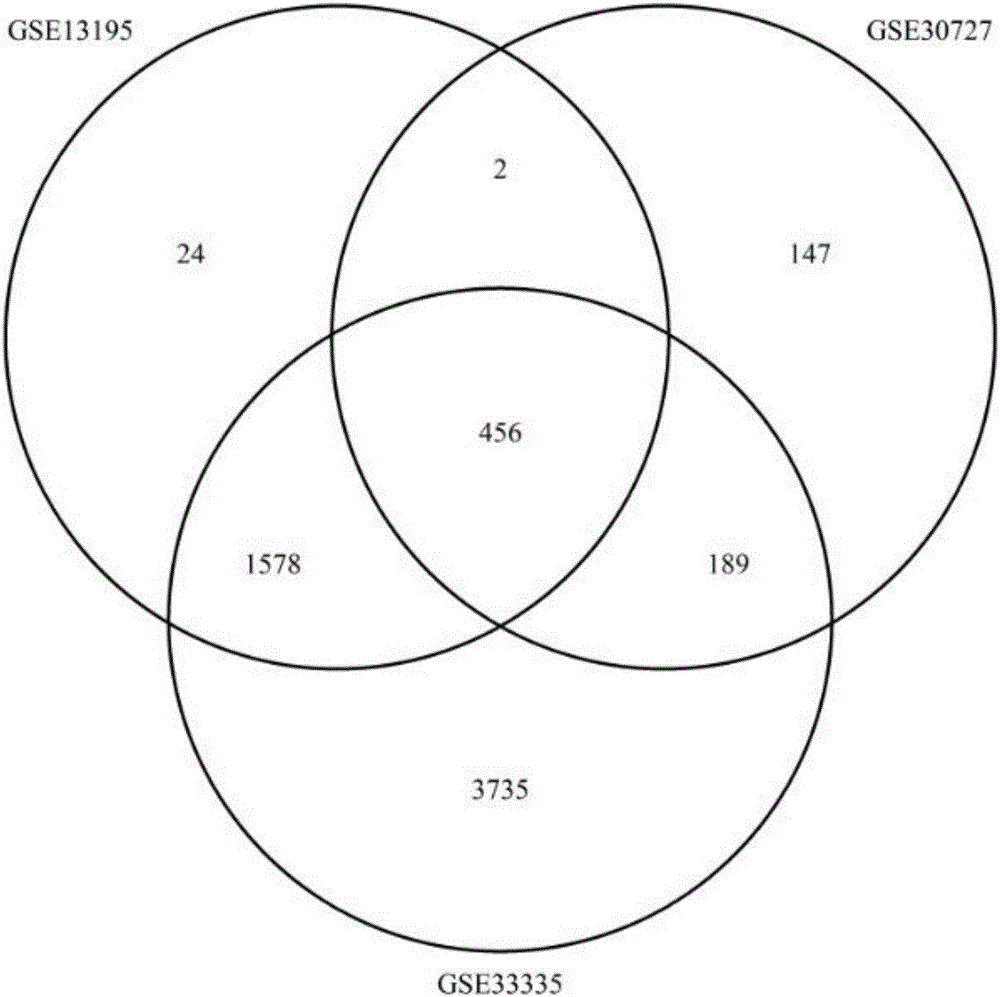
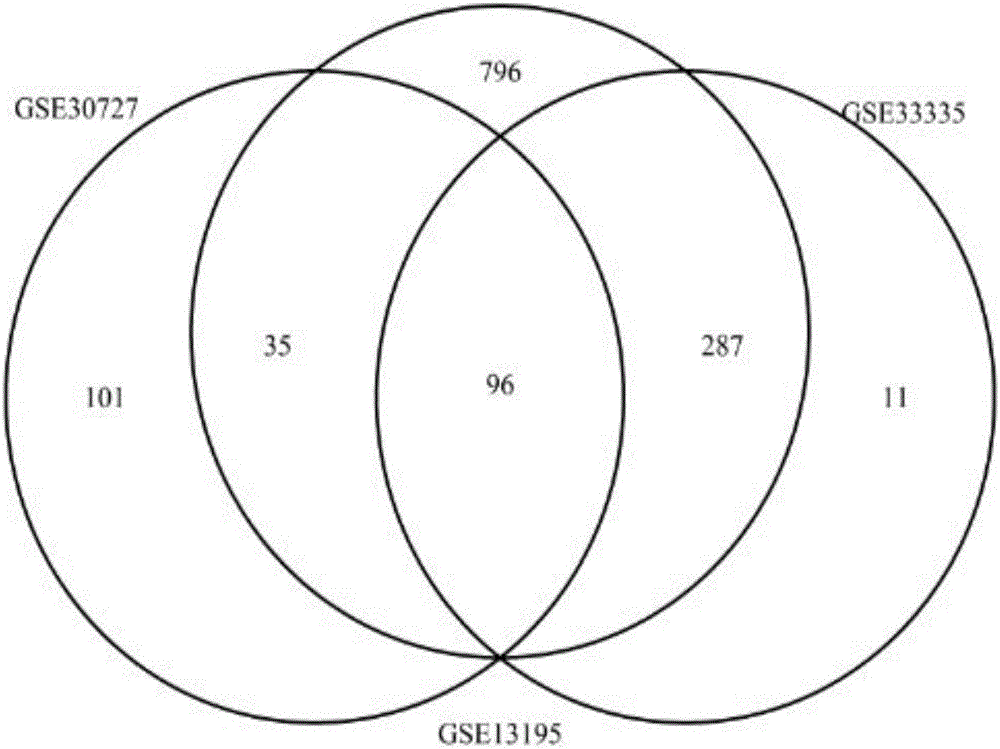
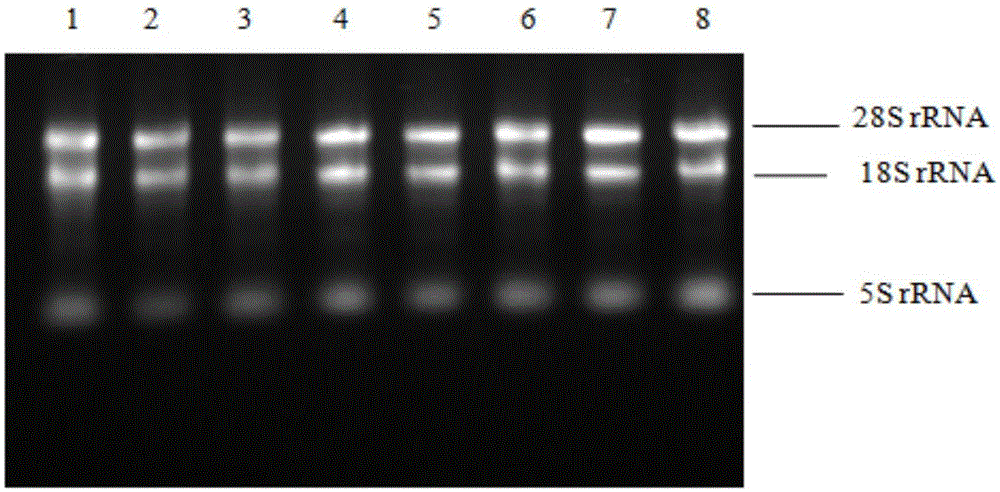
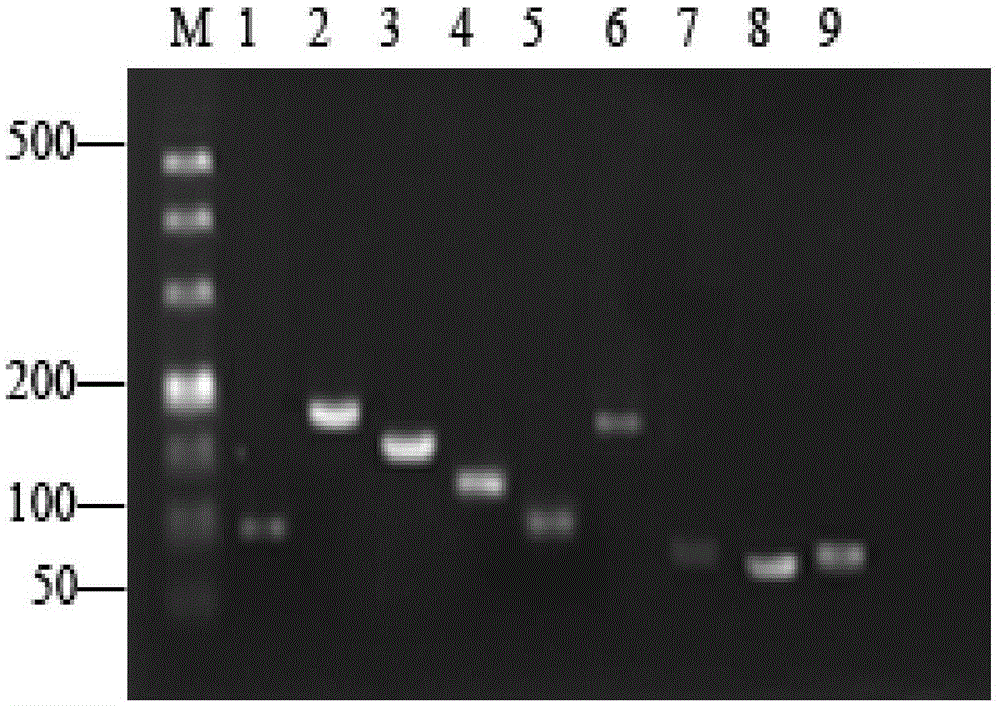
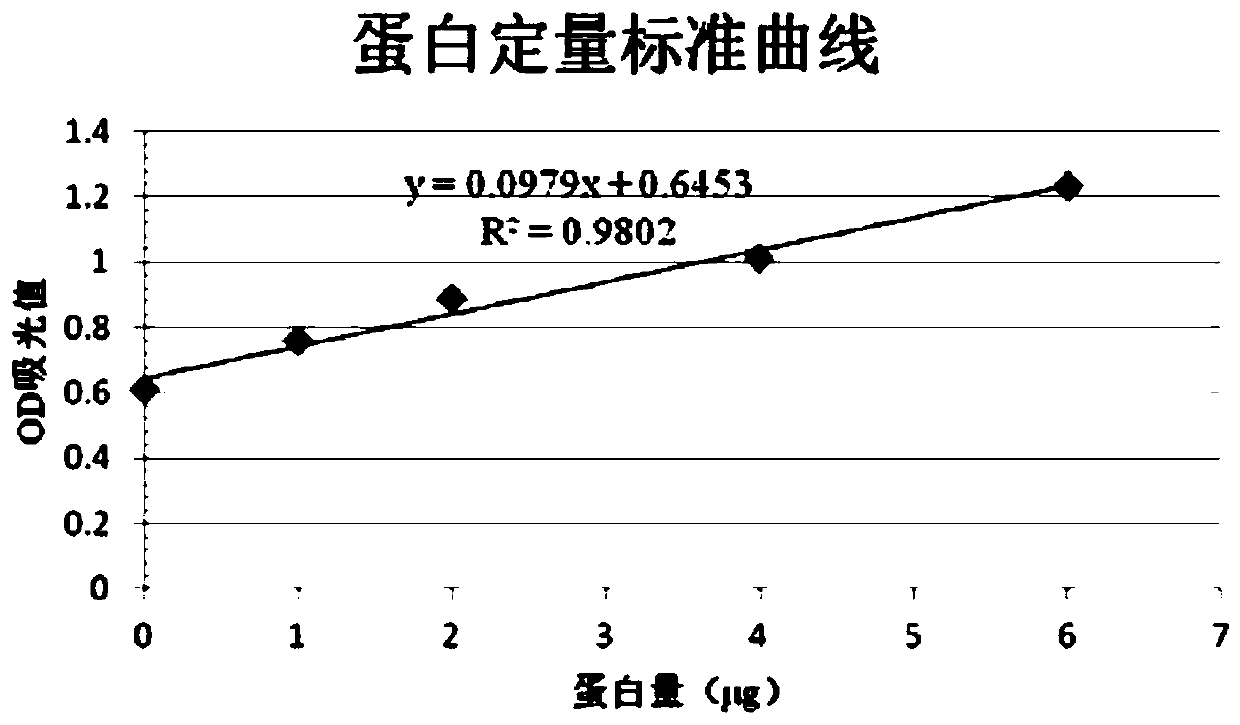
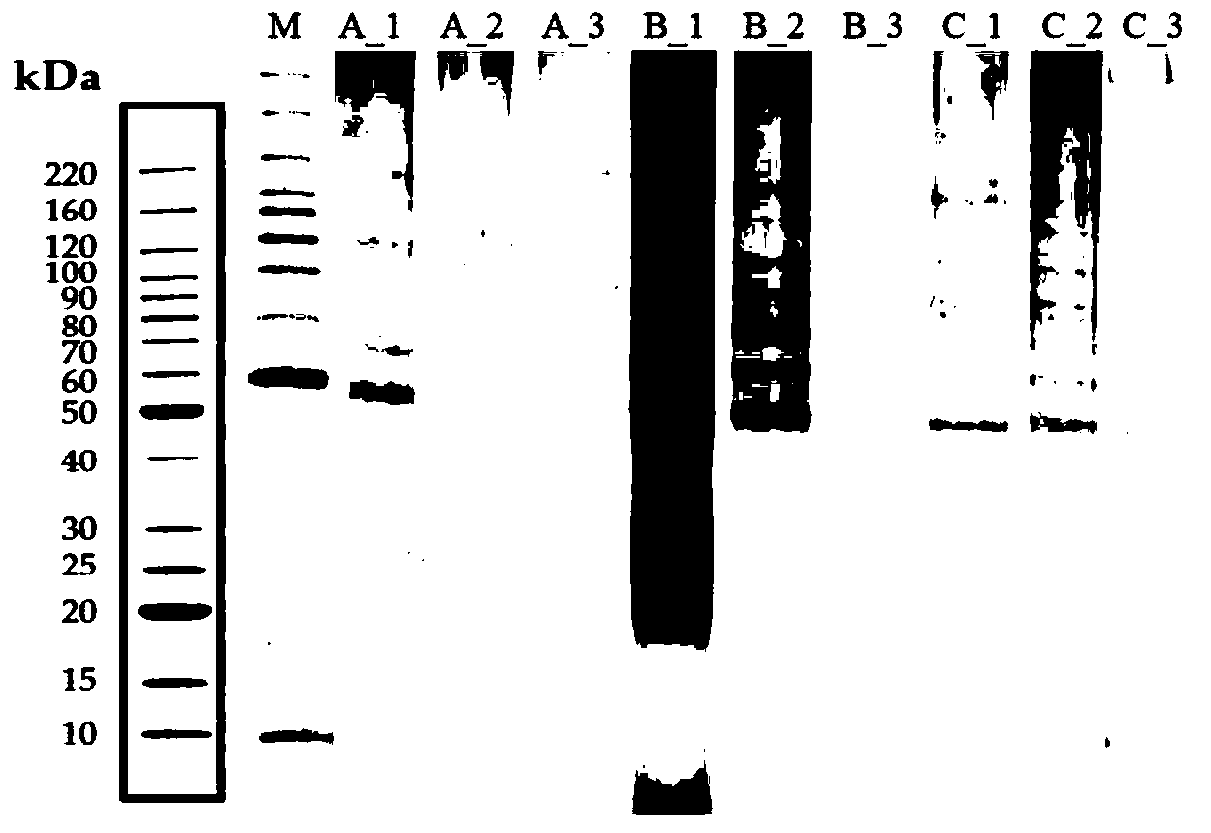
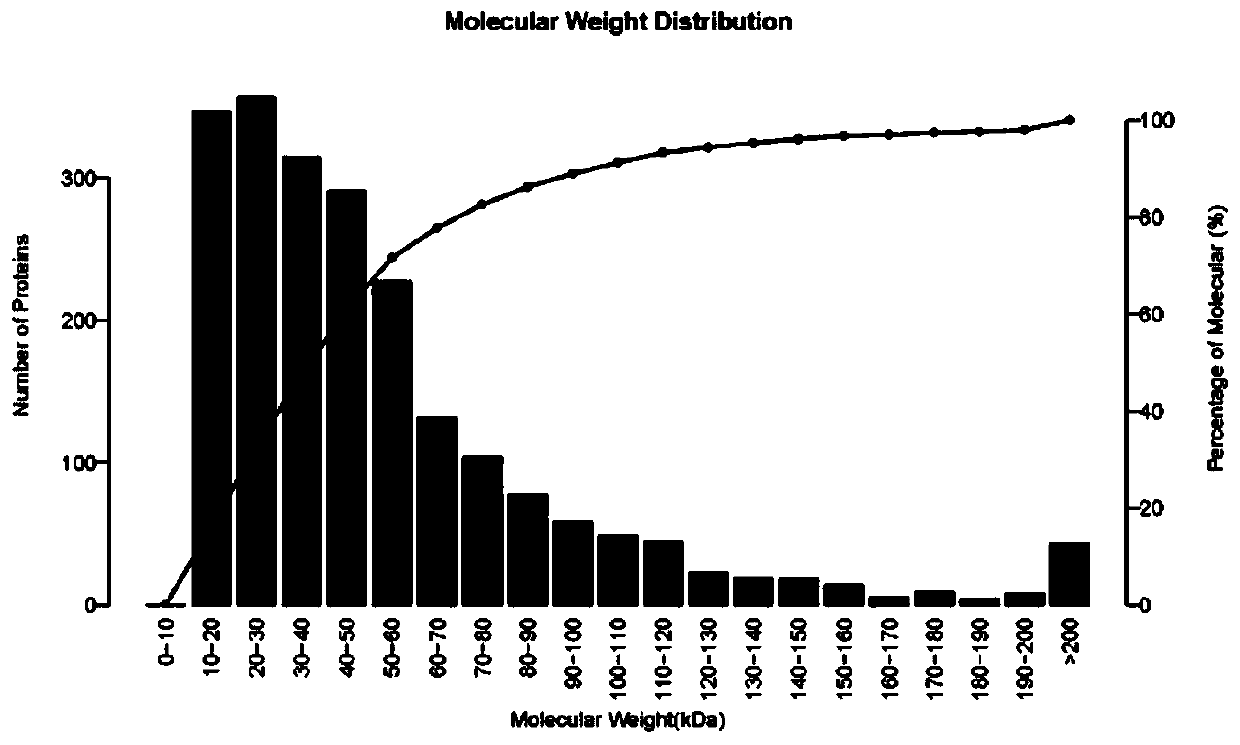
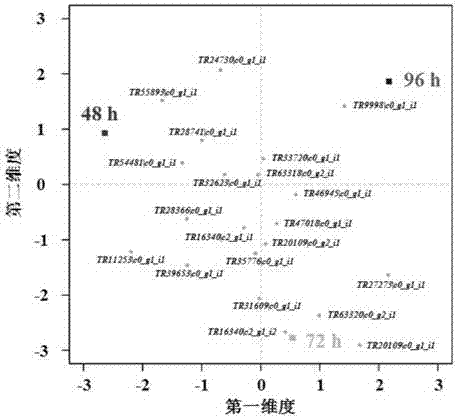
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
44 results about "Differential expression analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
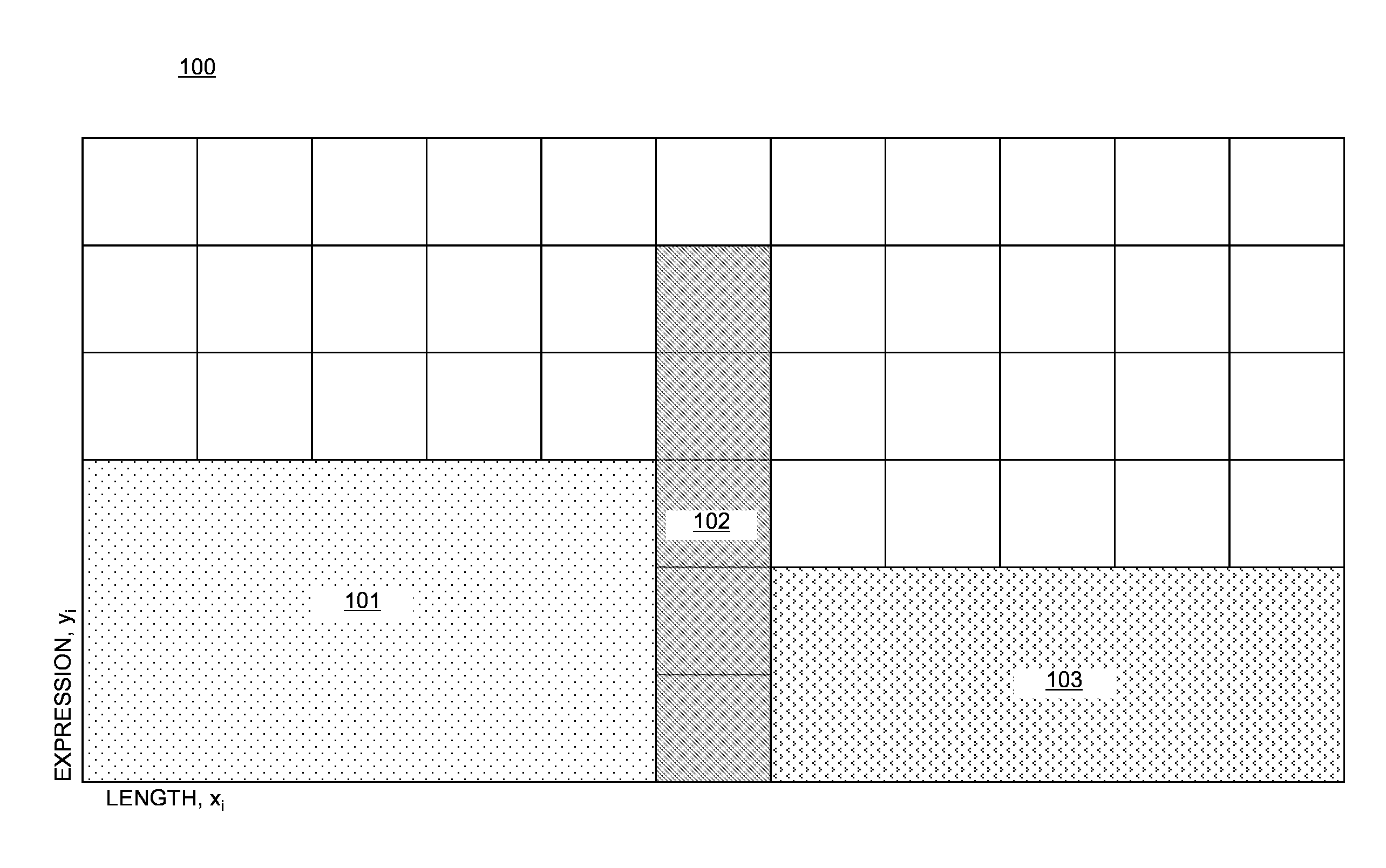
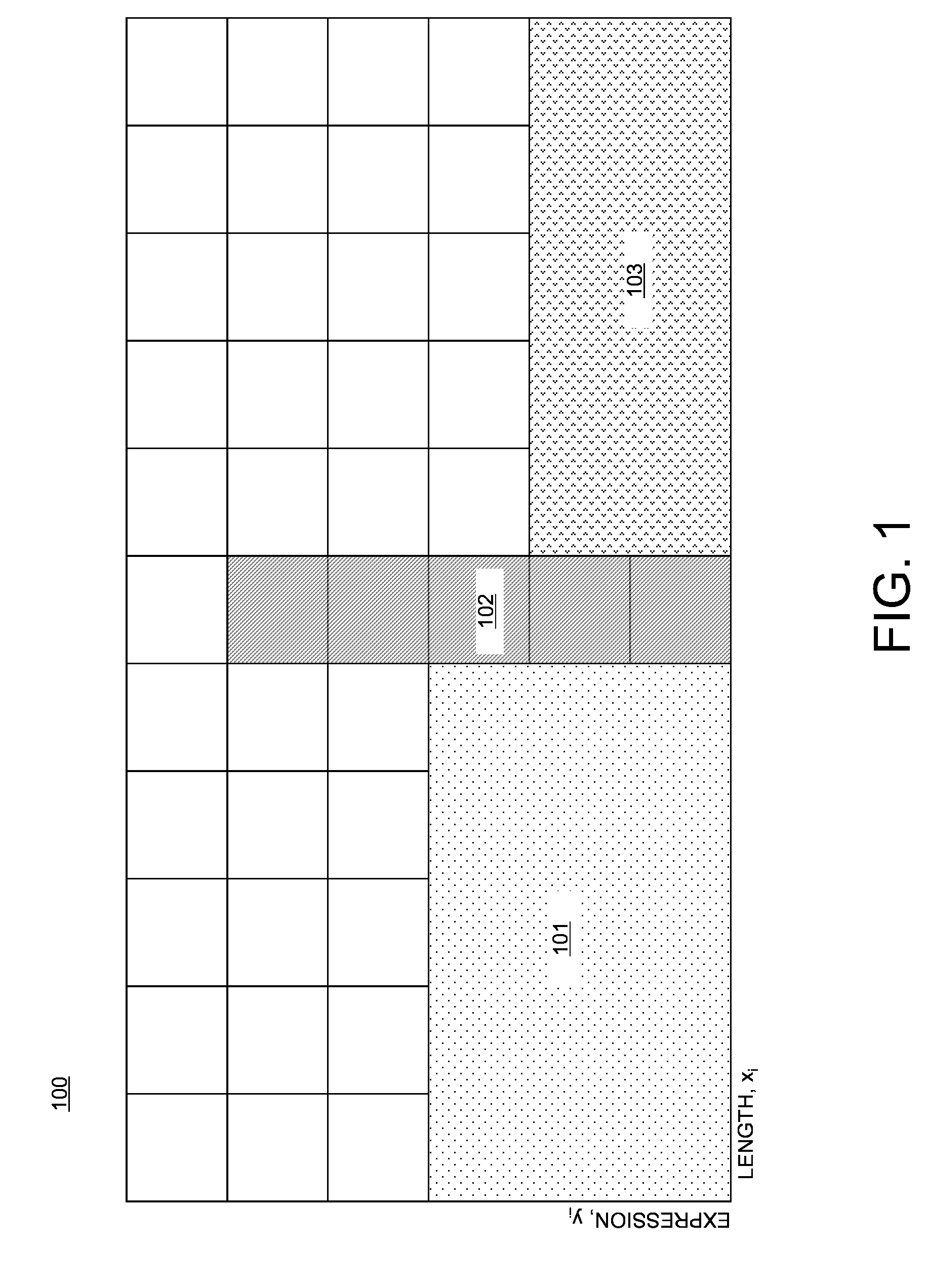
Differential expression analysis means taking the normalised read count data and performing statistical analysis to discover quantitative changes in expression levels between experimental groups.
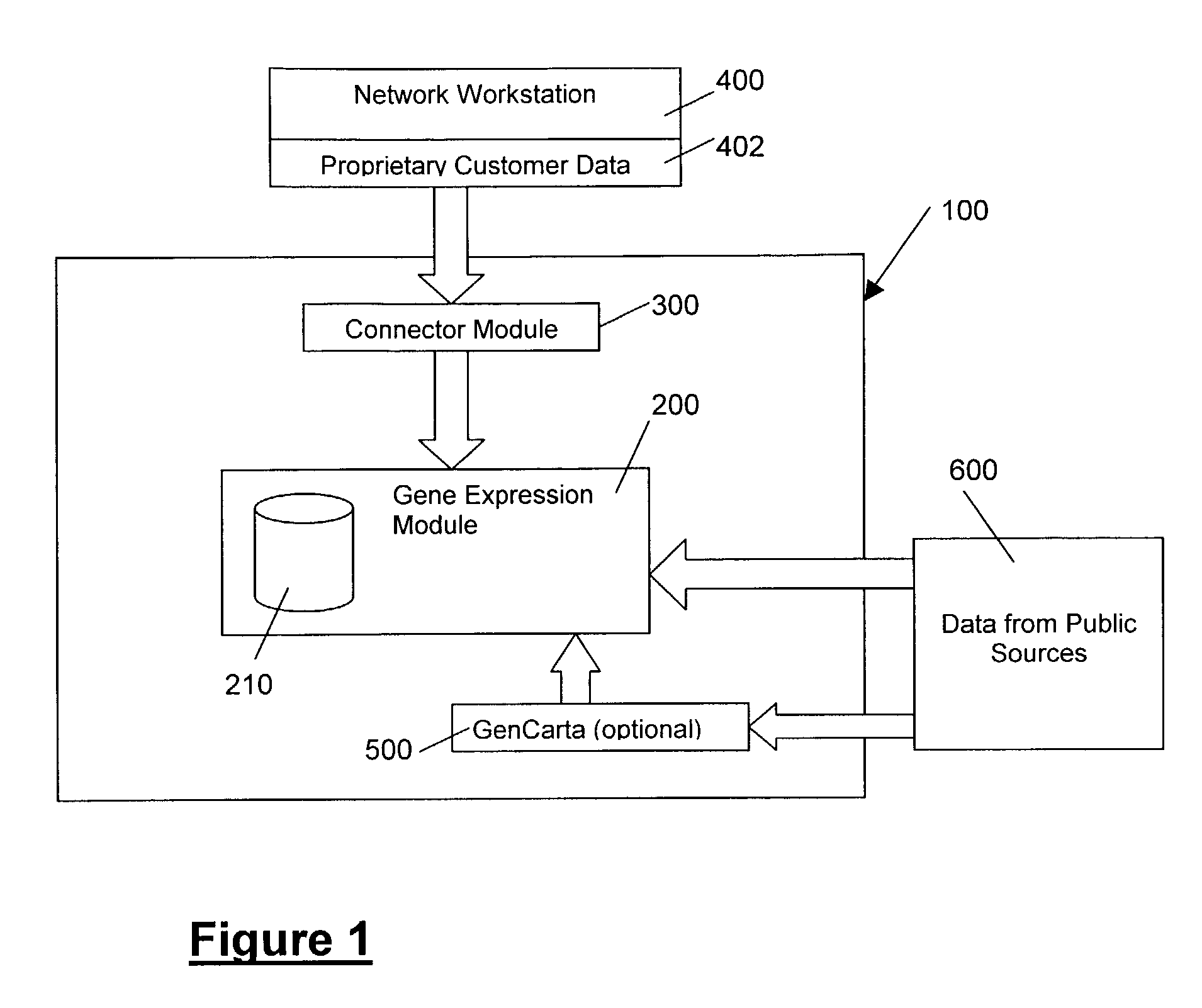
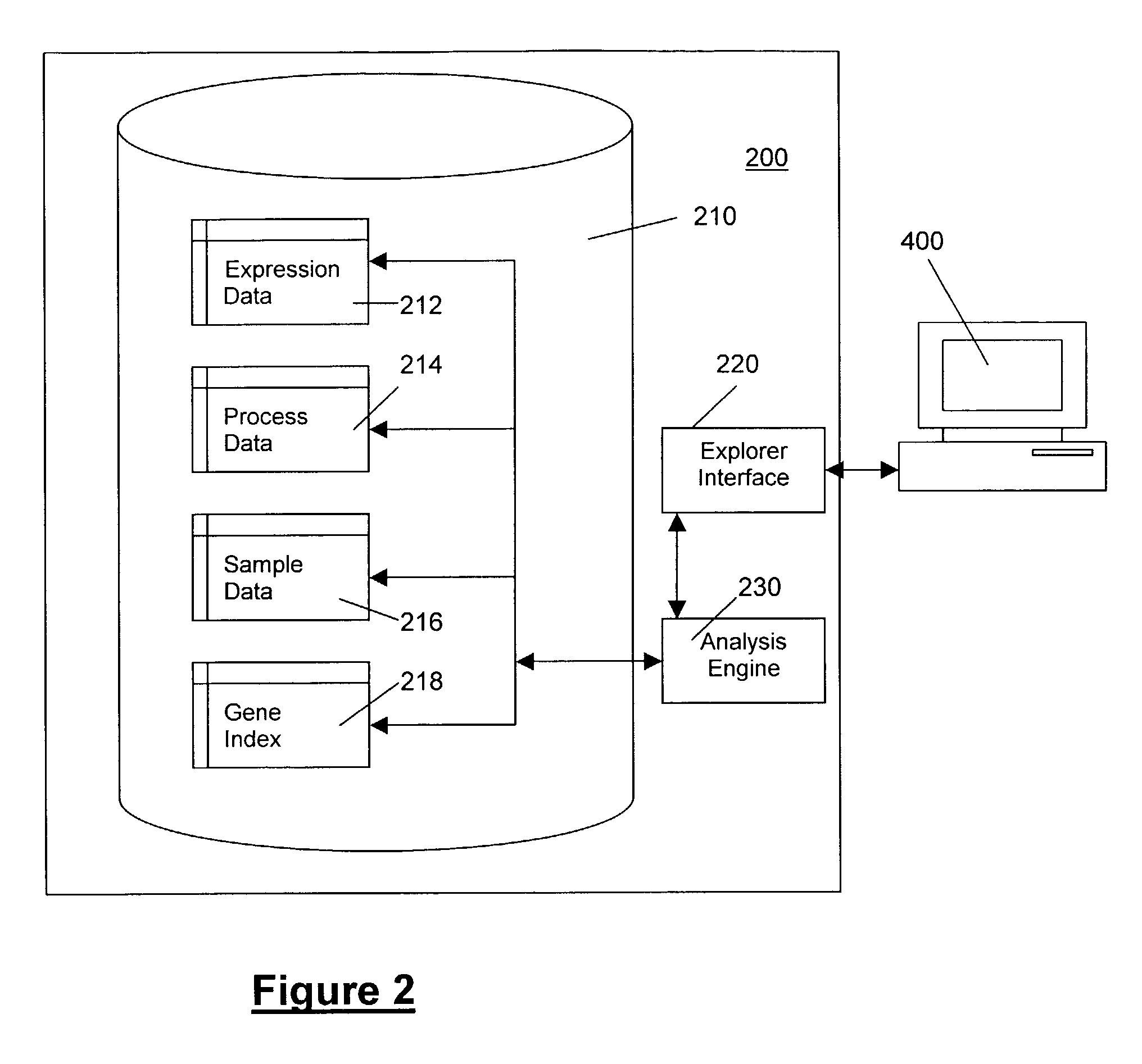
Methods and systems for efficient comparison, identification, processing, and importing of gene expression data
The present invention provides systems and methods for analyzing gene expression, gene annotation, and sample information in a relational format supporting efficient exploration and analysis, including comparative differential expression analysis, expression pattern matching, and mapping of external attributes or identifiers to internal representations of gene fragments or samples.
Owner:OCIMUM BIO SOLUTIONS
Tumor-related IncRNA and tumor-related IncRNA function predication
InactiveCN106295246ASimple processSimple methodBiostatisticsProteomicsDifferential codingPredictive marker
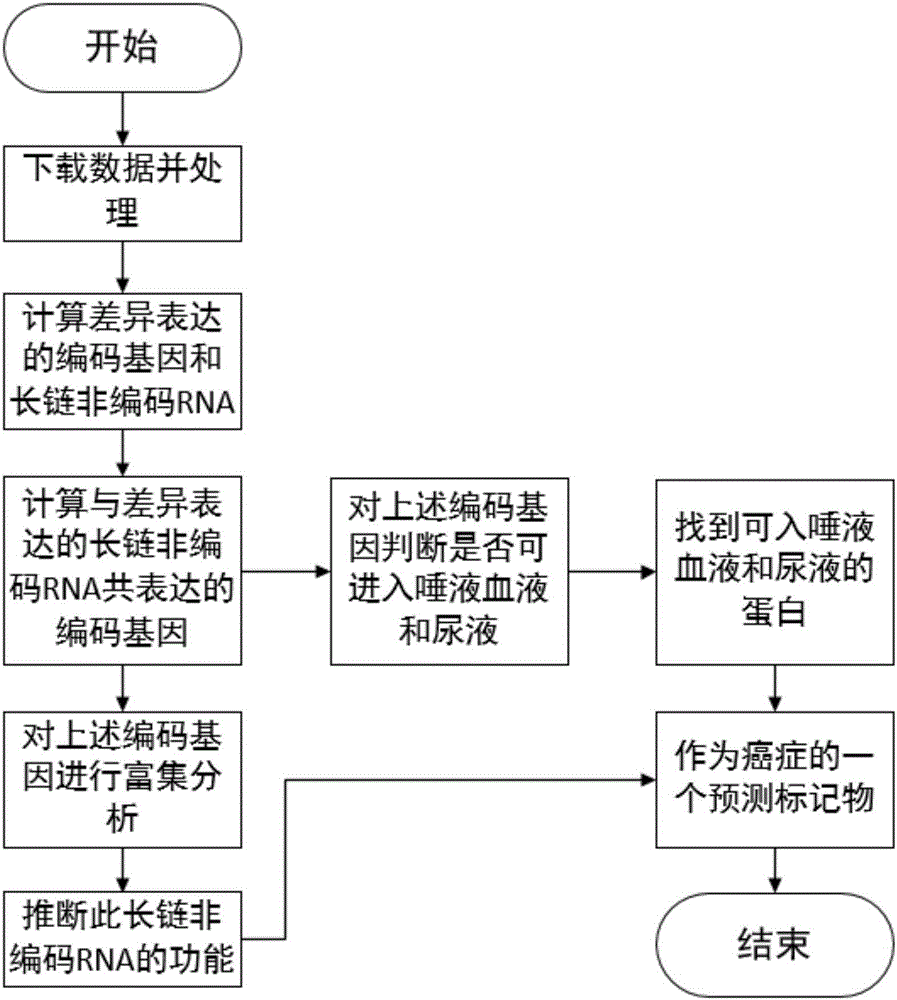
The invention relates to tumor-related IncRNA and tumor-related IncRNA function predication. By taking differential expression of IncRNA in tumors as diagnosis references, a process for finding a relation between IncRNA and tumors includes: step one, downloading data from GEO database, processing the data to obtain expression data of part of IncRNA and exons; step two, subjecting the processed expression data to differential expression analysis; step three, analyzing co-expression and differential coding genes and IncRNA for IncRNA in differential expression; step four, subjecting the coding genes to probe platform annotation; step five, further screening the IncRNA in differential expression to select out most obviously differential IncRNA; step six, performing enrichment analysis to obtain a GOBP process and pathyway, and speculating IncRNA functions through coding gene related biological processes; step seven, analyzing whether common coding genes obtained at the step six can enter blood, saliva and urine or not, analyzing genes which are allowed to enter, and taking the genes and IncRNA as potential predication marks of cancers.
Owner:JILIN UNIV
Tea tree miRNA fluorescent quantitative PCR reference gene under low temperature stress as well as screening method and application of reference gene
ActiveCN105132417AMicrobiological testing/measurementDNA/RNA fragmentationReference genesGene selection
The invention relates to a tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p, which has a nucleotide sequence as shown in SEQ ID NO: 1, as well as a screening method of the reference gene. According to the method, 6 common plant miRNA reference genes and 3 tea tree miRNAs are selected as candidate reference genes, and the expression stabilities of the candidate reference genes under different low temperature stresses and in different tea tree tissues are evaluated by virtue of qRT-PCR technology through two software, namely BestKeeper and geNorm, so as to provide a reference for the selection of reference genes for tea tree miRNA differential expression analysis under low temperature stress. The obtained tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p is stable in gene expression under different temperatures of treatment and in different tea tree tissues; therefore, the reference gene Pc-222-3p can be used as a reference gene for tea tree miRNA fluorescent quantitative PCR under low temperature stress.
Owner:ANHUI QIMEN COUNTY QIHONG TEA
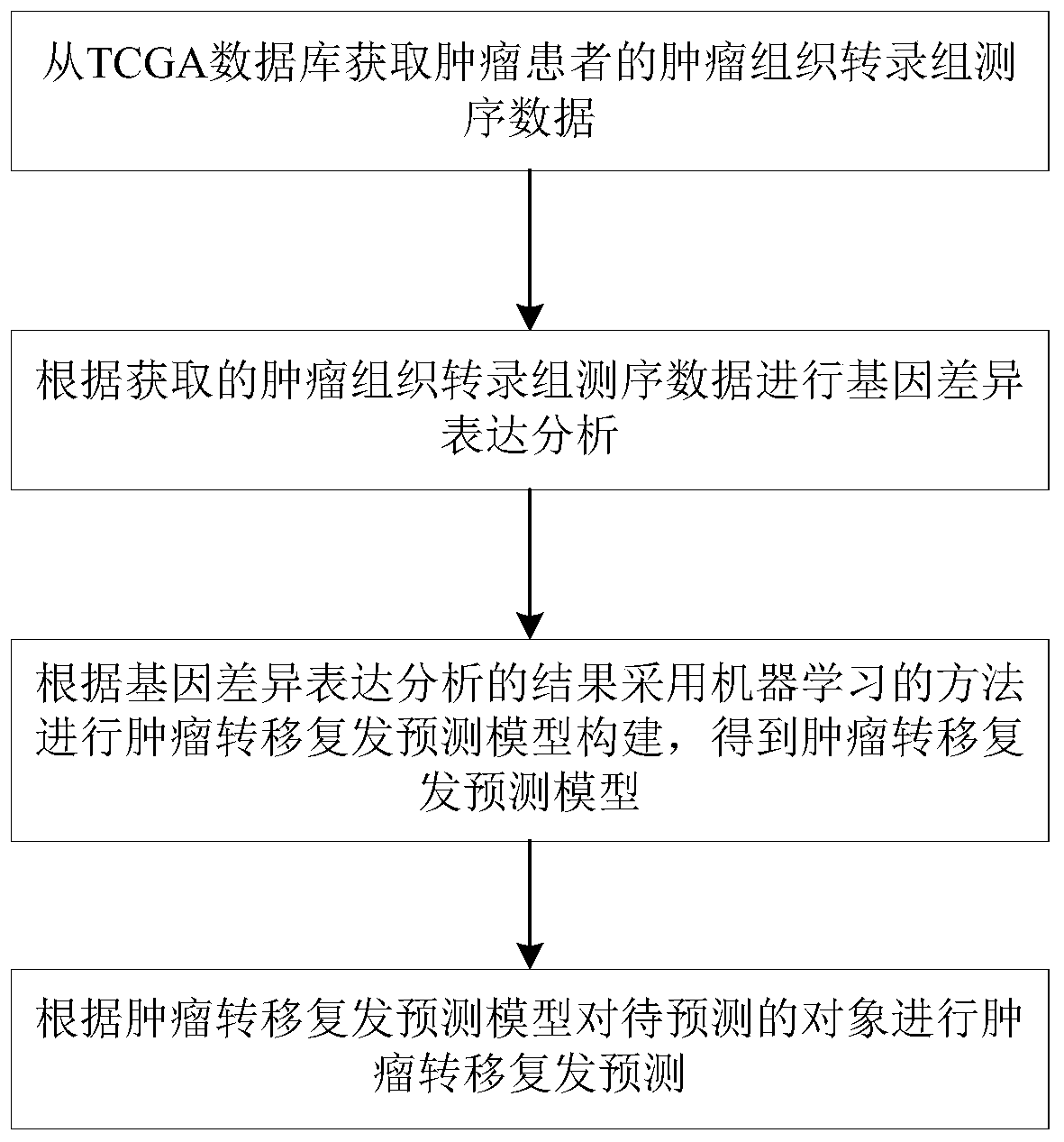
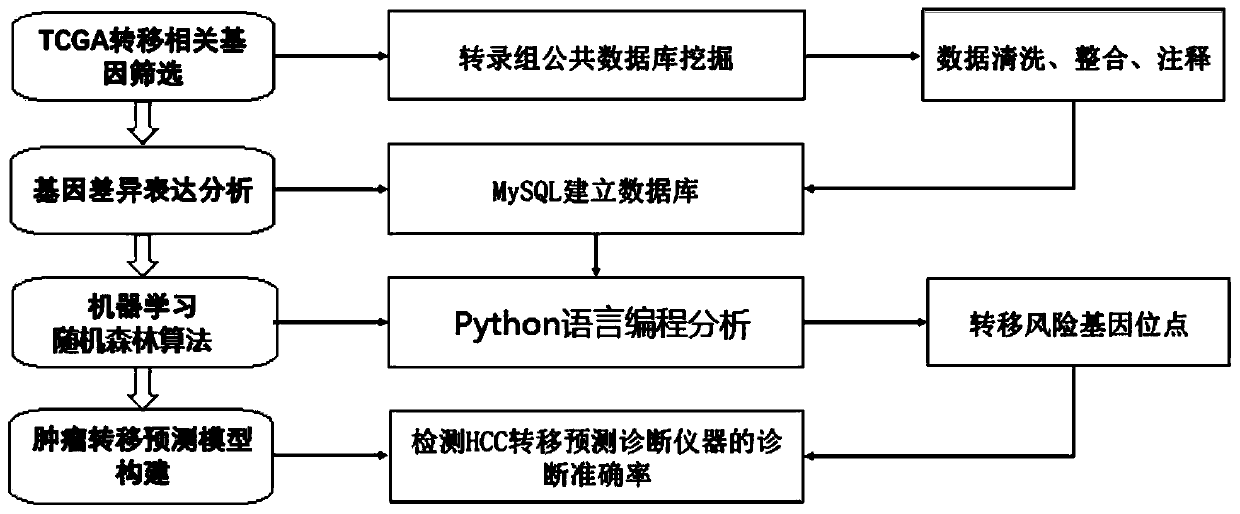
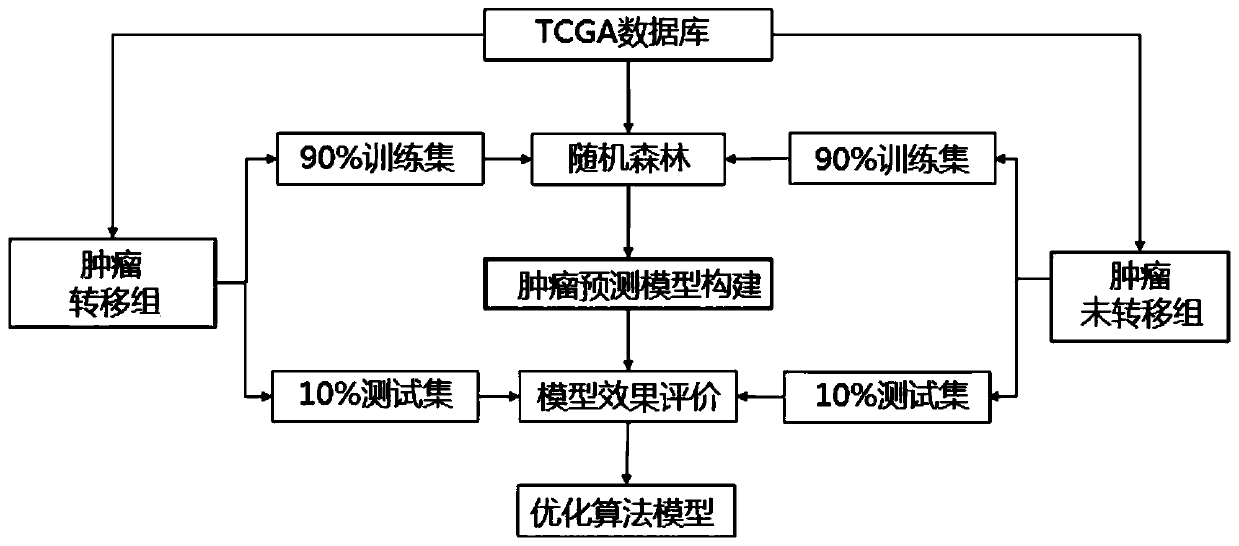
Tumor metastasis and recurrence prediction method and system based on TCGA database
ActiveCN109801680ARealize fully automated managementHealth-index calculationBiostatisticsCancer genomeRecurrence prediction
The invention discloses a tumor metastasis and recurrence prediction method and system based on a TCGA (The Cancer Genome Atlas) database. The tumor metastasis and recurrence prediction method includes the steps: obtaining transcriptome sequencing data of tumor tissues of tumor patients from the TCGA database; performing gene differential expression analysis according to the acquired transcriptomesequencing data of tumor tissues; performing construction of a tumor metastasis and recurrence prediction model by using a machine learning method according to results of gene differential expressionanalysis to obtain a tumor metastasis and recurrence model; and performing tumor metastasis and recurrence prediction on an object to be predicted according to the tumor metastasis and recurrence prediction model. The tumor metastasis and recurrence prediction method based on a TCGA database utilizes the machine learning method and the TCGA database to realize the fully automated management of the tumor metastasis and recurrence prediction, can directly provide a clear diagnosis and prognosis reference and guidance for tumor patients, and is more timely, accurate and efficient. The tumor metastasis and recurrence prediction method based on a TCGA database can be widely applied to the field of medical computer applications.
Owner:GUANGZHOU UNIVERSITY OF CHINESE MEDICINE
Acipenser dabryanus gonad differential expression gene GSDF and screening method thereof
InactiveCN107937394AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyScreening method
The invention discloses an acipenser dabryanus gonad differential expression gene expression gene GSDF and a screening method thereof, and belongs to the technical field of biology. A high-flux sequencing technique is used for performing male and female transcriptome comparative sequencing on acipenser dabryanus; transcriptome data is subjected to assembling, annotation and differential gene expression analysis. An FPKM method is used for analyzing the gene expression level in male and female gonad. EBSeq is used for performing gene differential expression analysis, so that a differential expression gene set between the male and female samples is obtained. In the screening process, FDR is selected as a key index of the differential expression gene screening; the conditions that the FDR issmaller than 0.05 and the differential multiple is greater than or equal to 2 are used as screening standards. Then, the differential gene is subjected to function annotation; the sex determination-related gene is screened; the basis is provided for the acipenser dabryanus sex determination and the sex determination gene and sex determination mechanism study in future.
Owner:FISHERIES INST SICHUAN ACADEMY OF AGRI SCI
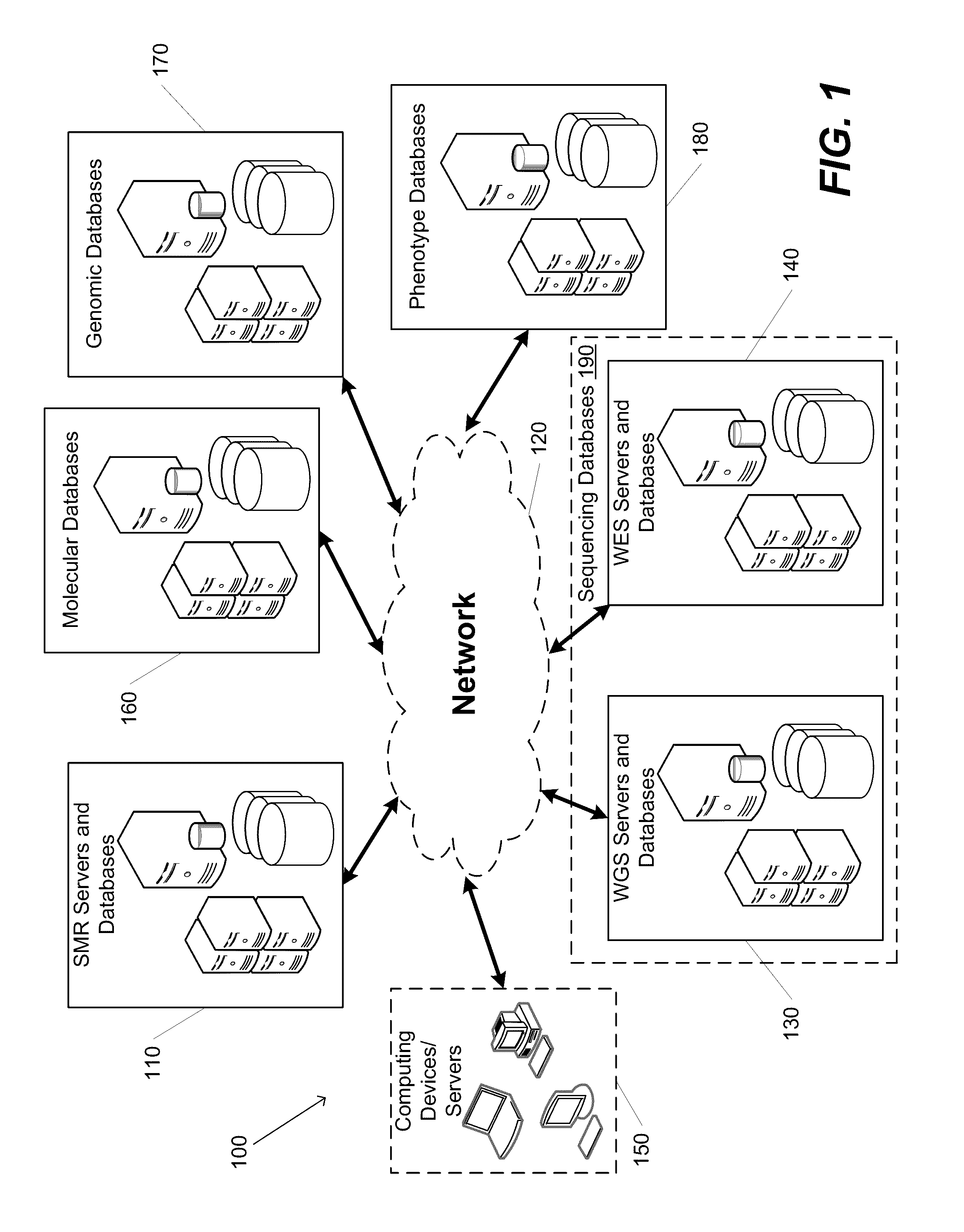
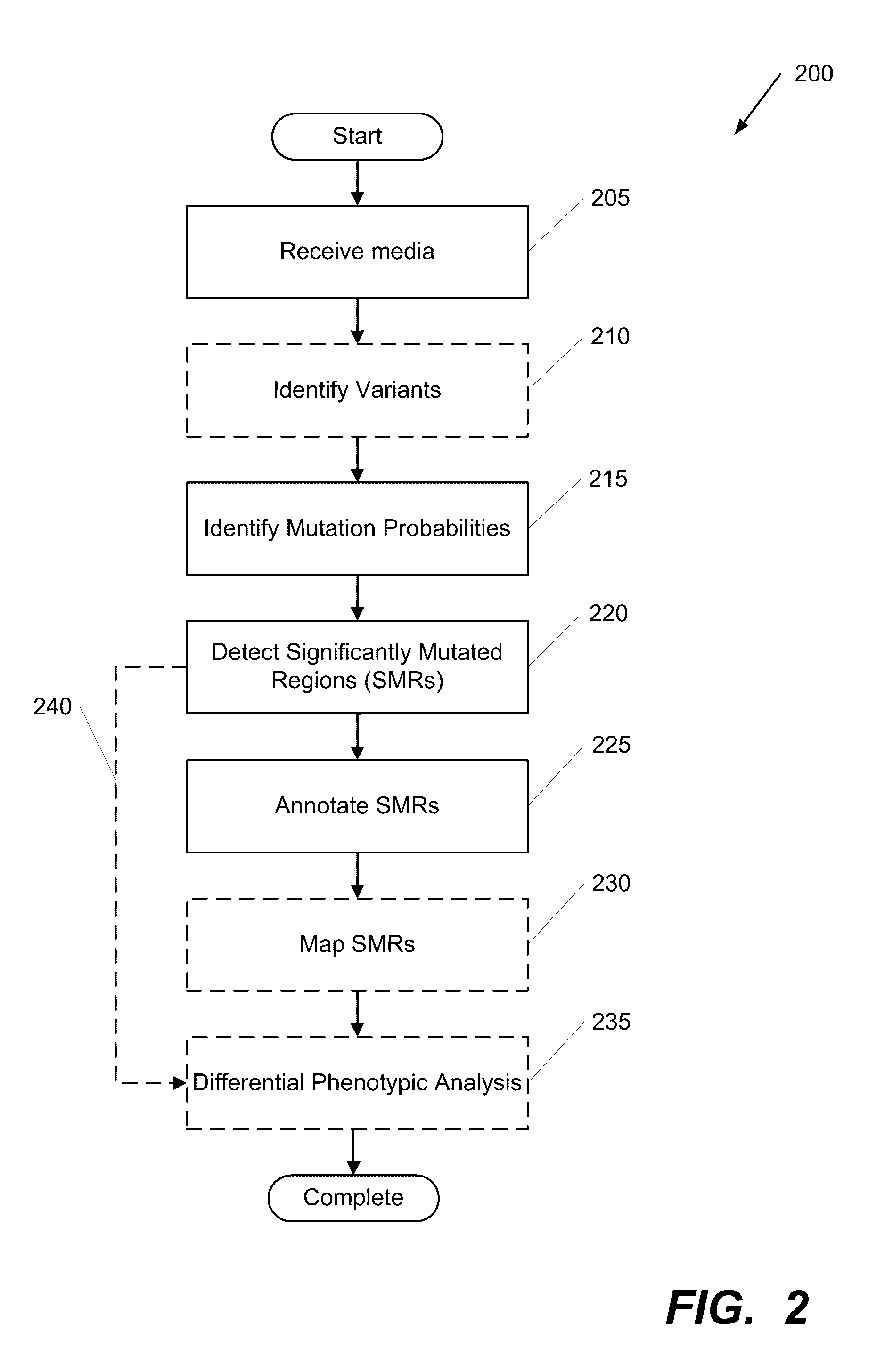
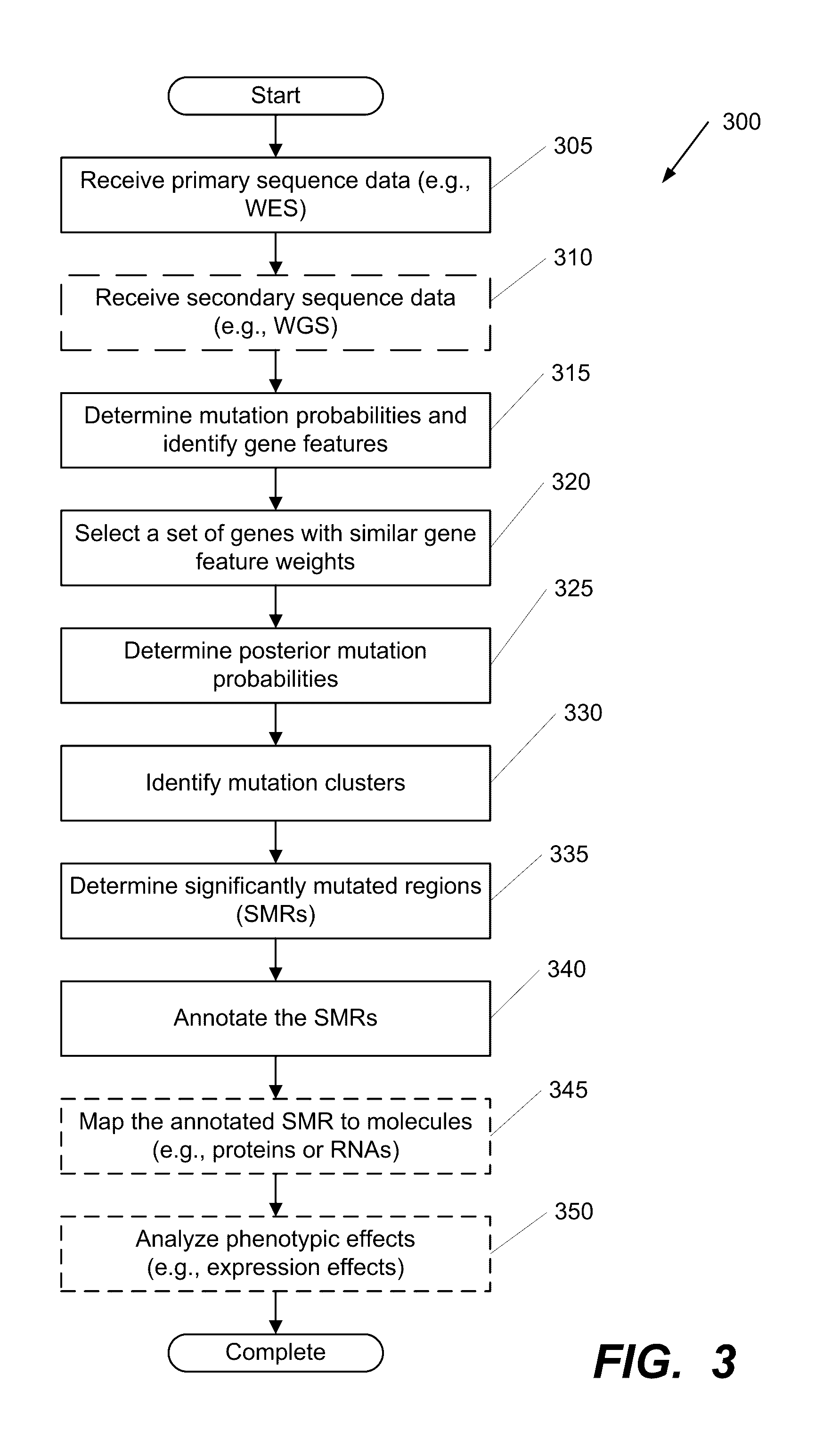
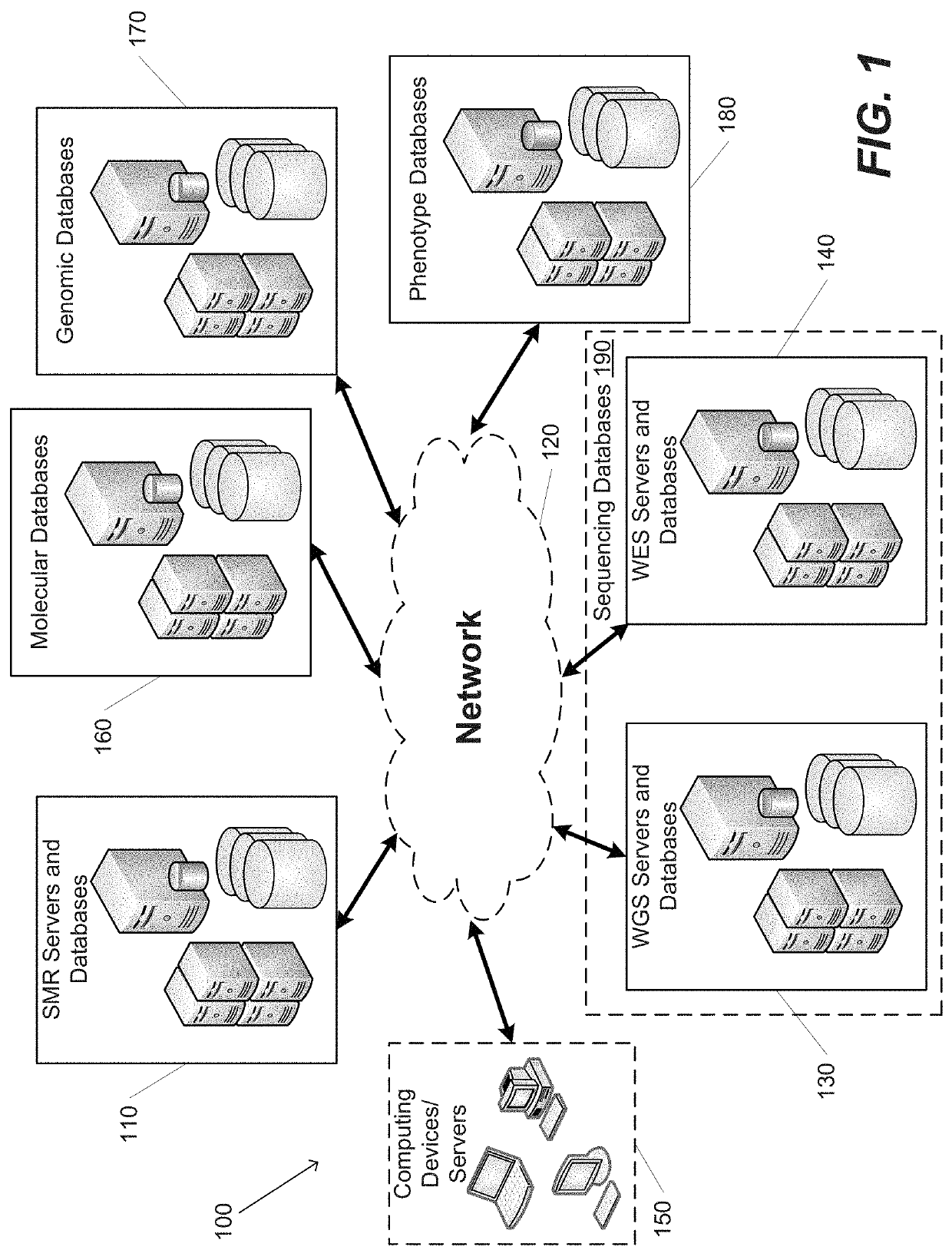
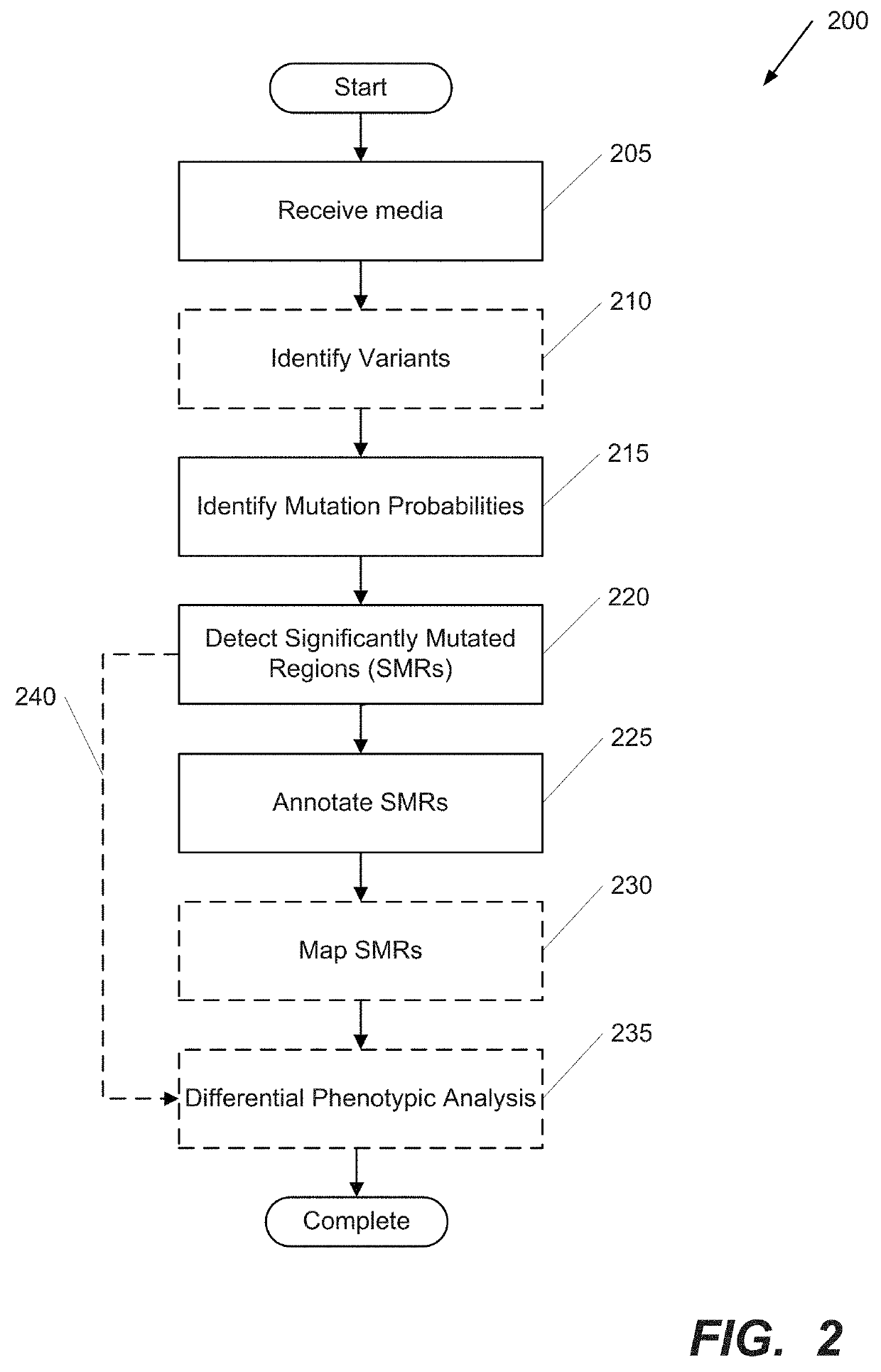
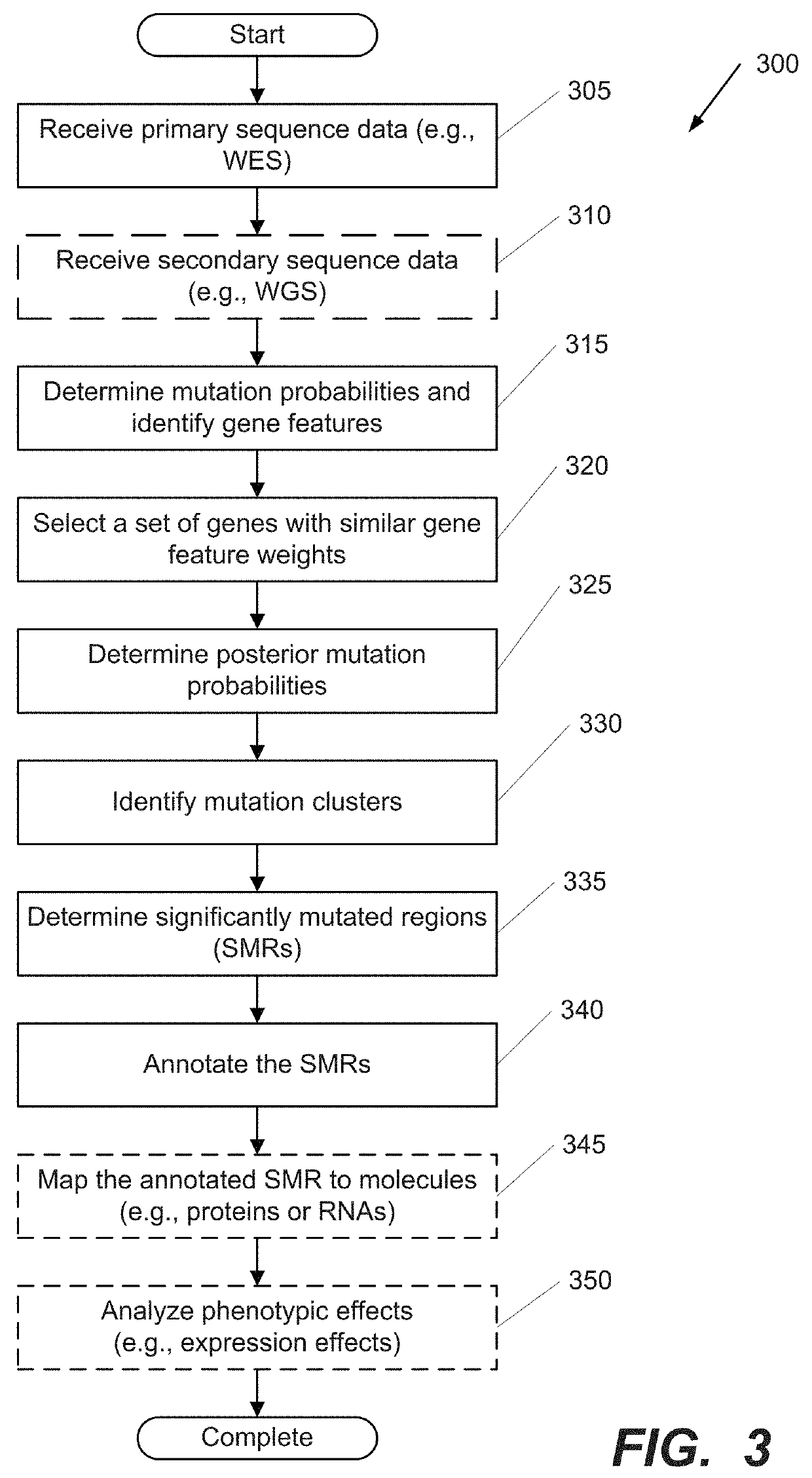
Systems and Methods for Multi-Scale, Annotation-Independent Detection of Functionally-Diverse Units of Recurrent Genomic Alteration
InactiveUS20160378915A1Useful in detectionMicrobiological testing/measurementBiostatisticsHuman DNA sequencingBinding site
The functional interpretation of somatic mutations remains a persistent challenge in the interpretation of human genome data. Systems and methods for detecting significantly mutated regions (SMRs) in the human genome permit the discovery and identification of multi-scale cancer-driving mutational hotspot clusters. Systems and methods of SMR detection reveal differentially mutated genetic regions across various cancer types. SMR detection and annotation reveals a diverse spectrum of functional elements in the genome, including at least single amino acids, compete coding exons and protein domains, microRNAs, transcription factor binding sites, splice sites, and untranslated regions. Systems and methods of SMR detection optionally including protein structure mapping uncover recurrent somatic alterations within proteins. Systems and methods of SMR detection optionally including differential expression analysis reveal previously unappreciated connections between recurrent and somatic mutations and molecular signatures.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
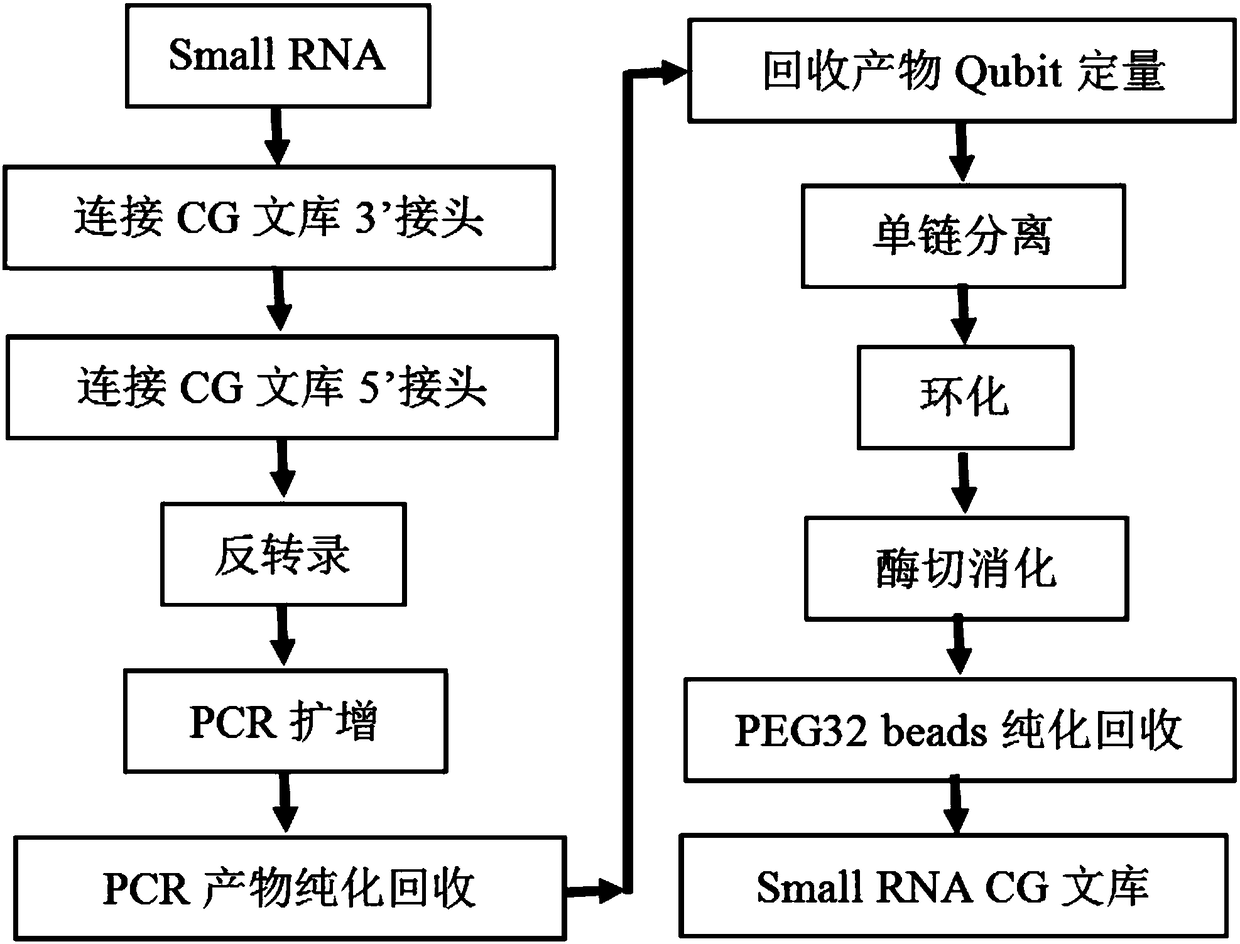
Library construction and sequencing method of small RNA (Ribonucleic Acid)
InactiveCN108060460AFavorable scientific basisMicrobiological testing/measurementLibrary creationSingle strand dnaDifferential expression analysis
The invention discloses a library construction and sequencing method of small RNA (Ribonucleic Acid). The construction method comprises the following steps: (1) connecting a small RNA segment with a 3' connector of a CG library; (2) connecting a product of step (1) with a 5' connector of the CG library; (3) carrying out reverse transcription on a product of step (2) to form cDNA (complementary Deoxyribonucleic Acid); (4) carrying out PCR (Polymerase Chain Reaction) amplification on the cDNA; purifying and recycling a PCR amplification product; (5) carrying out single-chain separation on the purified and recycled PCR amplification product and cyclizing single-chain DNA; (6) carrying out enzyme digestion on a cyclized product and recycling to obtain a small RNA CG library. According to the construction method disclosed by the invention, a novel CG library construction scheme is provided for the small RNA so that whole-genome level small RNA sequencing based on CG sequencing is realized and a foundation is laid for novel small RNA molecule digging, cancer target gene predication and identification, difference expression and analysis between samples, small RNA clustering, expression pattern analysis and the like and scientific evidence is also provided for cancer early diagnosis and targeting treatment and detection.
Owner:SHENZHEN HUADA GENE INST
Cloud computing-based method for quality control and management of gene sequence data
InactiveCN109584958AHigh precisionImprove experiment qualityProteomicsGenomicsGenomicsProcess quality
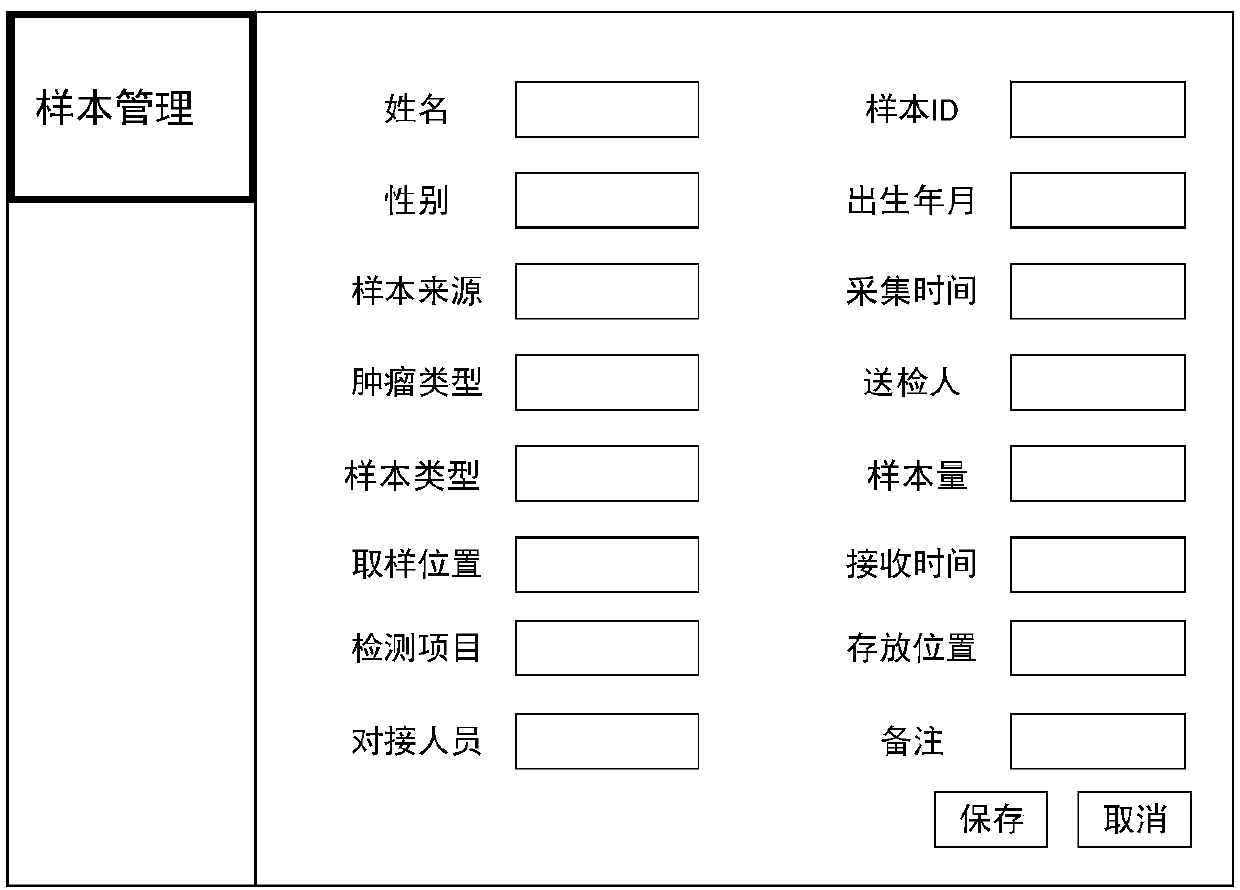
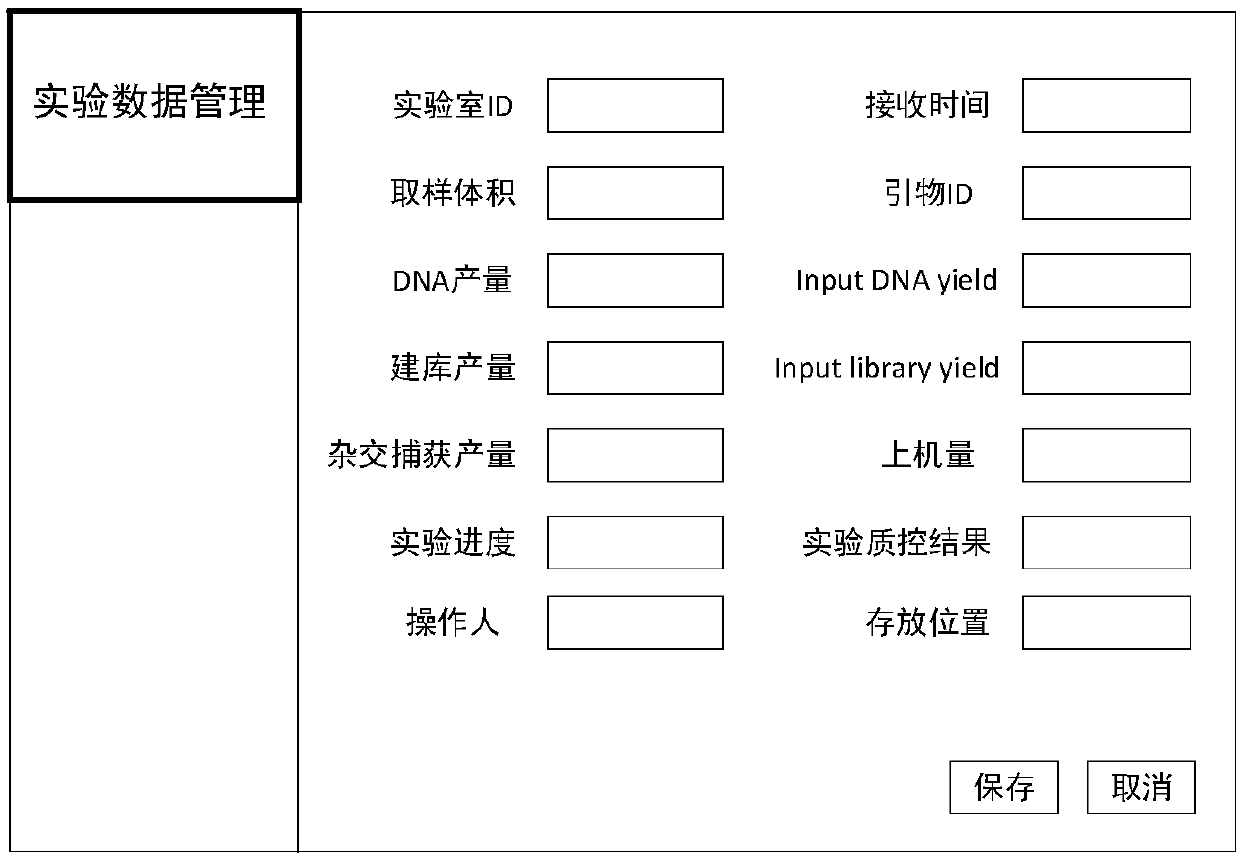
The invention relates to a cloud computing-based method for quality control and management of gene sequence data. The method includes sample clinical information management, experimental data management, data whole process quality control and analysis result management. Testing items of the sample clinical information management includes genomic analysis and transcriptomics analysis, and the genomic analysis includes whole genome sequencing, whole exome sequencing and whole exome sequencing; the transcriptomic analysis includes gene fusion, variable splicing and differential expression analysis. Experimental links are closely connected with analytical links, an analyst can comprehensively understand clinical information, sample information and experimental results, subsequent analysis is facilitated, the accuracy of the analysis result is improved, and the method can help experimenters to understand the quality of sequencing data and thus reflect on shortcomings in the experimental process, the experimental quality is thus improved, and the need of practical use can be applied well.
Owner:江苏医联生物科技有限公司
Exon conserved sequence amplified polymophic molecular marker and its analysis method
InactiveCN102443583AReduce design costLarge amount of amplified bandsMicrobiological testing/measurementDNA/RNA fragmentationConserved sequenceGenetic diversity
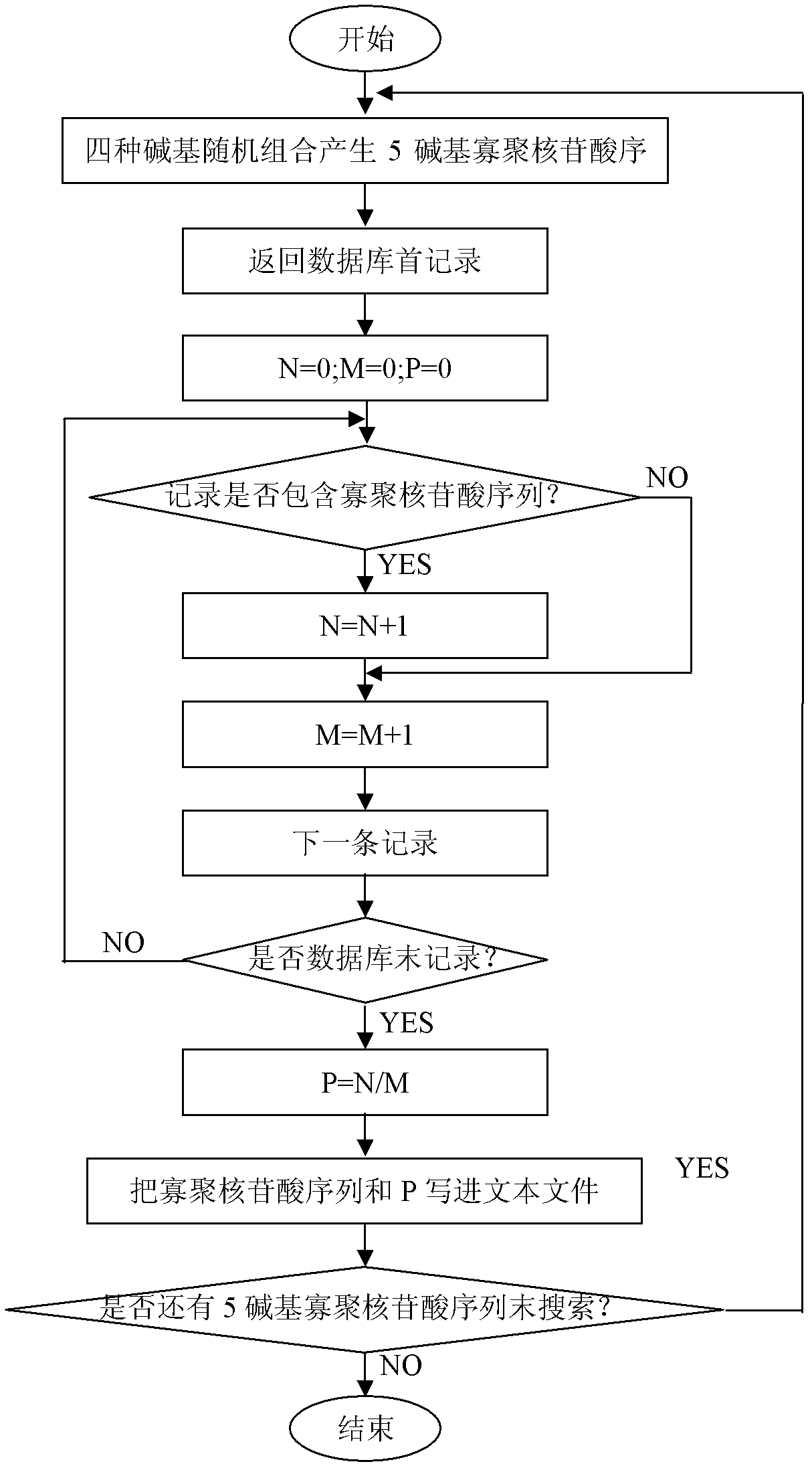
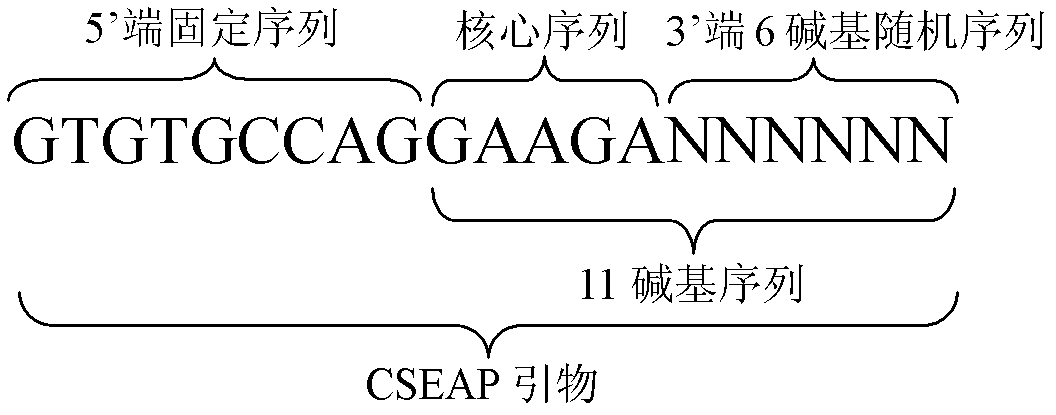
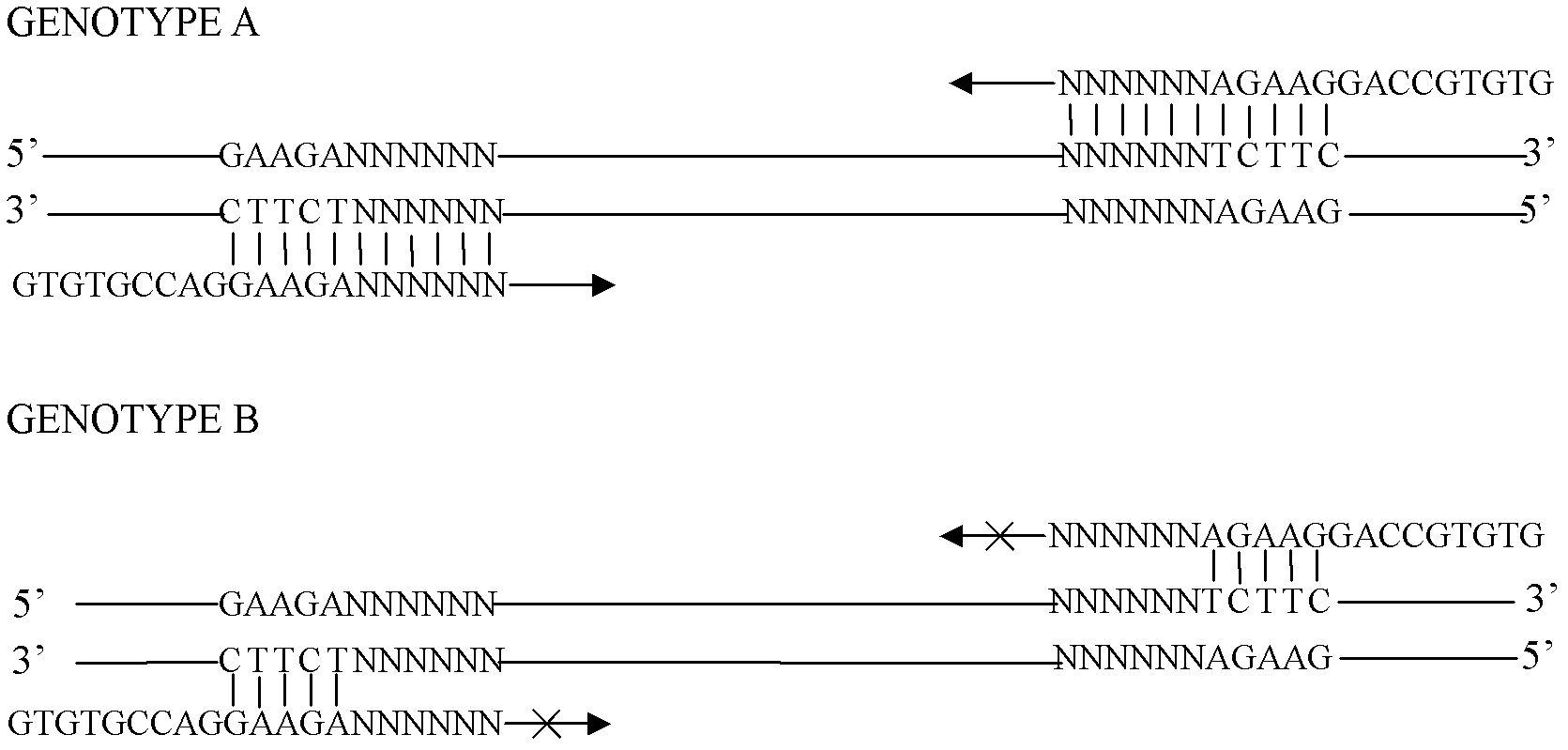
The invention discloses an exon conserved sequence amplified polymorphic molecular marker and its analysis method. According to the invention, mRNA sequence information of known species in public database can be utilized to find out five bases oligonucleotide conserved sequence of gene exon. The conserved sequence is taken as a core to link six bases random sequence to its 3' terminal to form an eleven bases sequence which makes up a primer of 20bp together with a fixed sequence of 5' terminal. Primers with higher occurrence frequency of the eleven bases sequence are preliminarily screened out by mRNA database. Then PCR is carried out for repeatability screening and the FCR based exon conserved sequence amplified polymorphic marker is obtained. The method provided by the invention has advantages of simple operation, high effectiveness, good repeatability, reduced time and cost of primer screening. It is a new effective molecular marking method for the genetic diversity analysis. The method can be widely applied in various fields of genetic map construction, gene or QTL location, marker-assisted breeding, genetic diversity analysis, genome research, mRNA differential expression analysis and the like and has a bright prospect.
Owner:GUANGXI UNIV
Application of miR-577 for preparing nephrosis diagnosis marker
InactiveCN108624693AMicrobiological testing/measurementAntineoplastic agentsNephrosisTarget analysis
The invention relates to application of miR-577 for preparing a nephrosis diagnosis marker, in particular to application of miR-577 and a target gene thereof in preparing a renal clear cell carcinomadiagnosis marker. Database renal clear cell carcinoma mRNA (Ribonucleic Acid) and miRNA data are retrieved, and miR-577 and a target gene regulated and controlled by the miR-577 are selected to carryout molecular verification through data integration, differential expression analysis and targeted analysis. A result indicates that the miR-577 and the target genes CD1D, TMEM133 and SLFN11 regulatedand controlled by the miR-577 can serve as the diagnosis marker of the renal clear cell carcinoma, and therefore good clinic application value is performed.
Owner:THE FIRST AFFILIATED HOSPITAL OF ARMY MEDICAL UNIV
Automated analysis method for prokaryotic transcriptome based on second generation sequencing
PendingCN110047560AConvenient error queryQuick check for errorsProteomicsGenomicsGenome alignmentStructure analysis
The invention discloses an automated analysis method for prokaryotic transcriptome based on second generation sequencing. The automated analysis method includes the steps: an original logout data filtering and quality control step; a genome comparison step; a transcript structure analysis step; a gene expression quantification and differential expression analysis step; and a result sorting step. The automated analysis method for prokaryotic transcriptome based on second generation sequencing has the advantages of: covering most of the analysis content required by the market, automatically sorting out all the analysis results, and after completing the analysis of each part, automatically counting, visualizing, and sorting the results, so that the results are arranged in order and can be directly used for generation of a report; being visible for all the operation steps, thus being convenient for error query; and when performing each step analysis, recording the used command lines and parameters and the log results generated during the operation, and once the program runs incorrectly, being able to quickly check the error.
Owner:南京派森诺基因科技有限公司
Ammopiptanthus mongolicus cold-resistant gene AmEBP1
InactiveCN101892242AImprove cold resistanceGrowth inhibitionMicrobiological testing/measurementFermentationNucleotideTransgene
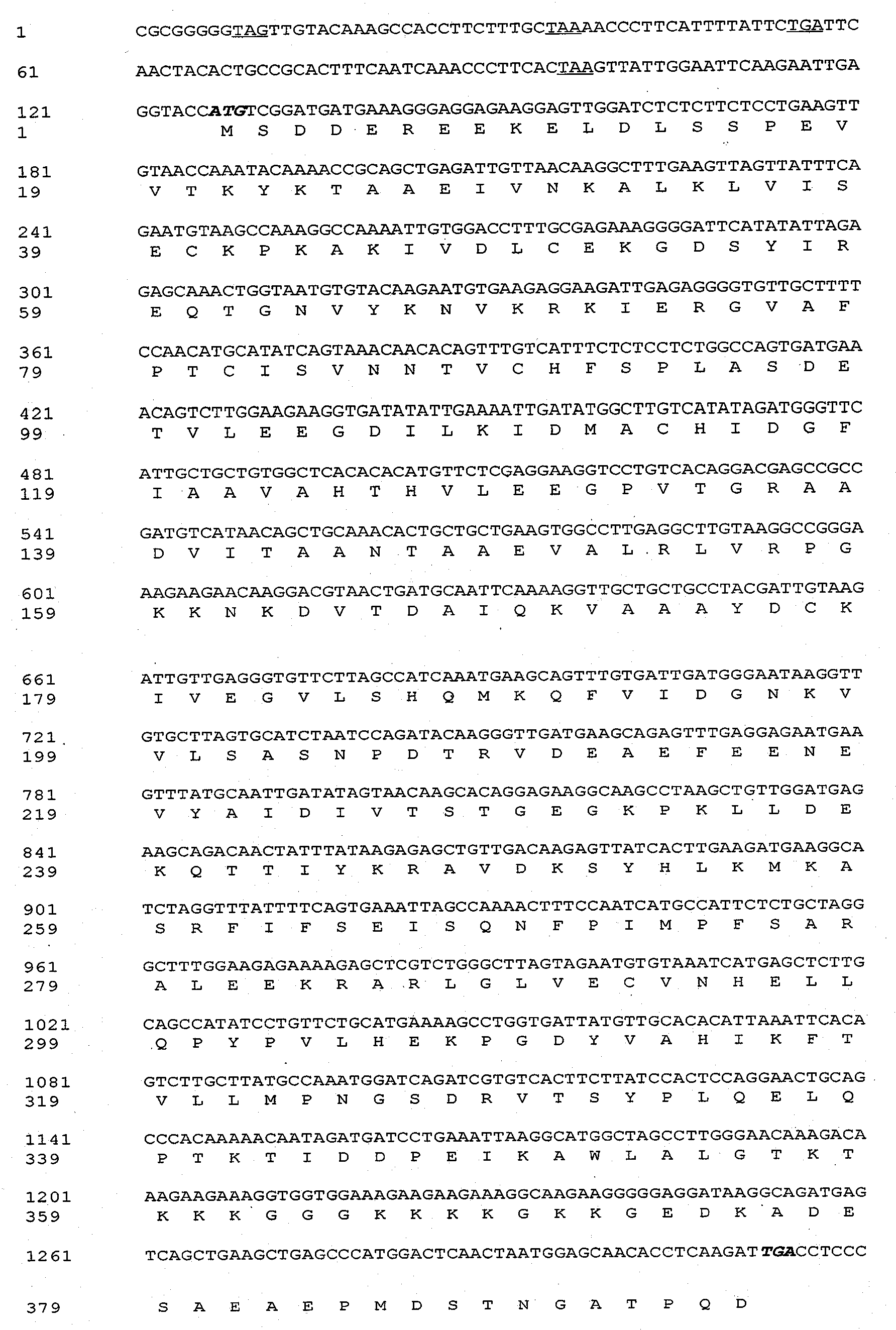
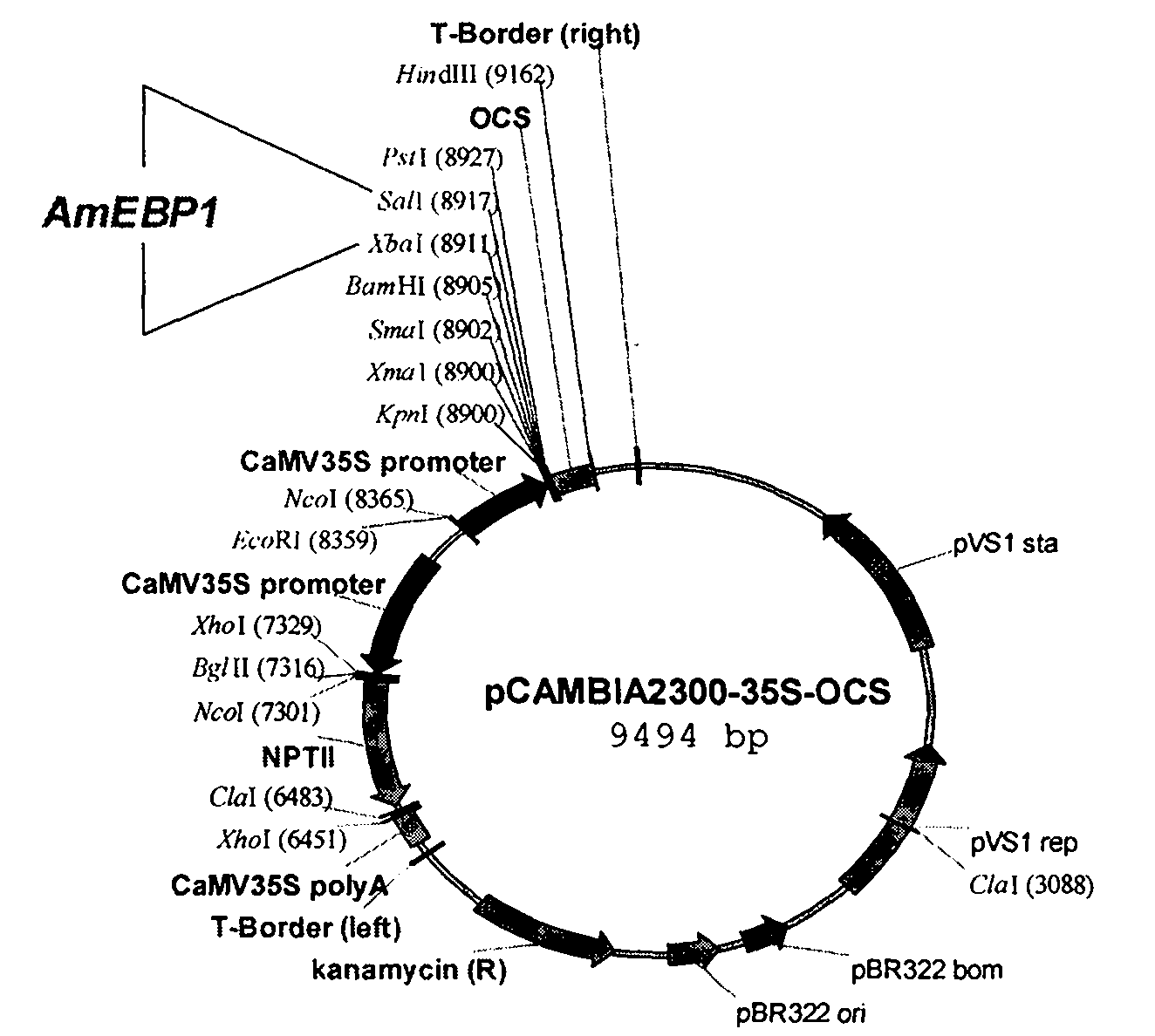
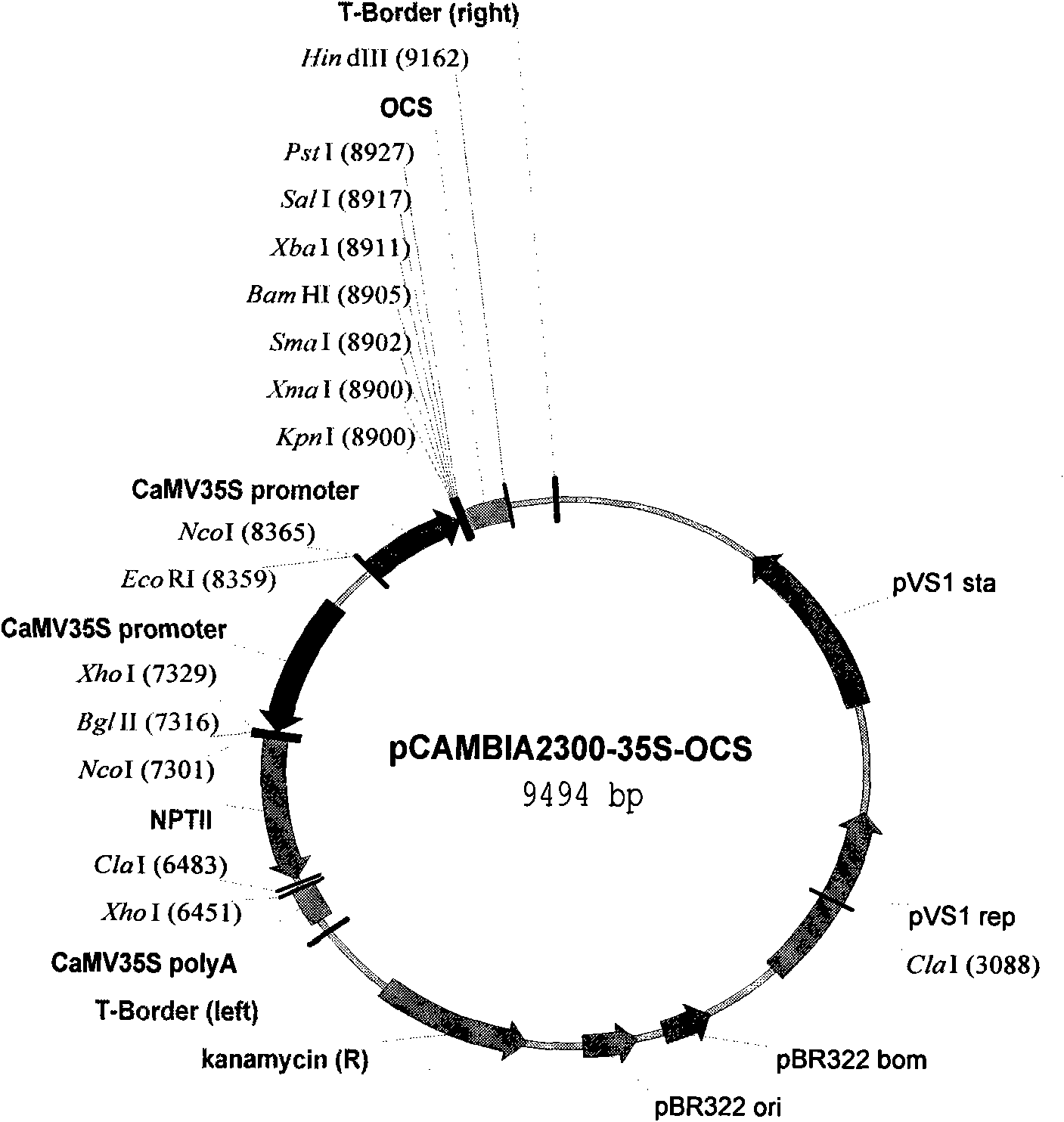
The invention discloses an ammopiptanthus mongolicus cold-resistant gene AmEBP1, and relates to a national secondary endangered and protected evergreen broad-leaved woody plant derived from the tertiary period in the desert regions of northwest China. The molecular cloning of the gene comprises the following steps of: obtaining a low temperature-induced and up regulation-expressed EST fragment through low temperature induction and differential expression analysis; and obtaining a full-length cDNA sequence of the gene through RACE amplification, and registering in a gene bank with the registration number of DQ519359. The AmEBP1 contains a whole ORF region with 1,188 nucleotides at the overall length, and the nucleotides code 395 amino acids and a terminator codon (TGA); a eukaryotic expression vector pCAMBIA2300-AmEBR1 is used to transform arabidopsis thaliana, and the obtained transgenic plant has improved cold resistance; and the ammopiptanthus mongolicus cold-resistant gene AmEBP1 is the first international woody plant cold-resistant gene which is cloned and passes genetic transformation verification.
Owner:TAISHAN RES INST OF FORESTRY +1
Specific protein marker for ovarian cancer diagnosis and preparation method of protein marker
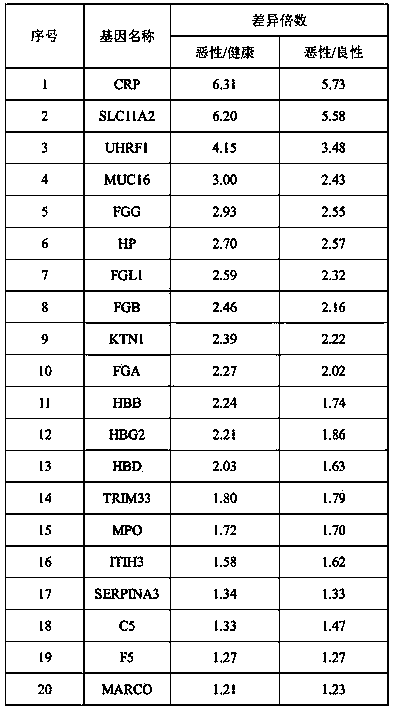
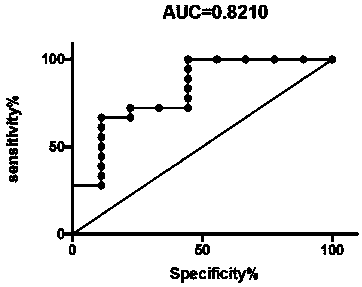
ActiveCN110596403AIncreased sensitivityImprove featuresBiological testingFermentationSolute transportersOvarian cancer
The invention relates to a specific protein marker for ovarian cancer diagnosis and a preparation method of the protein marker. The specific protein marker comprises the components including C reactive protein in serum exosome, solute transport protein family 11 member 2, ubiquitin-like PHD and ring finger domain protein 1. The preparation method comprises the following steps of protein extraction, pancreatin enzymolysis, TMT labeling, HPLC grading, liquid chromatography-mass spectrometry analysis, database search, protein identification, protein differential expression analysis, protein annotation and bioprotein marker screening. The protein marker has the advantages that (1) the marker has relatively high sensitivity and specificity; (2) ovarian malignant tumors can be distinguished frombenign tumors / healthy people; and (3) the preparation process is clear, the operability is high, and the repeatability is high.
Owner:THE FIRST AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
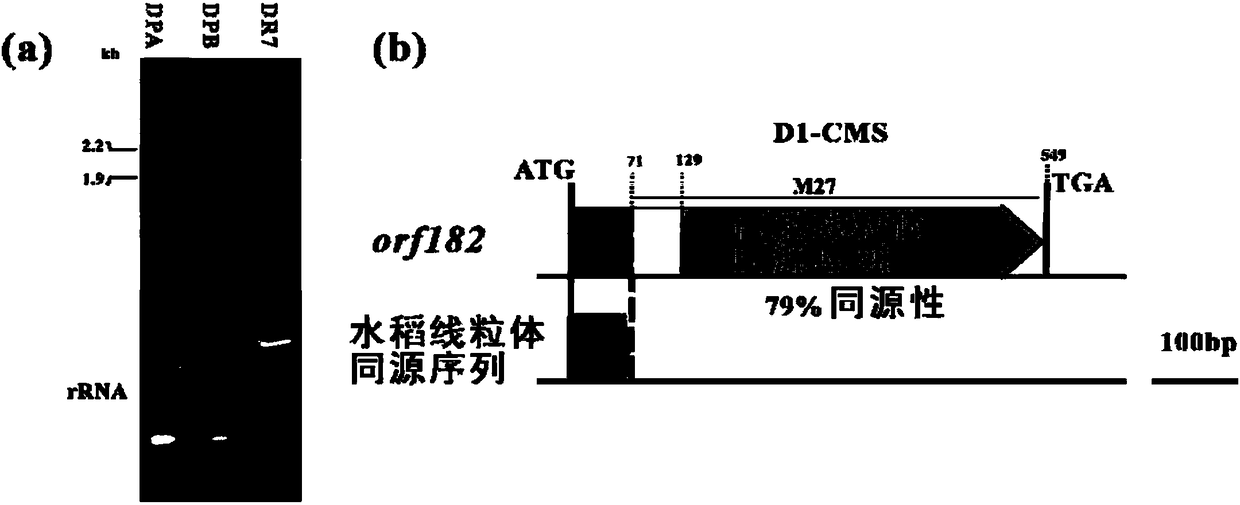
Rice mitochondria sterility gene and application thereof
The invention relates to the technical field of rice breeding, and specifically relates to a rice mitochondria sterility gene and the application thereof. The invention discloses a rice mitochondria sterility gene and a coding sequence thereof, and D1 type cytoplasm can be quickly and efficiently screened and authenticated from wild rice for cultivation of a D1 type cytoplasm sterile line by designing a specific molecular marker through the gene sequence. A mitochondrial genome specific sequence of the D1 type cytoplasm sterile is authenticated through comparative genomics, sterility related ORFs are authenticated through gene predication and differential expression analysis, and a plant transformation vecter is constructed for transforming maintenance line verification and sterility function.
Owner:JIANGXI SUPER RICE RES & DEV CENT (HAINAN RICE BREEDING CENT OF JIANGXI ACAD OF AGRI SCI) +1
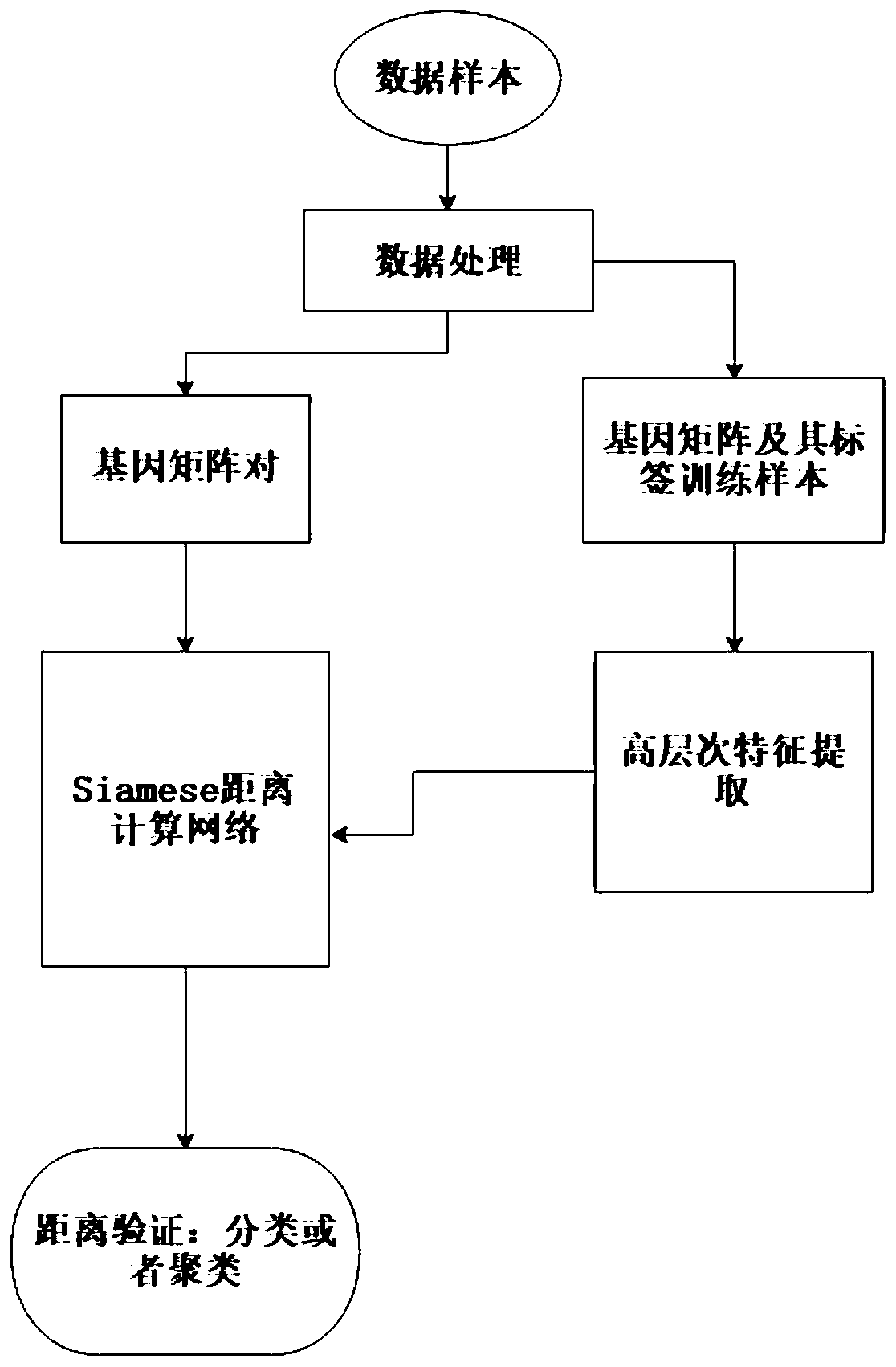
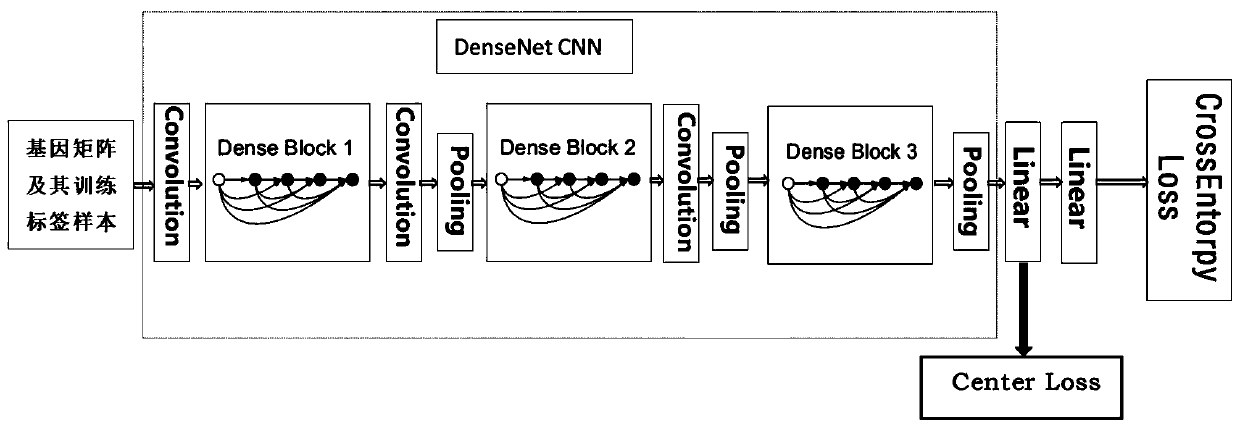
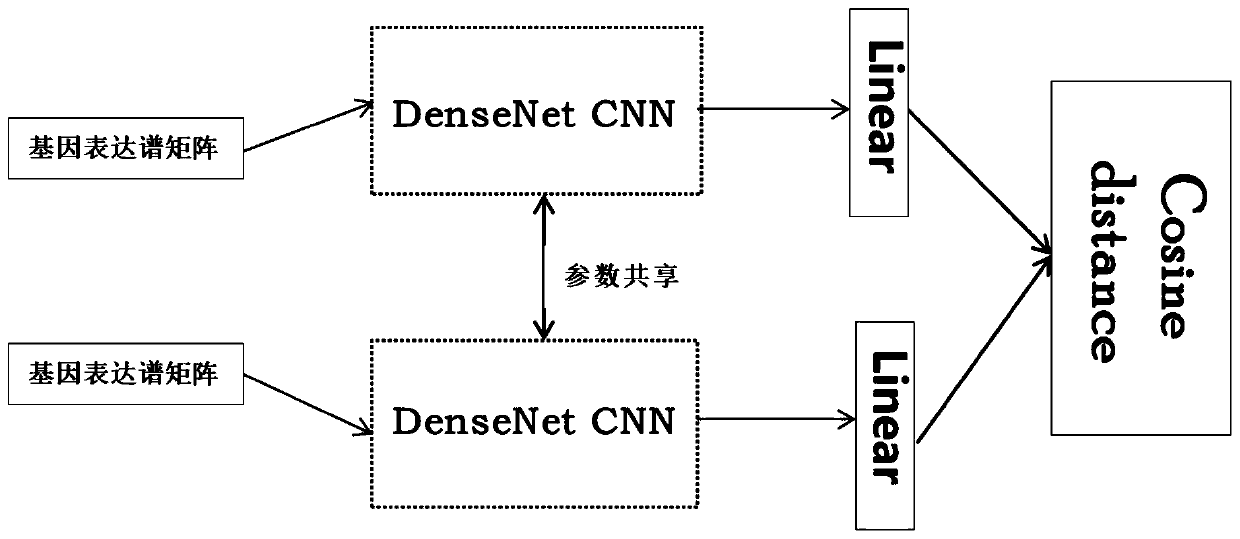
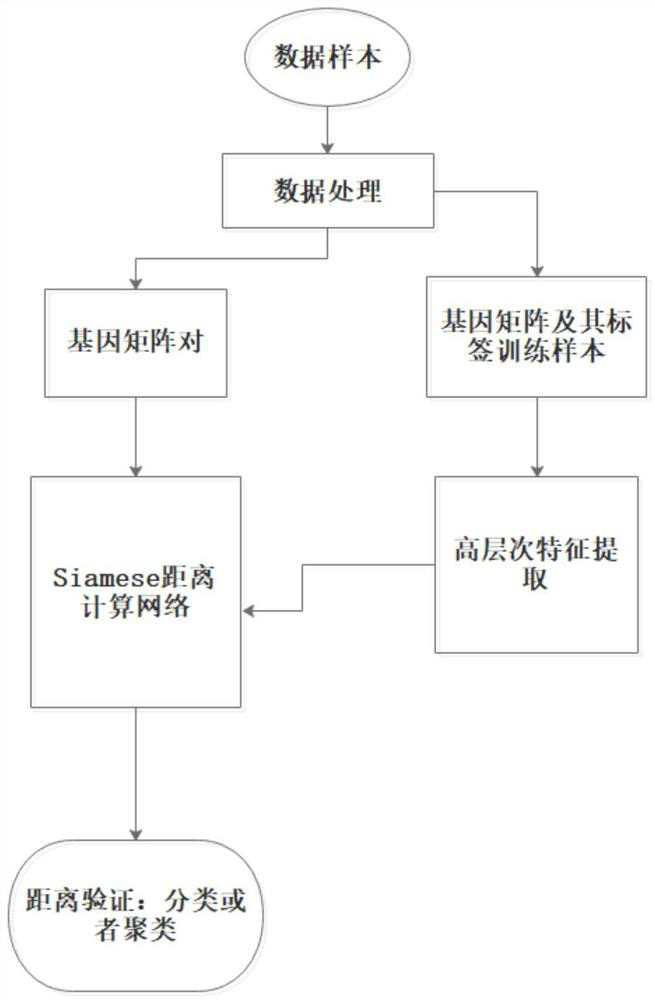
Gene expression profile distance measurement method based on deep learning
ActiveCN110033041AAccurate and fast calculationCharacter and pattern recognitionNeural architecturesGene EnrichmentProfile analysis
The invention belongs to the field of gene expression profile classification, discloses a gene expression profile distance measurement method based on deep learning, and belongs to mining and application of deep learning on biological big data. Firstly, a convolutional neural network model suitable for gene characteristic metric learning is designed to extract data characteristics, then the distance between the data is calculated by applying an improved cosine distance, and finally, the good performance of the method is measured through the classification effect of a classification algorithm.According to the method, the similarity between different gene expression profiles can be quickly and efficiently measured, and data is provided for subsequent researches such as gene classification,clustering, differential expression analysis and compound screening. Compared with a traditional gene enrichment method, the method has the advantages that the distance measurement effect between thedata is obviously improved, the manual intervention during gene expression profile analysis can be effectively reduced, the overfitting phenomenon easily generated by a conventional deep network is avoided, and the method has relatively high mobility.
Owner:HUNAN UNIV
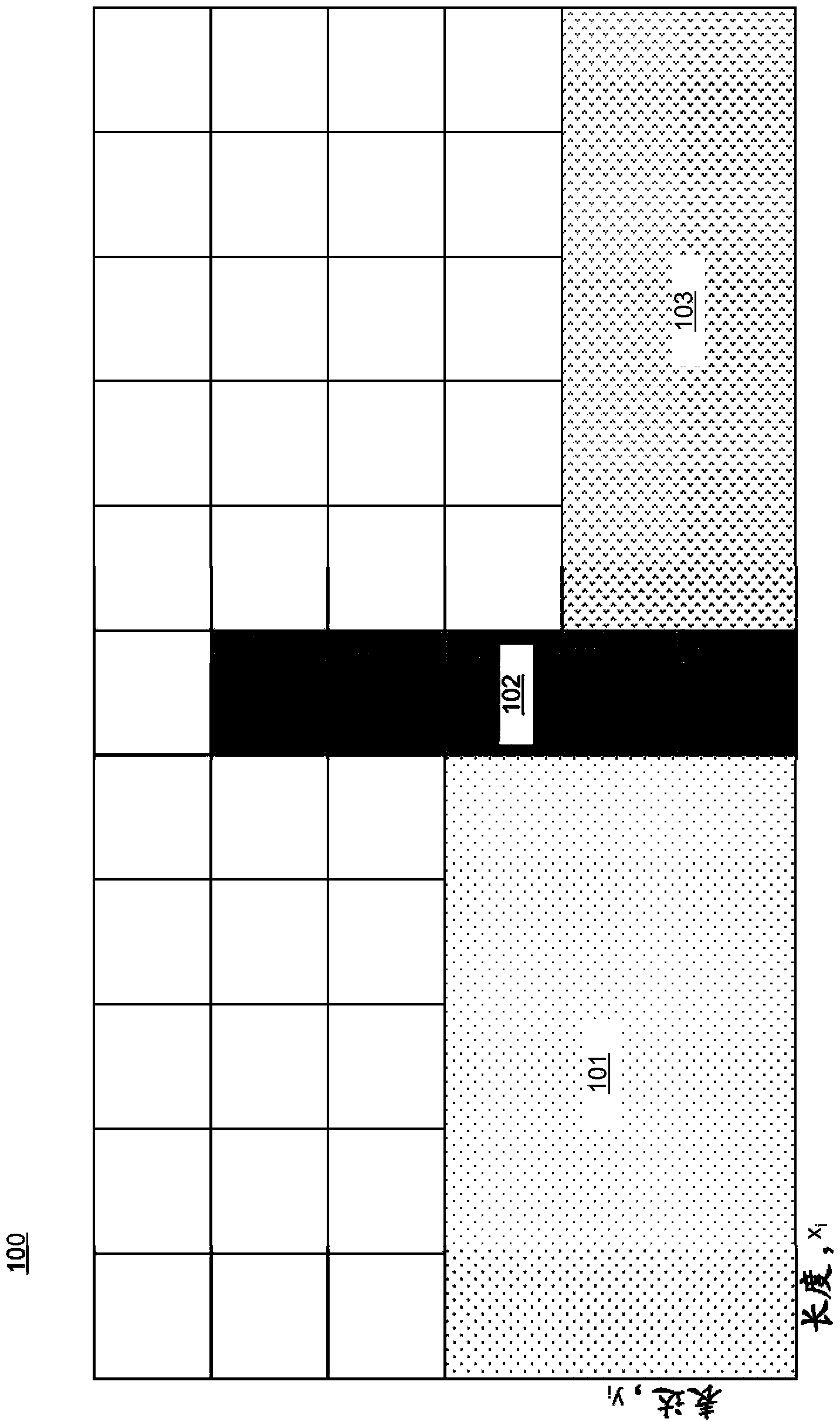
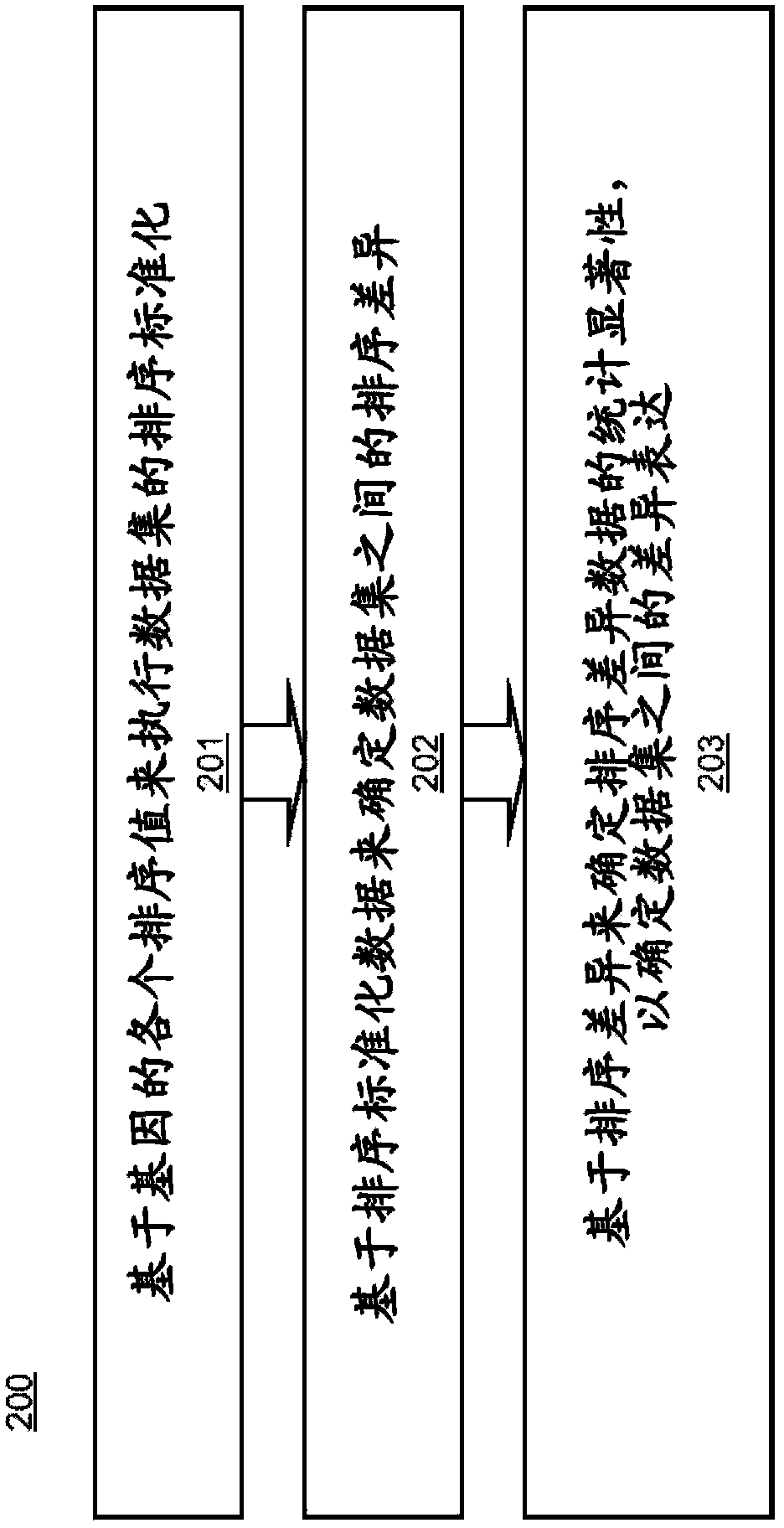
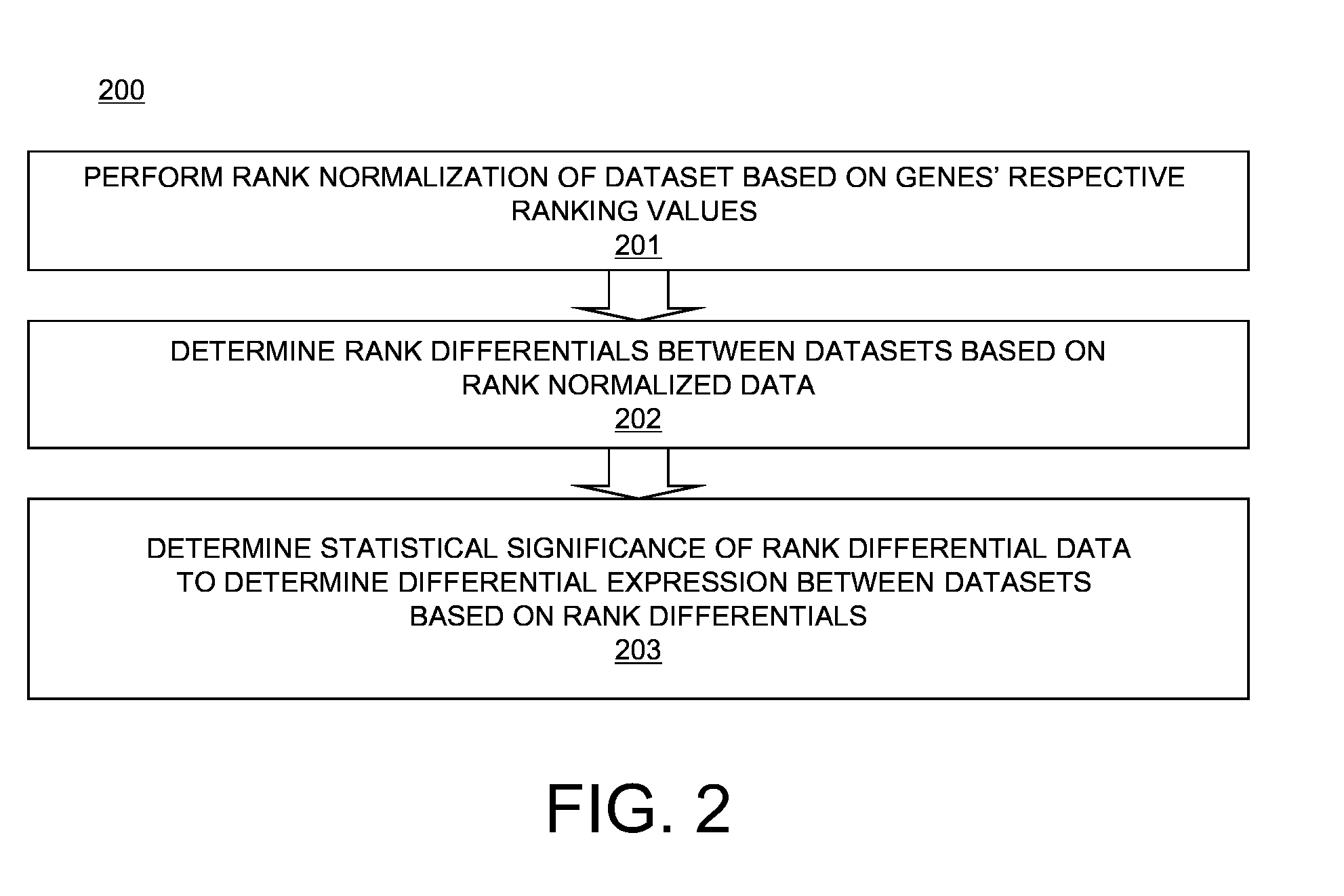
Computer-implemented method and computer system for rank normalization for differential expression analysis of transcriptome sequencing data
A computer-implemented method for rank normalization for differential expression analysis of transcriptome sequencing data includes receiving, by a computer, a first dataset comprising transcriptome sequencing data, the first dataset comprising a plurality of genes, and further comprising a respective ranking value associated with each of the plurality of genes; assigning a rank to each of the genes of the plurality of genes based on the ranking value to produce a first rank normalized dataset; determining a change between a first rank of a particular gene in the first rank normalized dataset, and a second rank of the particular gene in a second rank normalized dataset, the second rank normalized dataset being based on a second dataset comprising transcriptome sequencing data; and determining whether the particular gene is differentially expressed between the first dataset and the second dataset based on the determined change in rank.
Owner:IBM CORP
Rank Normalization for Differential Expression Analysis of Transcriptome Sequencing Data
A computer system for rank normalization for differential expression analysis of transcriptome sequencing data includes a processor; and a memory comprising a first dataset comprising transcriptome sequencing data, the first dataset comprising a plurality of genes and a respective ranking value associated with each of the plurality of genes, the system configured to perform a method including assigning a rank to each of the genes of the plurality of genes based on the ranking value to produce a first rank normalized dataset; determining a change between a first rank of a particular gene in the first rank normalized dataset, and a second rank of the particular gene in a second rank normalized dataset, the second rank normalized dataset being based on a second dataset comprising transcriptome sequencing data; and determining whether the particular gene is differentially expressed between the first and second datasets based on the determined change in rank.
Owner:IBM CORP
Transcriptomics and proteomics bio-information analysis method for liver cancer biological process
The invention discloses a transcriptomics and proteomics bio-information analysis method for a liver cancer biological process. The method comprises the steps of (1) performing basic analysis: performing Intersize length check; performing comparison information statistics; obtaining a randomness experimental check diagram; and performing data coverage degree and depth statistics; and (2) performing advanced analysis: performing gene optimization including new exons and 3', 5' UTR region identification; performing variable splicing including reserved introns, skipped exons, a variable first exon, a variable last exon, a variable splicing first exon, a variable splicing last exon and mutually exclusive exons; obtaining new genes or new transcripts; predicting annotations of the new transcripts; performing expression quantity analysis; performing differential expression analysis; performing differential expression clustering analysis; and performing function annotation analysis of differential expression genes: performing GO function annotation enrichment analysis. Clinical samples are subjected to located and quantitative expression verification; evidences of the clinical samples andclinical correlation are searched for; clinical values are evaluated; and a new clue is provided for liver cancer pathogenesis and liver cancer mechanism research.
Owner:南宁科城汇信息科技有限公司
Ammopiptanthus mongolicus cold resistant gene AmGS
InactiveCN102586269AImprove cold resistanceGrowth inhibitionFermentationVector-based foreign material introductionNucleotideApricot Tree
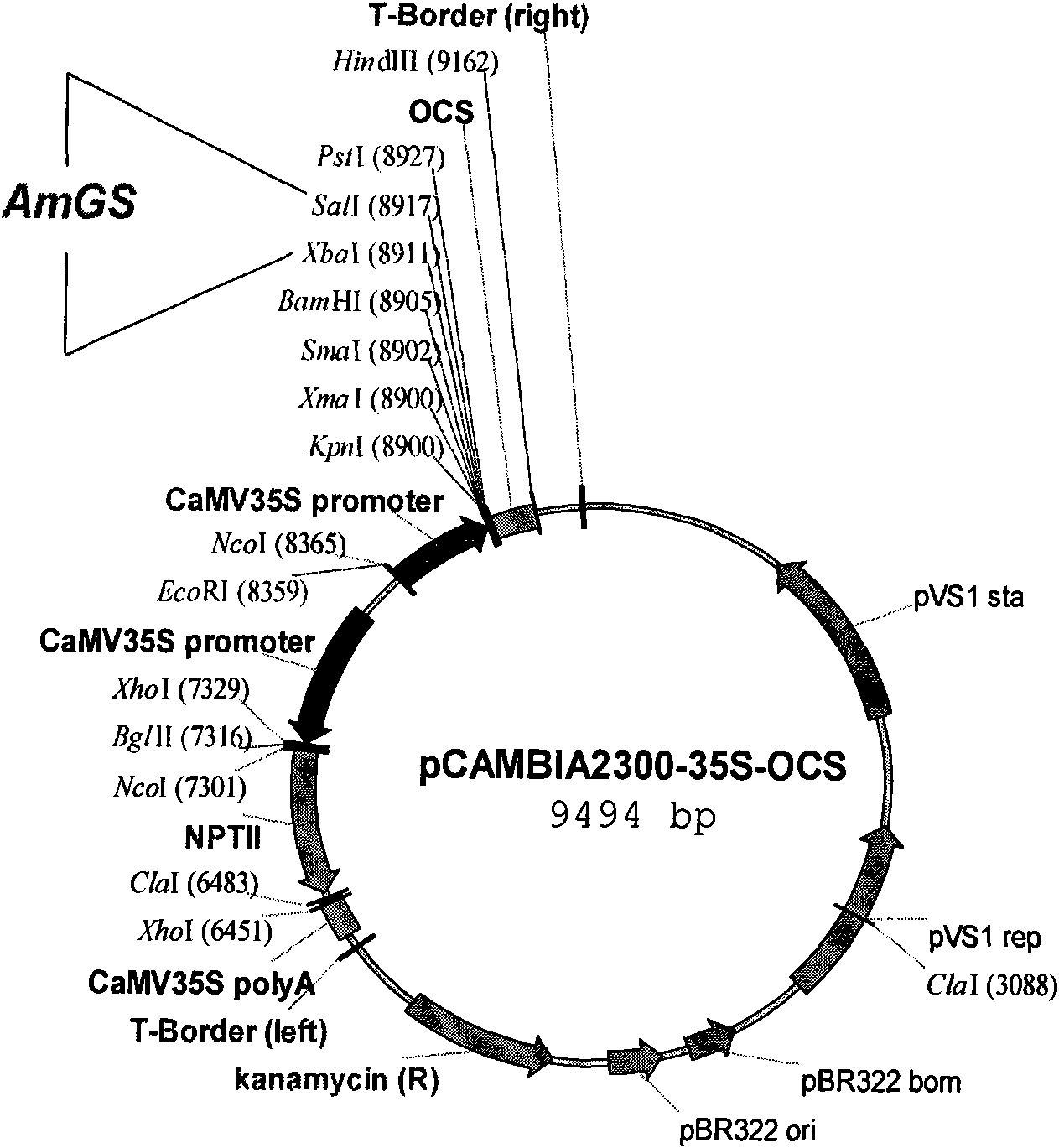
The invention provides an ammopiptanthus mongolicus cold resistant gene AmGS and relates to an endangered evergreen broad leaf woody plant considered as the second grade protection in the country and derived from the ancient tertiary period in the northwest desert region in China. According to molecular cloning the gene, the EST segment of the ammopiptanthus mongolicus cold resistant gene with low temperature induced up regulated expression is obtained through low temperature induction and differential expression analysis, and the full length cDNA sequence of the gene is obtained through RACE amplification. The AmGS comprises a complete ORF region; the ORF full length comprises 987 nucleotides; 328 amino acids and a termination codon (TAA) are coded; a plant expression vector pCAMBIA2300-AmGS of the AmGS gene is established; and the pCAMBIA2300-AmGS is used for transforming woody plants such as photinia fraseri and apricot trees, so that transgenic plants with improved cold resistance are obtained.
Owner:TAISHAN RES INST OF FORESTRY +1
Subarachnoid hemorrhage prediction model establishment method and system based on gene and cell signal pathways
InactiveCN111584085AEasy to analyzeImprove robustnessMedical simulationProteomicsCell signaling pathwaysMedicine
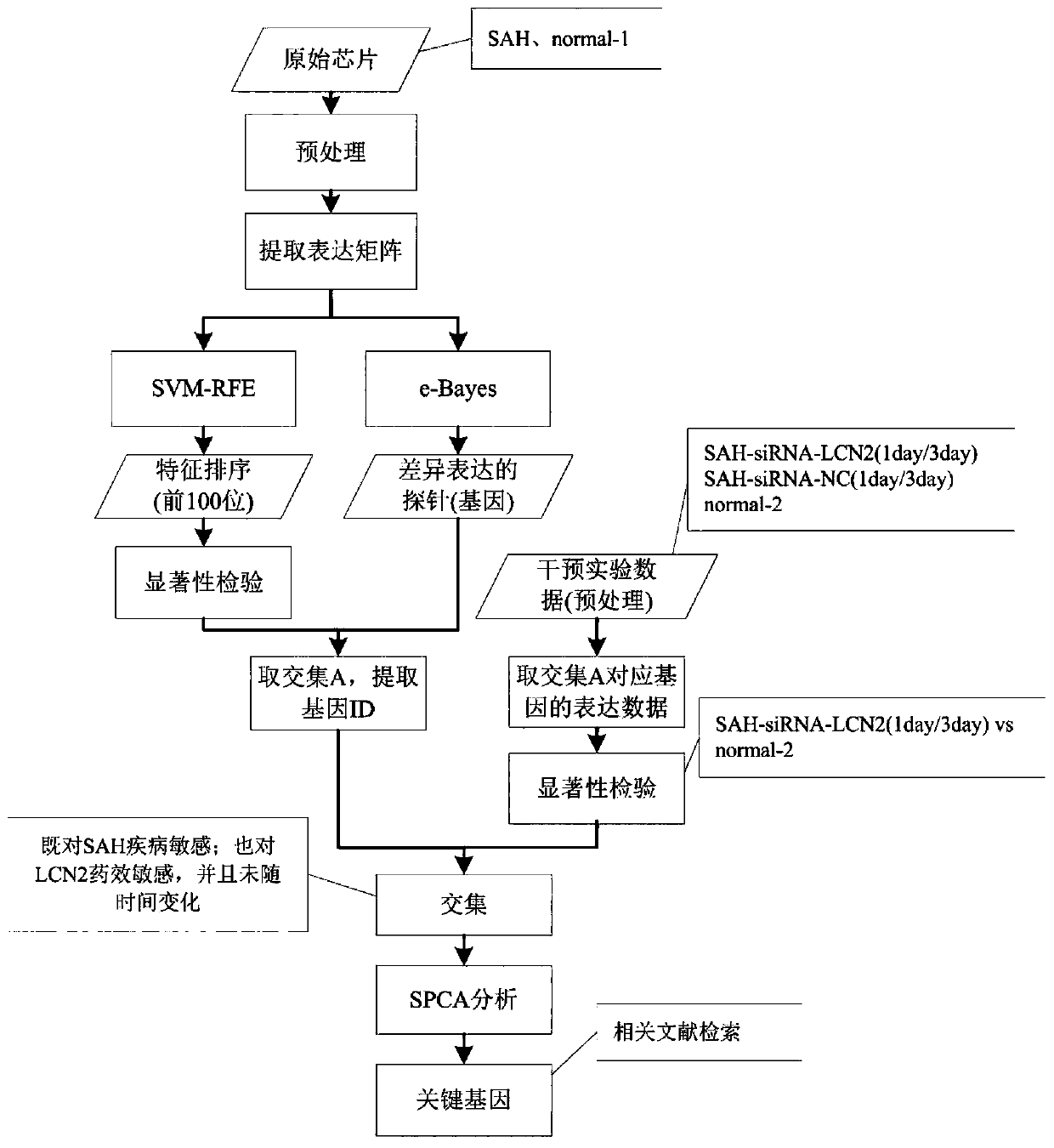
The invention provides a subarachnoid hemorrhage prediction model establishment method and system based on gene and cell signal pathways. The method comprises the steps: obtaining normal brain cell group 1 and SAH brain cell gene chip data, and preprocessing; carrying out differential expression analysis and signal path analysis on the data of the normal brain cell group 1 and the SAH brain cell gene chip; carrying out LCN2 intervention on a normal brain cell group 2 and SAH brain cells to obtain RNA-Seq data under different conditions, and preprocessing to form LCN2 data; carrying out differential expression analysis and signal path analysis on the LCN2 data; carrying out feature selection on the differentially expressed gene data obtained through the differential expression analysis to obtain feature gene data; and obtaining a training sample based on the feature gene data, training a plurality of classifiers based on the training sample, integrating the trained classifiers, and establishing a prediction model. According to the method, the subarachnoid hemorrhage model can be accurately established, and the treatment target can be obtained.
Owner:SICHUAN UNIV
Rank Normalization for Differential Expression Analysis of Transcriptome Sequencing Data
A computer-implemented method for rank normalization for differential expression analysis of transcriptome sequencing data includes receiving, by a computer, a first dataset comprising transcriptome sequencing data, the first dataset comprising a plurality of genes, and further comprising a respective ranking value associated with each of the plurality of genes; assigning a rank to each of the genes of the plurality of genes based on the ranking value to produce a first rank normalized dataset; determining a change between a first rank of a particular gene in the first rank normalized dataset, and a second rank of the particular gene in a second rank normalized dataset, the second rank normalized dataset being based on a second dataset comprising transcriptome sequencing data; and determining whether the particular gene is differentially expressed between the first dataset and the second dataset based on the determined change in rank.
Owner:IBM CORP
RNA analyzing method of paraffine section tissue
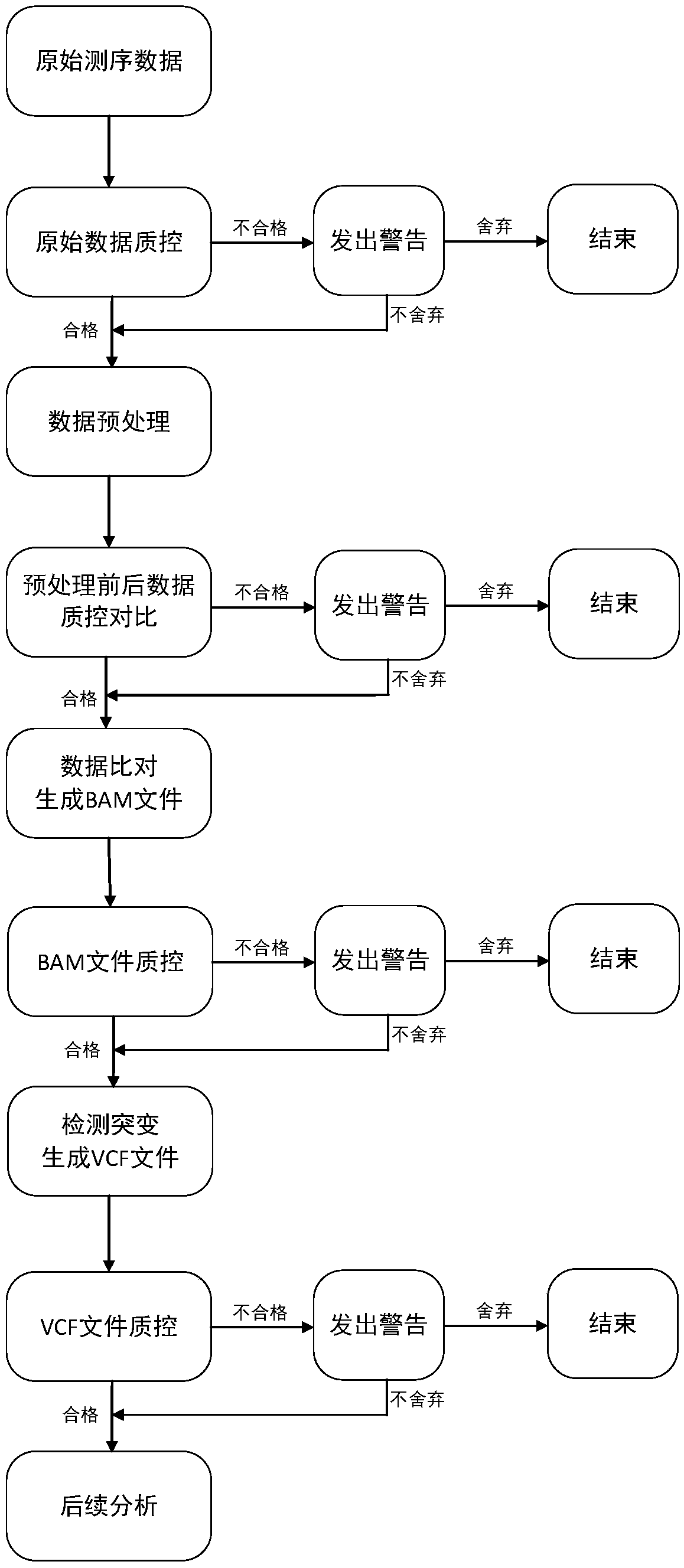
PendingCN110684830AUnderstand the reaction mechanismThe assessment results are accurateMicrobiological testing/measurementSequence analysisFusion geneExpression analysis
The invention provides an RNA analyzing method of paraffine section tissue. The analyzing method comprises the following steps of performing DNA degradation on the paraffine section tissue, and extracting RNA of samples; preparing a paraffine section sample nucleic acid library, and sequencing sample RNA; performing quality control on the sample data obtained by sequencing; comparing the sample data after quality control with a reference genome, and performing quality control on comparing results; and performing transcriptome assembling and transcript quantifying on the sample data after quality control of comparing results, and performing quantification analysis, gene difference expression analysis and fusion gene analysis on gene expression. The invention provides an index and detectionmethod for completely assessing RNA quality of the paraffine section tissue, so that RNA of the paraffine section tissue can be comprehensively assessed, and the assessment results are accurate and effective. An effective reference basis is provided for subsequent analysis accuracy.
Owner:深圳吉因加医学检验实验室
Parkinson disease evolution key module identification method based on miRNA sequencing data
The invention provides a Parkinson's disease key module identification method based on miRNA (micro Ribonucleic Acid) sequencing data. The method comprises the following steps: firstly, preprocessing high-throughput sequencing data; then, grouping the samples according to different stages of PD diseases, and carrying out differential expression analysis; then, carrying out hierarchical clustering according to correlation coefficients between differential expression miRNAs, and constructing a co-expression network and a module; and finally, constructing a module network and performing identifying to obtain a PD key module. By means of the method, key module identification in the PD evolution process can be carried out, the PD stage of the current patient is judged according to the key module, and help is provided for doctors to find early PD patients.
Owner:NORTHWESTERN POLYTECHNICAL UNIV
Protein biomarker for sepiella japonica in senescence process
InactiveCN110734485ALow costA large number of screeningComponent separationBiological material analysisPotential biomarkersCarrier protein
The invention provides a protein biomarker for sepiella japonica in the senescence process and belongs to the technical field of genetic engineering. The protein biomarker comprises a mitochondrial ATP / ADP (adenosine triphosphate / adenosine diphosphate) carrier protein SLC25A4S, has relative abundance of 1.82*10<7> before sexual maturation and oviposition, and has relative abundance of 0 during oviposition and articulo mortis after oviposition. A screening method of the protein biomarker comprises the following steps: extracting optic gland protein samples from individuals of sepiella japonicaat different reproductive stages; performing enzymolysis on the protein samples by using trypsin; analyzing the protein samples after enzymolysis by using liquid chromatogram-mass spectrum combination; and performing protein differential expression analysis, gene ontology analysis, signal pathway analysis and protein homologous cluster analysis through LC-MS (liquid chromatogram-mass spectrum) data processing. Through proteomic analysis and screening on the optic gland protein samples of the sepiella japonica at different reproductive stages, potential biomarkers can be obtained.
Owner:ZHEJIANG OCEAN UNIV
Method of analyzing and identifying DHA biosynthesis related fatty acid desaturase gene in crypthecodinium cohnii by means of De novo transcriptome
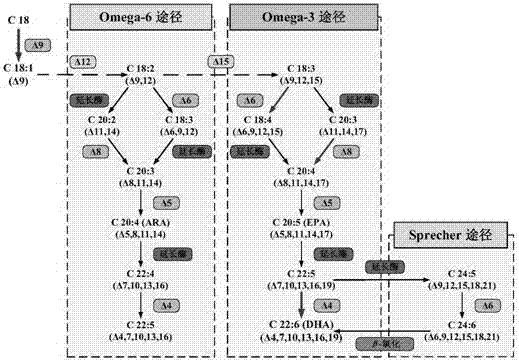
ActiveCN106916891AImprove synthesis abilityMicrobiological testing/measurementProteomicsRNA extractionOrganism
The invention discloses a method of identifying DHA (docosahexaenoic acid) biosynthesis related fatty acid desaturase gene in crypthecodinium cohnii through De novo transcriptome sequencing, transcript splicing and differential expression analysis. The method particularly includes the following steps: (1) performing DHA fermentation culture to the crypthecodinium cohnii; (2) collecting thallus at different growth and DHA accumulation stages, and performing RNA extraction and transcriptome Illumina deep sequencing; (3) analyzing and pre-processing sequenced data; (4) performing function annotation and correlation statistic analysis to the spliced transcriptome, identifying the DHA biosynthesis related fatty acid desaturase in the crypthecodinium cohnii and acquiring a whole gene sequence thereof; and (5) by means of qRT-PCR analysis technology, performing expression verification to the DHA biosynthesis related fatty acid desaturase gene in the crypthecodinium cohnii. The method provides important approaches and knowledge basis for researching biosynthesis mechanism of DHA in the crypthecodinium cohnii and improving DHA synthesis capability of the crypthecodinium cohnii.
Owner:KUNMING ZAONENG BIOTECH CO LTD
Method for predicting abnormity of triple negative breast cancer spindle assembly checkpoints by means of lncRNA-mRNA co-expression network
The invention relates to a method for predicting the abnormity of triple negative breast cancer spindle assembly checkpoints by means of an lncRNA-mRNA co-expression network. The method includes experimental grouping, RNA extraction, lncRNA chip data differential expression analysis, and lncrna target gene analysis and control network construction. The method for predicting the abnormity of triplenegative breast cancer spindle assembly checkpoints by means of the lncRNA-mRNA co-expression network provides a new way for finding a new molecular marker of the triple negative breast cancer and provides the reference basis for predicting the prognosis of the triple negative breast cancer.
Owner:范志民
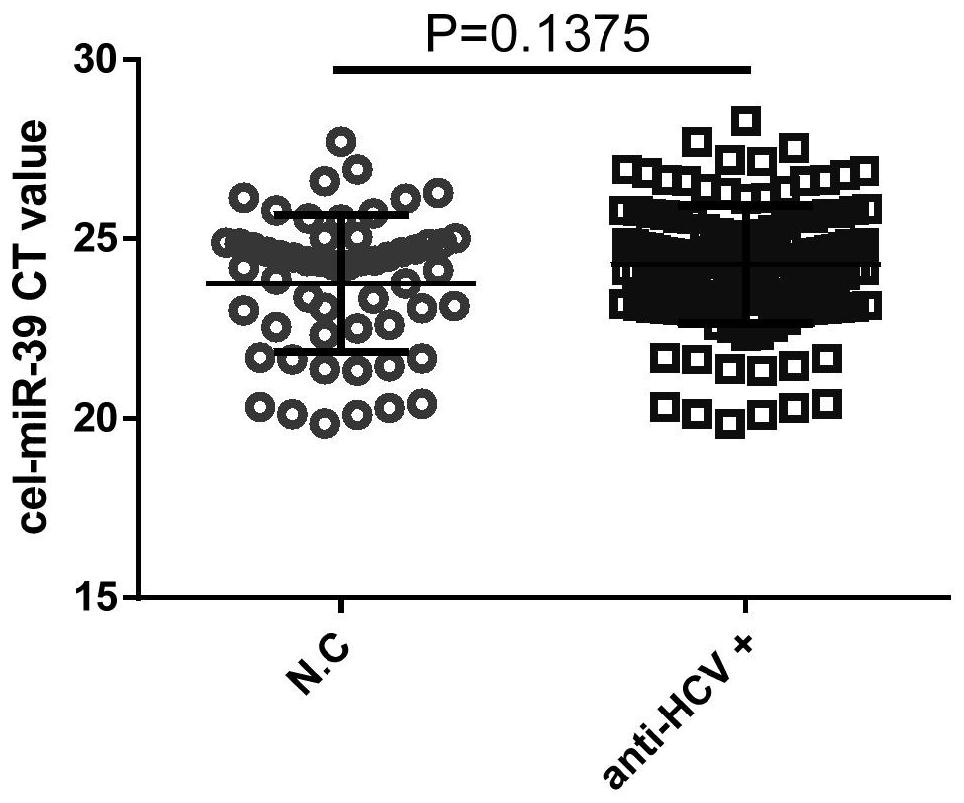
Plasma miRNA markers and application thereof
The invention provides a group of plasma miRNA (micro Ribonucleic Acid) markers, the plasma miRNA markers are used for determining HCV (Hepatitis C Virus) infection, and the markers comprise miR-20 and miR-122. Through differential expression analysis of miRNA in plasma, the inventor finds that the plasma miRNA marker combination is used for confirmation and diagnosis of anti-HCV+ / HCV RNA+ (anti-HCV positive and HCV RNA positive) blood donors, anti-HCV+ / HCV RNA-(anti-HCV positive, HCV RNA negative) blood donors, and anti-HCV- / HCV RNA+ (anti-HCV negative and HCV RNA positive) blood donors, and has good sensitivity, specificity and universality.
Owner:SHENZHEN BLOOD CENT
Systems and Methods for Multi-Scale, Annotation-Independent Detection of Functionally-Diverse Units of Recurrent Genomic Alteration
InactiveUS20190348151A1Microbiological testing/measurementBiostatisticsHuman DNA sequencingBinding site
The functional interpretation of somatic mutations remains a persistent challenge in the interpretation of human genome data. Systems and methods for detecting significantly mutated regions (SMRs) in the human genome permit the discovery and identification of multi-scale cancer-driving mutational hotspot clusters. Systems and methods of SMR detection reveal differentially mutated genetic regions across various cancer types. SMR detection and annotation reveals a diverse spectrum of functional elements in the genome, including at least single amino acids, compete coding exons and protein domains, microRNAs, transcription factor binding sites, splice sites, and untranslated regions. Systems and methods of SMR detection optionally including protein structure mapping uncover recurrent somatic alterations within proteins. Systems and methods of SMR detection optionally including differential expression analysis reveal previously unappreciated connections between recurrent and somatic mutations and molecular signatures.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
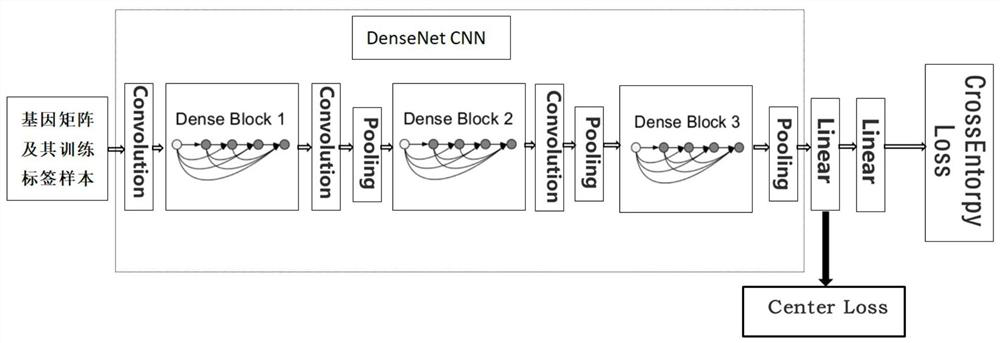
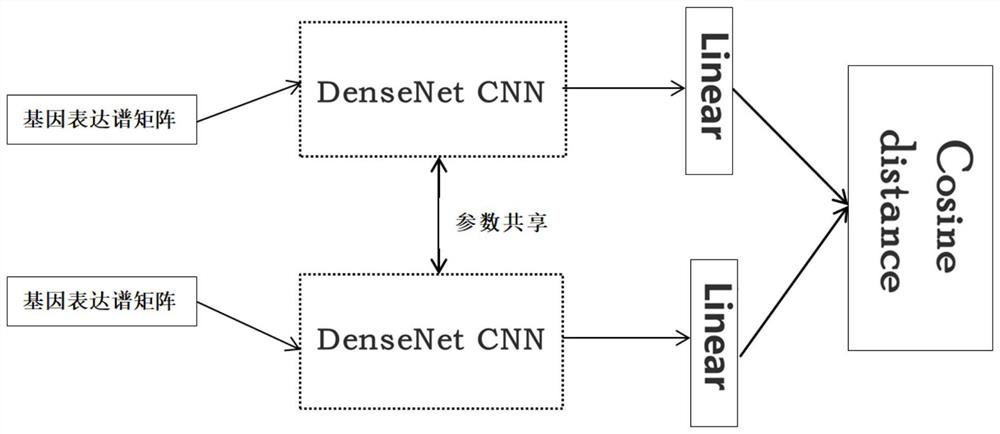
A distance measurement method for gene expression profiles based on deep learning
ActiveCN110033041BAccurate and fast calculationCharacter and pattern recognitionNeural architecturesGene FeatureExpression analysis
The invention belongs to the field of gene expression spectrum classification, discloses a gene expression spectrum distance measurement method based on deep learning, and belongs to the mining and application of deep learning on biological big data. First, a convolutional neural network model suitable for gene feature metric learning is designed to extract the characteristics of the data, and then the distance between the data is calculated by using the improved cosine distance, and finally the classification effect of the classification algorithm is used to measure the performance of the method excellent. This method can quickly and efficiently measure the similarity between different gene expression profiles, and provide data for subsequent studies such as gene classification, clustering, differential expression analysis, and compound screening. Compared with the traditional gene enrichment method, this method significantly improves the distance measurement effect between data, and can effectively reduce the manual intervention in gene expression profile analysis, avoiding the over-fitting phenomenon that is easy to occur in conventional deep networks. This method has strong transferability.
Owner:HUNAN UNIV
Method for screening genes related to synthesis of target compound and application
PendingCN113337589AImprove accuracyAchieve high quality and high yieldMicrobiological testing/measurementTranscriptome SequencingGenomic annotation
The invention discloses a method for screening genes related to synthesis of a target compound and application. The method in the invention comprises the following steps of: taking a third-generation transcript as a reference transcript, and comparing second-generation sequencing data to the transcript to obtain the abundance of each transcript; and quantifying the second-generation assembled transcript by using RSEM software, and carrying out differential expression analysis between genes to obtain a key gene in the terpenoid synthesis process. Compared with a second-generation transcriptome sequencing technology and a third-generation full-length transcriptome sequencing technology which are independently used, the invention has the advantages that: the advantage that the full-length transcriptome sequence can be obtained without splicing the third-generation transcriptome can be sufficiently utilized, so that the accuracy is high; the advantage of quantitative expression detection of genes by second-generation transcriptome sequencing data can be fully considered; the accuracy of genome annotation is greatly improved; and related genes in a synthetic route of the target compound are obtained through screening. According to the method, four genes related to cinnamomum burmannii monoterpenoid synthetase are obtained for the first time.
Owner:SOUTH CHINA UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com