Database establishing method suitable for plant transposase accessible chromatin analysis
A technology of transposase and proximity, applied in the field of molecular biology, can solve the problem of reducing the activity of Tn5 transposase and achieve the effect of reducing interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0029] A method for building a library suitable for plant transposase accessible chromatin analysis, comprising the following steps:
[0030] S1: After the plant tissue sample is treated with enzymatic solution TS2 and lysate TS1, cell nucleus suspension A is obtained;
[0031] S2: Staining and sorting the nuclei in the nuclei suspension A to obtain the nuclei B;
[0032] S3: Perform transposition reaction and purification on cell nucleus B to obtain transposition product C;
[0033] S4: performing PCR amplification on the transposition product C to obtain the amplified product D;
[0034] S5: Take part of the amplified product D and perform qPCR to determine the additional required number of PCR cycles, and continue to amplify the remaining amplified product D according to the determined additional required number of PCR cycles to obtain an amplified product E;
[0035]S6: Purify the amplified product E to obtain a single library, and perform fragment sorting on the single ...
Embodiment 1
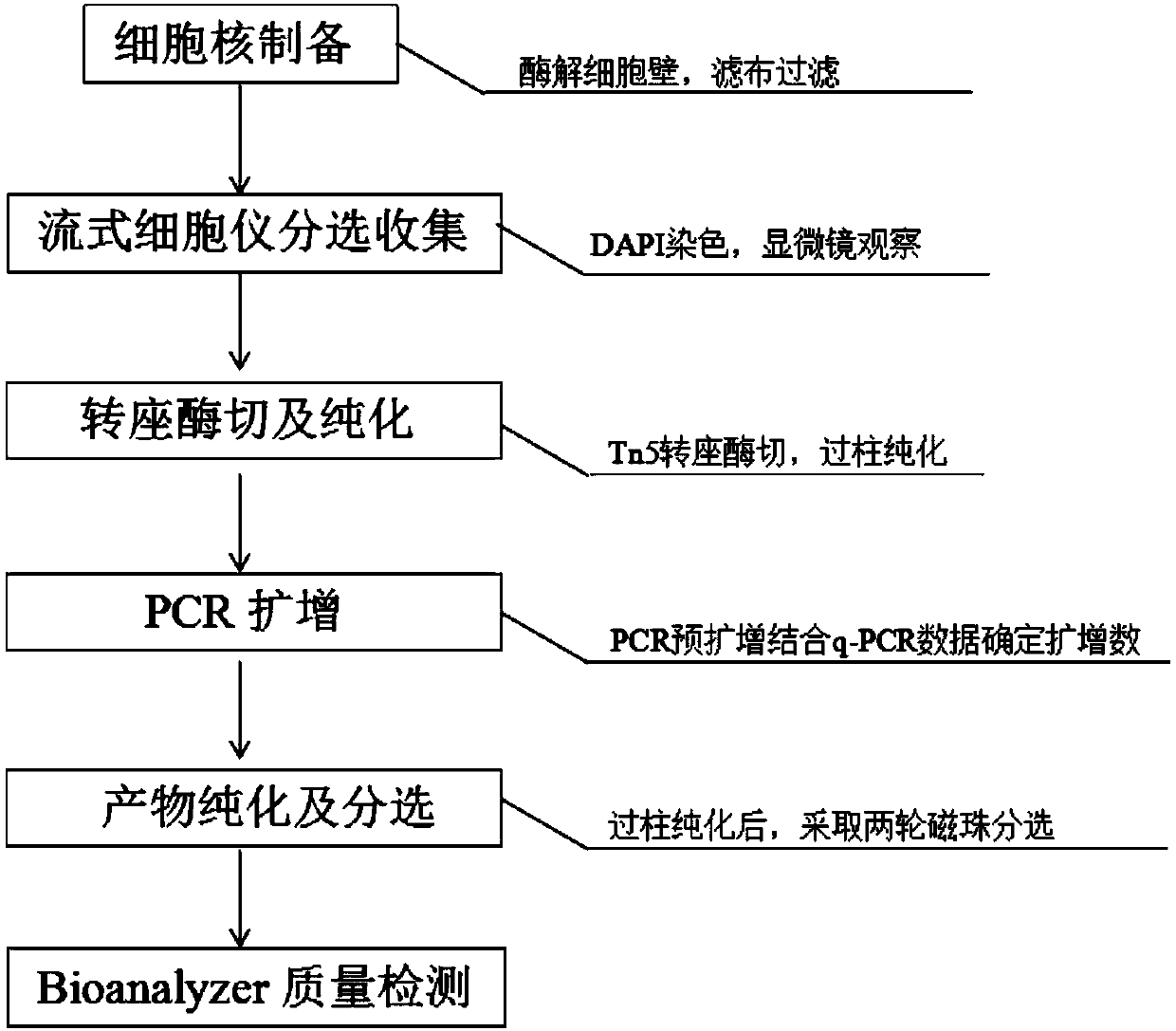
[0045] A library construction method suitable for the analysis of accessible chromatin of peony flower leaf transposase, refer to figure 1 The flow chart, the specific implementation steps are as follows:
[0046] (1) Separation of nuclei.
[0047] a. Take by weighing 200mg peony flowers and leaves.
[0048] b. Mince the tissue into a homogenate with a sharp blade in a Petri dish placed on ice.
[0049] c. Add 2.5mL TS2 enzymatic hydrolysis solution for 4h to remove the cell wall. The preparation of TS2 enzymolysis solution is: 0.5% cellulase, 0.4% pectinase, 0.3% hemicellulase, 0.5M mannitol, 20mmol / L KCl, 10mmol / L MES (2-(N-morpholine) Ethylsulfonic acid monohydrate, pH 5.7) the mixture was placed in a water bath at 56°C for 10 min, and after cooling to room temperature, 10 mmol / LCaCl was added 2 , 0.1% BSA, and filtered with a 0.45 μm filter.
[0050] d. After enzymatic hydrolysis, centrifuge at 4°C and 400rpm for 10min, and discard the supernatant.
[0051] e. Add 2....
Embodiment 2
[0153] Example 2: A library construction method suitable for tobacco leaf transposase accessible chromatin analysis, refer to figure 1 The flow chart, the specific implementation steps are as follows:
[0154] (1) Separation of nuclei.
[0155] a. Weigh 200 mg of tobacco leaves.
[0156] b. Mince the tissue into a homogenate with a sharp blade in a Petri dish placed on ice.
[0157] c. Add 2.5mL TS2 enzymatic hydrolysis solution for 4h to remove the cell wall. The preparation of TS2 enzymatic hydrolysis solution is: 1.3% cellulase, 0.4% isolated enzyme, 0.3% pectinase, 0.4M mannitol (pH 5.7) mixed solution was placed in 56 ° C water bath for 10 minutes, after cooling to room temperature, 10 mmol / L CaCl 2 , 0.1% BSA, and filtered with a 0.45 μm filter.
[0158] d. After enzymatic hydrolysis, centrifuge at 4°C and 400 rpm for 10 min, and discard the supernatant.
[0159] e. Add 2.5mL pre-cooled TS1 lysate to the precipitate, pipette repeatedly to resuspend, then transfer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com