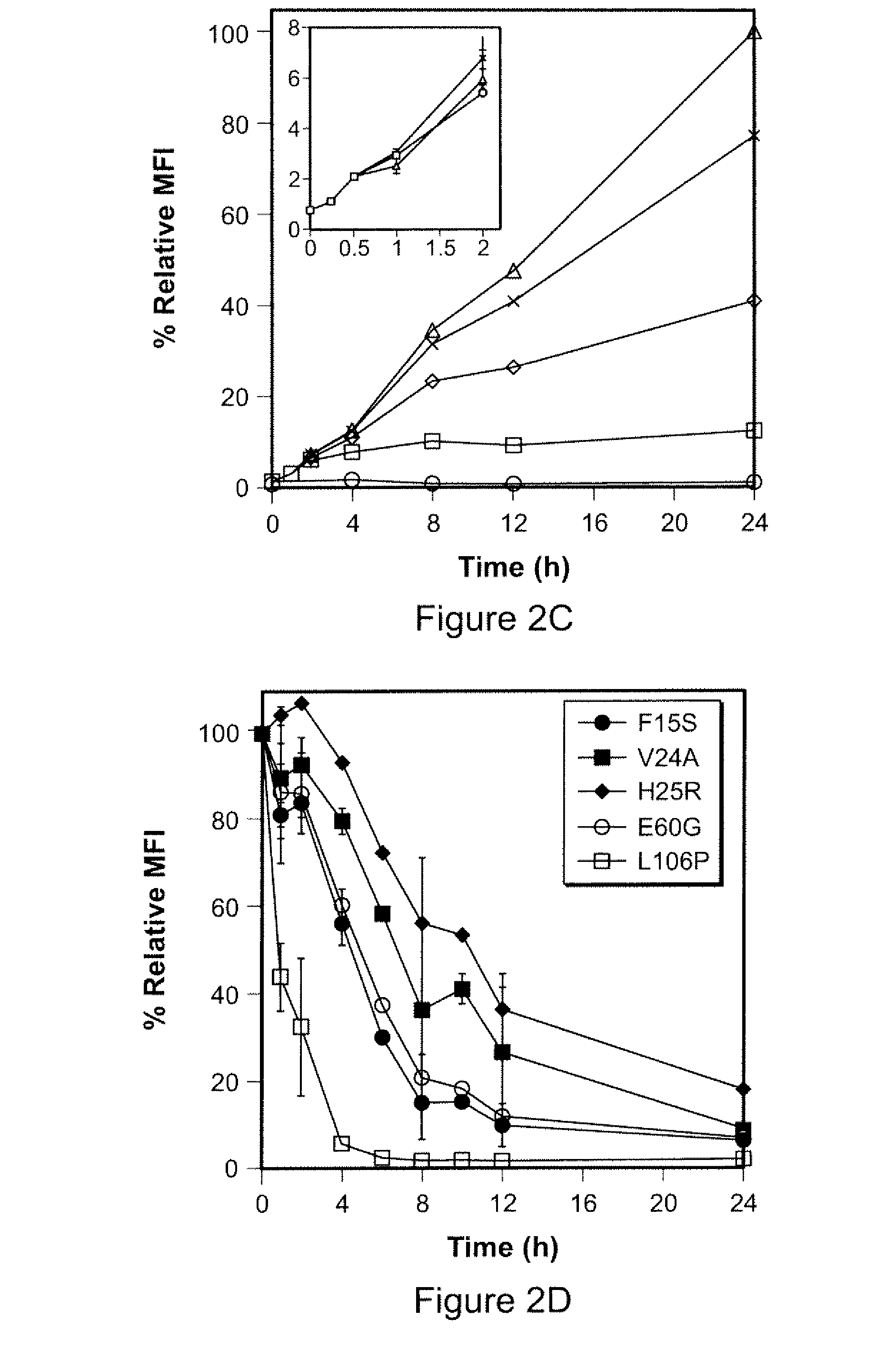
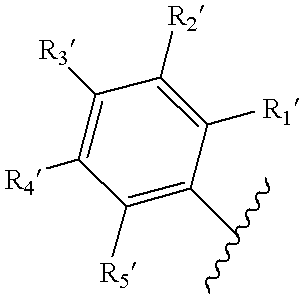
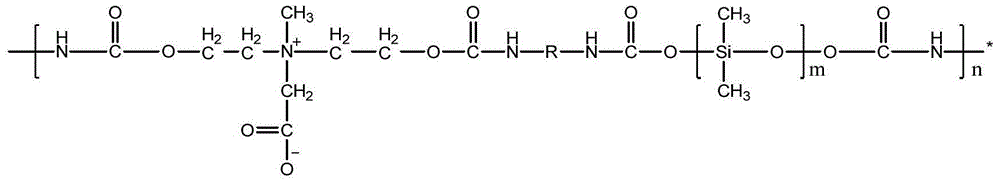
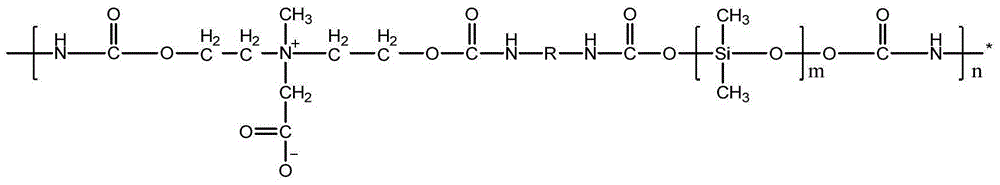
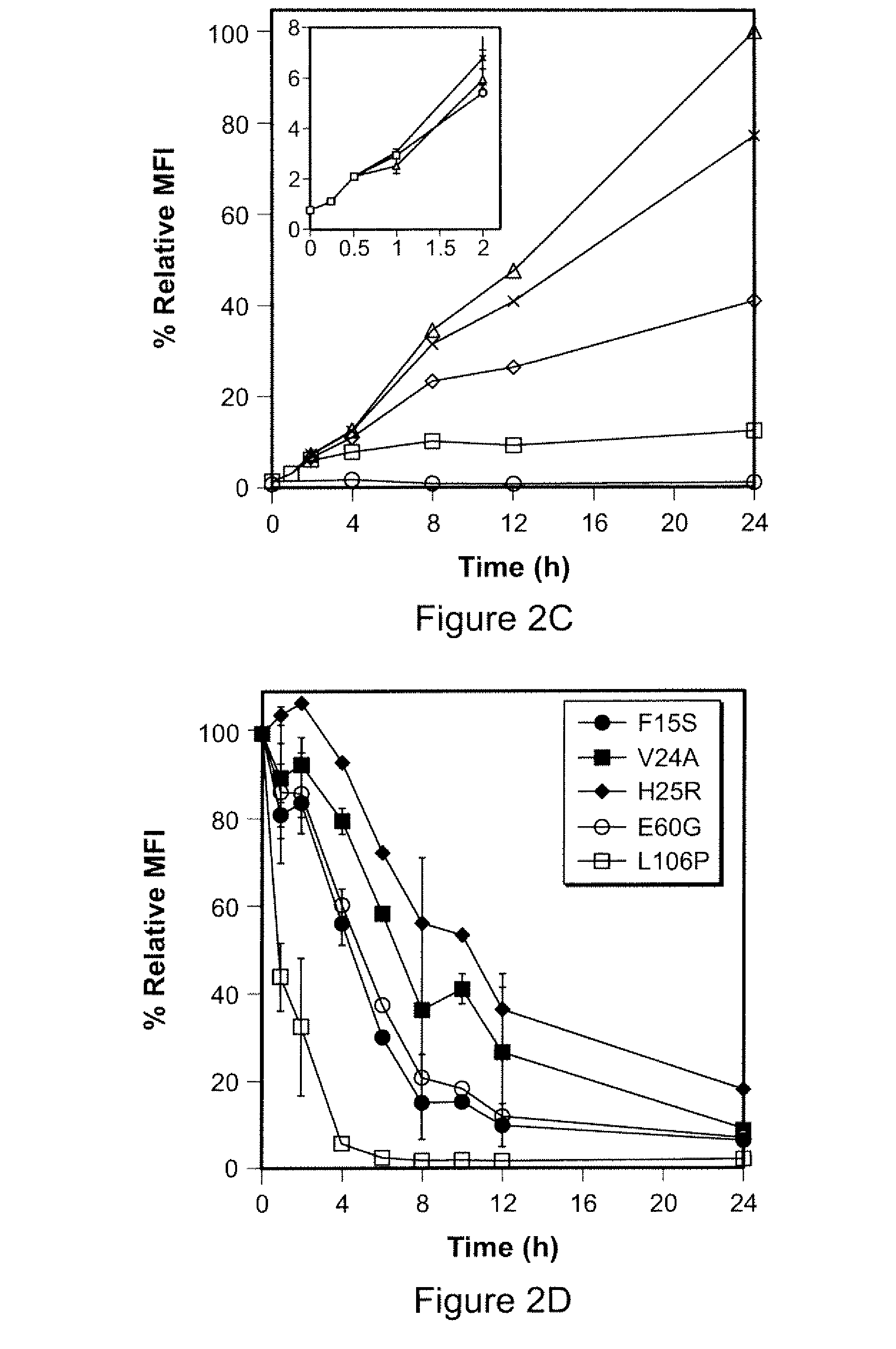
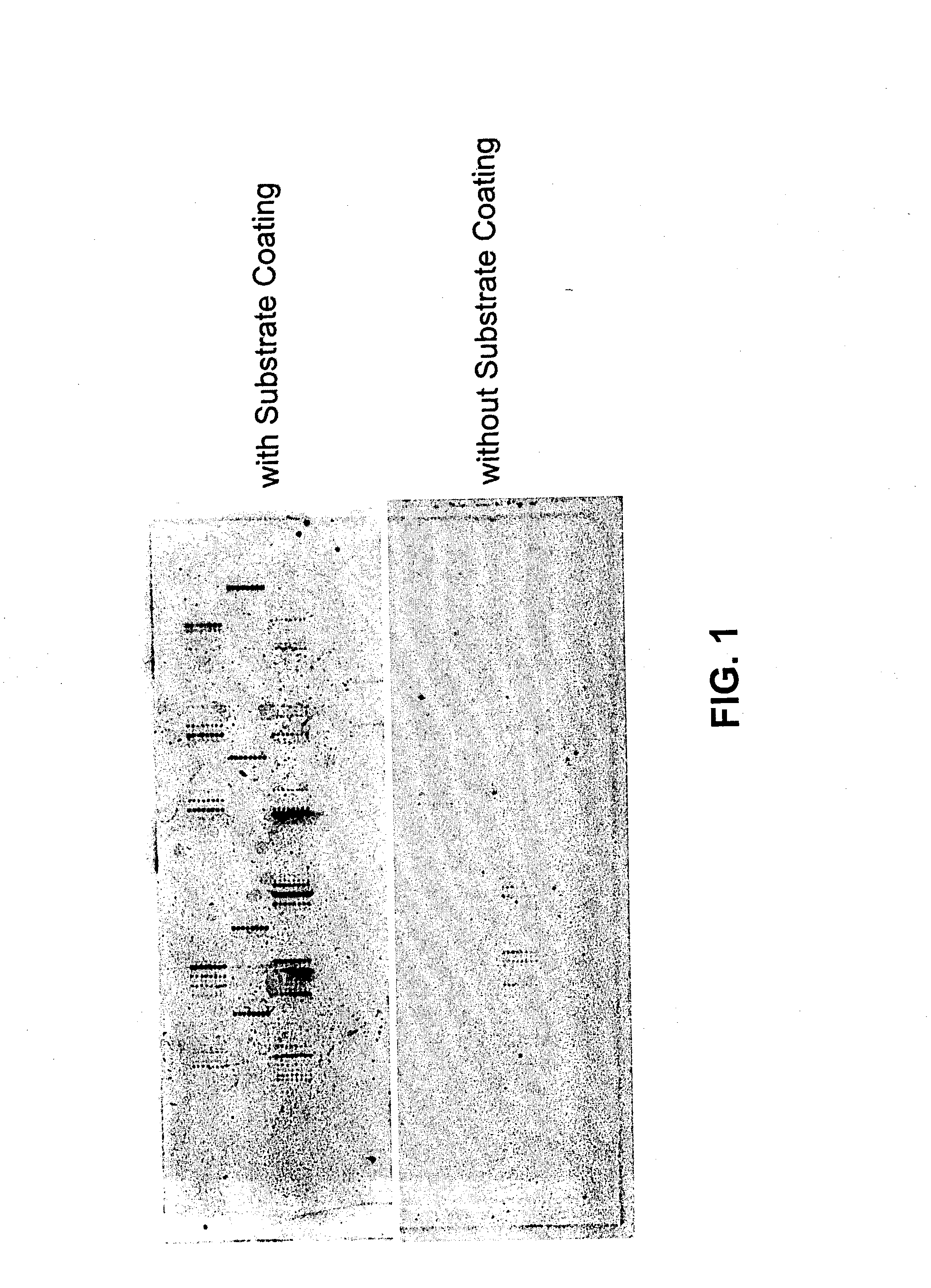
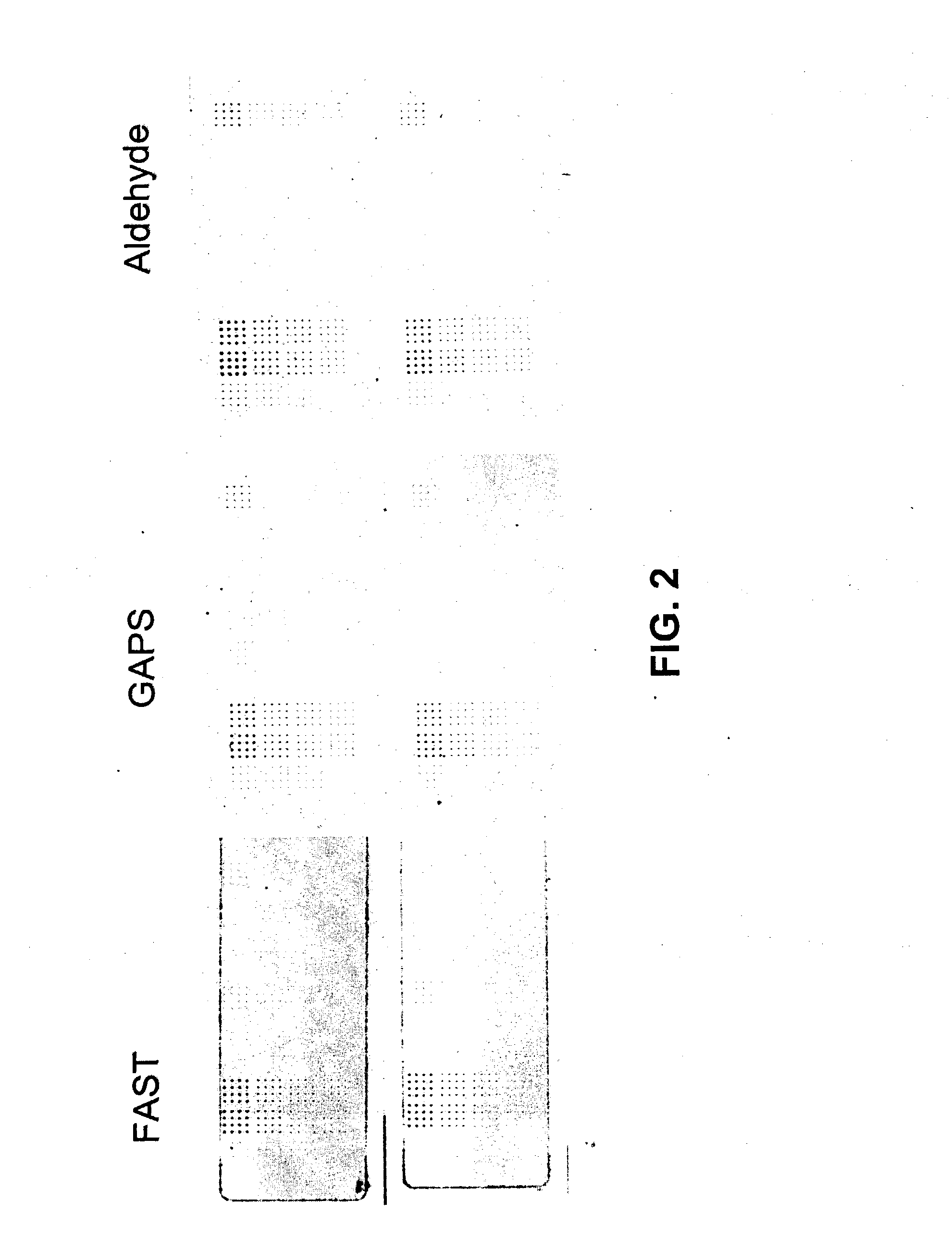
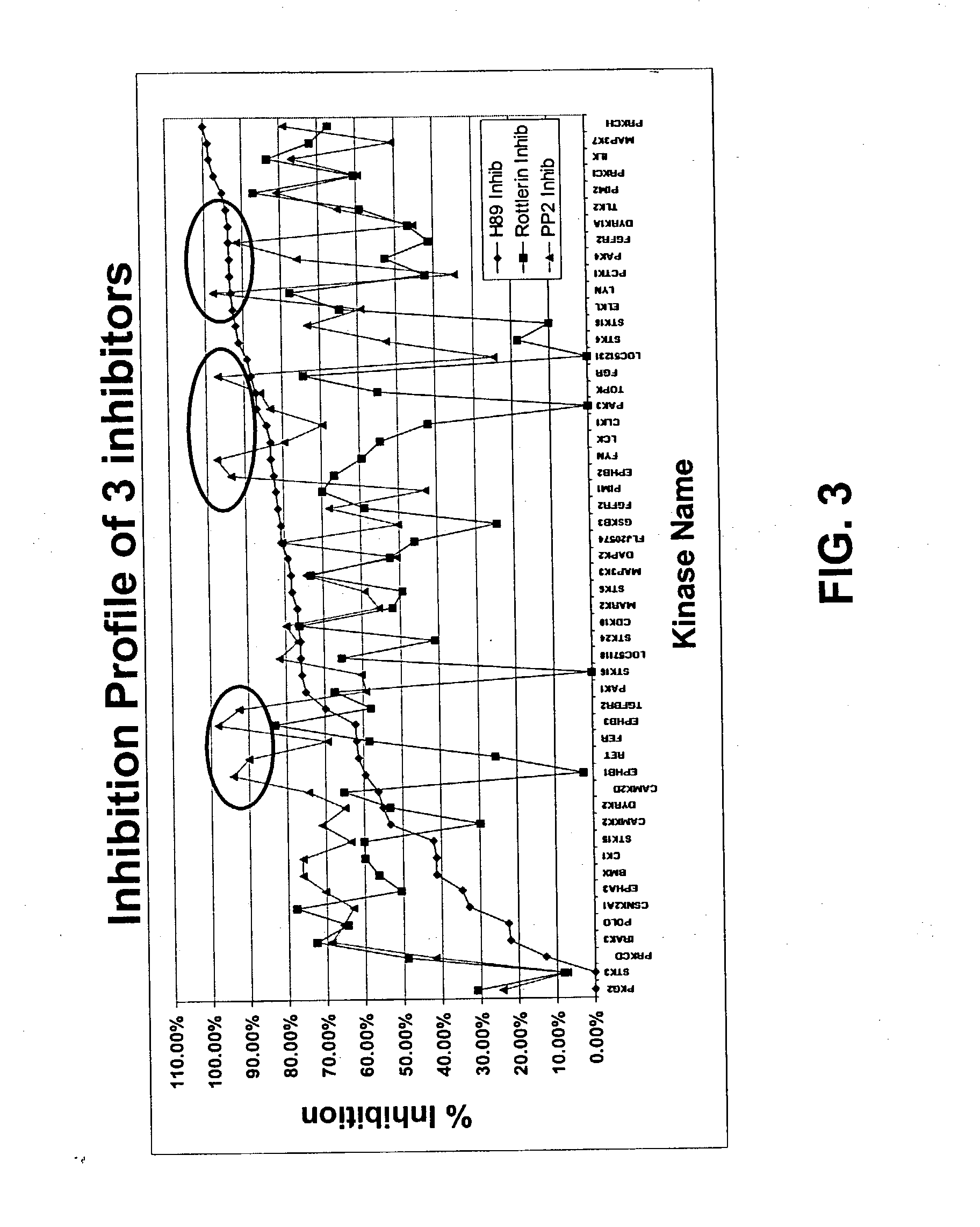
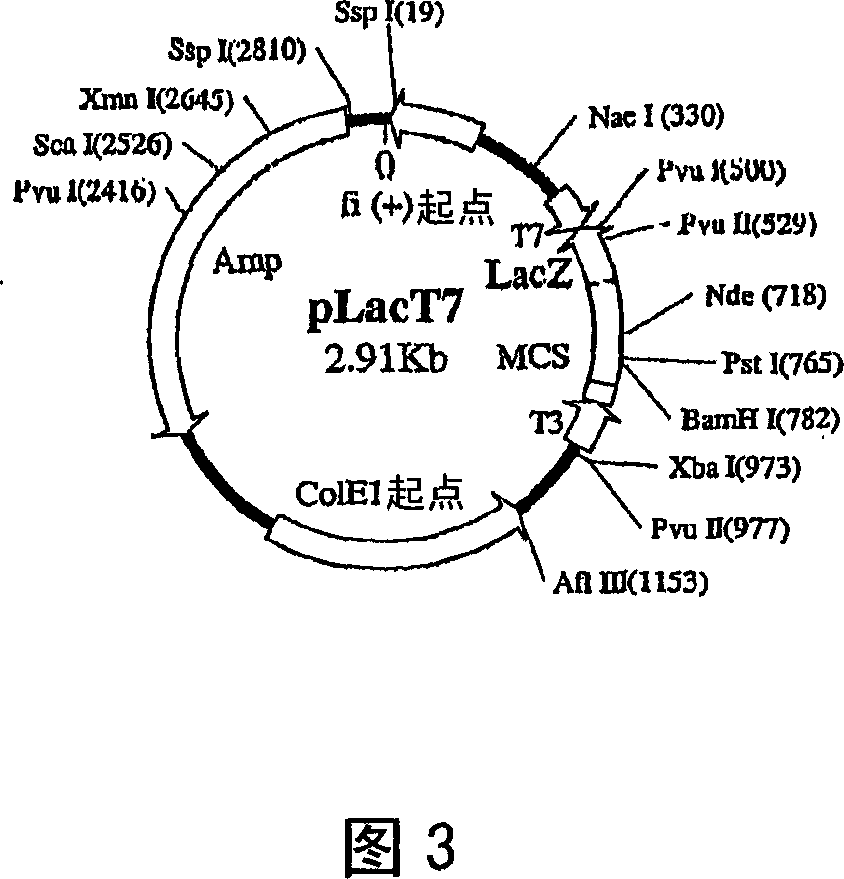
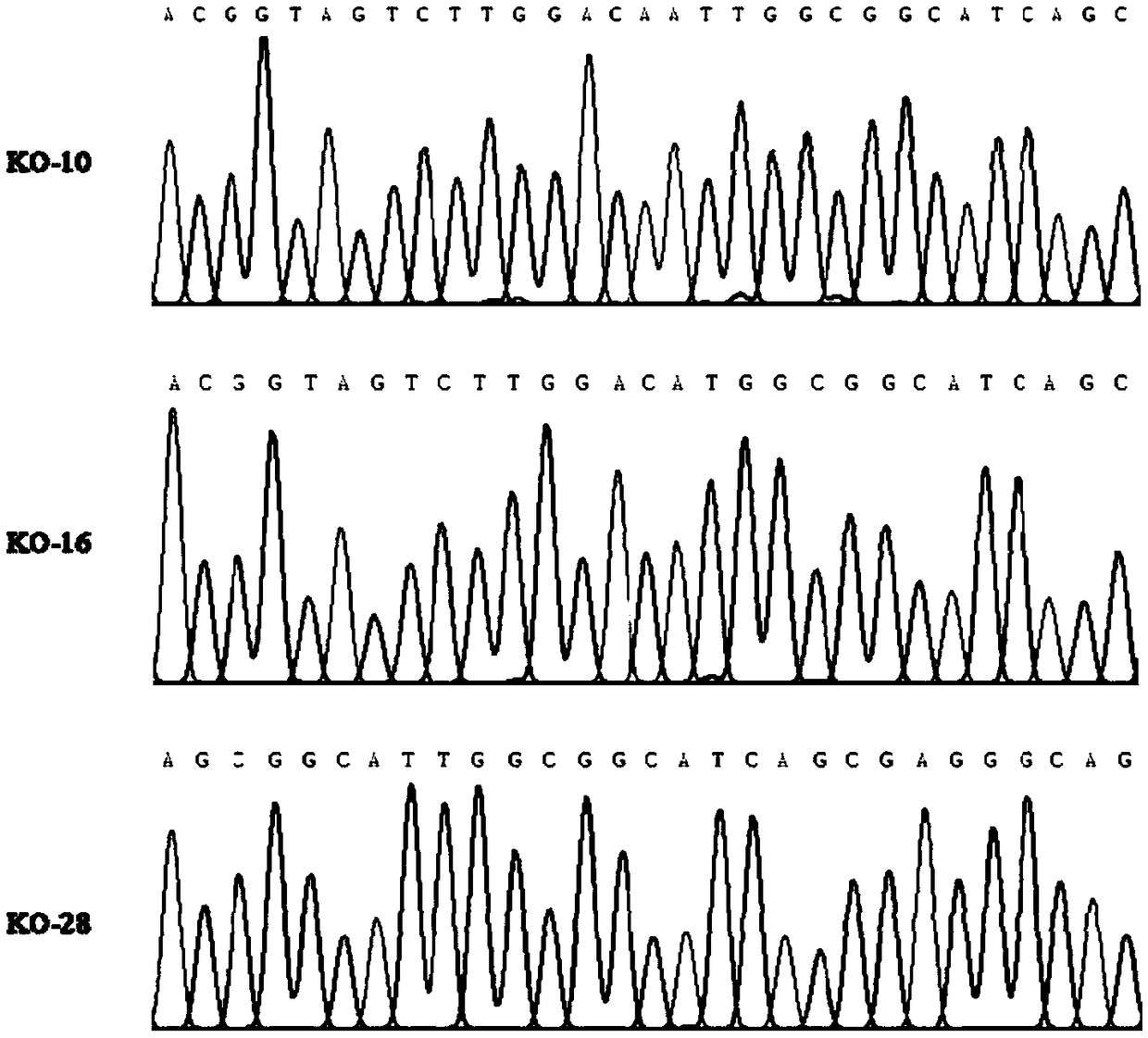
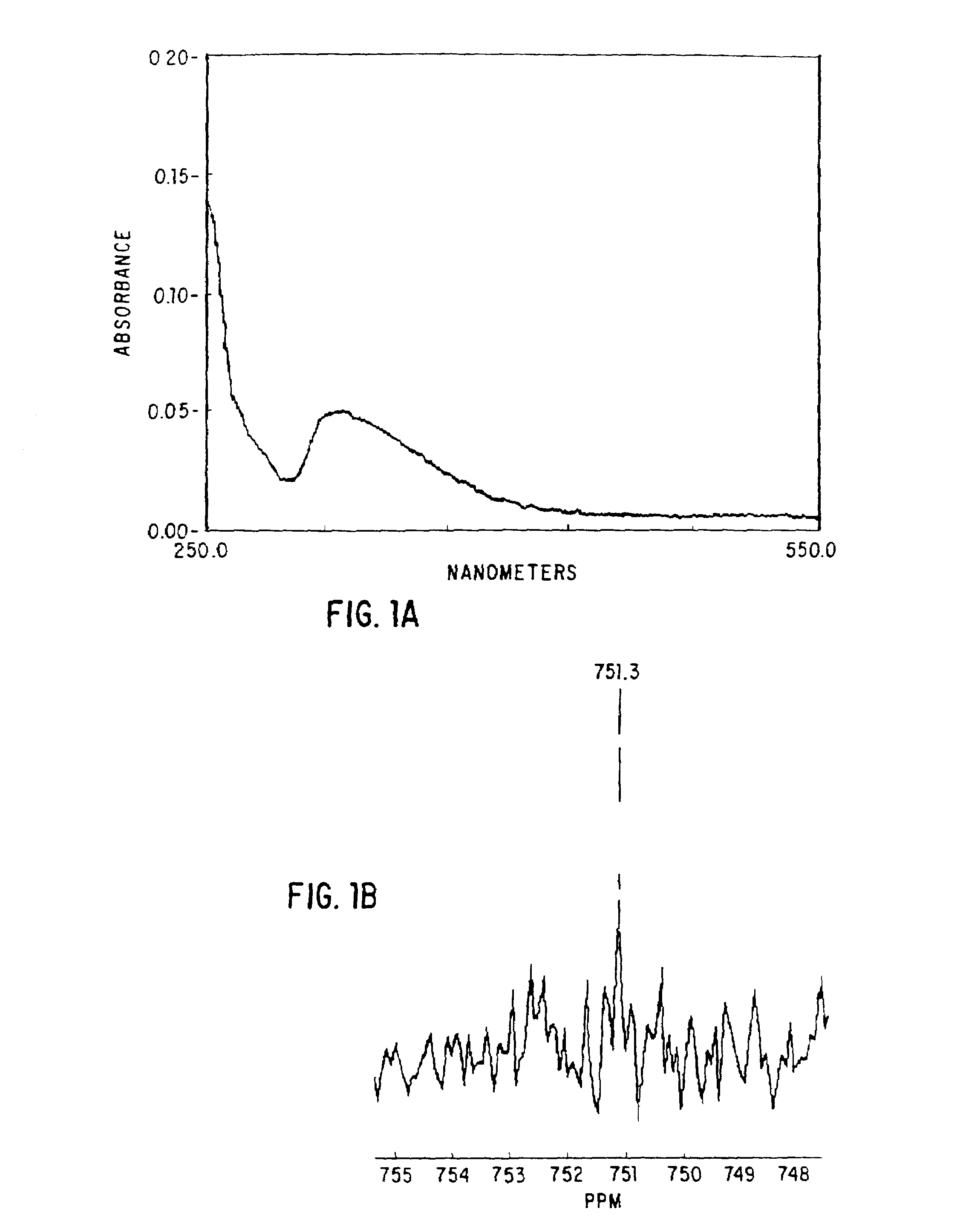
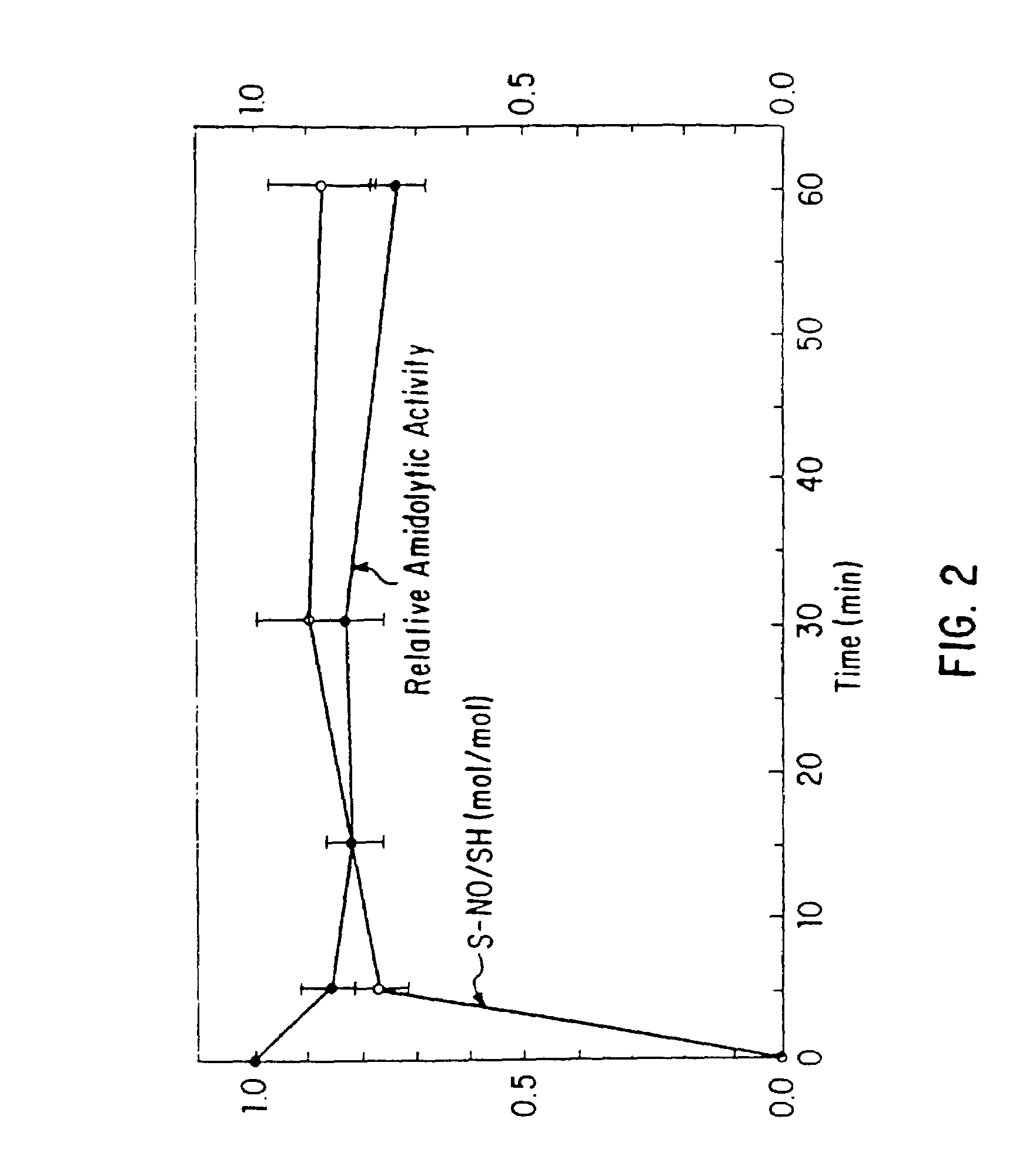
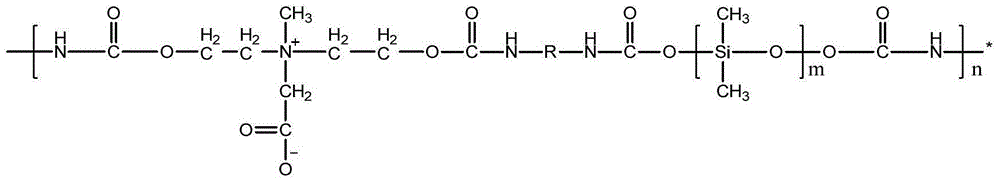Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
291 results about "Protein function" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Methods and products related to the improved analysis of carbohydrates
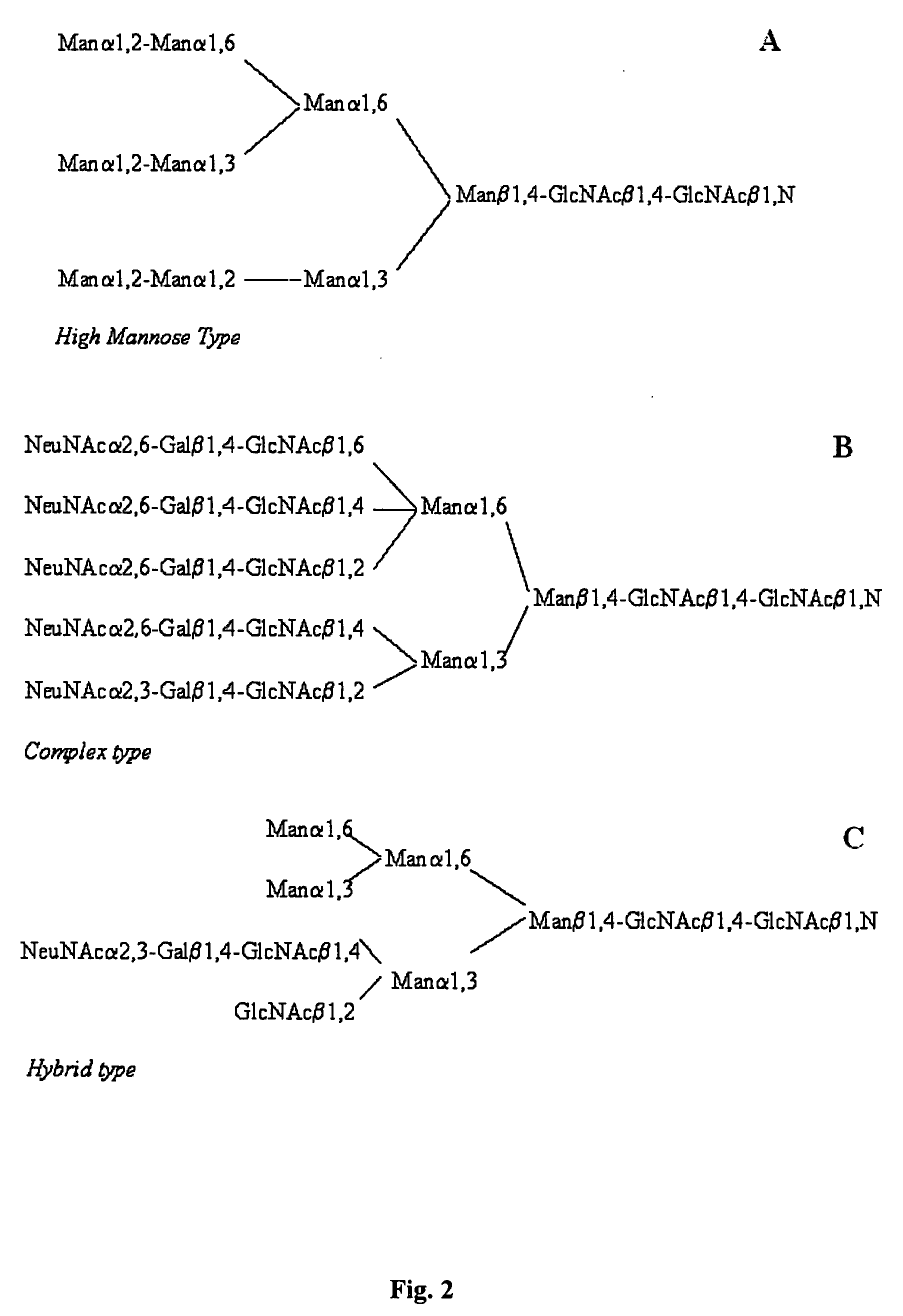
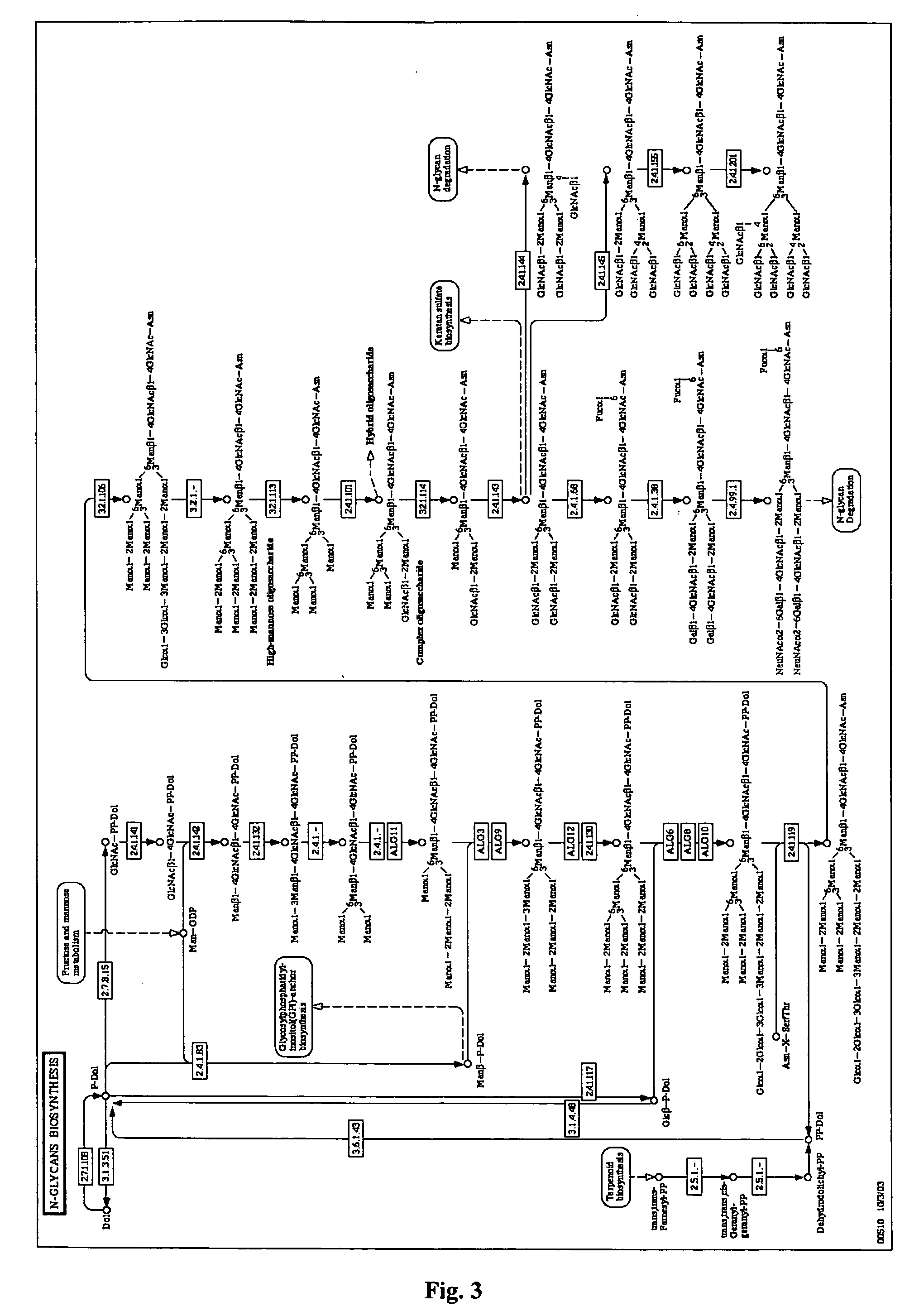
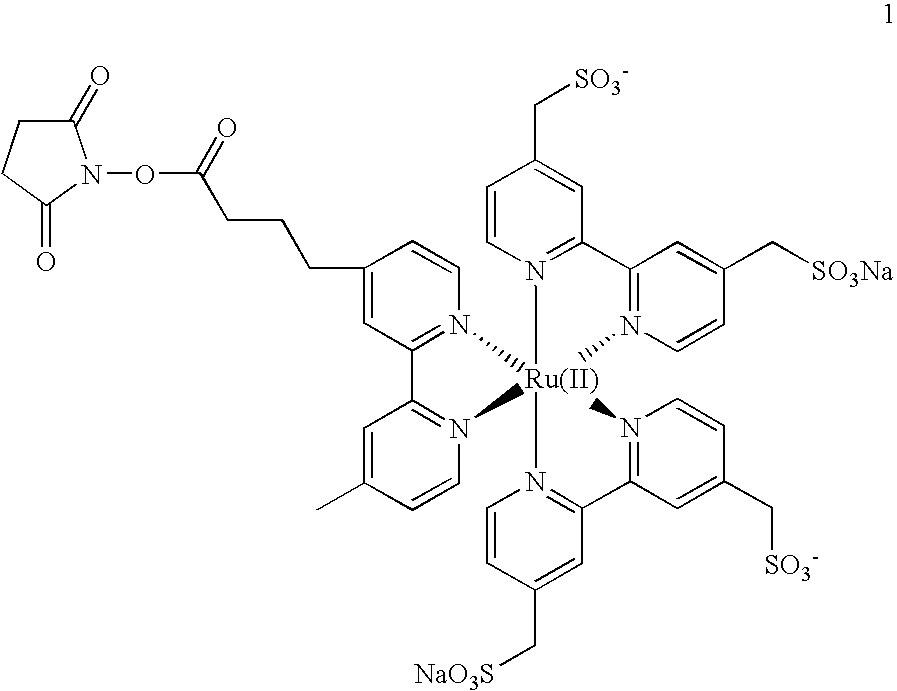
The invention relates, in part, to the improved analysis of carbohydrates. In particular, the invention relates to the analysis of carbohydrates, such as N-glycans and O-glycans found on proteins. Improved methods, therefore, for the study of glycosylation patterns on cells, tissue and body fluids are also provided. Information regarding the analysis of glycans, such as the glycosylation patterns on cells, tissues and in body fluids, can be used in diagnostic and treatment methods as well as for facilitating the study of the effects of glycosylation / altered glycosylation on protein function. Such methods are also provided. Methods are also provided to assess protein production processes, to assess the purity of proteins produced, and to select proteins with the desired glycosylation.
Owner:MOMENTA PHARMA +2
Method for in vitro molecular evolution of protein function
InactiveUS6989250B2Altered propertyIncreased complexitySugar derivativesMicrobiological testing/measurementNucleotideIn vivo
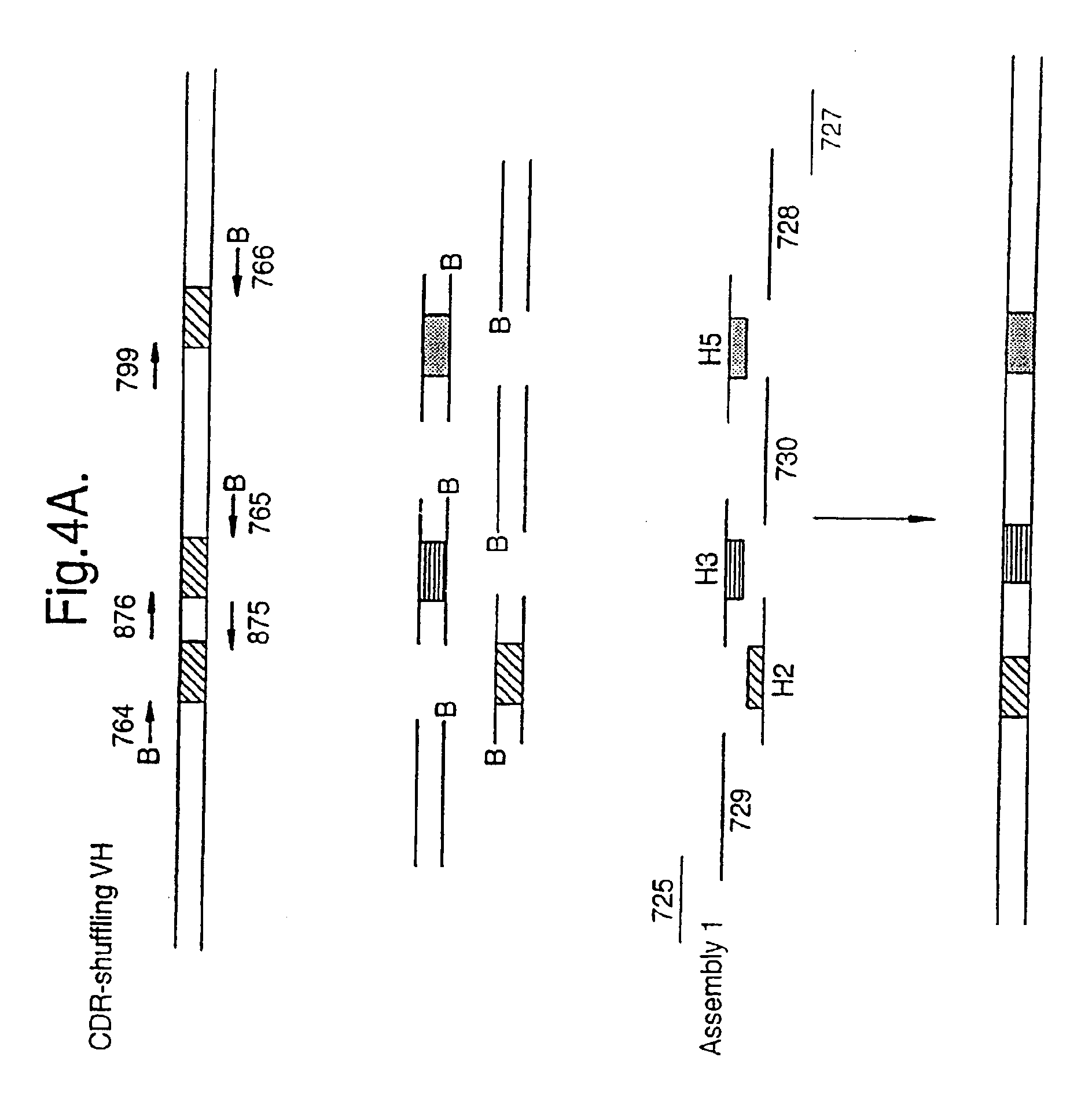
The present invention relates to a method for in vitro creation of molecular libraries evolution of protein function. Particularly, it relates to variability and modification of protein function by shuffling polynucleotide sequence segments. A protein of desired characteristics can be obtained by incorporating variant peptide regions (variant motifs) into defined peptide regions (scaffold sequence). The variant motifs can be obtained from parent DNA which has been subjected to mutagenesis to create a plurality of differently mutated derivatives thereof or they can be obtained from in vivo sequences. These variant motifs can then be incorporated into a scaffold sequence and the resulting coded protein screened for desired characteristics. This method is ideally used for obtaining antibodies with desired characteristics by isolating individual CDR DNA sequences and incorporating them into a scaffold which may, for example, be from a totally different antibody.
Owner:BIOINVENT INT AB
Methods, reagents, kits and apparatus for protein function analysis
InactiveUS20030207290A1Bioreactor/fermenter combinationsBiological substance pretreatmentsProtein functionEnzyme
Owner:MESO SCALE TECH LLC
Browsable database for biological use
The browsable database can allow for high-throughput analysis of protein sequences. One helpful feature may be a simplified ontology of protein function, which allows browsing of the database by biological functions. Biologist curators may have associated the ontology terms with Hidden Markov Models (HMMs), rather than individual sequences, so that they can be applied to additional sequences. To ensure accurate functional classification, HMMs may be constructed not only for families, but for curator-defined subfamilies, whenever family members have divergent functions or nomenclature. Multiple sequence alignments and phylogenetic trees, including curator-assigned information, can be available for each family. Various versions of the browsable database may include training sequences from all organisms in the GenBank non-redundant protein database, and the HMMs can be used to classify gene products across the entire genomes of human, and Drosophila melanogaster.
Owner:APPL BIOSYSTEMS INC
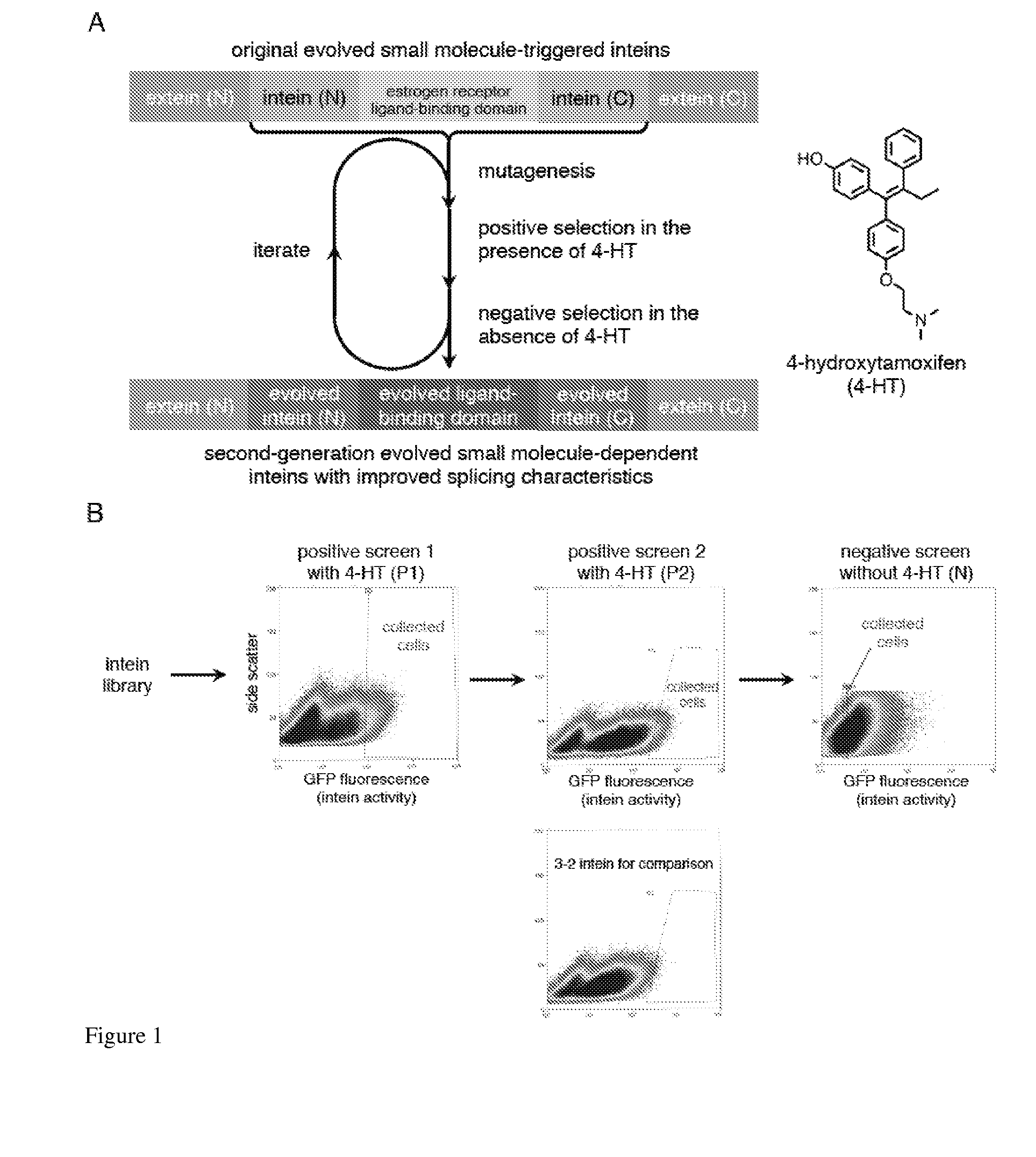
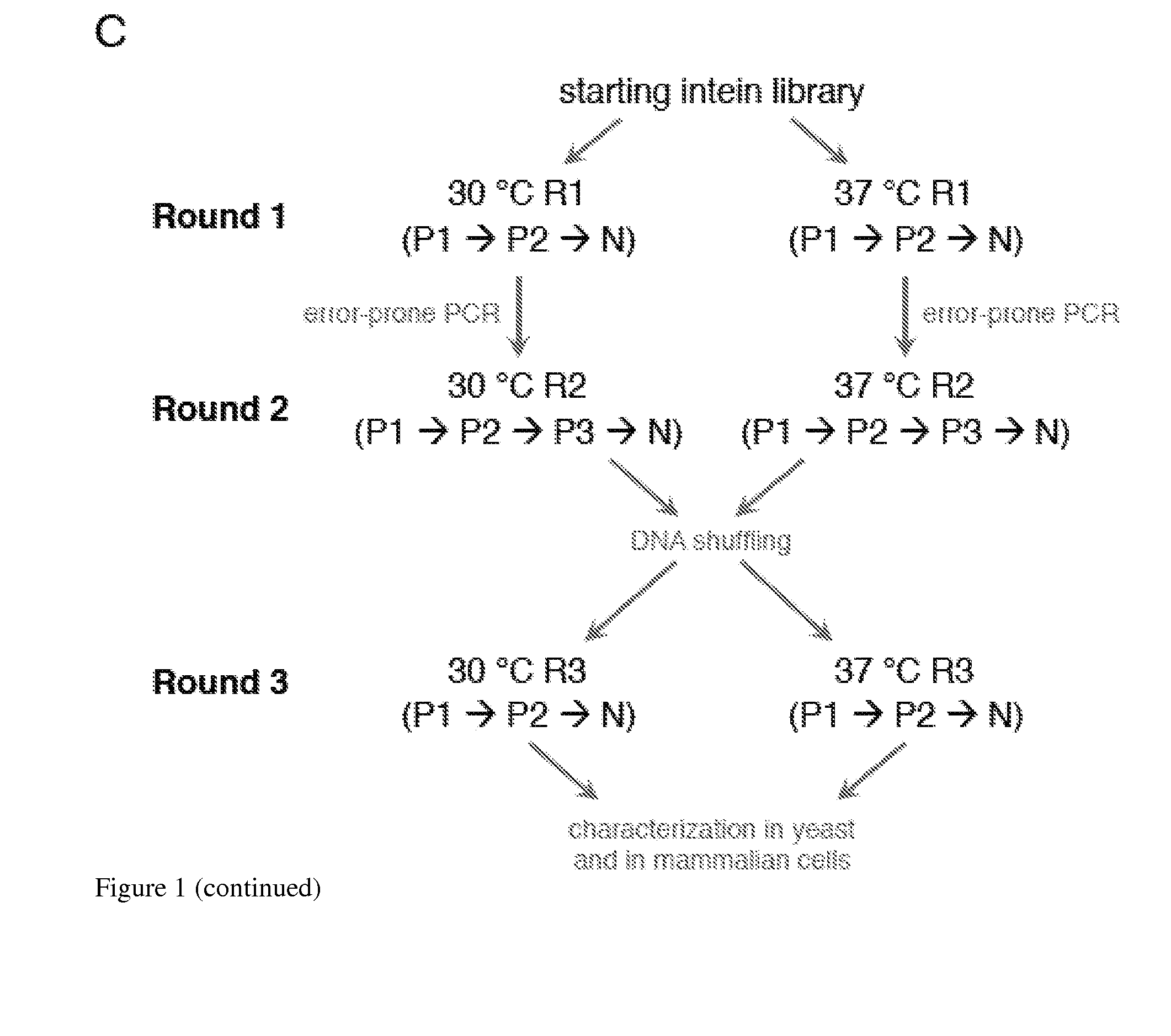
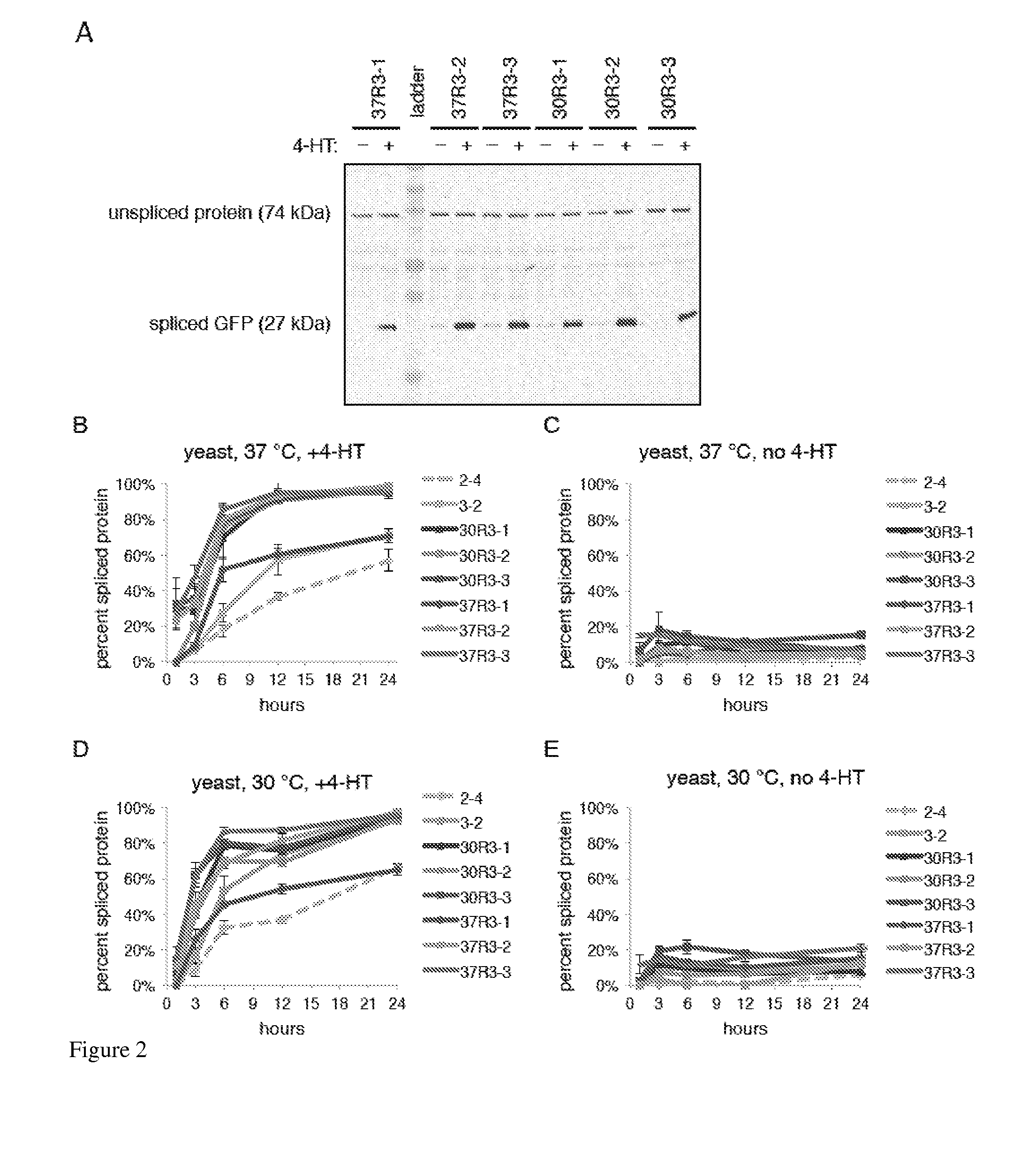
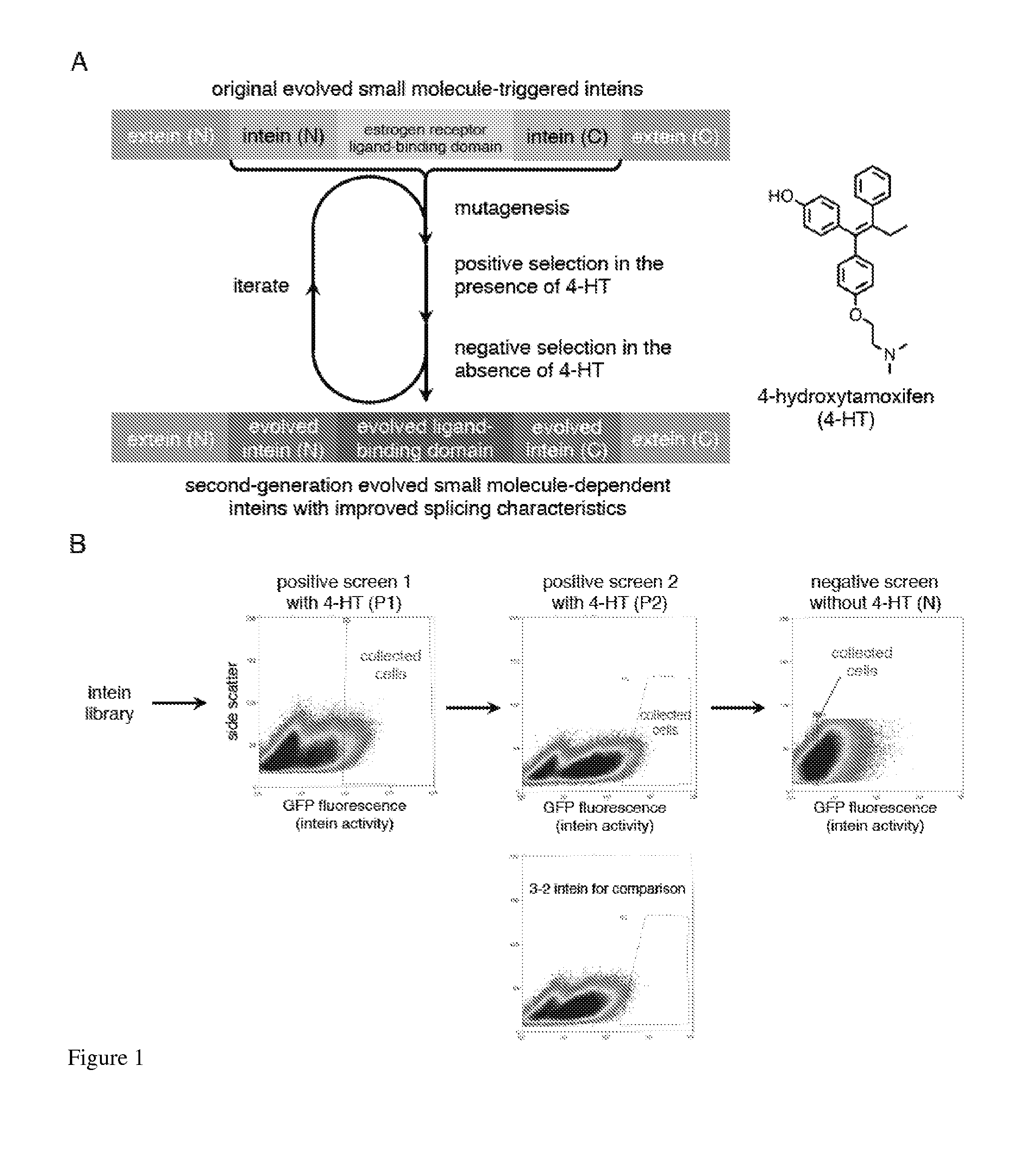
Small molecule-dependent inteins and uses thereof
ActiveUS20140065711A1Low splicing efficiencySlow splicingHydrolasesFusion with post-translational modification motifInteinNatural state
Elucidating the function of proteins in mammalian cells is particularly challenging due to the inherent complexity of these systems. Methods to study protein function in living cells ideally perturb the activity of only the protein of interest but otherwise maintain the natural state of the host cell or organism. Ligand-dependent inteins offer single-protein specificity and other desirable features as an approach to control protein function in cells post-translationally. Some aspects of this invention provide second-generation ligand-dependent inteins that splice to substantially higher yields and with faster kinetics in the presence of the cell-permeable small molecule 4-HT, especially at 37° C., while exhibiting comparable or improved low levels of background splicing in the absence of 4-HT, as compared to the parental inteins. These improvements were observed in four protein contexts tested in mammalian cells at 37° C., as well as in yeast cells assayed at 30° C. or 37° C. The newly evolved inteins described herein are therefore promising tools as conditional modulators of protein structure and function in yeast and mammalian cells.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Process for analyzing protein samples
InactiveUS7045296B2Reduce disulfide bondEasy labelingSamplingMicrobiological testing/measurementProtein expression profileUnit operation
Methods using gel electrophoresis and mass spectrometry for the rapid, quantitative analysis of proteins or protein function in mixtures of proteins derived from two or more samples in one unit operation are disclosed. In one embodiment the method includes (a) preparing an extract of proteins from each of at least two different samples; (b) providing a set of substantially chemically identical and differentially isotopically labeled protein reagents; (c) reacting the extract of proteins from different samples of step (a) with a different isotopically labeled reagent from the set of step (b) to provide two or more sets of isotopically differentially labeled proteins; (d) mixing each of the two or more sets of isotopically labeled proteins to form a single mixture of isotopically differentially labeled proteins; (e) electrophoresing the mixture of step (d) by an electrophoresing method capable of separating proteins within the mixture; (f) digesting at least a portion of one or more separated proteins of step (e) and (g) detecting the difference in the expression levels of the proteins in the two samples by mass spectrometry based on one or more peptides in the sample of labeled peptides. The analytical method can be used for qualitative and particularly for quantitative analysis of global protein expression profiles in cells and tissues, i.e. the quantitative analysis of proteomes.
Owner:DH TECH DEVMENT PTE +1
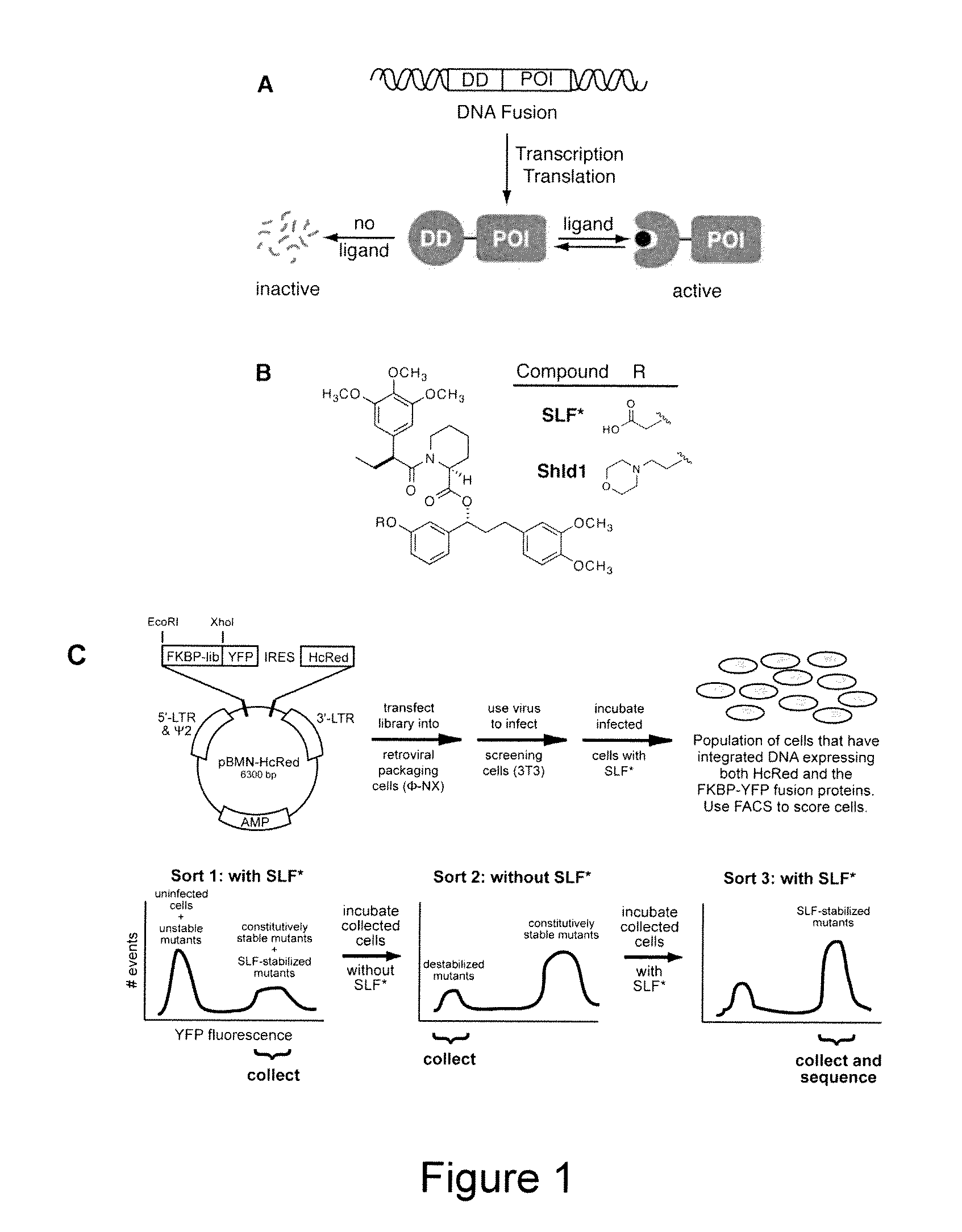
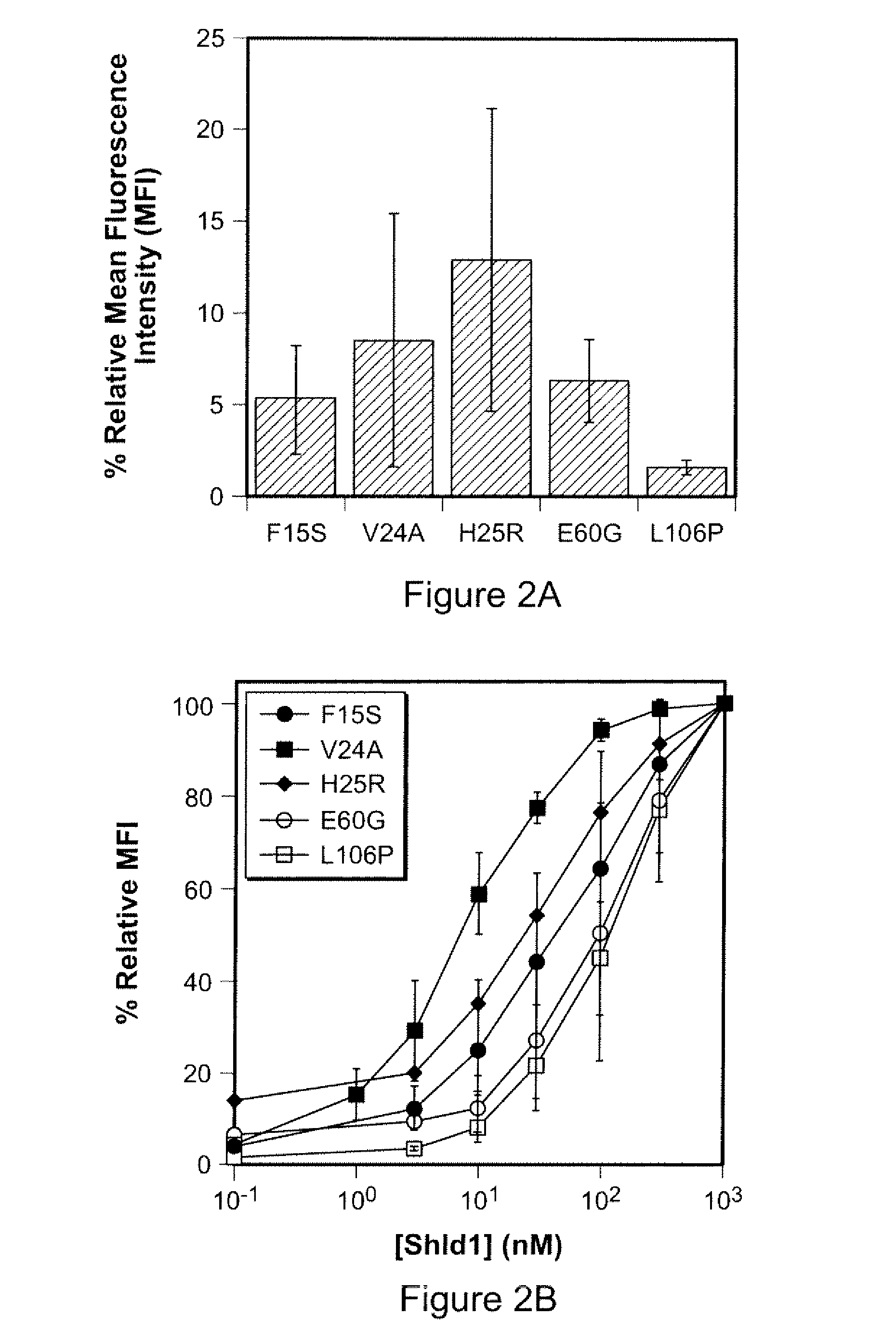
Method for regulating protein function in cells using synthetic small molecules
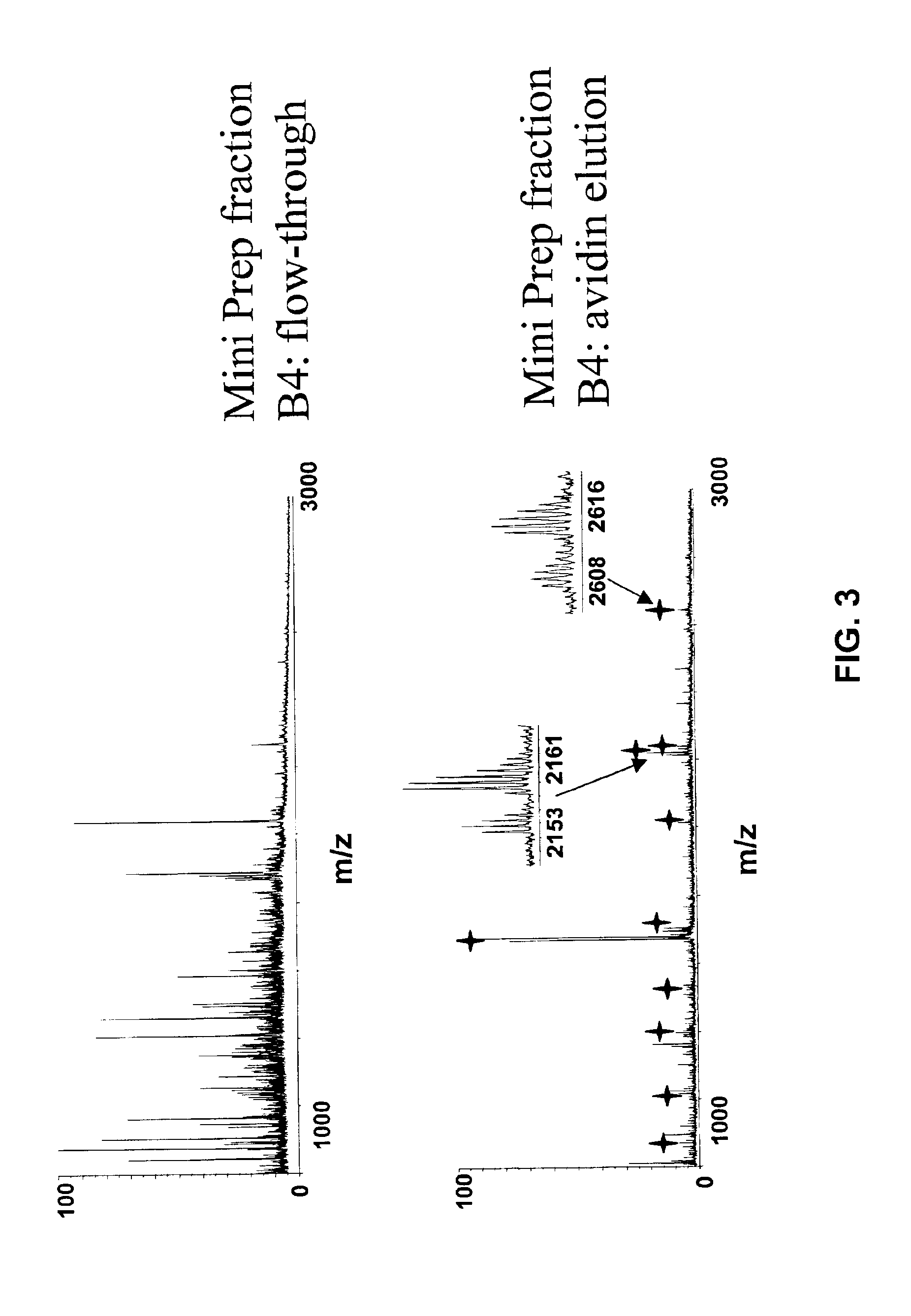
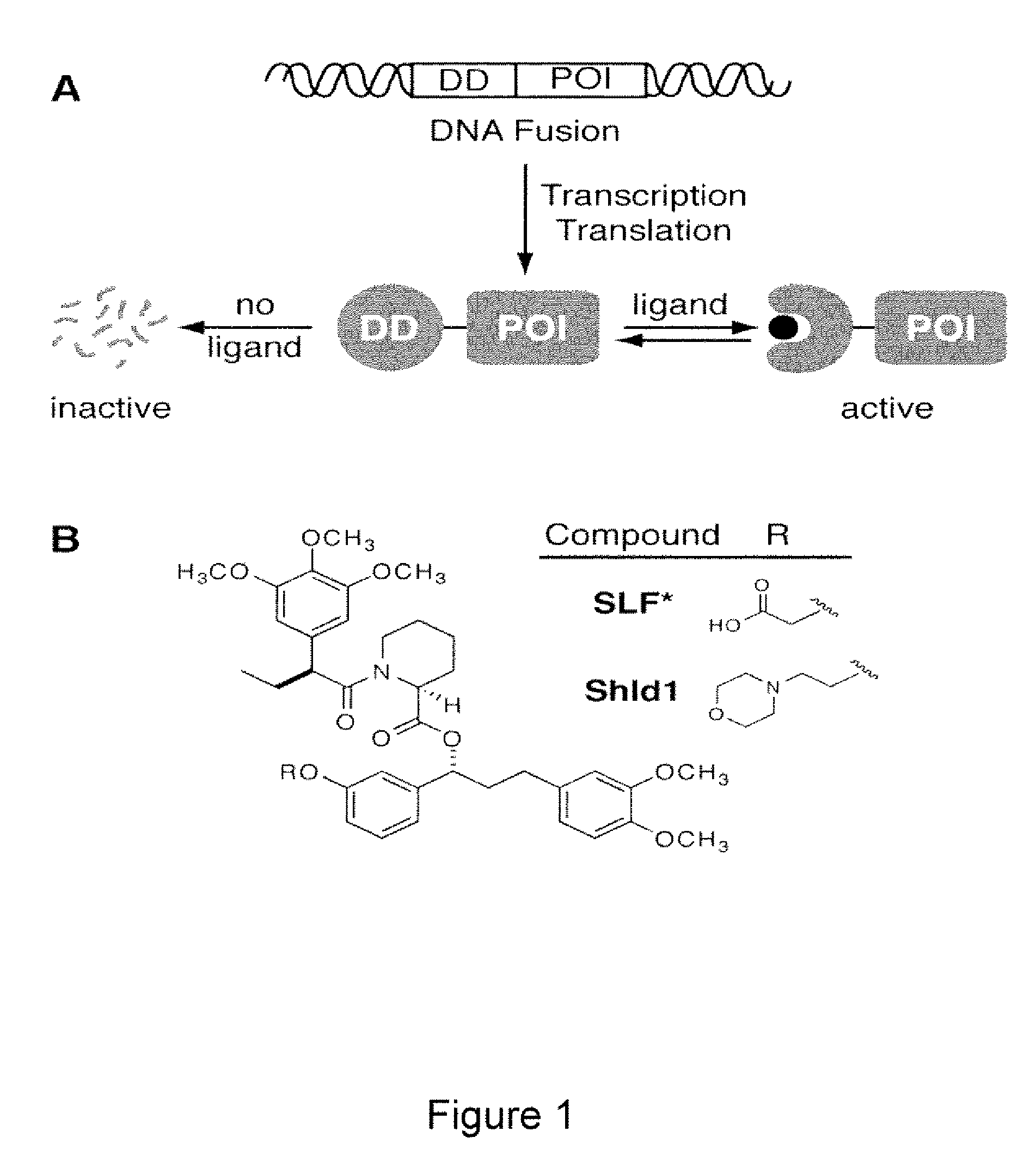
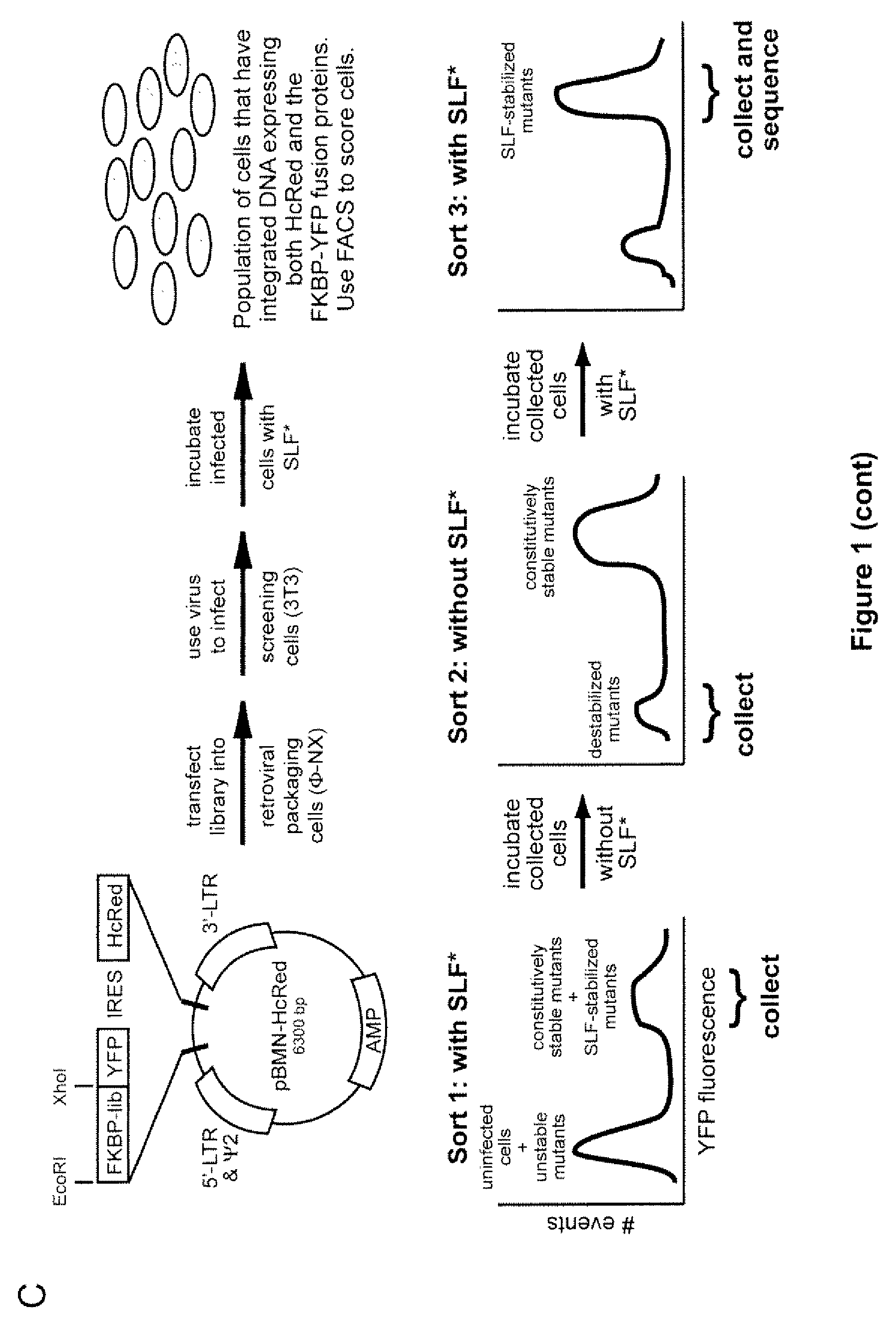
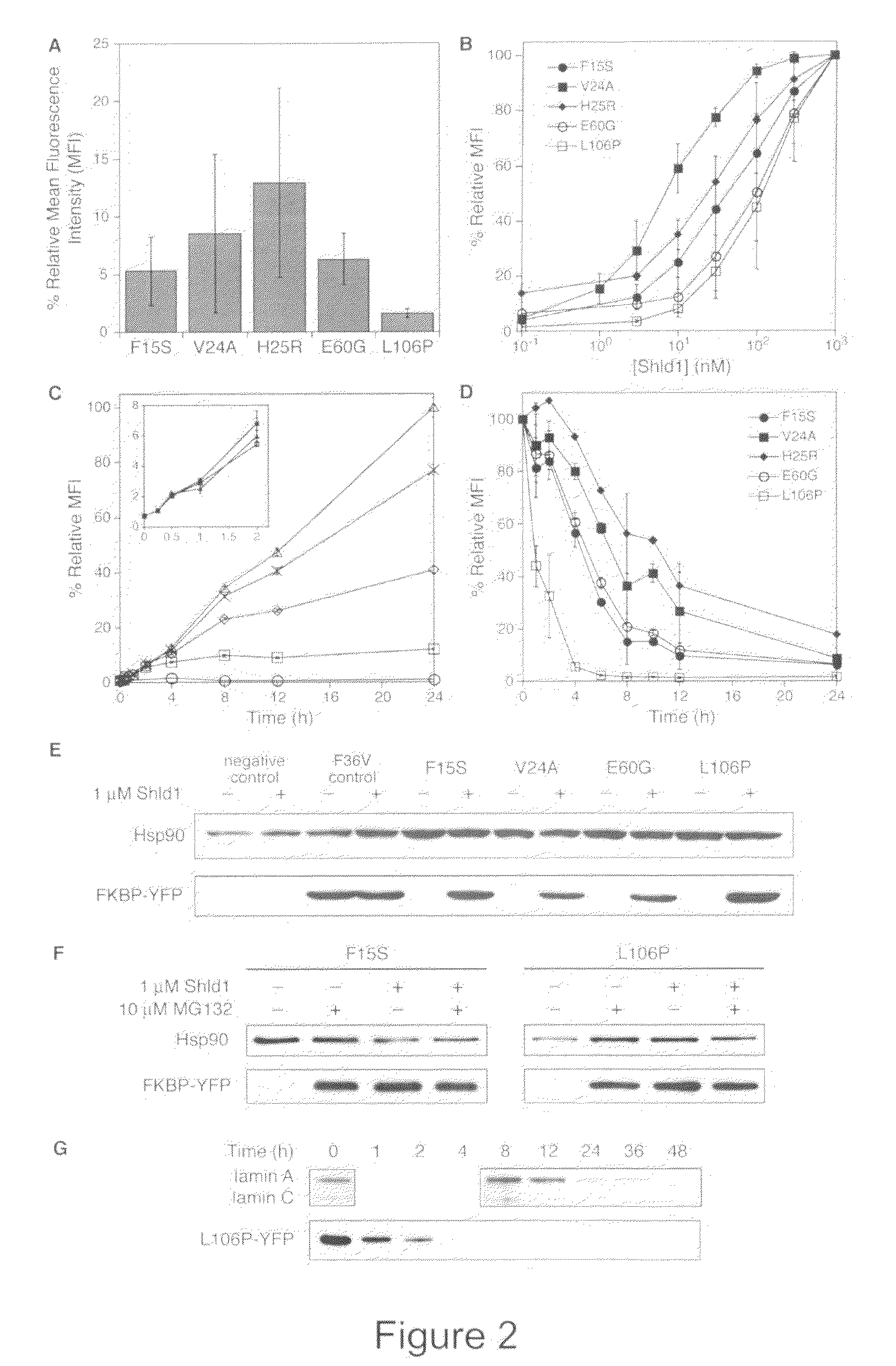
Methods and compositions for the rapid and reversible destabilizing of specific proteins using cell-permeable, synthetic molecules are described. Stability-affecting proteins, e.g., derived from FKBP and DHFR proteins are fused to a protein of interest and the presence or absence of the ligand is used to modulate the stability of the fusion protein.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
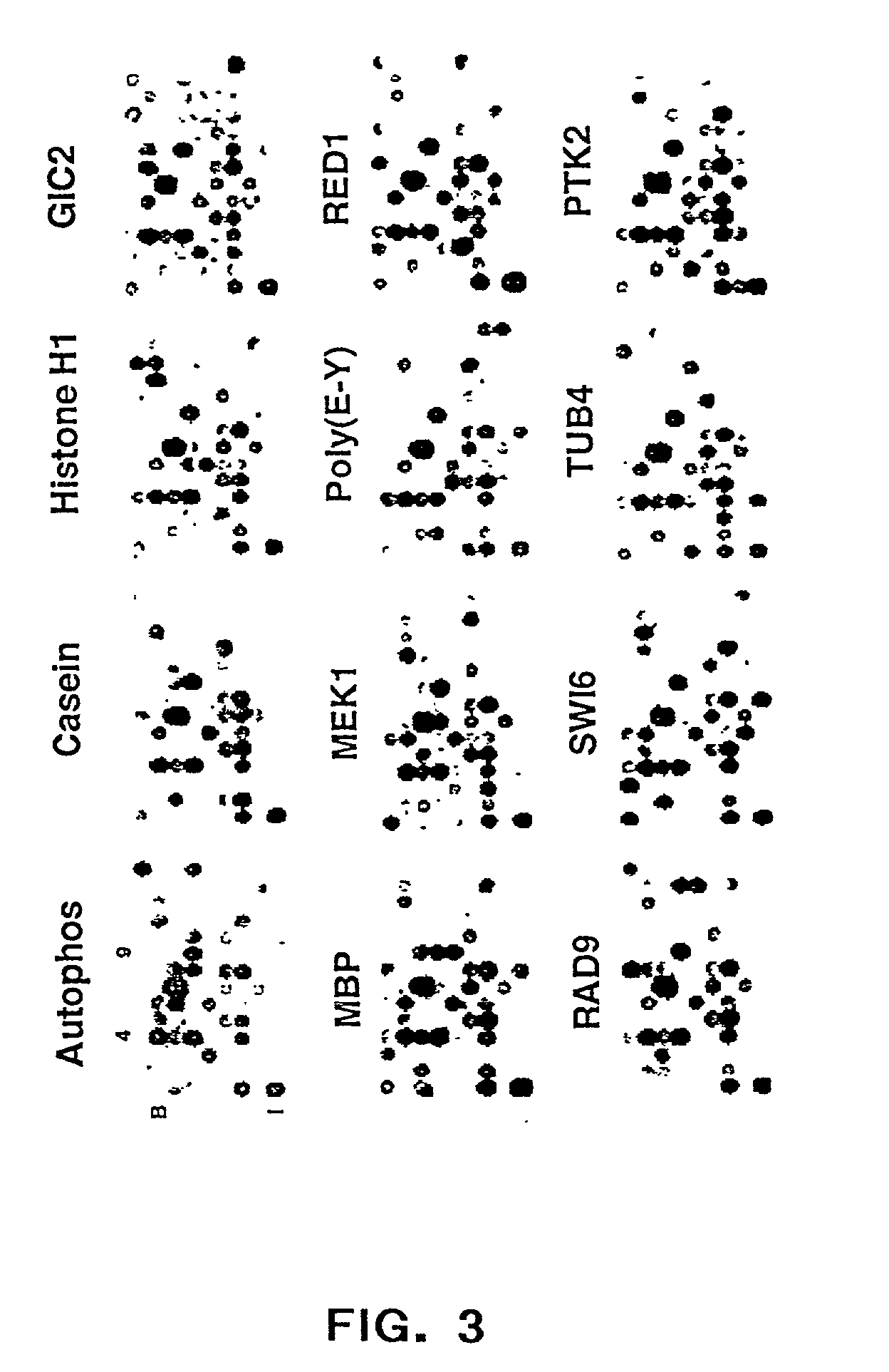
Methods for conducting assays for enzyme activity on protein microarrays
The present invention relates to methods of conducting assays for enzymatic activity on microarrays useful for the large-scale study of protein function, screening assays, and high-throughput analysis of enzymatic reactions. The invention relates to methods of using protein chips to assay the presence, amount, activity and / or function of enzymes present in a protein sample on a protein chip. In particular, the methods of the invention relate to conducting enzymatic assays using a microarray wherein a protein and a substance are immobilized on the surface of a solid support and wherein the protein and the substance are in proximity to each other sufficient for the occurrence of an enzymatic reaction between the substance and the protein. The invention also relates to microarrays that have an enzyme and a substrate immobilized on their surface wherein the enzyme and the substrate are in proximity to each other sufficient for the occurrence of an enzymatic reaction between the enzyme and the substrate.
Owner:LIFE TECH CORP
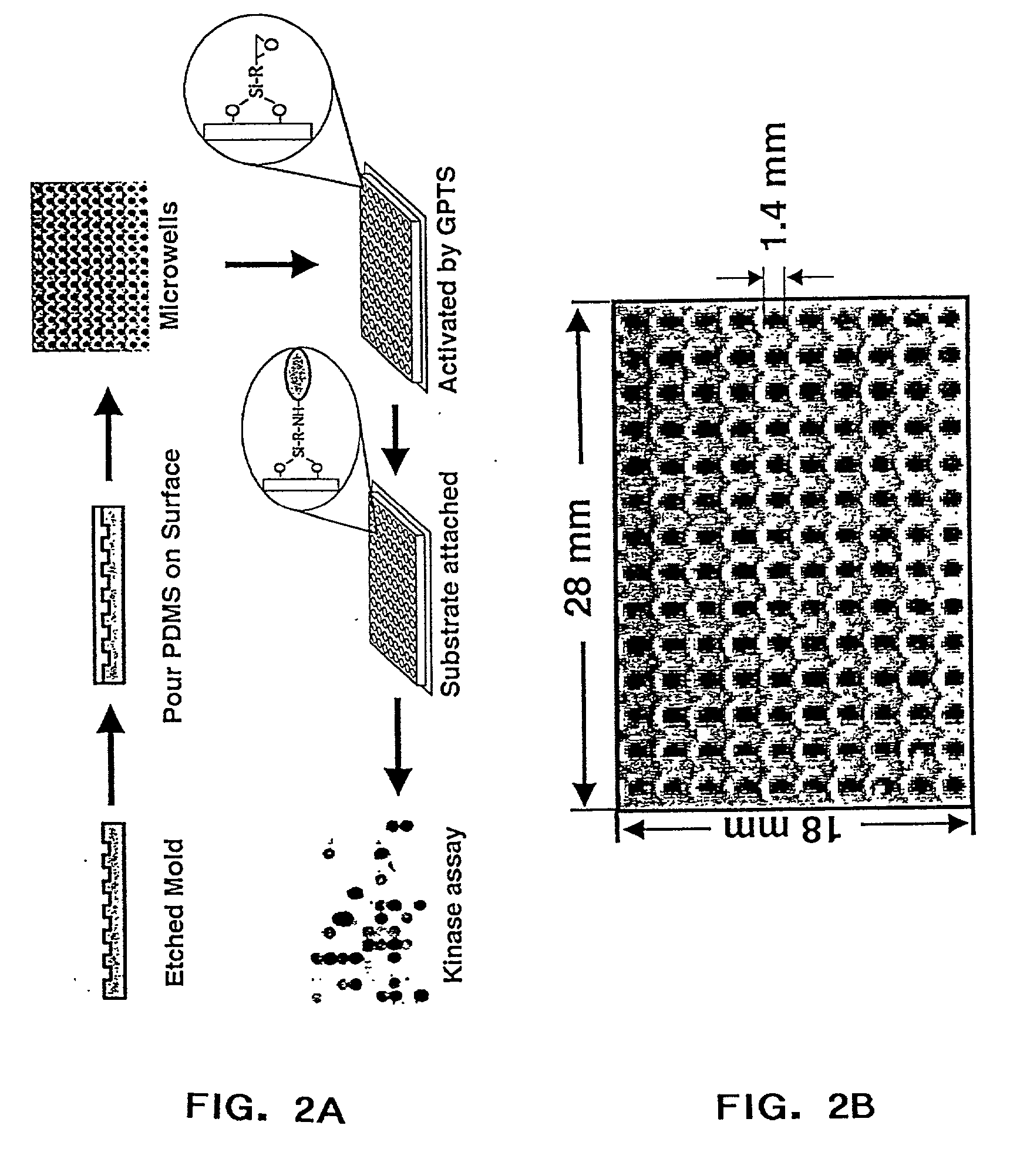
Protein chips for high throughput screening of protein activity
InactiveUS20030207467A1Material nanotechnologyBioreactor/fermenter combinationsHigh-Throughput Screening MethodsChemical reaction
The present invention relates to protein chips useful for the large-scale study of protein function where the chip contains densely packed reaction wells. The invention also relates to methods of using protein chips to assay simultaneously the presence, amount, and / or function of proteins present in a protein sample or on one protein chip, or to assay the presence, relative specificity, and binding affinity of each probe in a mixture of probes for each of the proteins on the chip. The invention also relates to methods of using the protein chips for high density and small volume chemical reactions. Also, the invention relates to polymers useful as protein chip substrates and methods of making protein chips. The invention further relates to compounds useful for the derivatization of protein chip substrates.
Owner:YALE UNIV
Systems and methods for silencing expression of a gene in a cell and uses thereof
InactiveUS20060178297A1Minimal disturbanceFast and nearly-complete uptakePeptide/protein ingredientsMicroencapsulation basedPrimary cellNeuron
The present invention provides RNA-based systems that are capable of silencing expression of a gene in a cell. Also provided are pharmaceutical compositions, cells, and kits that include the systems; cultures of primary cells that have been contacted with the systems; use of the systems in methods of studying protein function in one or more cells; and use of the systems in methods of studying interactions between neurons in culture; and use of the systems in a method of studying the function of mRNA. The present invention further provides methods for silencing expression of a gene in a cell, and methods for determining the function of a gene in a cell. Additionally, the present invention provides systems for use in genetic screening, and methods for performing genetic screening.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
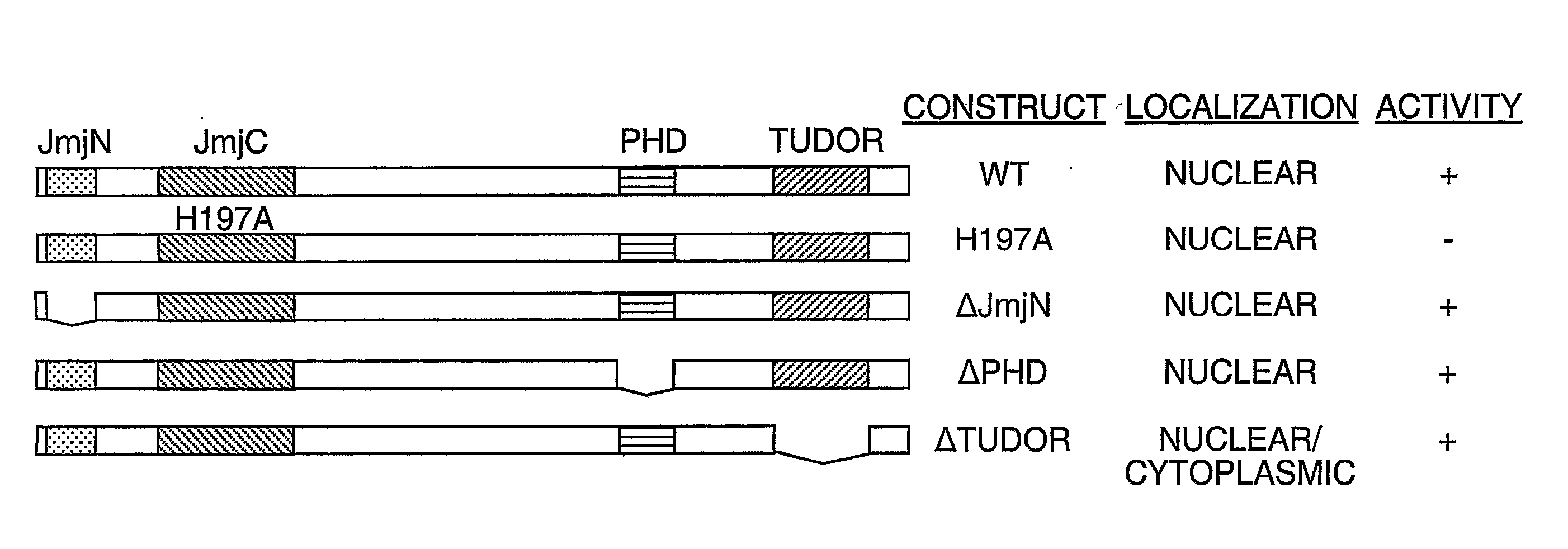
Protein demethylases comprising a jmjc domain
Post-translational modification, including protein methylation, plays an important role in regulating protein function. The present invention provides a novel assay for evaluating demethylase activity and the discovery of a family of protein demethylases comprising a novel demethylase motif.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
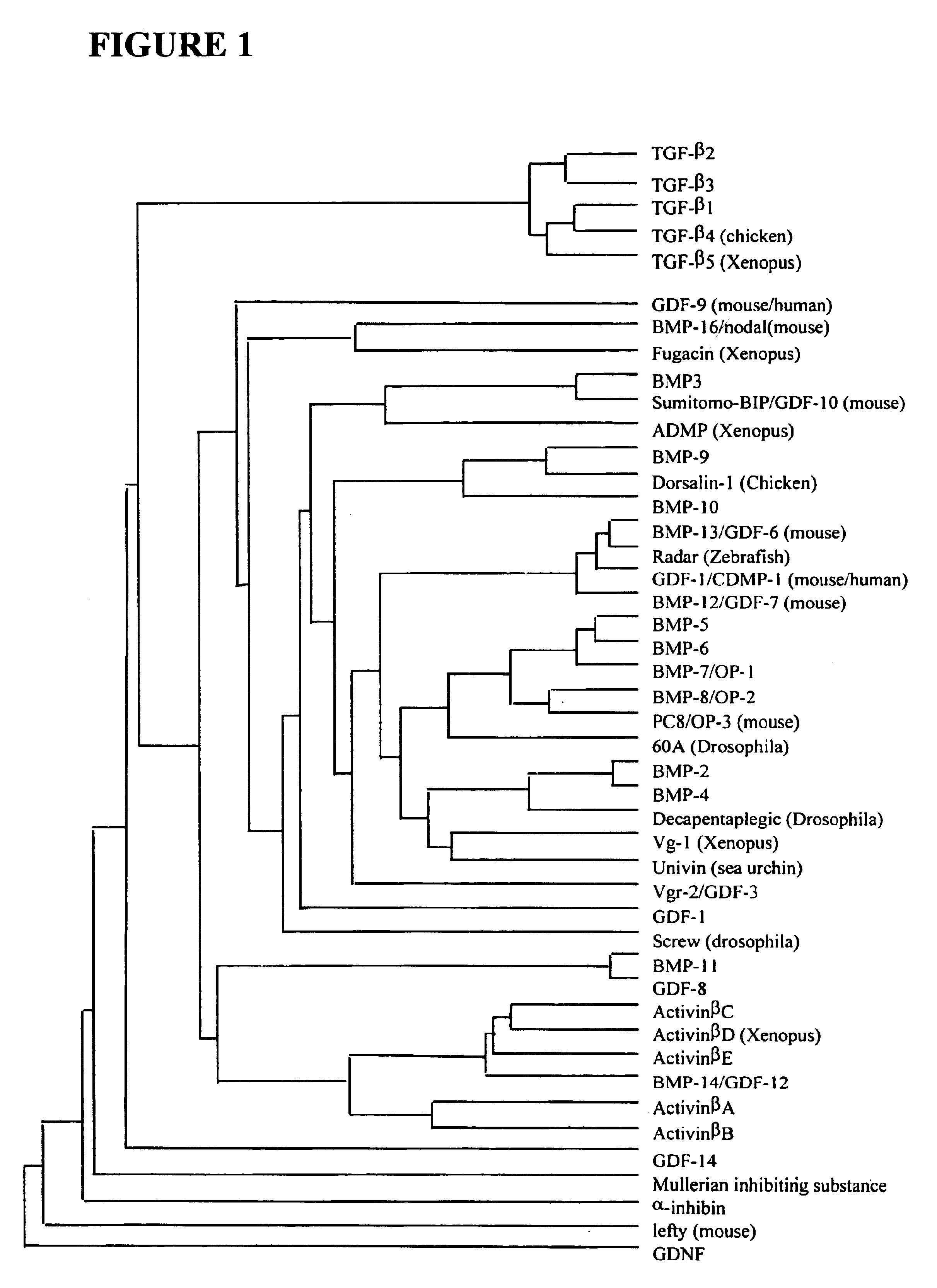
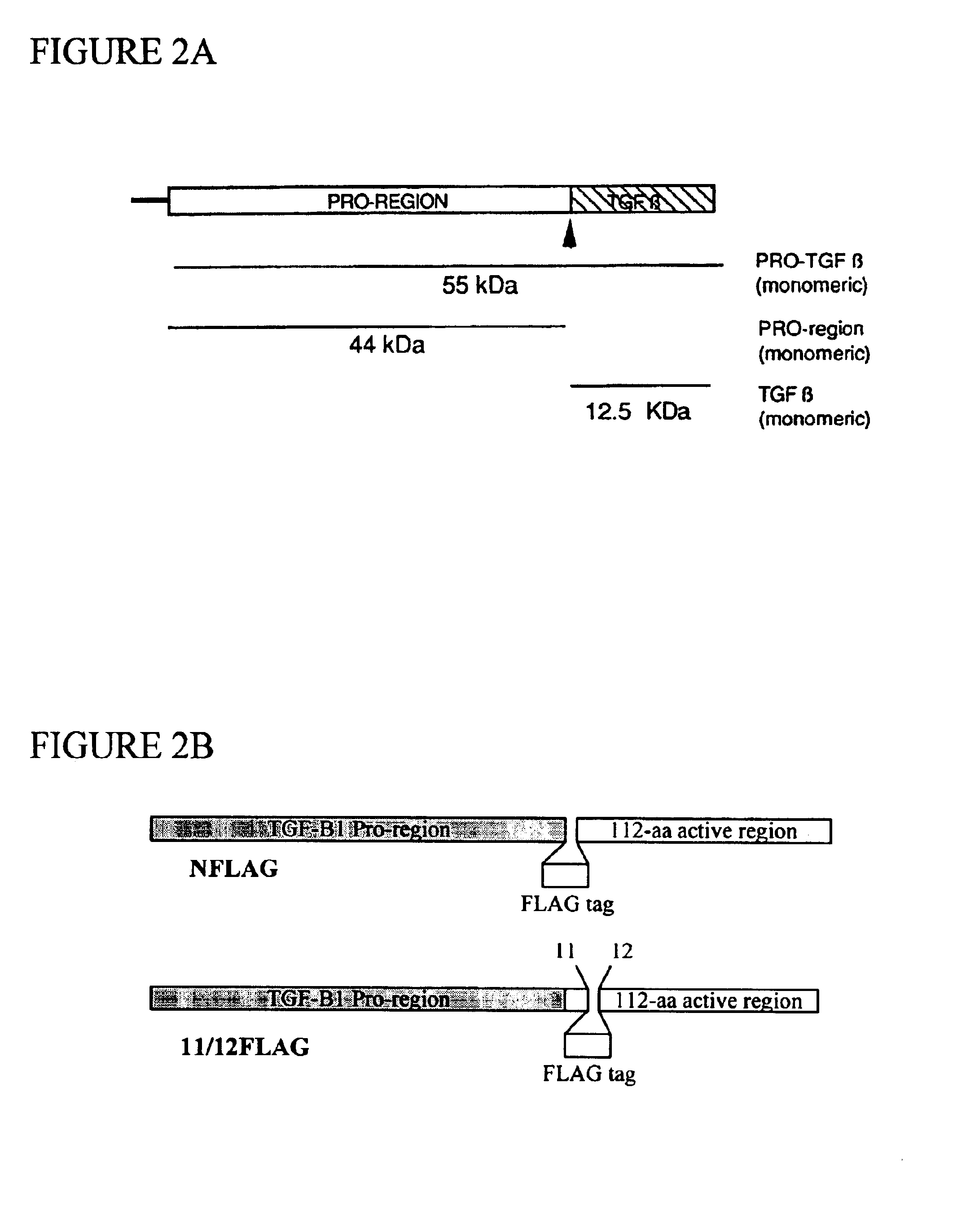
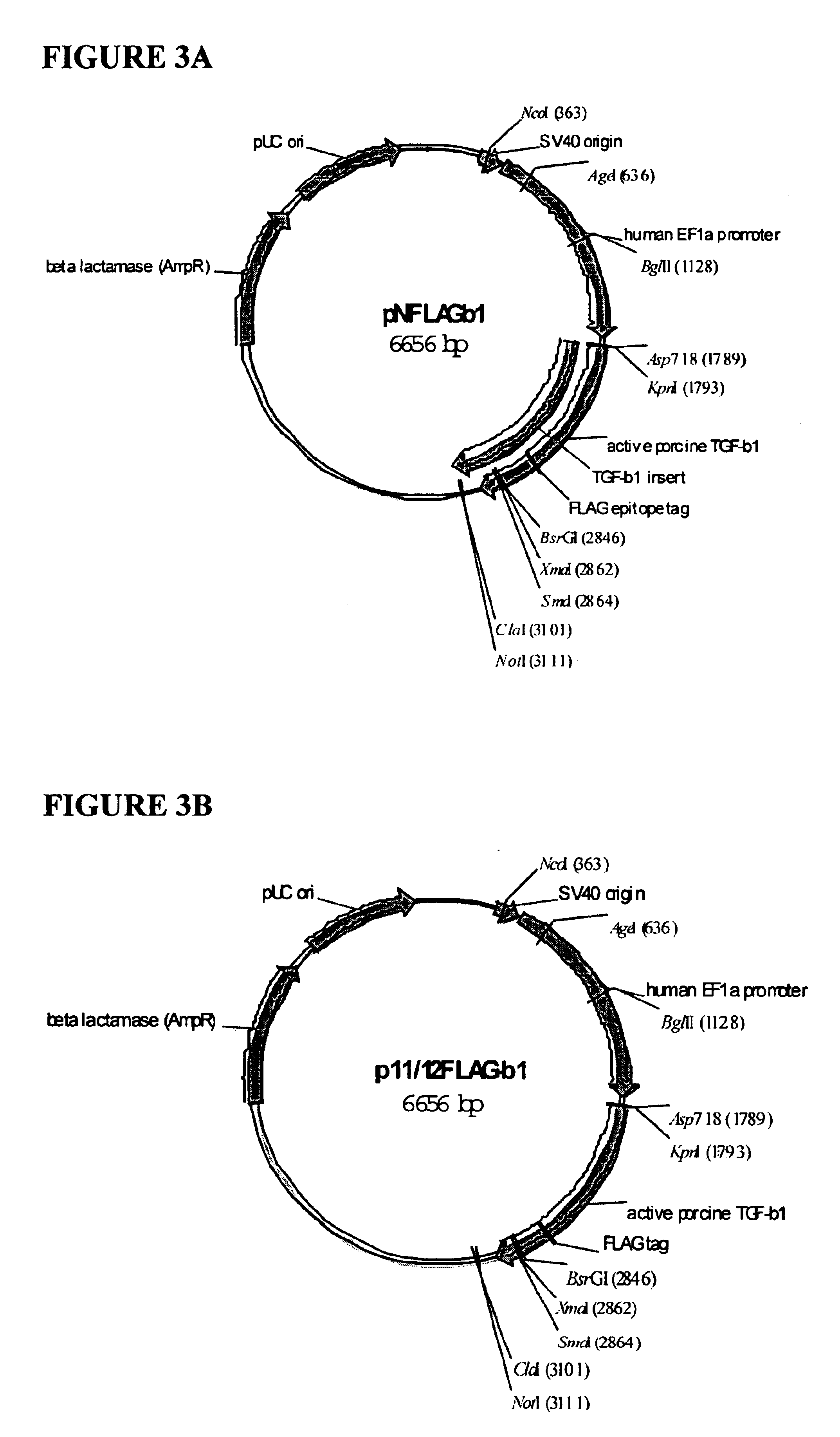
Functionalized TGF-beta fusion proteins
This disclosure relates to TGF-beta family protein fusions that display substantial native TGF-beta family protein function while also having an additional functionality conveyed by the addition of a functionalizing peptide domain. Such functionalizing peptide domain can be a tag peptide (e.g., an epitope tag, a purification tag, a molecular size differentiation tag, etc.) or a passenger or targeting protein. Also provided are methods of making these fusions, as well as methods of using them for diagnosis and diagnosis of various conditions, in measuring and monitoring levels of the fusion molecule in experimental systems and subjects, and in measuring and detecting receptor proteins.
Owner:GOVERNMENT OF THE UNITED STATE OF AMERICA AS REPRESENTED BY THE SEC OF THE DEPT OF HEALTH & HUMAN SERVICES THE
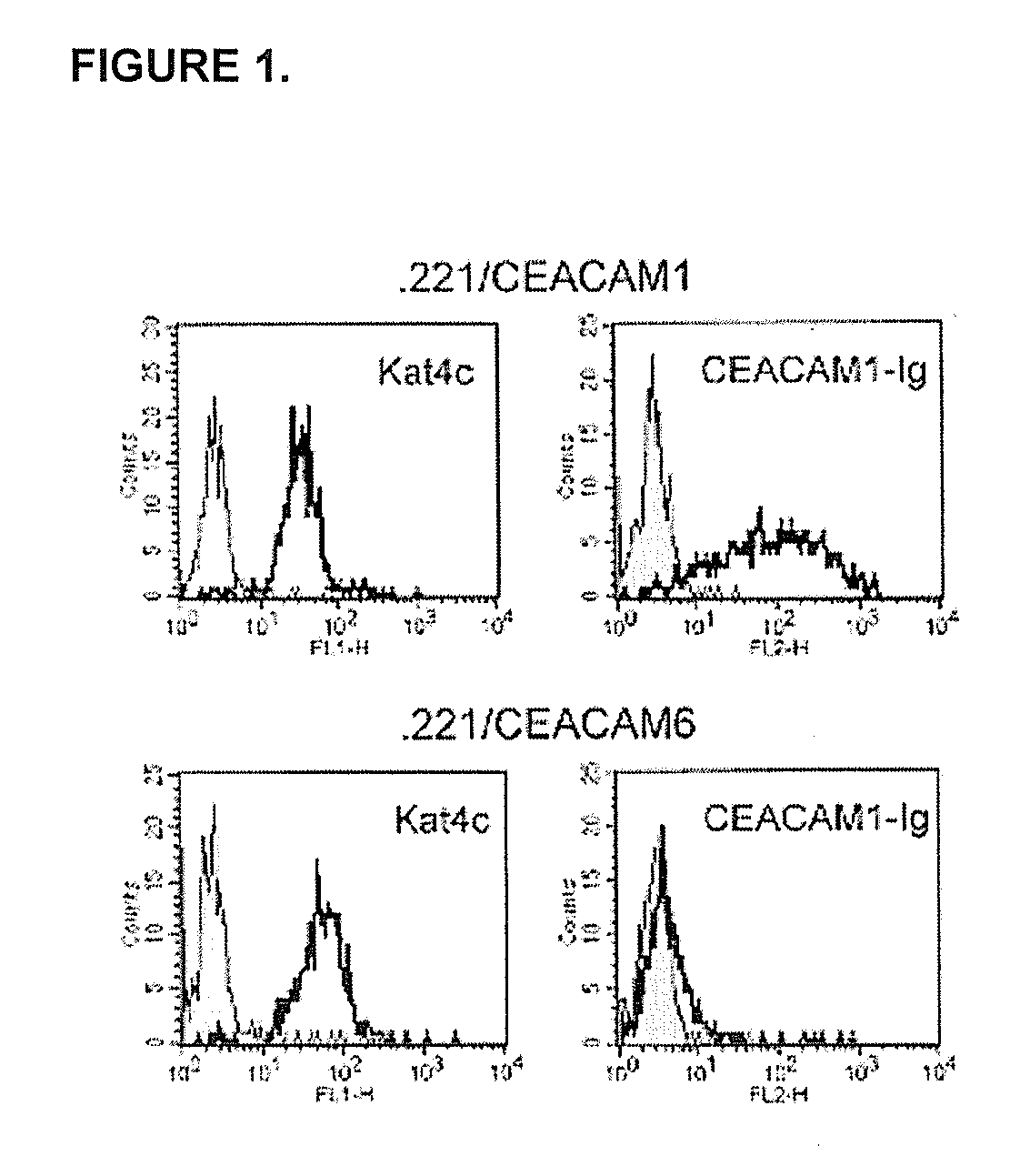
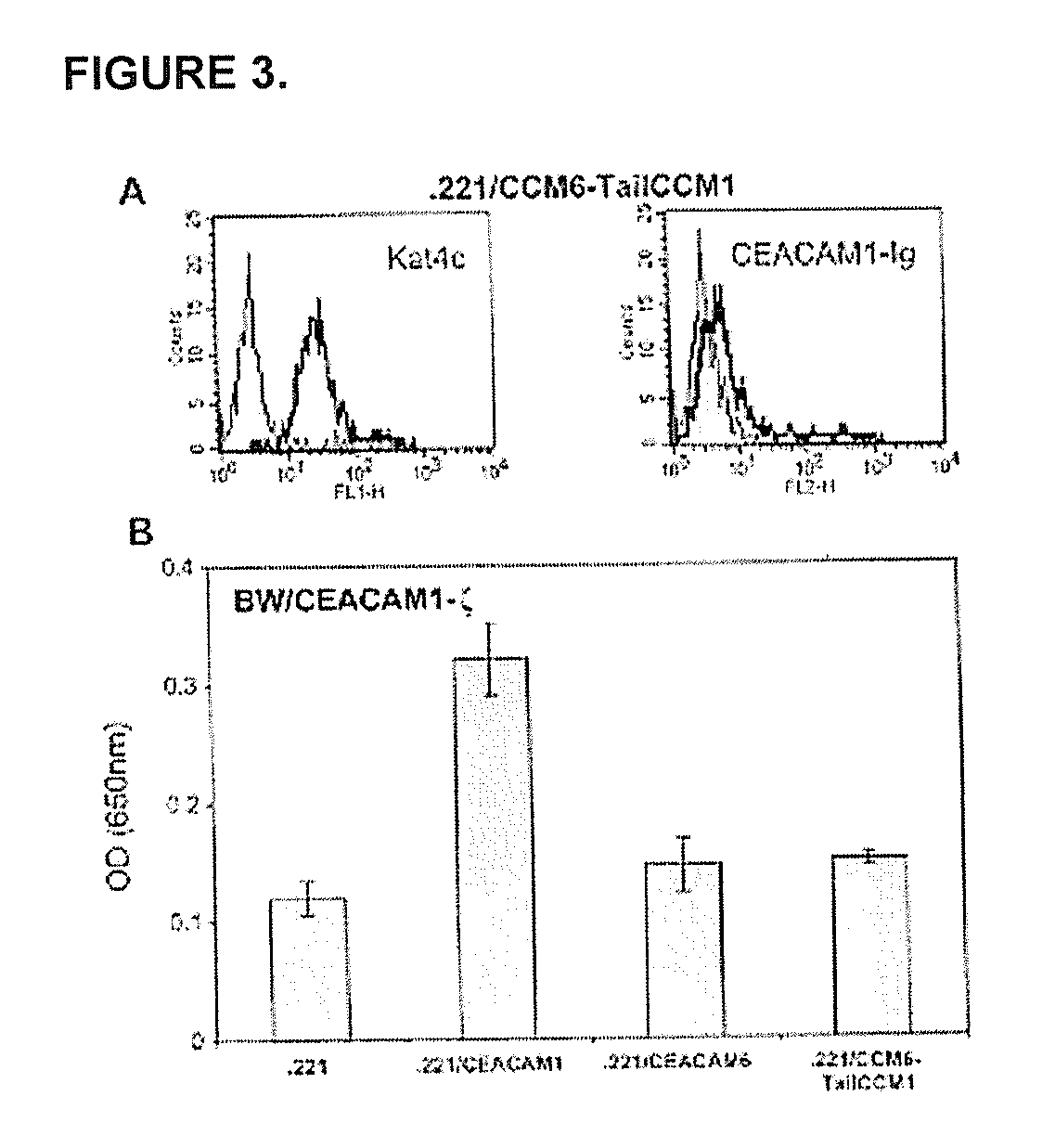
Modulation of Immunity and Ceacam1 Activity
ActiveUS20070110668A1Good curative effectCompounds screening/testingBiocideLymphocyteSpecific immunity
The present technology comprises methods for regulating an the immune system, and in particular methods for the regulation of a specific immune response, including the regulation of lymphocyte activity. Methods of the present technology comprise both the negative and positive modulation of CEACAM1 protein function.
Owner:MARKEL GAL
Rapid quantitative analysis of proteins or protein function in complex mixtures
InactiveUS7544518B2Facilitates quantitative determinationFacilitates quantitative determination of the absolute amountsComponent separationMaterial analysis by electric/magnetic meansIsotopic labelingProtein expression profile
Analytical reagents and mass spectrometry-based methods using these reagents for the rapid, and quantitative analysis of proteins or protein function in mixtures of proteins. The methods employ affinity labeled protein reactive reagents having three portions: an affinity label (A) covalently linked to a protein reactive group (PRG) through a linker group (L). The linker may be differentially isotopically labeled, e.g., by substitution of one or more atoms in the linker with a stable isotope thereof. These reagents allow for the selective isolation of peptide fragments or the products of reaction with a given protein (e.g., products of enzymatic reaction) from complex mixtures. The isolated peptide fragments or reaction products are characteristic of the presence of a protein or the presence of a protein function in those mixtures. Isolated peptides or reaction products are characterized by mass spectrometric (MS) techniques. The reagents also provide for differential isotopic labeling of the isolated peptides or reaction products which facilitates quantitative determination by mass spectrometry of the relative amounts of proteins in different samples. The methods of this invention can be used for qualitative and quantitative analysis of global protein expression profiles in cells and tissues, to screen for and identify proteins whose expression level in cells, tissue or biological fluids is affected by a stimulus or by a change in condition or state of the cell, tissue or organism from which the sample originated.
Owner:UNIV OF WASHINGTON
Method for identifying protein functions based on protein-protein interaction network and network topological structure features
InactiveCN105138866ARobustSignificant predictive advantageSpecial data processing applicationsNODALData set
The invention discloses a method for identifying protein functions based on a protein-protein interaction network and network topological structure features. Firstly, a node and side-weighted protein-protection interaction network is established, wherein the node represents protein while the edge represents the interaction; then the nodes and the sides in the network are weighted by protein first-grade structural description and protein-protein interaction trust scoring; protection functional annotation data is collected to establish a data set, and a new protein with overall and local information network topological structure features is provided based on a graph theory; and finally, the protein functions are predicated by choosing features through adopting a minimum-redundancy maximum-correlation method and by modeling through a support vector machine. The protein function predication method is greatly better than the prior art, and has robustness on sequence similarity and sampling; and meanwhile, information of three-dimensional structure and the like of protein is not required, so that the method is simple, rapid, accurate and efficient, and the method is expected to be applied in the research fields of proteomics and the like.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
Hierarchical multi-label classification method for protein function prediction
ActiveCN106126972ASolve the multi-label problemAchieve forecastBiostatisticsProteomicsData setProtein function prediction
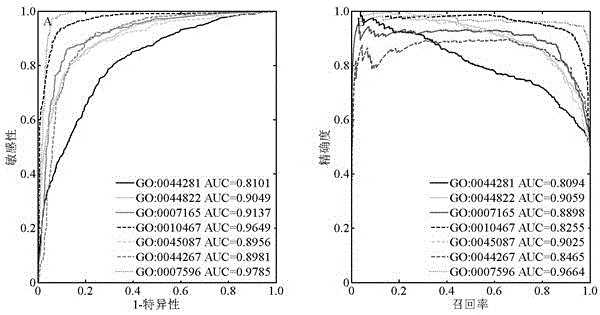
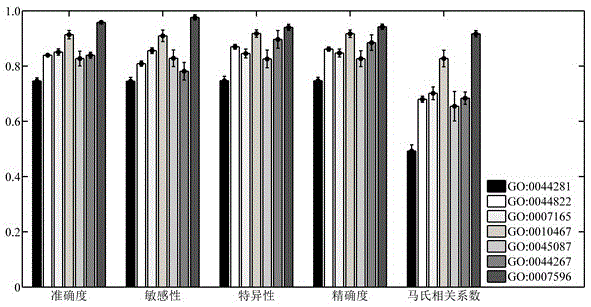
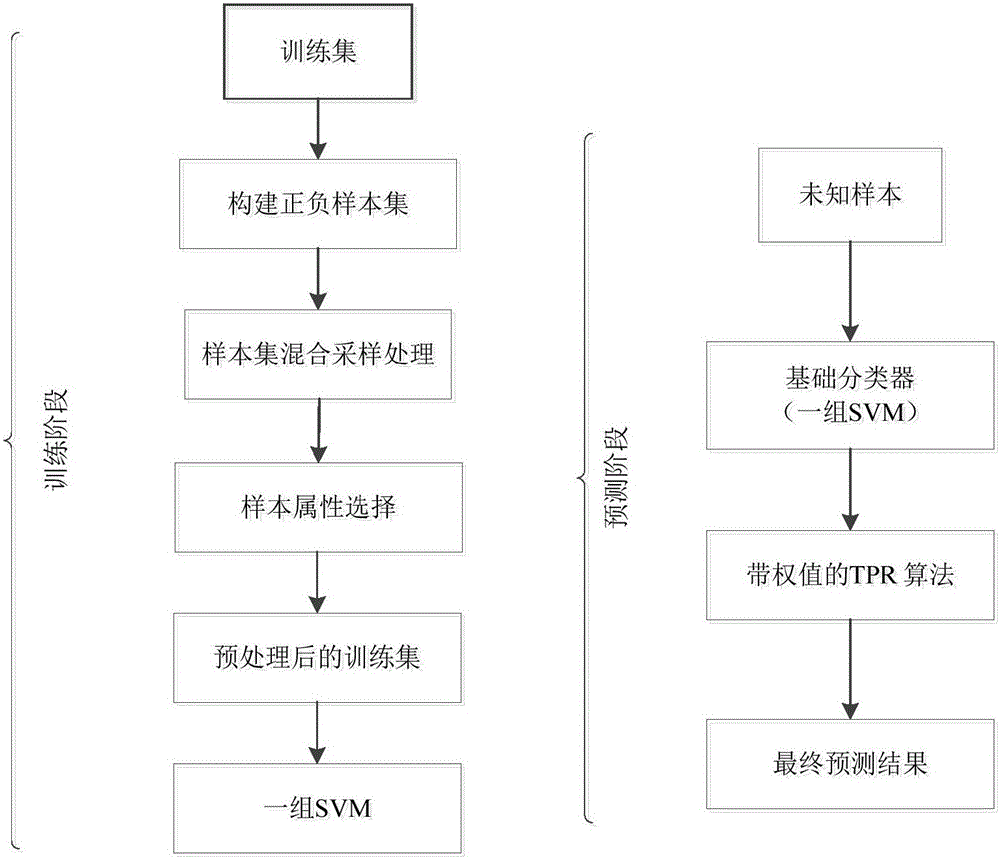
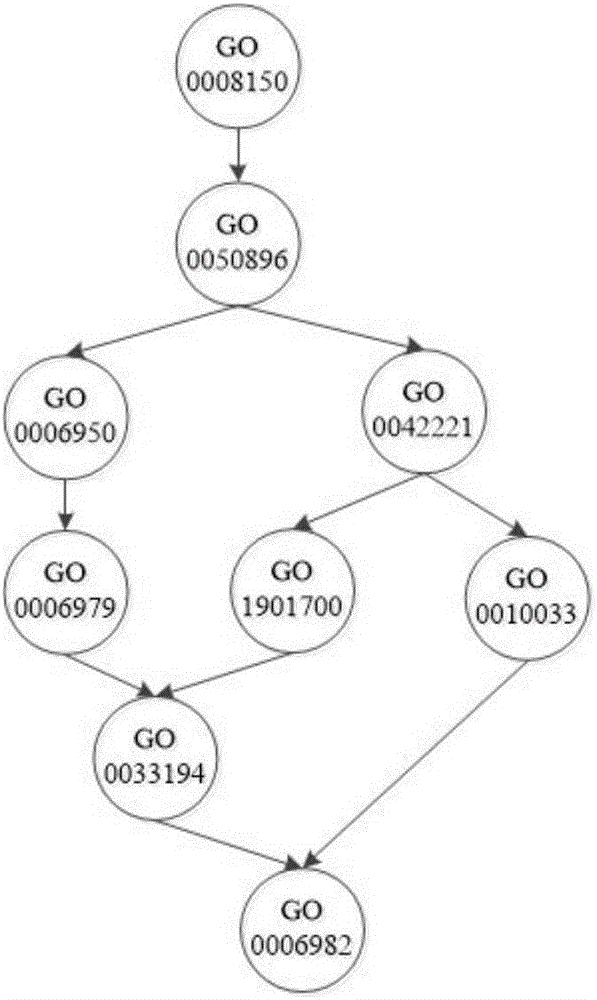
The invention relates to the field of bioinformatics and data mining, in particular to a hierarchical multi-label classification method for protein function prediction, and aims at solving the data set imbalance problem, multi-label problem and hierarchical constraint problem when the conventional classification methods are used for predicting protein functions. The method comprises the following steps of: 1, a training stage: training a data set of each node in a class label hierarchical structure by adopting an SVM classifier in the training stage so as to obtain a group of basic classifiers; and 2, a prediction stage: firstly obtaining preliminary results of unknown samples by using the group of basic classifiers obtained in the training stage in the prediction stage, and processing the results by adopting a TPR algorithm with a weight so as to obtain a final result which satisfies a hierarchical constraint condition and realize the prediction of the protein functions. The hierarchical multi-label classification method for protein function prediction is applied to the field of bioinformatics and data mining.
Owner:NAT INST OF ADVANCED MEDICAL DEVICES SHENZHEN
Small molecule-dependent inteins and uses thereof
ActiveUS9200045B2Low efficiencySlow splicingHydrolasesFusion with post-translational modification motifInteinNatural state
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Method for regulating protein function in cells in vivo using synthetic small molecules
ActiveUS8530636B2Improve survivalInhibit tumor growthOrganic active ingredientsBiocideIn vivoProtein function
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Mono- and bis-indolylquinones and prophylactic and therapeutic uses thereof
InactiveUS6376529B1Effective treatmentLower blood sugar levelsBiocideOrganic chemistryMedicineInsulin deficiency
The present invention relates to a class of indolylquinone compounds that inhibit GRB-2 adaptor protein function, pharmaceutical compositions comprising these compounds, and methods for ameliorating the symptoms of cell proliferative disorders associated with GRB-2 adaptor protein function using these compounds. The present invention further relates to methods for treating insulin-related disorders, such as diabetes, insulin resistance, insulin deficiency and insulin allergy, and for ameliorating the symptoms of insulin-related disorders, using certain indolylquinone compounds and pharmaceutical compositions thereof. The present invention also relates to novel synthetic methods for the preparation of mono- and bis-indolylquinone compounds.
Owner:SUGEN INC
Amphipathic polyurethane with anti-bacterial and anti-protein function as well as preparation method and application of amphipathic polyurethane
ActiveCN105199070AImprove stain resistanceGood biological stabilityAntifouling/underwater paintsPaints with biocidesCross-linkHydrolysis
The invention discloses amphipathic polyurethane resin based on polysiloxane and a preparation method of the amphipathic polyurethane resin, as well as an anti-bacterial anti-protein performance of the amphipathic polyurethane resin. The preparation method comprises the following steps: carrying out a reaction between tert-Butyl bromoacetate and N-methyldiethanolamine under a certain condition to obtain a quaternary ammonium diol; carrying out cross-link between the quaternary ammonium diol and hydroxymethyl-terminated polydimethylsiloxane; carrying out hydrolysis to obtain the amphipathic polyurethane of which a molecular chain segment has not only a hydrophilic ingredient but also a hydrophobic ingredient. The amphipathic polyurethane has all of the relatively high hydrophilicity of zwitter ions, the low surface energy and dirt discharging superiority of polysiloxane, and the stability of polyurethane, thereby having an excellent practical application value in the anti-bacterial and anti-protein field.
Owner:ZHEJIANG UNIV +1
Nitrosated and nitrosylated heme proteins
Nitrosylation of proteins and amino acid groups enables selective regulation of protein function, and also endows the proteins and amino acids with additional smooth muscle relaxant and platelet inhibitory capabilities. Thus, the invention relates to novel compounds achieved by nitrosylation of protein thiols. Such compounds include: S-nitroso-heme proteins, S-nitroso-t-PA, S-nitroso-cathepsin; S-nitroso-lipoproteins; and S-nitroso-immunoglobulins. The invention also relates to therapeutic use if S-nitroso-protein compounds for regulating protein function, cellular metabolism and effecting vasodilation, platelet inhibition, relaxation of non-vascular smooth muscle, and increasing blood oxygen transport by hemoglobin and myoglobin. The compounds are also used to deliver nitric oxide in its most bioactive form in order to achieve the effects described above, or for in vitro nitrosylation of molecules present in the body. The invention also relates to the nitrosylation of oxygen, carbon and nitrogen moieties present on proteins and amino acids, and the use thereof to achieve the above physiological effects.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC
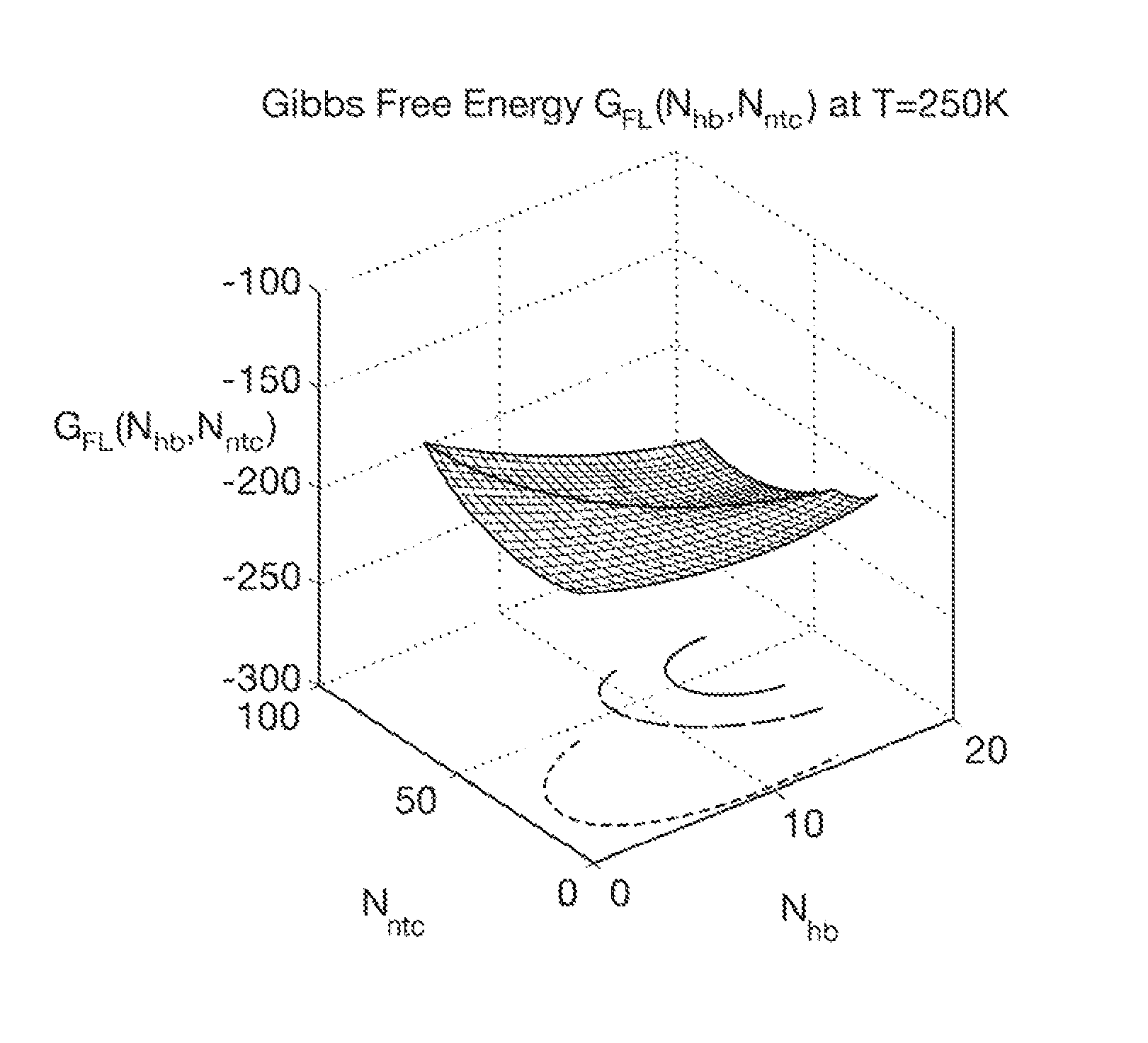
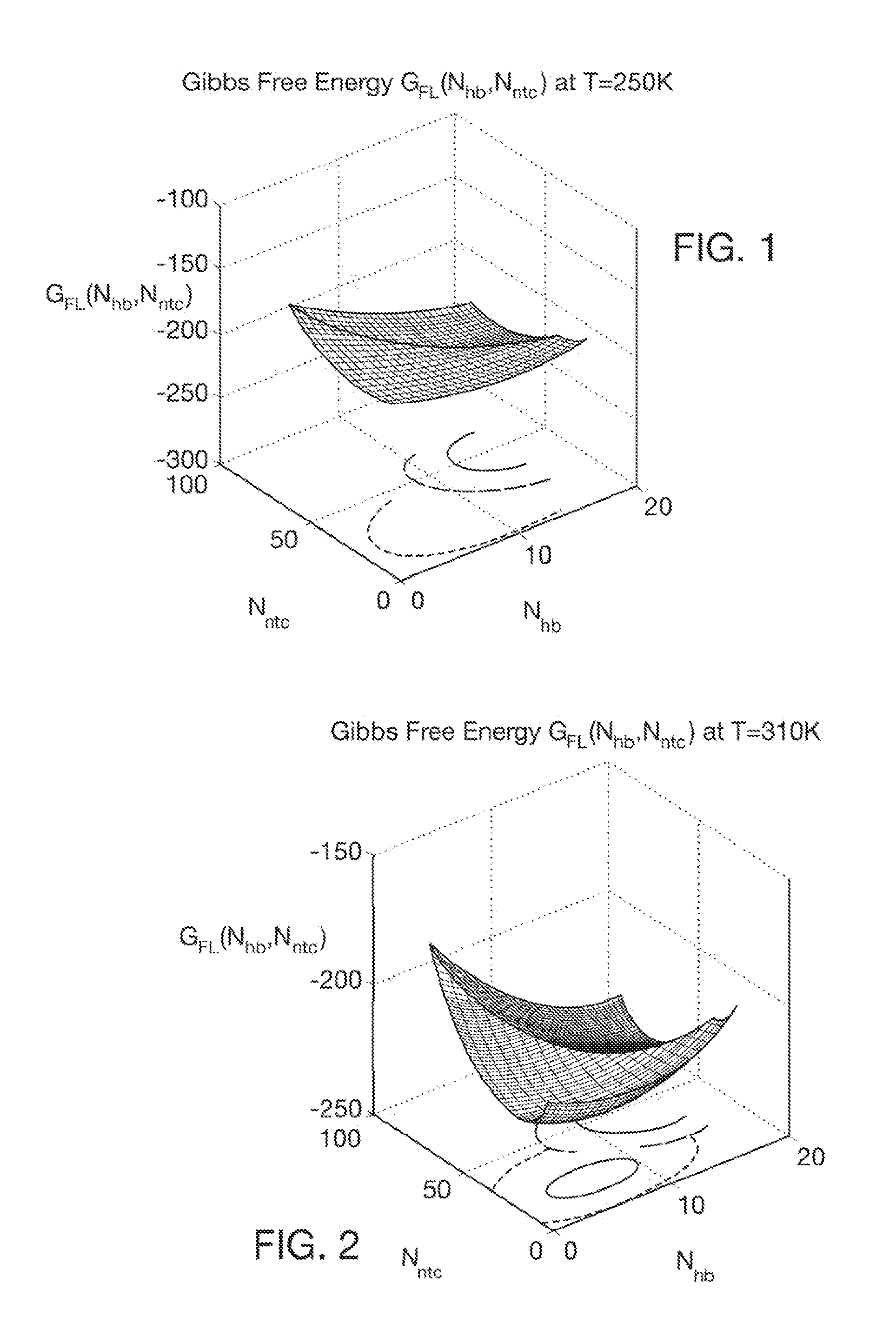
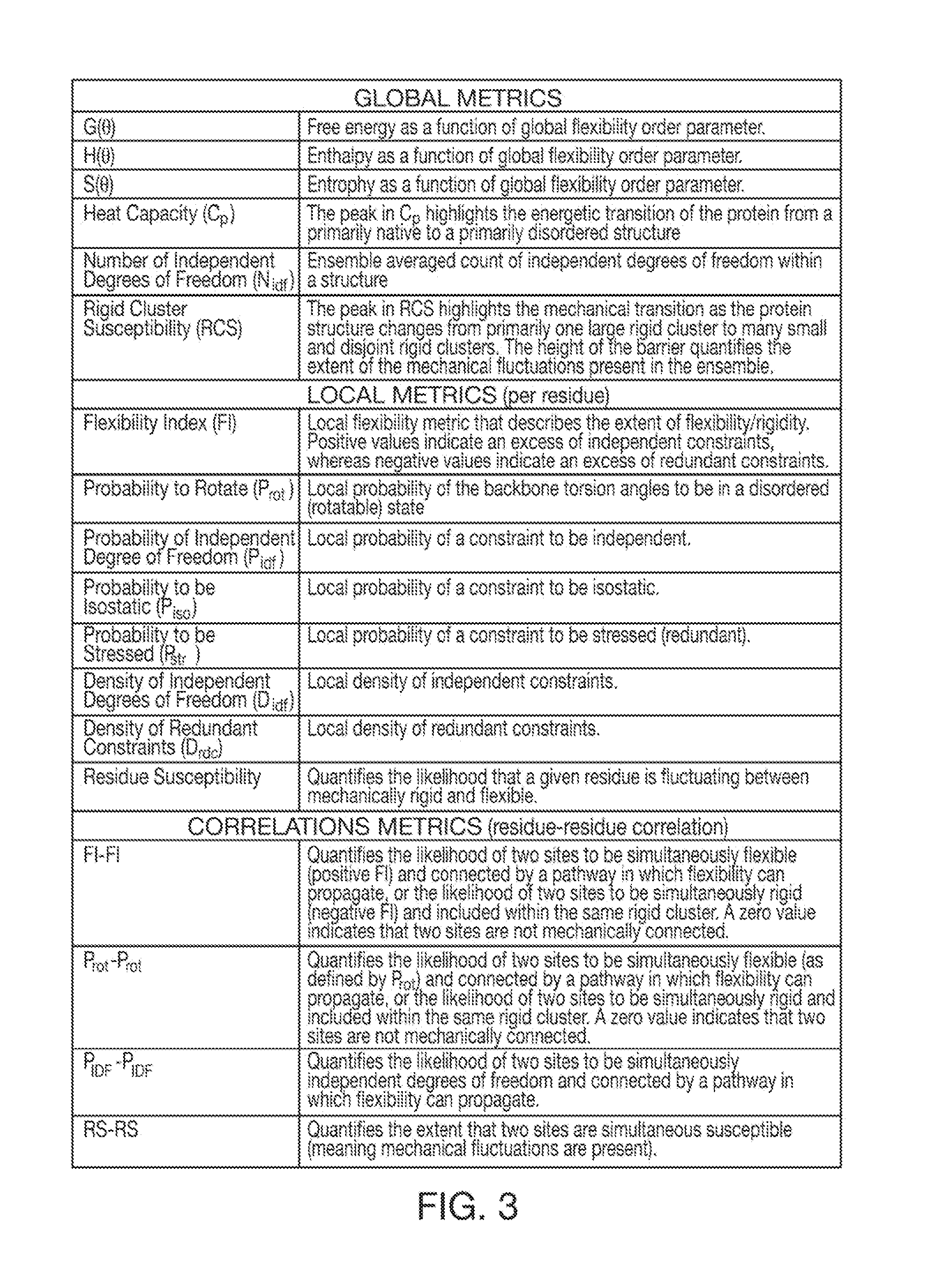
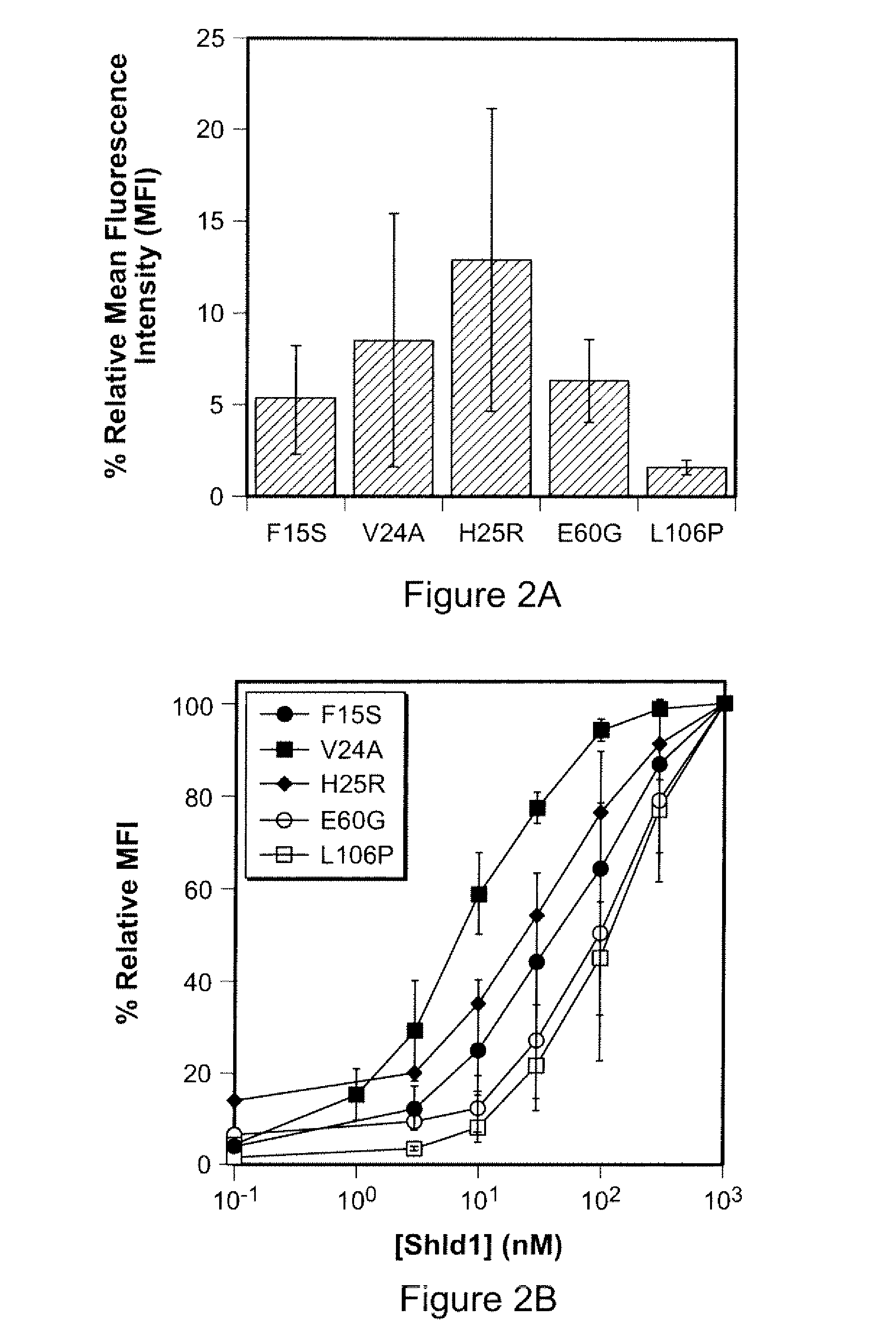
Computer implemented system for protein and drug target design utilizing quantified stability and flexibility relationships to control function
InactiveUS8374828B1High catalytic efficiencyFast computerMolecular designMicrobiological testing/measurementHigh dimensionalDrug target
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
Method for Regulating Protein Function in Cells In Vivo Using Synthetic Small Molecules
ActiveUS20100034777A1Improve survivalInhibit tumor growthBiocideOrganic active ingredientsIn vivoProtein function
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Methods for conducting assays for enzyme activity on protein microarrays
The present invention relates to methods of conducting assays for enzymatic activity on microarrays useful for the large-scale study of protein function, screening assays, and high-throughput analysis of enzymatic reactions. The invention relates to methods of using protein chips to assay the presence, amount, activity and / or function of enzymes present in a protein sample on a protein chip. In particular, the methods of the invention relate to conducting enzymatic assays using a microarray wherein a protein and a substance are immobilized on the surface of a solid support and wherein the protein and the substance are in proximity to each other sufficient for the occurrence of an enzymatic reaction between the substance and the protein. The invention also relates to microarrays that have an enzyme and a substrate immobilized on their surface wherein the enzyme and the substrate are in proximity to each other sufficient for the occurrence of an enzymatic reaction between the enzyme and the substrate.
Owner:PROTOMETRIX
Uses of spatial configuration to modulate protein function
ActiveCN101001644APeptide/protein ingredientsMedical atomisersRecombinant Super-compound InterferonInterferon alpha
This invention provides a method for preventing or treating Severe Acute Respiratory Syndrome in a subject comprising administering to the subject an effective amount of recombinant super-compound interferon or a functional equivalent thereof. This invention provides a method for inhibiting the causative agent of Severe Acute Respiratory Syndrome comprising contacting the agent with an effective amount of super-compound interferon or its equivalent.
Owner:SUPERLAB FAR EAST LTD
Rice bacterial blight resistance related proteins OsBBR1 and coding genes and application thereof
ActiveCN109369790AImprove disease resistanceReduced disease resistancePlant peptidesFermentationDiseaseBiotechnology
The invention discloses rice bacterial blight resistance related proteins OsBBR1 and coding genes and application thereof. The rice bacterial blight resistance related proteins OsBBR1 are following (A1), (A2) or (A3), wherein (A1) the proteins have the amino acid sequence of the 240-438 bits in the third sequence, (A2) the proteins are obtained in the process that the amino acid sequence shownin the 240-438 bits of the third sequence in a sequence table is subjected to substitution and / or deletion and / or addition of one or more amino acid residues, and have the same function, and (A3)the fusion proteins are obtained by connecting labels to the N ends or / and the C ends of the (A1) or the (A2). Experiments show that plants transfecting OsBBR1 genes can significantly improve diseaseresistance of plants, the disease resistance of the plants missing the OsBBR1 protein function after gene editing is lowered, which indicates that the OsBBR1 proteins and the coding genes thereof canbe used for improving the disease resistance of the plants and have great significance for cultivating the disease-resistant plants.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI +1
Nitrosylation of protein SH groups and amino acid residues as a therapeutic modality
InactiveUS7157500B2Organic active ingredientsHydrolasesVascular smooth muscle relaxationPlatelet inhibition
Nitrosylation of proteins and amino acid groups enables selective regulation of protein function, and also endows the proteins and amino acids with additional smooth muscle relaxant and platelet inhibitory capabilities. Thus, the invention relates to novel compounds achieved by nitrosylation of protein thiols. Such compounds include: S-nitroso-t-PA, S-nitroso-cathepsin; S-nitroso-lipoprotein; and S-nitroso-immunoglobulin. The invention also relates to therapeutic use of S-nitroso-protein compounds for regulating protein function, cellular metabolism and effecting vasodilation, platelet inhibition, relaxation of non-vascular smooth muscle, and increasing blood oxygen transport by hemoglobin and myoglobin. The compounds are also used to deliver nitric oxide in its most bioactive form in order to achieve the effects described above, or for in vitro nitrosylation of molecules present in the body. The invention also relates to the nitrosylation of oxygen, carbon and nitrogen moieties present on proteins and amino acids, and the use thereof to achieve the above physiological effects.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC
Methods for comparing functional sites in proteins
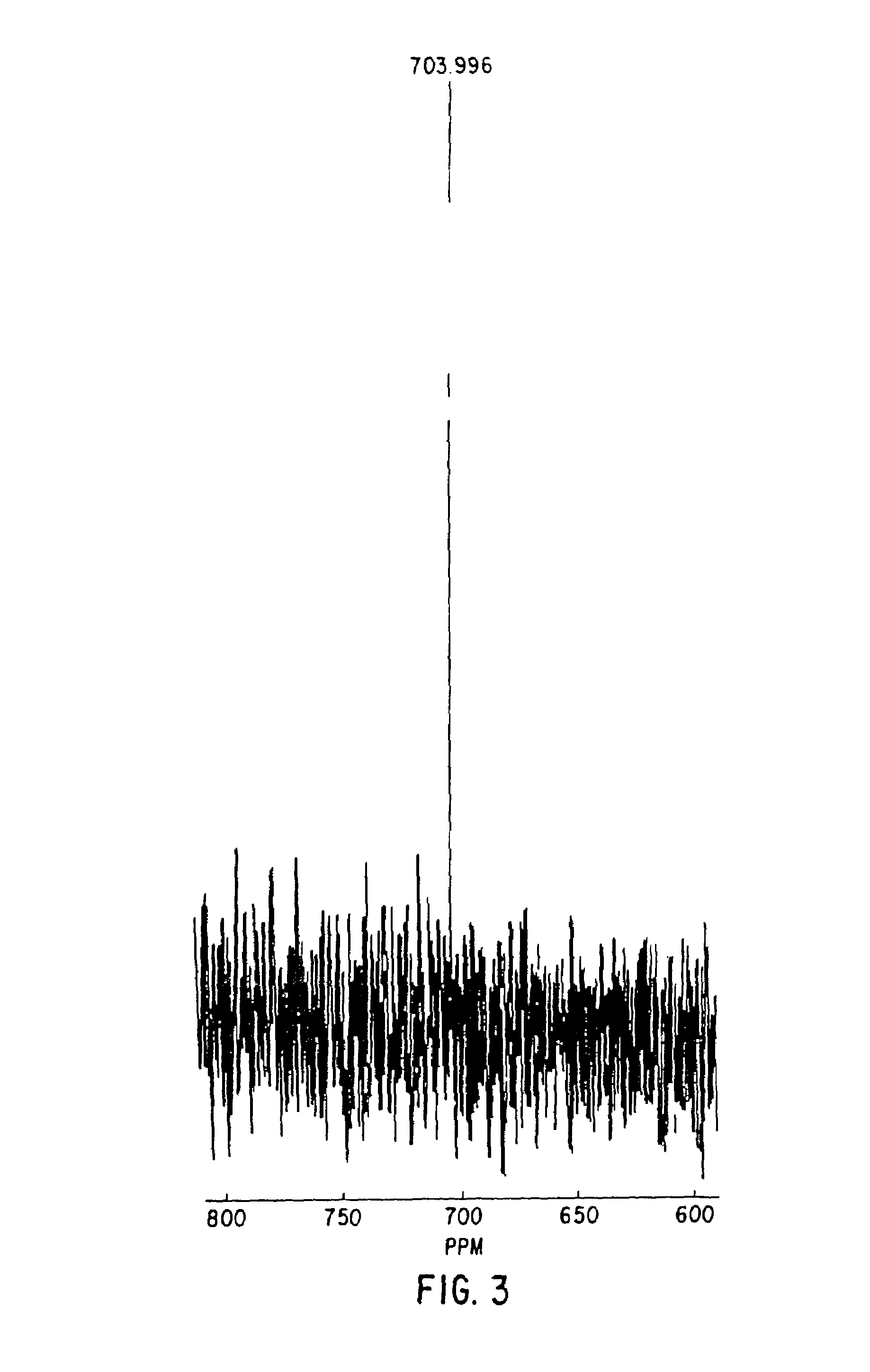
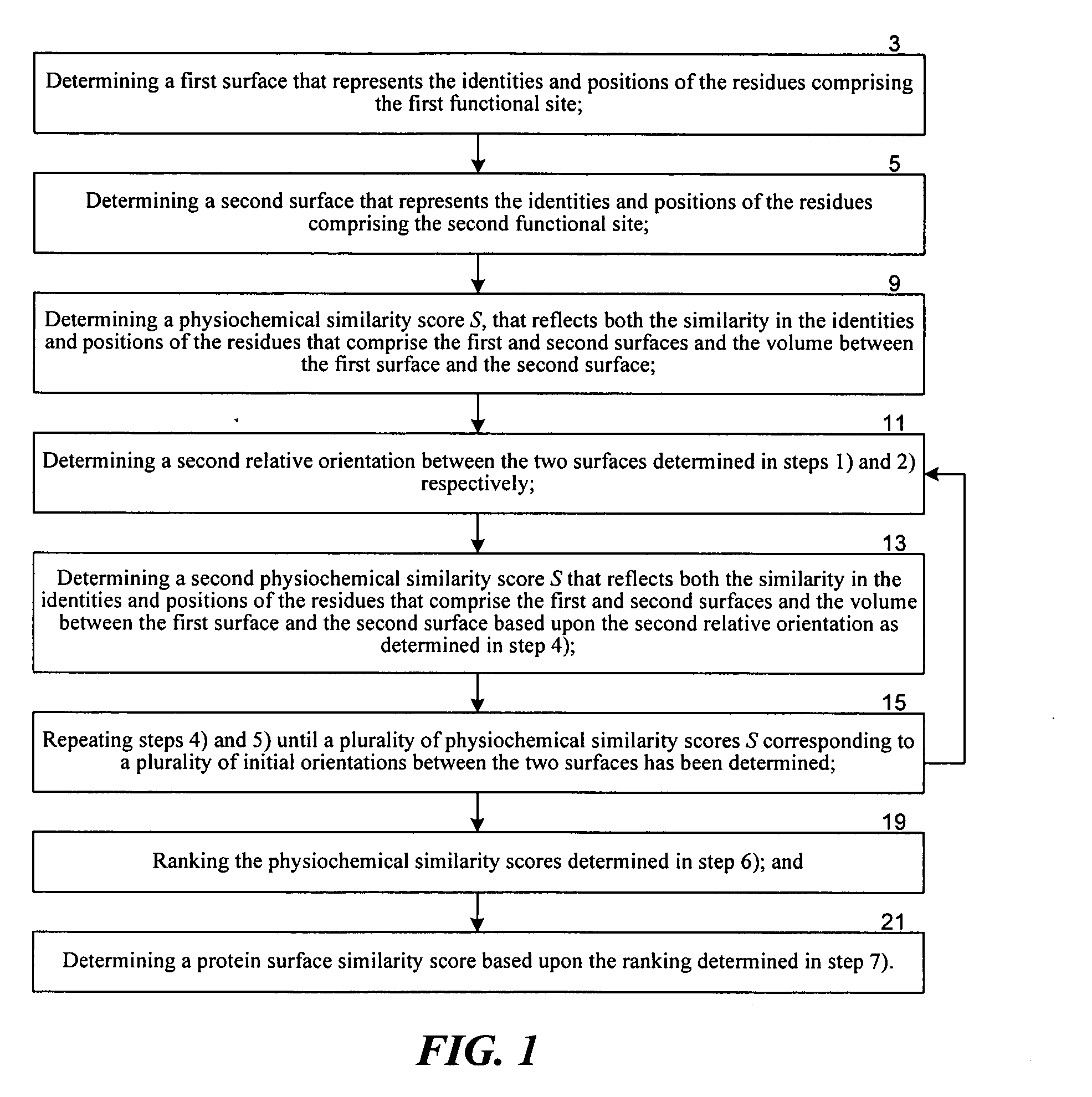
InactiveUS20050192758A1Effective calculationComputationally efficientProteomicsGenomicsTARP ProteinProtein surface
The present invention relates to methods and systems for representing and scoring the similarity of two protein by iteratively rotating and translating one protein surface representation relative to the other protein surface representation in order to maximize (or minimize) a score that represents both the volume between the two surface representations and the similarity in the identities and positions of the residues comprising the two protein surfaces. In another aspect of the invention, such methods and systems are used to compare and annotate a protein comprising a putative functional site of unknown function with a database of reference proteins of known function.
Owner:EIDOGEN SERTANTY INC
Hcv coreceptor and methods of use thereof
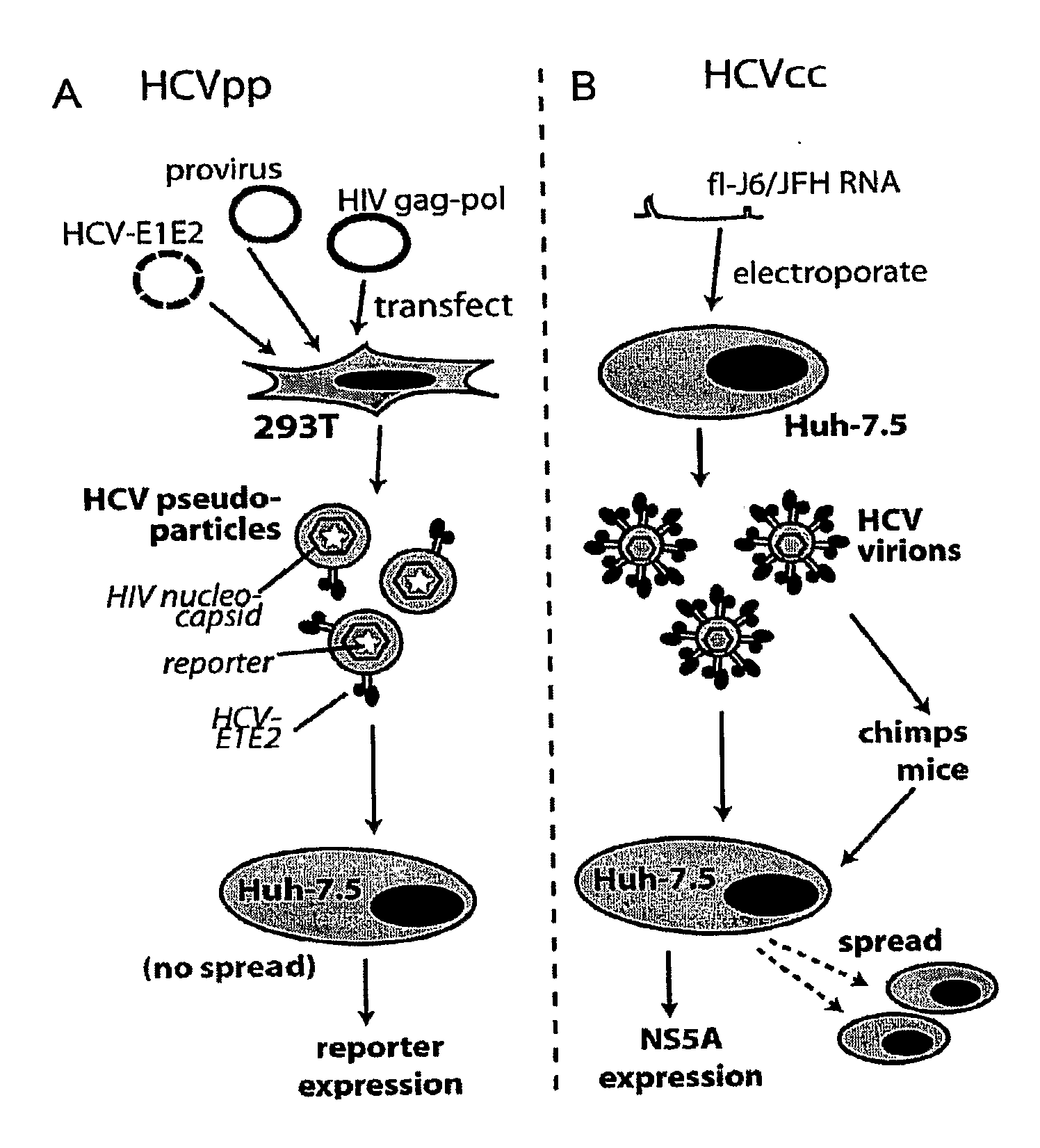
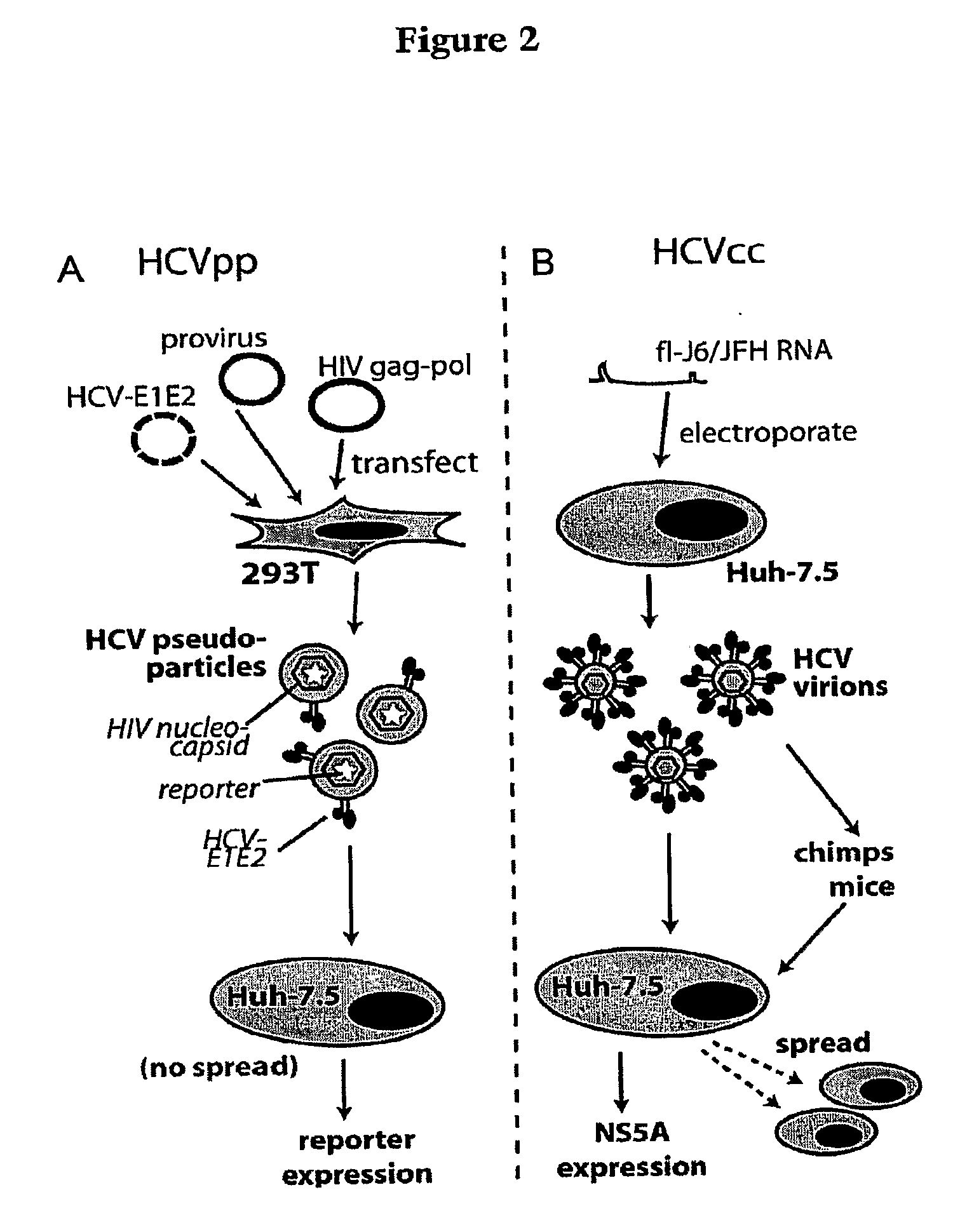
ActiveUS20090098123A1Prevents and mitigates interactionReduce retentionPeptide/protein ingredientsMicrobiological testing/measurementCo-receptorProtein function
The invention relates to the discovery that the Claudin-1 protein functions as a co-receptor for entry of HCV into cells. Methods of inhibiting, preventing or mitigating HCV infections by inhibiting HCV interactions with Claudin-1 are provided. Methods of identifying agents or compounds that interfere with HCV interactions with Claudin-1 are also provided. Finally, useful kits, cell culture compositions, agents, and compounds related to the inhibition of HCV interactions with Claudin-1 are also disclosed.
Owner:THE ROCKEFELLER UNIV
Marking method and system for protein functions
ActiveCN106599611AImprove labeling abilitySolve the costProteomicsGenomicsProtein insertionGene coexpression
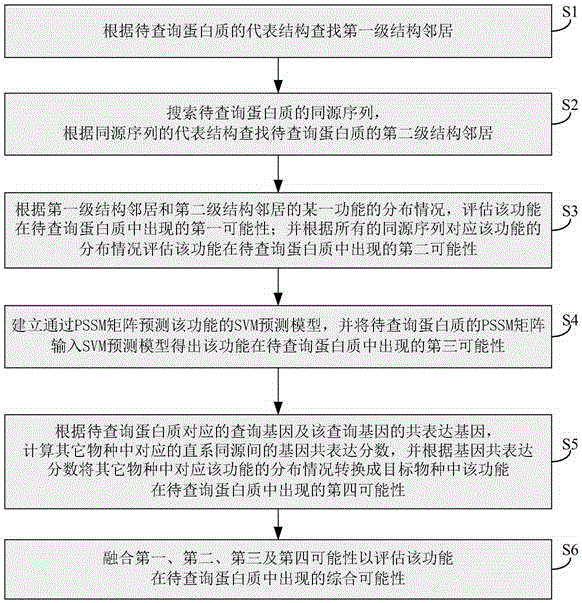
The invention relates to the technical field of bioinformation, and discloses a marking method and system for protein functions. Therefore, protein marking performance is improved, expensive cost of a bioexperiment method and poor efficiency are solved. The method comprises following steps: estimating the first possibility of a certain function in a to-be-inquired protein according to a first-stage structure neighborhood and a second-stage structure neighborhood; estimating the second possibility of the certain function in the to-be-inquired protein according to all homologous sequences; inputting a PSSM matrix of the to-be-inquired protein into an SVM prediction model to obtain the third possibility of the certain function in the to-be-inquired protein; converting the distribution of the function corresponding to other species according to the gene co-expression fraction into the fourth possibility of the function occurring in a target species in the to-be-inquired protein; and mixing the first possibility, the second possibility, the third possibility and the fourth possibility to estimate the comprehensive possibility of the function in the to-be-inquired protein.
Owner:CENT SOUTH UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com