Cell subset annotation method based on single cell transcriptome sequencing
A technology of transcriptome sequencing and cell subgroups, which is applied in the field of bioinformatics analysis, can solve problems such as annotation of single cell subgroups, and achieve the effect of comprehensive cell subgroups and avoiding subjective consciousness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
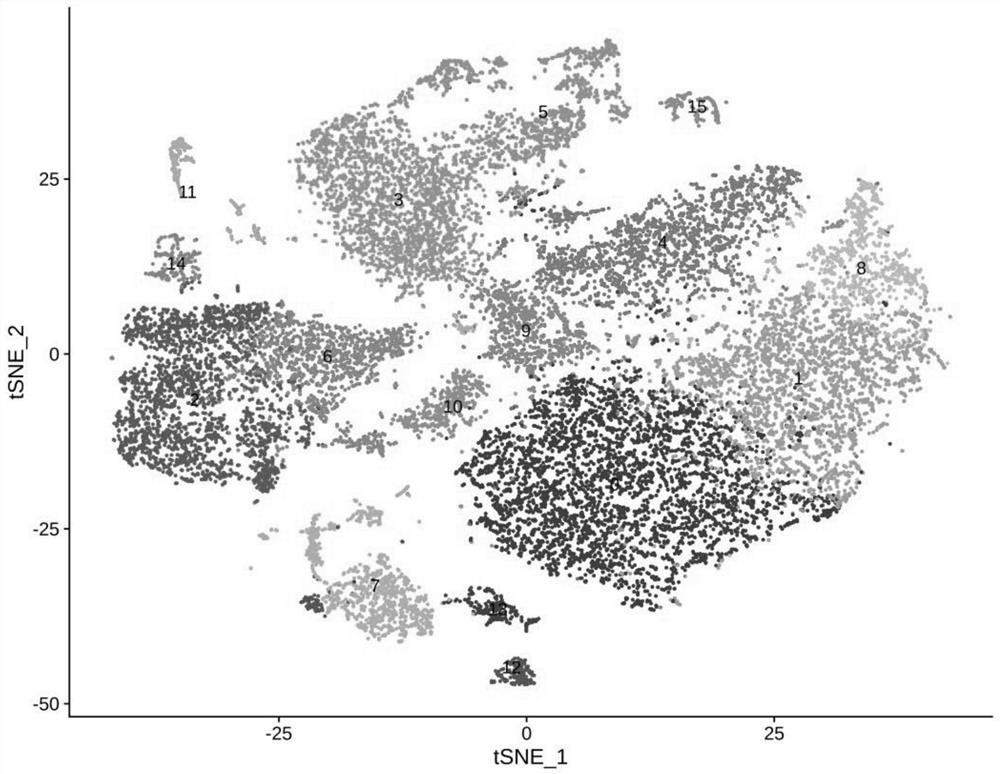
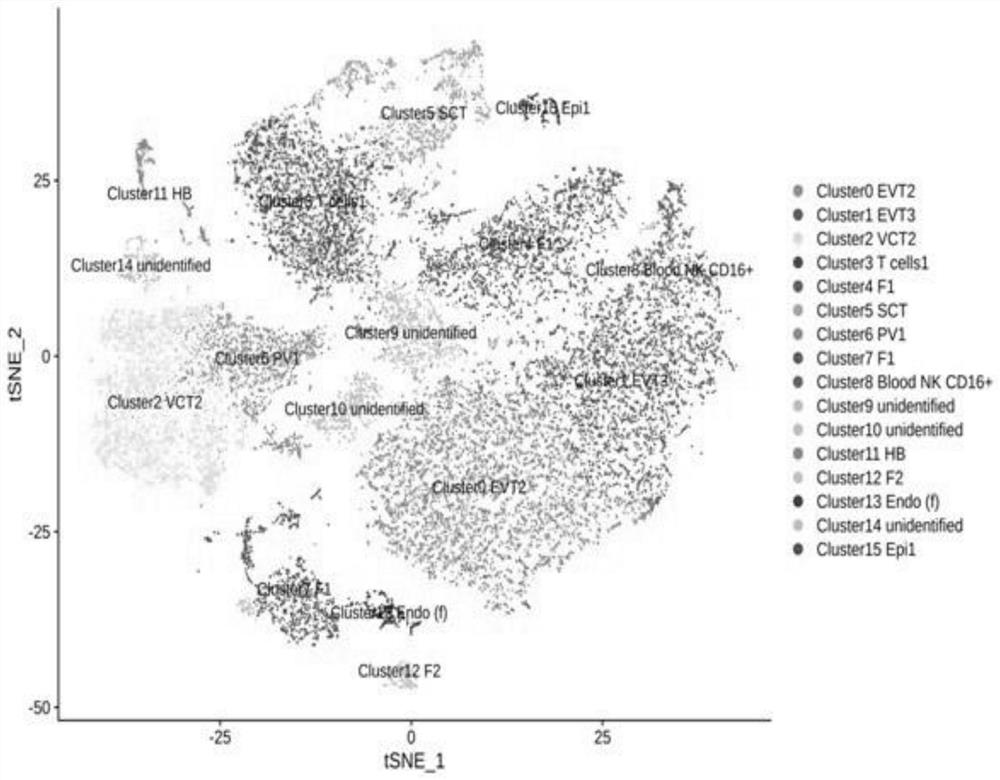
[0037] Example 1 Cell Segmentation and Annotation of Normal Chorionic Cells
[0038] A cell subpopulation annotation method based on single-cell transcriptome sequencing, including the following steps:
[0039] S1. 10x barcode UMI identification: The off-machine sequencing data of the 10x genomics platform database is a fastq sequence, and the fastq sequence with the same ID number includes 3 parts: barcode+UMI+mRNA sequence, use the software cellrangercount to distinguish the sequence through the barcode sequence Source cells, quantify gene expression through UMI sequence, and use 3' end mRNA sequence for gene identification;
[0040] S2. Genome comparison: use the STAR algorithm to compare the fastq sequence obtained by sequencing to the reference genome, and locate the measured sequence to the corresponding gene;
[0041] S3. Gene expression profile construction: including the following steps: 1) Data integration and data volume normalization: when samples from multiple li...
Embodiment 2
[0052] Example 2 Cell Segmentation and Annotation of Diseased Chorionic Cells
[0053] A cell subpopulation annotation method based on single-cell transcriptome sequencing, including the following steps:
[0054] S1. 10x barcode UMI identification: The off-machine sequencing data of the 10x genomics platform database is a fastq sequence, and the fastq sequence with the same ID number includes 3 parts: barcode+UMI+mRNA sequence, use the software cellrangercount to distinguish the sequence through the barcode sequence Source cells, gene expression quantification by UMI sequence, gene identification by 3' end mRNA sequence;
[0055] S2. Genome comparison: use the STAR algorithm to compare the fastq sequence obtained by sequencing to the reference genome, and locate the measured sequence to the corresponding gene;
[0056] S3. Gene expression profile construction: including the following steps: 1) Data integration and data volume normalization: when samples from multiple librarie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com