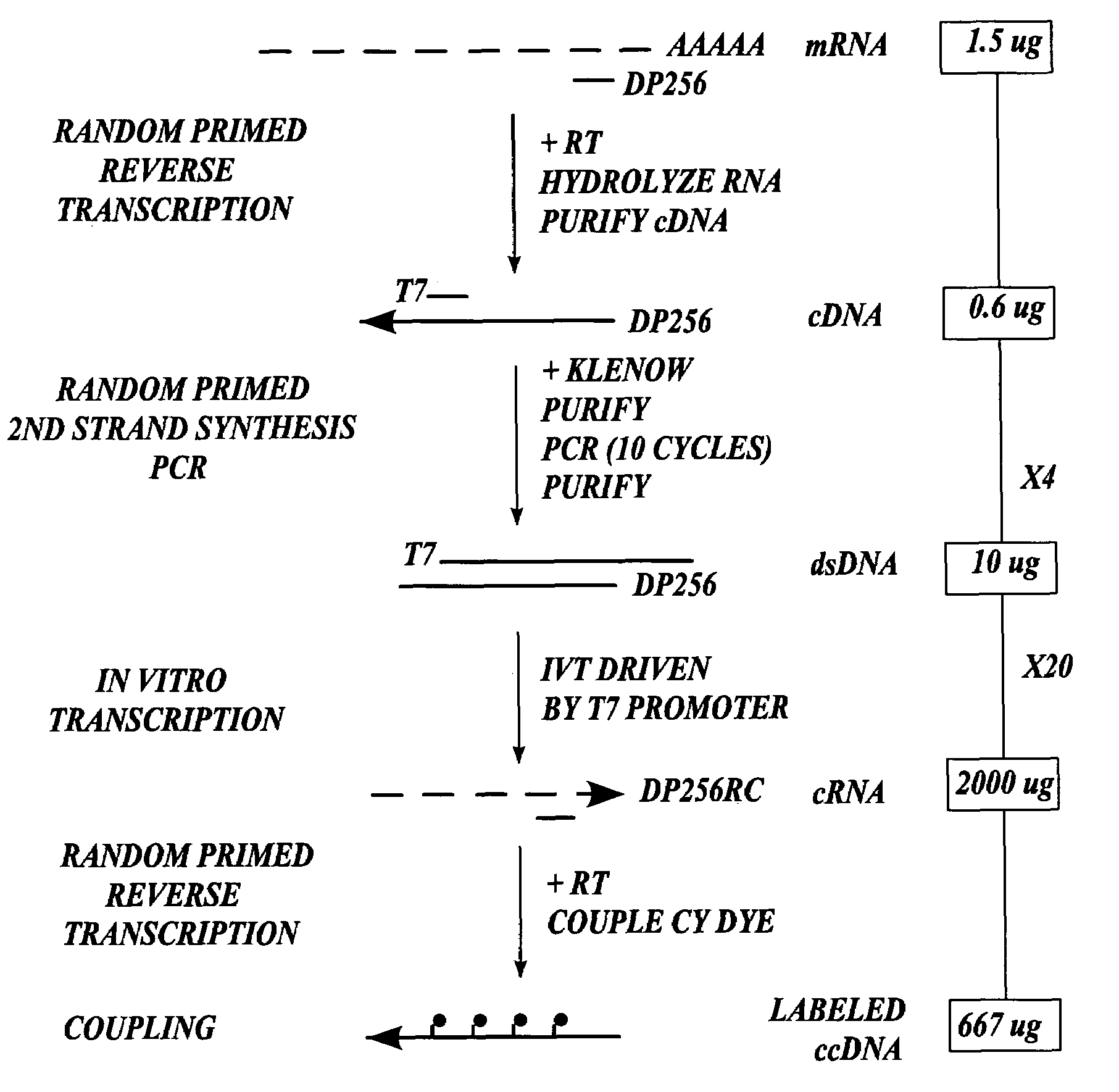
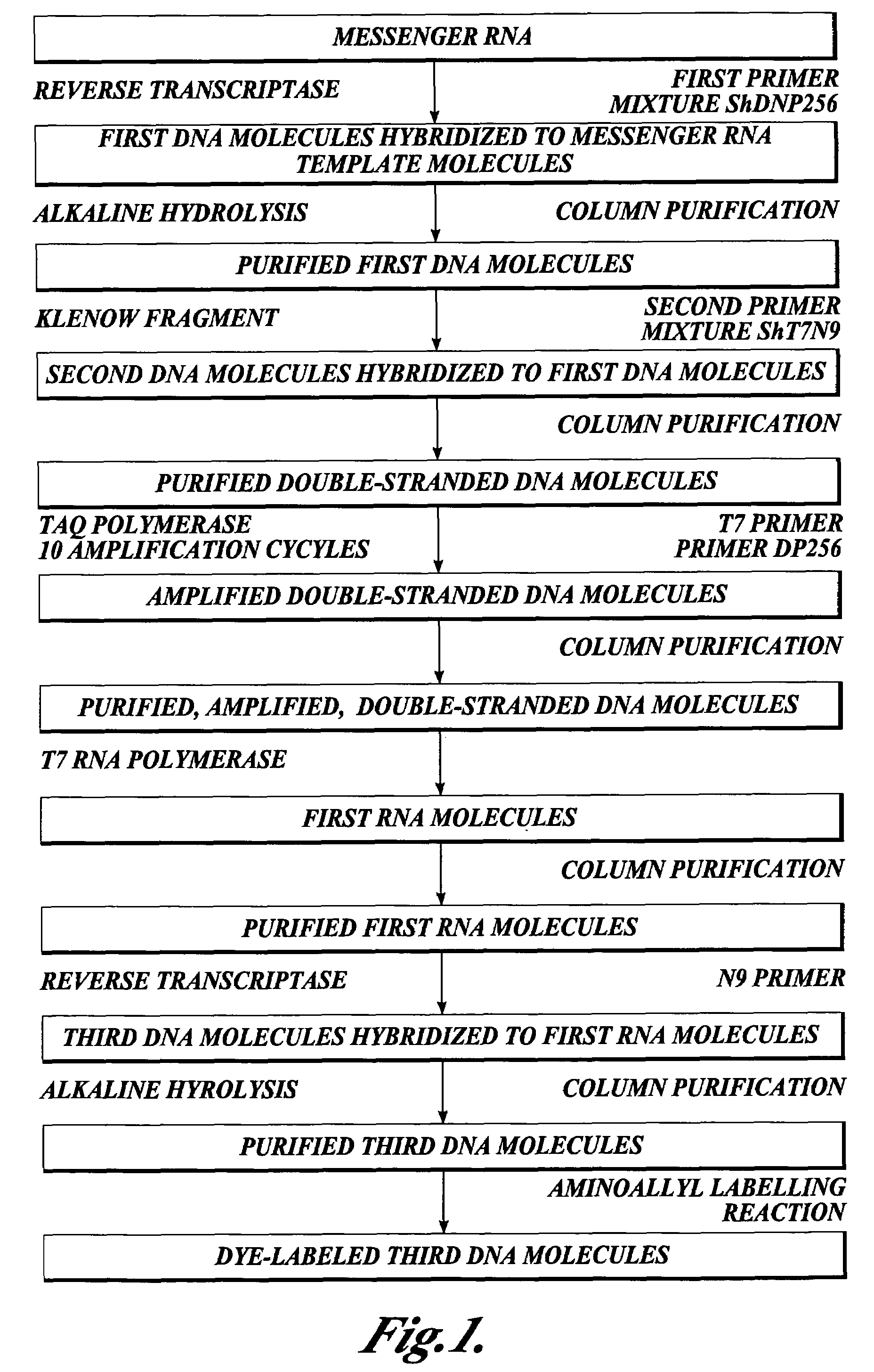
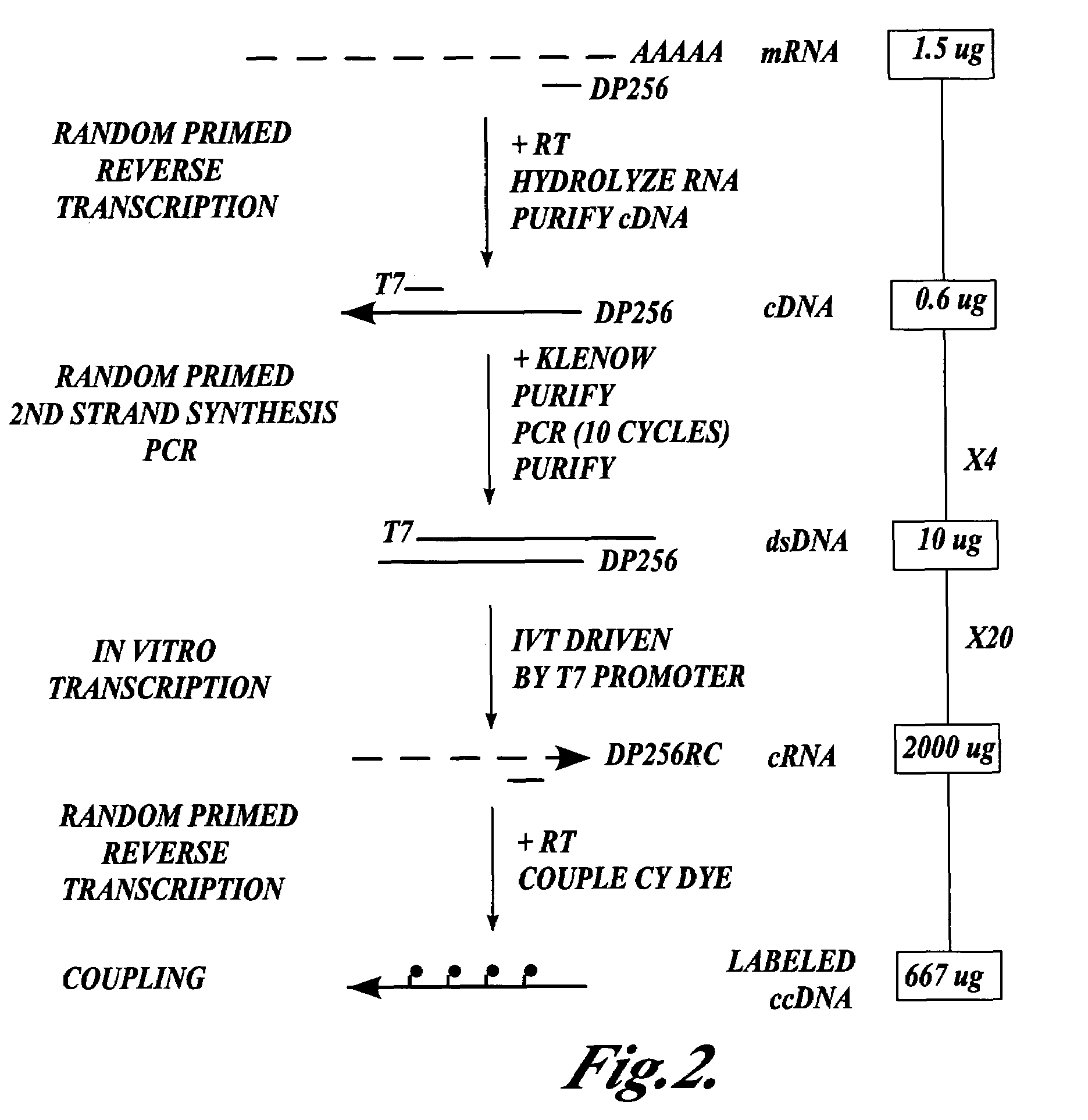
In one aspect the present invention provides methods of synthesizing a preparation of nucleic acid molecules, the methods comprising the steps of: (a) utilizing an RNA template to enzymatically synthesize a first DNA molecule that is complementary to at least 50 contiguous bases of the RNA template; (b) utilizing the first DNA molecule as a template to enzymatically synthesize a second DNA molecule, thereby forming a double-stranded DNA molecule wherein the first DNA molecule is hybridized to the second DNA molecule; (c) utilizing the first or second DNA molecule of the double-stranded DNA molecule as a template to enzymatically synthesize a first RNA molecule that is complementary to either the first DNA molecule or to the second DNA molecule; and (d) utilizing the first RNA molecule as a template to enzymatically synthesize a third DNA molecule that is complementary to the first RNA molecule. In another aspect, the present invention provides processed DNA samples prepared by a method of the invention for synthesizing a preparation of nucleic acid molecules. In another aspect, the present invention provides methods for hybridizing a processed DNA sample to a population of immobilized nucleic acid molecules.
Owner:LIFE TECH CORP
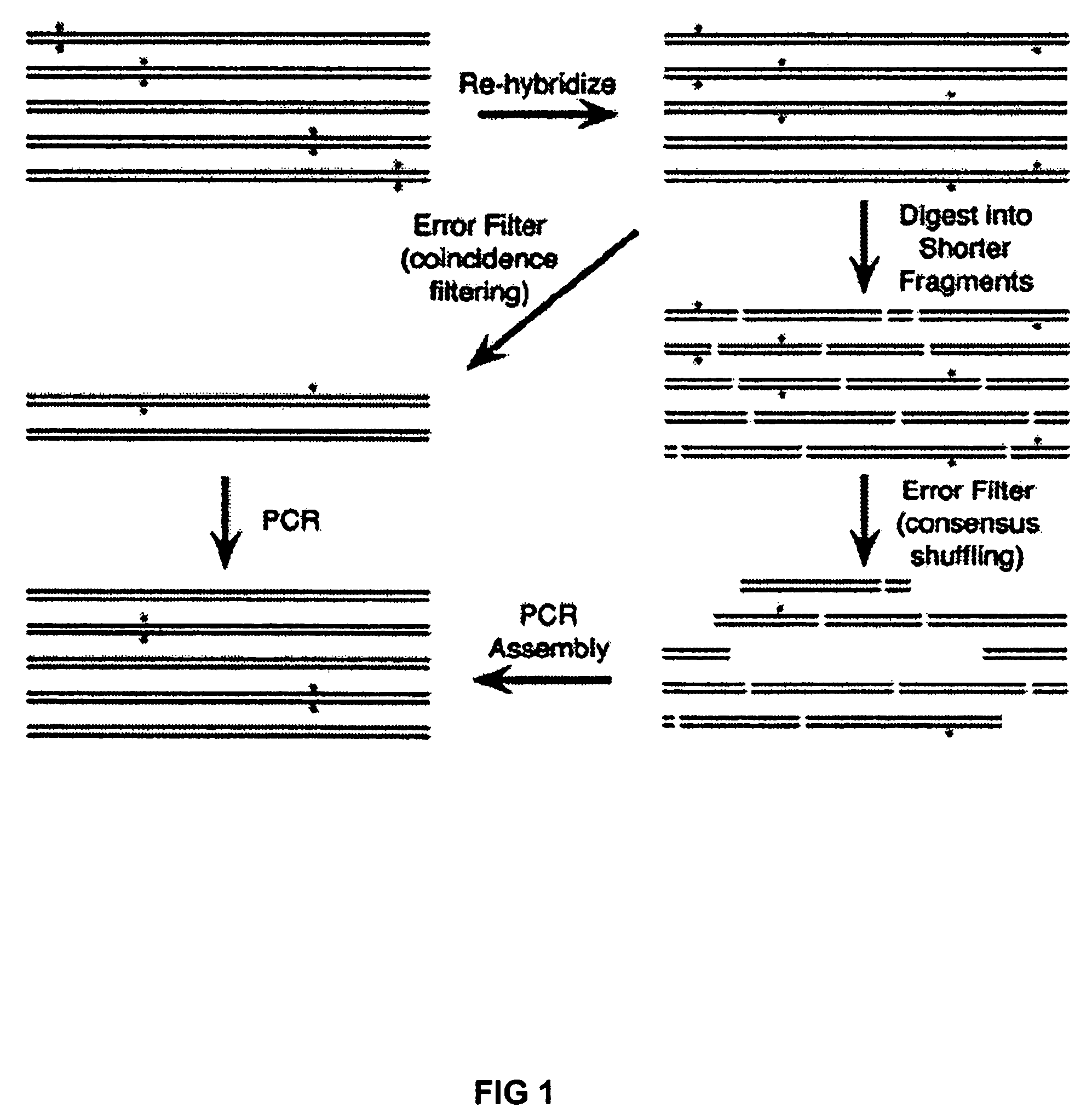
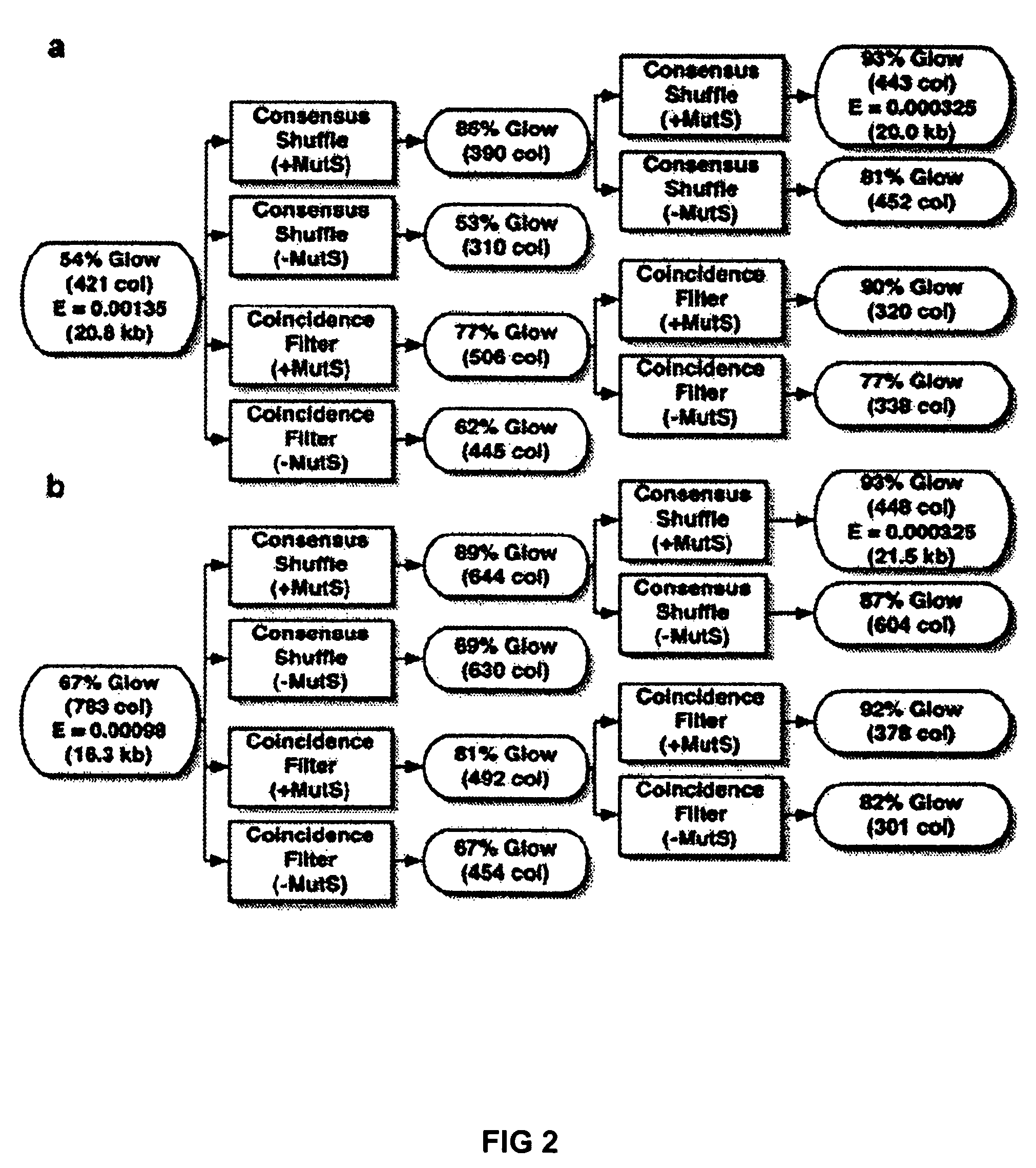
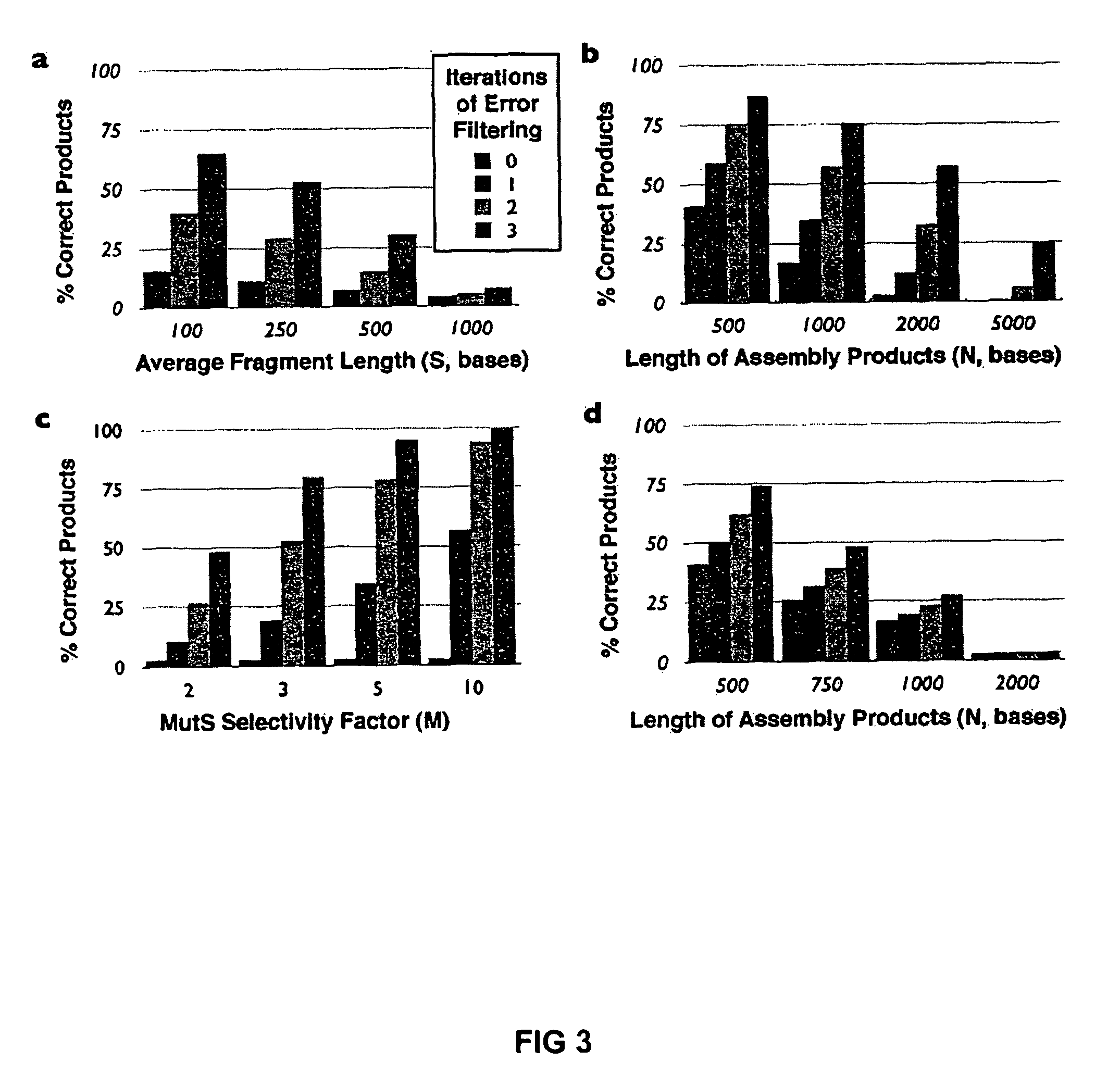
Method of error reduction in nucleic acid populations
A method is disclosed for the direct synthesis of double stranded DNA molecules of a variety of sizes and with any desired sequence. The DNA molecule to be synthesis is logically broken up into smaller overlapping DNA segments. A maskless microarray synthesizer is used to make a DNA microarray on a substrate in which each element or feature of the array is populated by DNA of a one of the overlapping DNA segments. The complement of each segment is also made in the microarray. The DNA segments are released from the substrate and held under conditions favoring hybridization of DNA, under which conditions the segments will hybridize to form duplexes. The duplexes are then separated using a DNA binding agent which binds to improperly formed DNA helixes to remove errors from the set of DNA molecules. The segments can then be hybridized to each other to assemble the larger target DNA sequence.
Owner:WISCONSIN ALUMNI RES FOUND
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com