A distance measurement method for gene expression profiles based on deep learning
A gene expression profile and distance measurement technology, which is applied in the field of gene expression profile distance measurement based on deep learning, can solve the problems of high time cost, poor performance of calculating gene expression profile distance, etc., and achieve the effect of low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
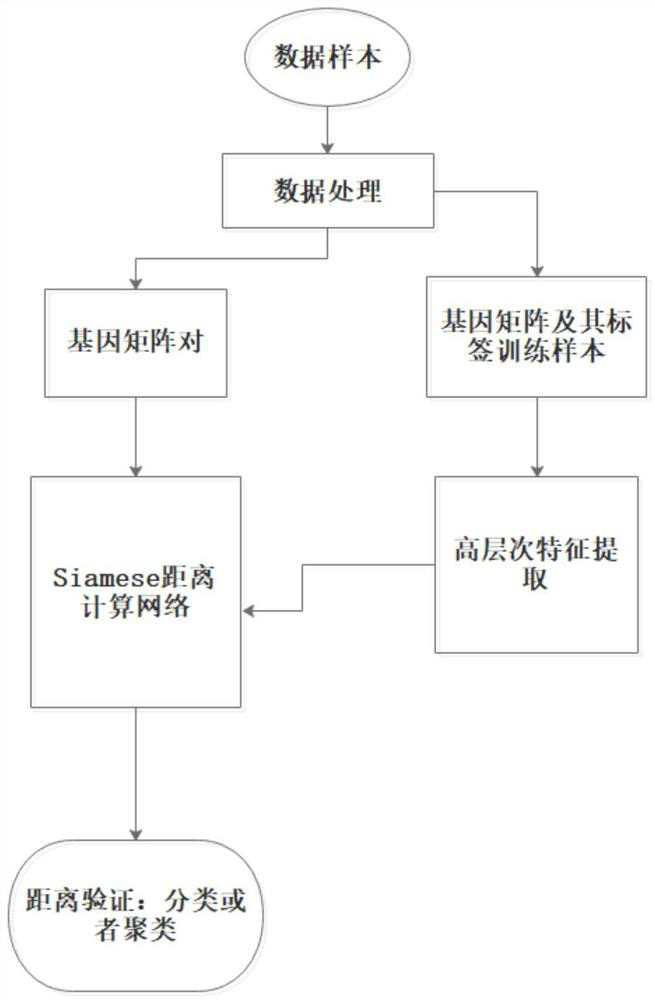
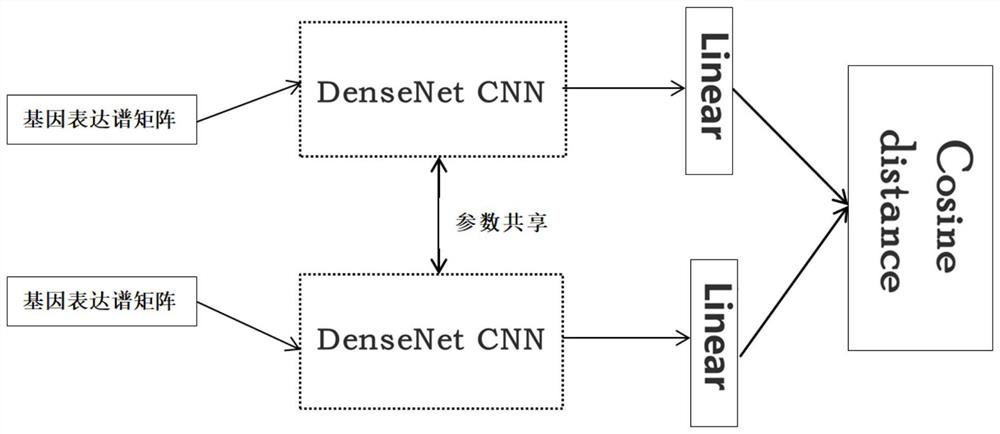
[0028] according to figure 1 The flow shown in the implementation mode includes the following four steps:
[0029] Step 1: data conversion processing, including the following steps,
[0030] 1.1. Convert the gene expression profile data into a square data matrix, and the length of the square matrix is calculated according to the dimension of the expression profile data. The specific calculation method is: convert the sample whose data dimension is N into a square matrix of x*x, where x is passed through the formula Obtained, the extra pixel position is filled to 0.
[0031] 1.2. Perform normalization and mean subtraction data preprocessing operations on the square matrix.
[0032] 1.3. Assign different category labels to the expression spectrum matrices of different categories, and divide the training, verification and test sample sets.
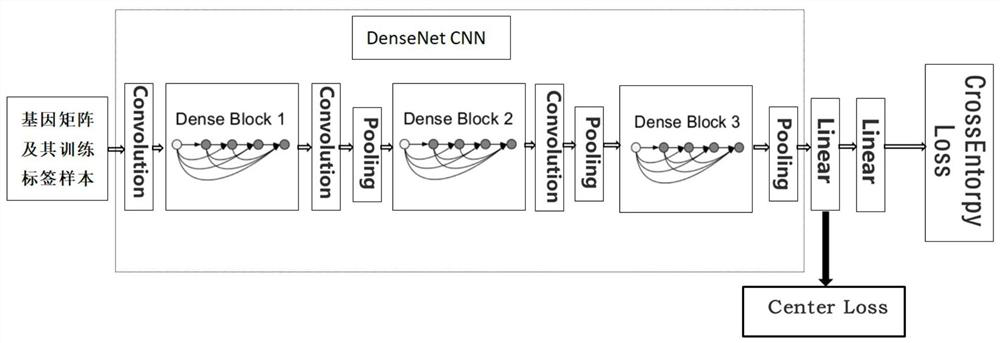
[0033] Step 2: extracting high-level features of training sample data, including the following steps,
[0034] 2.1. Pass the trainin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com