DNA barcode compositions and methods of in situ identification in a microfluidic device
A barcode, object technology, applied in the fields of biochemical equipment and methods, DNA preparation, chemical instruments and methods, etc., can solve the problems of difficulty and impossibility of specific phenotype association
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0467] Example 1. RNA capture, sequencing library preparation and sequencing results as demonstrated in OKT3 cells.
[0468] Cells: OKT3 cells, a murine myeloma hybridoma cell line, were obtained from ATCC ( Catalog Number CRL-8001 TM ). Cells are provided as suspension cell lines. By inoculating about 1 x 10 5 to about 2×10 5 viable cells / mL and maintain the culture by incubating at 37°C with 5% carbon dioxide in air as the atmosphere. Flask cells every 2-3 days. Count the OKT3 cell number and viability, and adjust the cell density to 5 × 10 5 / ml to load into the microfluidic device.
[0469] Culture medium: 1000ml Iskoff's modified medium ( Cat. No. 30-2005), 200ml fetal bovine serum ( Cat. No. 30-2020) and 10ml penicillin-streptomycin ( Cat. No. 15140-122) were combined to prepare media. Filter the complete medium through a 0.22 μm filter and store at 4 °C in the dark until use.
[0470] When perfusing during incubation, the medium was continuously conditio...
Embodiment 2
[0481] Example 2. T cell phenotyping, culture, measurement and RNA sequencing. Association of phenotypes with genomic information.
[0482] The microfluidic system, materials, and methods were the same as in Experiment 1, with the following differences:
[0483] Cells: Control cells were human peripheral blood T cells. Sample cells are human T cells derived from a human tumor sample.
[0484] Medium: RPMI 1640 medium (Gibco, #12633-012), 10% fetal bovine serum (FBS), (Seradigm, #1500-500); 2% human AB serum (Zen-bio, #HSER-ABP100ml), IL-2 (R&D Systems, 202-IL-010) 2U / ml; IL-7 (PeproTech, #200-07) 10ng / ml; IL-15 {PeproTech, #200-15) 10ng / ml, IX Pluronic F -127 (Life Tech Cat. No. 50-310-494).
[0485] Human T cells derived from human tumor samples were stained off-chip with antigen and then introduced into the microfluidic channels of the OptoSelect device at a density of 5×E6 cells / ml. Antigen-positive T cells (P-Ag) and antigen-negative cells (N-Ag) were moved by light-d...
Embodiment 3
[0493] Example 3. DNA capture, sequencing library preparation and sequencing results as demonstrated in OKT3 cells. Unless otherwise noted in this example, equipment, priming and perfusion protocols, cell sources and preparation were used / performed as in the general methods above. Media and OptoSelect devices were maintained at 37°C unless otherwise stated.
[0494] Table 4. Primers used in this experiment.
[0495]
[0496]
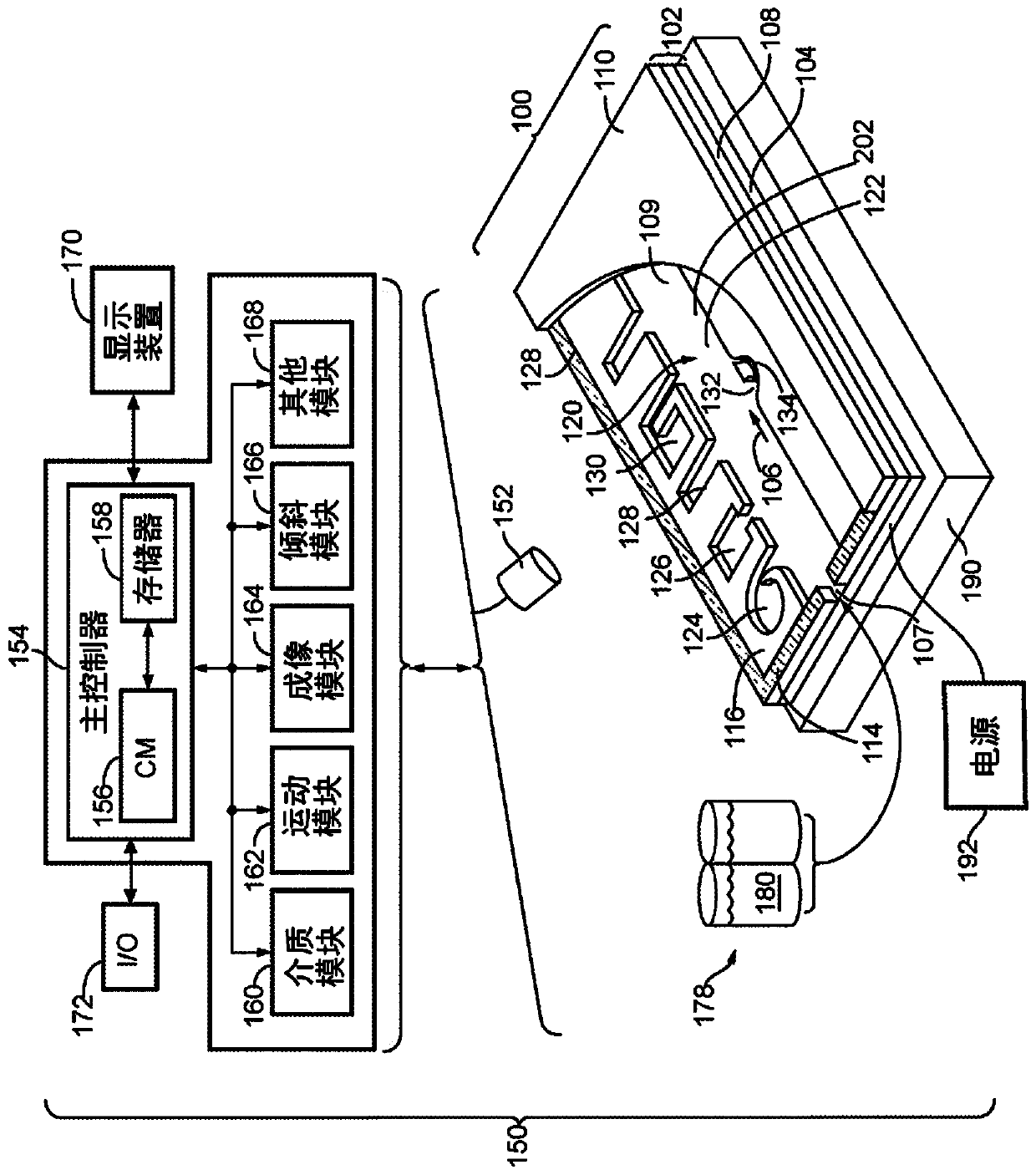
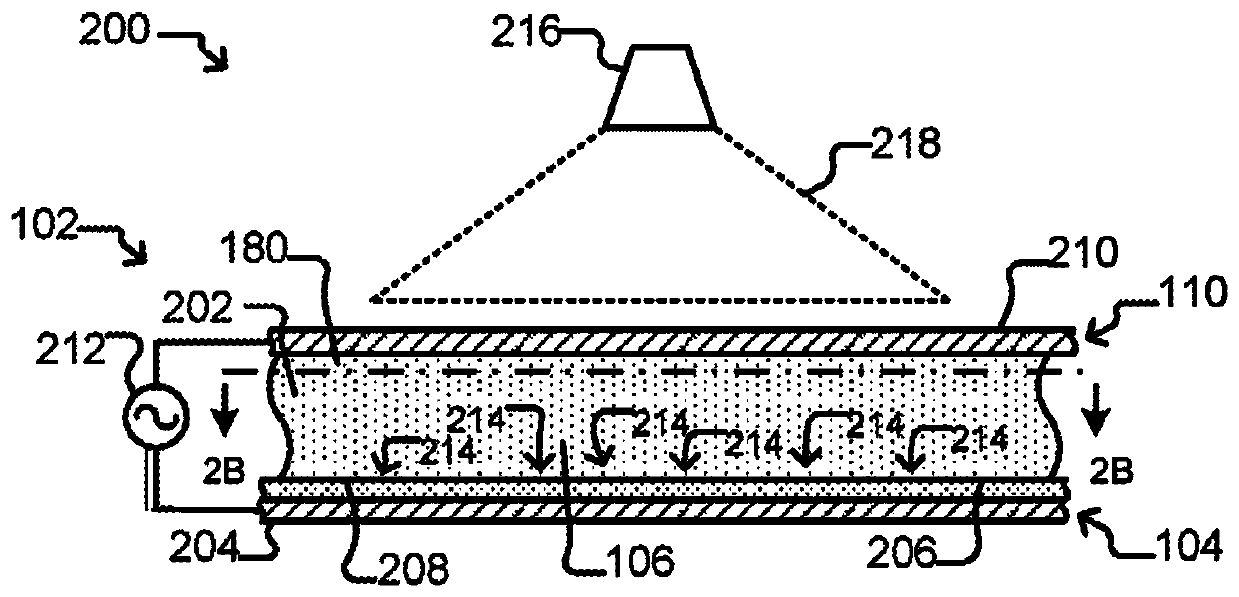
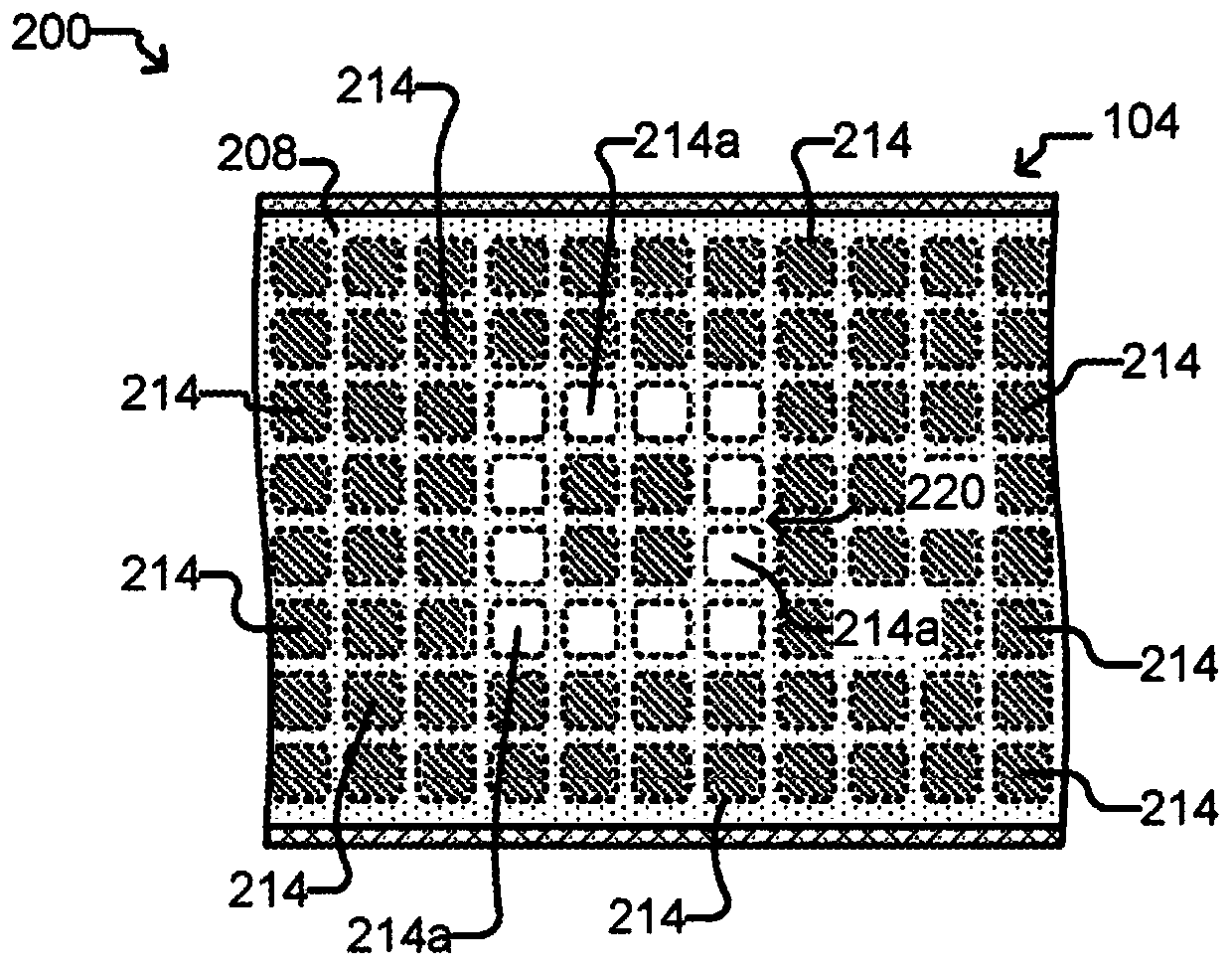
[0497] This experiment demonstrates that Nextera sequencing libraries (Illumina) can be generated using isothermal PCR with a biotinylated priming sequence (carrying the barcode) attached to a bead and a primer free in solution. OKT3 cells (150) were input into the OptoSelect device and loaded into the NanoPen chamber using light-driven dielectrophoretic force. Figure 24 Cells are shown after delivery to the NanoPen chamber. Light-driven dielectrophoretic force delivers one cell to each of the seven NanoPen chambers, where misses deliver one cell...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com