Method for capturing genome target sequence based on Crispr/cas9 and application thereof in high-throughput sequencing
A genome and high-throughput technology, applied in the biological field, can solve problems such as unsuitable for single-gene genetic disease detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
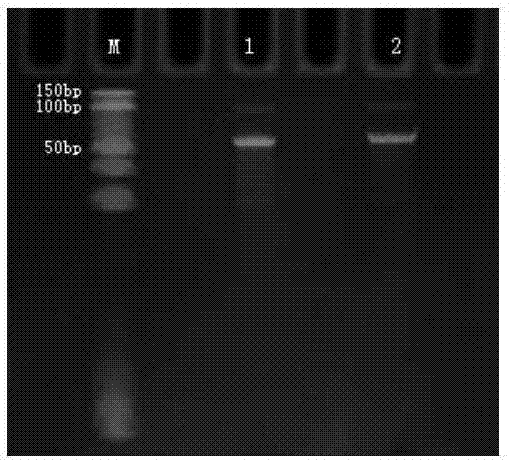
[0054] Example 1. Method for capturing target gene sequence in genome based on Crispr / cas9
[0055] The following example takes the BRCA1 and BRCA2 genes in the genomic DNA of human 293T cells as an example.
[0056] According to the exons of BRCA1 gene (GI: 262359905) and BRCA2 gene (GI: 568815585) as the target gene sequence.
[0057] 1. Preparation of transcription RNA template
[0058] 1. Design and synthesis of primers for transcription RNA template
[0059] According to the exons of BRCA1 gene (GI: 262359905) and BRCA2 gene (GI: 568815585) in the target genome, 63 BRCA1 gRNA (sequence 158-220) and 96 BRCA2 gRNA (sequence 221-313) were designed, and these gRNAs can guide CAS9 enzyme divides all exons of BRCA1 and BRCA2 into multiple fragmented products of 100-300bp in size.
[0060] Based on 63 BRCA1gRNA and 96 BRCA2gRNA, respectively design the amplification primers for 63 BRCA1 exon transcription RNA template and 93 BRCA2 exon transcription RNA template amplification primers as f...
Embodiment 2
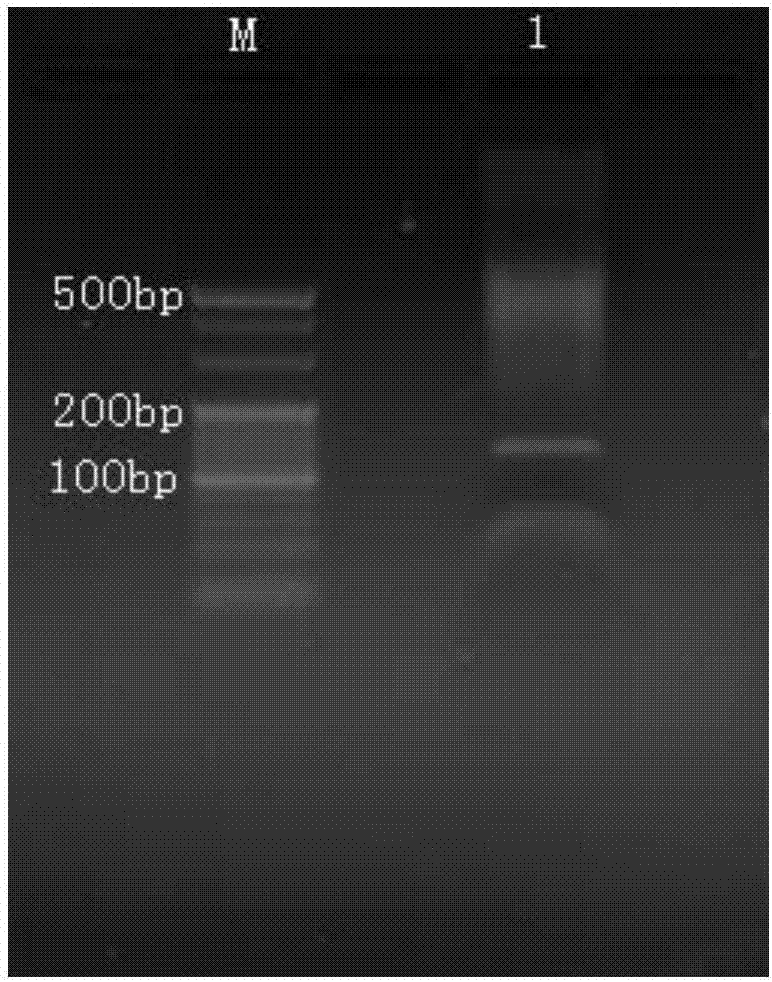
[0117] Example 2. High-throughput sequencing target sequence
[0118] 1. High-throughput sequencing library construction
[0119] 1. End repair, phosphorylation and adding dA tail
[0120] (1) Add the following reagents to the sterilized EP tube as shown in Table 4:
[0121] Table 4 shows the end repair, phosphorylation and dA tail system
[0122]
[0123] (2) Use a pipette to mix gently, and centrifuge briefly to collect the reaction solution to the bottom of the tube.
[0124] (3) Put the reaction tube in the PCR machine, and run the program in Table 5 to obtain the reaction product:
[0125] table 5
[0126]
[0127] 2. Connector connection
[0128] (1) Add the following components to the reaction product obtained in step 1:
[0129] Table 6
[0130]
[0131] T4 DNA Ligase, NEB (Beijing) Co., Ltd., NEB (Beijing) Co., Ltd., M0202L.
[0132] Y connector (UAF / AI1)
[0133] UAF AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T
[0134] AI1GATCGGAAGAGCACACGTCTGAACTCCAGTCACATCACGATCTCGTAT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com