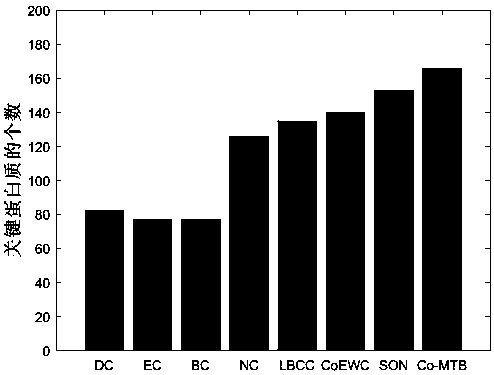
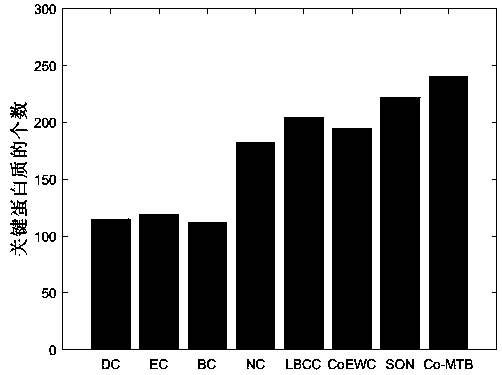
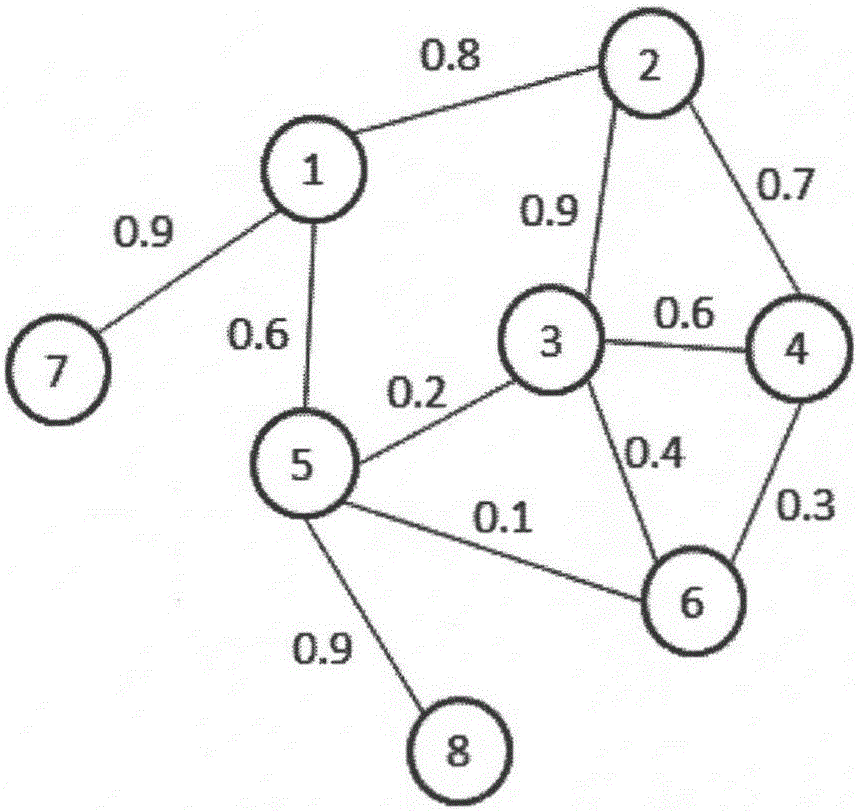
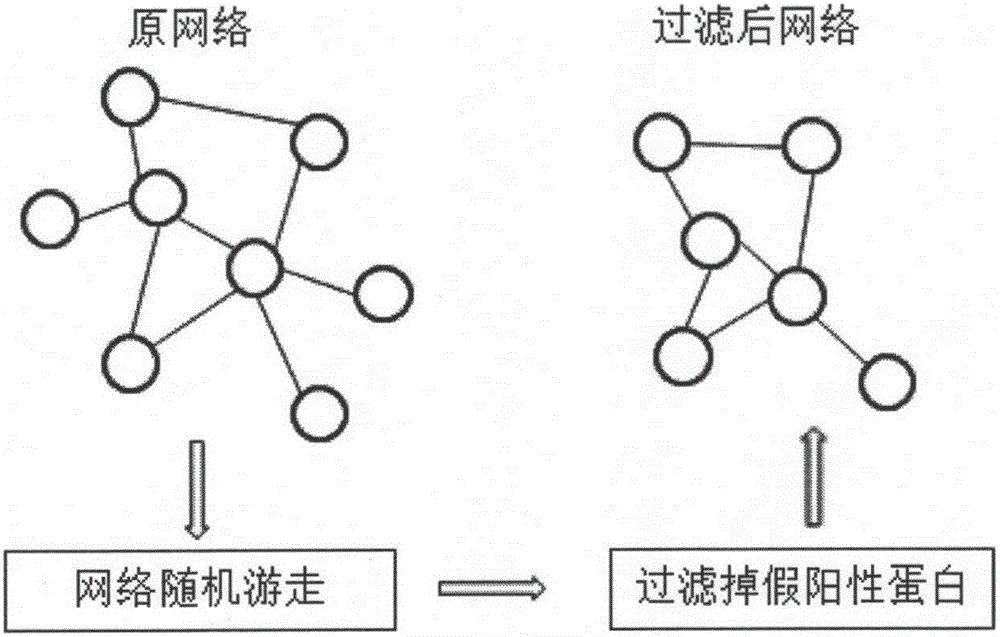
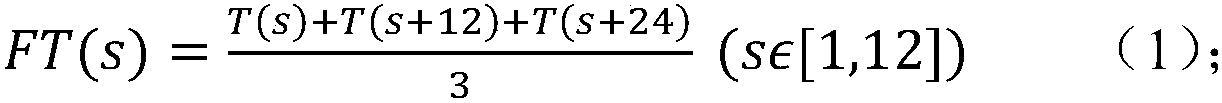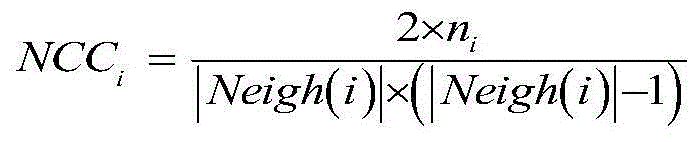Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
162 results about "Protein Interaction Networks" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for identifying protein functions based on protein-protein interaction network and network topological structure features
InactiveCN105138866ARobustSignificant predictive advantageSpecial data processing applicationsNODALData set
The invention discloses a method for identifying protein functions based on a protein-protein interaction network and network topological structure features. Firstly, a node and side-weighted protein-protection interaction network is established, wherein the node represents protein while the edge represents the interaction; then the nodes and the sides in the network are weighted by protein first-grade structural description and protein-protein interaction trust scoring; protection functional annotation data is collected to establish a data set, and a new protein with overall and local information network topological structure features is provided based on a graph theory; and finally, the protein functions are predicated by choosing features through adopting a minimum-redundancy maximum-correlation method and by modeling through a support vector machine. The protein function predication method is greatly better than the prior art, and has robustness on sequence similarity and sampling; and meanwhile, information of three-dimensional structure and the like of protein is not required, so that the method is simple, rapid, accurate and efficient, and the method is expected to be applied in the research fields of proteomics and the like.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
Method for identifying key proteins in protein-protein interaction network
ActiveCN105279397AImprove recognition accuracySolve the costSpecial data processing applicationsProtein insertionProtein protein interaction network
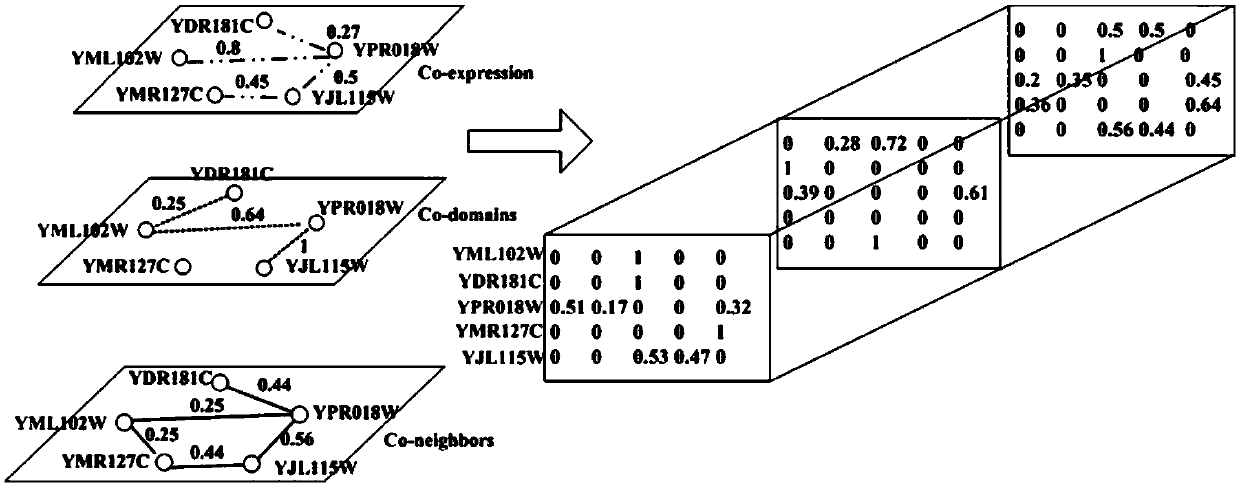
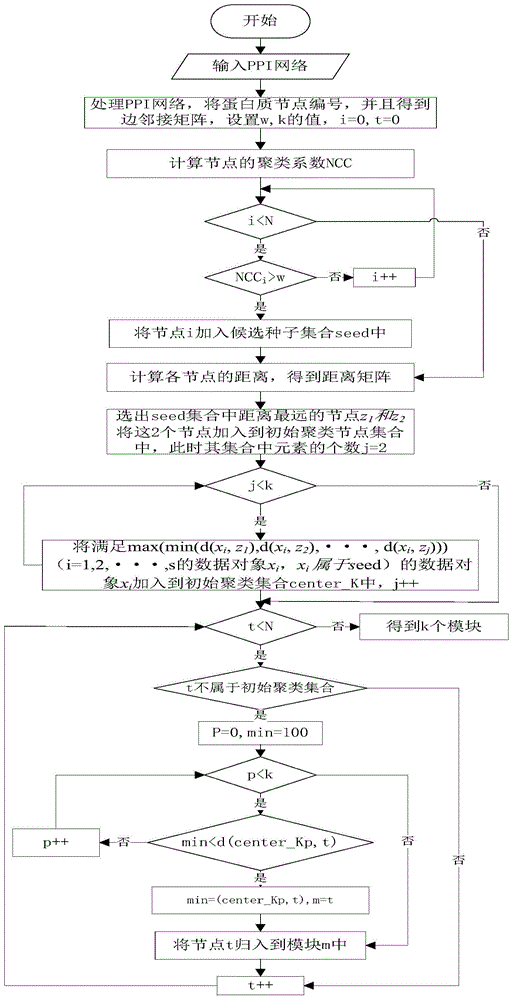
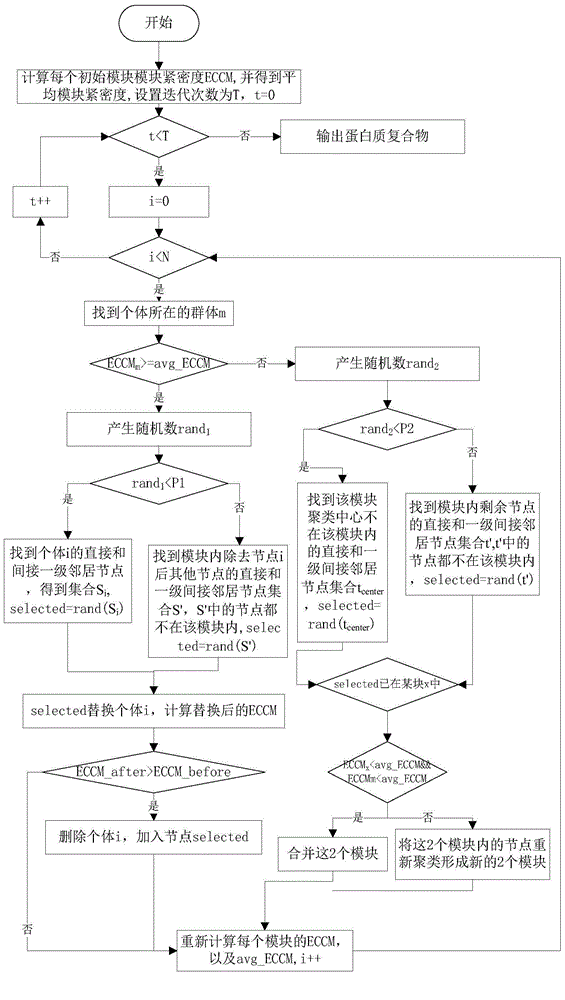
The invention discloses a method for identifying key proteins in a protein-protein interaction network. According to the method, an undirected graph G is constructed according to the protein-protein interaction data, and the edge clustering coefficient of the graph is calculated. Compared with the prior art, the method provided by the invention has the advantages of combining the gene expression profile data and the gene function annotation information data on the basis of considering the topological structure characteristics of the protein-protein interaction network, and integrating three groups of data to predict the key proteins, so that the influence caused by the data noise of a single data source on the prediction correctness can be effectively decreased, and the key proteins in the network can be predicted through the key protein characteristics embodied by three types of data, such as the edge clustering coefficient in the protein-protein interaction network, the Pearson's correlation coefficient of the gene expression value and the gene function similarity index. According to the method, the identification correctness of the key proteins in the protein-protein interaction network can be remarkably improved, and abundant key proteins can be predicted once, so that the problem that the biological experiment method is high in cost and time-consuming is solved.
Owner:EAST CHINA JIAOTONG UNIVERSITY
Protein functional module excavating method for multi-view data fusion
The invention belongs to the field of data excavation and discloses a protein functional module excavating method for multi-view data fusion. The method comprises the following steps: firstly performing quantifying description on strong and weak interaction of multiple data sources on protein and forming multi-view data; further performing uniform matrix decomposition on the multi-view data by utilizing a polymerization nonnegative matrix algorithm provided by the invention; determining the functional module of the protein by virtue of obtaining the optimal approximation of the multi-view information. The protein functional module excavating method for multi-view data fusion, provided by the invention, aims at simultaneously analyzing multiple biodata and comprises gene coexpression, GO annotation and PPIN and can be used for extracting the protein functional module with the most consistent polymerization characteristic from the multi-view. The method disclosed by the invention is especially suitable for interaction networks and biodata of the protein and meanwhile can be applied to community excavation problems of social complex networks and communication networks.
Owner:BEIJING UNIV OF TECH
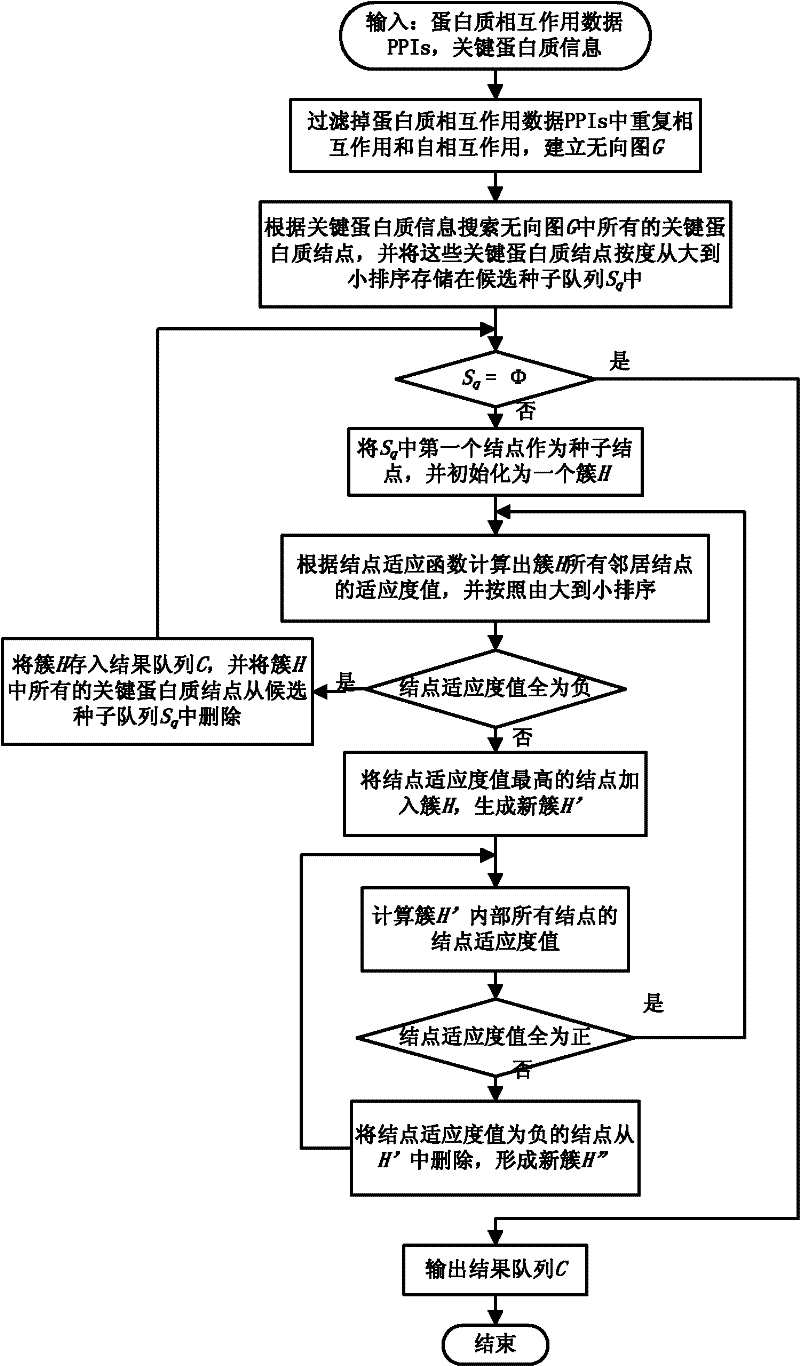
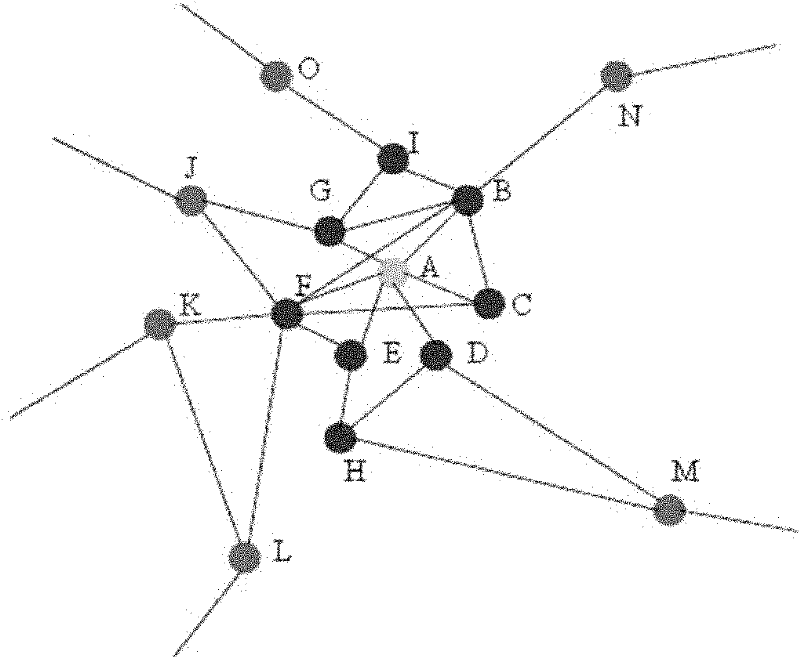
Protein complex identification method based on key protein and local adaptation
ActiveCN102176223AEfficient identificationAccurate identificationSpecial data processing applicationsLife activityProtein-protein complex
The invention discloses a protein complex identification method based on key protein and local adaptation. Based on the importance of the key protein to life activities of organisms and the topological property of a protein interaction network, the invention discloses the protein complex identification method (EPOF) based on key protein and locally adapted protein by using the key protein as a seed. The protein complex identification method not only can be applied to a non-weighted protein interaction network, but also can be applied to a weighted protein interaction network. The protein complex identification method can be used for recognizing the protein complex more accurately only according to the protein interaction information and the key protein information, and predicting a large quantity of protein complexes in one step, and solves the problems of high cost, high time consumption and the like of the chemical experiment method.
Owner:CENT SOUTH UNIV
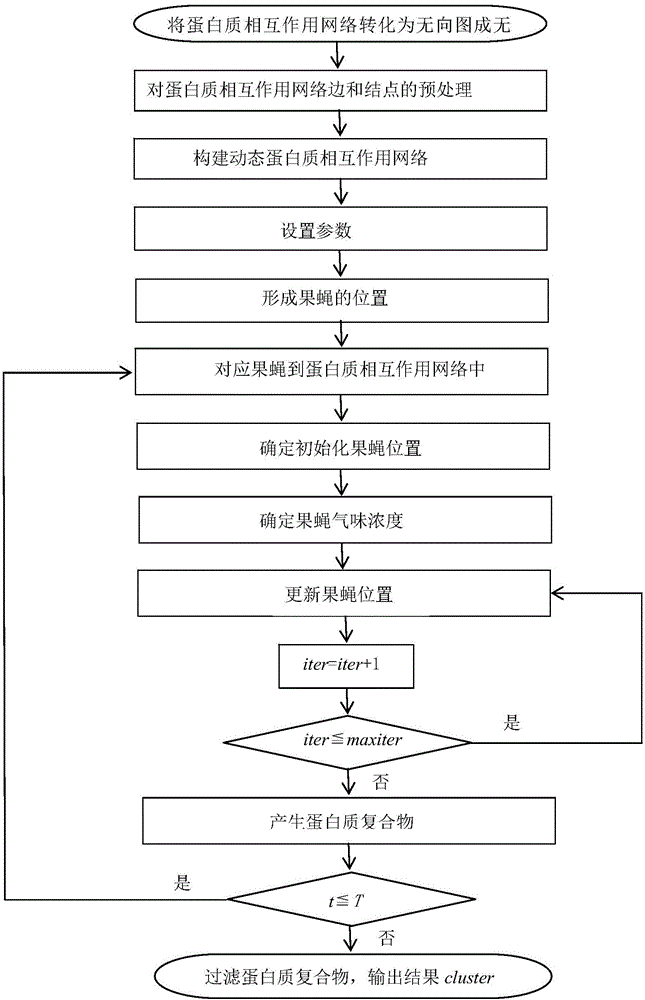
A method of identifying protein compounds by using a fruit fly optimization method
InactiveCN105868582AImprove accuracyRealistically simulate dynamicsProteomicsGenomicsProtein protein interaction networkProtein-protein complex
The invention provides a method of identifying protein compounds by using a fruit fly optimization method. The method comprises the steps of converting a protein-protein interaction network into a undirected graph, performing pretreatment on the edges and nodes of the protein-protein interaction network, establishing a dynamic protein-protein interaction network, setting parameters, forming fruit fly positions, matching fruit flies with the protein-protein interaction network, determining initialization fruit fly positions, determining the fruit fly odor concentration, updating the fruit fly positions, generating a protein compound, and filtering the protein compound. The method gives full consideration to the dynamic nature of the protein network, the protein compound inner core-attachment structure and the locality and wholeness of the protein-protein interaction network and can identify protein compounds accurately. The results of simulation experiments show that the performance of the indexes such as the accuracy and the recall ratio are excellent. Compared with other clustering methods, the method, based on the characteristics of the protein network and the protein compounds, realizes the protein compound identification process and improves the protein compound identification accuracy.
Owner:SHAANXI NORMAL UNIV
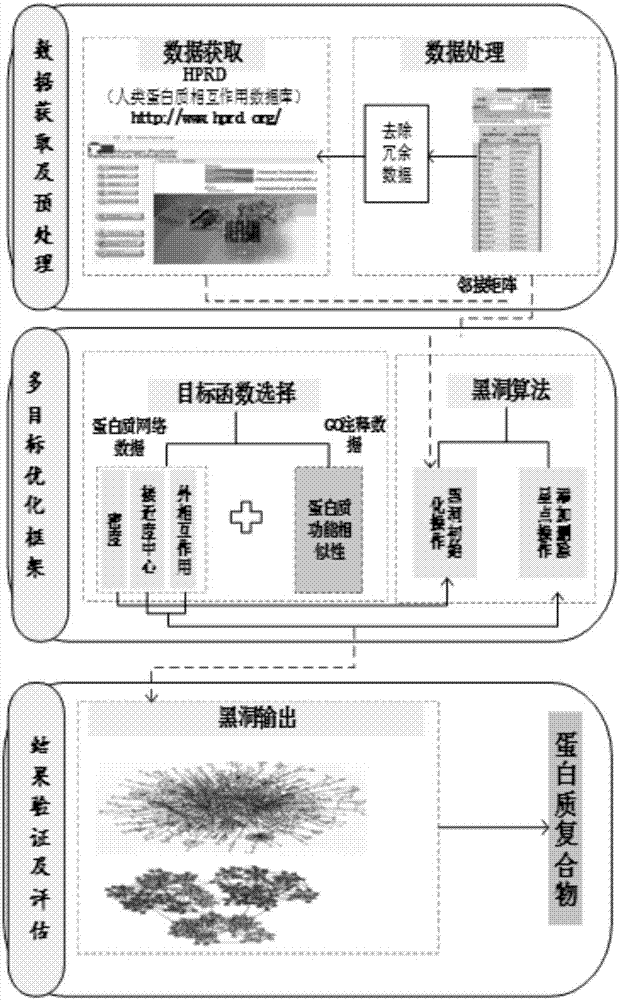
Protein complex recognizing method based on multi-source data fusion and multi-target optimization
InactiveCN108009403AImprove accuracyRecognition speed is fastSpecial data processing applicationsComplex network analysisAlgorithm
The invention discloses a protein complex recognizing method based on multi-source data fusion and multi-target optimization, comprising: preprocessing protein interaction network data to obtain adjacent matrixes; primarily clustering protein complexes to obtain an initial protein complex module; further optimizing the initial protein complex module, fusing topological structural features of the protein interaction network data and functional similar features of GO (gene ontology) annotation data during optimizing, and performing optimizing operation in conjunction with an adaptive multi-target blackhole optimization algorithm to obtain a more precise protein complex module; postprocessing to obtain a final optimal protein complex. The method of the invention has the advantages that protein complex recognition speed and precision are increased, the method is applicable to protein interaction networks and extensible to the analysis of other complex community networks, and the method isvery practical in complex network analysis.
Owner:CHINA UNIV OF GEOSCIENCES (WUHAN)
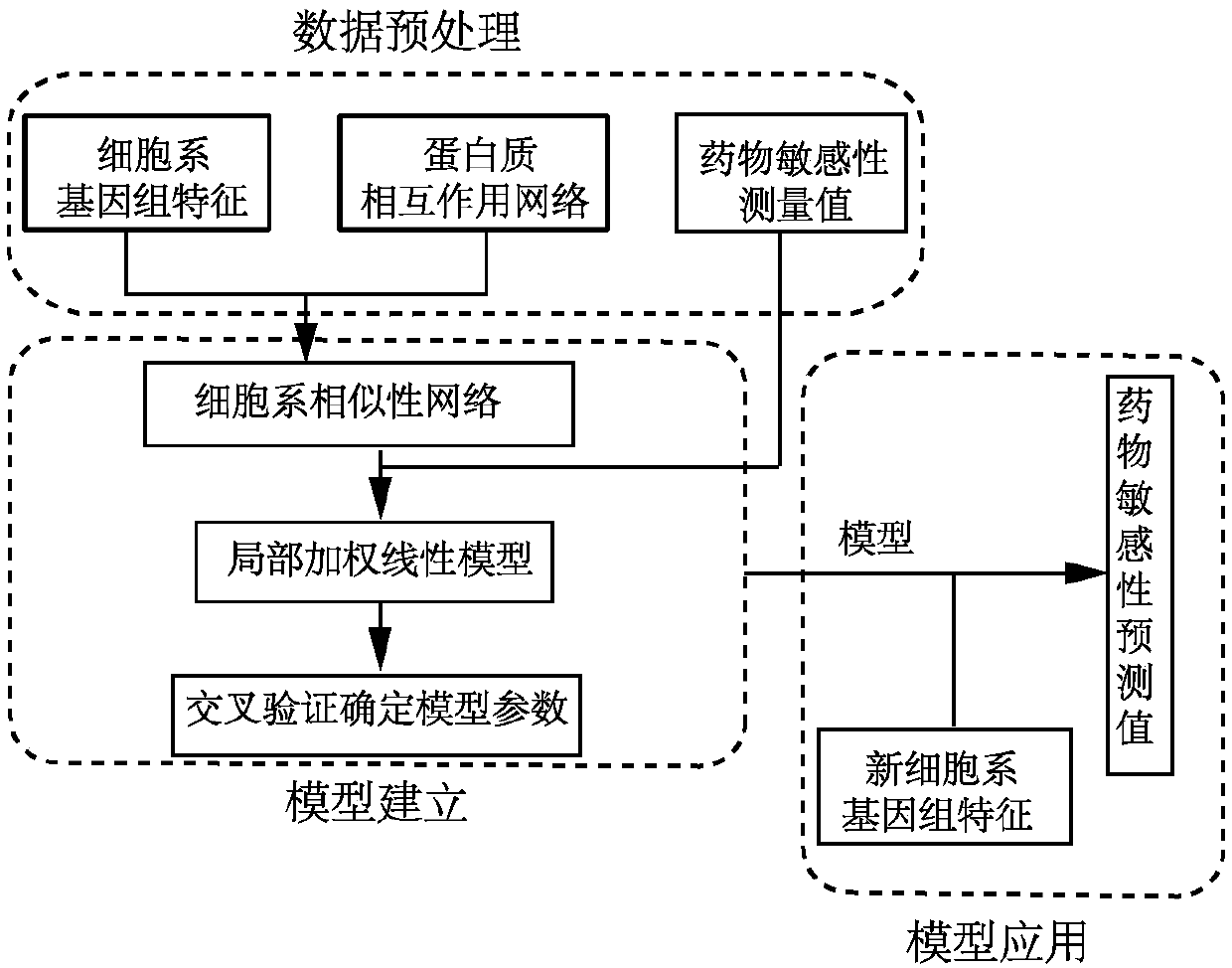
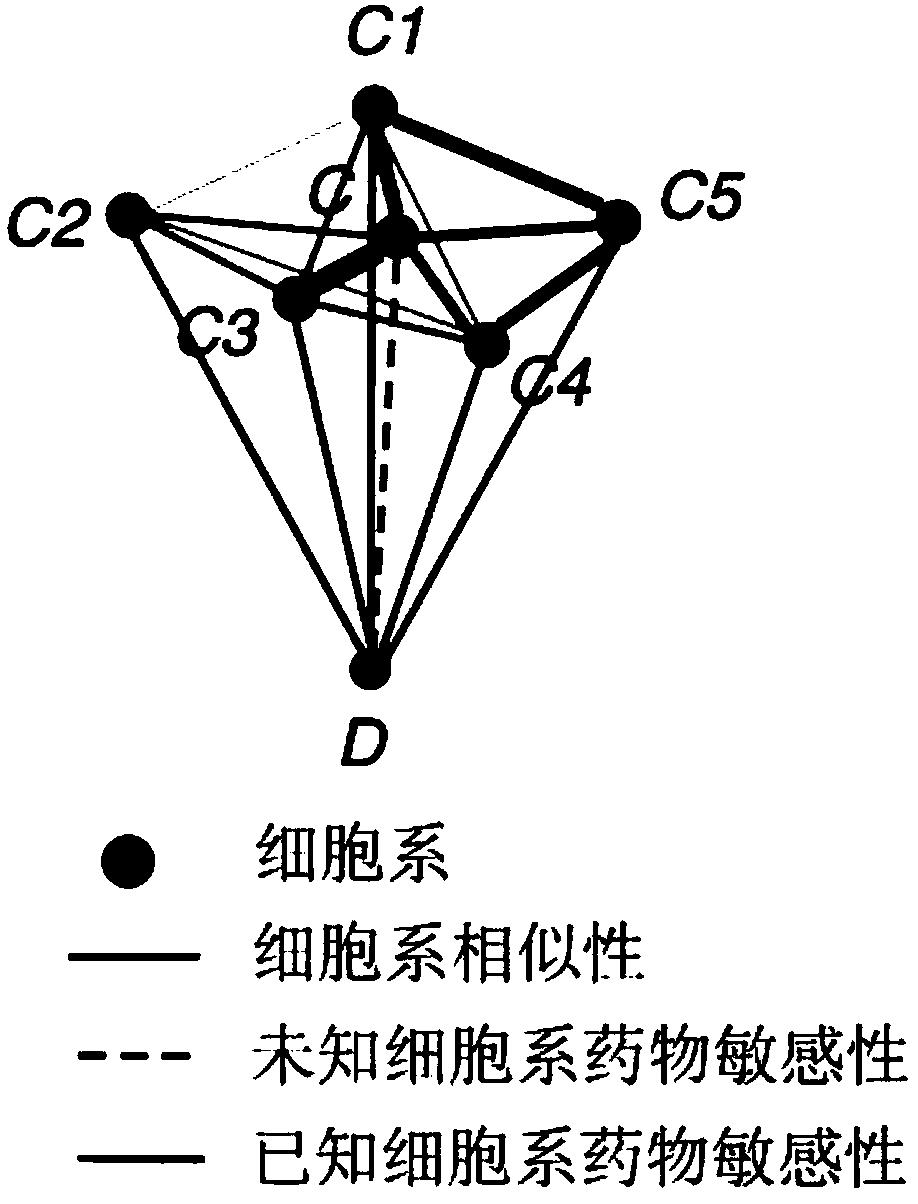
Drug sensitivity prediction method for cancer precision treatment
InactiveCN107609326ASolve the high latitude input problemLow running costSpecial data processing applicationsGenomicsGene expression profiling
The invention discloses a drug sensitivity prediction method for cancer precision treatment. Anti-cancer drug sensitivity prediction is performed by comprehensively utilizing genomics data of cell lines, so that theoretical medication guidance is provided for individual treatment. A calculation model is built for predicting drug sensitivity to individuals, thereby guiding clinical medication, which is one of core goals of precision treatment. According to the method, interaction terms among genes are added by utilizing a protein interaction network; a mutual relationship among the cell lines is described by utilizing the similarity of gene expression profiles of the cell lines; a local weighted linear model is built without depending on an independence assumption among genomic characteristics; and compared with a conventional linear model, a better prediction effect is achieved, the problem of high-latitude input caused by large-scale data characteristics is solved, and the operation cost of the model is reduced. The method can be widely applied to the drug sensitivity prediction of new cell lines, and provides the theoretical guidance for guiding the clinical medication.
Owner:TONGJI UNIV
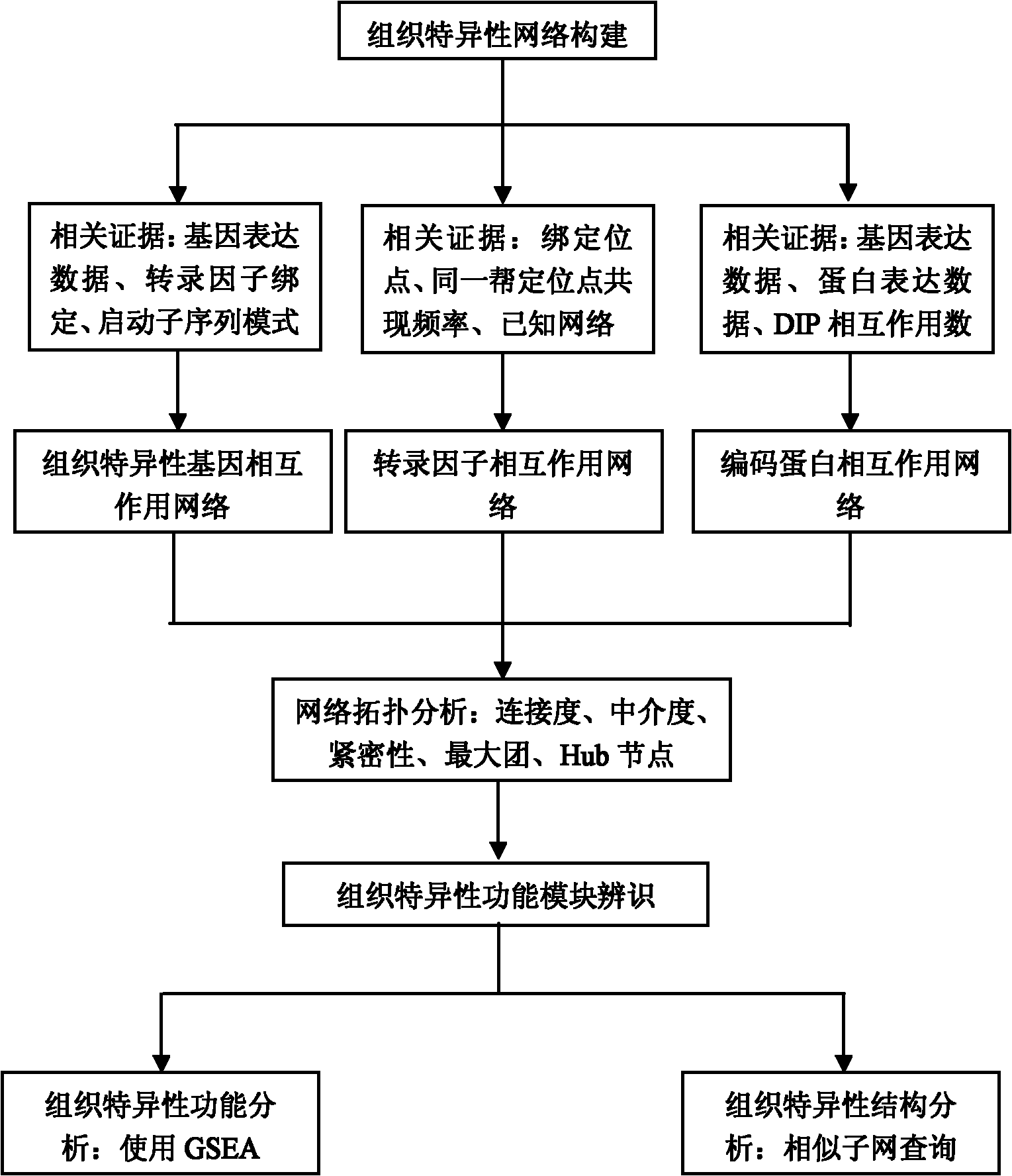
Method for constructing and analyzing tissue-specific interaction topology network
The invention discloses a method for constructing and analyzing a tissue-specific interaction topology network. The method comprises the following steps of: measuring similarity of integrated interaction network nodes; constructing a tissue-specific gene interaction network, a tissue-specific transcription factor interaction network and a tissue-specific encoded protein interaction network by a construction module of the tissue-specific interaction network; counting connection degree of each node in the network, counting betweenness of each node in the network, calculating closeness of each node in the network, calculating max clique in the interaction network and counting a Hub node in the interaction network by a topology analysis module of the interaction network; and acquiring related tissue-specific knowledge from a gene ontology database and a KEGG (Kyoto Encyclopedia of Genes and Genomes) biological field knowledge base by an identification module of the tissue-specific functional module. The invention provides an ideal method for constructing the interaction network from genetic information; and the method is a more ideal analysis tool for researching a tissue-specific intrinsic mechanism.
Owner:TIANJIN UNIV
Weight assembly clustering method for excavating protein complex
InactiveCN103235900AEnhance explanatoryReduce sensitivitySpecial data processing applicationsProtein containing complexUndirected graph
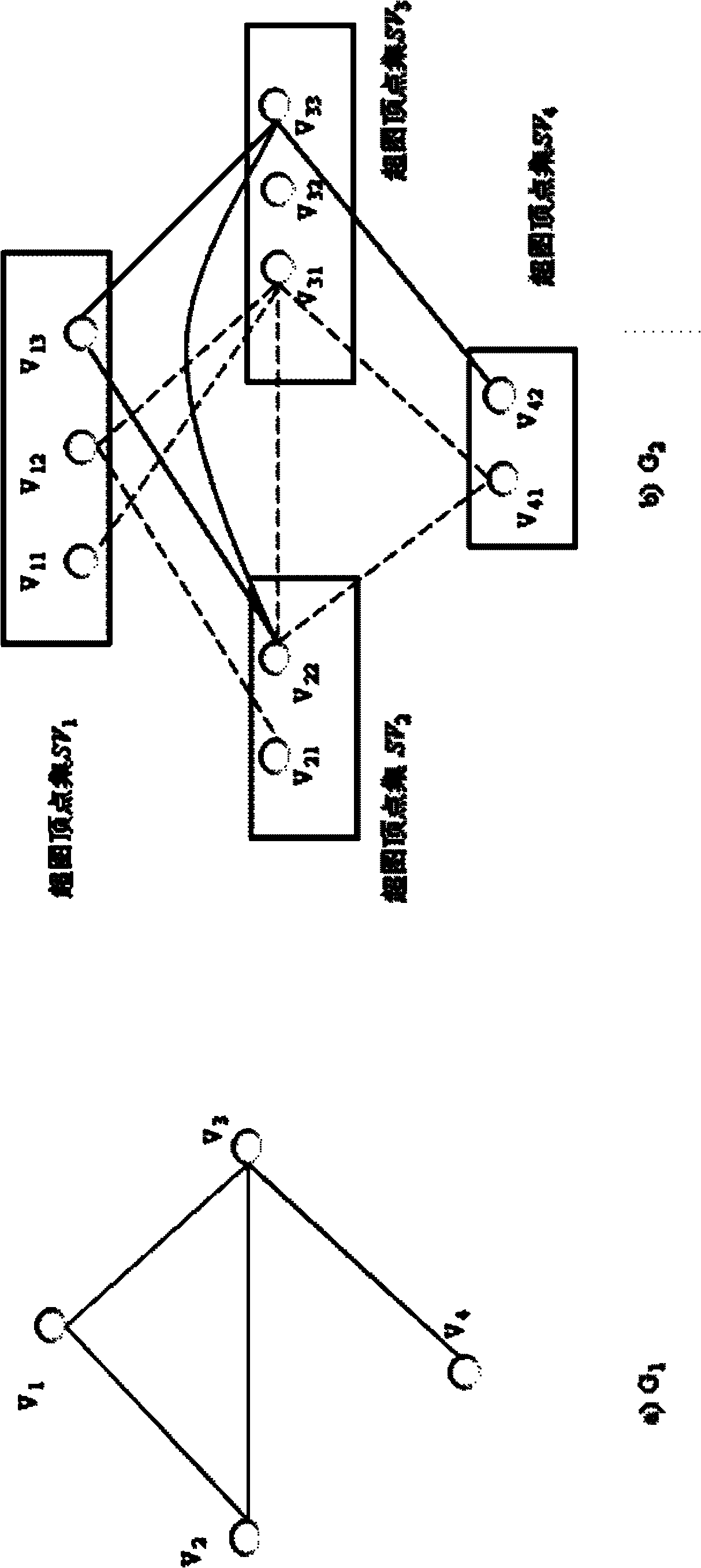
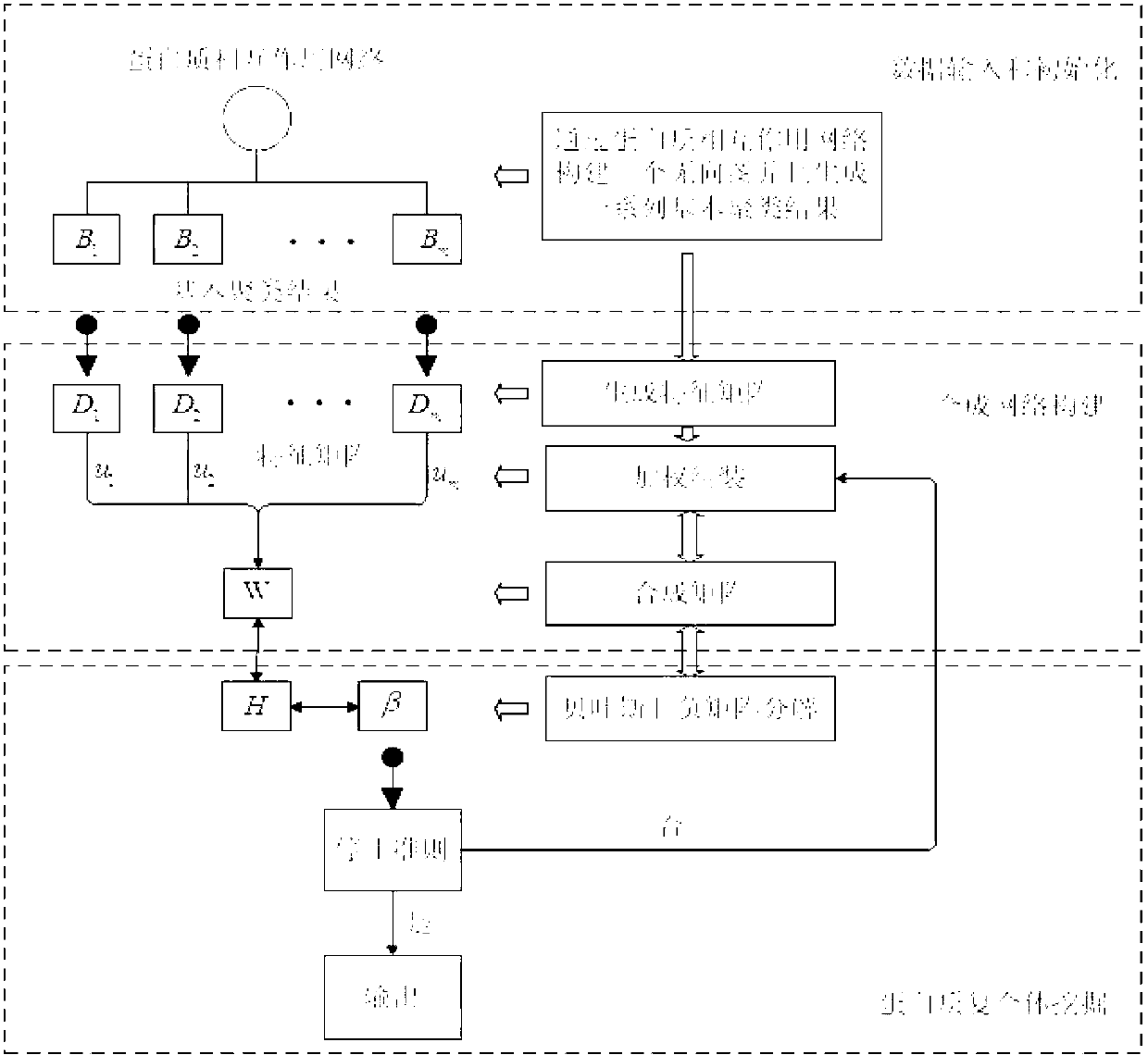
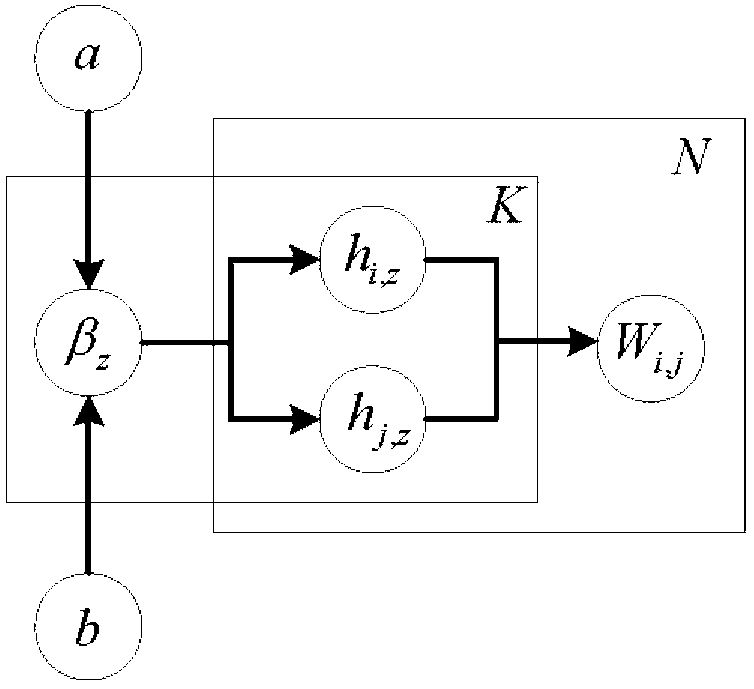
The invention discloses a weight assembly clustering method for excavating a protein complex. The method comprises the following steps: inputting a protein interaction network to produce an undirected graph, selecting m clustering methods to be applied to the network to obtain m clustering results; rebuilding characteristic networks for the base clustering results to obtain m characteristic networks, wherein the m characteristic networks correspond to m characteristic matrixes; combing the m characteristic matrixes to obtain a combined matrix W, wherein uq refers to weights of the qth characteristic network, and uq is larger than or equal to 0, the combined matrix W corresponds to a new network, and elements Wi and j are used for measuring the similar degree of the protein i and the protein j in the new network; excavating the clusters in the new network through adopting a Bayes non-negative matrix factorization algorithm; integrating the weight studying and the complex discovering into an optimal object, so that the weight is optimized through the clustering result, otherwise, the clusters are guided by the weight results; and obtaining the final protein complex excavating result after the optimizing is finished.
Owner:SUN YAT SEN UNIV
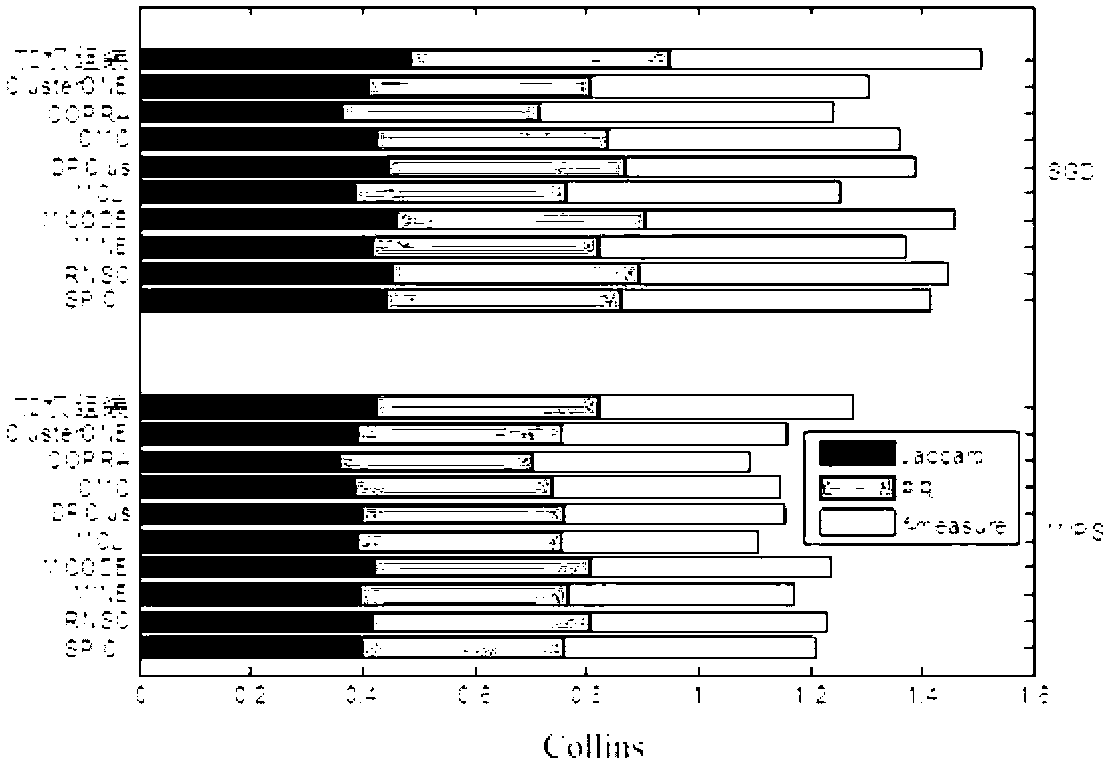
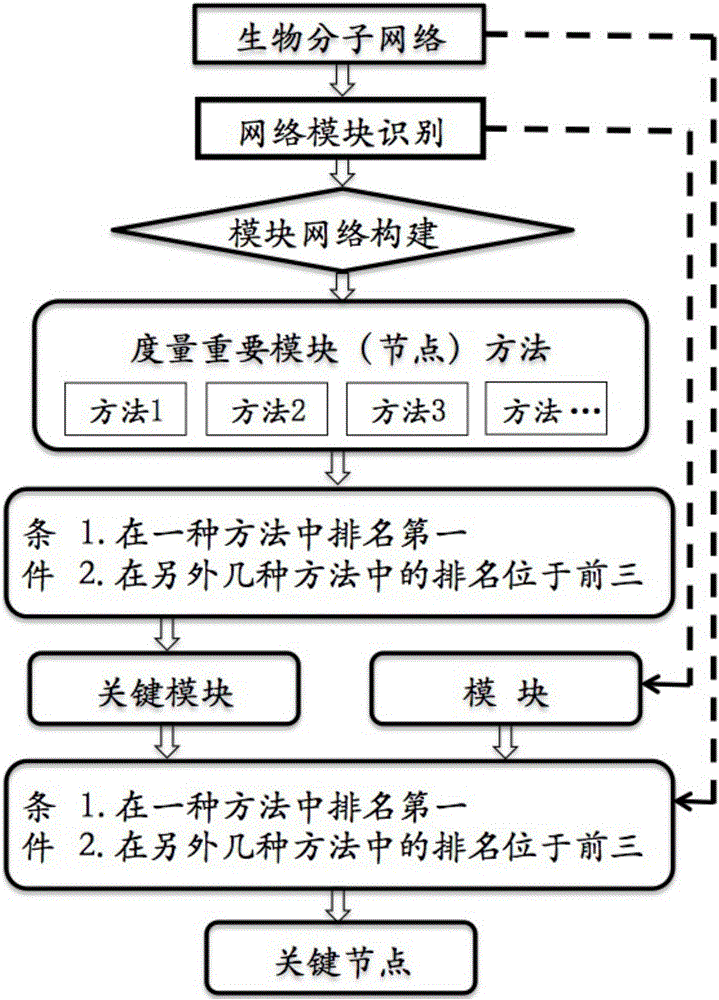
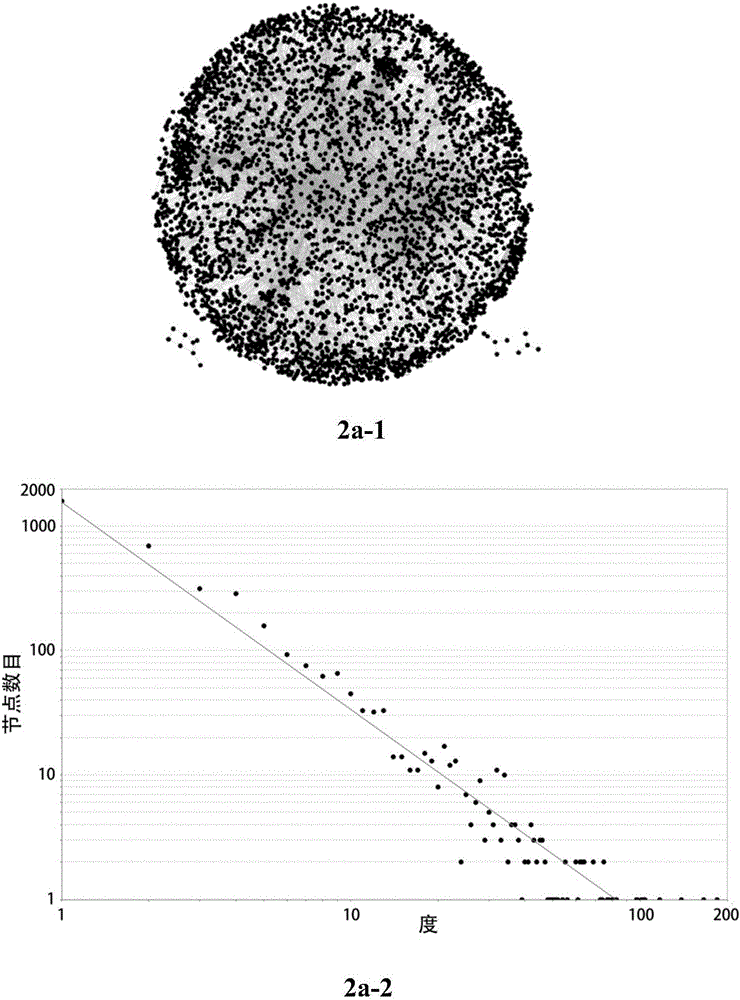
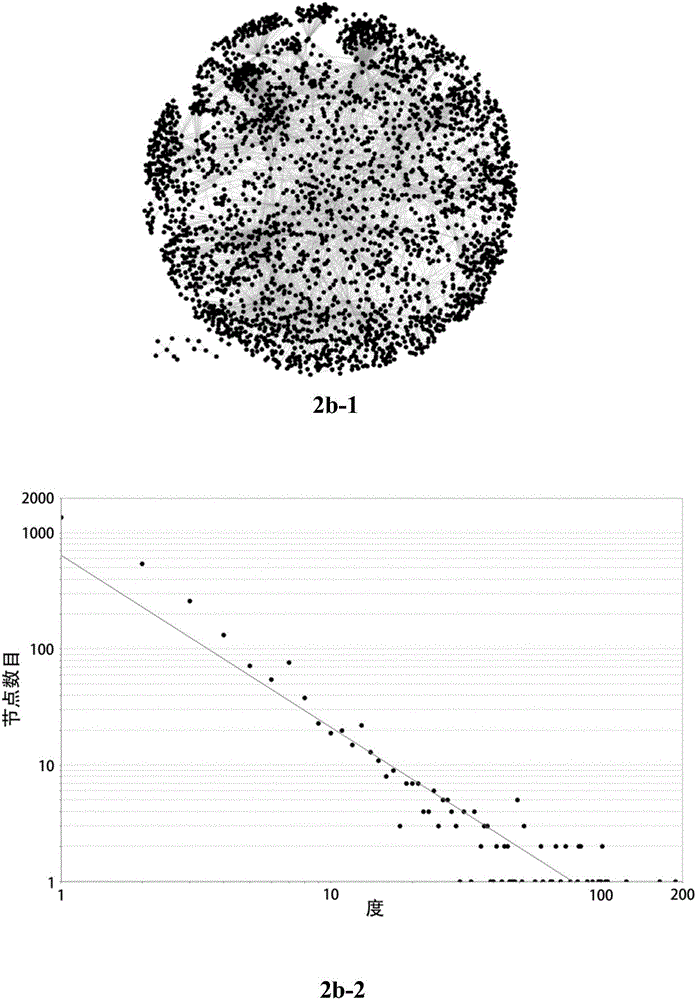
Method for identifying key module or key node in biomolecular network
The invention provides a method for identifying a key module and a key node by fusing multiple methods based on a topological structure of a biomolecular complex network such as a protein interaction network, a gene expression regulation network, a biological metabolism network, an epigenetic network, a phenotype network or a signal conduction network. The method comprises the following main steps of performing module division on the network based on the biomolecular network, and comprehensively and quantitatively identifying the key module and the key node from multiple angles by adopting multiple measurement methods based on the topological structure of the network for the network or each module.
Owner:王忠 +1
Plant protein interaction network constructing method based on deep learning
InactiveCN110136773AImprove forecast accuracyReduce the workload of parameter adjustmentSystems biologyInstrumentsAlgorithmInteractions protein
The invention relates to a plan protein interaction network constructing method based on deep learning. The method comprises the following steps of 1), acquiring 11 characteristic data of a protein interaction pair; 2), performing screening for obtaining a training set and a testing set; 3), constructing a deep learning classification model; 4), performing batch optimization on the parameter of the deep learning classification model, and obtaining a classification model of an optimal optimization parameter combination; 5), performing interaction relation predicting on all possible two-to-two interaction proteins of the whole genome according the classification model of the optimal optimization parameter combination; and 6), predicting the protein interaction network according to an interaction relation predicting result. Compared with the prior art, the method has advantages of high predicting accuracy, high modeling efficiency, etc.
Owner:SHANGHAI JIAO TONG UNIV
Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks
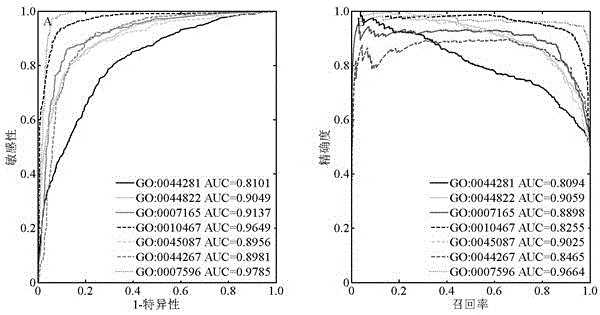
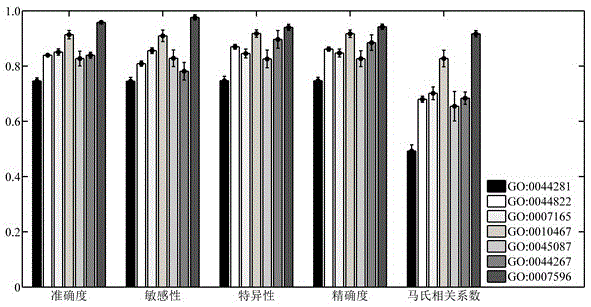
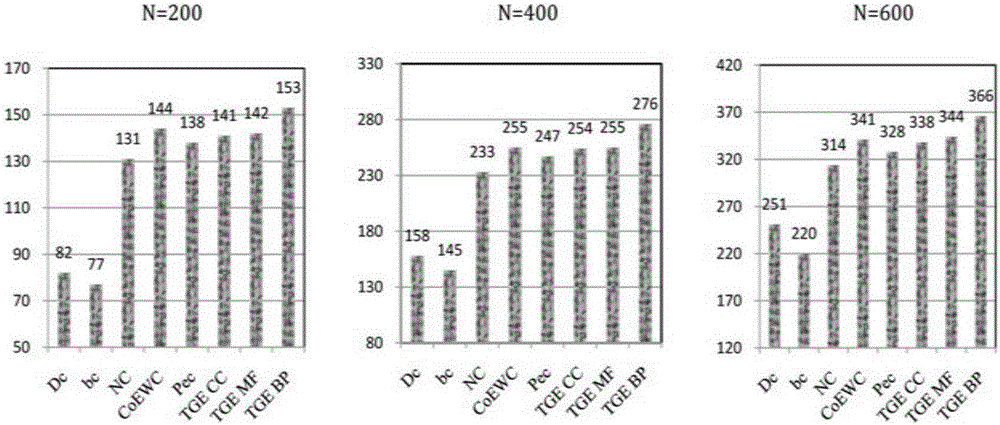
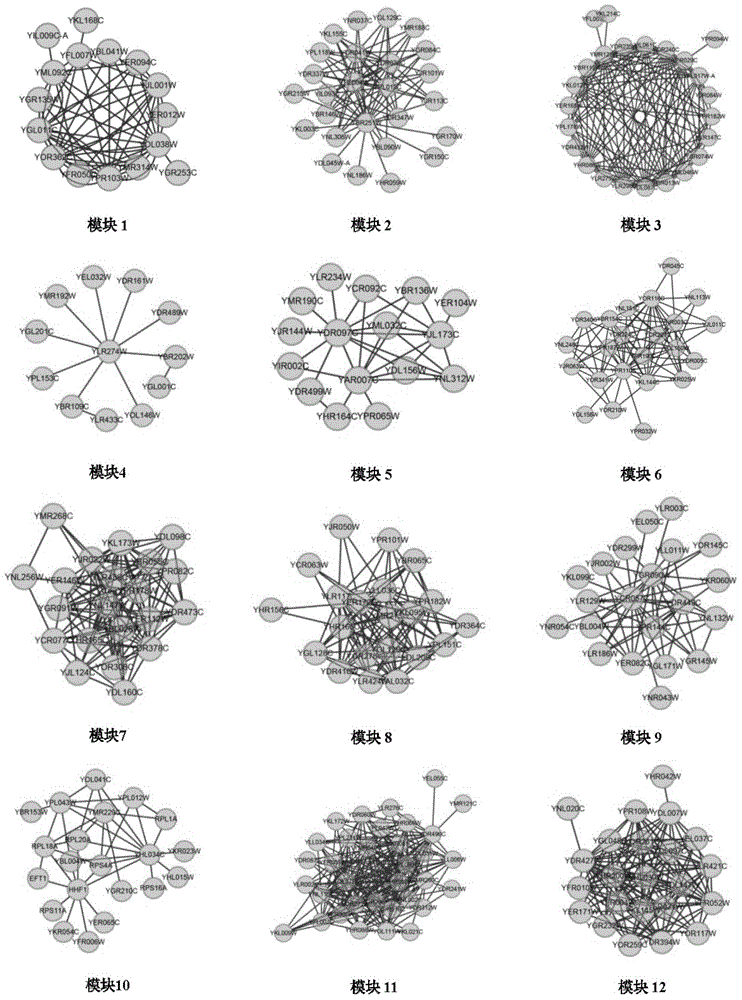
InactiveCN106529203AConvenient researchData visualisationSystems biologyProtein targetProtein insertion
The invention discloses a method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks. The method includes steps of building three sub-networks including a human protein-protein interaction network on the basis of HIPPIE, a miRNA-target protein network on the basis of mirTARbase and a miRNA-miRNA network on the basis of target protein overlapping structures; combining the three sub-networks with one another according to acquisition numbers of proteins and ID (identification) numbers of miRNA molecules in miRbase databases and building fusion miRNA-target protein association relationship networks; representing miRNA-target protein association features on the basis of guilt-by-association principles, building classification prediction models by the aid of random forest and predicting potential interaction association relationships between miRNA and the target proteins. The method has the advantages that research on many-to-many relationships of the miRNA regulation target proteins can be effectively carried out, and accordingly the method has high application value.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
Protein complex recognizing method based on range estimation
InactiveCN101246520AEfficient identificationImprove robustnessSpecial data processing applicationsShortest distanceProtein-protein complex
The invention discloses a protein compound recognition method based on the range estimation, which takes the most short distance between protein apexes as a key parameter to recognize the protein compound, and controls the dense degree of the recognized the protein compound by the function probability between the protein apexes and the protein compound based on finding that the most short distance between the protein apexes in the known protein compound generally does not surpass 2. The realization of the invention is simply; a plurality of known proteins compound with the biological significance can be recognized through the protein interaction network, the invention has the very good toughness for the high proportion false positive and false negative which universally exist in the protein interaction large-scale data, and solves the chemical experiment cost to be expensive and the biology difficult problems that the quantity of single recognition is small and the dynamic compound is very difficult to recognized effectively.
Owner:CENT SOUTH UNIV
Key-protein recognition method based on fusion of biological and topological features
ActiveCN108733976AImprove accuracyImprove recognition efficiencySpecial data processing applicationsProtein Interaction NetworksEuclidean vector
The invention belongs to the field of bioinformatics technology, and particularly relates to a key-protein recognition method based on fusion of biological and topological features. The method includes: assigning a score, which indicates an importance degree of each protein vertex, to the protein vertex, wherein the scores of all the vertices constitute a vector of n columns; giving initial valuesof the scores, constituting the attribute values of the protein vertices according to biological information and values of topological characteristics, and constituting an attribute matrix; and finally, carrying out descending arrangement according to the scores, and outputting k proteins corresponding to the scores, namely final results. Combination of the topological characteristics of a protein interaction network and the protein biological-attributes facilitates identification of key proteins, and improves efficiency of key-protein recognition.
Owner:YANGZHOU UNIV
Protein composite identification method based on random walking model
InactiveCN106355044ASolve the noiseOvercome limitationsProteomicsGenomicsExperimental validationProtein insertion
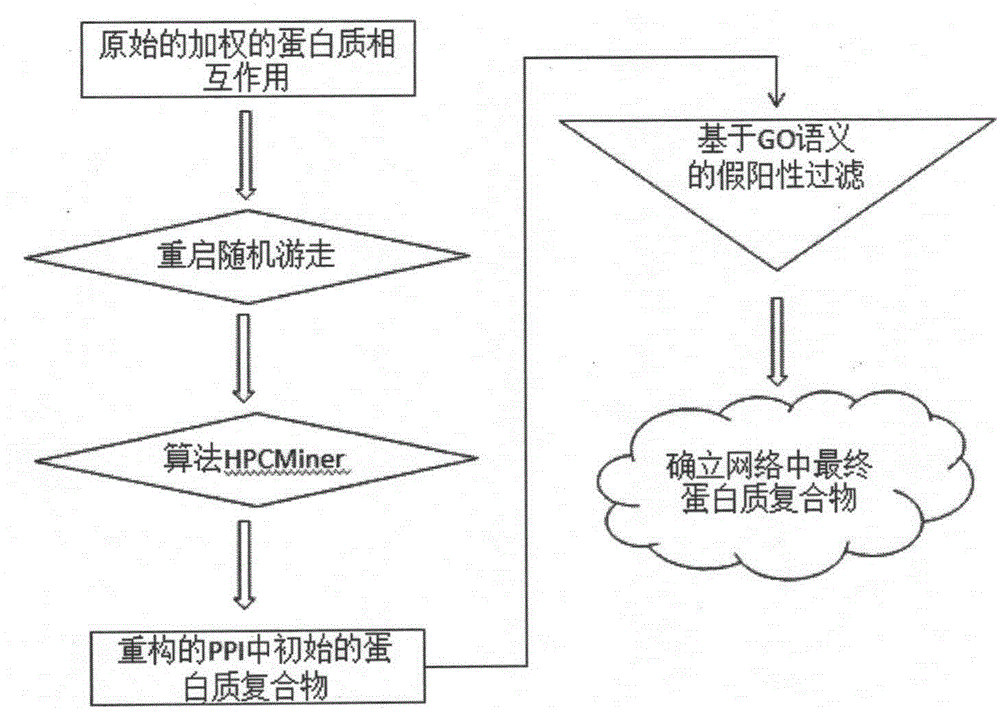
The invention provides a protein composite identification method based on a random walking model. Interaction data and false-negative or false-positive noisy data truly existing on a protein network are forecasted through the random walking algorithm. On the protein interaction network obtained after false-negative data and false-positive noisy data are removed, protein composites with the biological significance are identified through a H-index graph model, the semantic similarity between the protein composites is calculated according to a GO body, and the identified protein composites are finally determined. According to the protein composite identification method based on the random walking model, the algorithm is insensitive to input parameters, and the effectiveness of the provided algorithm is verified through experiments.
Owner:SHANGHAI DIANJI UNIV
Method for recognizing protein network compound based on semantic density
InactiveCN104992078AReduce time complexityShort timeSpecial data processing applicationsData setGene ontology
The invention discloses a method for recognizing a protein network compound based on a semantic density, which is specifically implemented at the steps that all the protein properties in a network data set are searched in a gene ontology base GO as for a protein-protein interaction network data set without a weight; based on searching results, similarity of connected proteins in the network data set is calculated by a semantic similarity calculation method based on gene ontology; according to obtained similarity results, the given protein-protein interaction network data set is converted into an undirected network data set with a weight, wherein nodes represent the proteins, borders stand for interactions among the proteins, and the similarity among the proteins is the weight of the border; and the protein compound can be recognized from a protein-protein interaction network, wherein recognition accuracy is high and time complexity is low.
Owner:XIAN UNIV OF TECH
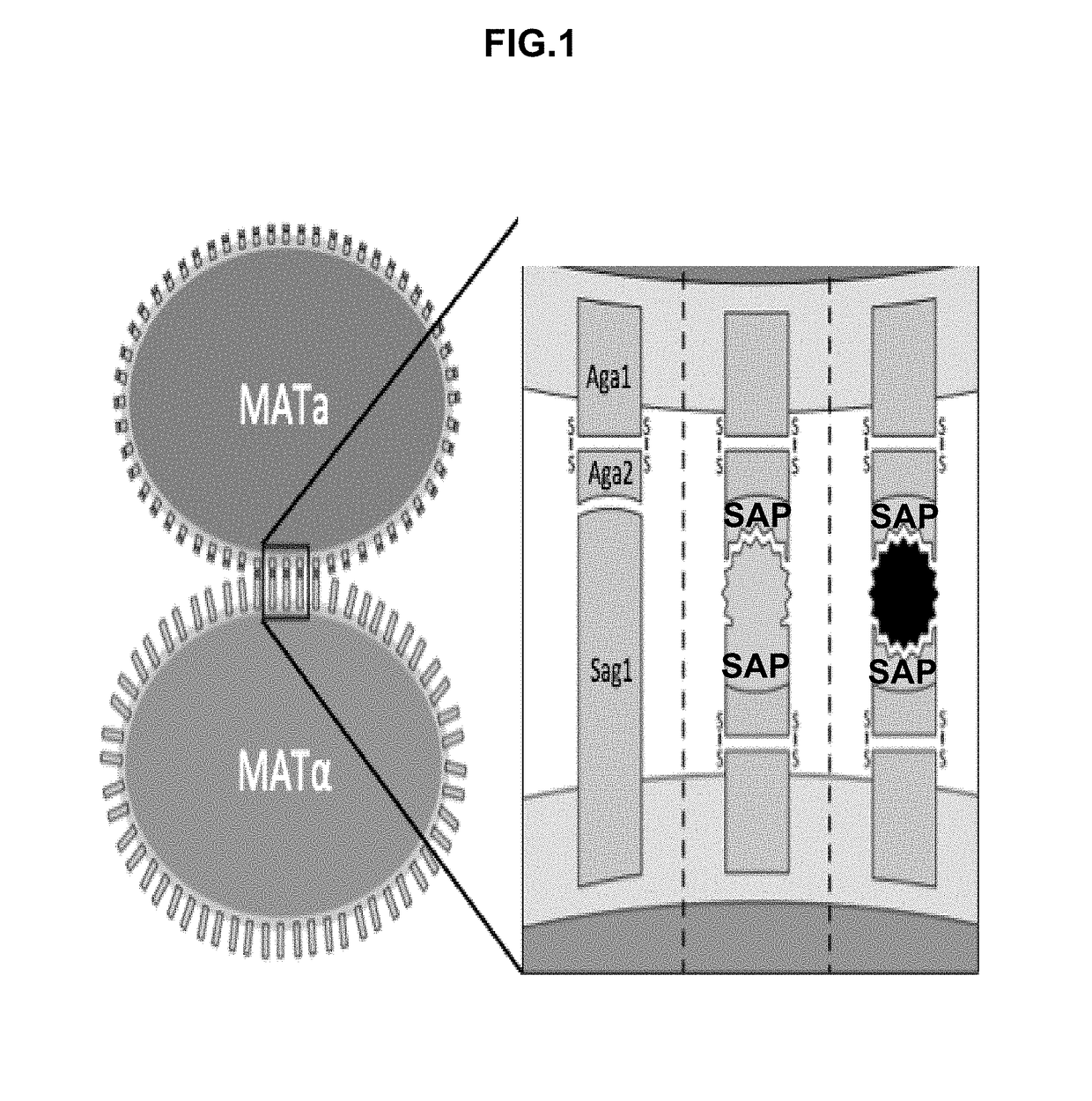
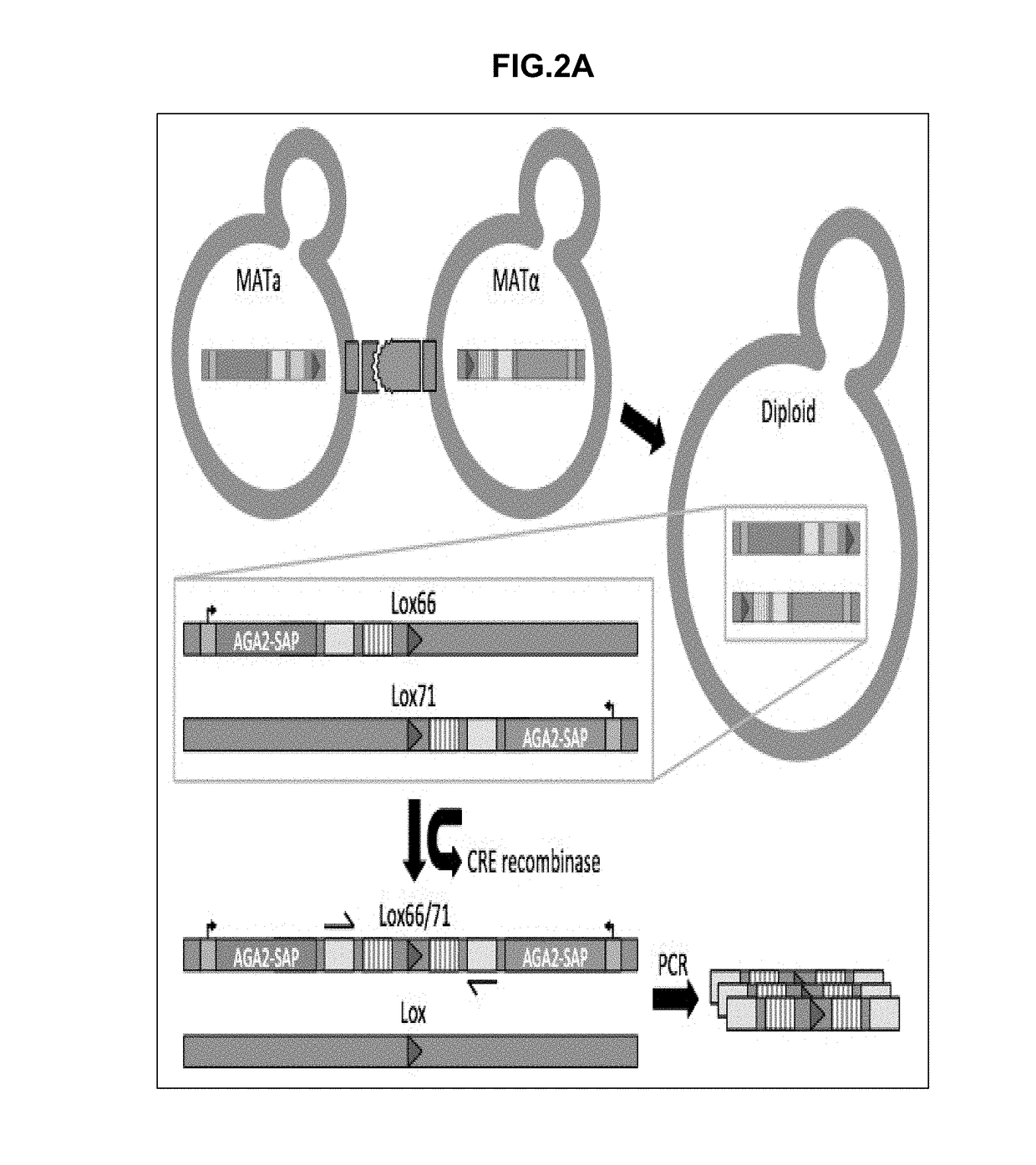
Synthetic yeast agglutination
InactiveUS20170205421A1High throughput screeningHydrolasesMicrobiological testing/measurementProtein protein interaction networkAgglutination
The present invention provides methods and compositions that can be applied to screening protein-protein interaction networks, screening drug candidates that modulate protein-protein interactions for on- and off-target effects, detecting extracellular targets for which no native S. cerevisiae receptor exists, and re-engineering yeast agglutination in order to answer biological questions about yeast speciation and ecological dynamics
Owner:UNIV OF WASHINGTON
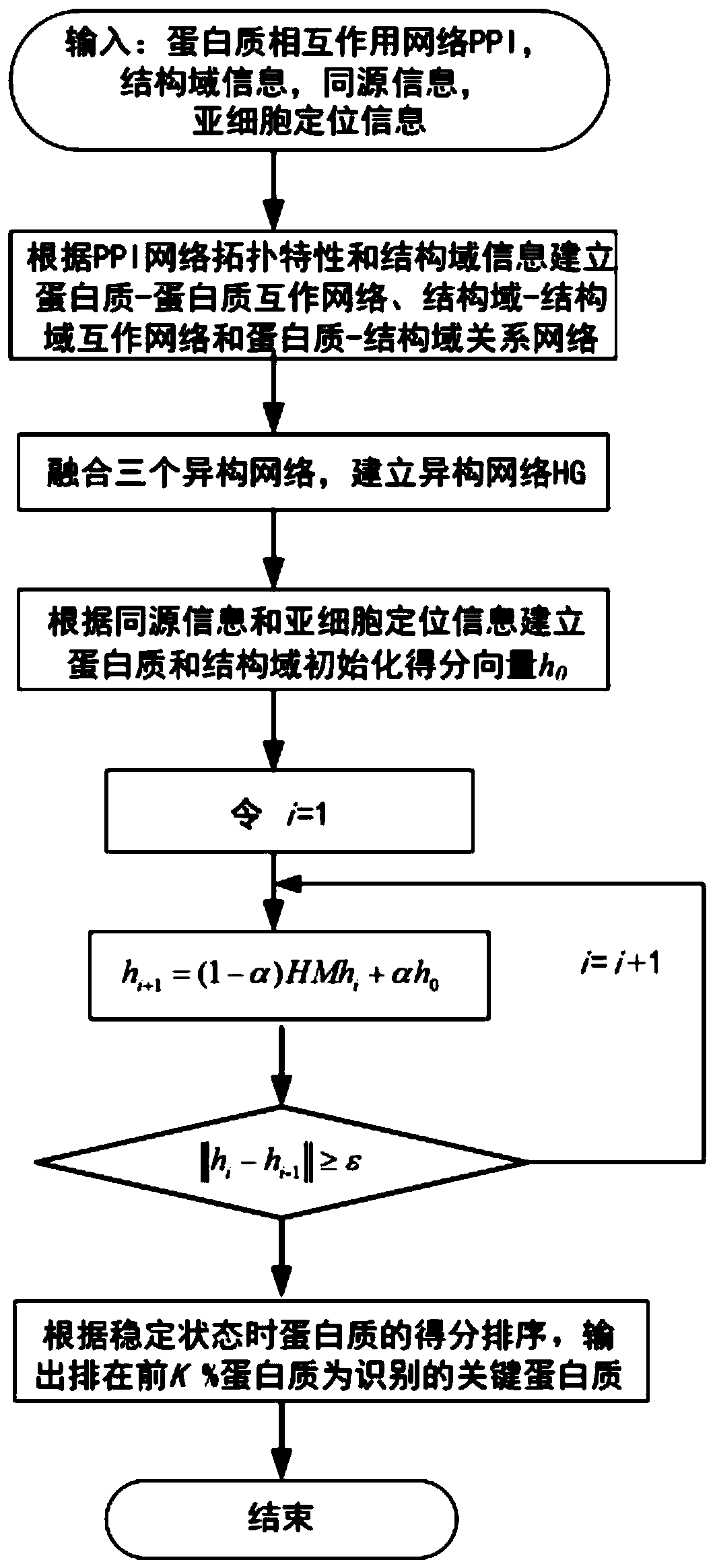
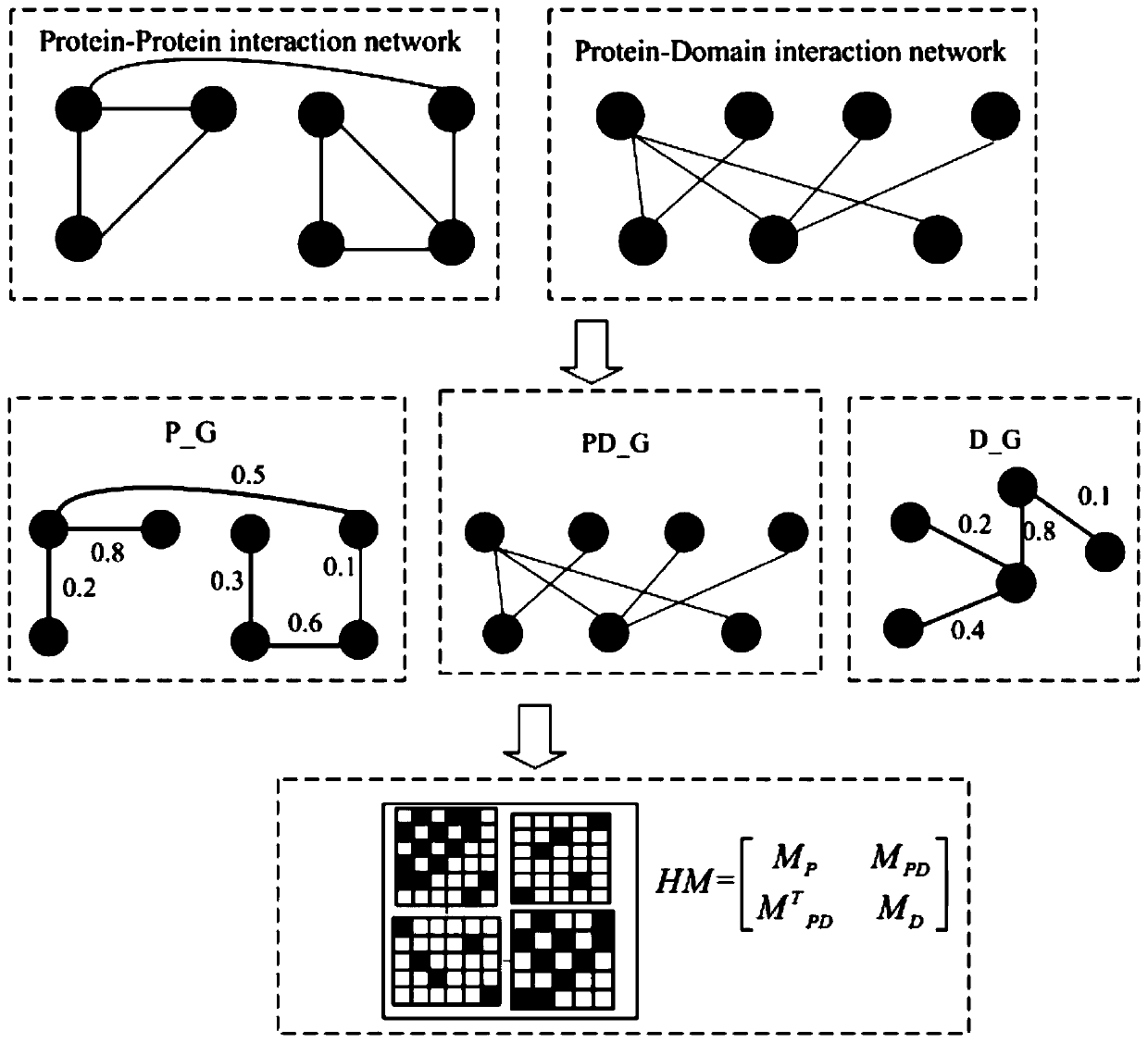
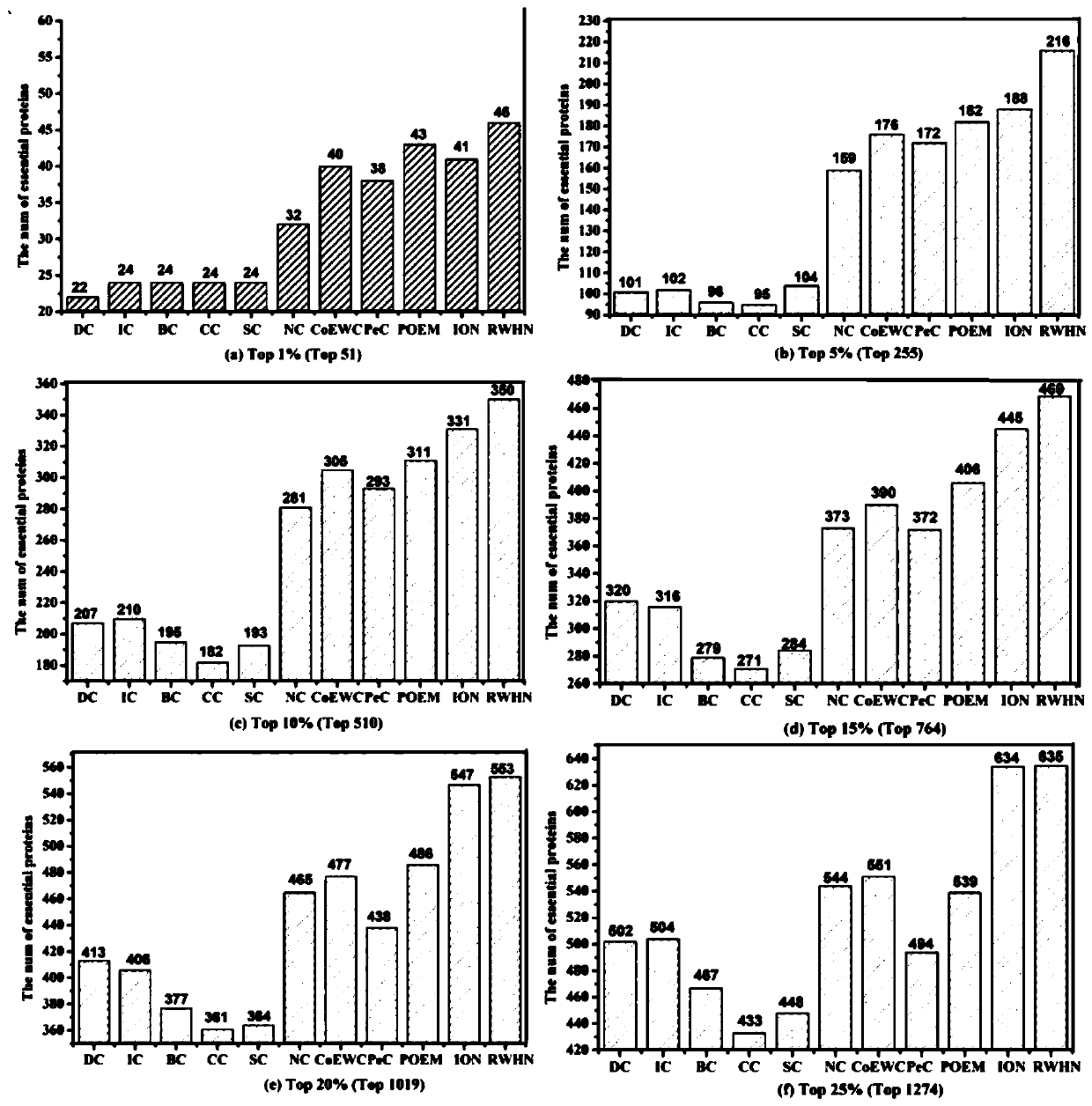
Key protein recognition method based on heterogeneous biological network fusion
ActiveCN109801674AImprove recognition accuracyBiostatisticsInstrumentsYeast ProteinsHeterogeneous network
The invention discloses a key protein recognition method based on heterogeneous biological network fusion. The key protein recognition method includes the steps: obtaining a yeast protein interactionnetwork topology, protein domain information, protein homology information, and protein subcellular localization information; respectively establishing a protein-protein interaction network P_G, a structural domain-structural domain interaction network D_G and a protein-structural domain network PD_G; fusing the three networks PD_G three networks, and establishing a heterogeneous network HG; establishing an initialized score vector h0 of the protein and structural domain; iteratively calculating the score vector h(i)t( / i) of the protein and structural domain based on a random walk model untila steady state is achieved; and sorting according to the score of the protein in the steady state, and identifying the key protein. The key protein recognition method based on heterogeneous biologicalnetwork fusion improves the fusion mode of multi-source biological data in the research of key protein recognition methods, and greatly improves the recognition accuracy of key proteins.
Owner:CHANGSHA UNIVERSITY
Method for identifying key proteins based on cuckoo search algorithm
ActiveCN108319812AImprove accuracyImprove reliabilityArtificial lifeProteomicsProtein-protein complexPredictive value
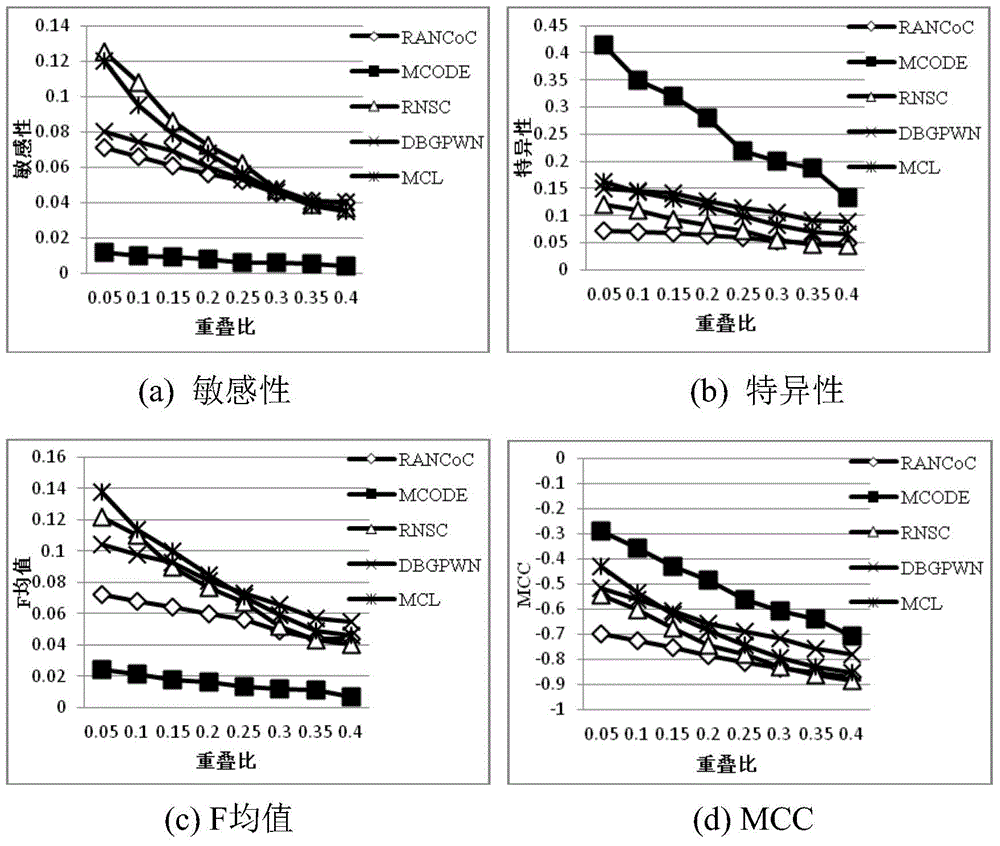
The invention discloses a method for identifying key proteins based on a cuckoo search algorithm, and belongs to the technical field of biological information. A protein interaction network is converted into an undirected graph, denoising processing is conducted on the protein interaction network to acquire gene expression data, subcellular localization information, standard protein complex information and GO annotation information corresponding to the protein, weighting processing is conducted on edges and nodes of the purified protein interaction network to generate a bird nest and cuckoos and update a bird nest adaptive value to generate the key proteins. The method can accurately identify the key proteins; simulation results show that the indexes such as sensitivity, specificity, F measure, a positive predictive value, a negative predictive value, correct rate and the like are superior; compared with other existing identification methods, the optimizing characteristics of the cuckoo search algorithm and topological characteristics of the protein interaction network are combined to identify the key proteins, and the recognition accuracy of the key proteins is improved.
Owner:SHAANXI NORMAL UNIV
Novel integrated pharmacology method and application thereof in treatment of breast cancer by astragalus membranaceus
InactiveCN110556166AEasy to identifyComprehensive detectionChemical property predictionMicrobiological testing/measurementNODALFishing
The invention provides a novel integrated pharmacology method. The method comprises the steps of differential expression gene identification and analysis, traditional Chinese medicine component database construction and ADME screening, identification of related compound targets, introduction and annotation of the compound targets, protein interaction network construction and analysis and moleculardynamics simulation and target fishing. The invention also provides an application of the novel integrated pharmacology method in the treatment of breast cancer by astragalus membranaceus. Multi-target effective components and targets are comprehensively modeled on the biological path and interaction network level, and the complex and multi-level interaction network is analyzed, so the interaction between the medicine and the specific node or module can be discovered, and a new target can be discovered; the detected target is more comprehensive and high in accuracy, and the multi-target characteristic of the traditional Chinese medicine intervention disease is effectively associated with the complex disease system.
Owner:刘存
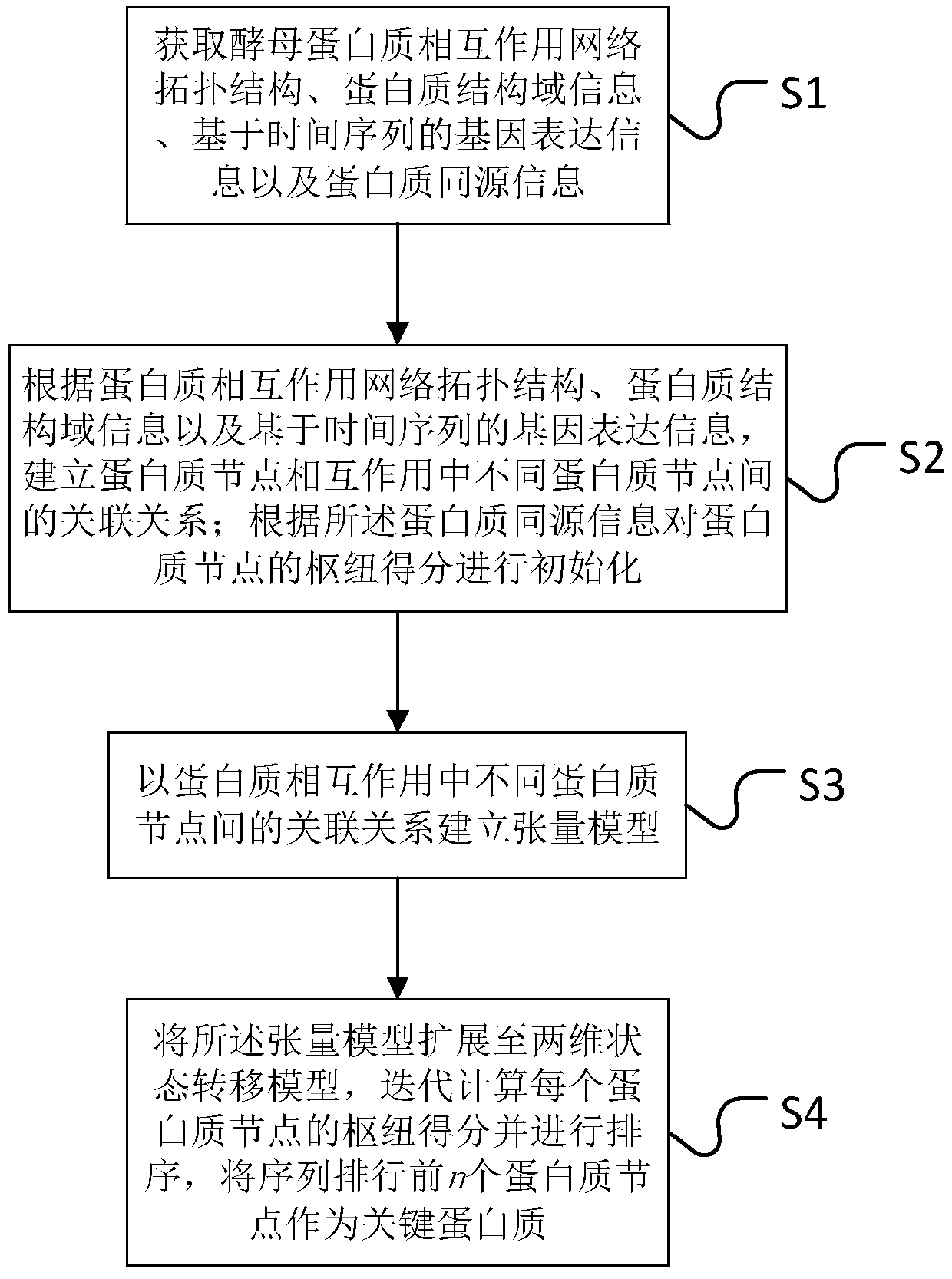
Key protein recognition method based on tensor random walking
The invention discloses a key protein recognition method based on tensor random walking. The method comprises the steps that a protein mutual impacting network topological structure, protein structural domain information, genetic expression information based on time sequence and protein homologous information are obtained; a correlation relationship of different protein nodes in the mutual impacting of protein nodes is constructed according to the above information; hub scores of protein nodes are initialized according to the protein homologous information; a tensor model is established according to the correlation relationship of different protein nodes in the protein mutual impacting; the hub scores of each protein nodes which are obtained by iterative computation based on the tensor model are sorted, and the top n protein nodes are taken as key proteins. The method has the advantages that the method is simple and effective, and compared with other methods, tests on multiple data sets show that the method has better prediction performance on key protein recognition.
Owner:CHANGSHA UNIVERSITY
Method for identifying key protein based on genetic algorithm in PPI network
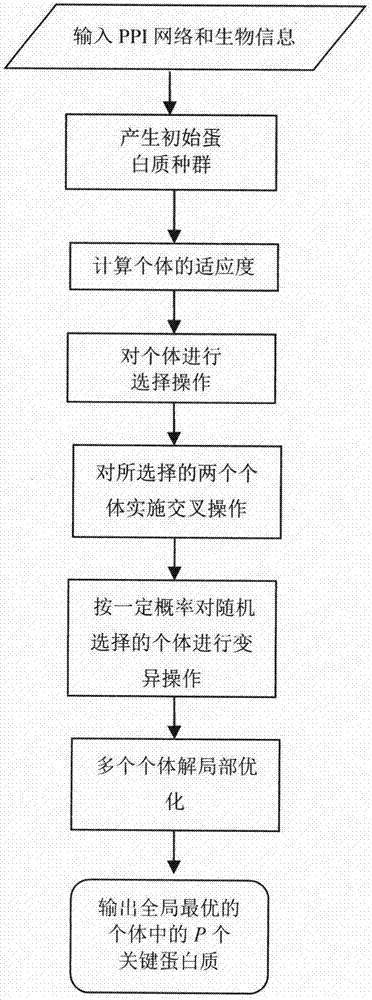
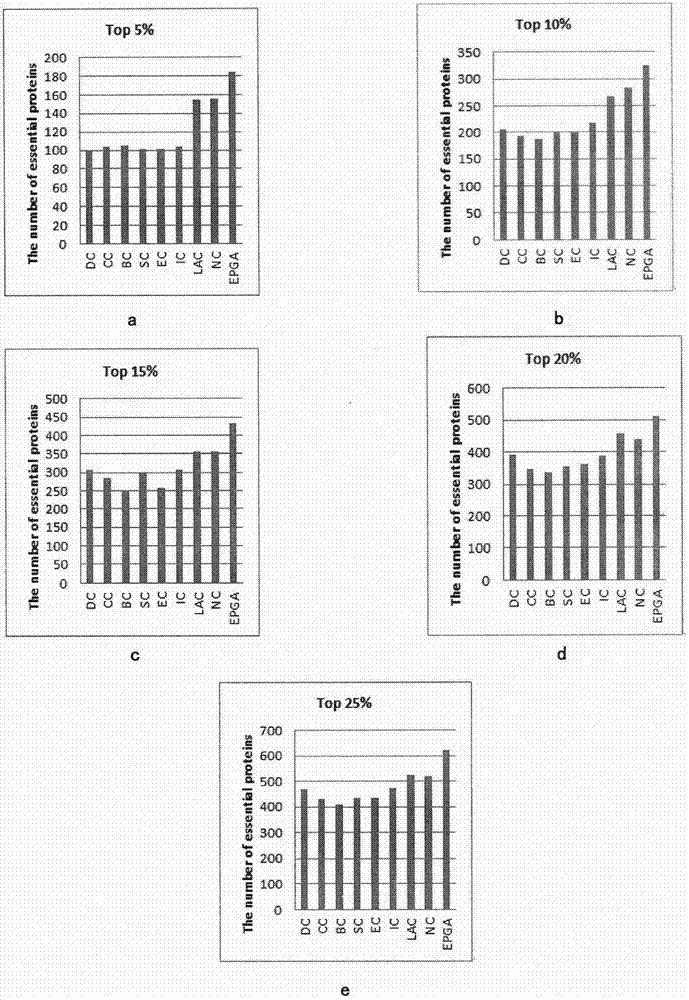
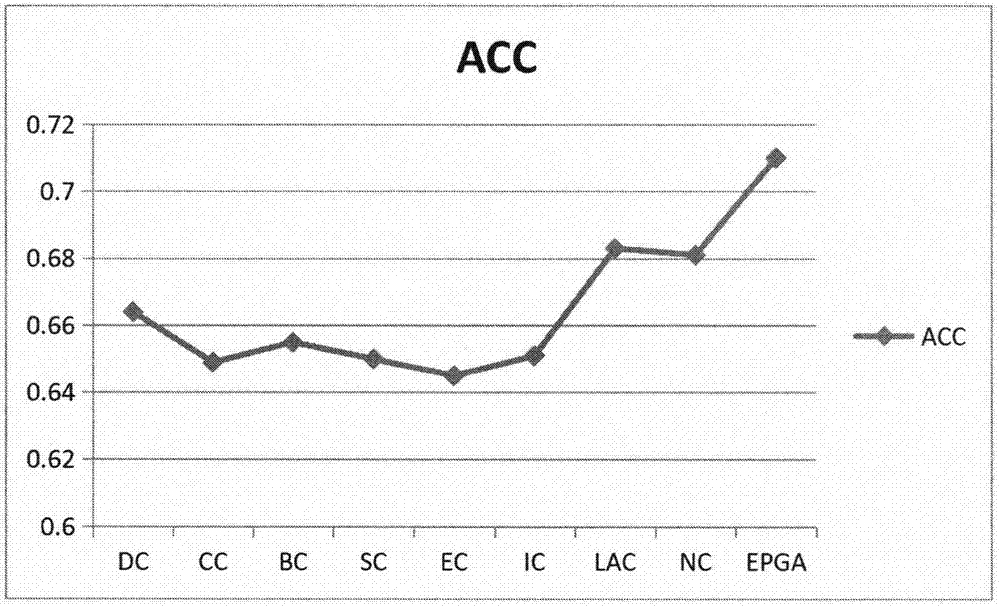
ActiveCN107092812APrediction is accurateImprove reliabilityProteomicsGenomicsAlgorithmMethod selection
The invention relates to an algorithm for identifying a key protein based on a genetic algorithm in a PPI network. The algorithm comprises the steps of generating an initial population in a protein interaction network; calculating the fitness of individuals; performing selection operation by a roulette wheel method; performing crossing operation and mutation operation among the randomly selected individuals; and performing local optimization on multiple individual solutions. According to the algorithm, the defects of existing methods are overcome; indexes are optimized and bio-information is fused, so that the reliability is higher and lots of unnecessary calculations are reduced; and the predicted key protein can be locally optimized, so that the efficiency of key protein identification is improved, and the application range and practicality of the technology in the field of the bio-information are expanded and improved.
Owner:YANGZHOU UNIV
Method for analyzing anti-glandular cystitis action mechanism of pachymaran based on network pharmacology
PendingCN112201365AReduce R&D costsImprove screening efficiencyDrug referencesSystems biologyPharmacometricsNetwork pharmacology
The invention belongs to the field of biomedicine, discloses a method for analyzing an anti-glandular cystitis action mechanism of pachymaran based on network pharmacology, and reports detailed targets and specific pharmacological mechanisms of pachymaran for preventing and treating glandular cystitis for the first time. The method comprises the following main points: obtaining a pachymaran pharmacological target and a glandular cystitis pathogen target through online database analysis of TCMSP, DisGeNET and the like; taking an intersection of the pachymaran and the glandular cystitis target to obtain a drug disease intersection target gene target, and constructing a related protein interaction network to screen a core target; further performing gene ontology GO biological process and KEGGpathway enrichment analysis on the core target by utilizing the R language related packet; and finally, constructing a drug target gene ontology functional pathway disease visualization diagram for deep analysis of a treatment mechanism. The method provided by the invention provides a new idea for explaining a research mechanism of pachymaran for exerting glandular cystitis resistance, and also provides an early-stage research basis for clinical application of pachymaran in glandular cystitis.
Owner:NANNING SECOND PEOPLES HOSPITAL
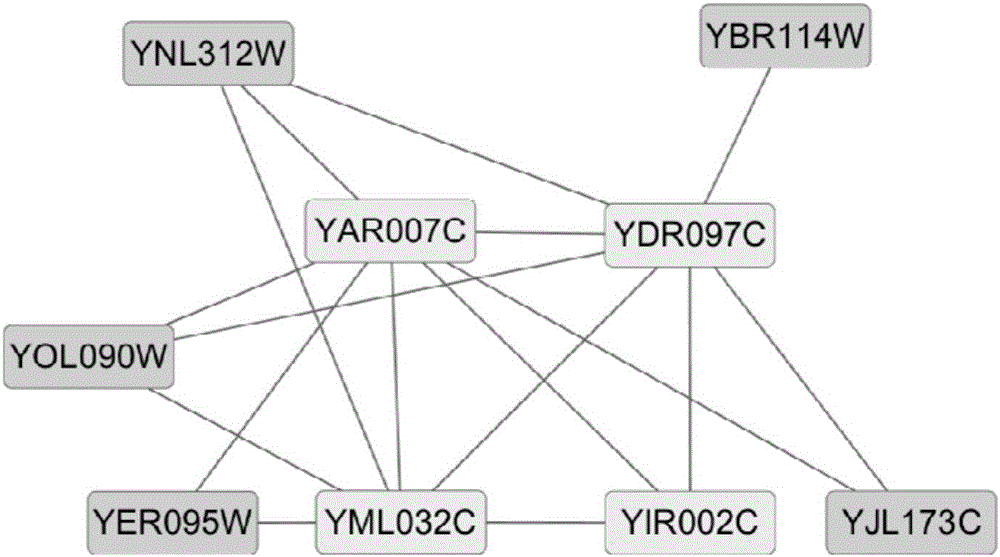
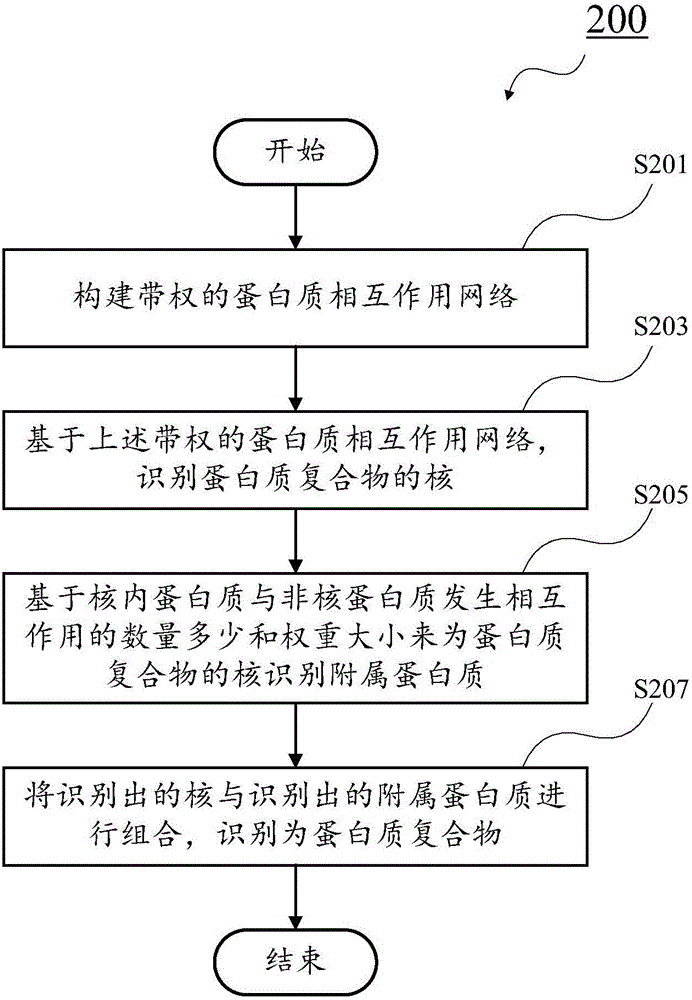
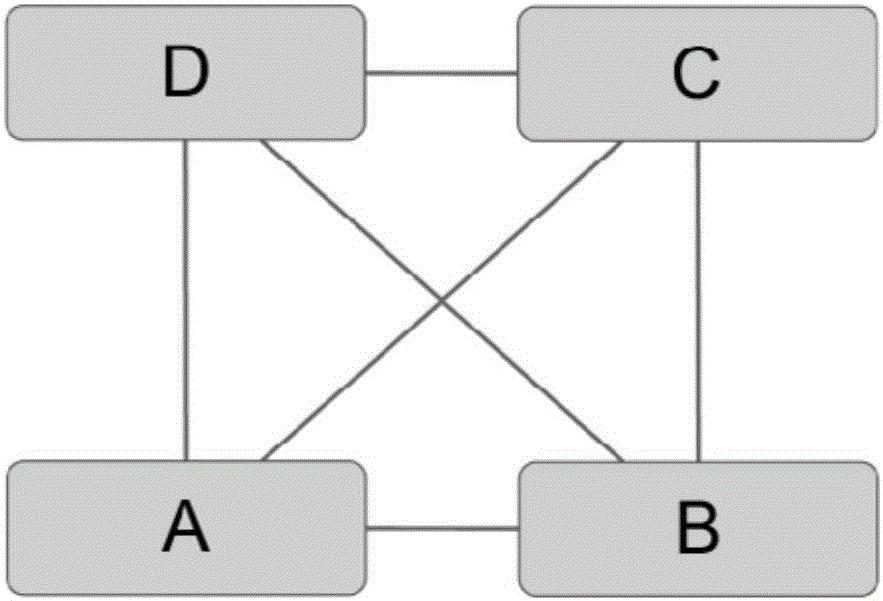
Recognition method of protein complexes
The invention discloses a recognition method of protein complexes. The method comprises following steps: constructing a weighted protein-protein interaction network, wherein weight value represents similarity between gene representing modes corresponding to proteins with codes interacting each other; recognizing nucleuses of protein complexes based on the weighted protein-protein interaction network; recognizing accessory proteins for the nucleuses of protein complexes based on the number of nuclear proteins interacting with non-nuclear proteins and weight; and combining recognized nucleuses and recognized accessory proteins to recognize the protein complexes. The method does not take internal structures of protein complexes into consideration and utilizes the feature of code interaction with high protein gene expression. Therefore, the method is capable of recognizing protein complexes with overlapping structures and the recognized protein complexes reflect actual internal structures of protein complexes.
Owner:HENAN UNIV OF URBAN CONSTR
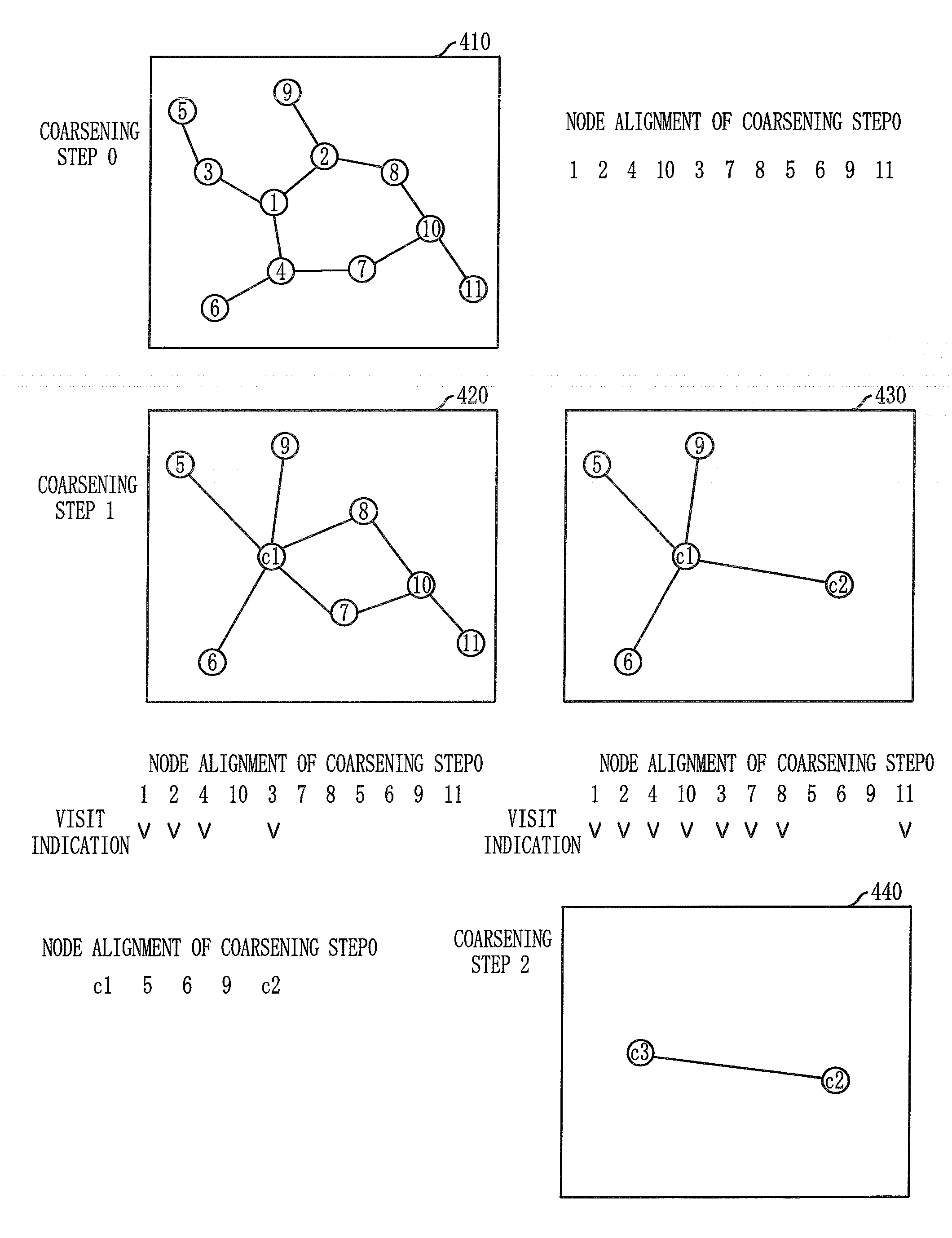
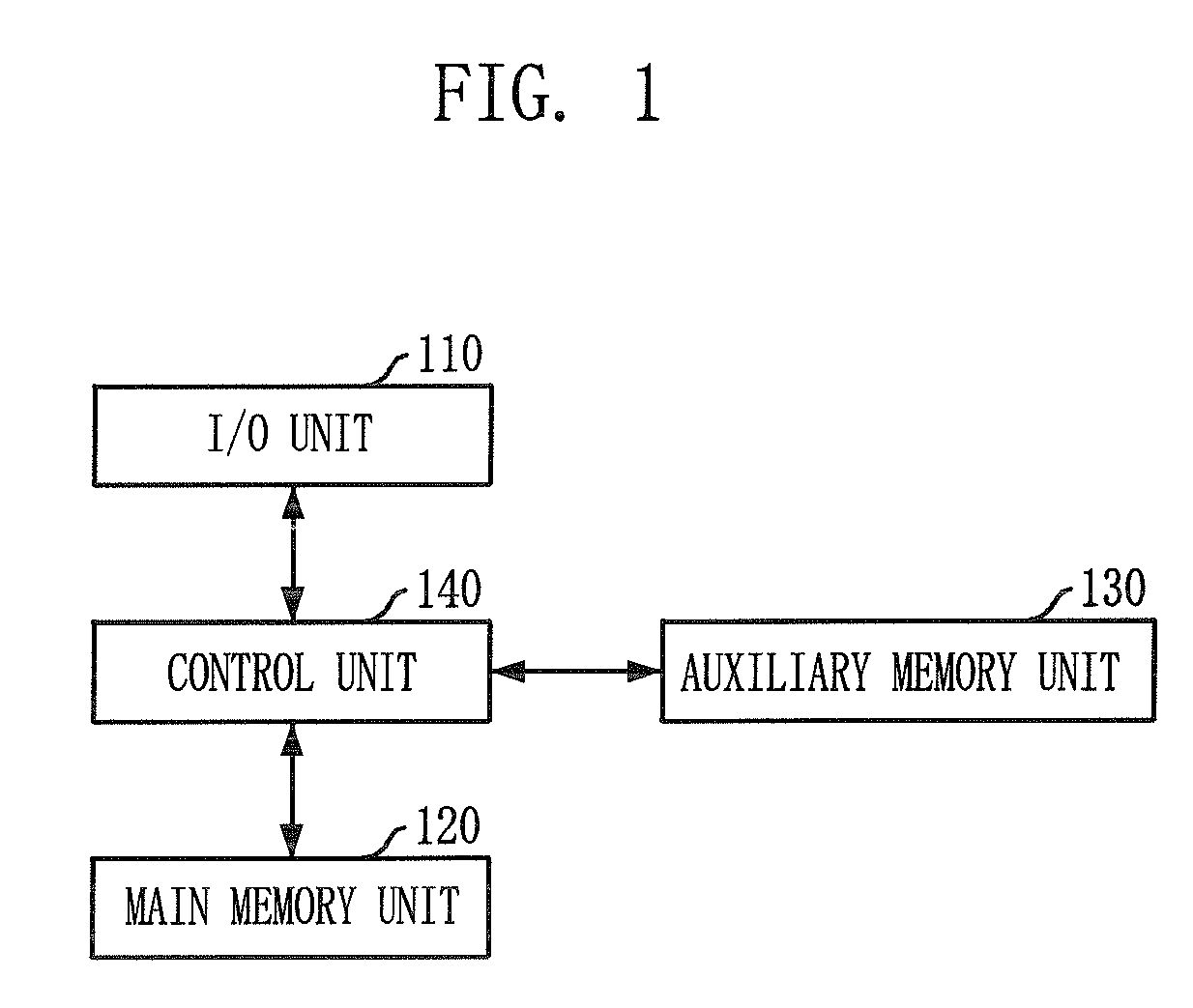
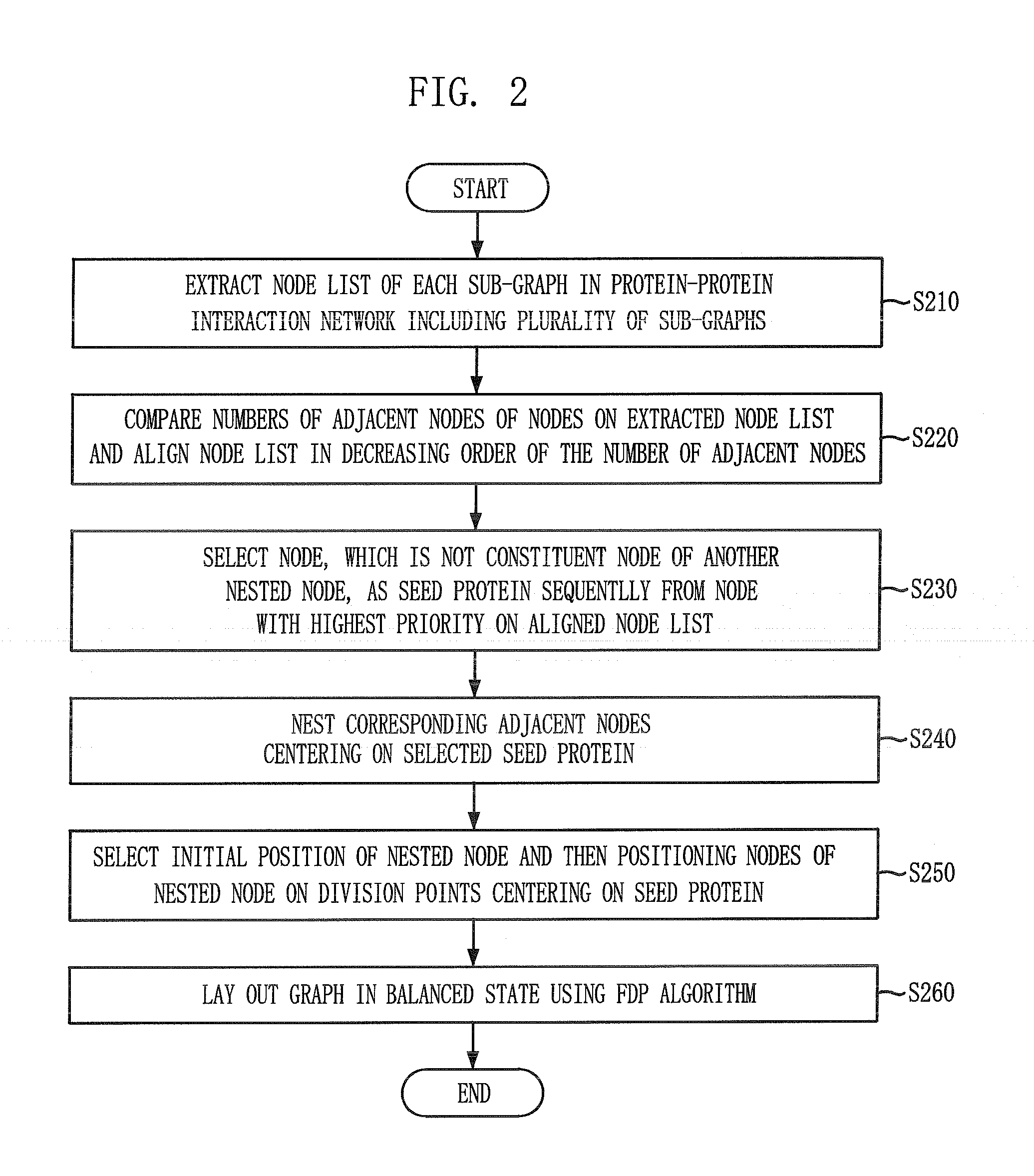
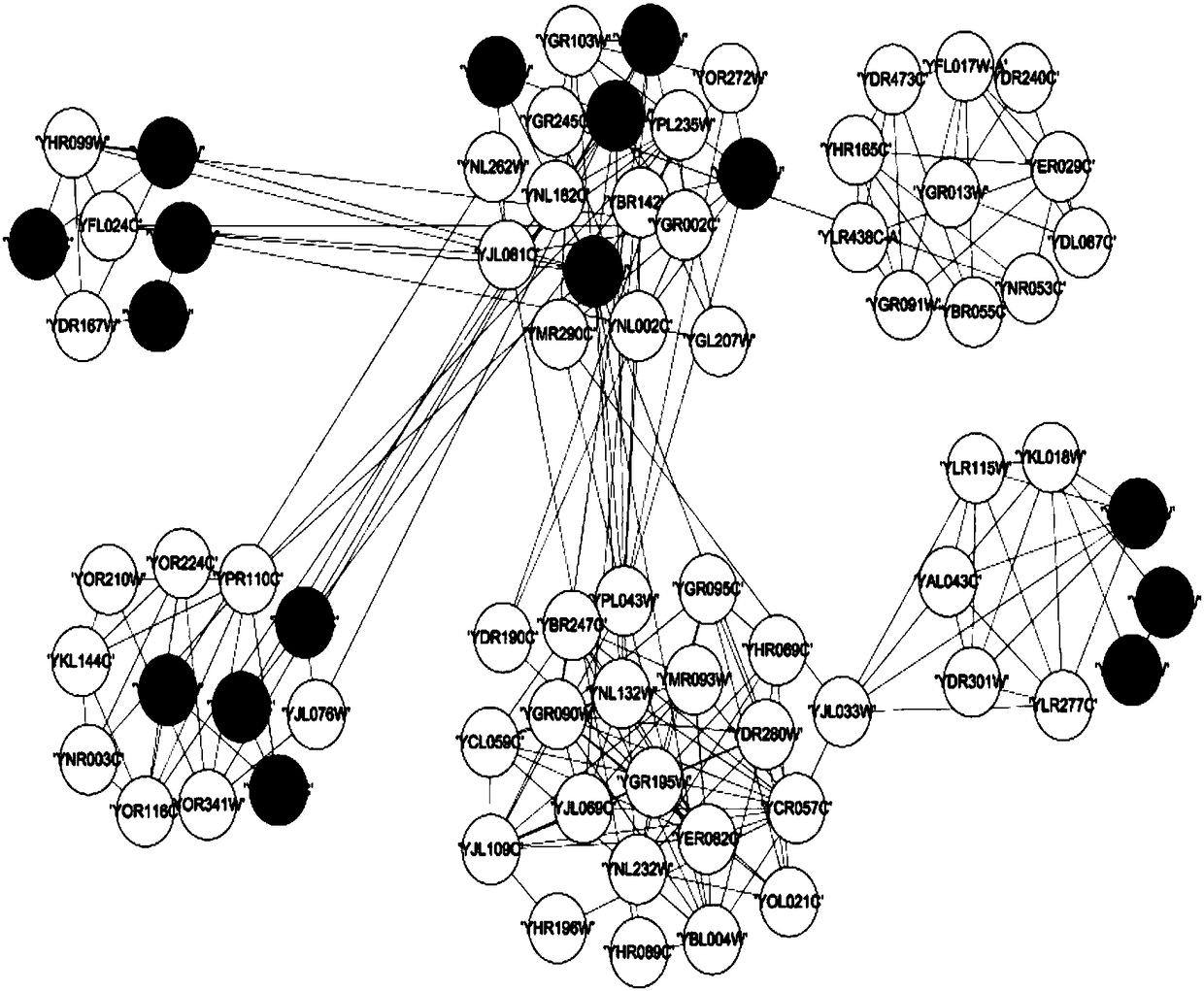
Layout method for protein-protein interaction networks based on seed protein
InactiveUS20080133197A1Increase speedLayout balanceData visualisationAnalogue computers for chemical processesNODALAlgorithm
Provided is a layout method for protein-protein interaction networks based on a seed protein, which is for performing multiple stages of nesting centered on a node having a high degree of physical relationship, and performing multiple stages of extension and force directed placement (FDP) with respect to a final nest graph. The layout method includes the steps of: a) extracting a node list of each sub-graph constituting a protein-protein interaction network, and aligning the node list according to adjacency of nodes; b) selecting a seed protein from the aligned node list according to node priority and nest relationship with another node; c) nesting adjacent nodes centered on the selected seed protein to generate a nested node; and d) selecting an initial position of the generated nested node, placing the nodes of the nested nodes on respective division points, centered on the seed protein, and then performing layout.
Owner:ELECTRONICS & TELECOMM RES INST
Method for identifying key protein through fruit fly optimization algorithm
ActiveCN108229643AEmbodies biological significanceImprove recognition efficiencyArtificial lifeUndirected graphProtein protein interaction network
The invention discloses a method for identifying key protein through a fruit fly optimization algorithm. The method includes the steps that a protein-protein interaction network is converted into an undirected graph, a dynamic protein-protein interaction network is constructed, the edges and nodes of the dynamic protein-protein interaction network are preprocessed, the position of a fruit fly group is randomly initialized, the food random direction and distance are searched for with the sense of smell, the taste concentration judgment value of each fruit fly individual is calculated, the odorconcentration value of each fruit fly individual is calculated, the highest odor concentration value in the current group is worked out, the fruit flies fly to the food with the sense of sight, and key protein is generated. By means of the method, the key protein can be accurately identified; the simulation experiment result shows that the method has good performance indexes including sensitivity,specificity, positive predictive value, negative predictive value, accuracy rate and recall rate harmonic value, precise value and the like; compared with other methods for identifying key protein, the method adopting the fruit fly optimization algorithm for identifying the key protein has certain advantages.
Owner:SHAANXI NORMAL UNIV
Method for identifying protein complex based on BSO (Brain Storm Optimization)
InactiveCN105590039ASignificant bioaccumulationStrong global optimization abilitySpecial data processing applicationsNODALProtein protein interaction network
The invention provides a method for identifying a protein complex based on BSO (Brain Storm Optimization); the method comprises the following steps: by utilizing strong global optimization searching capability of a BSO algorithm, regarding a protein-protein interaction network as a full network connected graph, combining gene ontology annotation function information of the protein with a topological structure of the protein-protein interaction network to define a distance among protein nodes, and carrying out preliminary clustering according to an improved k-means algorithm; then, according to four optimization searching principles of the BSO algorithm, generating a new fitness value, respectively carrying out module internal and module external optimization searching operations on a protein module which is formed preliminarily, iterating in a circulative manner and searching a most optimal global solution; and at last, carrying out post processing process. The method disclosed by the invention can keep the diversity of a group in the optimization searching process, thereby avoiding getting into local optimization; the global optimization module division is obtained, and the protein complex with remarkable biological enrichment is obtained.
Owner:HUAZHONG NORMAL UNIV
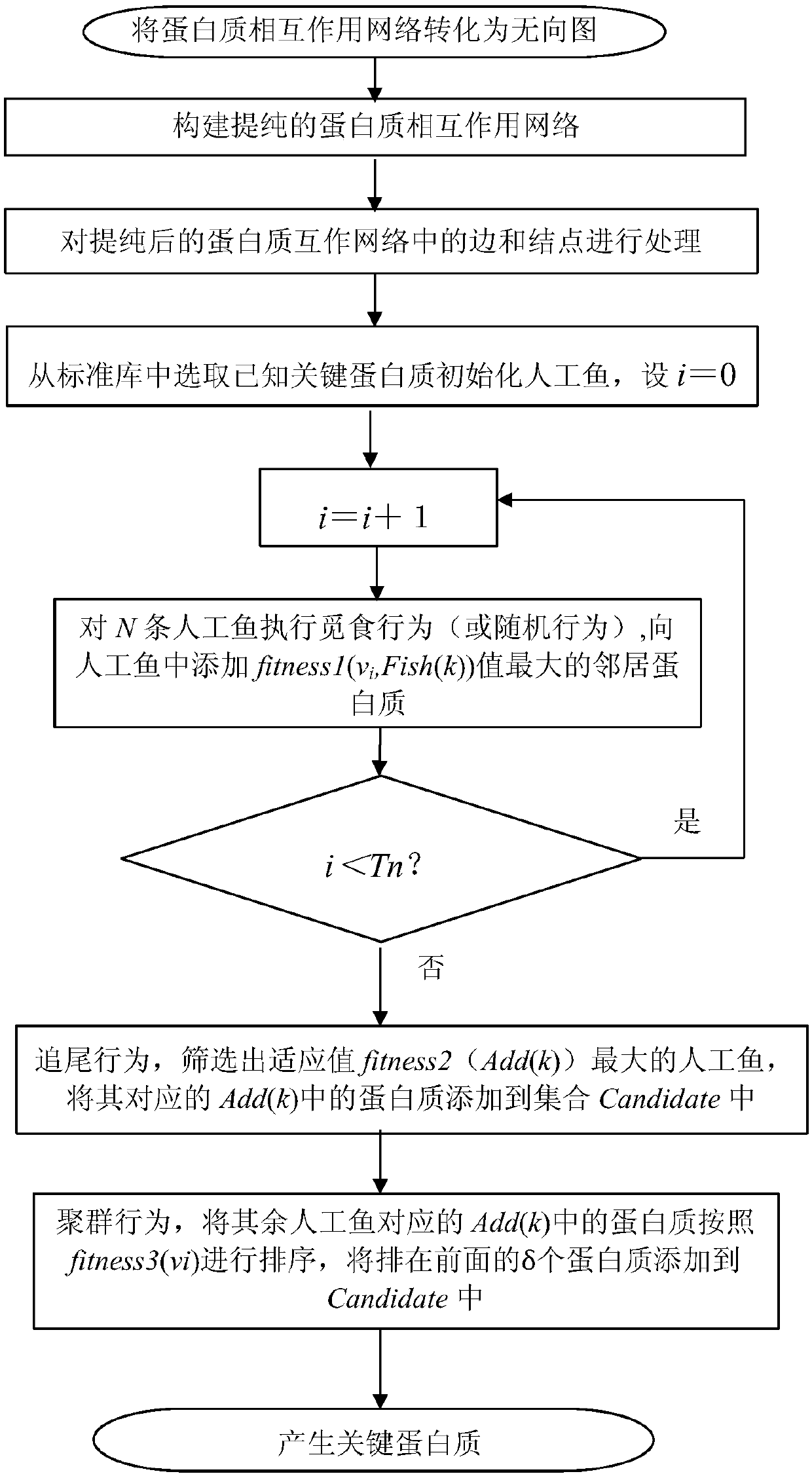
Method for identifying key proteins with AFSO (artificial fish school optimization) algorithm
ActiveCN107784196AFully consider the topological characteristicsAccurate identificationArtificial lifeProteomicsUndirected graphProtein protein interaction network
The invention discloses a method for identifying key proteins with an AFSO (artificial fish school optimization) algorithm. The method comprises steps as follows: a protein-protein interaction networkis converted into an undirected graph, a purified protein-protein interaction network is constructed, RNA gene expression values corresponding to proteins, GO comment information and degrees of proteins in known compounds are obtained, edges and nodes of the purified protein-protein interaction network are treated, known key proteins are selected as initial artificial fishes, the artificial fishes execute foraging behavior, random behavior, following behavior and swarm behavior, and the key proteins are produced. According to the method, the key proteins can be identified accurately; a simulation experiment result indicates that performance of indexes such as sensitiveness, specificity, a positive predictive value, a negative predictive value and the like is better; compared with other methods for identifying the key proteins, the method has the advantages that optimizing characteristics of artificial fish schools are combined with topological characteristics of the protein-protein interaction network to realize the key protein identification process, and the accuracy rate of the key protein identification is increased.
Owner:SHAANXI NORMAL UNIV
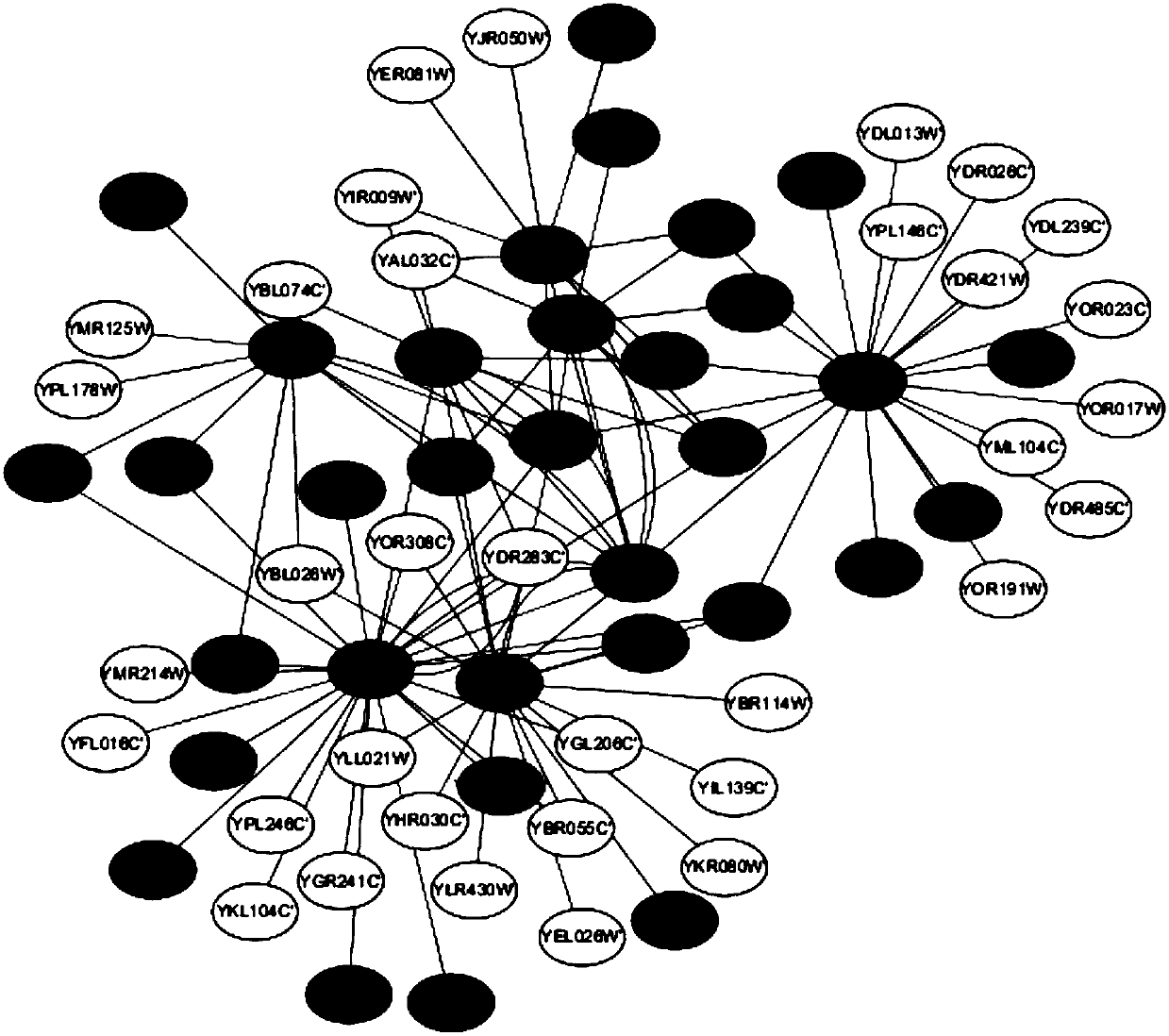
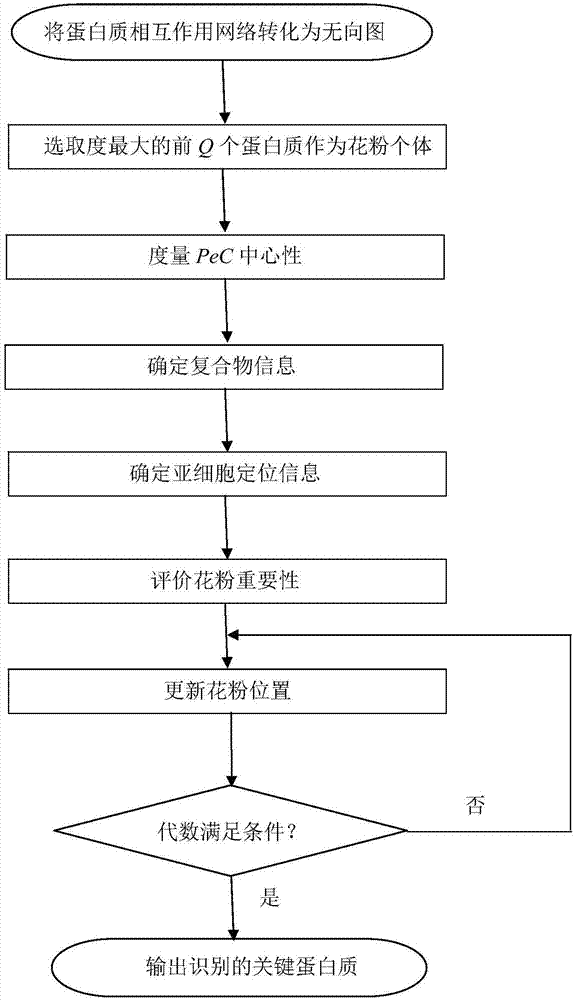
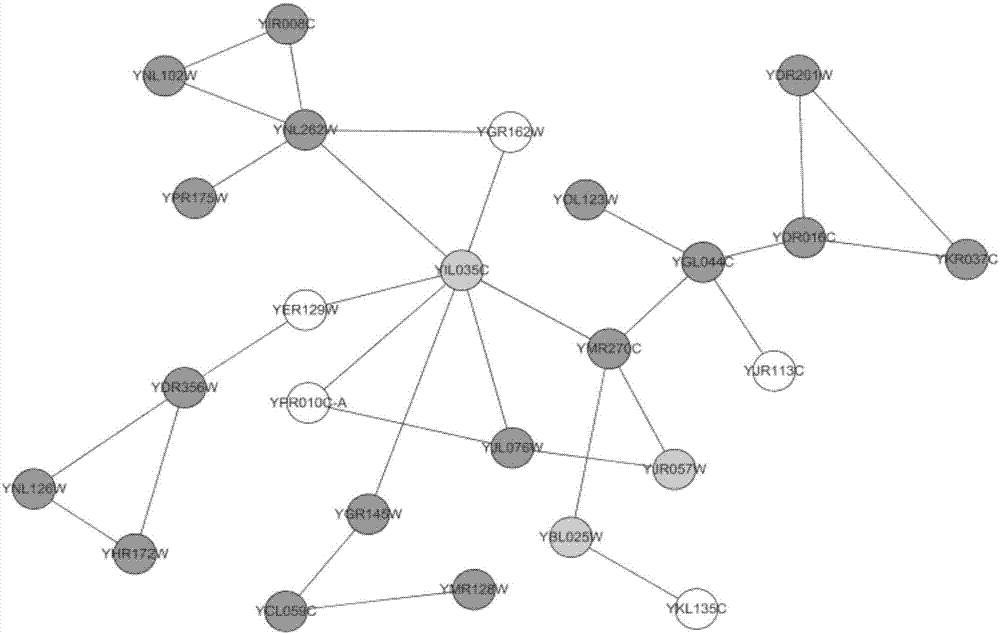
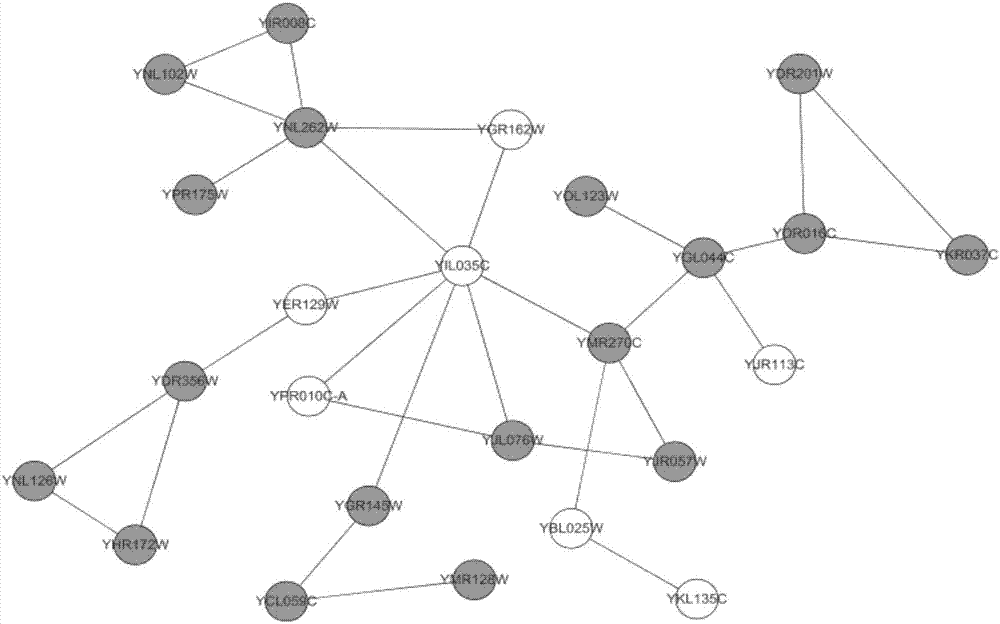
Method for identifying key protein by adopting improved flower pollination algorithm
ActiveCN107885971AImprove accuracyUnderstanding Growth Regulatory ProcessesSystems biologySpecial data processing applicationsUndirected graphPollen
The invention discloses a method for identifying a key protein by adopting an improved flower pollination algorithm. A protein-protein interaction network is converted into an undirected graph, previous Q proteins having the largest choice degree serve as pollen individuals, the PeC centrality is measured, compound information is determined, subcellular localization information is determined, pollen significance is evaluated, a pollen position is updated, and an identified key protein is output. The topological attributes of the protein network are considered and the biological characteristicsof the protein network are fused during evaluation of the pollen significance, and the key protein can be accurately identified. A simulation experiment result shows that the index properties such asaccuracy, specificity and sensitivity are more excellent. Compared with other key protein identifying methods, the key protein identifying process is achieved by combining with the topological attributes and biological characteristics of the protein network, and the key protein identifying accuracy is improved.
Owner:SHAANXI NORMAL UNIV
Method of adopting firework algorithm to recognize protein complex
InactiveCN106228036AImprove accuracyEfficient identificationBiostatisticsSpecial data processing applicationsAccessory structureFireworks
The invention provides a method of adopting a firework algorithm to recognize a protein complex. The method includes following steps: converting a protein interaction network into an undirected graph; pre-treating sides and nodes of the protein interaction network; building a dynamic protein interaction network; setting parameters; initializing positions of fireworks; simulating firework explosion to generate spark; selecting part of good points from the spark to serve as fireworks, wherein all fireworks form a class; filtering undesirable classes; outputting a finally-acquired class. The method takes dynamic nature of a protein network and locality and globality of a core-accessory structure inside the protein complex and the protein interaction network into consideration, and the protein complex can be recognized accurately. Simulation experiment results show that performance of indexes like accuracy and recall ratio is excellent. Compared with other clustering methods, the method has the advantages that characteristics of the protein interaction network and the protein complex are combined to recognize the protein complex, and recognition accuracy of the protein complex is improved.
Owner:SHAANXI NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com










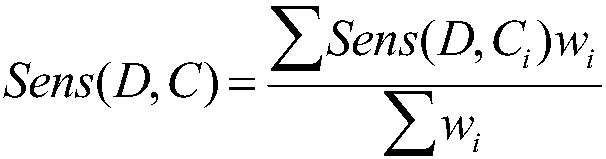
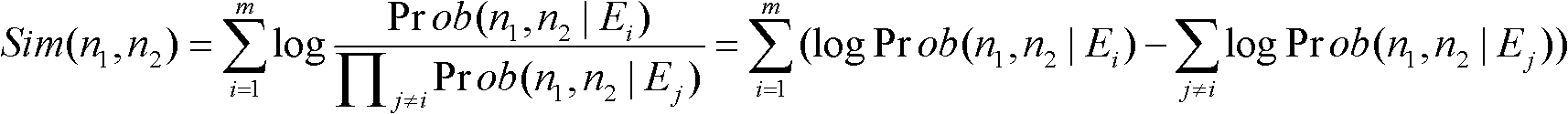



![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE001.png)
![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE002.png)
![Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks Method for predicting miRNA [micro-RNA (ribonucleic acid)] target proteins of miRNA regulation protein interaction networks](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/2ceb1e3a-4328-4313-b6c3-95d3a50590fe/DEST_PATH_IMAGE003.png)