Method for screening genes related to synthesis of target compound and application
A compound and purpose technology, applied in the field of genes related to compound synthesis, can solve the problems of less chimeras, differentially expressed genes, and unobtainable genes, etc., and achieve the effect of improving accuracy and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
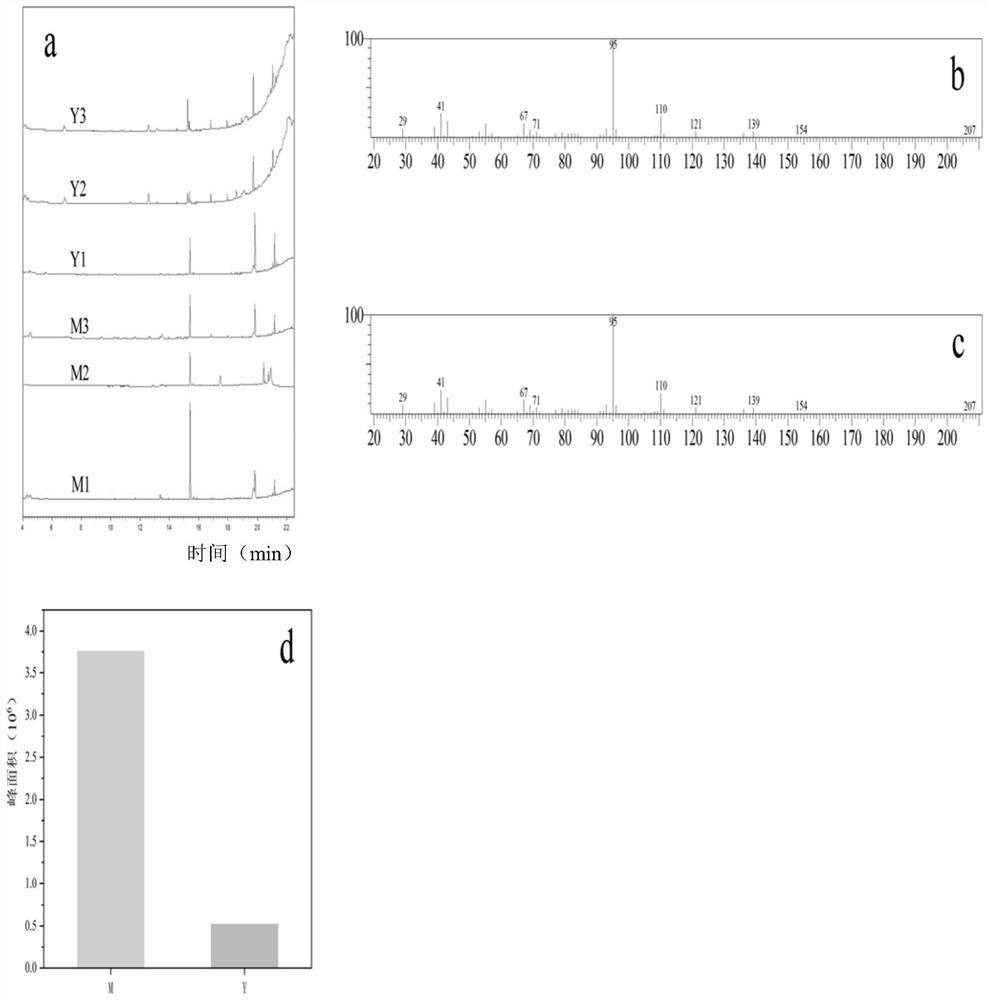
[0061] 1. Extraction of secondary metabolites in Cinnamomum burmanii
[0062] Weigh 1g of fresh Cinnamomum burmanii leaves and remove the veins and petioles and cut them into pieces and place them in 60mL of absolute ethanol. The ratio of fresh leaf samples to absolute ethanol is 1:60 (g / mL). At a temperature of 80°C, the extraction time was 6h, and then the volatile oil was dilute to 100mL.
[0063] 2. GC-MS analysis of compound composition in two chemical types of Cinnamomum burmanii
[0064] Gas chromatography analysis using Agilent 7890A gas chromatograph (column: Agilent 19091N-113, 30m × 320μm × 0.25μm). The carrier gas was nitrogen with a flow rate of 2 mL / min. GC program: 70°C / min, rising to 100°C at 3°C / min, rising to 250°C at 15°C / min, and maintaining for 1 min. The injection volume was 1.0 μL without splashing. The injection port temperature is 220°C, and the detection temperature is 230°C.
[0065] Gas chromatography-mass spectrometry uses Shimadzu QP2010 PLUS...
Embodiment 2
[0068] 1. Extraction of RNA
[0069] TaKaRa MiniBEST Plant RNA Extraction Kit was used.
[0070] (1) Quickly transfer fresh or cryopreserved plant tissue samples to a mortar pre-cooled with liquid nitrogen, and grind the tissue with a pestle, during which liquid nitrogen is continuously added until it is ground into powder (no obvious visible particles , if it is not ground thoroughly, it will affect the yield and quality of RNA). Add the powdered sample (20-50 mg) into a 1.5 mL sterilized tube containing 450 μL Buffer PE, and pipette repeatedly until there is no obvious precipitation in the lysate.
[0071] (2) Centrifuge the lysate at 12,000 rpm at 4°C for 5 minutes.
[0072] (3) Pipette the supernatant carefully into a new 1.5mL sterile tube. Add 1 / 10 volume of Buffer NB to the supernatant (precipitation will appear at this time), shake and mix.
[0073] (4) Centrifuge at 12,000 rpm at 4°C for 5 minutes.
[0074] (5) Pipette the supernatant carefully into a new 1.5mL s...
Embodiment 3
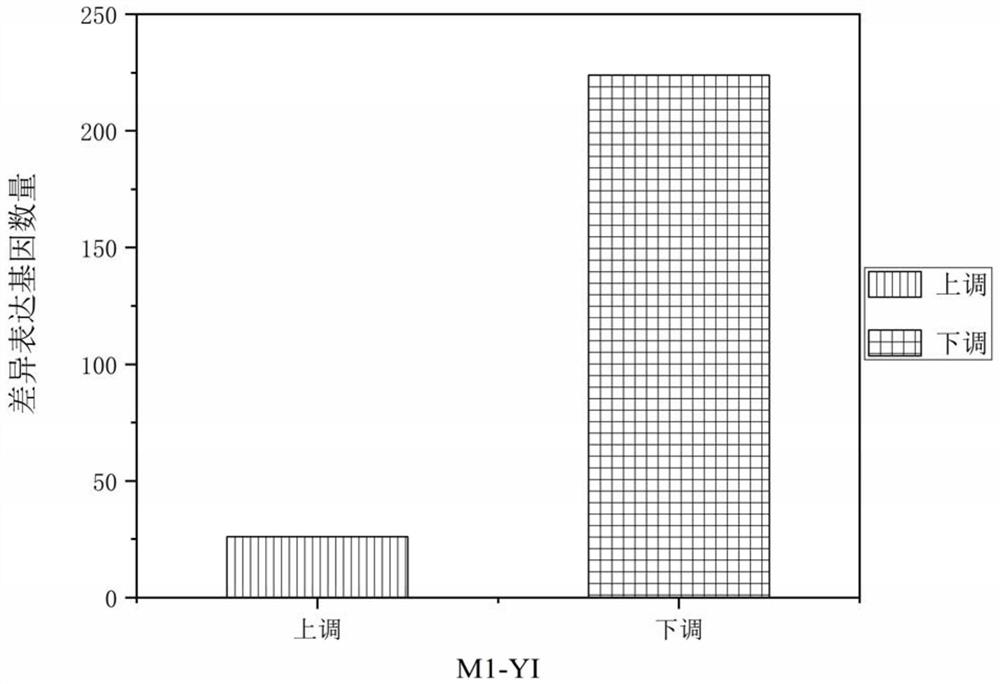
[0093] 1. Three-generation full-length transcriptome sequencing
[0094](1) Synthesis of first-strand cDNA: The qualified RNA (or poly A+RNA) is reverse-transcribed into first-strand cDNA using Clontech SMARTerPCR cDNA Synthesis Kit; PCR amplification is used to synthesize double-stranded cDNA; PCR product purification: use AMPure PB Bead purifies PCR amplification products; SMRTbell library construction: including DNA damage repair, end repair, ligation of aptamers, and library quality assessment; Sequencing on the machine: annealing of SMRTbell library combined with primers and polymerase, using MagBead Loading on the machine sequencing.
[0095] (2) Using SMRT Link v5.0.1 supported by Pacific Biosciences to pair the raw sequencing data (reads) of the cDNA library
[0096] Sorted and clustered into consensus transcripts. Simply put, CCS (circular consensus sequence) is extracted from the BAM file. Then according to the cDNA primers and polyA tail signal, the CCS data were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com