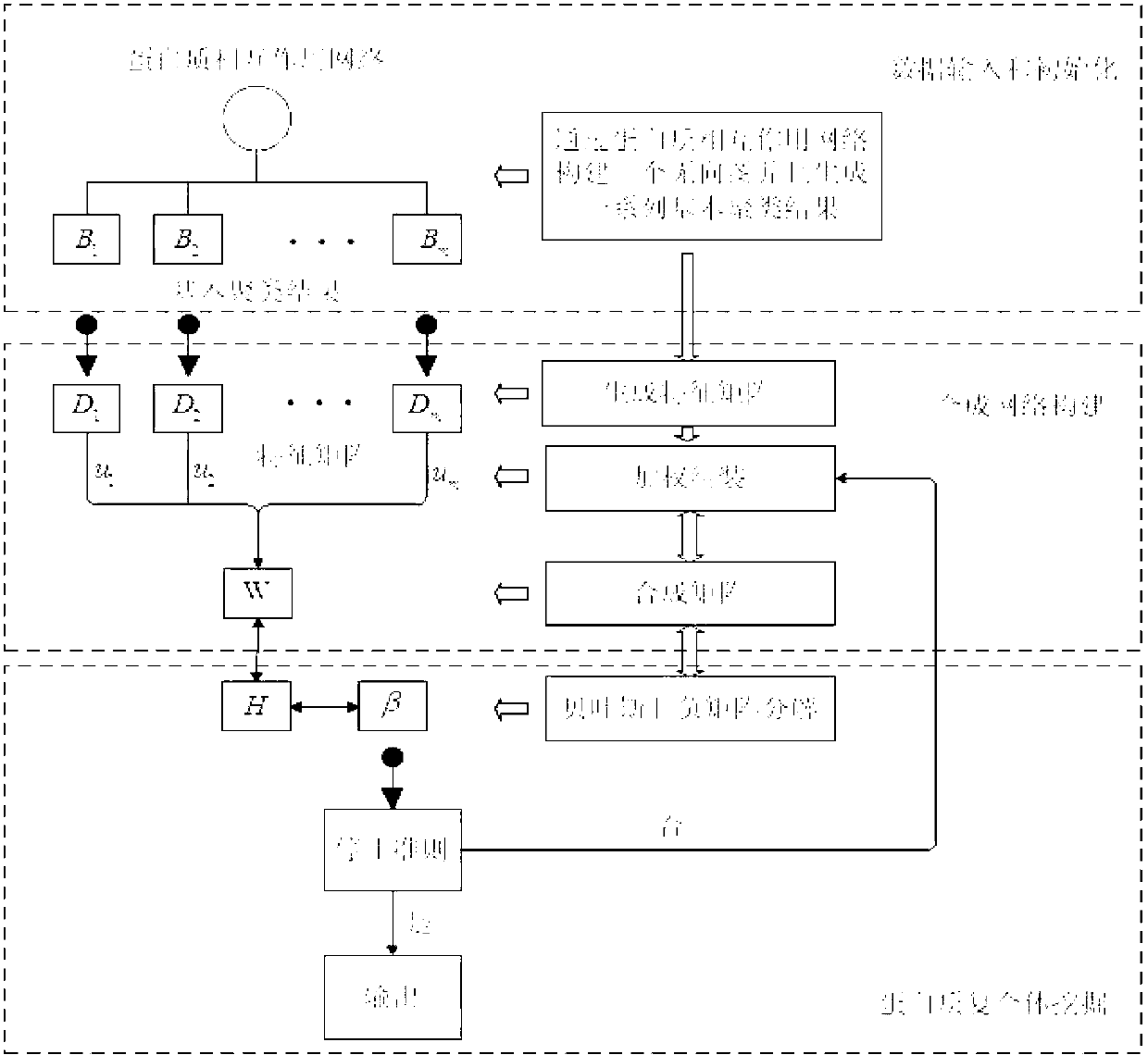
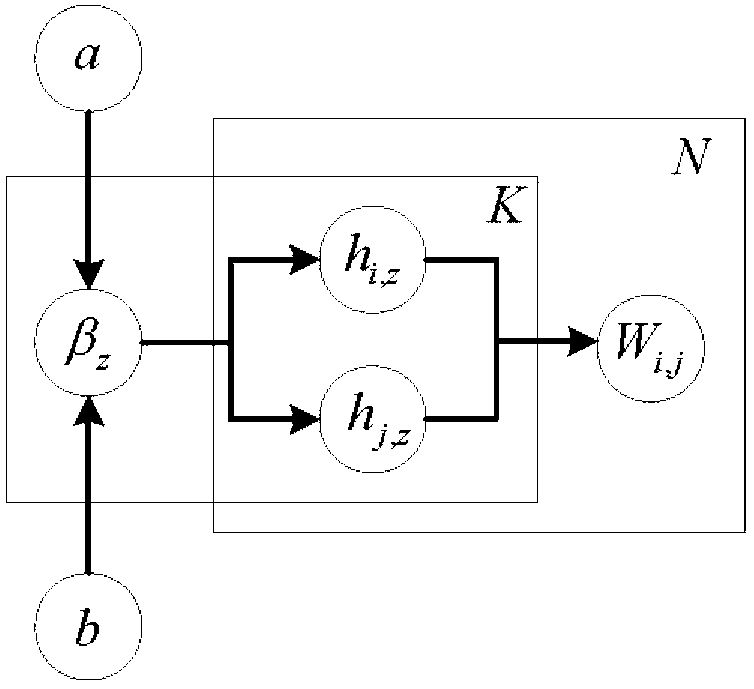
Weight assembly clustering method for excavating protein complex
A clustering method and protein technology, applied in the field of systems biology, to reduce sensitivity and dependence and enhance interpretability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
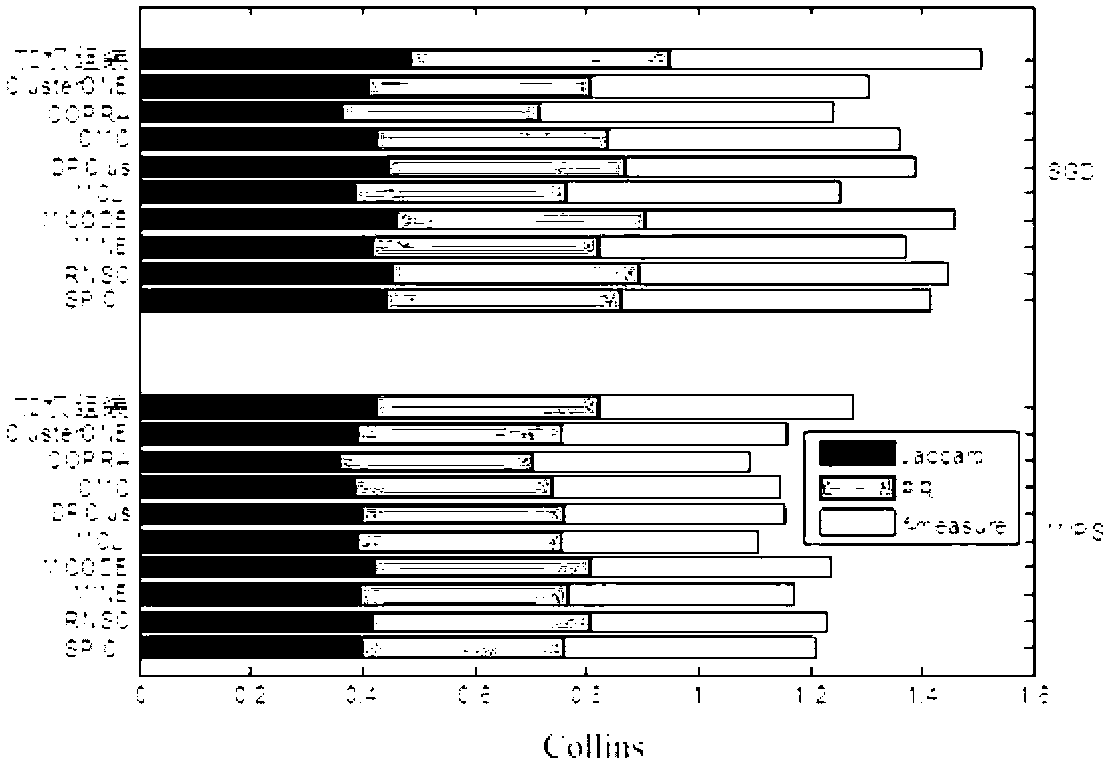
[0136] Nine classic protein complex mining algorithms (ClusterONE, CMC, COPRA, DPClus, MCL, MCODE, MINE, RNSC, SPICi) were selected to work on three yeast protein interaction network databases (Collins, Gavin and BioGRID). Two reference protein complex databases (MIPS and SGD) and three evaluation criteria (f-measure, Jaccard and PR) were used to verify the accuracy of the results of different algorithms. The statistical characteristics of the three protein interaction network databases and two reference databases corresponding to these three networks are shown in Table 2 and Table 3. Among the three evaluation criteria, f-measure measures the similarity between the predicted complex and the complex in the reference library from the protein complex level. Jaccard and PR measured the match between the predicted complex and the complex in the reference library at the complex-protein level.
[0137] Before describing these evaluation criteria, we give some notation explanations....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com