SNP sites related to number of pig nipples and detection method and application thereof
A locus and nipple technology, applied in the field of molecular genetics, can solve the problems of inability to achieve large-scale sequencing, limited labeling density, limited application, etc., and achieve excellent economic value, increase the number of nipples, and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
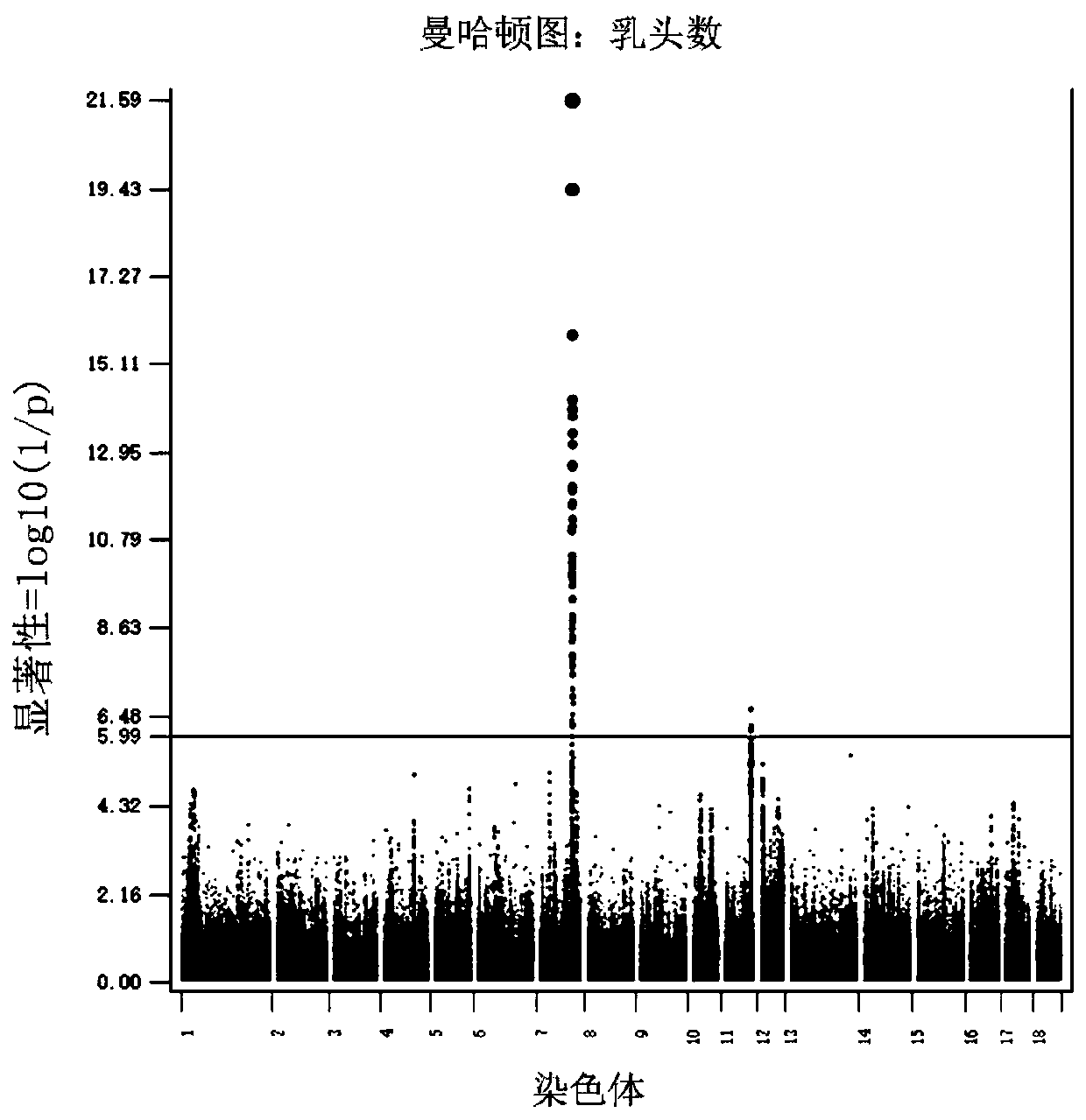
[0033] Example 1 Genome-wide association analysis of pig teat number traits
[0034] The performance records of 33,960 individuals (12,987 boars and 20,973 sows) born between August 2007 and January 2016 were collected for this study.
[0035] 2. Test method
[0036] 2.1 Teat number phenotype: the number of left teats, right teats and total teats of each pig were recorded within one week after the individual was born.
[0037] 2.2 Tn5 transposase library construction:
[0038] (1) The genome was uniformly diluted to 40ng / μl, and 50ng of each sample was taken as the initial amount. Tn5 proenzyme and specific Tn5ME-A / Tn5Merev and Tn5ME-B / Tn5MErev linkers were embedded at 72°C for 2h to obtain Tn5 working enzyme with cut-paste activity. The linker structure is the same as the Tn5ME-A, Tn5ME-B, and Tn5Merev structures disclosed in the document "Tn5 transposase and tagmentation procedures formatively scaled sequencing projects".
[0039] (2) Dilute Tn5 working enzyme to 16.5ng / ...
Embodiment 2
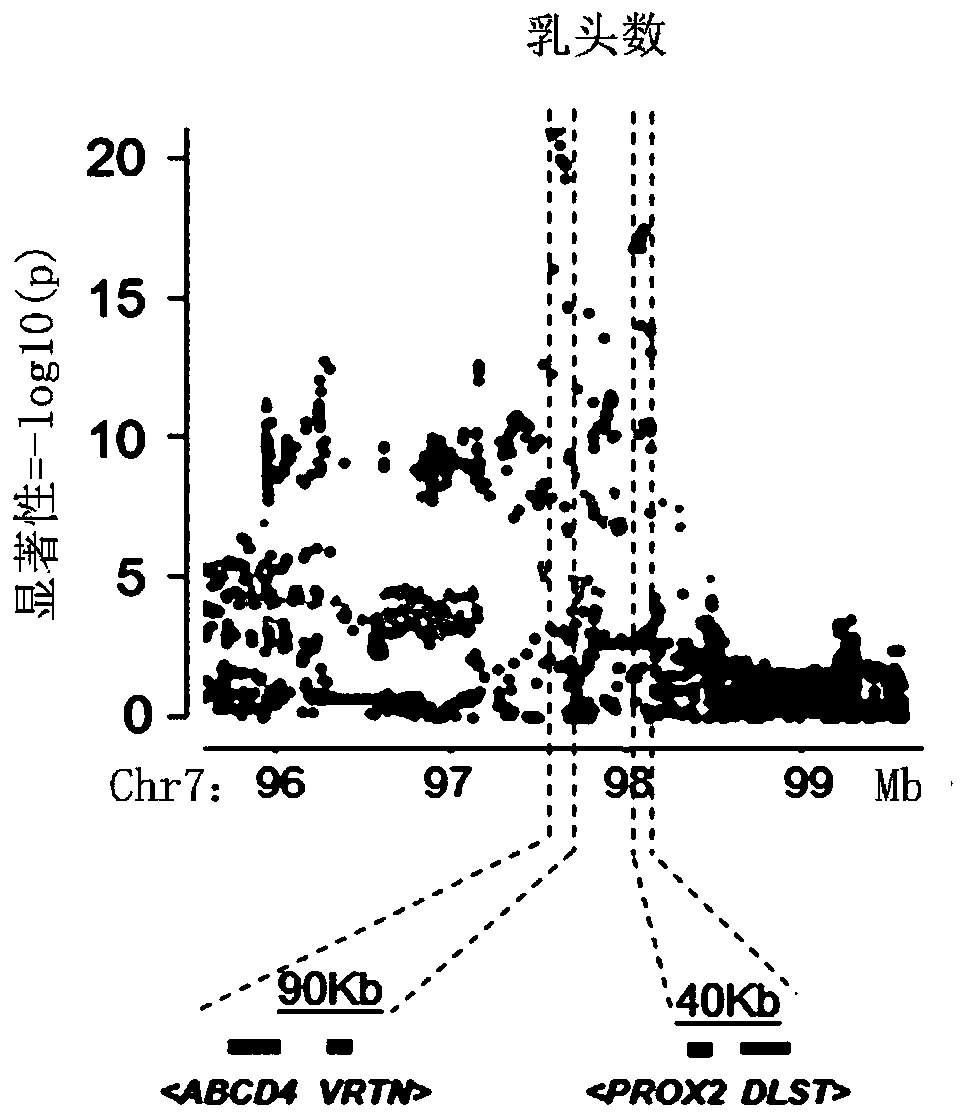
[0051] Example 2: Breeding application of SNP locus (SSC7:97570528) in pig teat number traits
[0052] 1. Test material
[0053] A population of Duroc purebred pigs with the teat number trait.
[0054] 2. Test method
[0055] Association analysis of missense mutations and teat number traits: 7 missense mutation loci were genotyped, and association analysis was performed between genotypes and teat number traits of Duroc pigs.
[0056] Table 2 shows the results of the association analysis between the genotypes of the 7 missense mutation sites and the number of papillae traits. SSC7:97570528 site, AA is the dominant genotype. SSC7: 97574214 site, CC is the dominant genotype; SSC7: 98073927 site, AA is the dominant genotype; SSC7: 98074438 site, TT is the dominant genotype; SSC7: 98074524 site, TT is the dominant genotype; SSC7: At site 98074744, CC is the dominant genotype; at site SSC7:98101170, GG is the dominant genotype.
[0057] Table 2 - Least squares mean and standard...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com