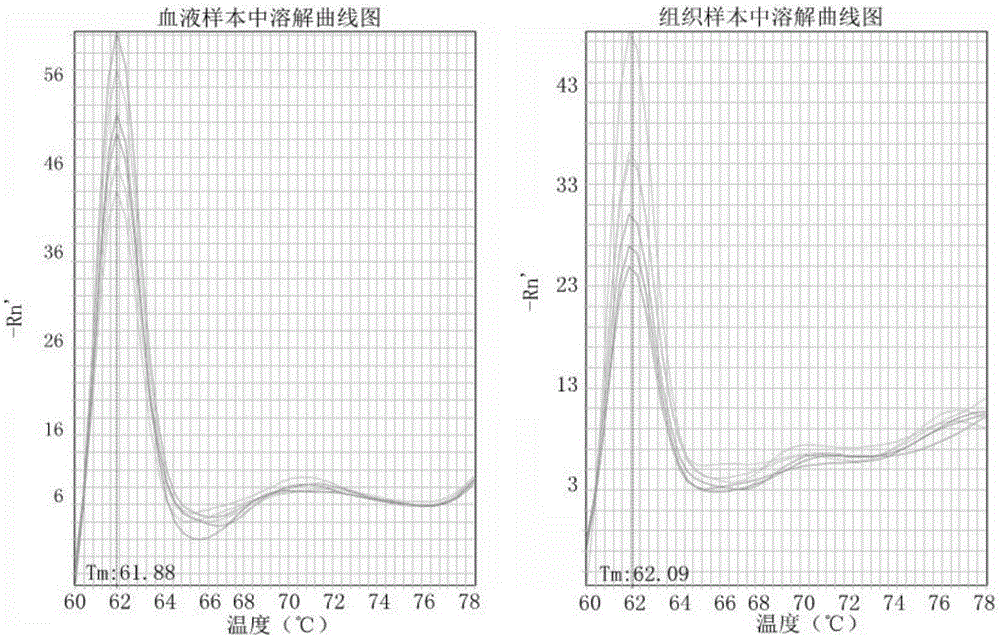
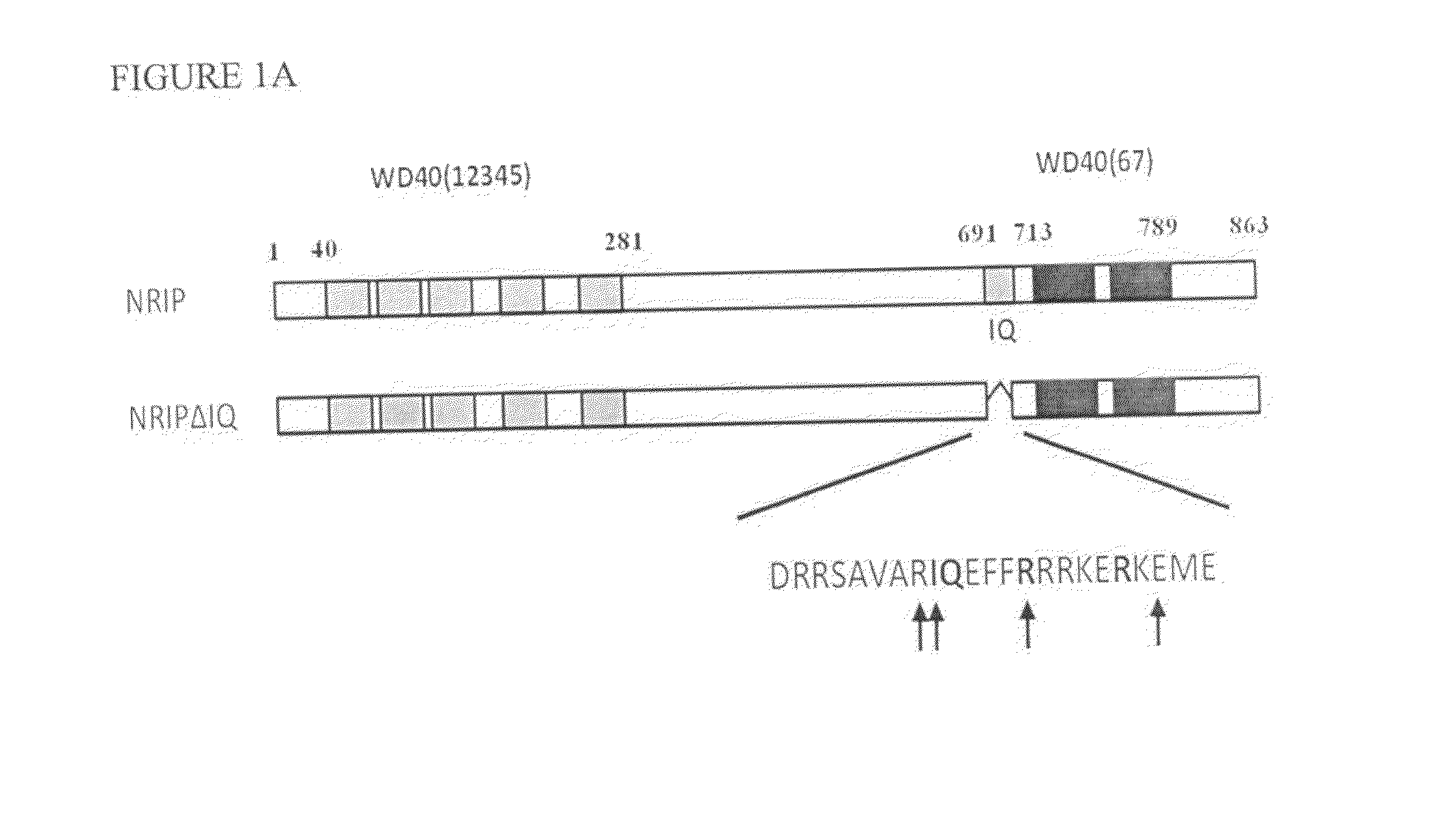
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
42 results about "Interactions protein" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
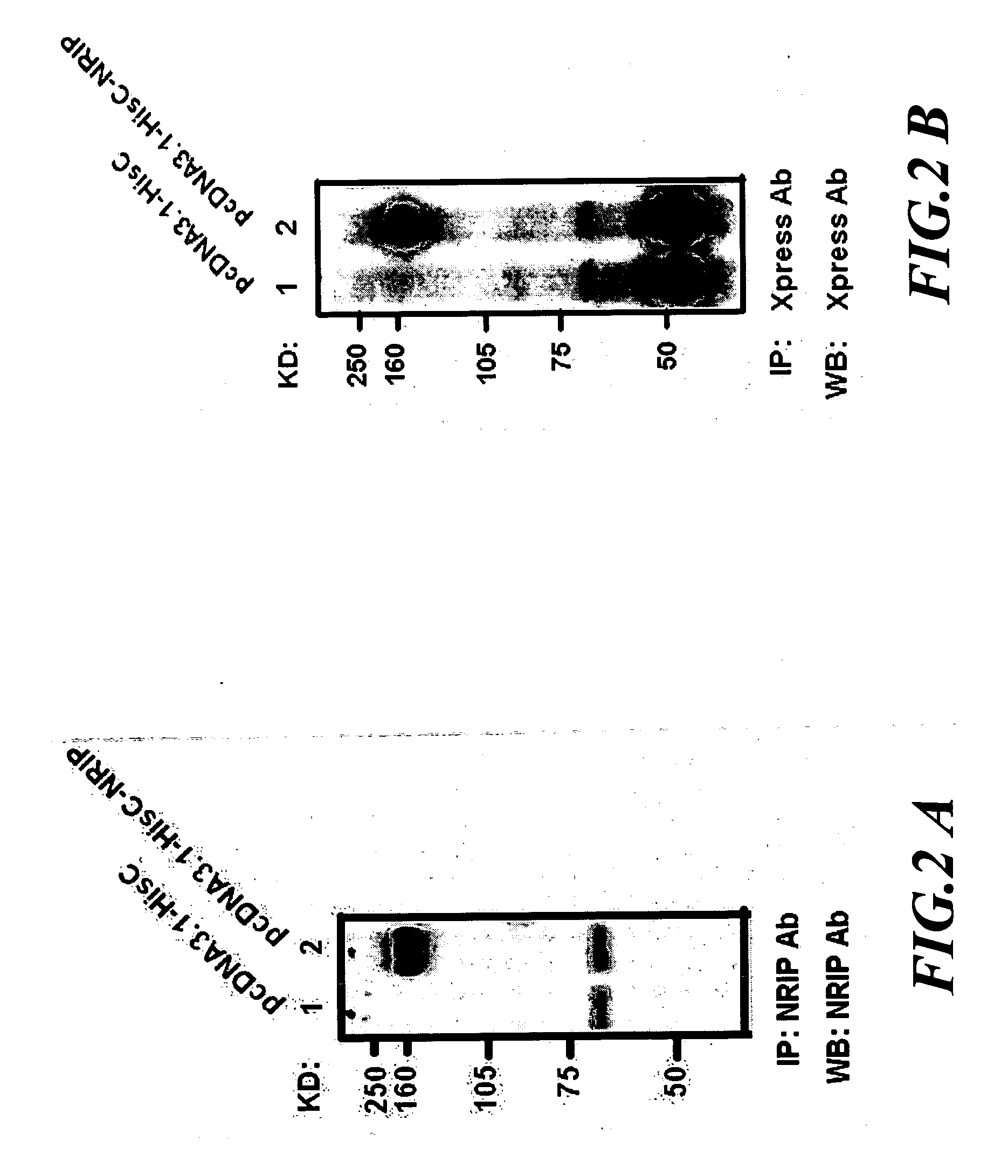
Novel diagnosis & treatment tools for cancer using the NRIP antibody and RNA interference
The invention provides an isolated nucleic acid encoding NRIP (Nuclear Receptor Interaction Protein) gene that highly expressed in tumorigenic development of human neoplasms in cervical cancer. The invention provides an antibody against the NRIP protein, and methods of diagnosing the NRIP related cancer. The invention also features RNA interferences of the NRIP gene, and the methods of treatment for the NRIP related cancer using the RNA interferences.
Owner:CHEN SHOW LI +2
Plant protein interaction network constructing method based on deep learning
InactiveCN110136773AImprove forecast accuracyReduce the workload of parameter adjustmentSystems biologyInstrumentsAlgorithmInteractions protein
The invention relates to a plan protein interaction network constructing method based on deep learning. The method comprises the following steps of 1), acquiring 11 characteristic data of a protein interaction pair; 2), performing screening for obtaining a training set and a testing set; 3), constructing a deep learning classification model; 4), performing batch optimization on the parameter of the deep learning classification model, and obtaining a classification model of an optimal optimization parameter combination; 5), performing interaction relation predicting on all possible two-to-two interaction proteins of the whole genome according the classification model of the optimal optimization parameter combination; and 6), predicting the protein interaction network according to an interaction relation predicting result. Compared with the prior art, the method has advantages of high predicting accuracy, high modeling efficiency, etc.
Owner:SHANGHAI JIAO TONG UNIV
Method for detecting protein interaction by immunological coprecipitation based on protein chip and reagent kit for detecting protein interaction
InactiveCN101105493AOvercoming difficult-to-obtain bottlenecksEfficient detection of interactionsFluorescence/phosphorescenceFluorescenceInteractions protein
Protein-protein interaction (PPI) is important for all biological processes; therefore, the research of protein interaction is always a research hot in the fields of cell biology and molecular biology. The co-immunoprecipitation (CoIP) is a classic method which is used to research protein interaction based on the specific action between antibody and antigen and is the commonest and most effective method to find and testify PPI; the traditional CoIP steps based on a resin are complicated and labor consuming and are hard to realize the flux detection. The key point in the invention is that CoIP is implemented on glass aldehyde sheet of the fixed anti-label antibody by utilizing the epitope label of recombinant protein; protein-protein interaction (PPI) is detected by the antibody labeled by fluorescence. The method greatly simplifies the operation steps of the traditional CoIP, greatly improves the flux of sample detection and is a novel technique platform to research PPI simply, conveniently and swiftly and with high flux.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
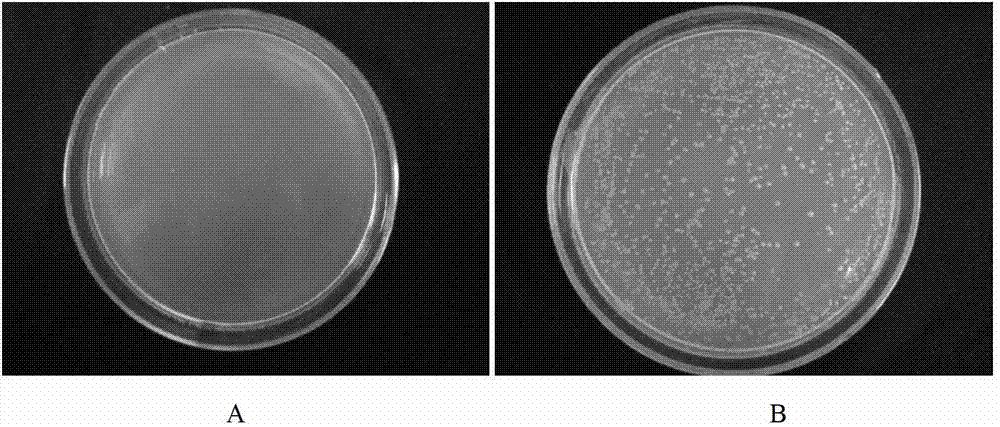
Library versus library yeast two-hybrid massive interaction protein screening method
InactiveCN103774241AReduce masking effectLow costLibrary member identificationLibrary creationCDNA libraryGenome scale
The invention discloses a library versus library yeast two-hybrid massive interaction protein screening method. The method takes a genome-grade cDNA as the bait to screen the genome-grade cDNA library. In the application process of the method, the cDNA library is subjected to a homogenization treatment so as to reduce the masking effect of the high-abundance interaction proteins to the low-abundance interaction proteins, and thus the screening efficiency is improved; and moreover the characteristic of yeast URA3 gene is utilized so as to automatically remove the sequences, which have the self-activating function, in bait cDNA. The invention establish a library versus library yeast two-hybrid massive interaction protein screening method, which has the advantages of high feasibility, low cost, low requirements on experiment conditions, and low workload; and successfully applies the method to a screening of pathogen-host plant interaction proteins.
Owner:HUAZHONG AGRI UNIV
Application of HsCIPK17 of Hordeum spontaneum C. Koch in Qinghai Tibet Plateau to improvement in rice resistance to abiotic stress
InactiveCN108588117AGood adversity tolerancePromote growth and developmentHydrolasesFermentationGenetically modified riceNucleotide
The invention discloses application of a calcineurin B protein interaction protein kinase gene HsCIPK17 of annual Hordeum spontaneum C. Koch in the Qinghai-Tibet Plateau: the HsCIPK17 is used for constructing Oryza sativa L.; the Oryza sativa L. has heavy metal stress tolerance and also has salt tolerance and abscisic acid stress tolerance; heavy metals are mercury, cadmium and chromium; a nucleotide sequence of the gene HsCIPK17 is that the GenBank accession number is JN655677.
Owner:LANZHOU UNIVERSITY
Method for predicting the direction and regulation of protein interactions
ActiveCN109086569AAccurate predictionEfficient forecastingNeural architecturesSpecial data processing applicationsProtein containing complexInteractions protein
The invention relates to a prediction method of protein interaction direction and regulation relationship, which comprises the following steps: homologous modeling of the interaction protein; comparing and superposing the homologous spatial structure of each interacting protein and each subunit of the protein complex, according to the structure of the stable complex, a protein-protein interactionmodel with spatial structure resolution being constructed, and the interaction direction and frequency of amino acid residues in the protein-protein interaction interface being calculated, and the directional contact map of amino acid residues in the protein-protein interaction interface being generated; a convolution neural network being used to extract the target characteristics of the directional contact map, and the recognition model being trained to obtain the training model, and the protein interaction direction and regulation relationship being predicted. The prediction method of protein interaction direction and regulation relationship is a prediction method based on depth learning, which can achieve accurate and efficient prediction of protein interaction direction and regulationrelationship.
Owner:南京深佰科技有限公司
Key protein identification method in network based on dynamic weighting interaction
PendingCN109686402AOvercome incompletenessImprove accuracyBiostatisticsProteomicsCorrelation coefficientGene ontology
The invention discloses a key protein identification method use principle in a network based on dynamic weighting interaction. A key protein identification method comprises the steps of calculating aprotein activity time point and a protein activity probability, constructing a dynamic PPI network, then calculating a protein interaction weight according to the protein activity probability, and constructing a dynamic weighted PPI network; based on the established dynamic weighted PPI network, according to the topological characteristic and biological attribute of the protein network, calculating an edge clustering coefficient, a gene body similarity and a Pearson correlation coefficient between the interaction protein pairs; afterwards, obtaining an importance score, and performing arrangement according to the score value from largest to lowest, and outputting k proteins which correspond with the scores as a final result. The method according to the invention improves key protein identification efficiency and expands application range and practicability of the technique in a bioinformation field.
Owner:YANGZHOU UNIV
Screening methods for inhibitors of protein-protein binding interactions
Protein-protein interactions represent a large and important group of drug targets involved in the development and progression of human diseases. However, their utilization in drug discovery has been hampered by the low probability of identifying small-molecule inhibitors able to disrupt protein binding with desirable potency and selectivity. Therefore, the capability for rapid screening of large compound libraries has been critical for the exploration of this target class. The present invention relates to a homogeneous time-resolved fluorescence assay for identification of inhibitors of Cks1-Skp2 binding that plays a critical role in the ubiquitin-dependent degradation of p27. The assay was implemented in a 1536-well format using the new Zeiss uHTS robot and achieved a throughput in excess of 100,000 data points per day. A protocol for a fully automated high throughput IC50 determination was developed for hit validation. The basic 1536 well screening platform reported here is simple, robust and cost effective. It is widely applicable to any protein-protein interaction of therapeutic interest.
Owner:HUANG KUO SEN +1
Method for screening MKL-1 protein interaction proteins via yeast two-hybrid screening
InactiveCN107034286AQuick filterGood for verification experimentsMicrobiological testing/measurementVector-based foreign material introductionBiotechnologyCDNA library
The invention relates to the technical field of genetic biology, in particular to a method screening MKL-1 protein interaction proteins via yeast two-hybrid screening. A recombinant yeast capable of expressing bait protein MKL-1 is prepared, cDNA library is transformed to the recombinant yeast, the two nutrient defects, His and Ade, and chromogenic reaction are used alone or in combination, and positive clones having different interactive intensities to the protein MKL-1 are acquired by further screening. By means of gradually increasing the selective pressure, it is convenient to quickly screen out proteins having different interactive degrees to the protein MKL-1, subsequent validation experiments are facilitated, and the method is significant to further elaboration of the functionality and regulatory effect of MKl-1 in the genesis and development of leukemia.
Owner:WUHAN UNIV OF SCI & TECH
Protein identification method based on protein interaction network and proteomics
ActiveCN104156603AHigh precisionEfficient identificationSpecial data processing applicationsInteractions proteinProtein Interaction Networks
The invention discloses a protein identification method based on a protein interaction network and proteomics. The method is based on the phenomenon that the existence probabilities between interaction proteins are influenced mutually. Protein interaction network information is fused with shotgun method proteomics data, a new protein identification graph model is defined, the probability of peptide mapped to the proteins is adjusted by means of the existence probability of the proteins in the graph model and the support degree of neighbor protein nodes obtained in the graph model, and therefore the existence probability of the proteins is adjusted. Most proteins can be identified through the method. Compared with other identification methods, the protein identification method based on the protein interaction network and the proteomics has high accuracy. Valuable reference information is provided for experiments for inference and identification of proteins of biologists through proteomics data and further study.
Owner:CENT SOUTH UNIV
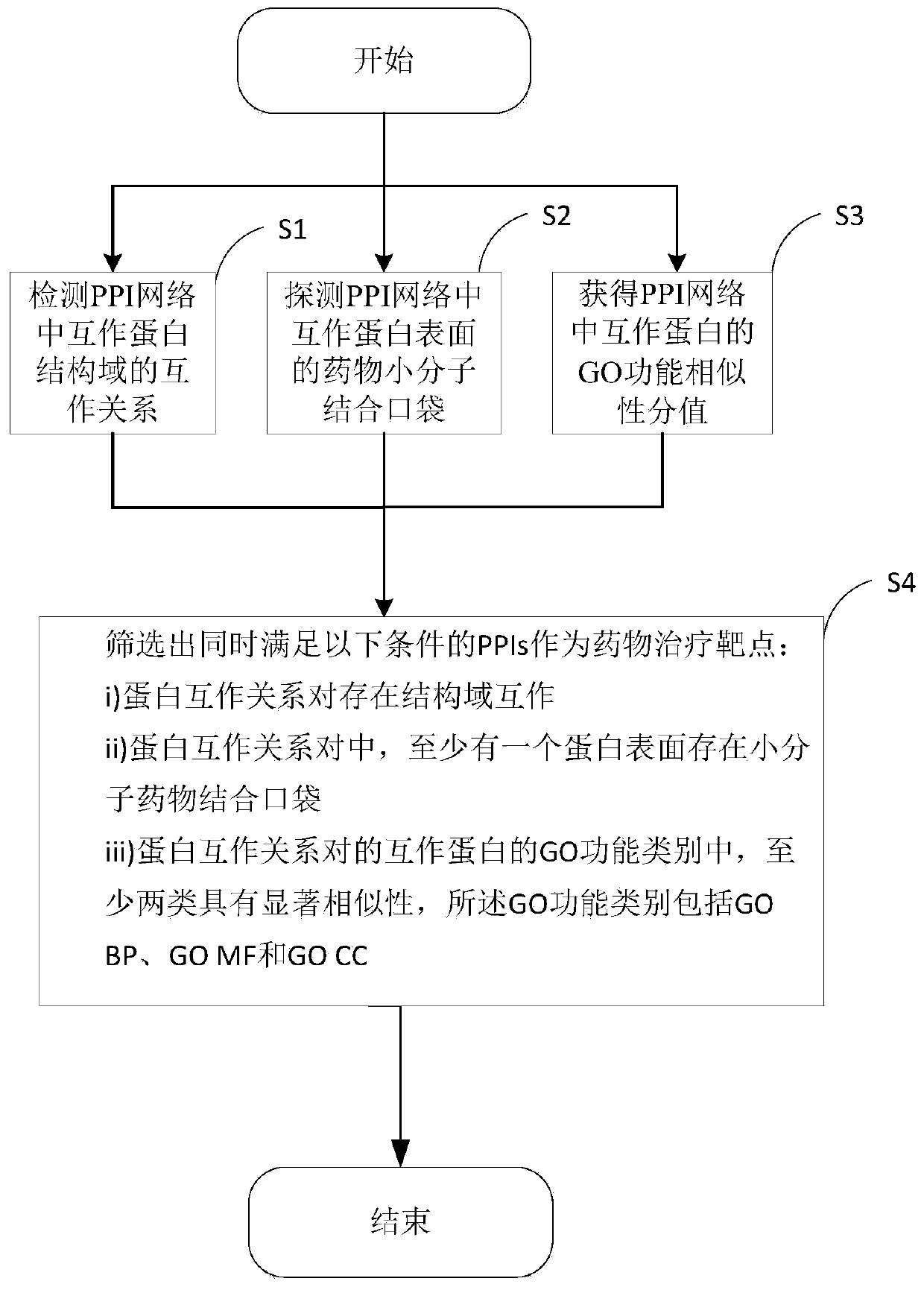
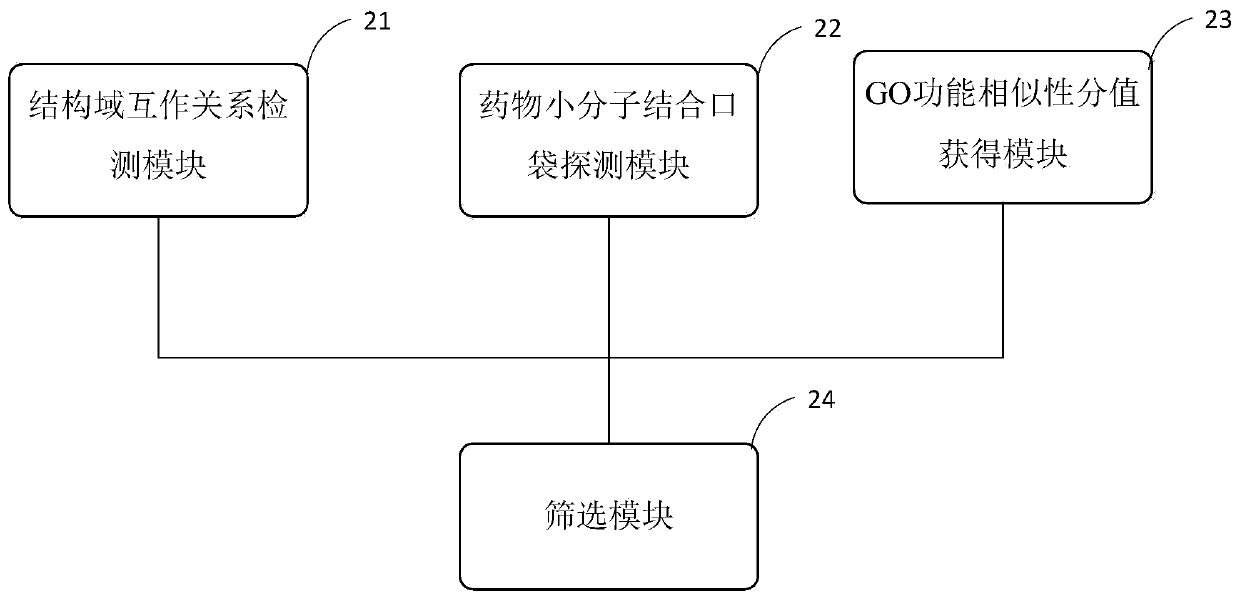
Target PPIs drug property prediction method and device based on protein interaction network
ActiveCN110544506AReliable interactionReduce false positivesProteomicsGenomicsPositive interactionInteractions protein
The invention provides a target PPIs drug property prediction method based on a protein interaction network. The method at least comprises the following steps: S1, detecting the interaction relationship of interaction protein structural domains in the PPI network; S2, detecting a drug small molecule binding pocket on the surface of interaction protein in the PPI network; S3, obtaining a GO function similarity score of the interactive protein in the PPI network; S4, screening out PPIs meeting the following conditions at the same time to serve as drug therapy targets: protein interaction relationship pairs have structural domain interaction; in the protein interaction relationship pair, at least one protein surface has a small molecule drug binding pocket; at least two of the GO function categories of the interaction proteins of the protein interaction relationship pair have significant similarity, and the GO function categories comprise GO BP, GO MF and GO CC. According to the method, three strict mutually independent standards are adopted to comprehensively explore and discover the target PPI, false positive interaction is systematically eliminated, more reliable PPIs are selectedas drug targets, and the calculation result better conforms to objective reality.
Owner:上海源兹生物科技有限公司
Yeast two-hybrid method
ActiveCN110004171AEnhanced interactionImprove the detection rateHybrid cell preparationVector-based foreign material introductionBiotechnologyYeast
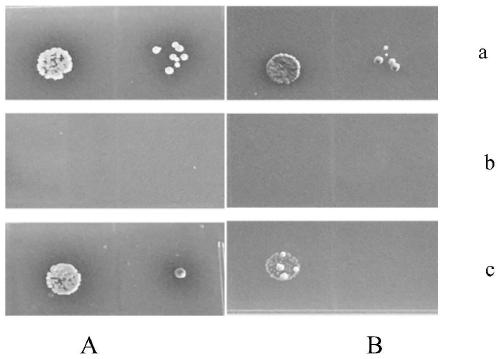
The invention provides a yeast two-hybrid method. The yeast two-hybrid method is characterized by comprising the steps that 1, library yeast is activated, and an AD-Y fusion vector is transformed intothe library yeast; 2, bait plasmids BD-X are transformed into yeast cells Y2H Gold; 3, recombined yeast of BD-X and recombined yeast containing the AD-Y are fused into a diploid, the diploid yeast iscultured on an amplification culture medium containing zinc ions to obtain a bacterial colony, and then positive clone screening is conducted. GAL4 protein in the yeast cells Y2H Gold used in the method is a dependent transcription factor of the zinc ions, therefore, the zinc ions are added into the amplification culture medium used in the method, interactions between low-abundance proteins are improved, the detection rate of low-interaction proteins is improved, and the positive clone rate is improved.
Owner:FOSHAN UNIVERSITY
Known molecule and protein interaction detection system based on covalent linkage and identification or verification method thereof
ActiveCN112904017AAccurate detectionMaintain structureBiological testingMicrobiological testing/measurementInteractions proteinOligopeptide
The invention discloses a known molecule and protein interaction detection system based on covalent linkage and an identification or verification method thereof. The detection system comprises: a) a streptavidin-oligopeptide tetramer; b) a PafA enzyme; and c) biotin-modified known molecules. Known molecules are labeled with biotin, after the known molecules interact with proteins, interaction proteins of the known molecules are efficiently captured through the streptavidin-oligopeptide tetramer under mild conditions, and then under catalysis of PafA enzyme, oligopeptide and the known molecules are in covalent linkage with the interaction proteins, therefore, non-covalent binding between the interacting known molecules and the protein is converted into covalent linkage between the streptavidin and the protein, and analysis, separation and identification are carried out. The method can realize capture of weak interaction and instantaneous interaction on the basis of keeping the natural structure of known molecules, and can be used for verification and discovery of known molecules and interaction proteins.
Owner:SHANGHAI JIAO TONG UNIV
Luciferase fluorescence-complementary system, and preparation method and application thereof
ActiveCN109957604AIncrease typeIncrease concentrationMicrobiological testing/measurementBiological testingBinding siteFluorescence
The invention provides a luciferase fluorescence-complementary system as well as a preparation method and application thereof. The luciferase fluorescence-complementary system comprises Rab1, SidM / DrrA, a luciferase C-terminal, a luciferase N-terminal, linker peptides and a reducer TCEP; the C terminal of the Rab1 is connected with the luciferase N-terminal by a linker peptide; and the SidM / DrrA is connected with a luciferase C-terminal fragment by a linker peptide. By introducing specific interaction protein pairs, designing appropriate flexible peptides and binding sites as well as optimizing type and concentration of a reducer in a buffer solution, the luciferase fluorescence-complementary system finally has significantly increased fluorescence complementary efficiency; and the fluorescence complementary efficiency can be increased to up to 30 times. Moreover, the preparation method of the luciferase fluorescence-complementary system provided by the invention is simple and highly efficient, so that, the preparation method is convenient to operate, easy to promote, wide in application prospects and high in market values.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
Kit for diagnosing cardiomyopathy and method for predicting cardiomyopathy
The present invention provides a kit for diagnosing the cardiomyopathy comprising an antibody or oligonucleotide for measuring an expression level of nuclear receptor interaction protein (NRIP). The present invention also provides a method of predicting a risk of cardiomyopathy for a subject. The present invention further provides a method of screening an agent for treating cardiomyopathy.
Owner:NAT TAIWAN UNIV
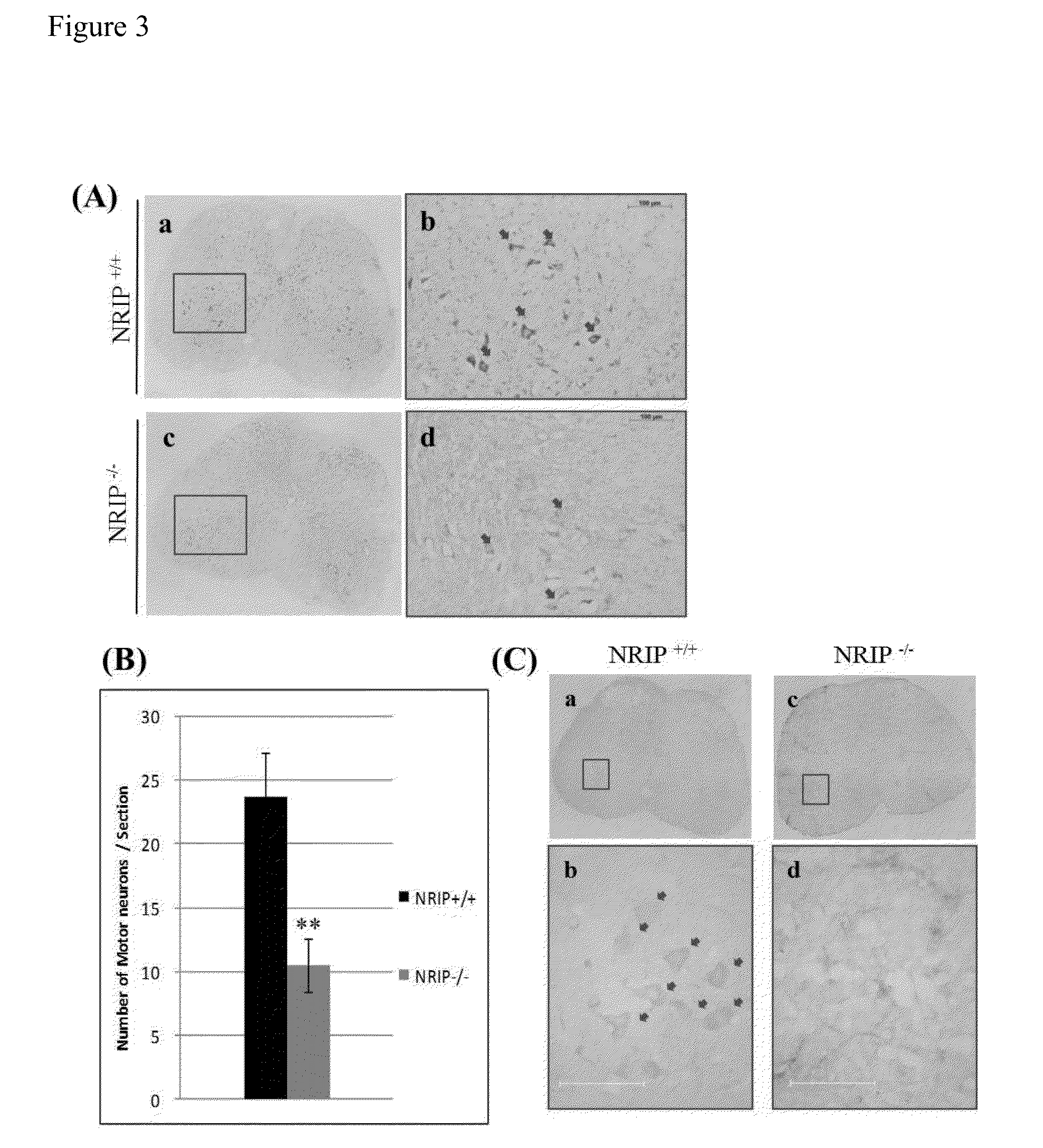
Nrip as a biomarker of abnormal function of motor neurons
InactiveUS20150226750A1Reduced expression levelLibrary screeningDisease diagnosisInteractions proteinPrimary motor neuron
The present invention provides a method for evaluating physiological state of motor neurons in a subject. The method comprises detecting an expression level of a nuclear receptor interaction protein (NRIP) in a biological sample from the subject; and comparing the expression level of the NRIP to a control value for NRIP; wherein detecting a decrease of the expression level of the NRIP in the biological sample from the subject as compared to the control value for NRIP indicates an abnormal function of the motor neurons of the subject
Owner:NAT TAIWAN UNIV
Library versus library yeast two-hybrid massive interaction protein screening method
InactiveCN103774241BReduce masking effectLow costLibrary member identificationLibrary creationCDNA libraryProtein insertion
The invention discloses a library versus library yeast two-hybrid massive interaction protein screening method. The method takes a genome-grade cDNA as the bait to screen the genome-grade cDNA library. In the application process of the method, the cDNA library is subjected to a homogenization treatment so as to reduce the masking effect of the high-abundance interaction proteins to the low-abundance interaction proteins, and thus the screening efficiency is improved; and moreover the characteristic of yeast URA3 gene is utilized so as to automatically remove the sequences, which have the self-activating function, in bait cDNA. The invention establish a library versus library yeast two-hybrid massive interaction protein screening method, which has the advantages of high feasibility, low cost, low requirements on experiment conditions, and low workload; and successfully applies the method to a screening of pathogen-host plant interaction proteins.
Owner:HUAZHONG AGRI UNIV
Construction method of yeast two-hybird bait vectors for screening ASICs interaction proteins
The invention discloses a construction method of yeast two-hybird bait vectors for screening ASICs interaction proteins, belonging to the technical field of biology. According to the construction method, subunit extracellular loop genes ASIC1a and ASIC2a in acid-sensitive ion channels (ASICs) are respectively cloned to a yeast two-hybird bait vector pGBKT7 by virtue of a gene engineering technique so as to successfully construct two yeast two-hybird bait vectors pGBKT7-ASIC1a and pGBKT7-ASIC2a, the expression and identification of the constructed vectors are realized, and the recombinant bait vectors are proved to be applicable to a yeast two-hybird system, so that an important experimental foundation is laid for screening the ASICs interaction proteins by virtue of the yeast two-hybird system and further researching and establishing an ASICs regulation and control machine, and a new direction is opened up for the treatment of inflammatory pain.
Owner:湖南甲骨文生物医药有限公司
Method for detecting protein interaction by immunological coprecipitation based on protein chip and reagent kit for detecting protein interaction
InactiveCN101105493BOvercoming difficult-to-obtain bottlenecksEfficient detection of interactionsFluorescence/phosphorescenceFluorescenceInteractions protein
Protein-protein interaction (PPI) is important for all biological processes; therefore, the research of protein interaction is always a research hot in the fields of cell biology and molecular biology. The co-immunoprecipitation (CoIP) is a classic method which is used to research protein interaction based on the specific action between antibody and antigen and is the commonest and most effectivemethod to find and testify PPI; the traditional CoIP steps based on a resin are complicated and labor consuming and are hard to realize the flux detection. The key point in the invention is that CoIPis implemented on glass aldehyde sheet of the fixed anti-label antibody by utilizing the epitope label of recombinant protein; protein-protein interaction (PPI) is detected by the antibody labeled byfluorescence. The method greatly simplifies the operation steps of the traditional CoIP, greatly improves the flux of sample detection and is a novel technique platform to research PPI simply, conveniently and swiftly and with high flux.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
Application of IPCEF1 (Interaction Protein For Cytohesin Exchange Factors 1) in diagnosing and treating osteosarcoma
The invention relates to application of IPCEF1 (Interaction Protein For Cytohesin Exchange Factors 1) in diagnosing and treating osteosarcoma. In recent years, the treatment of the osteosarcoma encounters the bottleneck, and particularly in an aspect of early diagnosis of osteosarcoma transfer, a related molecular tumor marker is clinically in urgent need. Osteosarcoma case samples are collected for carrying out high-throughput sequencing by an inventor, genes IPCEF1 which are closely related to the osteosarcoma are picked out by adopting bioinformatics analysis, the relevance of the IPCEF1 and the osteosarcoma is verified in a large sample through a molecular biology experiment, the molecular biology experiment shows that the genes IPCEF1 are expected to be a diagnosis molecular marker of the osteosarcoma, and an ideal is provided for early diagnosis of the osteosarcoma.
Owner:BEIJING MEDINTELL BIOMED CO LTD
Complete set of reagents for detecting interaction between protein subjected to post-translational modification and ligand thereof
The invention discloses a complete set of reagents for detecting interaction between protein subjected to post-translational modification and ligands thereof. The complete set of reagents consists offour reagents A, B, C and D, wherein A is formed by connecting a biomolecule named as R and a protein named as X; B contains a biomolecule named as L; R and L are the same or different and have interaction, and phase change occurs after the R and L interact; C is a polymer formed by a monomer C, the monomer C is a molecule obtained by connecting a monomer named as mc, a reporter group named as I and a biomolecule named as YC, and two or more mc can form the polymer; D is formed by connecting a modified protein named as XL and a biomolecule named as YD; and the YC and the YD have interaction. According to the invention, high enrichment of the interaction protein and the ligand in the phase-change liquid drop is realized, and the weak interaction signal is amplified, so that the detection iseasy.
Owner:抟相医药(杭州)有限公司
Method for identifying and analyzing specific locus interaction proteins on basis of CRISPR/cas9 and peroxidase APEX2 system
InactiveCN109507429AAchieve highly specific enrichmentEasy to operateBiological testingMagnetic beadInteractions protein
The invention belongs to the technical field of biology, and relates to a method for identifying and analyzing specific locus interaction proteins on the basis of a CRISPR / cas9 and peroxidase APEX2 system. The method comprises the following steps: plasmid transformation; biotin labeling of specific locus binding protein complex; enrichment of biotin-labeled protein complex by utilizing streptomycin magnetic beads; and assay and analysis of enriched proteins. An experimental analysis result shows that the method is applicable to the research and analysis of protein at any given chromosomal location, and meanwhile, the idea that the method combines the CRISPR gene editing system with APEX2 to carry out specific locus protein analysis is also applicable to the combination of other genome editing methods (such as TALEN) and the APEX2. The method has the advantages of simplicity in operation, high sensitivity, high specificity, good repeatability and adaption to the requirement of the research of local chromatin interaction at any given chromosomal location.
Owner:FUDAN UNIV
Protein interaction network construction method
InactiveCN105389483AComprehensive and extensive storageLarge storageSpecial data processing applicationsBioinformaticsProtein targetInteractions protein
The present invention discloses a protein interaction network construction method. The method comprises the following steps: acquiring a protein set that is interested in; constructing a first-level interaction protein database of a target protein, and drawing a first-level protein interaction network of the target protein according to the first-level interaction protein database; and constructing a second-level interaction protein database of the target protein, and in conjunction with the first-level protein interaction network of the target protein, drawing an interaction protein database of the target protein. Compared with the prior art, according to the method disclosed by the present invention the candidate database used when constructing the protein interaction network is comprehensive and extensive, and the idea of integrated use and expanded application of an existing database is novel; the protein interaction database can comprehensively and extensively store existing protein interaction relationships, the source is reliable, and the query is convenient; and a reliable basis is provided for experimental verification of unknown protein functions.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
Experimental method for improving corn amylose content by knocking out SIP1 gene
ActiveCN110144366AIncrease contentImprove qualityClimate change adaptationPlant peptidesExperimental methodsInteractions protein
The invention discloses an experimental method for improving the corn amylose content by knocking out an SIP1 gene, and belongs to the field of genetic engineering.The method mainly comprises the following steps: constructing a CRISPR / Cas9-SIP1 gene knockout vector; and transforming target corn by agrobacterium to obtain a corn plant with the SIP1 gene knockout. According to the genetic engineering method, the content of amylose in corn kernels is improved by interfering the starch branching action of SIP1 in the participation of the main gene ZmSBEII. The realization of the method has important practical guiding significance for developing a new high-amylose corn variety by utilizing ZmSBEII interaction protein.
Owner:陈超
Methods for modulating slow myosin
InactiveUS20130178410A1Peptide/protein ingredientsGenetic material ingredientsMyosinInteractions protein
The present invention provides a method for modulating an expression level of a gene encoding slow myosin in a subject in need thereof, comprising administering to said subject a pharmaceutically effective amount of a nuclear receptor interaction protein (NRIP) and a pharmaceutically acceptable carrier. The present invention also provides a method for modulating an expression level of a gene encoding slow myosin in a subject in need thereof, comprising administering to said subject a pharmaceutically effective amount of an expression vector comprising a gene encoding a nuclear receptor interaction protein (NRIP) and a pharmaceutically acceptable carrier. In a preferred embodiment, the expression vector is an adenoviral vector.
Owner:NAT TAIWAN UNIV
Nrip knockout mice and uses thereof
The present invention directs to a transgenic NRIP knockout mouse, the genome of which is manipulated to comprise a disruption of a nuclear receptor interaction protein (NRIP) gene, wherein the NRIP gene is disrupted by deletion of exon 2, the mouse exhibits a phenotype comprising abnormal muscular function. The present invention also directs to a method for making a transgenic NRIP knockout mouse whose genome comprises a homozygous disruption of the NRIP gene, the mouse exhibits abnormal muscular function.
Owner:NAT TAIWAN UNIV
Protein interaction prediction method
PendingCN112259157AImprove robustnessPredictableHybridisationSystems biologyInteractions proteinMolecular function
The invention discloses a protein interaction prediction method based on a sampling strategy of non-interaction protein pairs fused with biological semantics. Protein pairs in different molecular functions, biological processes and cell components are sampled and combined based on GO term semantic similarity to obtain an NIPs subset. With a negative set sampling strategy, a non-protein interactiondata set with higher quality and low selection deviation is obtained, so that a protein interaction prediction model with better robustness and better prediction performance is obtained through training.
Owner:HANGZHOU NORMAL UNIVERSITY
Research method for analyzing Klotho interaction protein in non-small cell lung cancer
ActiveCN109100514AIncrease the level of distributionEasy to transportMicrobiological testing/measurementBiological testingInteractions proteinStudy methods
The invention discloses a research method for analyzing a Klotho interaction protein in non-small cell lung cancer which comprises the three steps, a screening of Klotho interaction protein, a verification of Rab8 and Klotho gene binds to one another, and the mechanism analysis of Rab8 and Klotho interaction. The present invention firstly finds that the presence of Rab8 and Klotho in non-small cell lung cancer cells binds to one another, and a co-location exist in the subcellular, Rab8 binds to Klotho through a protein synthesis pathway and promotes a transport thereof to the membrane surfacein the form of vesicles, so that the level of the surface of the membrane is increased.
Owner:JIANGSU PROVINCE HOSPITAL THE FIRST AFFILIATED HOSPITAL WITH NANJING MEDICAL UNIV
Yeast transformant for screening protein with interaction with G protein beta-gamma dimer and screening method of yeast transformant
The invention relates to yeast transformant for screening a protein with interaction with G protein beta-gamma dimer and a screening method of the yeast transformant. Yeast transformant has encoding genes of a beta-subunit and a gamma-subunit of mulberry, a mulberry cDNA (Complementary Deoxyribonucleic Acid) library is converted into the yeast transformant capable of simultaneously expressing thebeta-subunit and the gamma-subunit, two types of nutrition defect screening of His and Met are carried out to obtain positive cloning with interaction with beta-gamma dimer, and finally the protein with interaction with the G protein beta-gamma dimer of the mulberry can be successfully screened. By adopting the method, a protein dimer interaction protein can be efficiently screened, and the methodcan be successfully applied to screening on a mulberry G protein beta-gamma dimer interaction protein.
Owner:SOUTHWEST UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com