Protein complex recognizing method based on range estimation
A technology of protein complexes and recognition methods, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as insurmountable efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
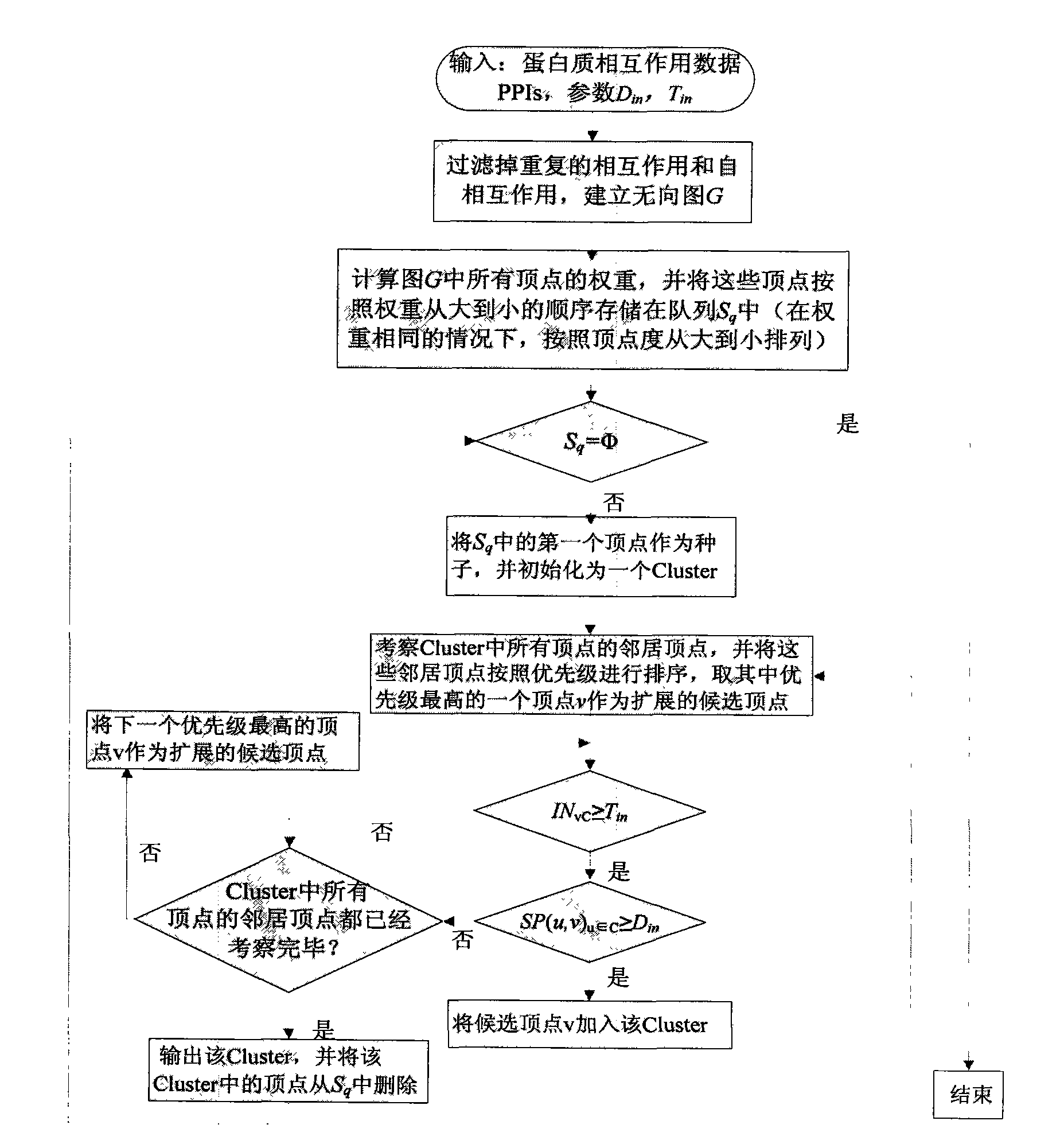
[0020] 1. Statistical analysis of topological features of known protein complexes
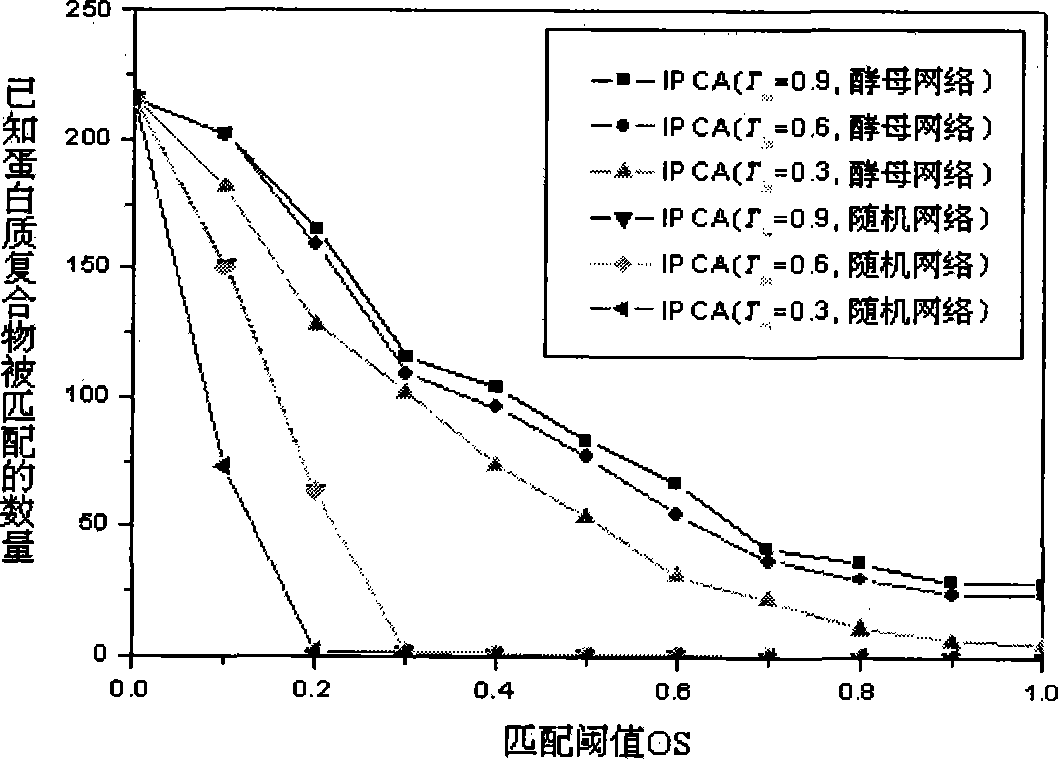
[0021] The most widely studied species is yeast, and there are already a number of experimentally determined yeast protein complexes. The present invention downloads the known yeast protein complex and yeast protein interaction network data from the MIPS (Munich Information center for Protein Sequences) database. The interaction data were removed from self-interactions and redundant interactions, and the final protein interaction network consisted of 4546 yeast proteins and 12319 pairs of interactions. The average clustering coefficient of the entire network is 0.4, the network diameter is 13, and the characteristic path length (ie, the average value of the shortest path length between any two vertices in the network) is 4.42. The protein complex data set has a total of 216 after removing complexes with only one protein. The smallest complex includes 2 proteins, the largest complex includes 81...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com