Tumor antigen expression examination primer and kit based on high-throughput sequencing method
A technique for detecting tumor antigens and primers, which is applied in the biological field and can solve problems such as low throughput and limited detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0152] Example 1 Identification of Individualized Tumor Antigen Peptide Library Establishment
[0153] The workflow for identifying individualized tumor antigen peptide libraries using fresh tumor tissue or FFPE sections is as follows: figure 1 shown.
[0154] 1. Total RNA Extraction
[0155] 1) Use RNA extraction kit or DNA and RNA co-extraction kit to extract total RNA.
[0156] 2) Use Qubit TM RNA HS Assay Kit for total RNA quantification.
[0157] 3) Agilent RNA 6000 Pico Kit was used to determine the integrity of RNA.
[0158] 4) At least 10 ng of DNase-treated total RNA (≥1.43 ng / μL) is required for each reverse transcription reaction. For samples with low extraction concentration (concentration lower than 1.43ng / μL), take the largest volume (7μL) for reverse transcription, and the initial amount of library construction is <10ng, which belongs to high-risk library construction.
[0159] 5) For samples with DV 200%30% of samples, use 10-50ng input for reverse transc...
Embodiment 2
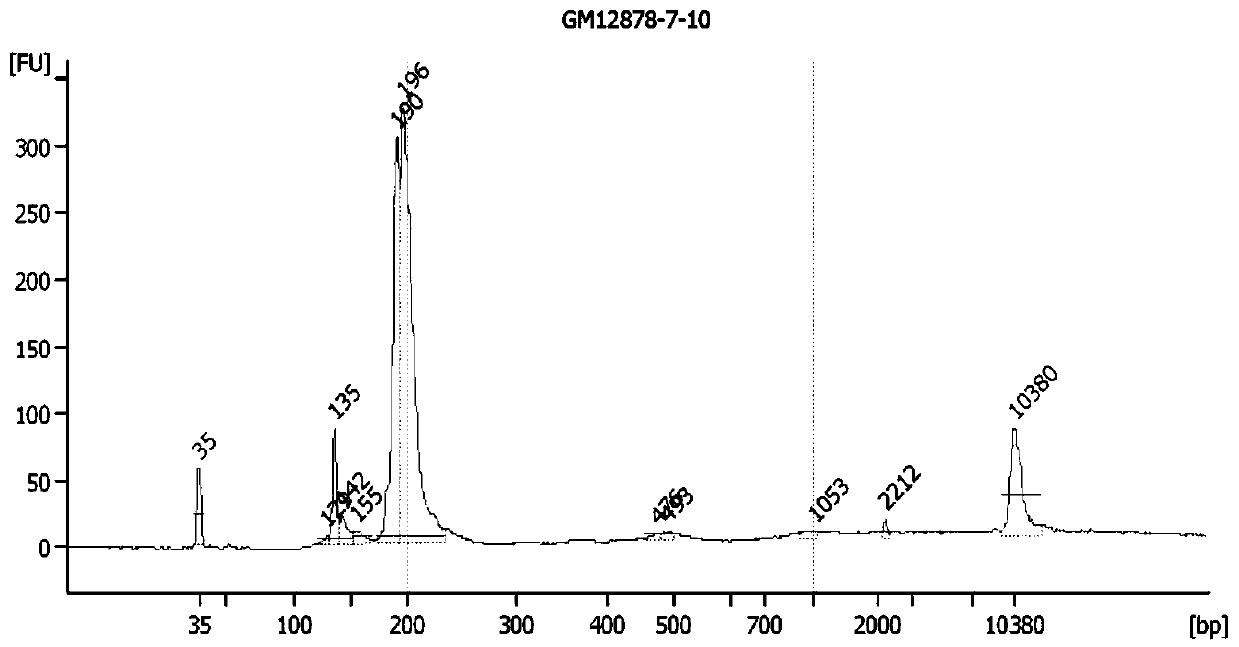
[0250] The different RNA starting amount of embodiment 2FFPE samples and repetition in batch
[0251] One case of FFPE sample with good RNA extraction quality was selected (RIN value 2.1, DV200 80, Figure 2A ), using 10ng, 20ng, and 50ng of total RNA as the starting amount, perform multiplex PCR amplification of the target sequence, build a library, and sequence it. It can be seen that when the total RNA quality is good, the total RNA can be as low as 10ng A better quality library was obtained ( Figure 2B -D). Each initial amount was repeated 3 times for library construction, and a total of 9 libraries were obtained with 3 initial amounts, and the sequencing was completed in the same batch. After obtaining the expression level (nrpm) of the target sequence in each library, Pearson correlation analysis was performed on the paired libraries. The correlation coefficient R was above 0.9, and the consistency was very high (see Figure 2E ).
Embodiment 3
[0252] Repeatability between batches of embodiment 3 fresh tumor cell samples
[0253] One fresh sample was selected, and under the same experimental conditions, the same initial amount of library construction (10ng) was used to construct the library, and 3 replicate libraries were obtained, which were repeated 2 times on the machine, and a total of 6 sequencing data were obtained. The expression level (nrpm) of the target sequence in each library was obtained, and the Pearson correlation analysis was carried out twice for the three libraries. The correlation coefficients between the two repetitions were all 0.99 or above, and the consistency was extremely high (see image 3 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com