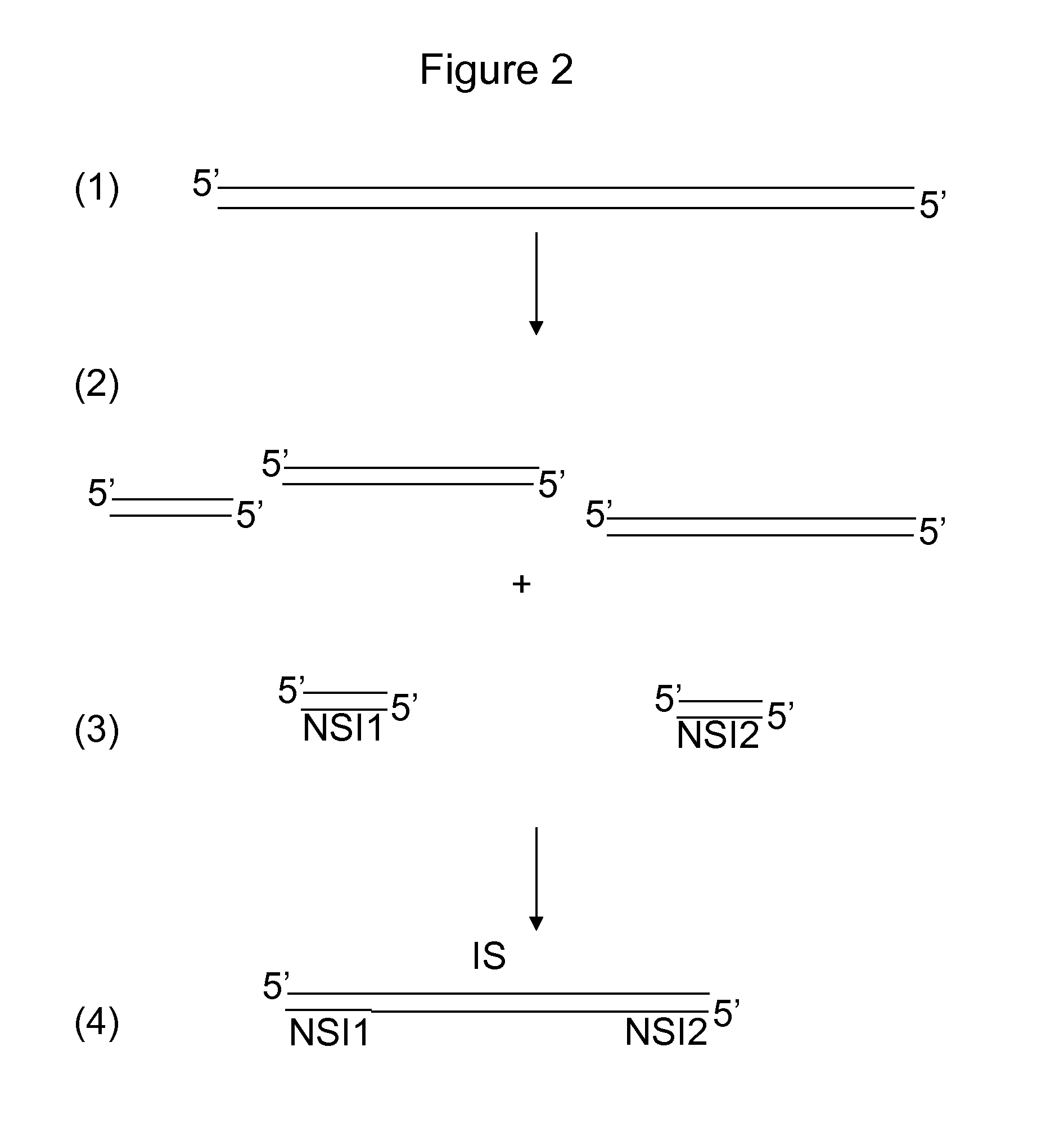
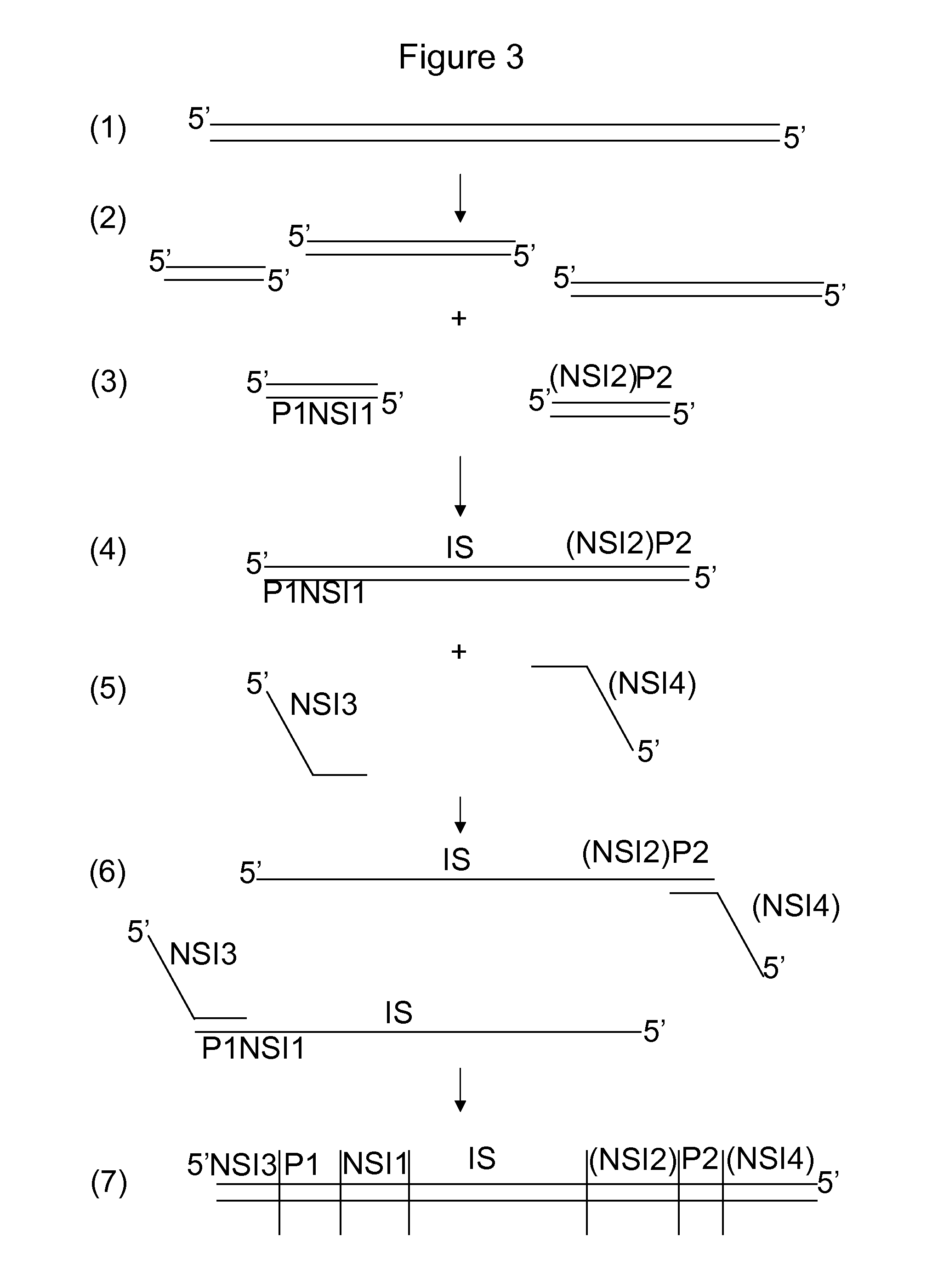
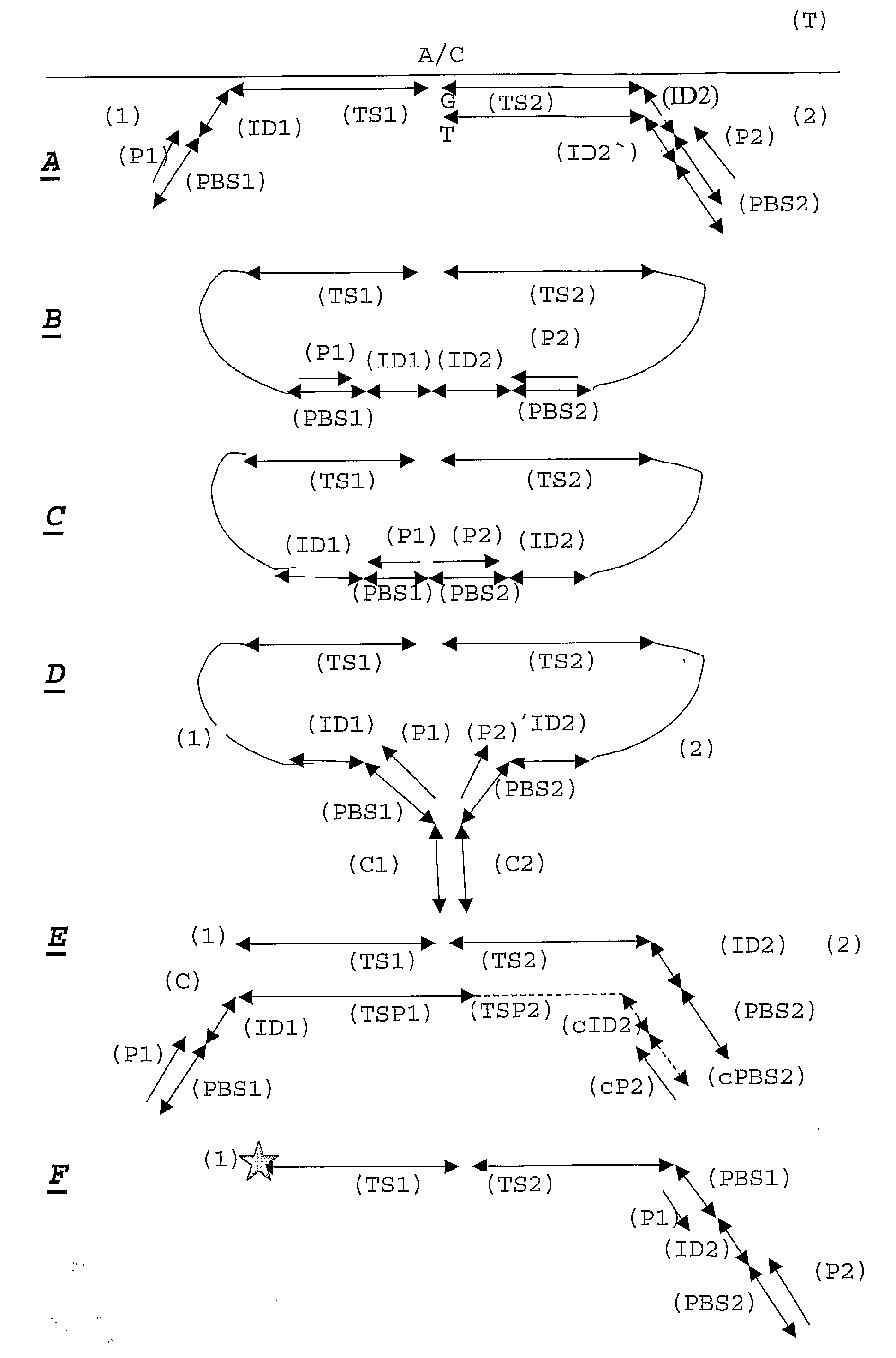
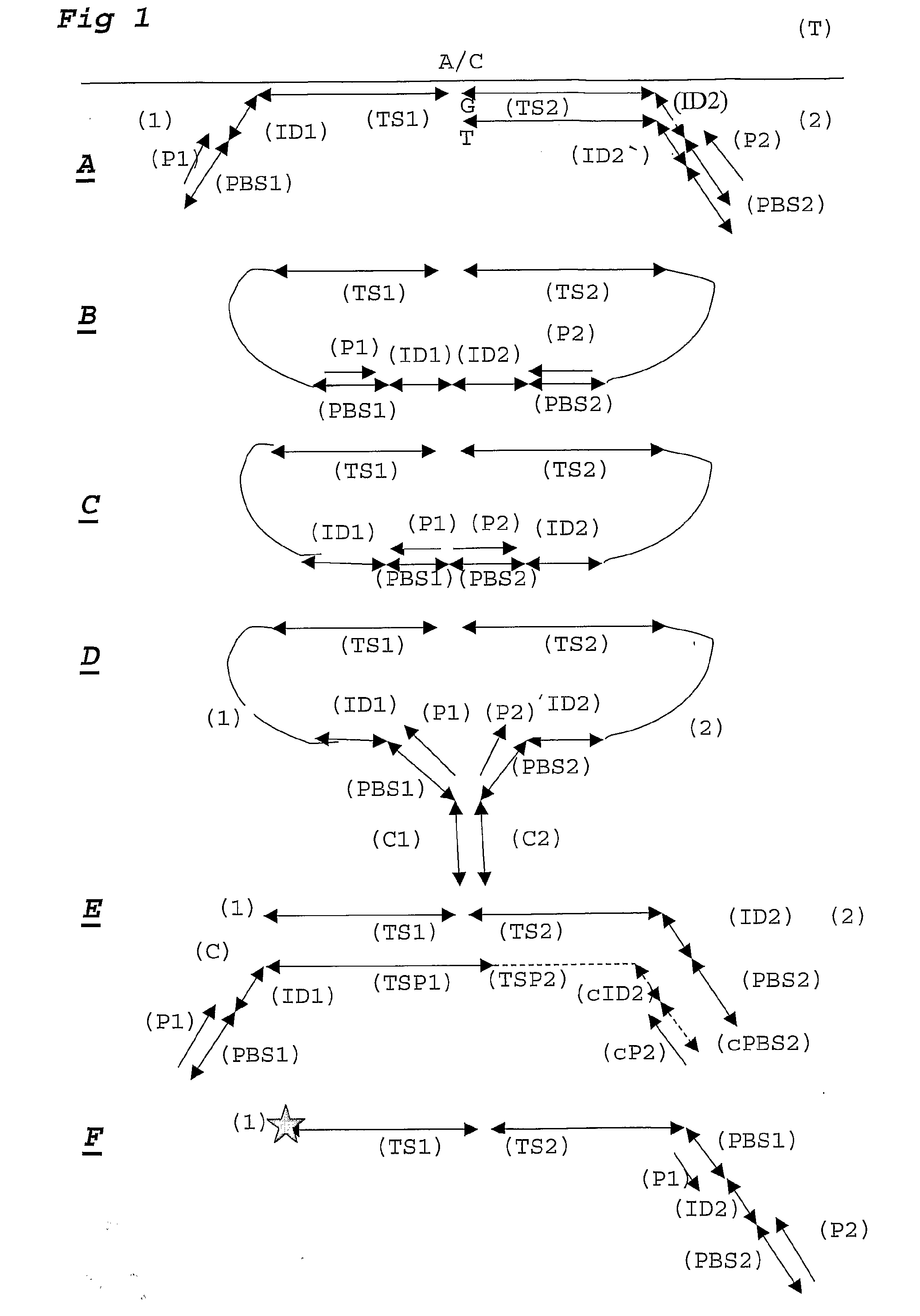
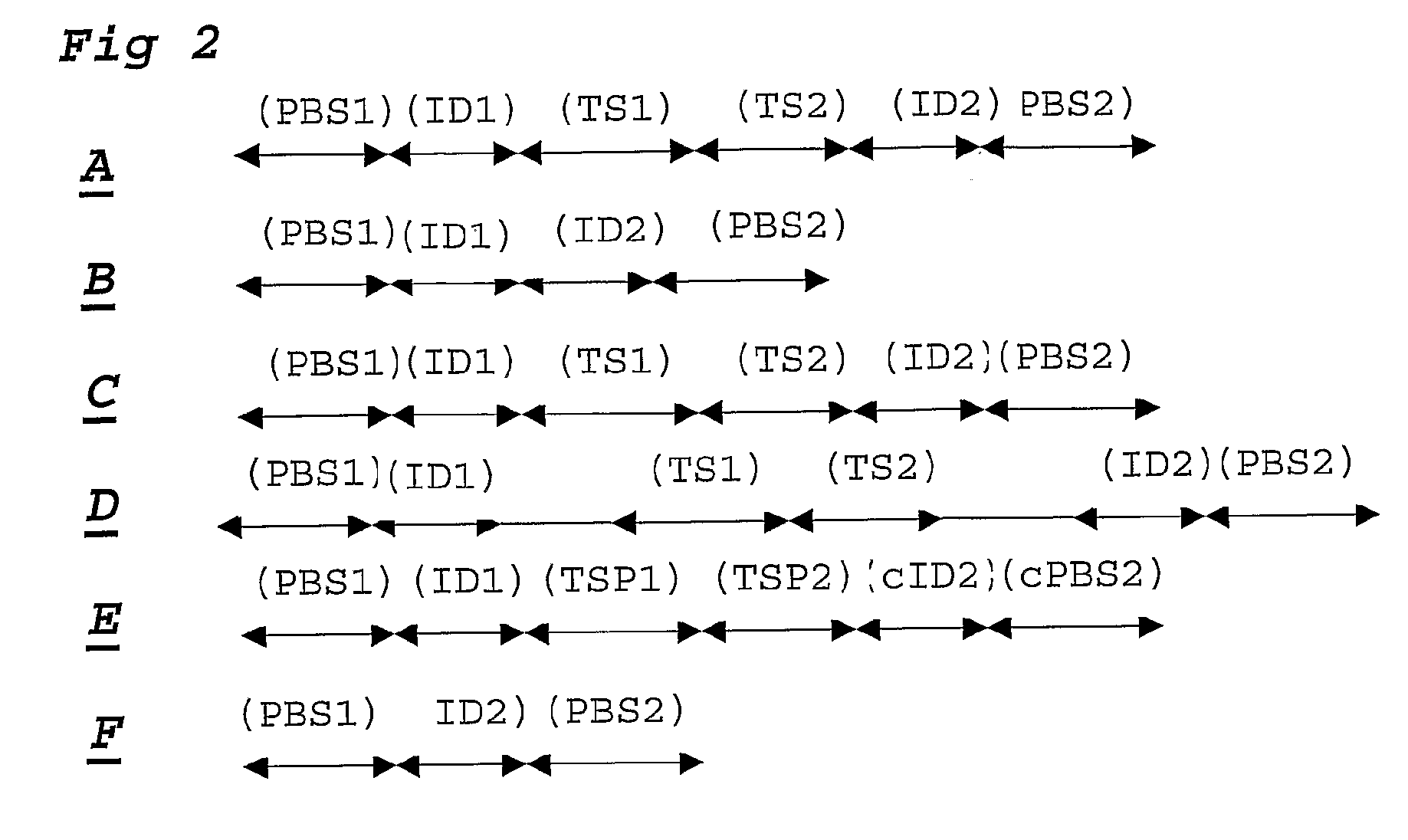
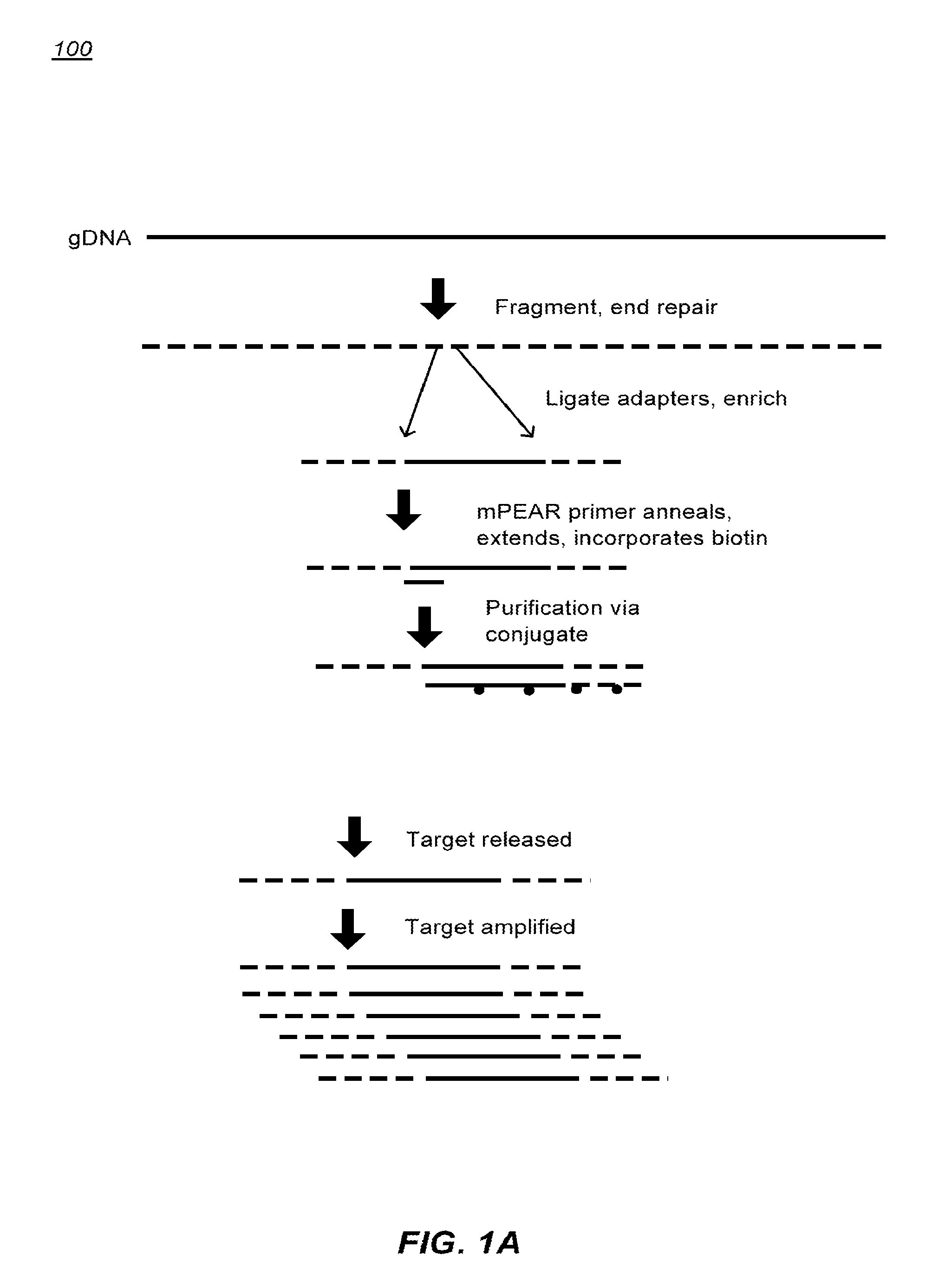
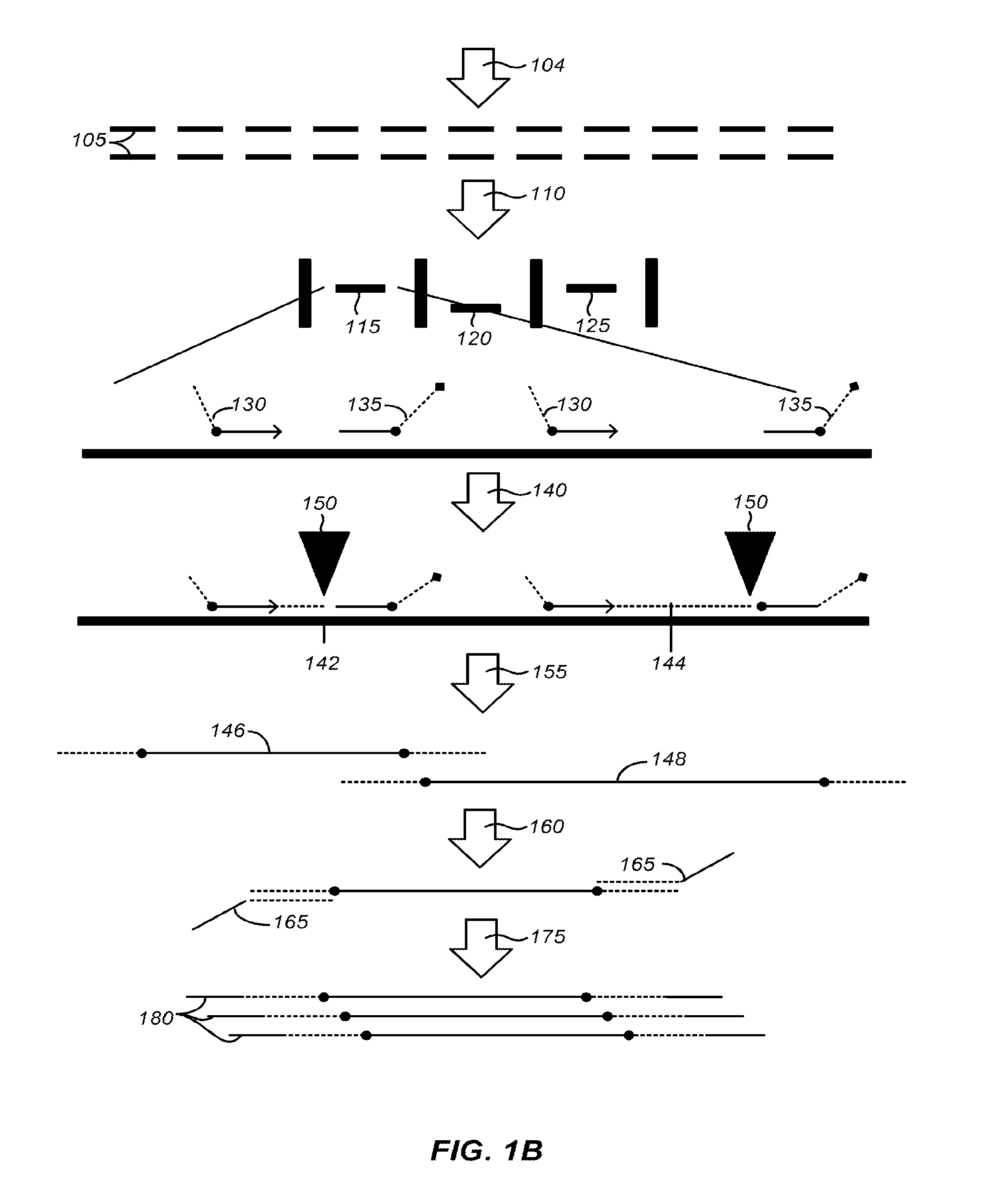
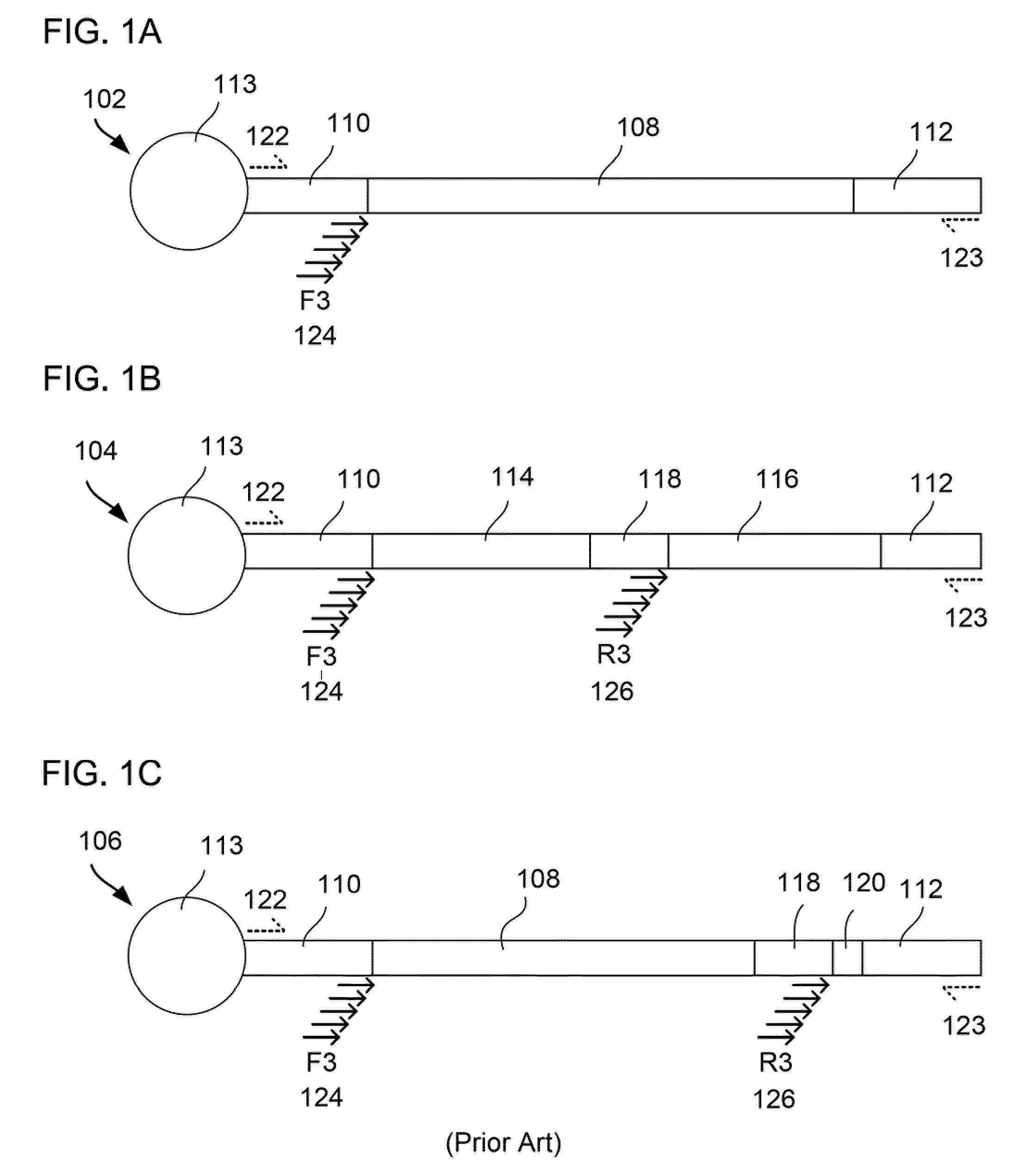
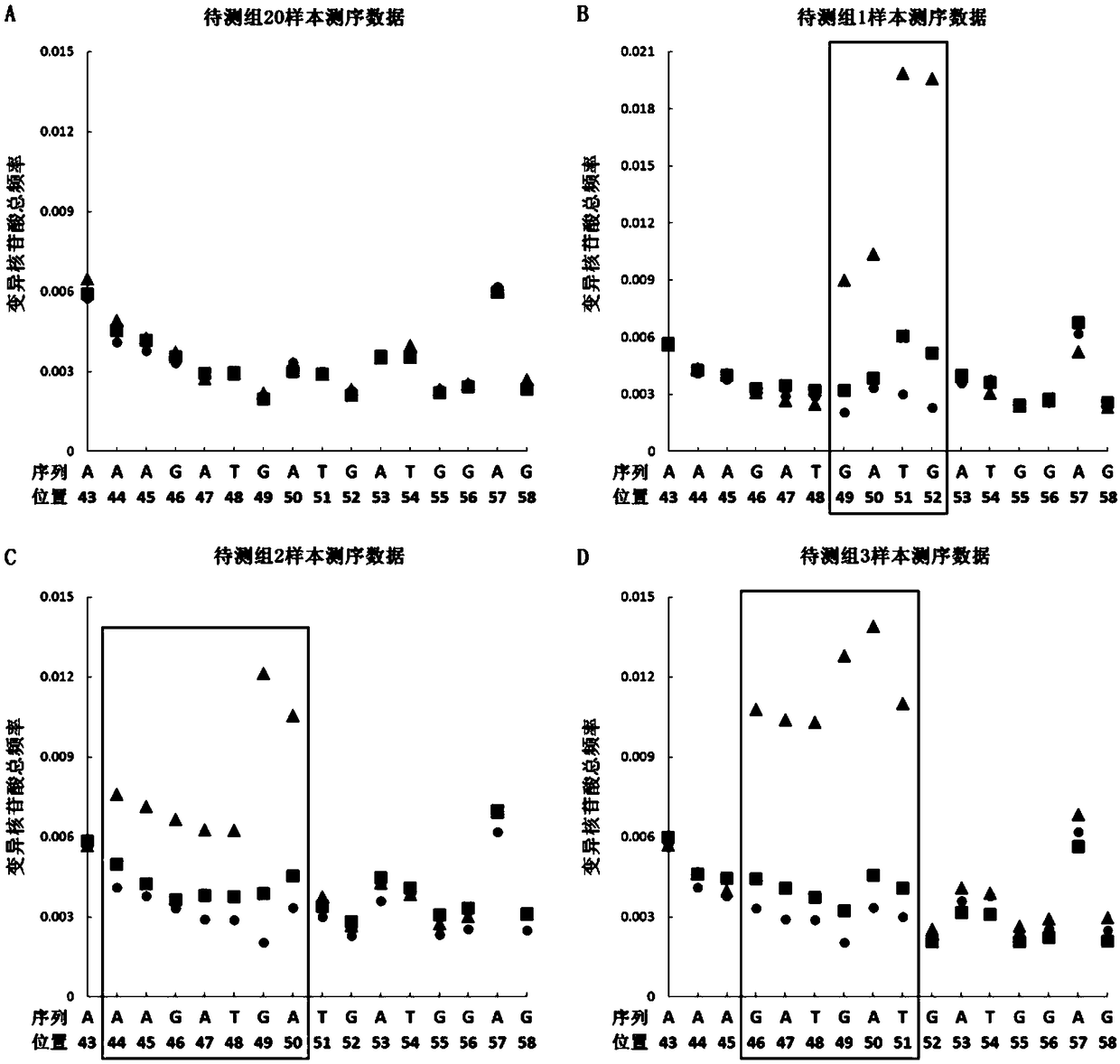
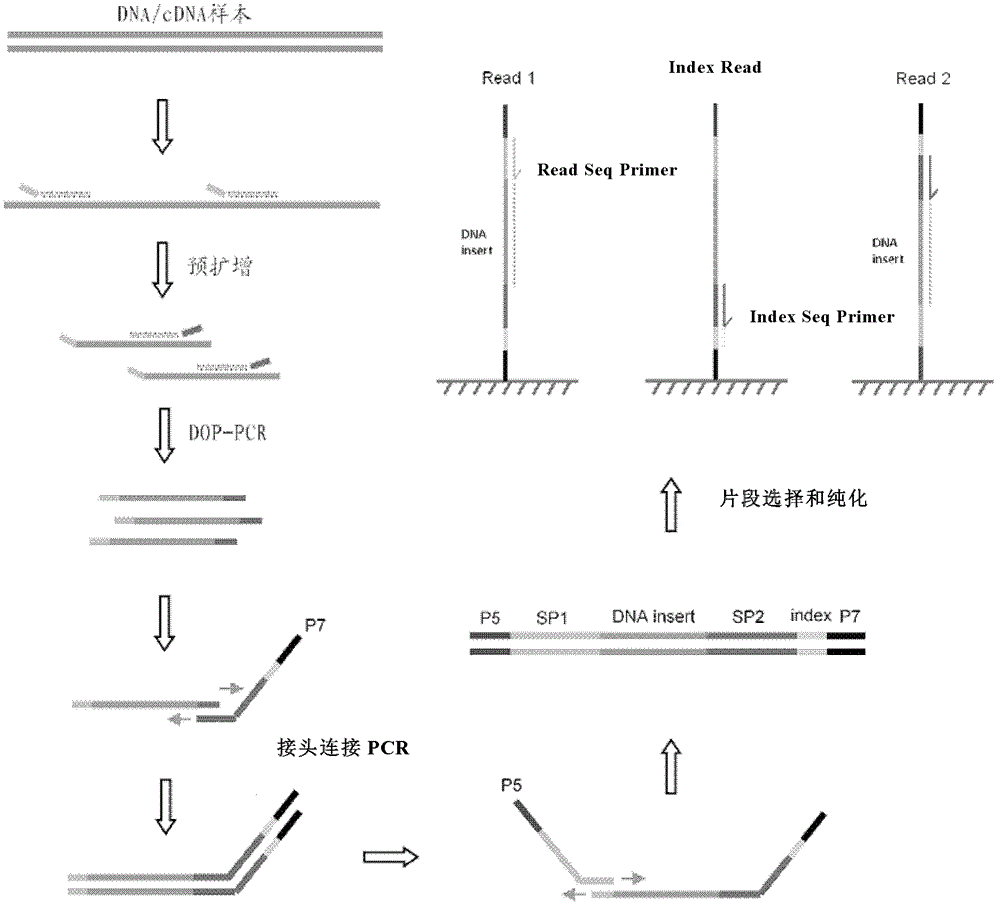
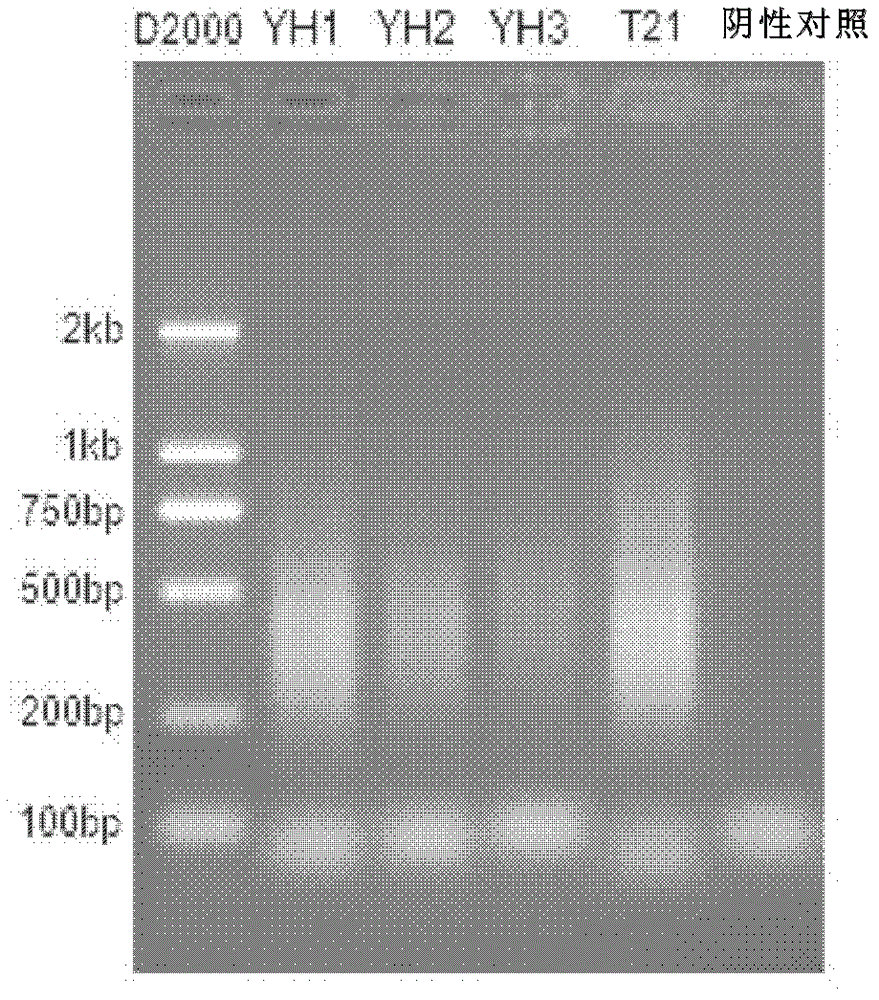
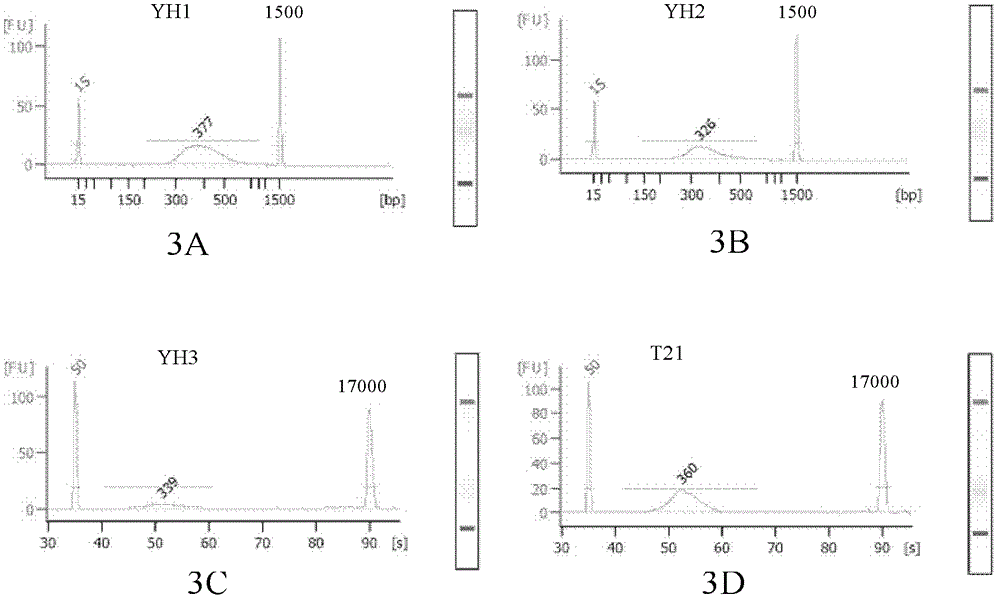
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
1158 results about "High throughput sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
4 Answers 4. High-throughput sequencing specifically refers to sequencing techniques like Illumina that allow you to sequence massive amounts of DNA at once (hundreds of thousands of strands), as opposed to older techniques such as cloning the cDNA in plasmids, followed by sequencing.
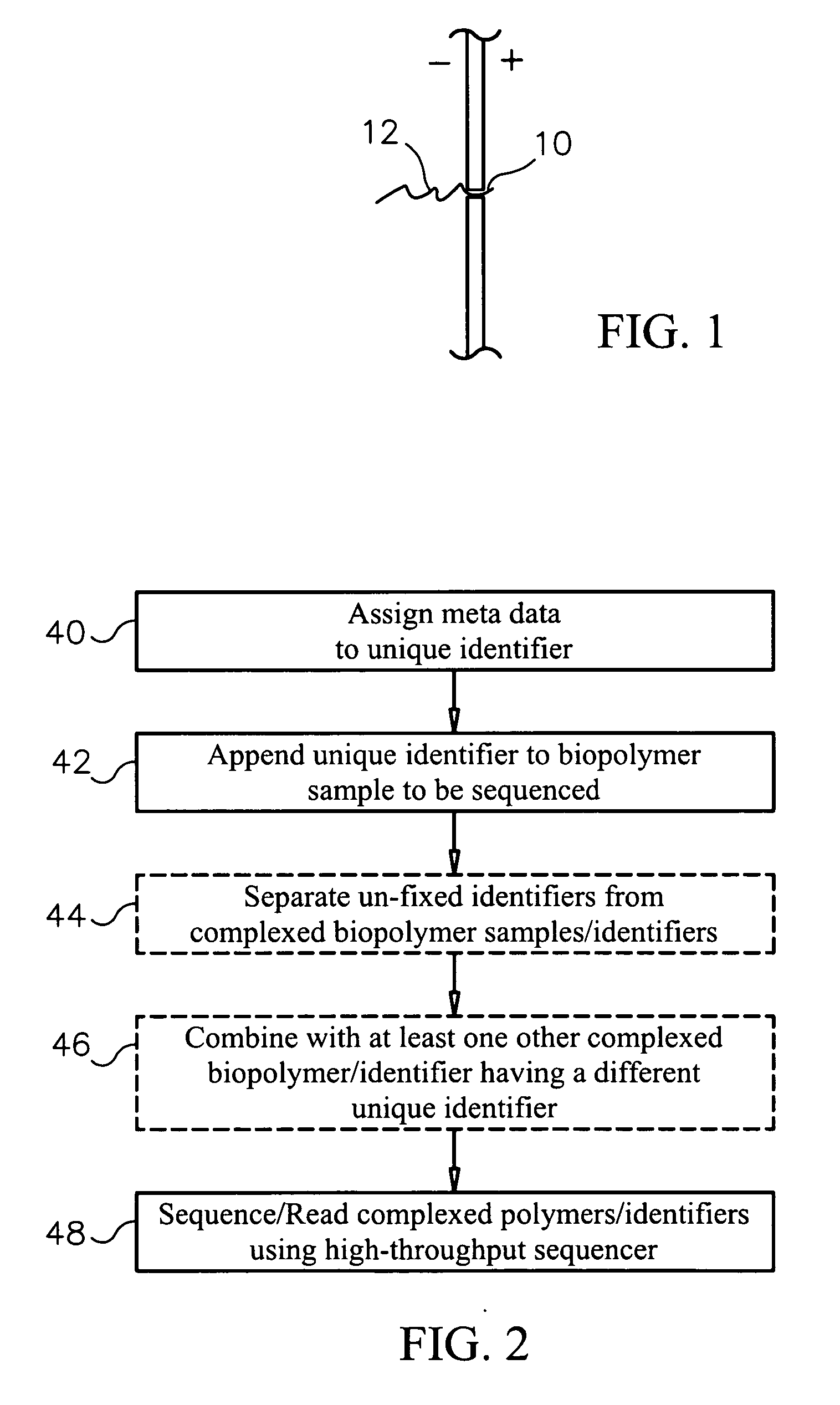
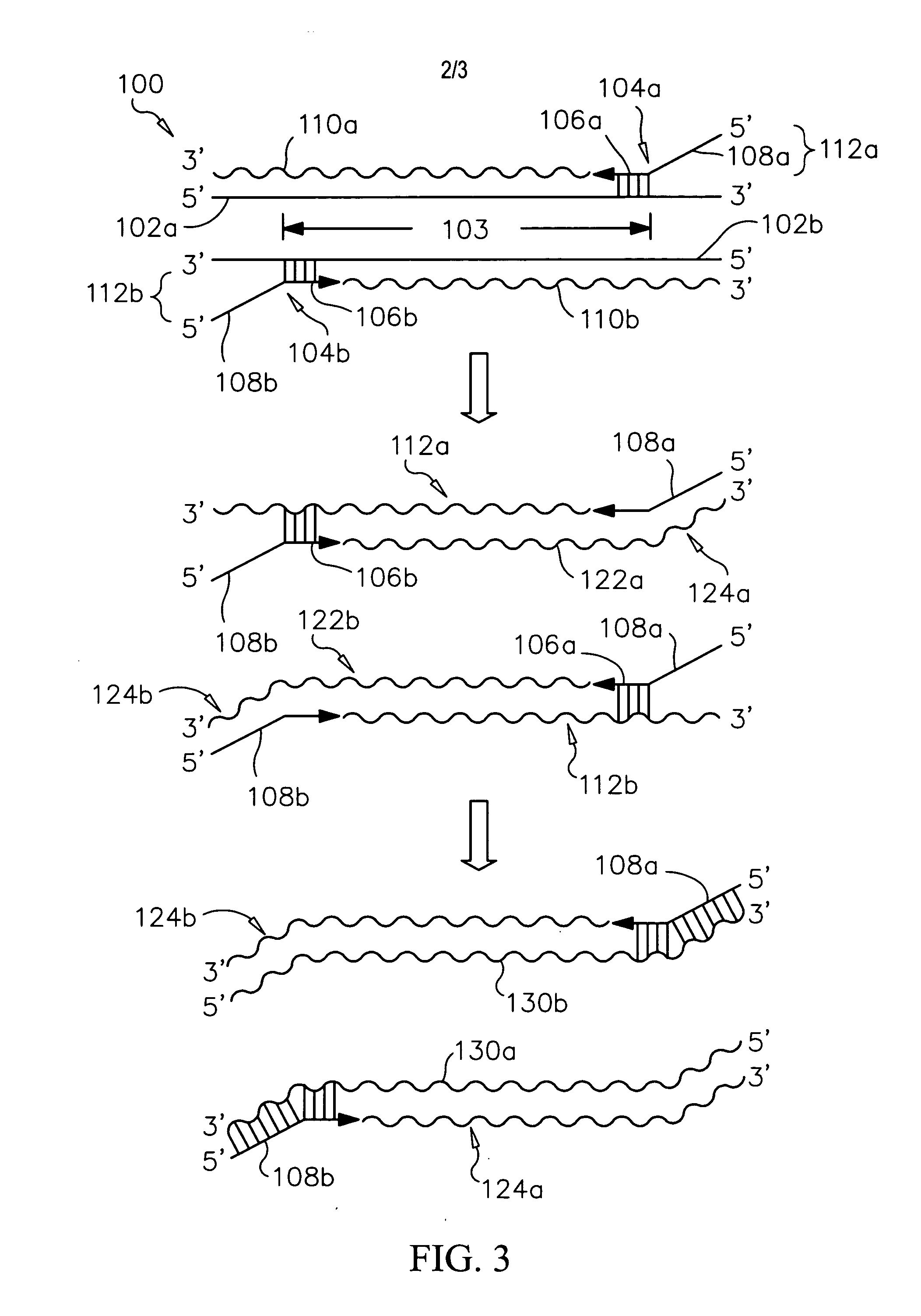
Unique identifiers for indicating properties associated with entities to which they are attached, and methods for using
InactiveUS20060263789A1Efficiently sequencingMicrobiological testing/measurementBiological testingBiopolymerUnique identifier
Methods, systems and computer readable media for sequencing a biopolymer specimen and tracking a source from which the specimen was derived. Methods, systems and computer readable media for multiplex sequencing biopolymer samples. Methods, systems and computer readable media for efficiently sequencing biopolymeric specimens through a high-throughput sequencer. Methods, systems and computer readable media for performing ratio-based analysis with a high throughput sequencer.
Owner:AGILENT TECH INC
High throughput screening using combinatorial sequence barcodes
ActiveUS9080210B2Reduce in quantityLarge capacityMicrobiological testing/measurementFermentationNucleotideBarcode
Owner:KEYGENE NV
High throughput sequence-based detection of snps using ligation assays
ActiveUS20090215633A1Compromise reliabilityEasy to detectSugar derivativesMicrobiological testing/measurementHigh throughput sequenceOligonucleotide
Method for the detection the presence or absence of one or more target sequences in one or more samples based on oligonucleotide ligation assays with a variety of ligatable probes containing identifiers and the subsequent identification of the identifiers in the amplicons or ligated probes using high throughput sequencing technologies.
Owner:KEYGENE NV
Methods of sample preparation
ActiveUS20150265995A1Sequential/parallel process reactionsNucleotide librariesHigh throughput sequenceComputational biology
The present disclosure provides methods, compositions, and kits for methods that can improve techniques nucleic acid analysis, and can allow for more reliable and accurate targeted, multiplexed, high throughput sequencing. The methods, compositions, and kits can be used for sequencing target loci of nucleic acid. The methods, compositions, and kits disclosed herein can be used for assisted de novo targeted sequencing. The methods, compositions, and kits disclosed herein can also be used for library labeling for de novo sequencing and phasing.
Owner:THE SCRIPPS RES INST
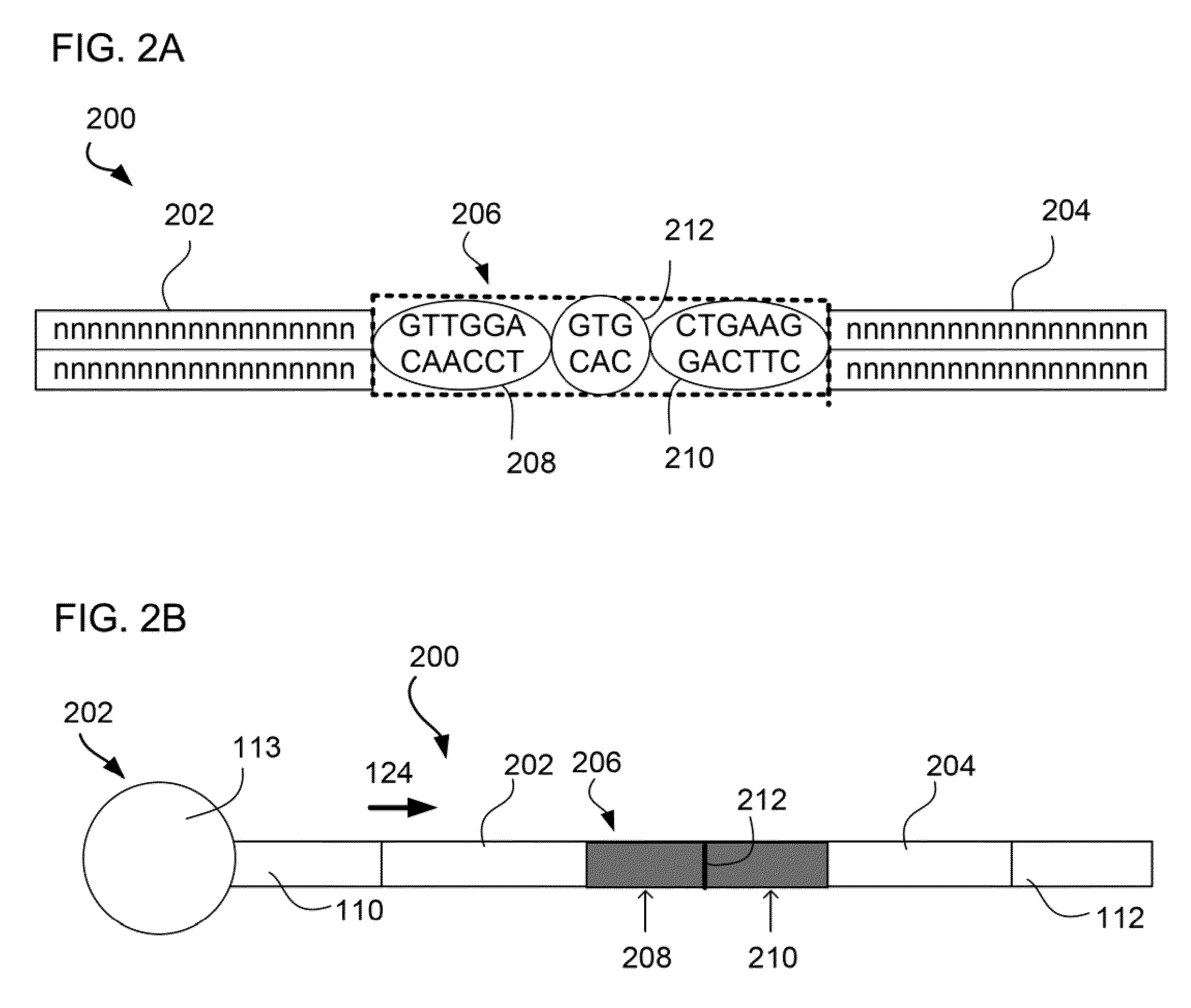
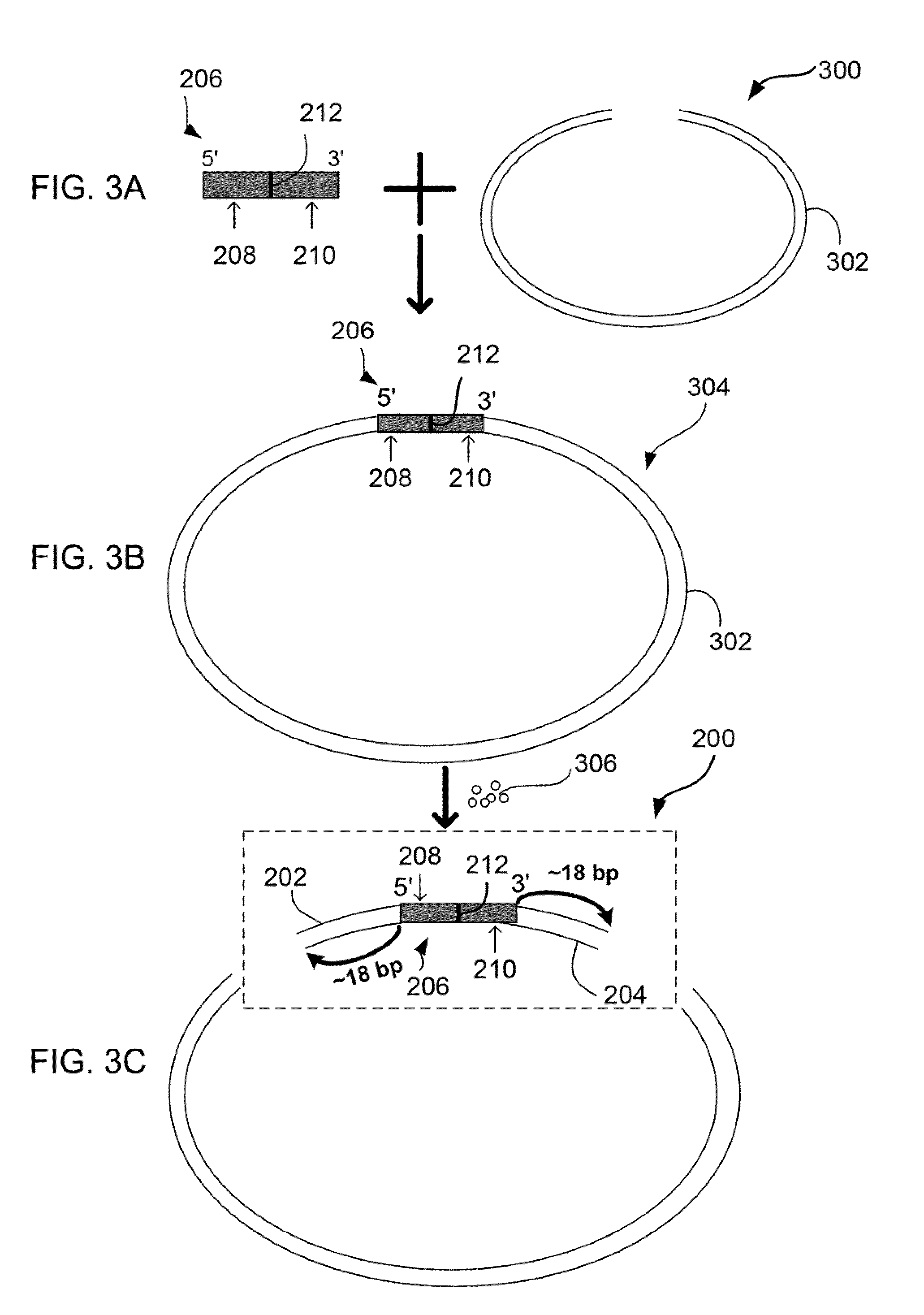
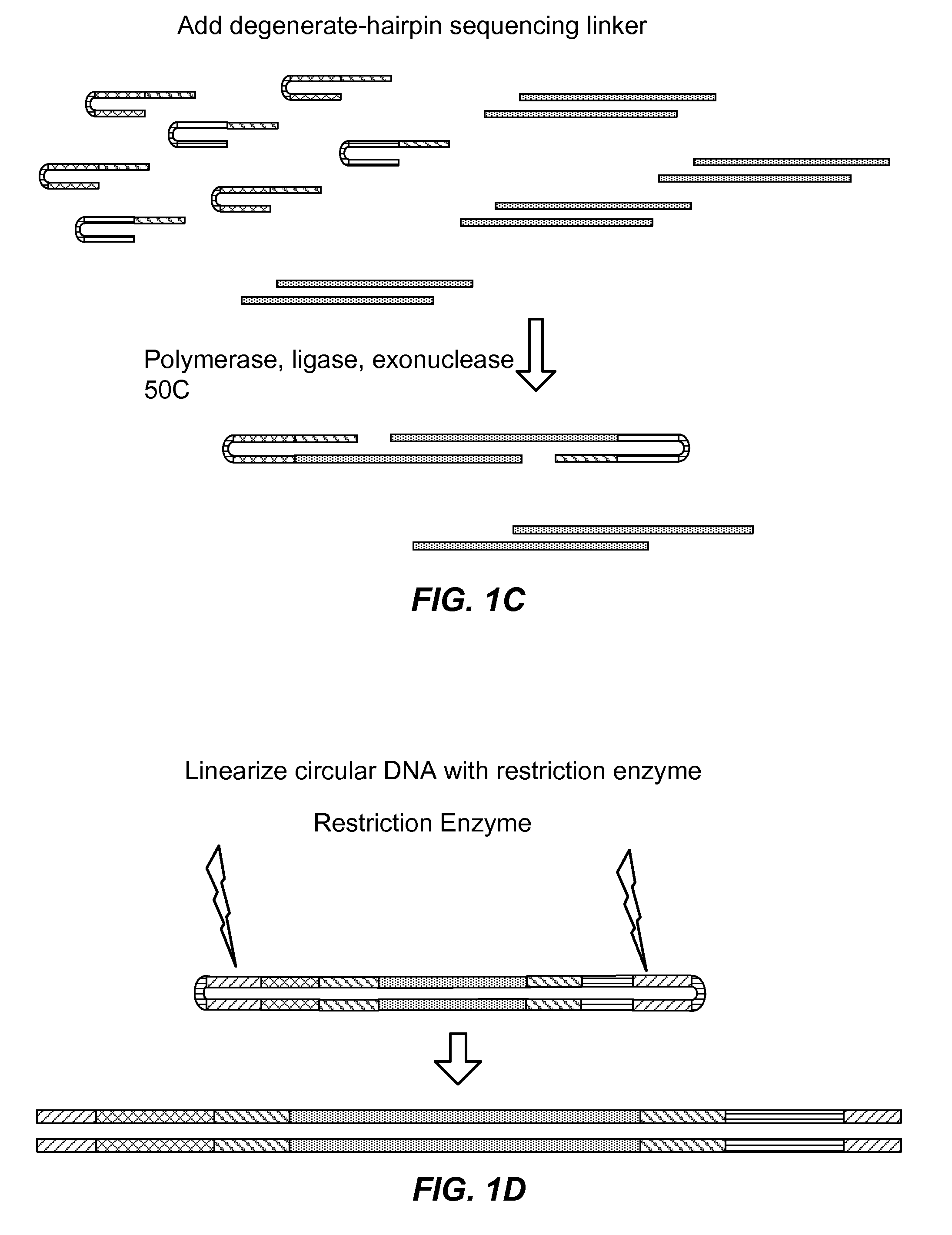
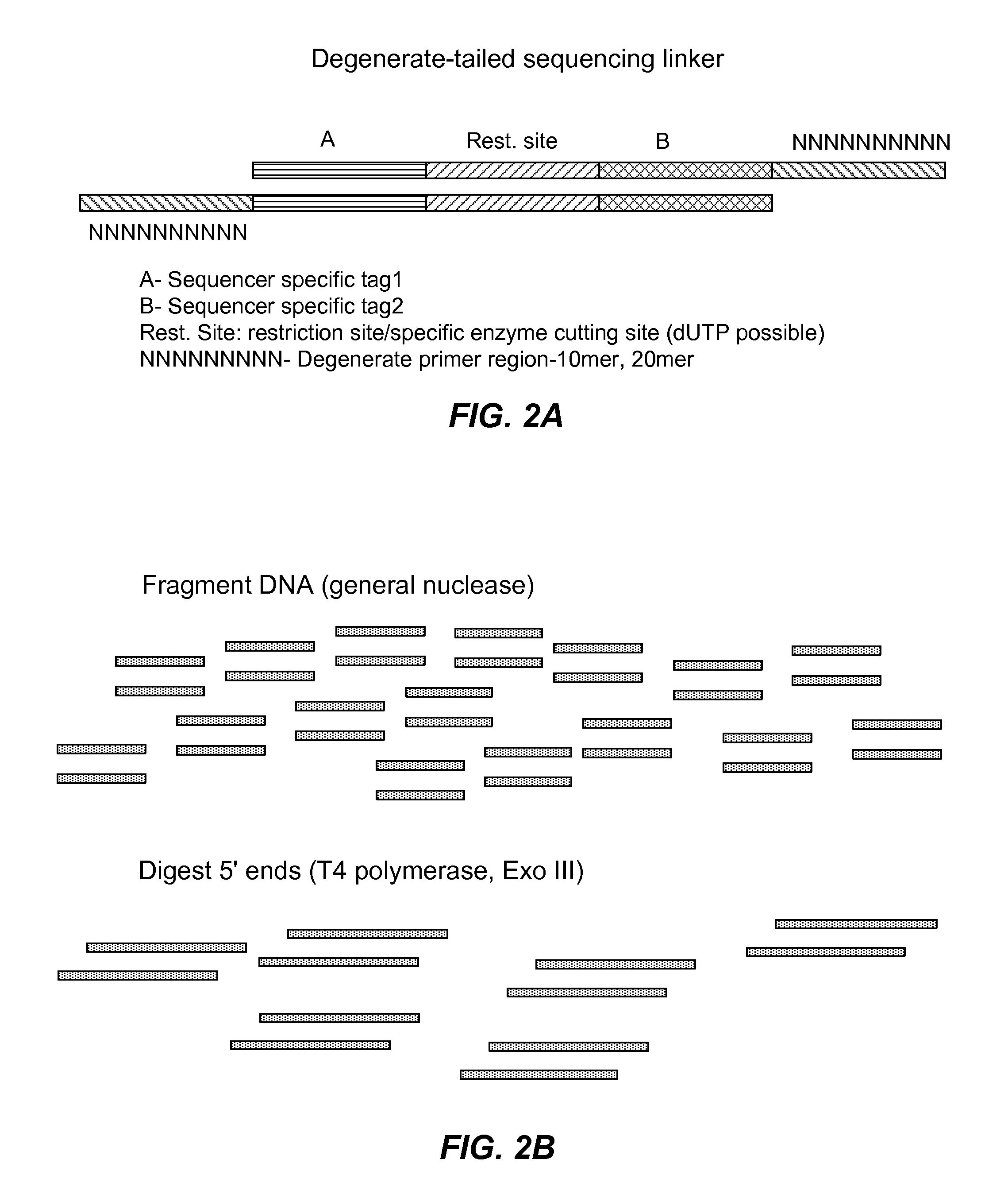
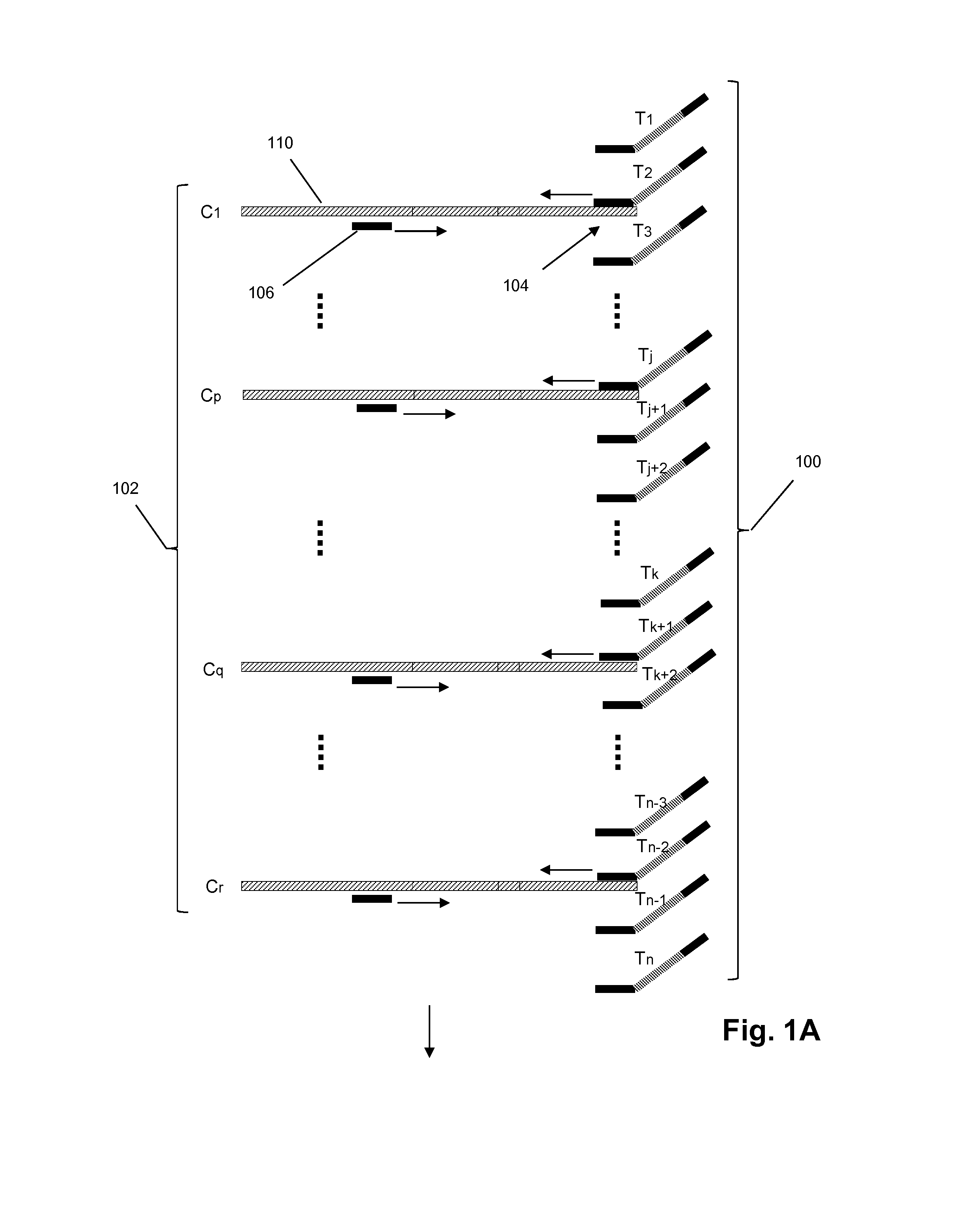
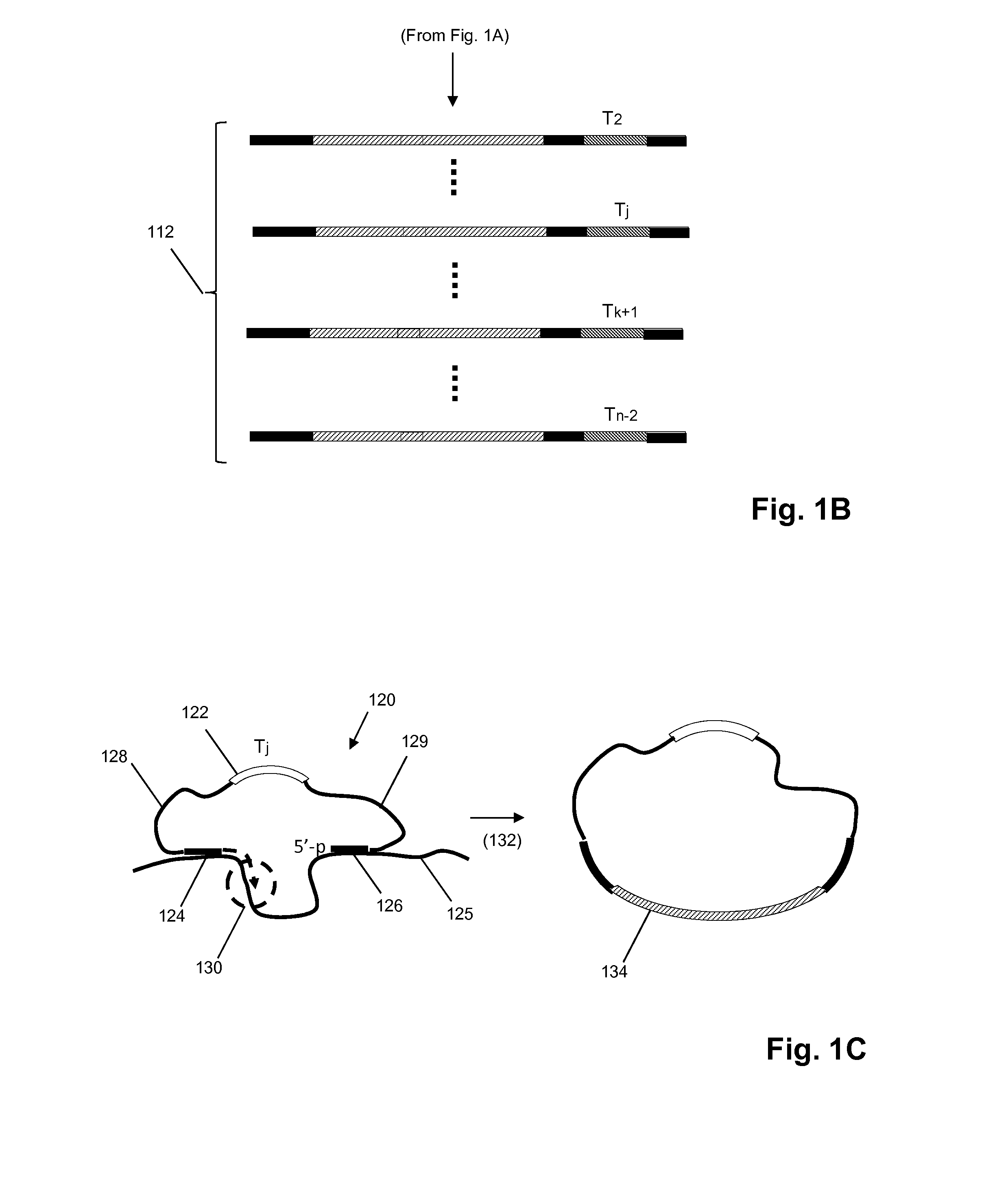
MULTIPLEX BARCODED PAIRED-END DITAG (mbPED) LIBRARY CONSTRUCTION FOR ULTRA HIGH THROUGHPUT SEQUENCING
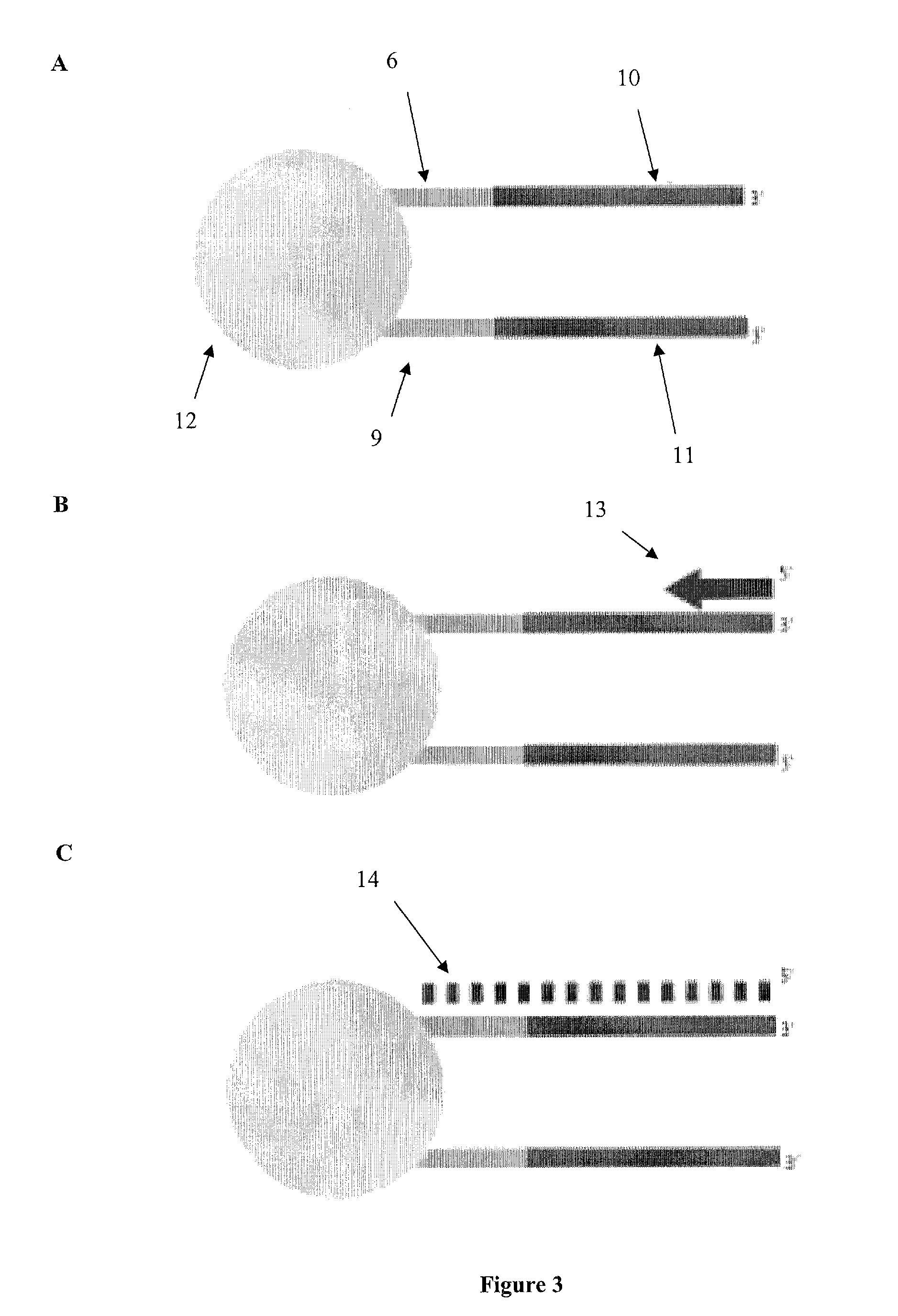
Multiplex barcoded Paired-End Ditag (mbPED) library construction for ultra high throughput sequencing is disclosed. The mbPED library comprises multiple types of barcoded Paired-End Ditag (bPED) nucleic acid fragment constructs, each of which comprises a unique barcoded adaptor, a first tag, and a second tag linked to the first tag via the barcoded adaptor. The two tags are the 5′- and 3′-ends of a nucleic acid molecule from which they originate. The barcoded adaptor comprises a barcode, a first polynucleotide sequence comprising a first restriction enzyme (RE) recognition site, and a second polynucleotide sequence comprising a second RE recognition site and covalently linked to the first polynucleotide sequence via the barcode. The two REs lead to cleavage of a nucleic acid at a defined distance from their recognition sites. The length of the adaptor is set so that the bPED nucleic acid fragment fits one-step sequencing.
Owner:ACAD SINIC
Nucleic acid encoding reactions
ActiveUS20160208322A1Microbiological testing/measurementLibrary member identificationNucleotideBarcode
Described herein are methods useful for incorporating one or more adaptors and / or nucleotide tag(s) and / or barcode nucleotide sequence(s) one, or typically more, target nucleotide sequences. In particular embodiments, nucleic acid fragments having adaptors, e.g., suitable for use in high-throughput DNA sequencing are generated. In other embodiments, information about a reaction mixture is encoded into a reaction product. Also described herein are methods and kits useful for amplifying one or more target nucleic acids in preparation for applications such as bidirectional nucleic acid sequencing. In particular embodiments, methods of the invention entail additionally carrying out bidirectional DNA sequencing. Also described herein are methods for encoding and detecting and / or quantifying alleles by primer extension.
Owner:FLUIDIGM CORP
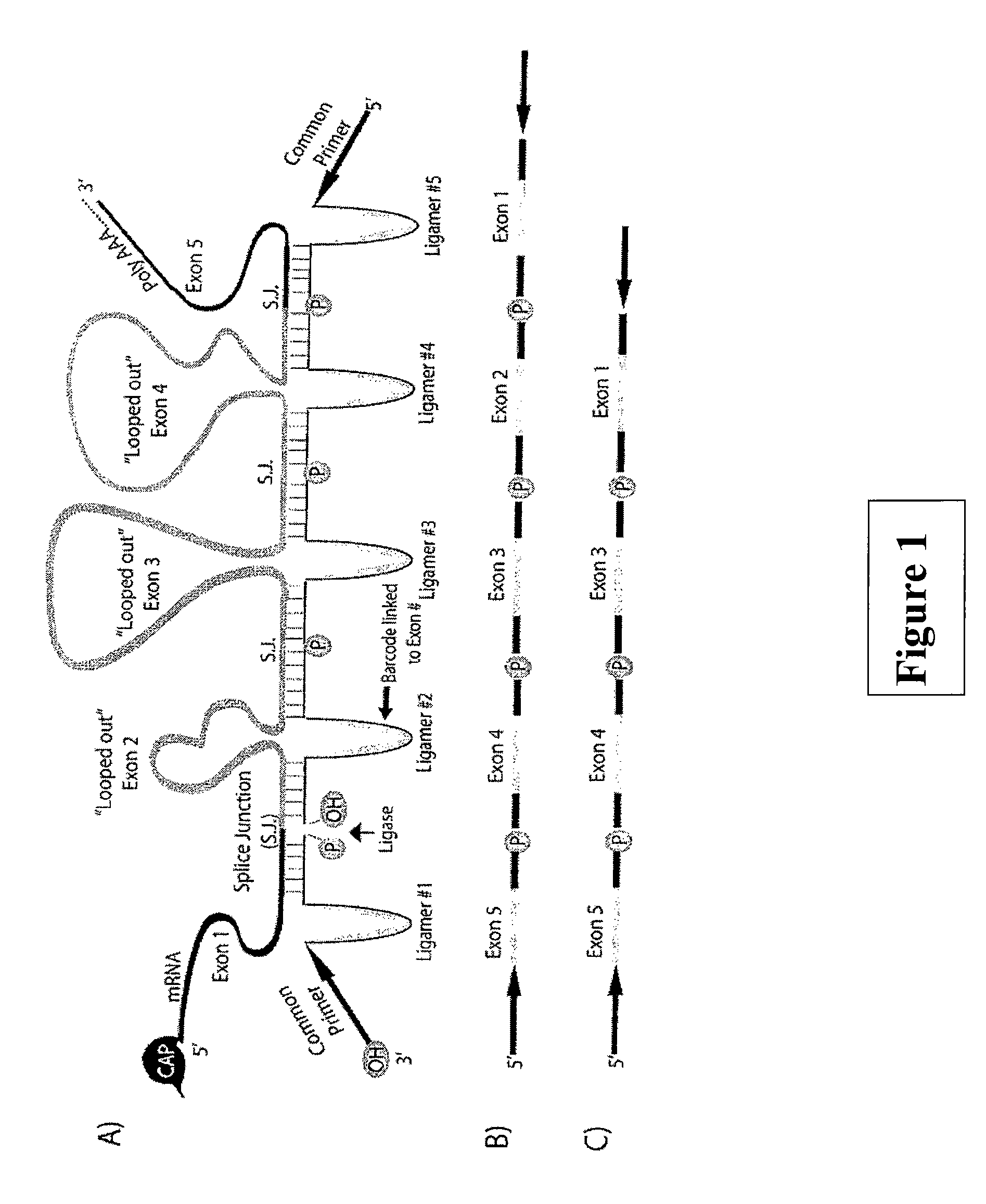
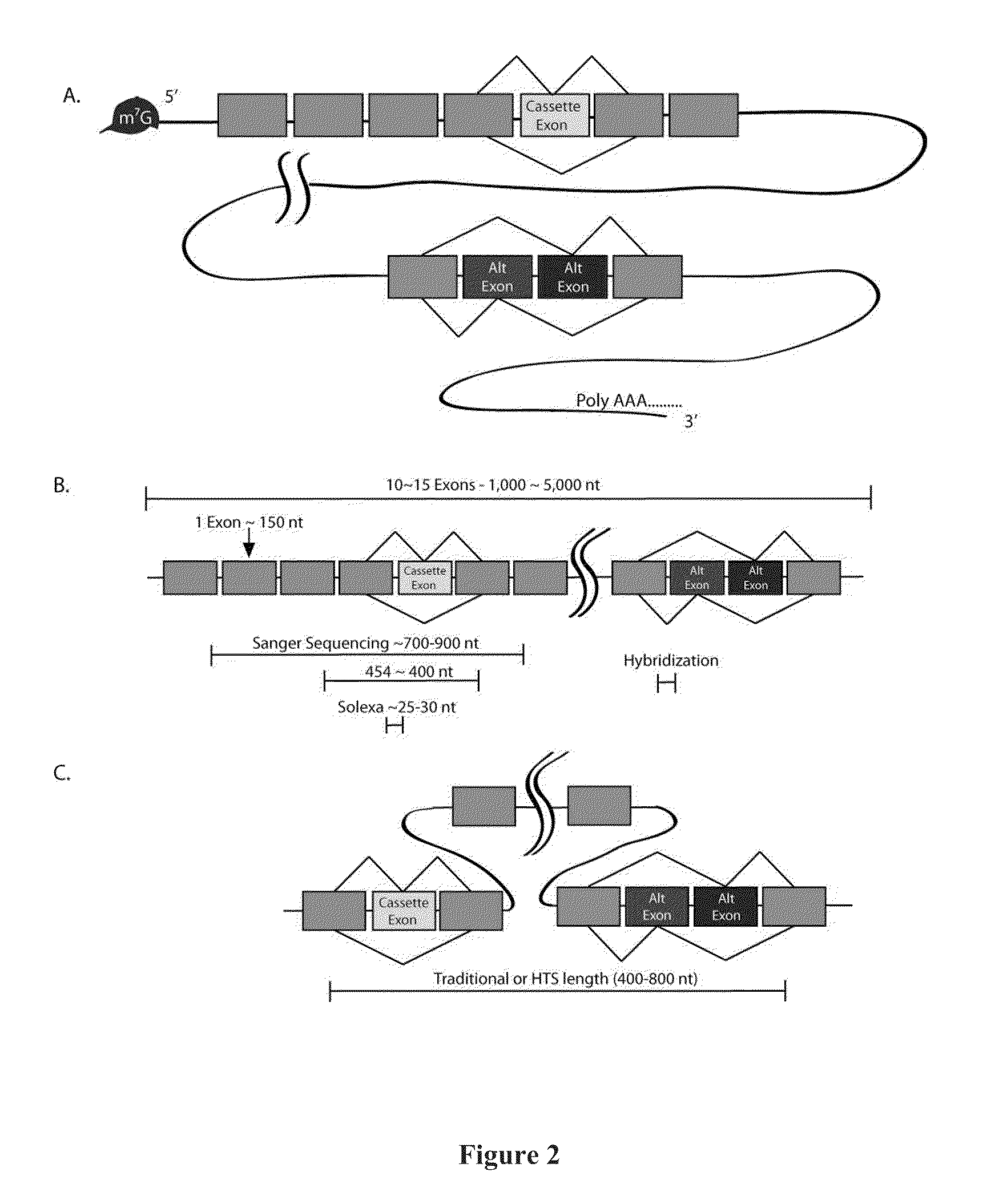
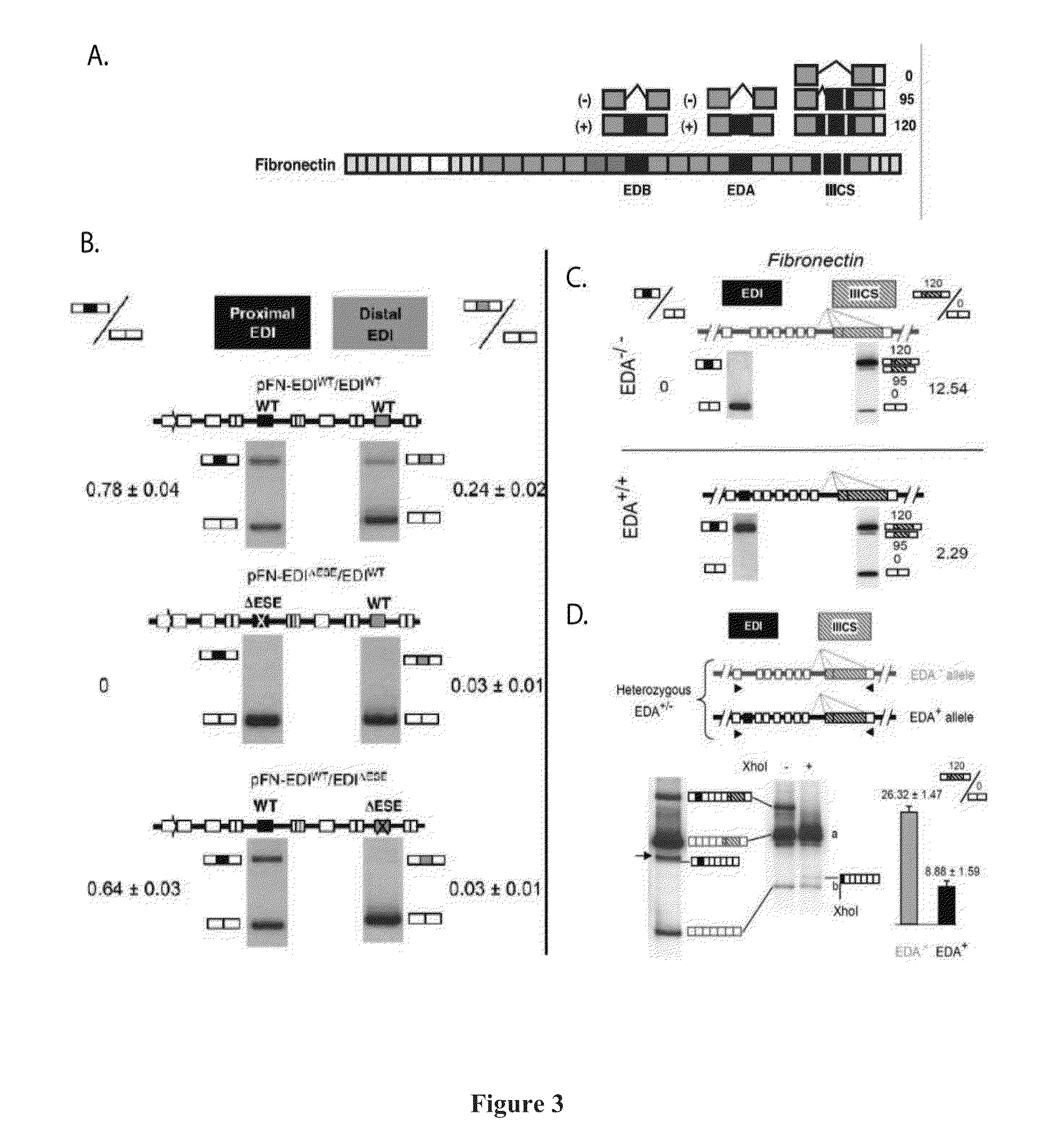
Deducing Exon Connectivity by RNA-Templated DNA Ligation/Sequencing
A technology is described that is capable of generating high-throughput sequencing (HTS) read length DNA products to accurately and reliably provide exon connectivity information for alternatively spliced isoforms. The method is not limited by the initial size of the isoform as the technology removes the template oligonucleotide sequence and a newly formed full length ligated product provides an HTS-compatible read length sequence that comprises information that corresponds to the consecutive order of the exons in the original template oligonucleotide.
Owner:UNIV OF MASSACHUSETTS MEDICAL SCHOOL
Comparative gene transcript analysis
InactiveUS6114114ASugar derivativesMicrobiological testing/measurementSequence analysisDisease cause
A method and system for quantifying the relative abundance of gene transcripts in a biological sample. One embodiment of the method generates high-throughput sequence-specific analysis of multiple RNAs or their corresponding cDNAs (gene transcript imaging analysis). Another embodiment of the method produces a gene transcript imaging analysis by the use of high-throughput CDNA sequence analysis. In addition, the gene transcript imaging can be used to detect or diagnose a particular biological state, disease, or condition which is correlated to the relative abundance of gene transcripts in a given cell or population of cells. The invention provides a method for comparing the gene transcript image analysis from two or more different biological samples in order to distinguish between the two samples and identify one or more genes which are differentially expressed between the two samples.
Owner:INCYTE PHARMA INC
Digital protein quantification
PendingUS20170192013A1Sequential/parallel process reactionsLibrary tagsQuantification methodsQuantitative proteomics
Methods and compositions are described for single cell resolution, quantitative proteomic analysis using high throughput sequencing.
Owner:BIO RAD INNOVATIONS SAS +1
Determining paired immune receptor chains from frequency matched subunits
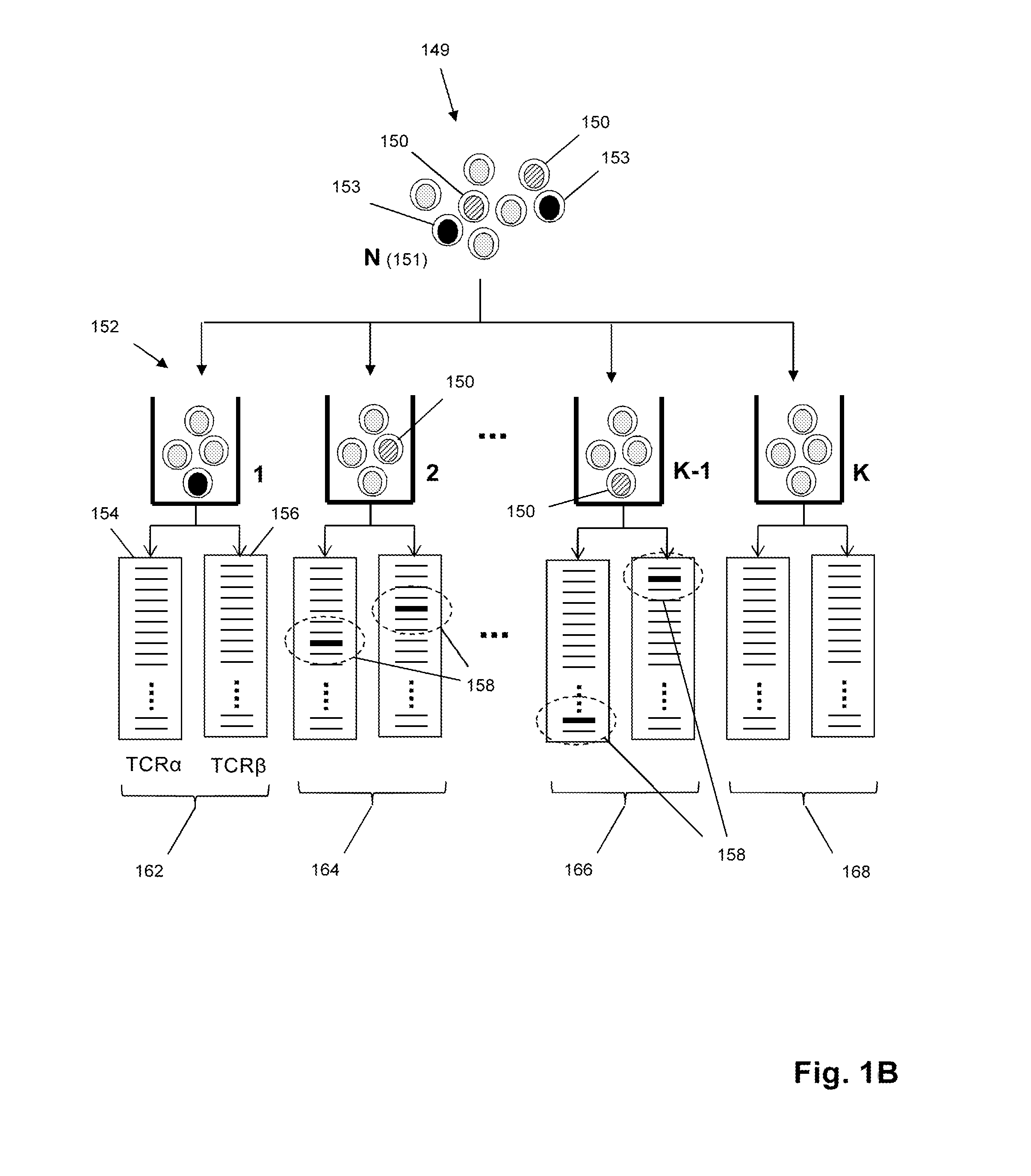
The invention is directed to methods for determining nucleic acids that encode immune receptor chains originating from the same cell, that is, paired immune receptor chains. Methods of the invention comprise high-throughput sequencing of rearranged nucleic acids encoding immune receptors from one or more samples of lymphocytes. In one aspect, from a plurality of subsets of a sample, nucleic acids encoding separate chains of a pair are separately sequenced, wherein the size of the sample and the number of subsets are selected so that the distribution of lymphocytes approximates a binomial model. Paired chains are determined by identifying pairs that appear together or that are entirely absent in the subsets.
Owner:ADAPTIVE BIOTECH
Compositions and methods for co-amplifying subsequences of a nucleic acid fragment sequence
ActiveUS20140243242A1Nucleotide librariesMicrobiological testing/measurementEmulsionEmulsion polymerization
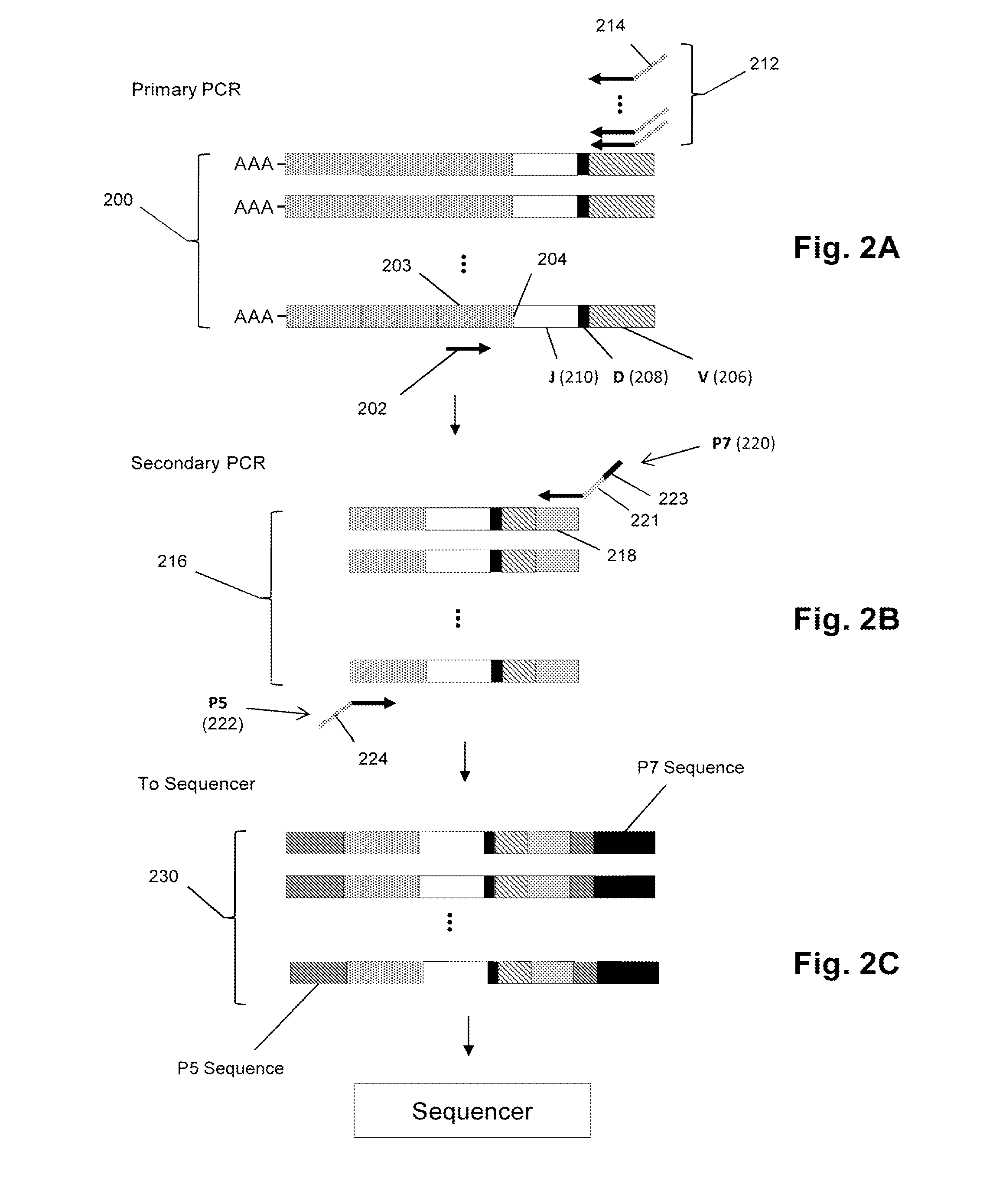
The present invention is related to genomic nucleotide sequencing. In particular, the invention describes a single reaction method to co-amplify multiple subsequences of a nucleic acid fragment sequence (i.e., for example, at least two read pairs from a single library insert sequence). Nucleic acid fragment sequences may include, but are not limited to, localizing library insert sequences and / or unique read pair sequences in specific orientations on a single emulsion polymerase chain reaction bead. Methods may include, but are not limited to, annealing, melting, digesting, and / or reannealing high throughput sequencing primers to high throughput sequencing primer binding sites. The compositions and methods disclosed herein contemplate sequencing complex genomes, amplified genomic regions, as well as detecting chromosomal structural rearrangements that are compatible with massively parallel high throughput sequencing platforms as well as ion semiconductor matching sequencing platforms (i.e., for example, Ion Torrent platforms).
Owner:THE BROAD INST INC
Method for capturing genome target sequence based on Crispr/cas9 and application thereof in high-throughput sequencing
PendingCN107190008AMicrobiological testing/measurementStable introduction of DNAEnzymeSpecific population
The invention discloses a method for capturing a genome target sequence based on Crispr / cas9 and application thereof in high-throughput sequencing. The method for capturing a target gene sequence from a genome DNA on the basis of a Crispr / cas9 system comprises the steps that 1, a plurality of gRNAs are designed and synthesized according to the target gene sequence; 2, the multiple gRNAs and a cas9 enzyme are adopted for performing a cleavage reaction on the genome DNA, and a cleavage product containing multiple 100-300 bp fragmentation products is obtained; 3, the multiple 100-300 bp fragmentation products in the cleavage product are captured. On the basis of the CRISPR gene, the target sequence is edited and captured, and the method can be used for high-throughput sequencing and is applicable to screening of specific population with family heredity, the public physical examination market and the like.
Owner:SUZHOU GENESCI CO LTD
High throughput sequencing of multiple transcripts of a single cell
ActiveUS20150141261A1Nucleotide librariesMicrobiological testing/measurementHigh throughput sequenceImmune receptor
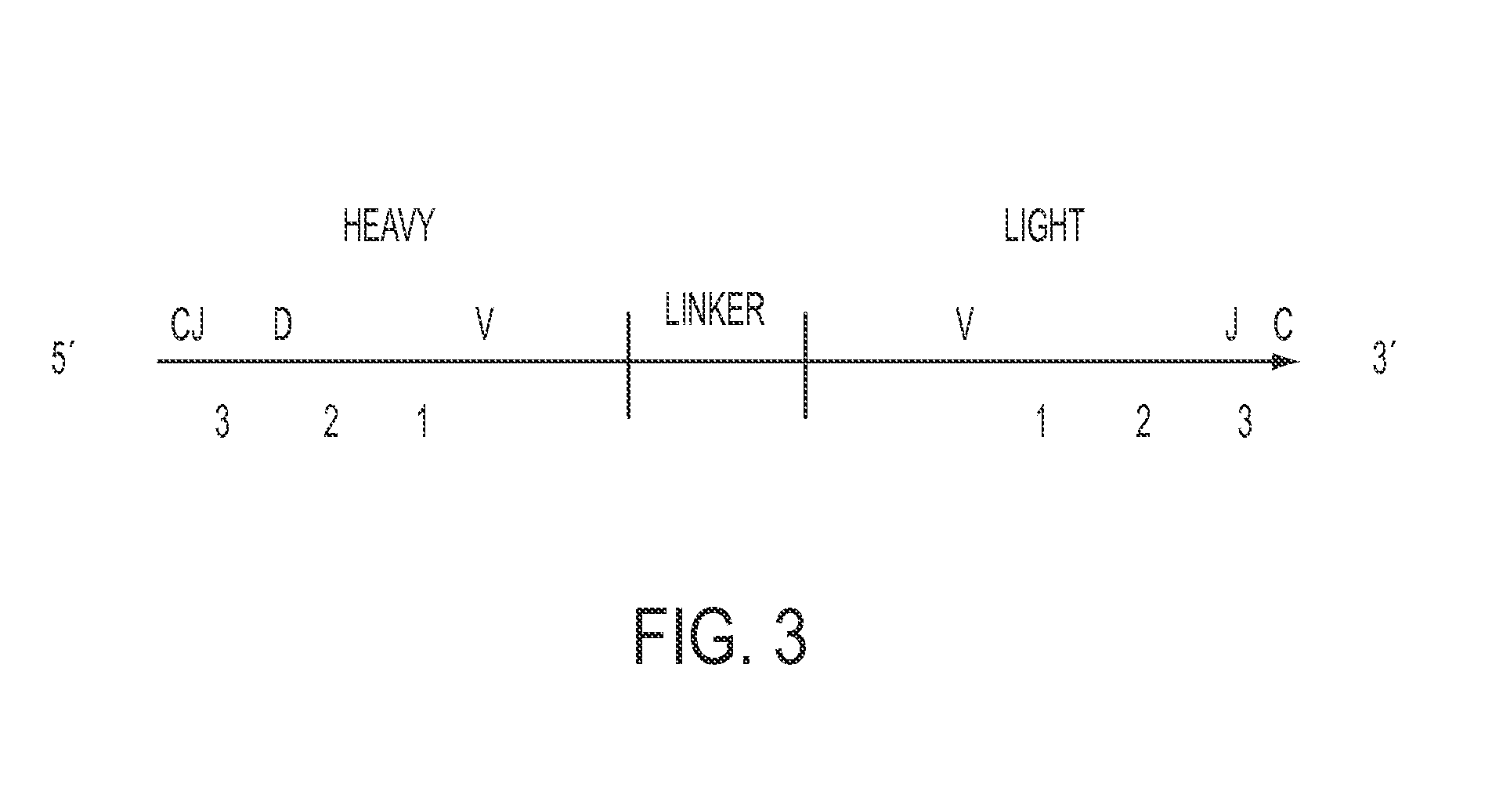
The present disclosure generally relates to sequencing two or more genes expressed in a single cell in a high-throughput manner. More particularly, the present disclosure relates to a method for high-throughput sequencing of pairs of transcripts co expressed in single cells (e.g., antibody VH and VL coding sequence) to determine pairs of polypeptide chains that comprise immune receptors.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Spatial and cellular mapping of biomolecules in situ by high-throughput sequencing
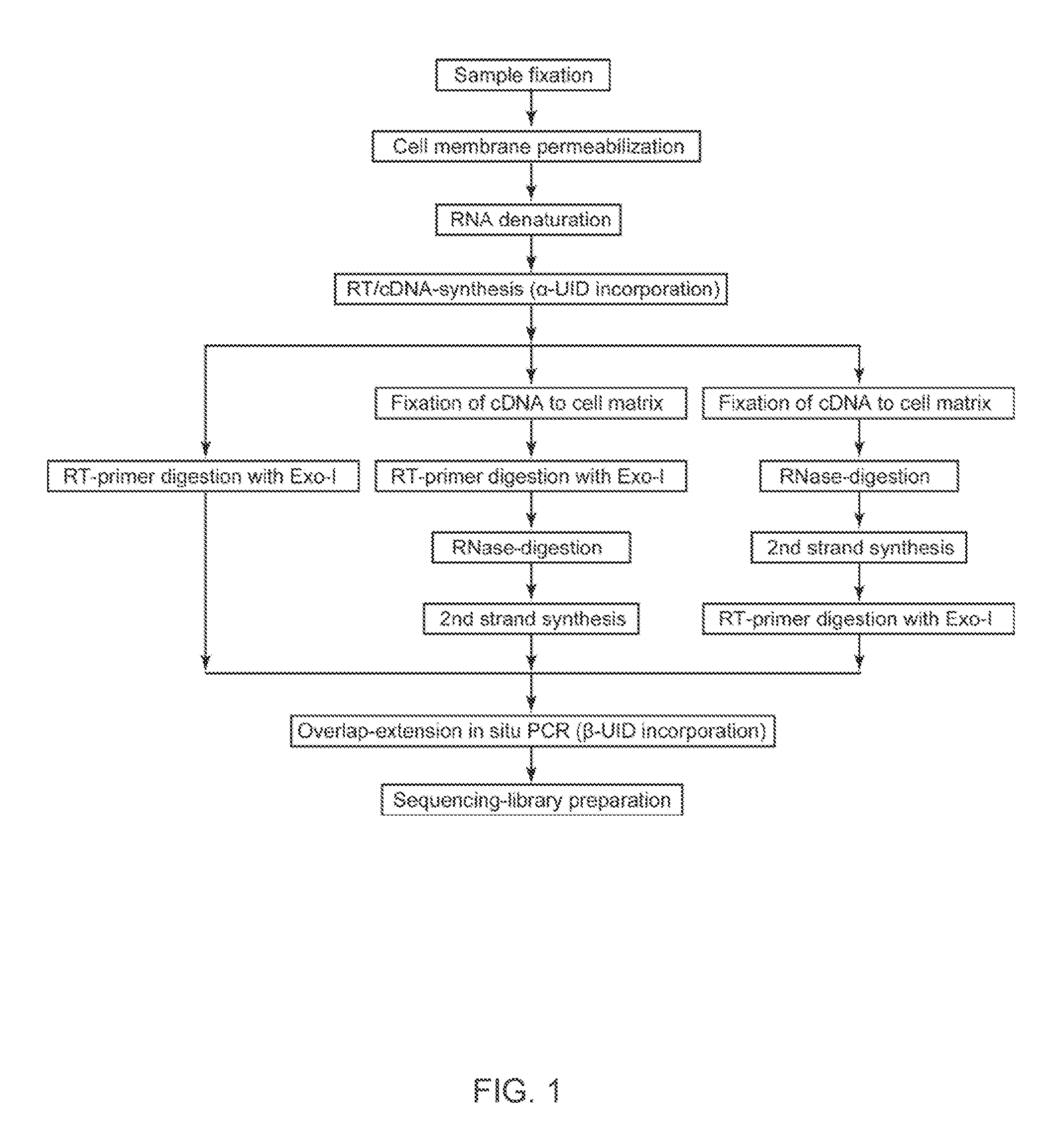
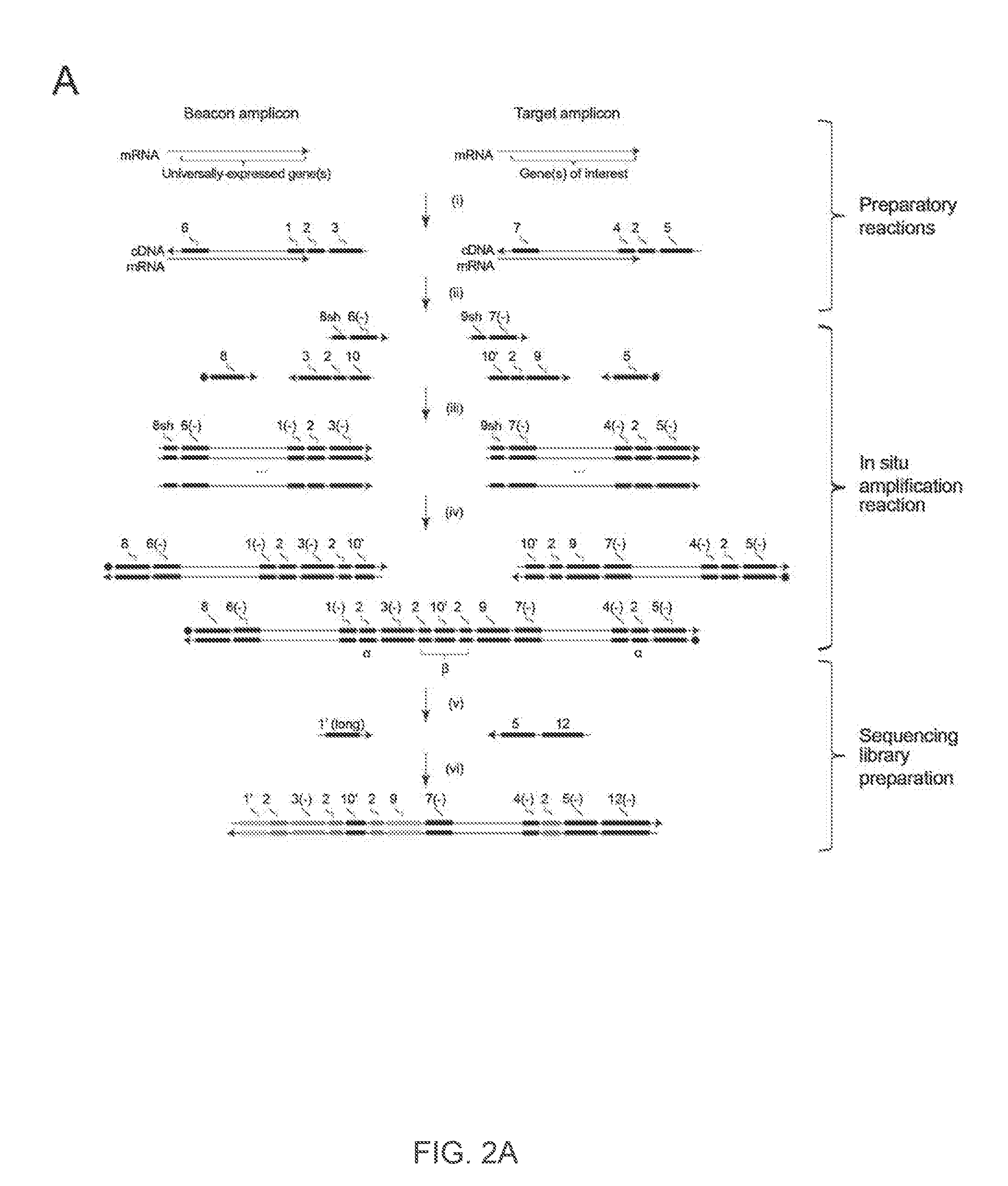
The present invention relates to molecular microscopy or volumetric imaging by proximal unique molecular identifiers (“UID”) reaction (“VIPUR”) microscopy methods to record the cellular co-localization and / or spatial distributions of arbitrary nucleic acid sequences, or other biomolecules tagged with nucleic sequences. The method involves one or both of two DNA sequence-components such as an α-UID, which may identify the targeted sequences-of-interest themselves and / or spatial beacons relative to which their distances are measured, and a −β-UID, which labels α-UID association events.
Owner:THE BROAD INST INC +1
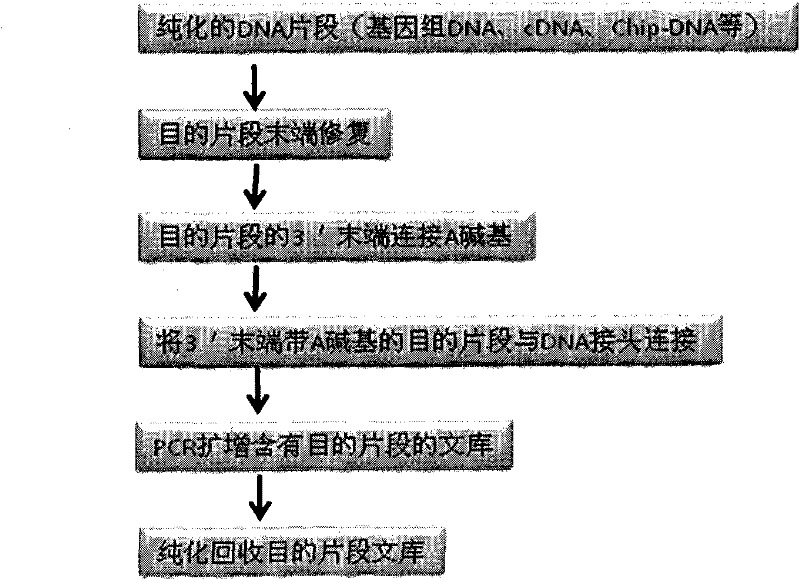
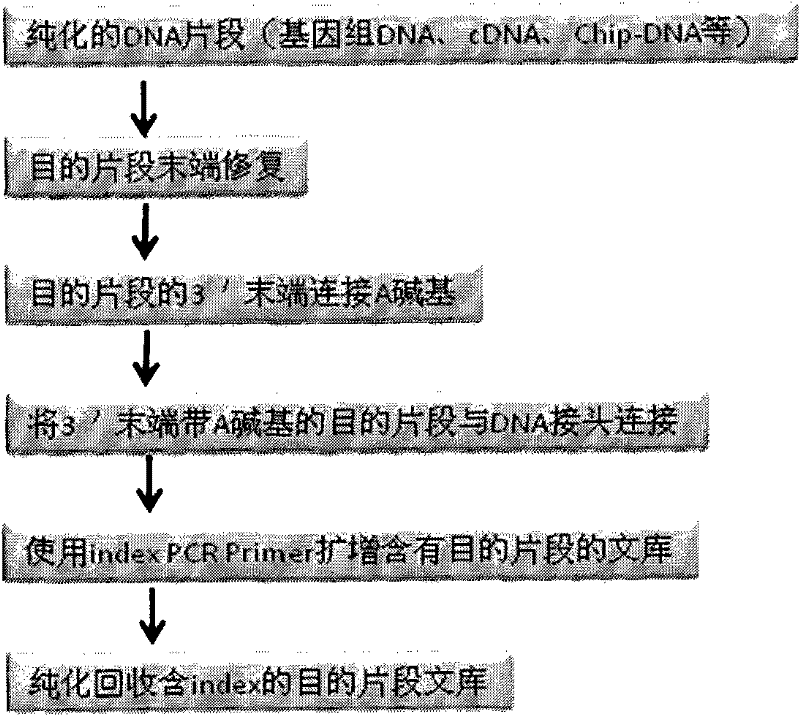
DNA index library building method based on high throughput sequencing
ActiveCN102409048AImprove production efficiencyHigh sequencing throughputNucleotide librariesMicrobiological testing/measurementSingle sampleA-DNA
The invention provides unique index sequences with a length of 7bp. The index sequences can be respectively imported into a DNA(deoxyribonucleic acid) index library through adapter link and PCR (polymerase chain reaction). The invention provides a method for building the DNA index library by using the index sequences based on a solexa sequencing platform of the current illumina company, and the method is applied to solexa DNA sequencing and has the effects on improving the preparation efficiency of the DNA index library, increasing the sequencing throughput of the DNA samples and lowering thesolexa sequencing cost of a single sample.
Owner:BGI TECH SOLUTIONS
High sensitivity mutation detection using sequence tags
InactiveUS20160115532A1Improve accuracyHigh sensitivityMicrobiological testing/measurementNucleotideMutation detection
The invention is directed to methods for increasing the sensitivity of high throughput sequencing, particularly for distinguishing true rare mutations from amplification, sequencing and other sample processing errors that occur in sequencing techniques. In one aspect, methods of the invention includes steps of (a) preparing templates from nucleic acids in a sample; (b) labeling by sampling the templates to form tag-template conjugates, wherein substantially every template of a tag-template conjugate has a unique sequence tag; (c) linearly amplifying the tag-template conjugates; (d) generating a plurality of sequence reads from the linearly amplified tag-template conjugates; and (e) determining a nucleotide sequence of each of the nucleic acids based on the frequencies, or numbers, of each type of nucleotide at each nucleotide position of each plurality of sequence reads having identical sequence tags.
Owner:ADAPTIVE BIOTECH
Methods and compositions for high-throughput sequencing
InactiveUS20140024541A1Microbiological testing/measurementLibrary screeningBarcodeHigh throughput sequence
The invention provides methods, apparatuses, and compositions for high-throughput amplification sequencing of specific target sequences in one or more samples. In some aspects, barcode-tagged polynucleotides are sequenced simultaneously and sample sources are identified on the basis of barcode sequences. In some aspects, sequencing data are used to determine one or more genotypes at one or more loci comprising a causal genetic variant.
Owner:COUNSYL INC
Preparation method of non-transgenic CRISPR mutant
InactiveCN108611364AReduce frequencyImprove accuracyNucleic acid vectorVector-based foreign material introductionScreening methodSexual reproduction
The invention provides a preparation method of a non-transgenic mutant based on a CRISPR-Cas9 technology. Specifically, the preparation method comprises a construction method and a screening method. The construction method is characterized in that CRISPR-Cas9 and sgRNA are preassembled and are located at T-DNA of agrobacterium tumefaciens; site-directed change of a target gene of a target plant can be realized by infection of the target plant by the agrobacterium tumefaciens and coculture without integrating the sequences of the CRISPR-Cas9 and the sgRNA into a genome of the target plant, so that an obtained site-directed mutant material of the target gene does not need the processes of sexual reproduction, segregation posterity, population screening and the like or does not contain any exogenous gene sequence. The obtained regeneration seedlings are screened by a high-throughput sequencing and high-resolution melting curve technology provided by the invention; mutant plants of which target genes are mutated can be obtained by identifying the regeneration seedlings efficiently and quickly at the current generation of transgene, even if low-proportion mutants of which the proportionis 1 / 100 of a mixed population can be screened. The screening method provided by the invention still can ensure excellent accuracy and sensitivity.
Owner:NANJING AGRICULTURAL UNIVERSITY
Library preparation method of trace nucleic acid sample and application thereof
ActiveCN103060924ANucleotide librariesMicrobiological testing/measurementNucleotideLibrary preparation
The invention relates to a library preparation method of a trace nucleic acid sample and application thereof. The method comprises: using a DOP (Degenerate Oligonucleotide Primed) primer to perform first amplification on DNA in the sample, with a DOP primer sequence having at least a 5' terminal nondegenerate nucleotide zone and a 3' terminal degenerate nucleotide zone; employing a DOP-Amp primer to conduct second amplification on a first PCR product so as to obtain a second PCR amplification product; and carrying out adaptor-ligation PCR on the second PCR amplification product to obtain a third PCR product, and taking the third PCR product with sequencing adaptors on both ends as the nucleic acid library. The library can be used for fragment size selection and high-throughput sequencing to obtain gene information related to diseases in the sample. The invention also provides a kit and its application in preparation of the trace nucleic acid sample library.
Owner:BGI BIOTECH WUHAN CO LTD
Lung cancer somatic mutation detection and analysis method based on high-throughput sequencing technique
InactiveCN107391965AEfficient detectionAnalytical results are reliableSequence analysisSpecial data processing applicationsMutation detectionLung cancer
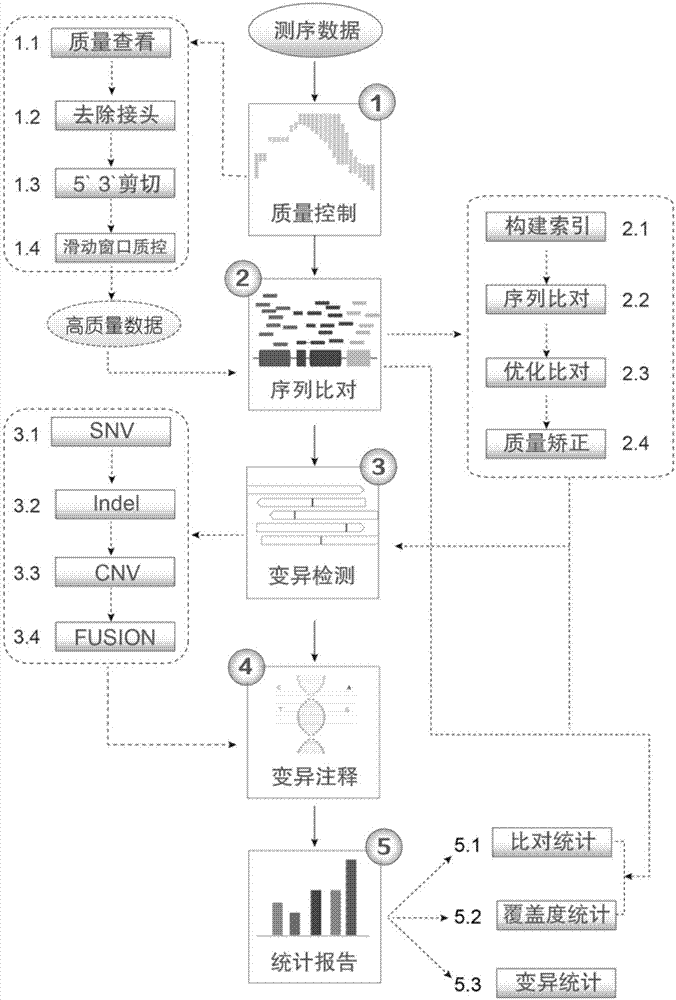
The invention discloses a lung cancer somatic mutation detection and analysis method based on a high-throughput sequencing technique. The method comprises the steps of (1) controlling quality; (2) comparing sequences; (3) detecting mutations; (4) annotating the mutations; (5) reporting the results. Compared with the prior art, the method of the invention has the advantages that the detection results are reliable; the detection content is diversified; the detection method is flexible; the data results are visualized.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Construction method of high-flux sequencing library and reagent kit for library construction
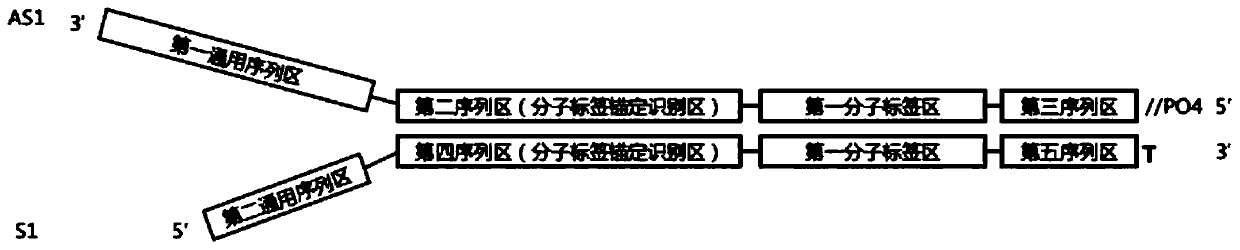
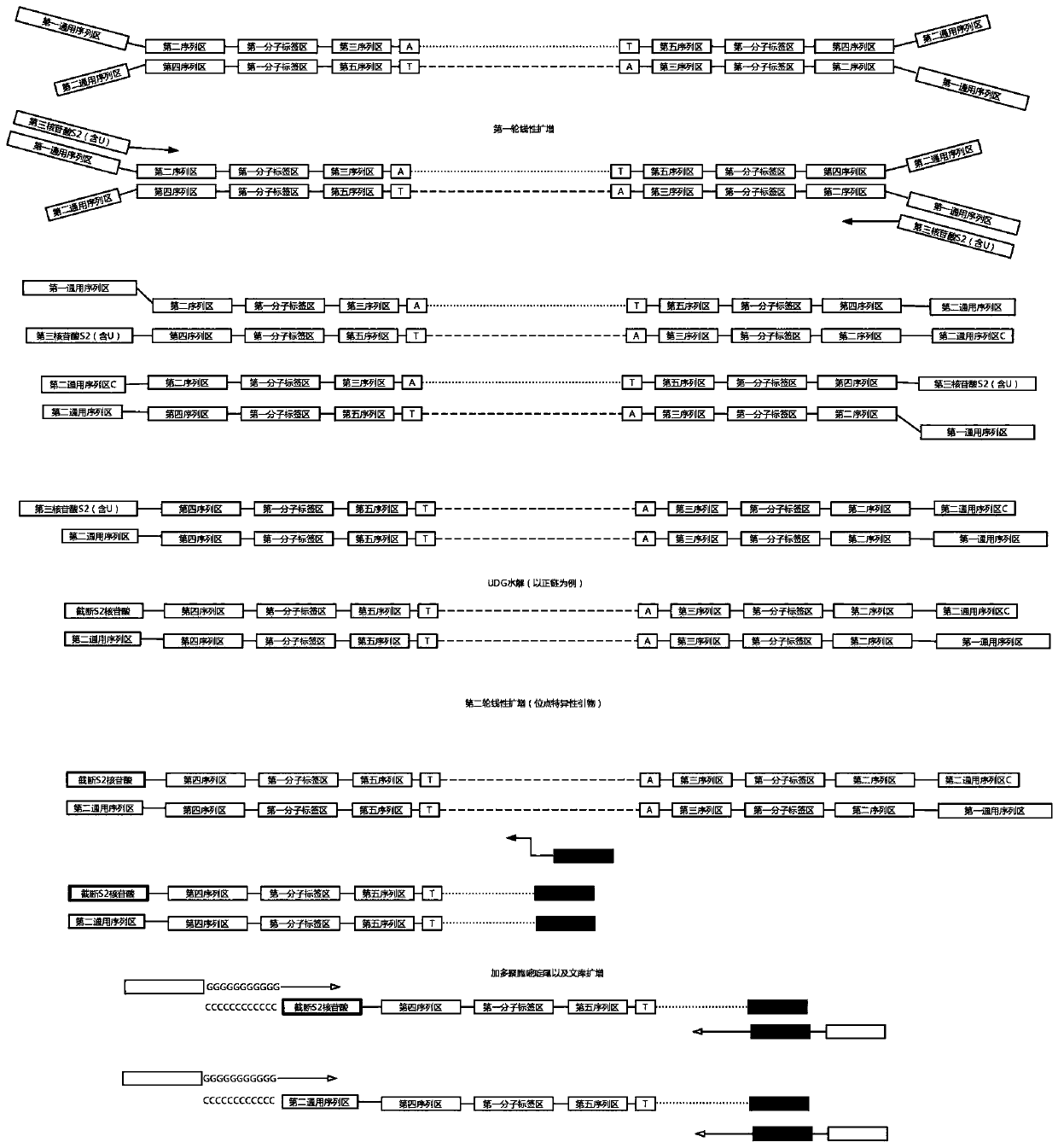
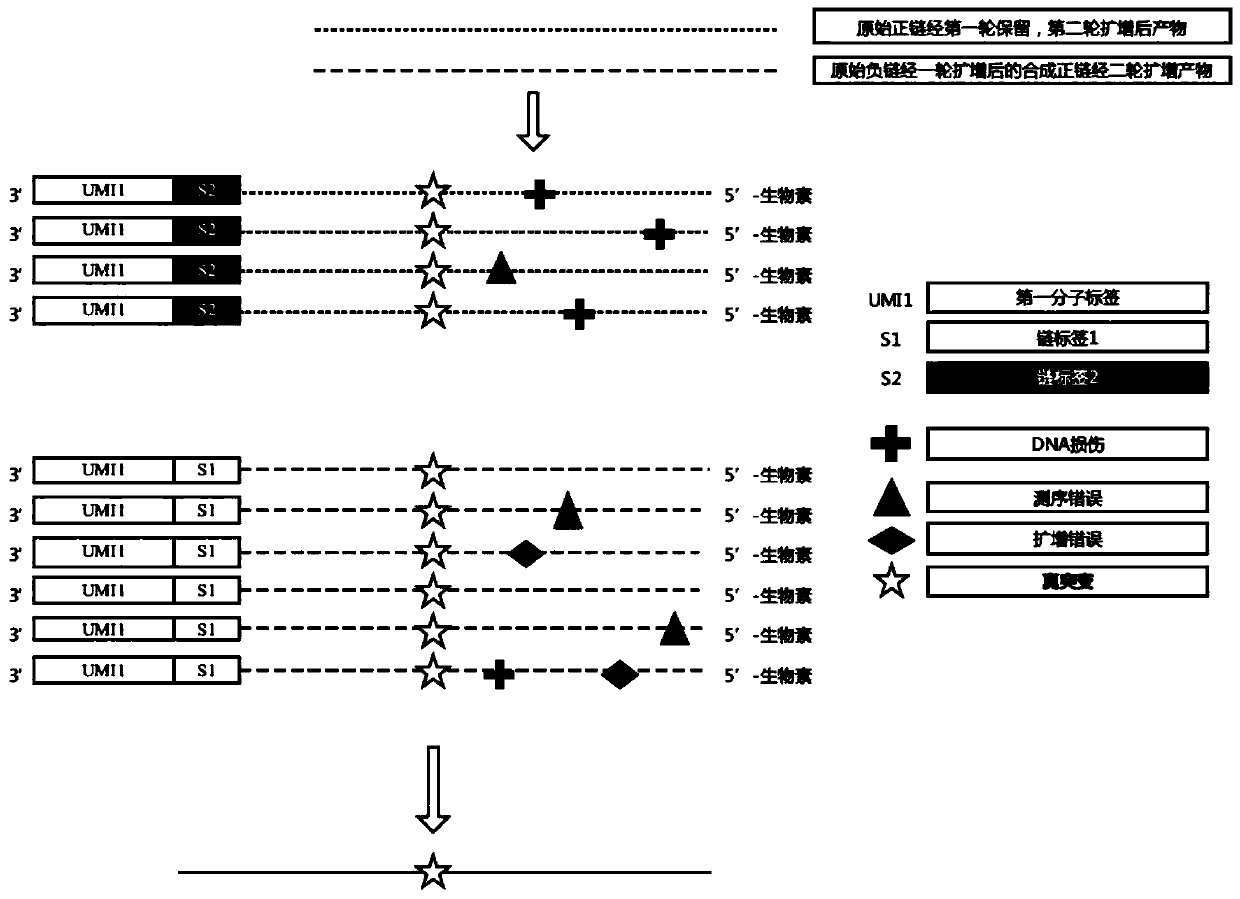
ActiveCN110734908AAchieve rearrangementAvoid the phenomenon of heterogeneous amplificationNucleotide librariesMicrobiological testing/measurementMultiplexMutation detection
The invention provides a construction method of a high-flux sequencing library and a reagent kit for library construction. The reagent kit comprises one or more of the following components: a high-flux sequencing Y-shaped joint, a universal primer for single-end linear PCR amplification, a biotin labeling specific primer for single-end linear multiplex PCR amplification, forward and backward library amplification primers, a UDG enzyme and the like. The invention relates to a method for constructing a targeting dimolecular identifier (UMI (unique molecular identifier) and chain unique molecularidentifier) high-flux sequencing library based on the reagent kit. According to the method, double error correcting mechanisms of a random UMI and a chain unique molecular identifier having sequencepolymorphism in a targeting sequencing system based on multiplex PCR amplification are realized, and disadvantages of most of conventional multiplex PCR amplification targeting sequencing systems areavoided, so that all false positives and false negatives in mutation detection are avoided, and high-sensitivity high-accuracy high-depth detection can be performed on low-frequency nucleic acid mutation in samples.
Owner:福州福瑞医学检验实验室有限公司
Construction method of single cell transcriptome sequencing library and application of construction method
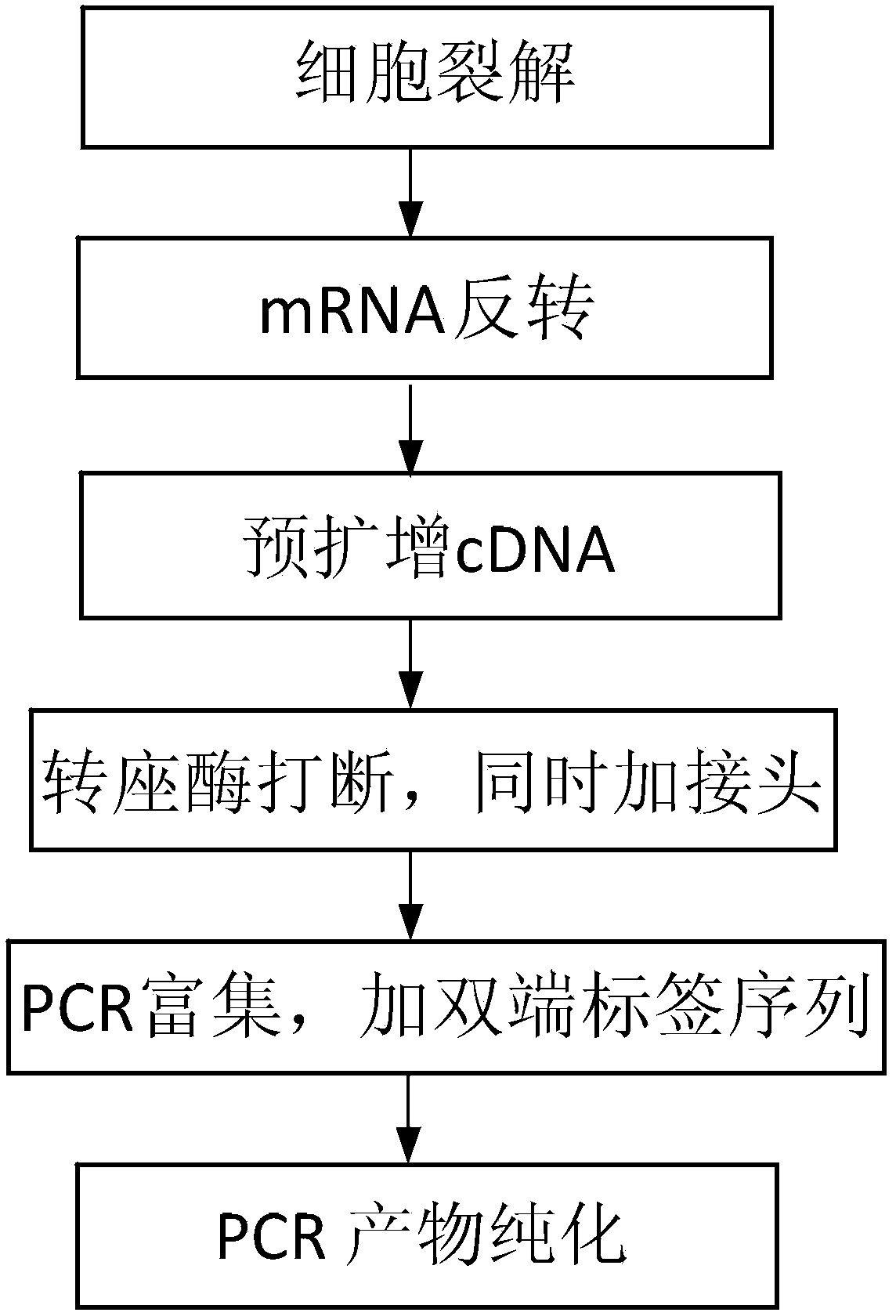
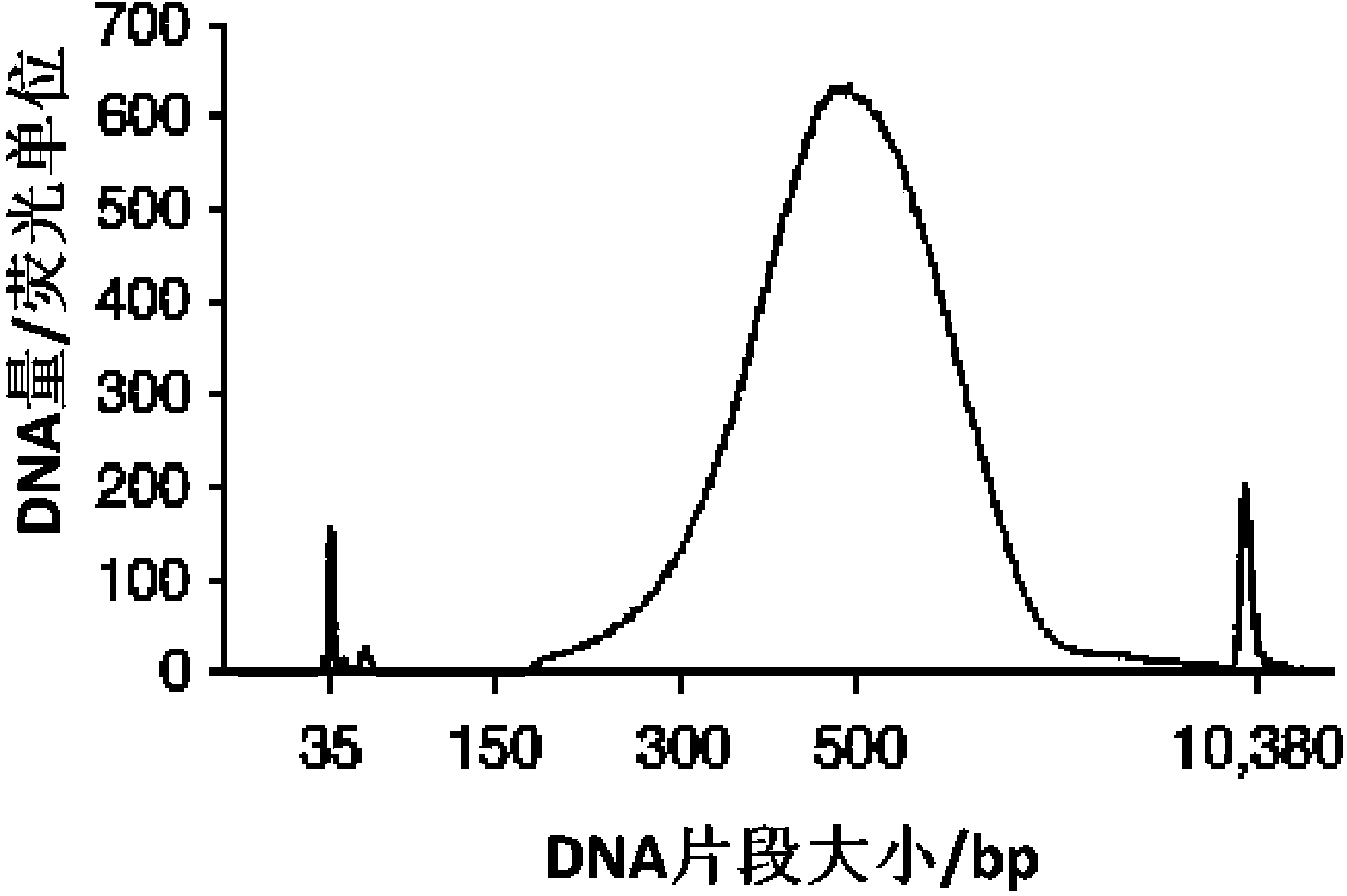
InactiveCN104032377AReduce sample lossImprove stabilityMicrobiological testing/measurementLibrary creationTranscriptome SequencingThroughput
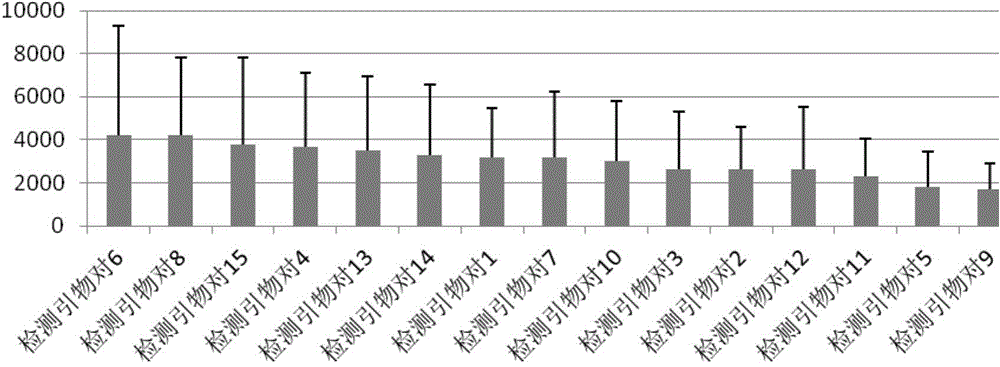
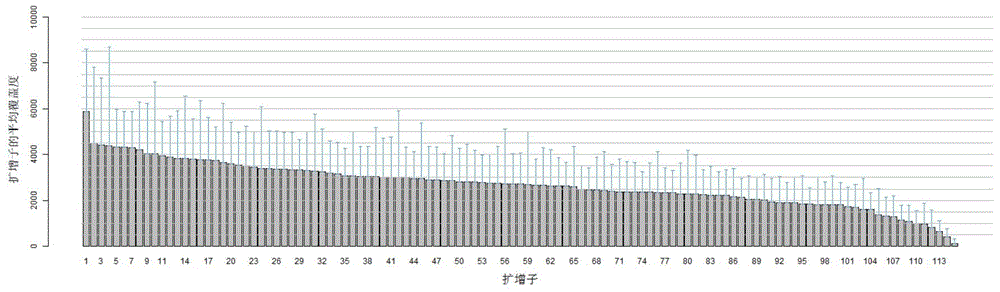
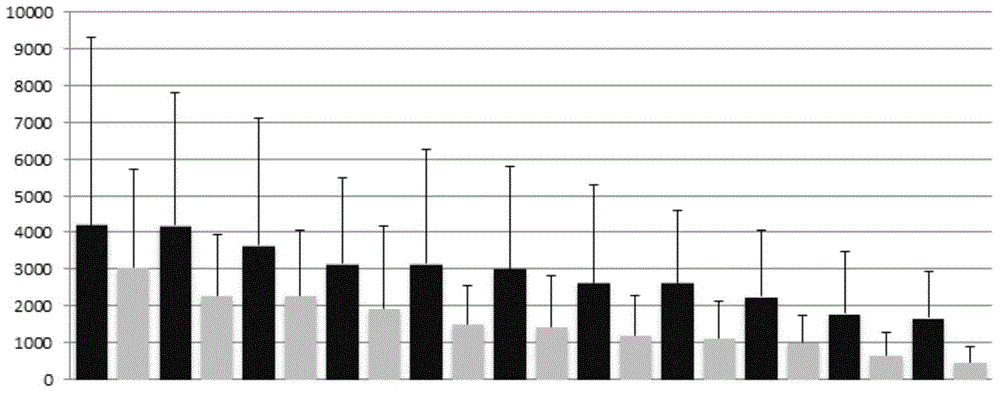
The invention discloses a construction method of a single cell transcriptome sequencing library and application of the construction method. The construction method includes the following steps of preparing a cDNA sample from a single cell and conducting fragmentized library construction on the cDNA sample to obtain the transcriptome sequencing library of the single cell, wherein fragmentation is conducted on the cDNA sample with a physical breaking method in the step of constructing the fragmentized library, and the step for purifying the fragmentized cDNA sample is not needed. According to the method, fragmentation is conducted on the sample of the single cell with the physical breaking method, and therefore fragments obtained through breaking are small and relatively uniform, and the peak width of the constructed single cell transcriptome sequencing library is concentrated; the purification step is not needed for the fragmentized sample, sample loss is reduced, construction of the single cell transcriptome sequencing library of the micro-sample can be achieved, stability of output of data volume can be improved, and therefore the reliability of large-scale and high-throughput sequencing of the single cell transcriptome sequencing library is improved.
Owner:BEIJING NOVOGENE TECH CO LTD
Cancer gene mutation and gene amplification detection
ActiveCN104630375AAmplified equalizationImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationMultiplexMedicine
The invention provides a primer set for simultaneously detecting mutation of a plurality of regions of two or more exons of one or more cancer drive genes in a sample on the basis of multiplex PCR (polymerase chain reaction) and high-flux sequencing techniques, a kit comprising the primer set, and a method for amplifying the cancer drive gene or detecting the cancer drive gene by using the primer set and / or kit. The mutation detection result can be used for instructing cancer clinic medication, auxiliary diagnosis or prognosis judgment on certain cancers.
Owner:北京圣谷智汇医学检验所有限公司
Mitochondria genome amplified universal primer mixture as well as design and amplification method thereof
ActiveCN105506103AEfficient and specific enrichmentAchieve deep coverageMicrobiological testing/measurementDNA/RNA fragmentationGenomic informationMitophagy
The embodiment of the invention discloses a universal primer mixture for animal mitochondria genome amplification as well as a design and amplification method thereof, and a kit comprising the universal primer mixture. By utilizing the primer mixture or the kit and the animal mitochondria genome amplified method and combining the novel high throughput sequencing technique, the animal mitochondria genome information can be efficiently obtained in low cost.
Owner:TIANJIN UNIV OF TRADITIONAL CHINESE MEDICINE
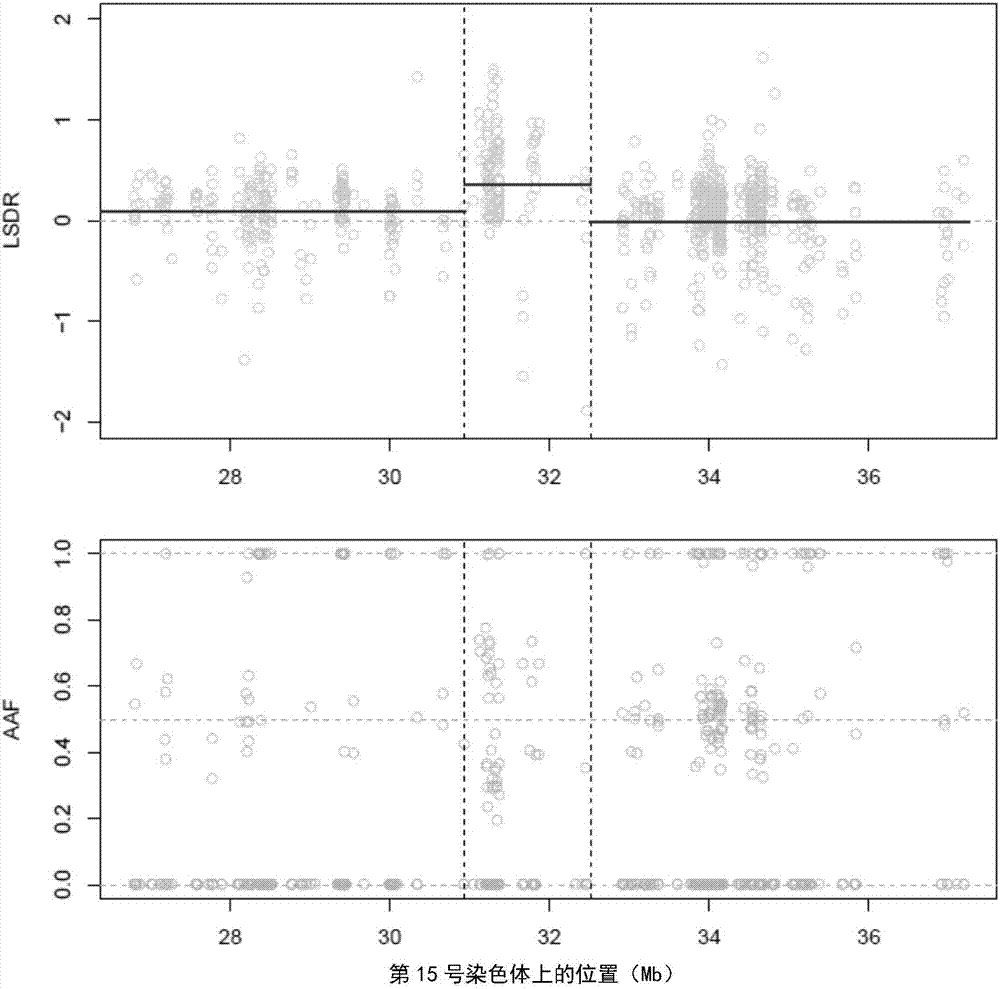
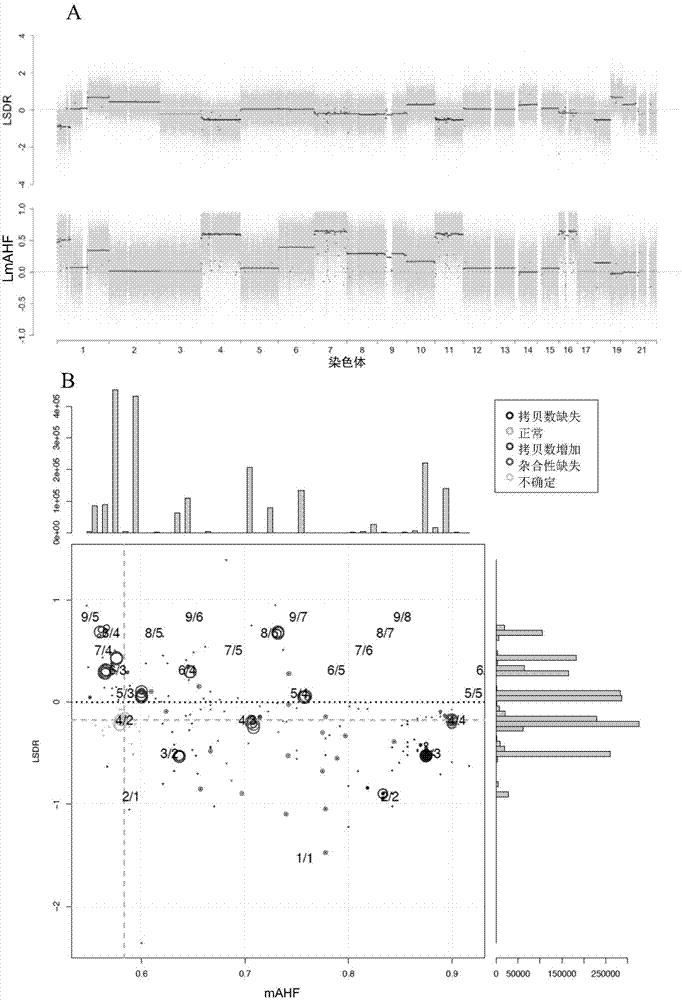
Detection method and system for copy number variation in genome
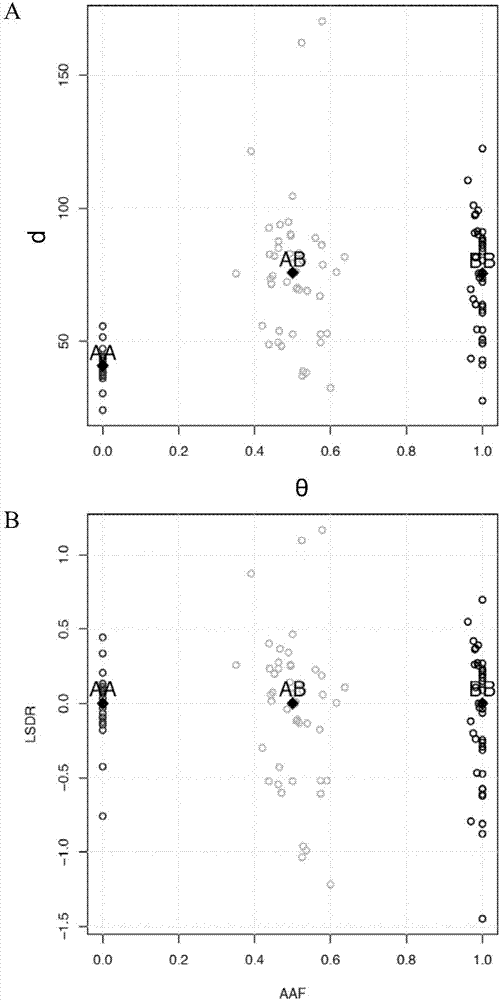
The invention provides a method for analyzing copy number variation in a genome of a testee. According to the method, mathematical manipulation is creatively performed on variables extracted from sequencing data, new two-dimensional variables, namely a sequencing depth ratio (SDR) and alternate allele frequency (AAF) or alternate allelic haploid frequency (AHF) are derived, the signal / noise ratio is further increased, detection sensitivity and precision are improved, and information contained in the high-throughput DNA sequencing data is utilized more fully. Besides, the method is used for processing free DNA in a body fluid sample of the testee, wherein when a CNV feature signal of a certain locus of the genome is extracted from the sequencing data, the information close to the locus is introduced, so that signal strength is greatly improved, and the copy number variation in the free DNA in urine or serum can be effectively detected. The invention furthermore provides a system for implementing the method.
Owner:郝柯 +1
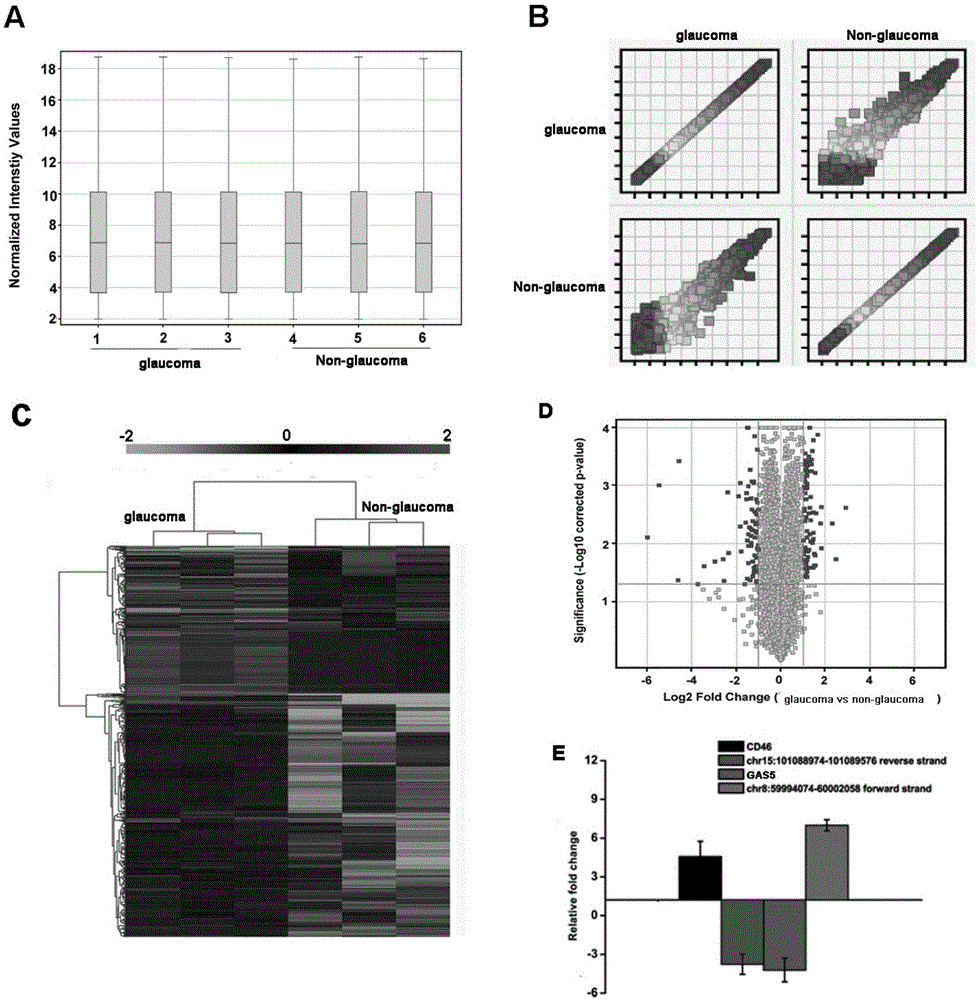
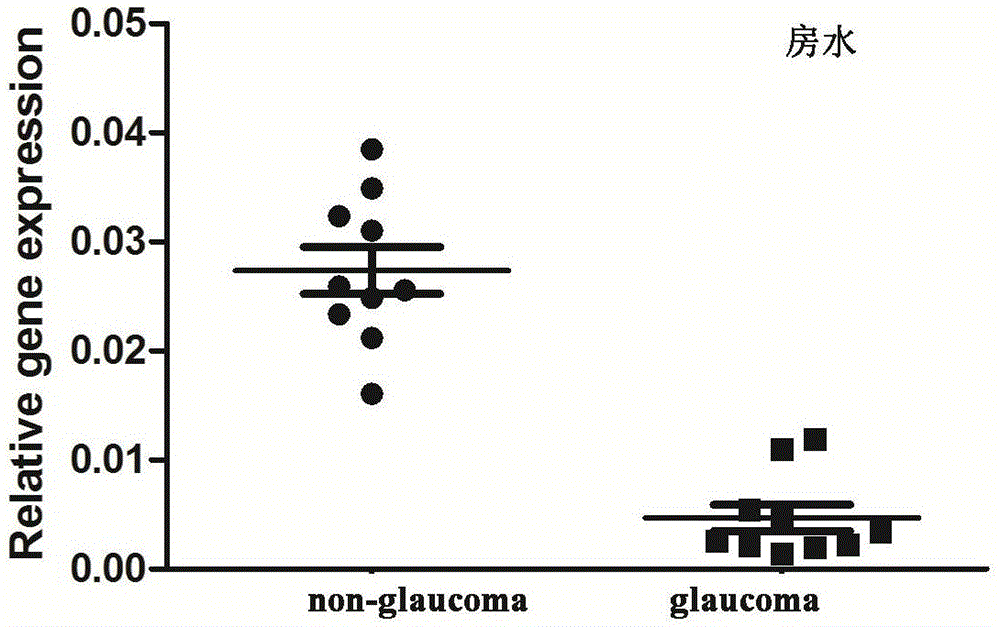
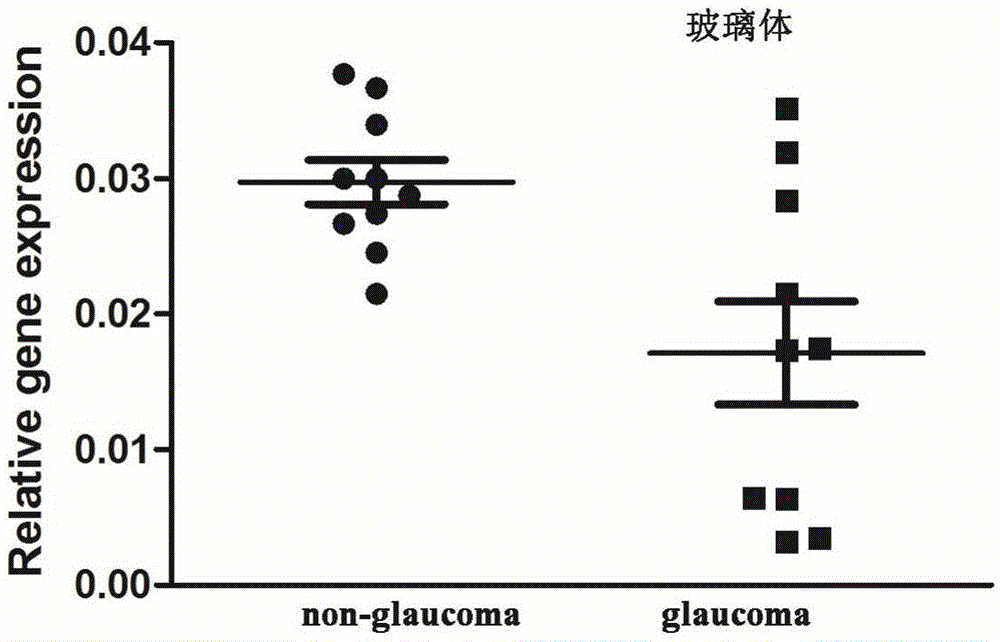
Application of LncRNA-GAS5 in preparing glaucoma diagnosis reagent
ActiveCN105002182AGood correlationReduce healingSenses disorderGenetic material ingredientsGlaucomaOcular disease
The invention discloses the application of LncRNA-GAS5 in preparing a glaucoma diagnosis reagent and a glaucoma lesion diagnosis reagent kit. Through high-throughput sequencing and quantitative PCR validation, it is found that the LncRNA-GAS5 has a significant correlation with the occurrence of glaucoma. The LncRNA-GAS5 can be used for screening clinical glaucoma patients and providing a theoretical basis for early intervention in glaucoma.
Owner:南京医科大学眼科医院
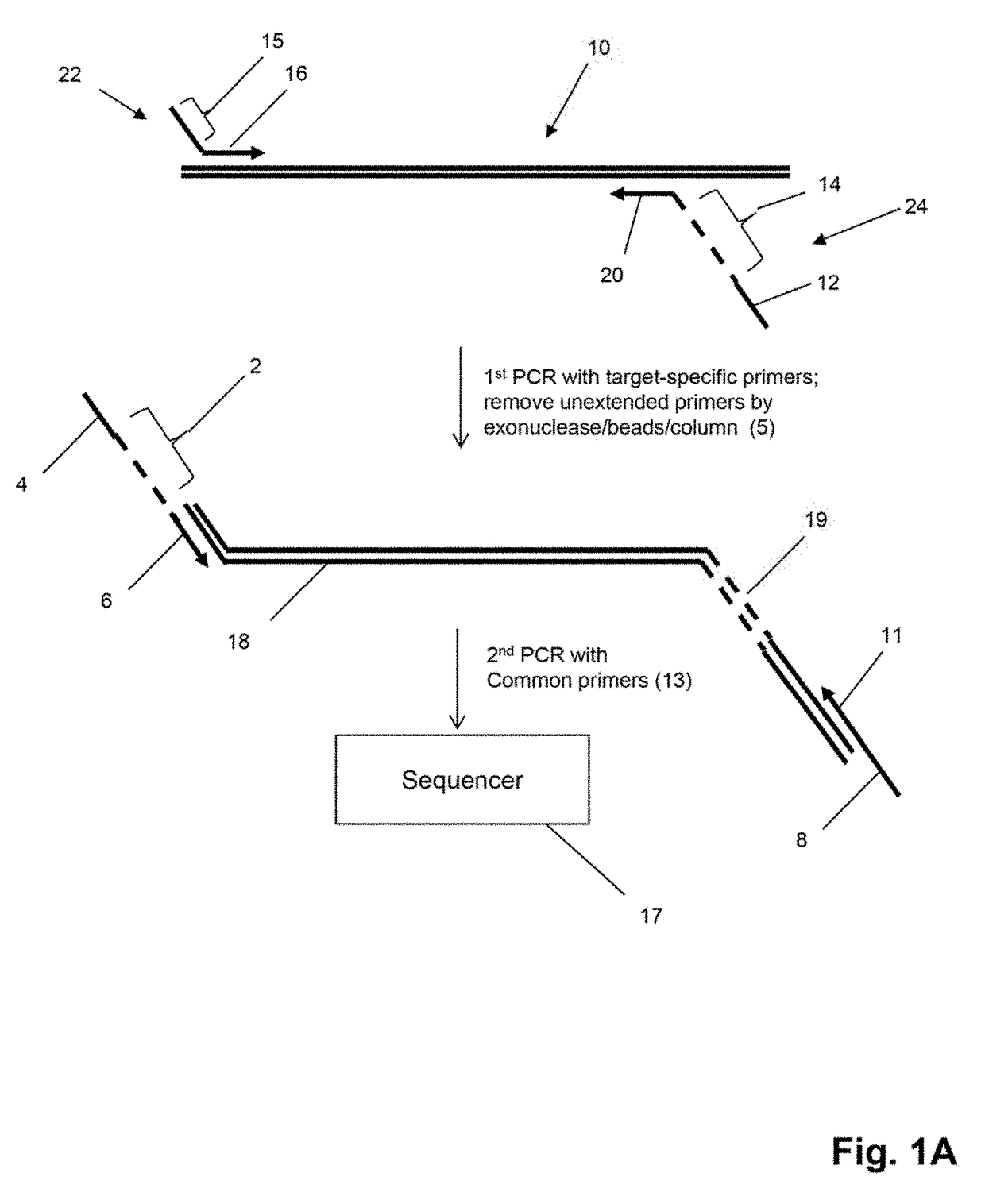
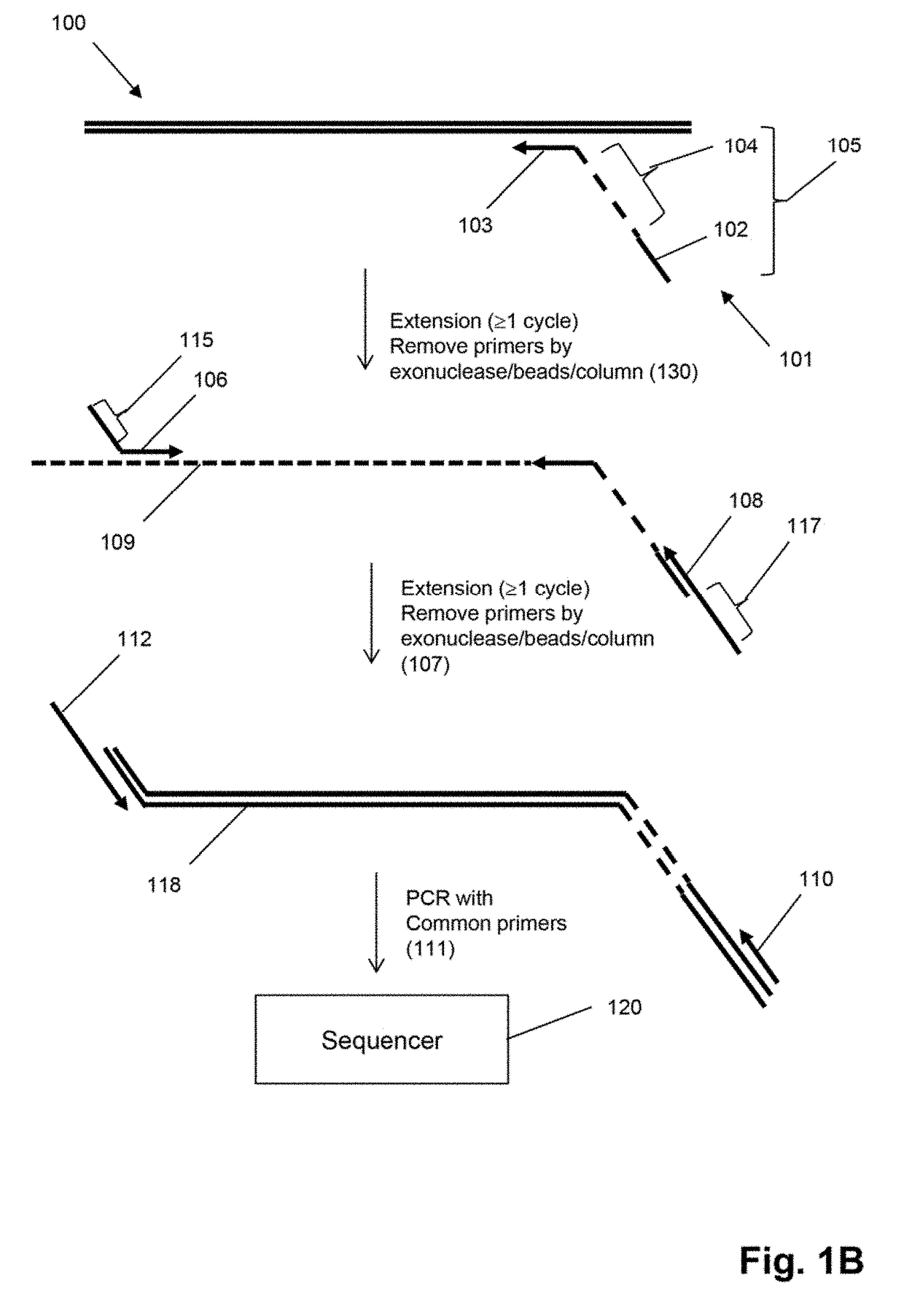
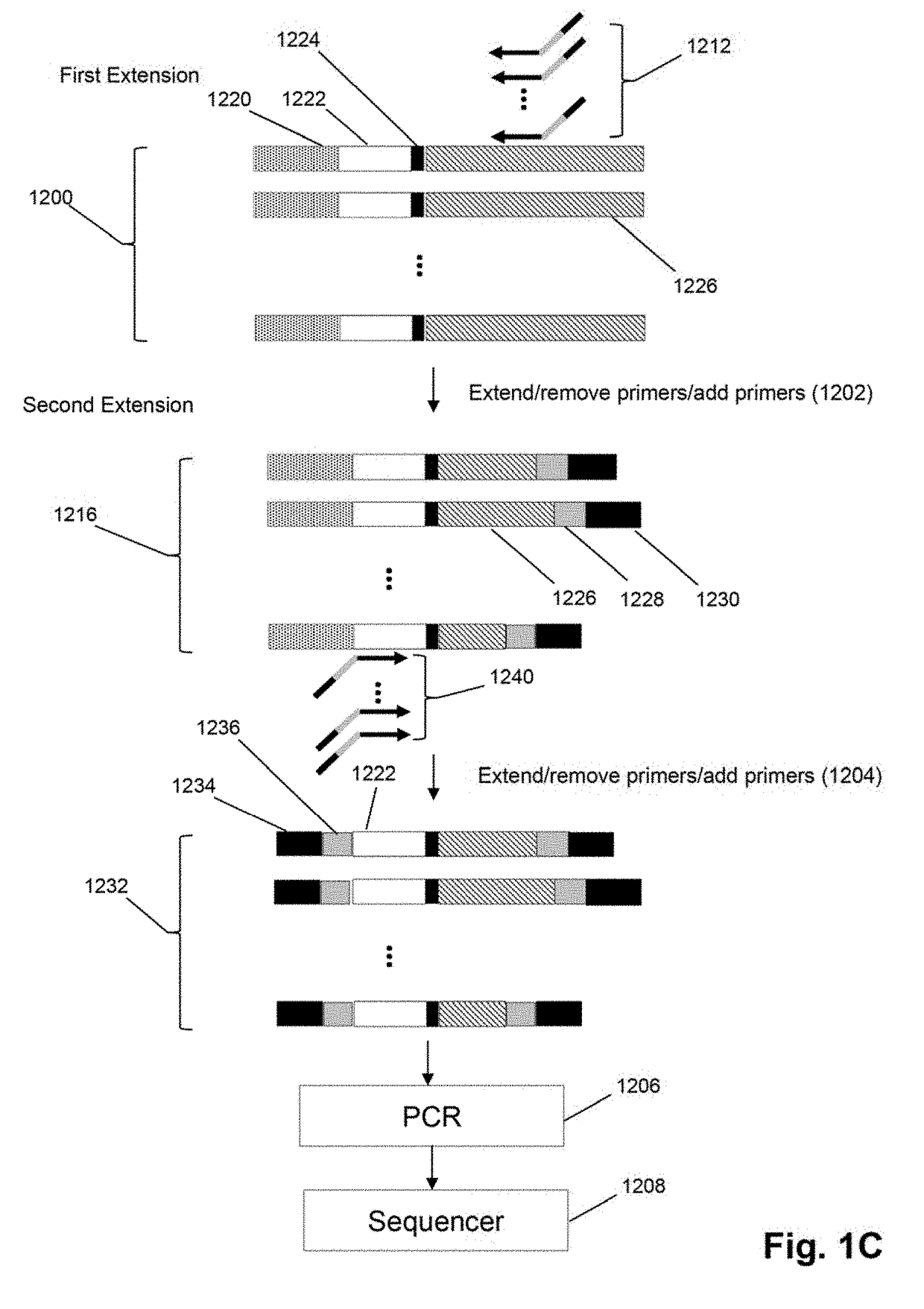
Method for generating clonotype profiles using sequence tags
ActiveUS9708657B2Health-index calculationMicrobiological testing/measurementHigh throughput sequenceContamination
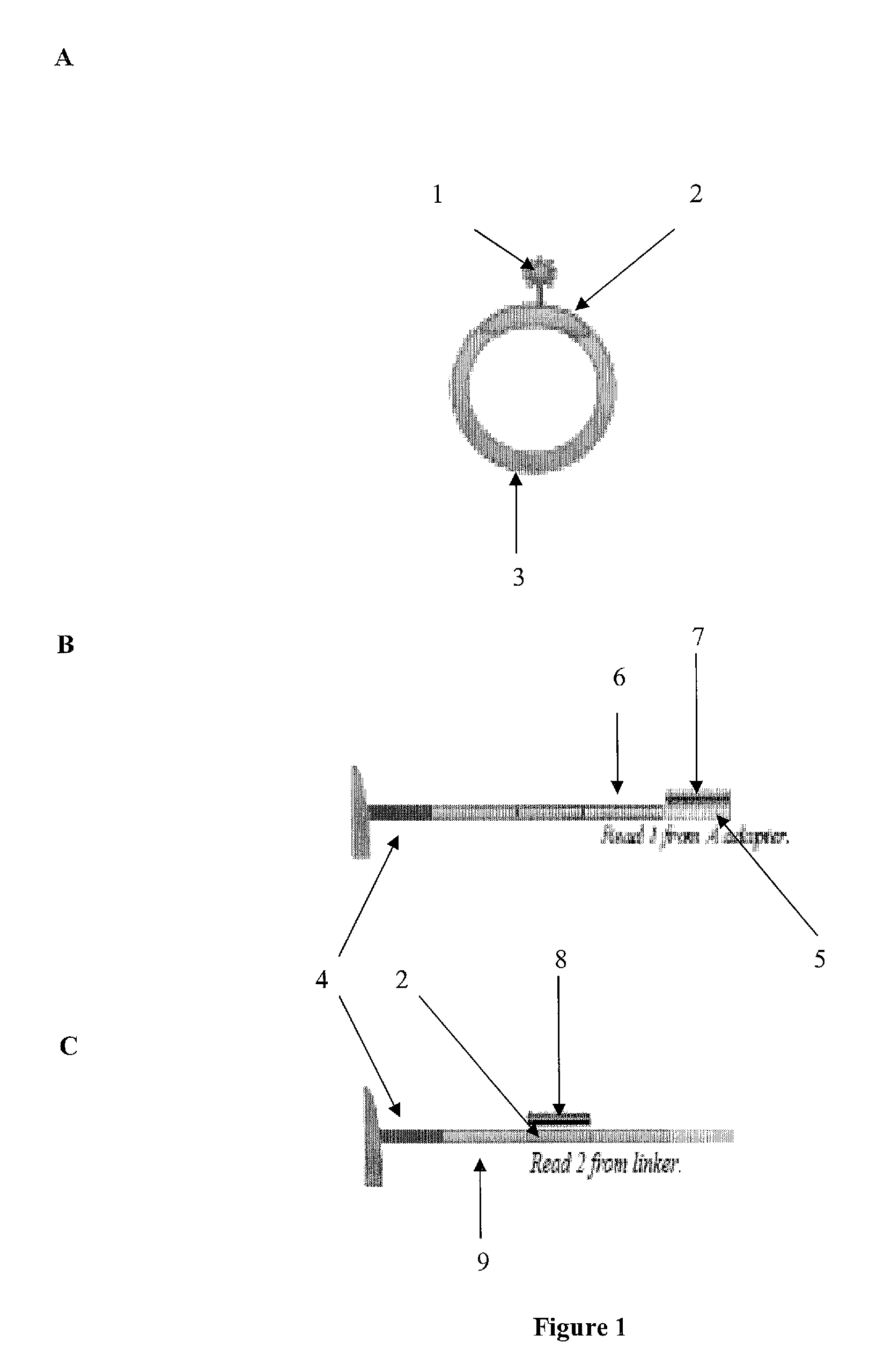
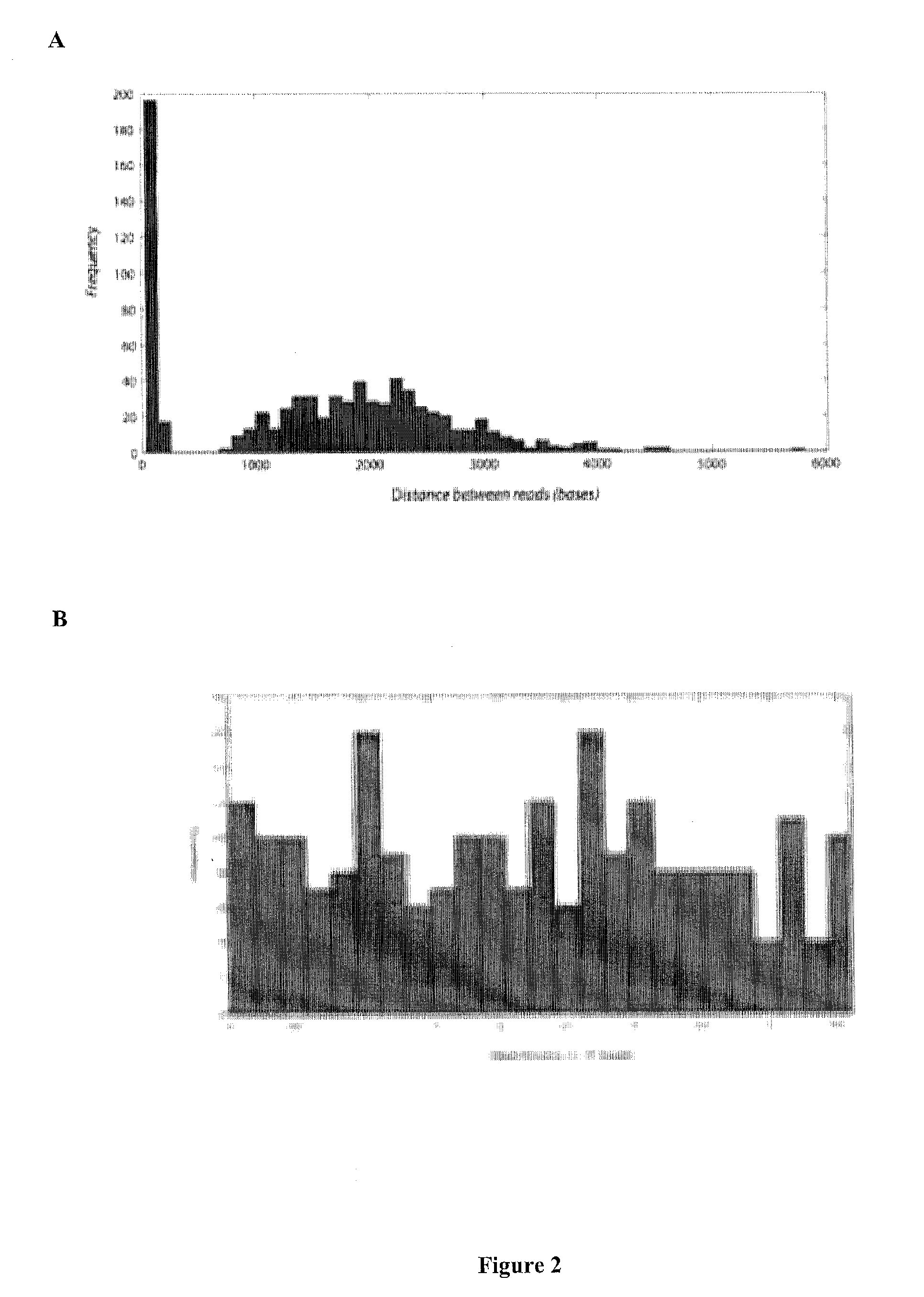
The invention is directed to sequence-based profiling of populations of nucleic acids by multiplex amplification and attachment of one or more sequence tags to target nucleic acids anchor copies thereof followed by high-throughput sequencing of the amplification product. In some embodiments, the invention includes successive steps of primer extension, removal of unextended primers and addition of new primers either for amplification (for example by PCR) or for additional primer extension. Some embodiments of the invention are directed to minimal residual disease (MRD) analysis of patients being treated for cancer. Sequence tags incorporated into sequence reads provide an efficient means for determining clonotypes and at the same time provide a convenient means for detecting carry-over contamination from other samples of the same patient or from samples of a different patient which were tested in the same laboratory.
Owner:ADAPTIVE BIOTECH
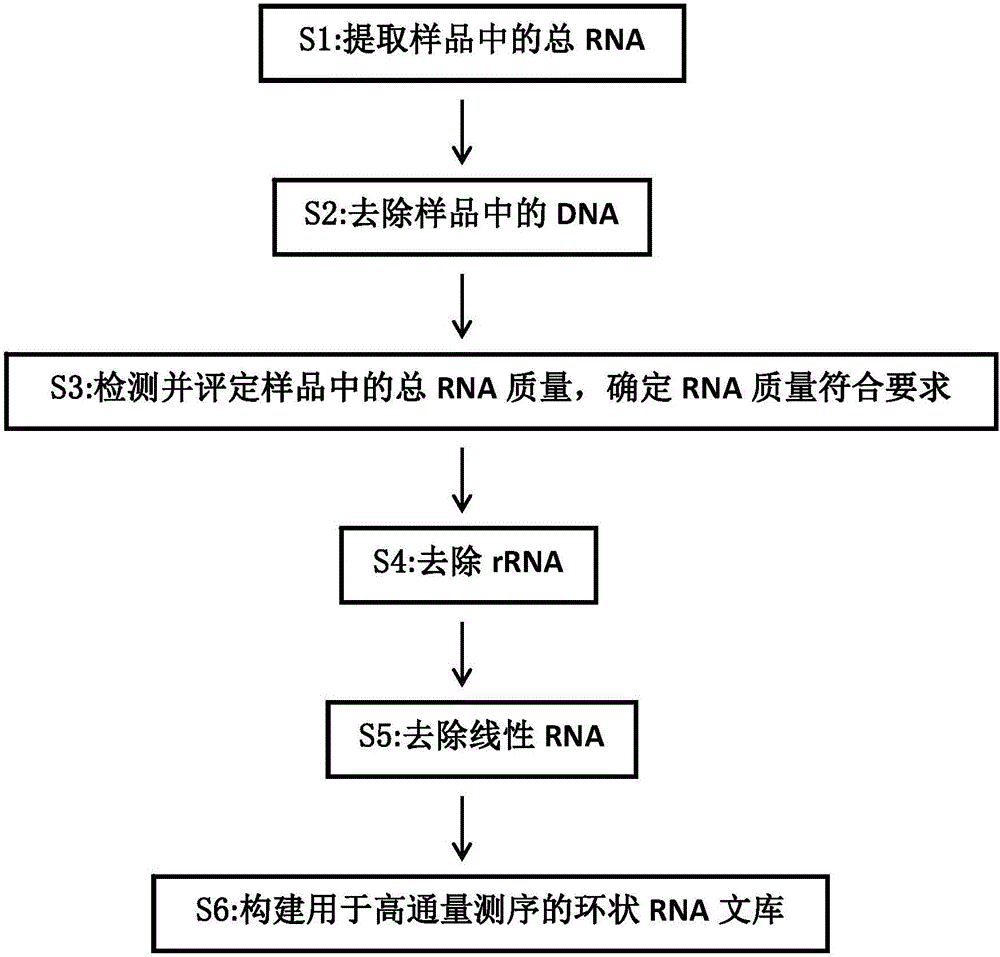
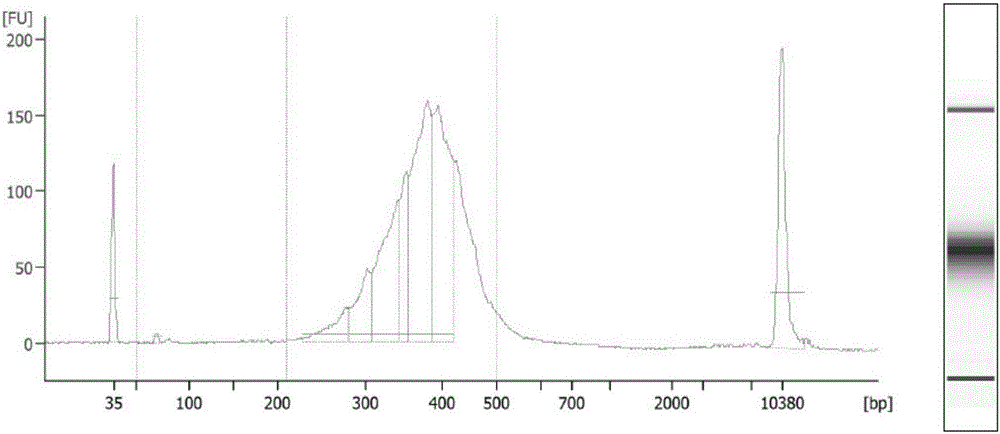
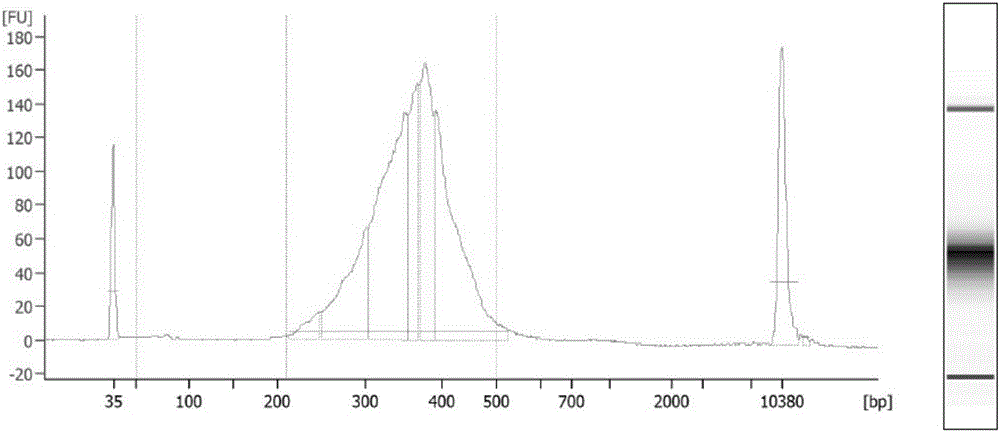
Establishing method of circular RNA high-throughput sequencing library and kit thereof
ActiveCN106801050AImprove detection efficiencyGood data repeatabilityMicrobiological testing/measurementLibrary creationTotal rnaRepeatability
The invention discloses an establishing method of a circular RNA high-throughput sequencing library and a kit thereof. The establishing method sequentially comprises the following steps: (S1) extracting total RNA from a sample; (S2) removing DNA in the sample; (S3) detecting and evaluating the quality of the total RNA in the sample, and determining that the RNA quality meets a requirement; (S4) removing rRNA; (S5) removing linear RNA; (S6) constructing the circular RNA library for high-throughput sequencing. The establishing method of the circular RNA high-throughput sequencing library, provided by the invention, is high in efficiency, stable, low in residual ratio of the rRNA, high in circular RNA detection efficiency, good in data reproducibility, and high in sequencing result verifying success rate, and is especially applicable to an FFPE tissue and other samples with poor RNA quality.
Owner:GUANGZHOU FOREVERGEN BIOTECH CO LTD +1
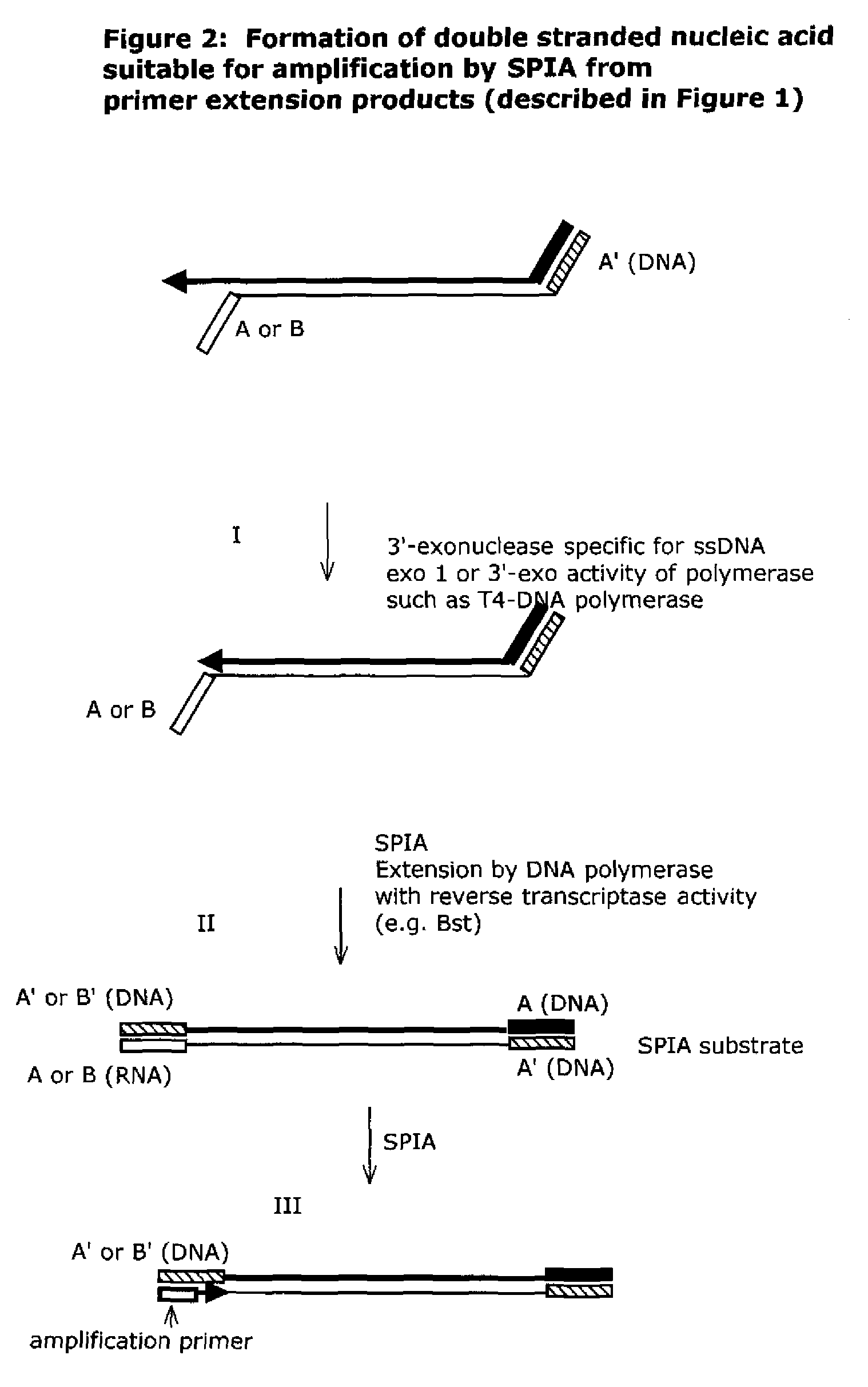
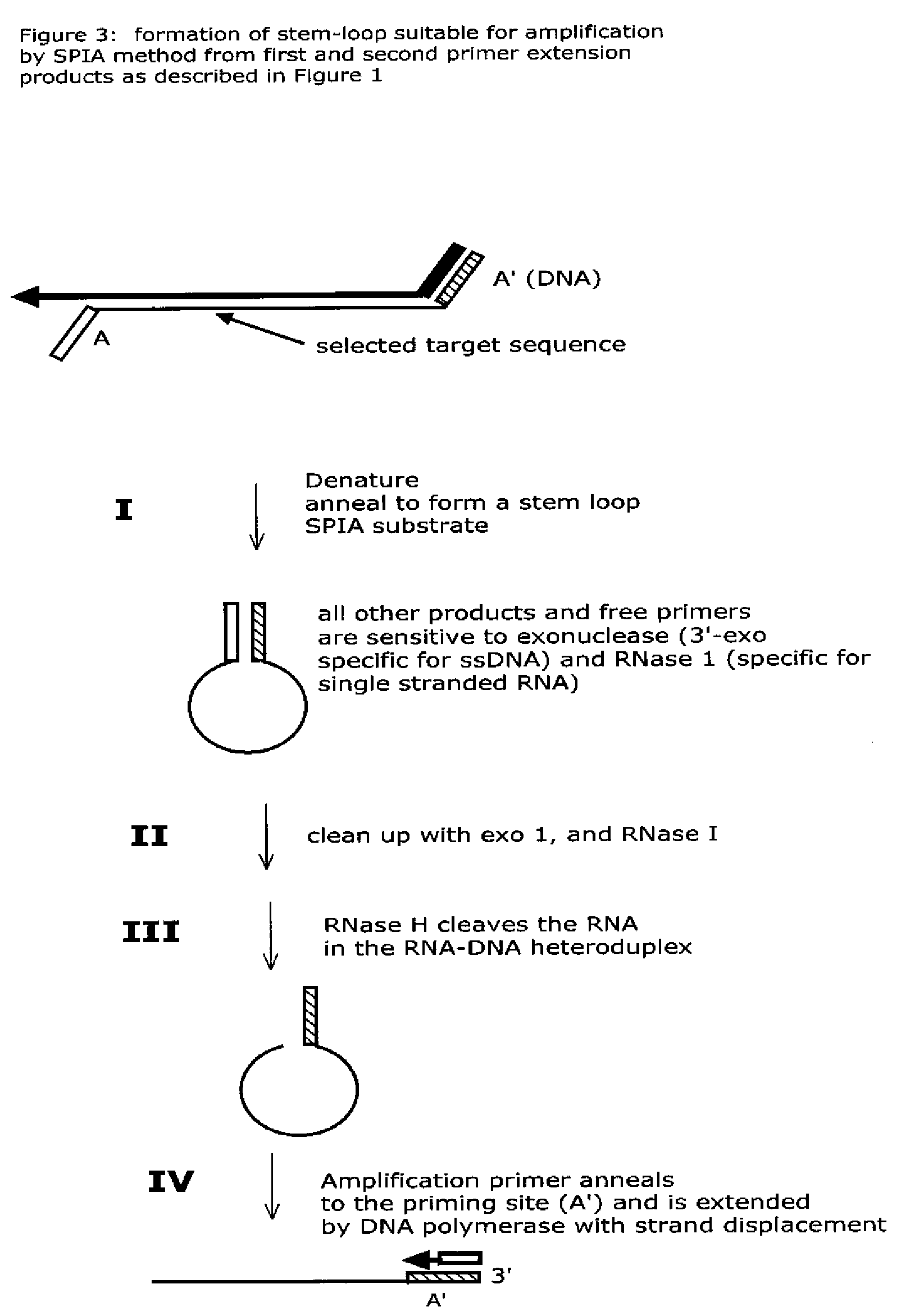
Isothermal nucleic acid amplification methods and compositions
Methods and compositions are provided related to the amplification of target polynucleotide sequences as well as total RNA and total DNA amplification. In some embodiments, the methods and compositions also allow for the immobilization and capture of target polynucleotides with defined 3′ and or 5′ sequences to solid surfaces. The polynucleotides attached to the solid surfaces can be amplified or eluted for downstream processing. In some cases, nucleotides attached to solid surfaces can be used for high throughput sequencing of nucleotide sequences related to target DNA or target RNA.
Owner:NUGEN TECH
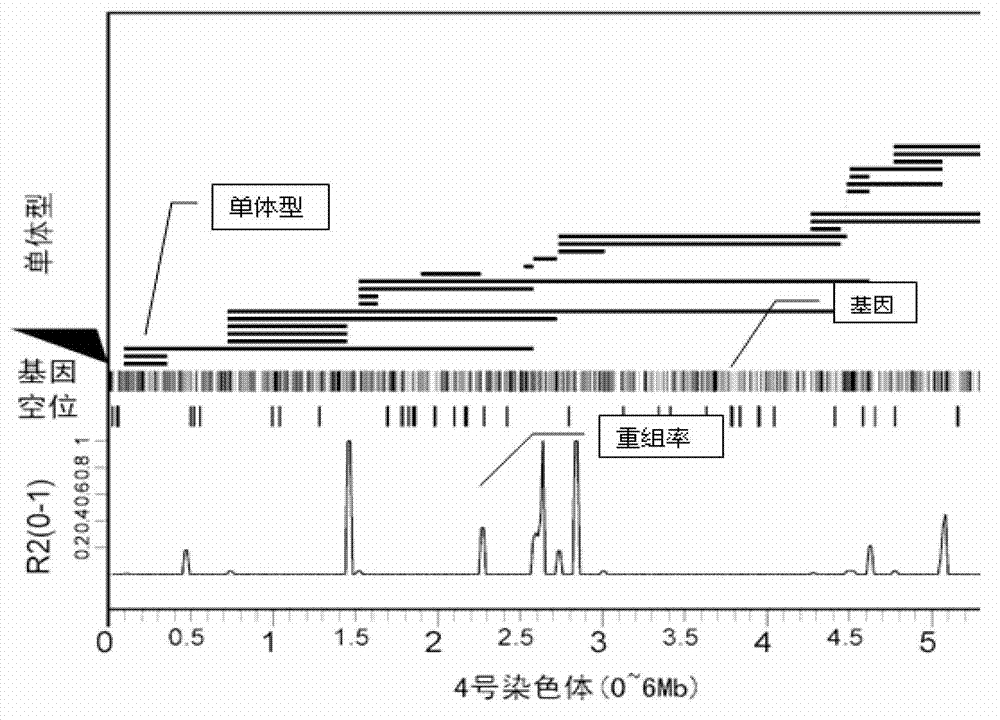
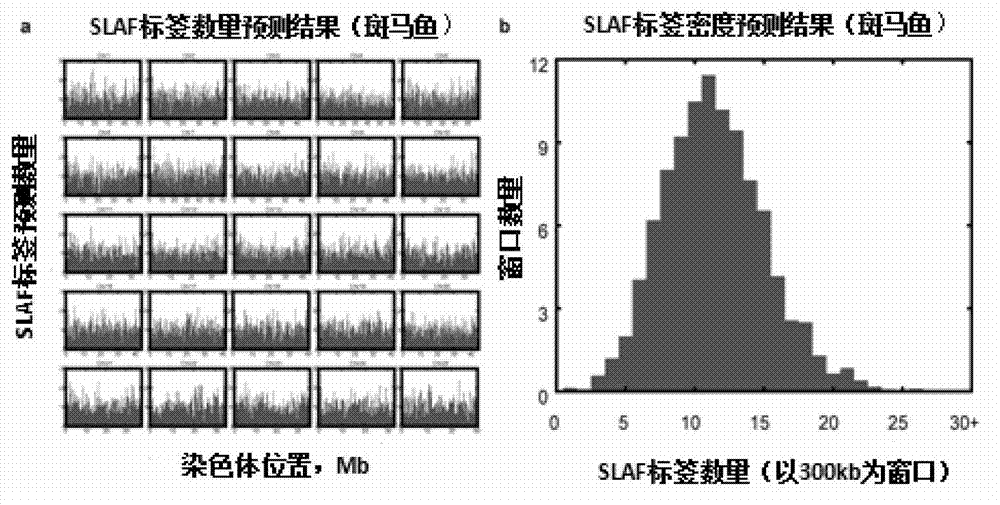
Large-scale genetic typing method based on SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology
ActiveCN103088120AGuarantee label qualityQuality assuranceMicrobiological testing/measurementGenotyping by sequencingGenome
The invention provides a method for carrying out large-scale gene typing based on an n SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology. Complexity of the genome is reduced by utilizing the SLAF-seq technology, and genetic typing is carried out on large-scale products. High-throughput sequencing is carried out on the genome, marker-developing, genetic map drawing and whole genome association analyzing are carried out on the samples by utilizing the technology. Compared with the conventional method, the large-scale genetic typing method disclosed by the invention has the advantages that the throughput is greatly improved, and the cost is greatly reduced. The method is mainly applied to marker-developing, genetic map drawing and whole genome association analyzing.
Owner:BIOMARKER TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com