MRNA capture sequences, synthesis method of capture carrier and production method of high-throughput single-cell sequencing library
A technology for capturing sequences and synthesizing methods, used in DNA/RNA fragments, chemical libraries, recombinant DNA technology, etc. In order to achieve the effect of optimizing the microporous structure, facilitating the reaction and purification, and facilitating the specific assembly
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] The present invention provides an mRNA capture sequence. The mRNA capture sequence is an oligonucleotide and includes a universal primer, a rare enzyme cutting site, a cell tag, a random molecular tag and a PolyT sequence.
[0056] Further, the structure of the mRNA capture sequence is universal primer-rare restriction site-cell tag-random molecular tag-PolyT sequence, that is, the mRNA capture sequence is composed of parts in the following order: universal primer, rare restriction site Dots, cell tags, random molecular tags and PolyT sequences, the sequence length is 38-180bp, preferably 57-100bp; among them,
[0057] The universal primer sequence is used to initiate cDNA double-strand PCR to realize the replication of the cDNA chain, and the sequence length is 10-30 bp, preferably 15-25 bp;
[0058] The rare enzyme cleavage site can be specifically recognized and cut by restriction enzymes, including but not limited to Acc65I, AccI, AciI, AclI, AcuI, AflII, AhdI, AluI, AlwNI...
Embodiment 2
[0063] Such as Figure 8 As shown, the present invention also provides a method for synthesizing a capture carrier for mRNA capture, including the following steps:
[0064] Step 1: Select a base material containing several micro-reaction units, and form surface functional groups for grafting oligonucleotides in the micro-reaction units by chemical modification methods;
[0065] Step two, starting with the surface functional group formed in step one for grafting oligonucleotides as a starting point, in situ synthesizing several mRNA capture sequences as described above in each micro-reaction unit to prepare mRNA Captured capture carrier.
[0066] The base material includes, but is not limited to, micropore arrays, microspheres, magnetic beads, gel microspheres, and resin fillers.
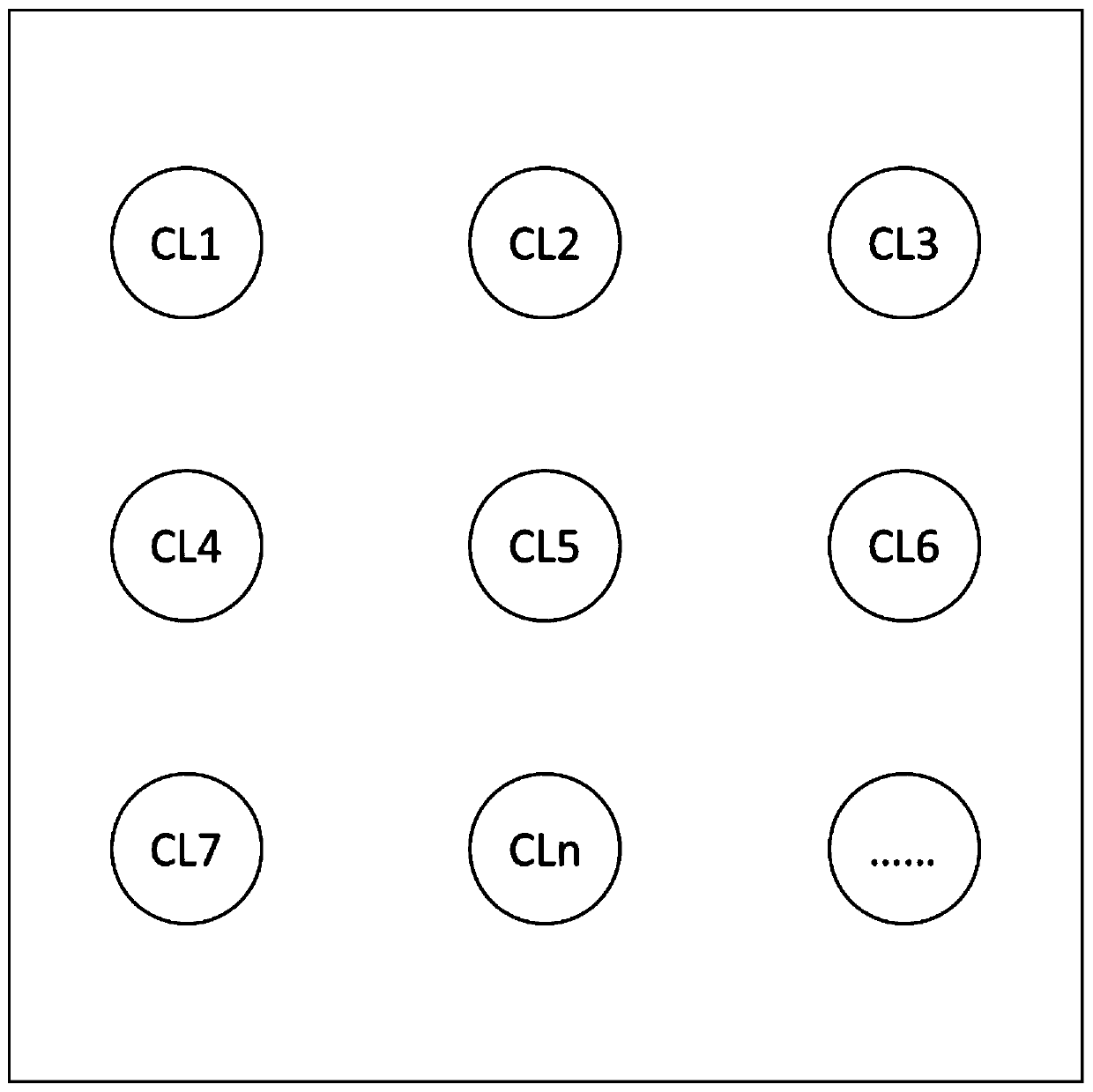
[0067] The substrate material is a micropore array, such as image 3 As shown in the figure, CL1, CL2 to CLn, etc. are all microwells on the microwell array shown, that is, microreaction units. The microwell...
Embodiment 3
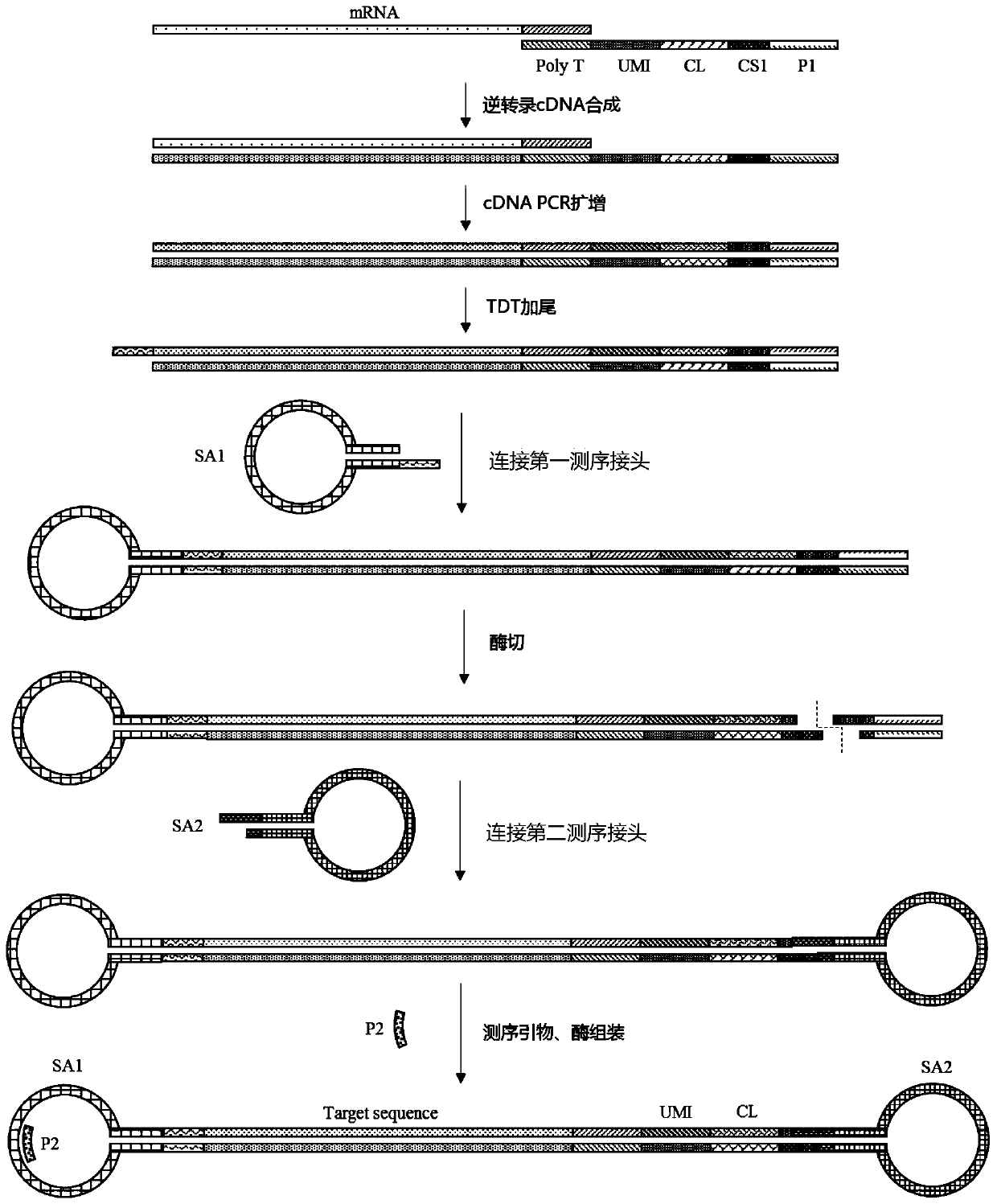
[0084] Such as figure 1 , figure 2 , Picture 10 As shown, the present invention also provides a method for preparing a high-throughput single-cell sequencing library, which includes the following steps:
[0085] S1. Capturing single cells by using the micro-reaction unit of the capture carrier for mRNA capture prepared by the synthetic method described above;
[0086] S2. The single cell is lysed to release mRNA, PolyT in the mRNA capture sequence in the micro-reaction unit specifically binds to PolyA on the mRNA, and cDNA is synthesized by reverse transcription;
[0087] S3. Replace buffer, add microspheres carrying replication primers, perform PCR amplification reaction to form a complete cDNA double-stranded template, wherein the replication primer and the universal primer sequence on the mRNA capture sequence in the microreaction unit the same;
[0088] S4. Recover the microspheres and combine them, wash the replacement buffer, and connect the first sticky end to the end of the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com