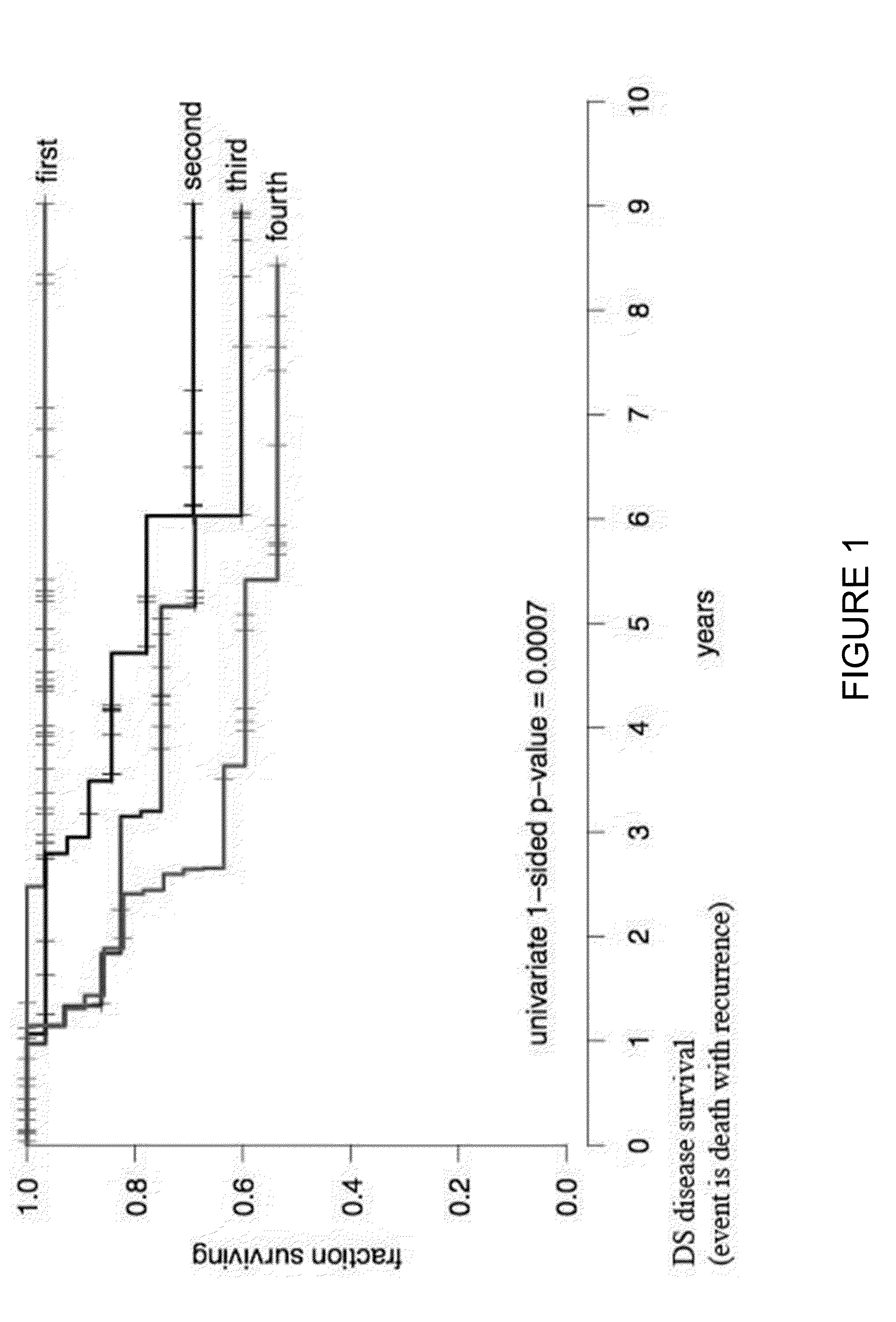
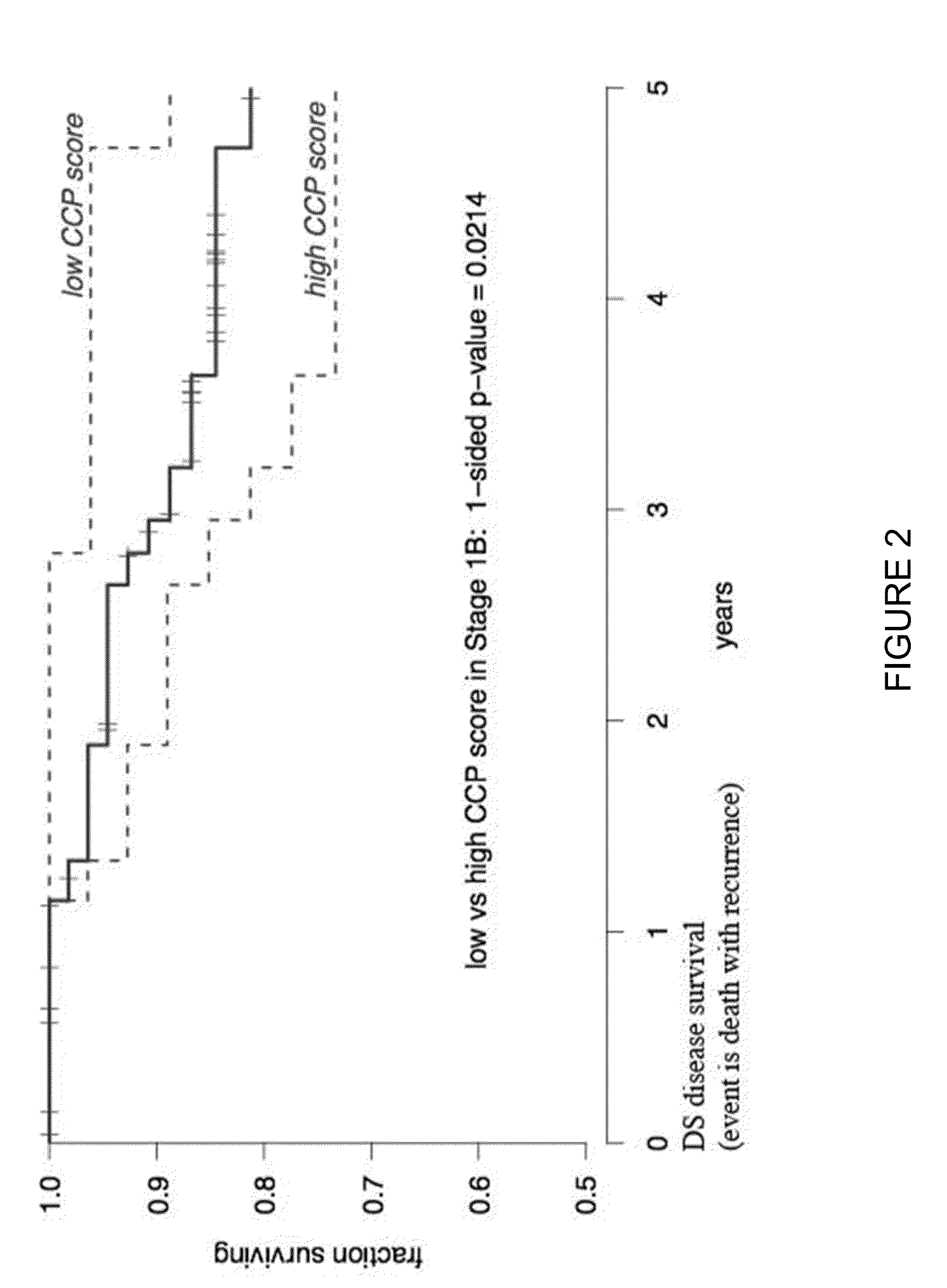
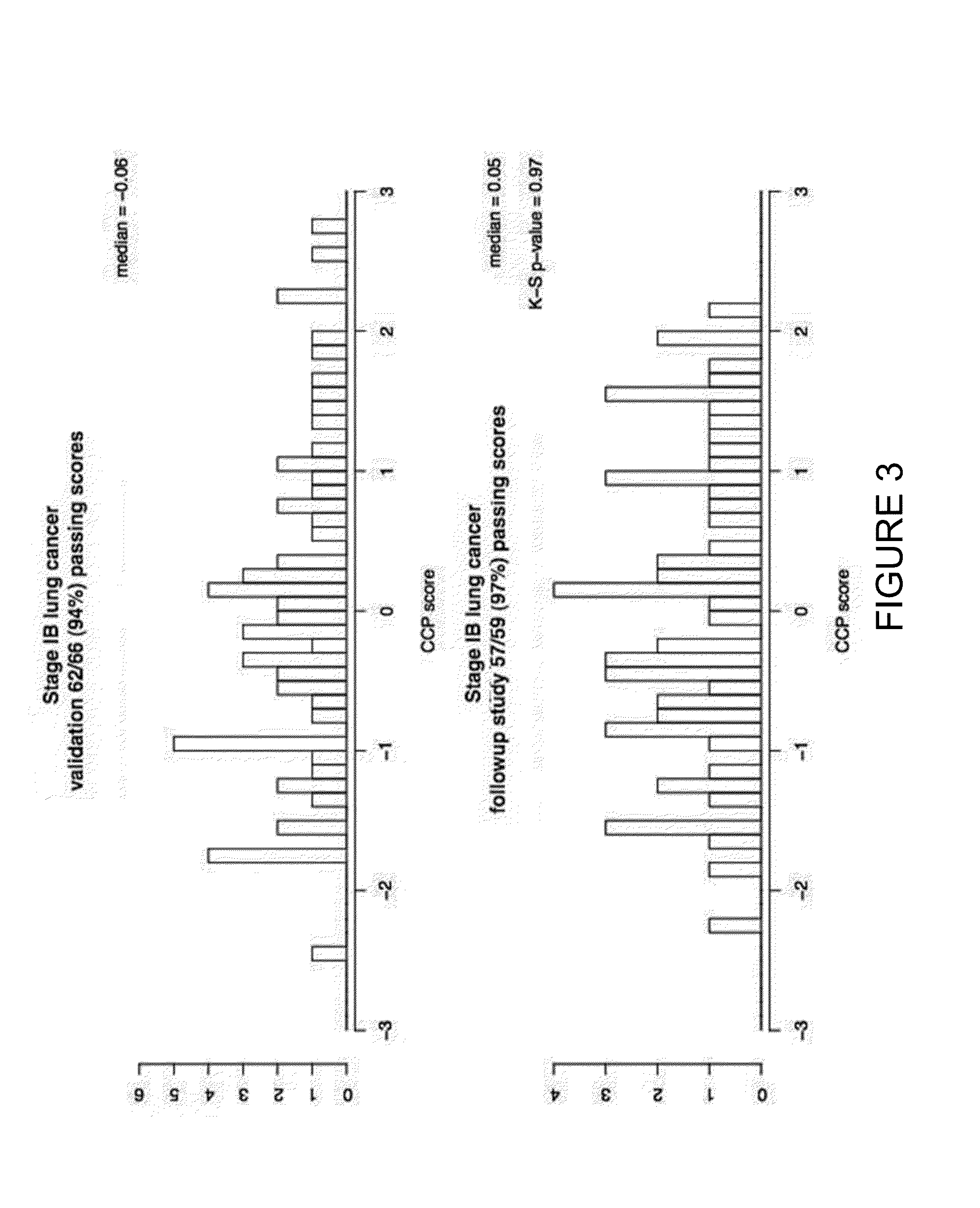
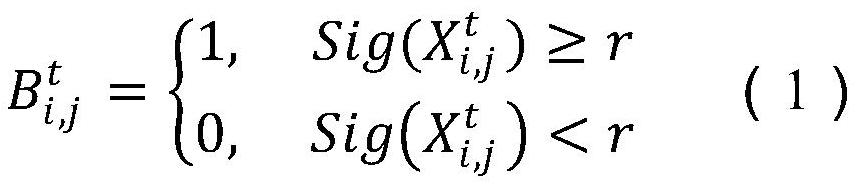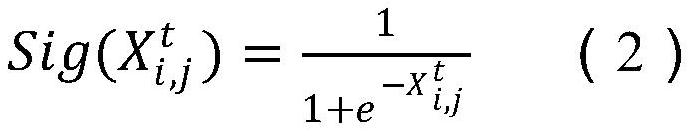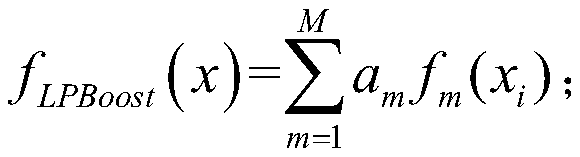Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
91 results about "Gene Feature" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Gene Feature Identification. Abstract. The identification of all genes is one of the goals of any genome‐sequencing project. Apart from laboratory techniques, genes can also be identified by using computational, homology‐based or ab initio (model‐based) methods, which differ in their performance according to the sequence being analysed.
Method for carrying out high-throughput sequencing on TCR (T cell receptor) or BCR (B cell receptor) and method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing tag sequences
InactiveCN103710454AFully functionalReduce sequencing errorsMicrobiological testing/measurementDNA/RNA fragmentationV regionPcr ctpp
The invention provides a method for carrying out high-throughput sequencing on a TCR (T cell receptor) or a BCR (B cell receptor). The method is characterized by designing upstream primers according to gene features of a V region of the TCR or the BCR and designing downstream primers according to gene features of a C region or a J region of the TCR or the BCR and obtaining sequences of the of the TCR or the BCR in combination with the multiplex PCR (polymerase chain reaction) technology and high-throughput sequencing, thus analyzing the rearrangement information of the TCR or the BCR. Compared with 25-30 cycles of existing multiplex PCR, two cycles of the multiplex PCR technology provided by the invention can conduce to greatly reducing the sequencing errors caused by primer amplification preference. Besides, the invention also provides a method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing DNA (deoxyribonucleic acid) tag sequences, thus further reducing the sequencing errors caused by primer amplification preference and intrinsic sequencing errors of high-throughput sequencing.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA +1
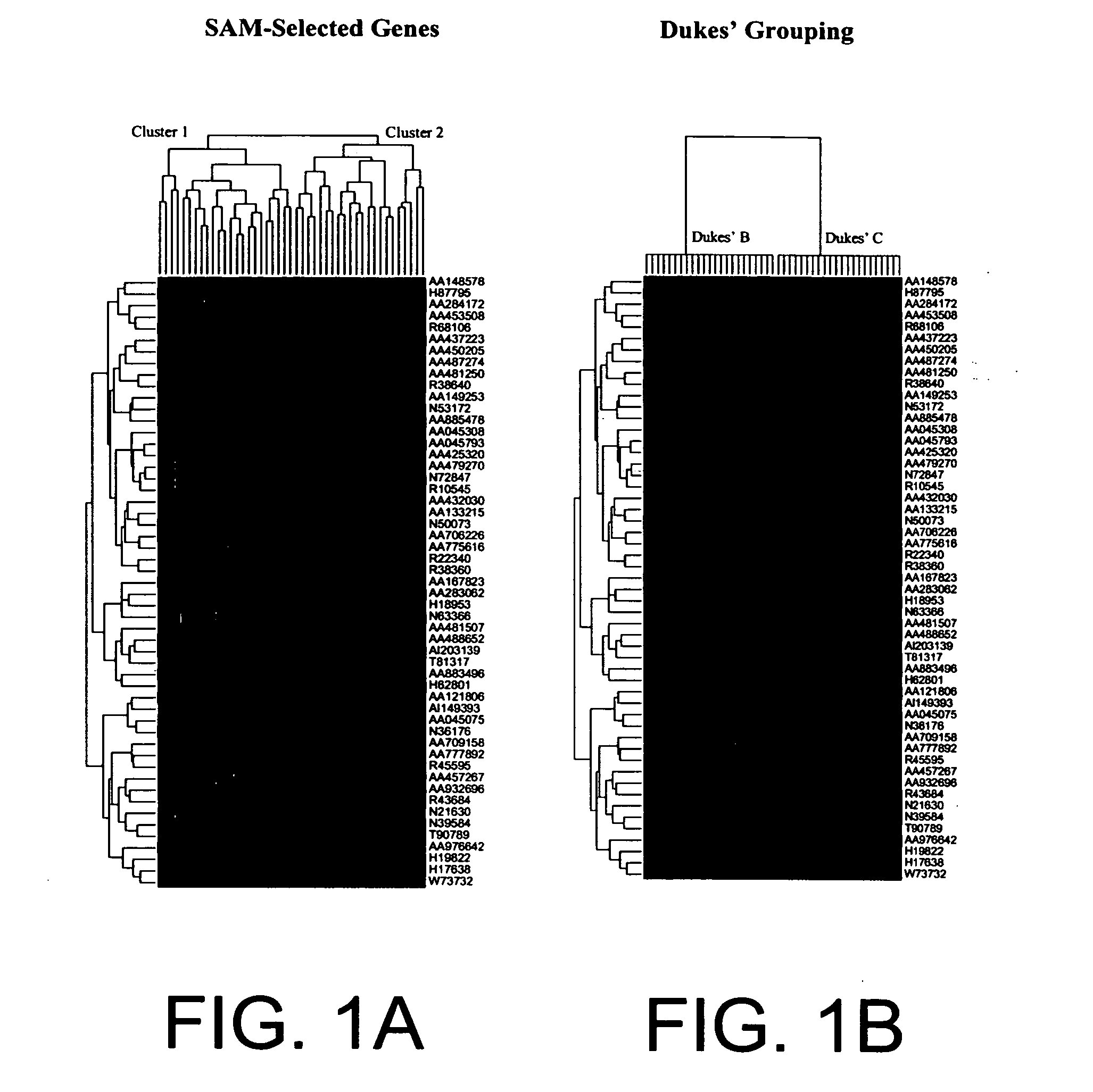
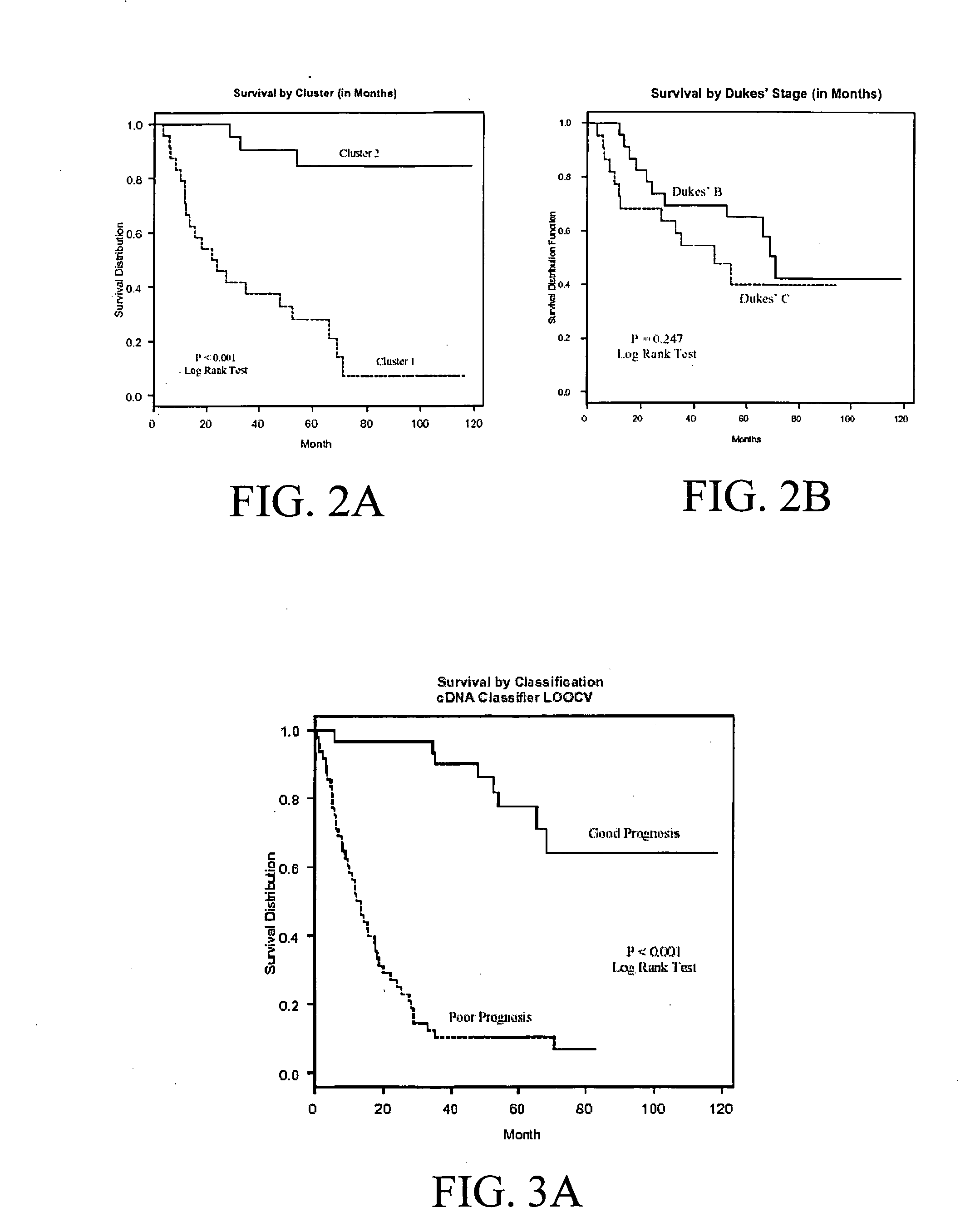
Methods for predicting cancer outcome and gene signatures for use therein
InactiveUS20060195266A1Accurate predictionMicrobiological testing/measurementBiostatisticsSupport vector machineGene Feature
The present invention pertains to specific gene signatures for cancer that are used to predict survival and novel processes for identifying such gene signatures. In one embodiment, gene signatures for human colorectal cancer are identified and outcomes are linked to the specific gene signatures using significance analysis of microarrays (SAM) and support vector machines (SVM) to provide a prognosis / survival classifier.
Owner:YEATMAN TIMOTHY
Methods and gene expression signature for assessing ras pathway activity
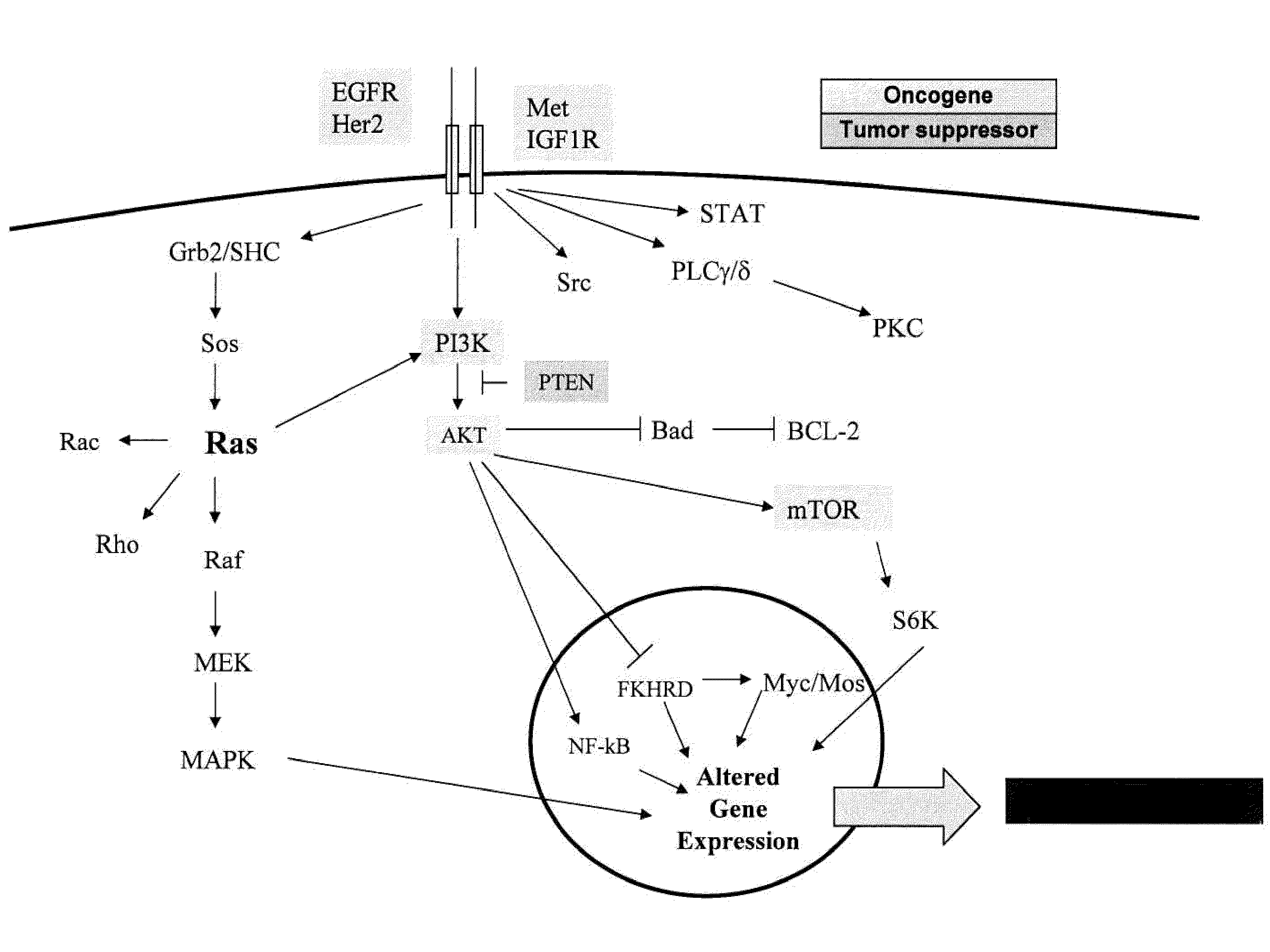
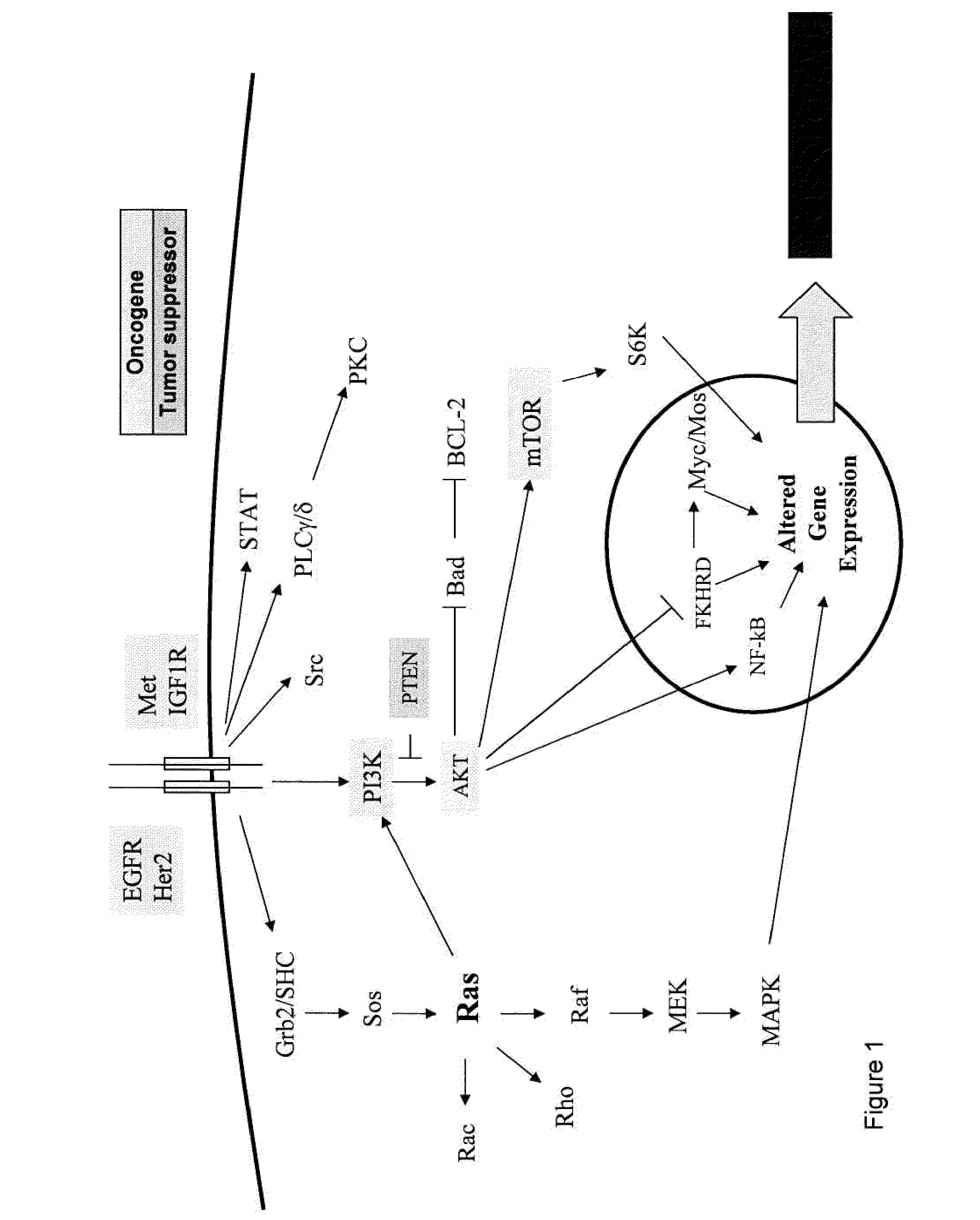
Methods, biomarkers, and expression signatures are disclosed for assessing the regulation status of RAS pathway signaling in a cell sample or subject. More specifically, several aspects of the invention provide a set of genes which can be used as biomarkers and gene signatures for evaluating RAS pathway deregulation status in a sample; classifying a cell sample as having a deregulated or regulated RAS signaling pathway; determining whether an agent modulates the RAS signaling pathway in sample; predicting response of a subject to an agent that modulates the RAS signaling pathway; assigning treatment to a subject; and evaluating the pharmacodynamic effects of cancer therapies designed to regulate RAS pathway signaling.
Owner:MERCK SHARP & DOHME CORP
Gene expression data classification method and system
ActiveCN105825081AReduce redundancyReduce computing timeKernel methodsRelational databasesCluster algorithmData set
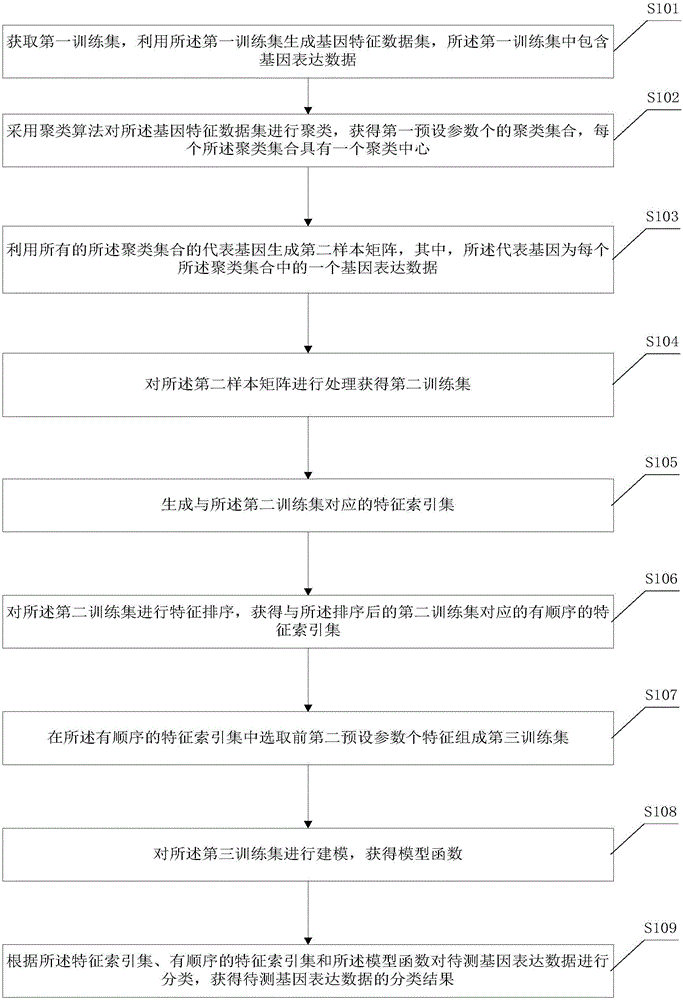
The invention discloses a gene expression data classification method and system. The gene expression data classification method comprises the following steps: after obtaining a gene characteristic data set, clustering the gene characteristic data set by adopting a clustering algorithm to obtain a first pre-set parameter clustering set; and processing the clustering set to obtain a second sample matrix, a second training set and a characteristic index set, and reducing dimensions of gene characteristic data so as to reduce the redundancy of the gene characteristic data, and reduce occupied computational resources and consumed computation time in a process of carrying out characteristic selection on the second training set to the very great extent. The occupied computational resources and the consumed computation time in a process of carrying out clustering operation on the gene characteristic data set by adopting the clustering algorithm are less, so that the occupied computational resources and the consumed computation time in a process of clustering gene characteristic data to be detected through adopting the gene characteristic data classification method are less.
Owner:SUZHOU UNIV
Fungus virulence new gene MgKIN17 coming from pyricularia gisea and its use
The present invention provides one new gene MgKIN17 from Pyricularia gisea, and the gene features that it has the amino acid sequence from the site 1 to the site 3544 in the SEQ ID No. 1, the cDNA with the nucleotide sequence of SEQ ID No. 2, the promoter with the nucleotide sequence of SEQ ID No. 4, and coded protein with the amino acid sequence of SEQ ID No. 3. The insertion mutation and gene complementation of the present invention shows that the destruction of the gene leads the formation of Pyricularia gisea infecting organ and reduced lesion capacity of rice. Wheat bakanae fungus, corn stalk rot and other pathogenic fungus have homogenous protein with homogeneity over 40 % to MgKIN17 and maybe similar biological function. The gene of the present invention and its protein expression and modification may be used as the target site for designing and screening antifungal medicines.
Owner:CHINA AGRI UNIV
Gene signatures for detection of potential human diseases
A process to select signature gene by performing statistical analyses on gene datasets of various types of diseases for identifying signature genes for at least one of the diseases followed by categorizing and establishing gene expression table with the signature genes. The signature genes in the gene expression table are tested and verified by applying additional datasets to finalize and confirm the signature genes. The step of performing the statistical analyses on the gene datasets of various types of diseases further comprising a step of performing a total background normalization (TBN) of a relative gene expression (RGE) ratio then carried a two-tail T-test of the RGE ratios between the various diseases. The step of identifying the signature gene further comprising a step of carrying out a false positive elimination (FPE) by identifying differently expressed genes and removing overlapping genes among different diseases from a list of the signature genes
Owner:YANG YIH SHENG
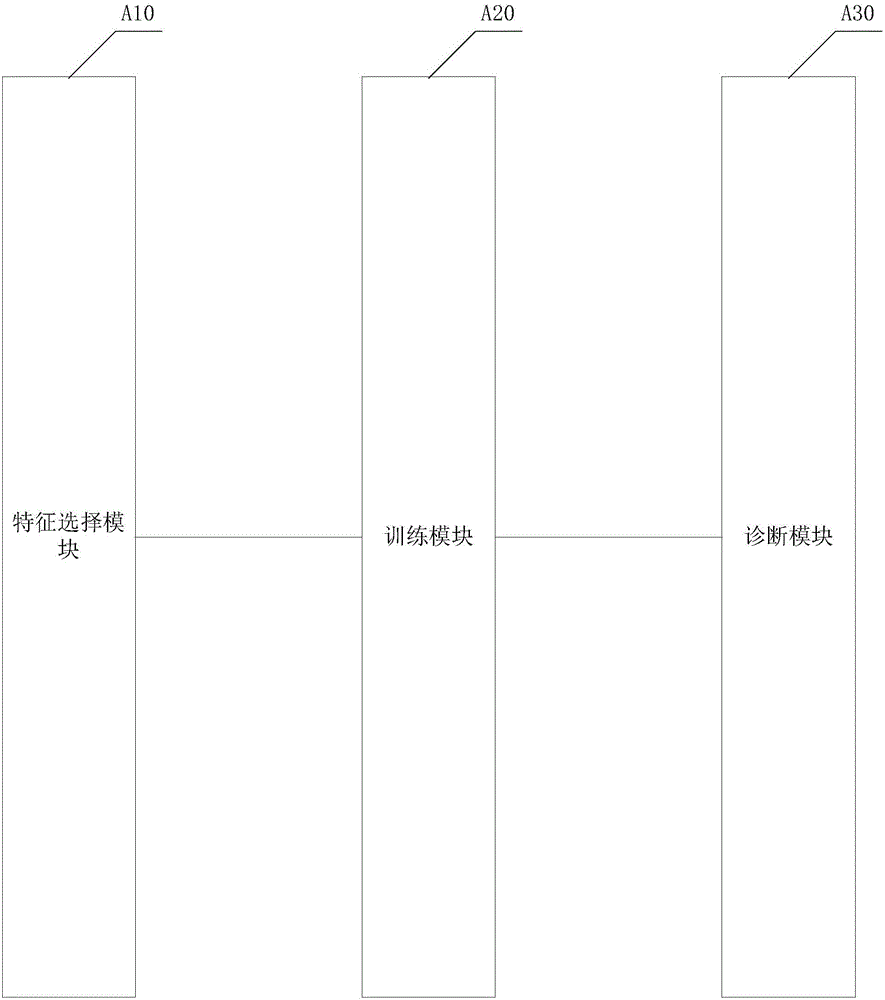
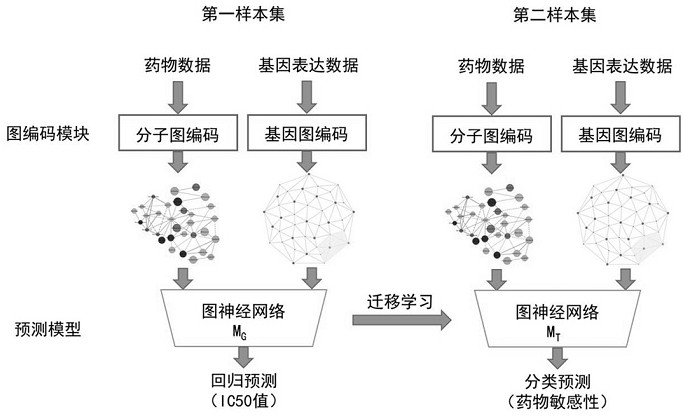
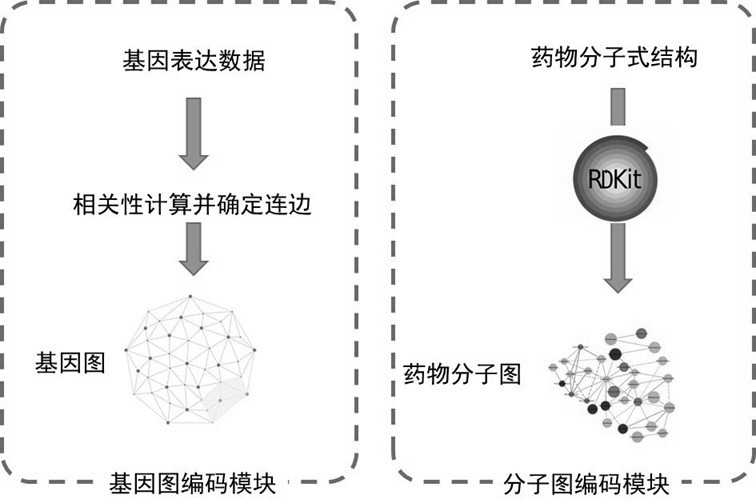
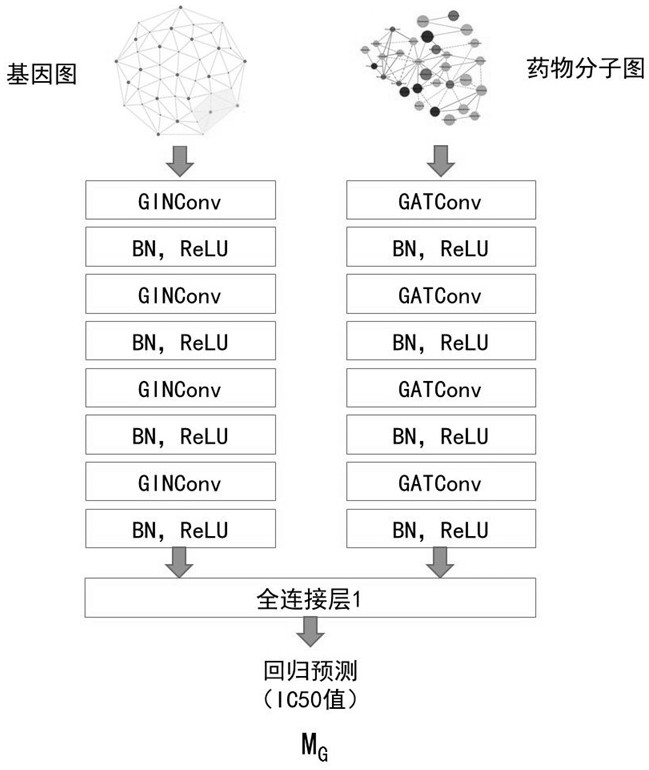
Drug sensitivity prediction method and device based on transfer learning and graph neural network
The invention discloses a drug sensitivity prediction method and device based on transfer learning and a graph neural network, and belongs to the technical field of drug sensitivity detection and evaluation. The method specifically comprises the steps: performing parameter optimization on a semi-inhibitory concentration prediction module through first sample data containing the semi-inhibitory concentration of a drug to a cell line; migrating the network parameters of a feature extraction unit of a semi-inhibitory concentration prediction module to a feature extraction unit of a drug sensitivity prediction module through migration learning, and on this basis, performing parameter optimization on the drug sensitivity prediction module through second sample data containing drug sensitivity category data; in this way, cross-dataset feature migration is effectively achieved, the defect that the sample size of the datasets is insufficient is overcome, the drug sensitivity prediction module is optimized by means of the datasets with large differences, the drug sensitivity prediction module can effectively extract gene features and drug features of individuals, and the drug sensitivity is predicted with high accuracy.
Owner:ZHEJIANG UNIV
Gene signature to predict homologous recombination (HR) deficient cancer
Methods for identifying and treating cancers that are homologous recombination (HR)-repair defective. In some aspects, HR defective cancers are treated with a PARP inhibitor therapy. Methods for sensitizing cancers to a PARP inhibitor therapy are also provided.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Producing, cataloging and classifying sequence tags
InactiveUS7618778B2Overcome limitationsShortening the linear chimeric nucleic acid intermediatesSugar derivativesMicrobiological testing/measurementCatalogingDouble strand
Owner:KAUFMAN JOSEPH C
Gene signatures for cancer diagnosis and prognosis
ActiveUS20130302242A1In-vivo radioactive preparationsNucleotide librariesPrognosis biomarkerCancers diagnosis
Owner:MYRIAD GENETICS
Gene signatures for lung cancer prognosis and therapy selection
InactiveUS20140170242A1Heavy metal active ingredientsNucleotide librariesDiseaseMolecular classification
The invention provides for molecular classification of disease and, particularly, molecular markers for lung cancer prognosis and therapy selection and methods and systems of use thereof.
Owner:MYRIAD GENETICS
False gene data bank construction method of rice genome
The invention relates to a method for constructing pseudogene database of rice global genome, which comprises constructing a local data base of known rice global genome sequences in a computer system, proceeding search and comparison to the data base through BLAST program, obtaining standard BLAST format comparison result, using the SeqIO module in the Bioperl to analyze the comparison result, obtaining the information file recording characteristic value data for the pseudogenes and genes, removing redundant pseudogenes and genes data, screening and sorting pseudogenes, building data base of the pseudogenes by using characteristic value corresponding to pseudogenes as the data item marker.
Owner:ZHEJIANG UNIV
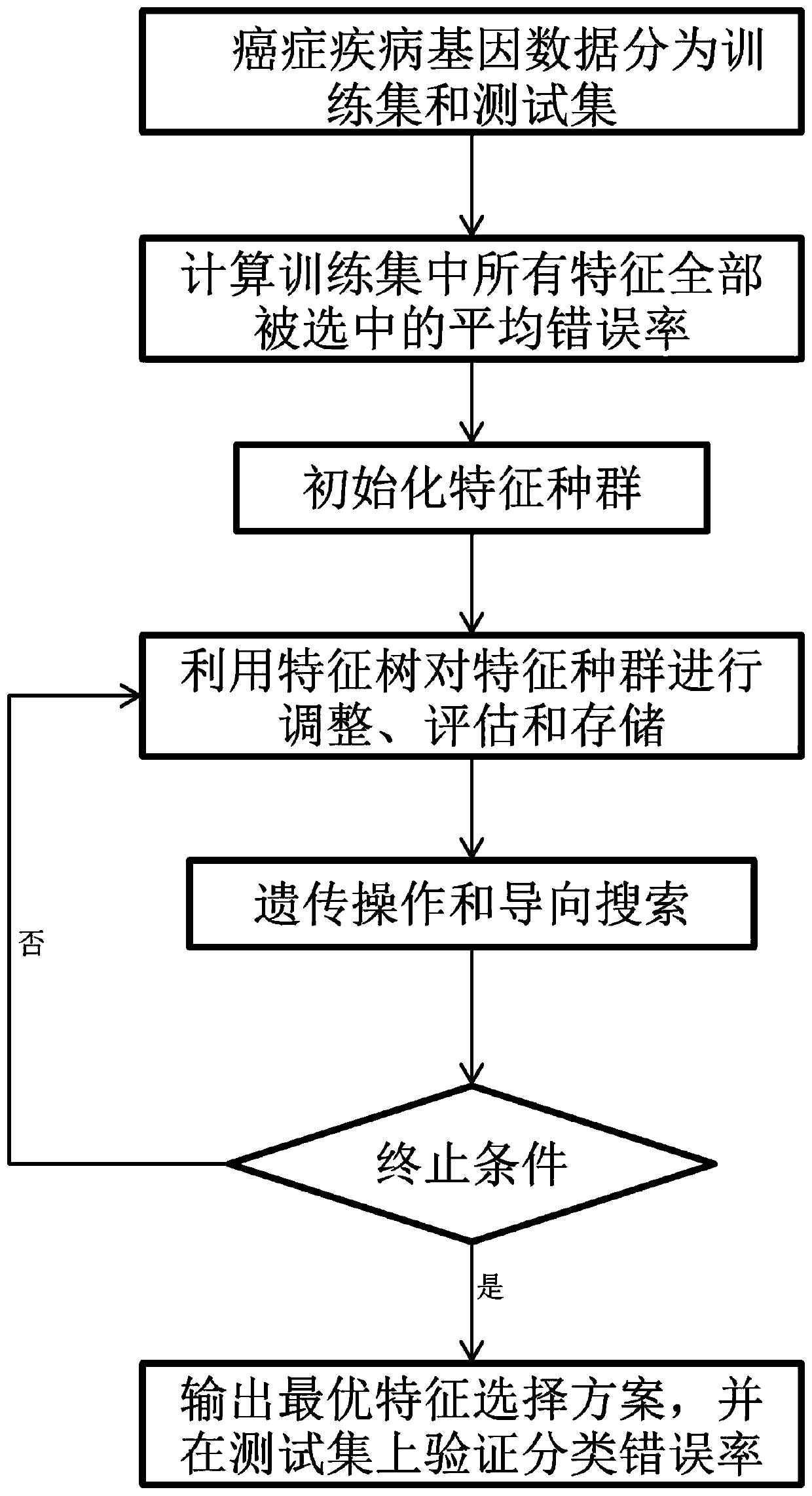
Cancer disease gene characteristic selection method based on historical data
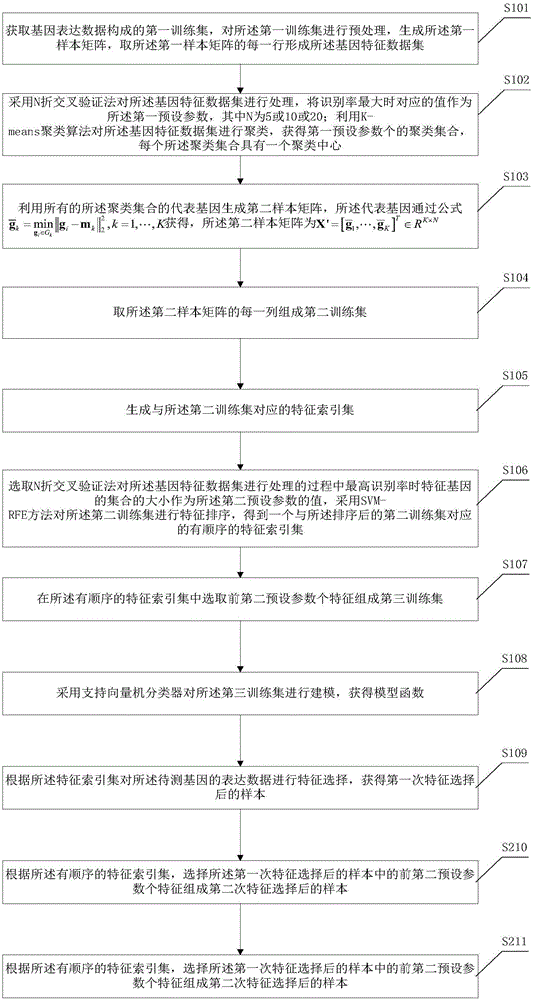
ActiveCN110070916AReduce dimensionalityImprove forecast accuracyBiostatisticsCharacter and pattern recognitionAlgorithmGene Feature
The invention discloses a cancer disease gene characteristic selection method based on historical data. The cancer disease gene characteristic selection method based on historical data includes the following steps: A, dividing cancer disease gene data into a training set and a test set; b, calculating a total average error rate after all characteristics on the training set are selected; C, generating an initial population, and constructing a fitness function; D, recording all characteristic selection schemes into a characteristic tree, adjusting the distribution of the characteristic selectionschemes, taking the characteristic selection scheme with the minimum fitness value as the optimal characteristic selection scheme, and returning a result to a genetic operator and a guide search operator; E, guiding the evolutionary direction of the characteristic population; and F, judging a termination condition, if the termination condition is not met, repeating the steps D-F, and if the termination condition is met, outputting an optimal solution. The cancer disease gene characteristic selection method based on historical data has the advantages that the data dimension can be effectivelyreduced; the prediction accuracy is improved; the related genes of diseases such as cancer are screened through the characteristic tree in combination with the genetic algorithm; and assistance is provided for diagnosis and treatment.
Owner:ANHUI UNIVERSITY
Deep learning method for predicting prognosis risk of cancer patient based on multi-omics data
PendingCN112820403AImprove robustnessEpidemiological alert systemsMedical automated diagnosisData setGene Feature
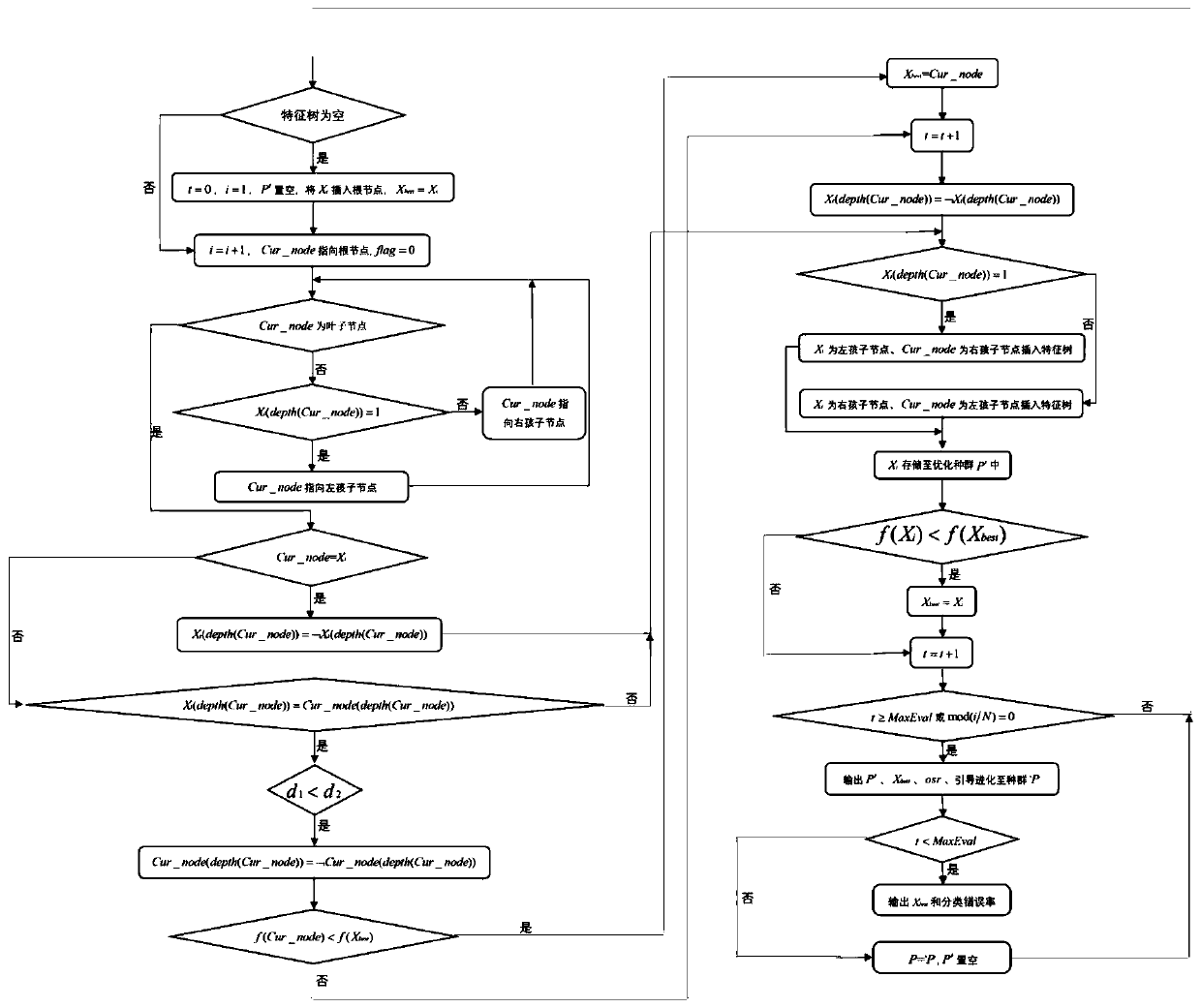
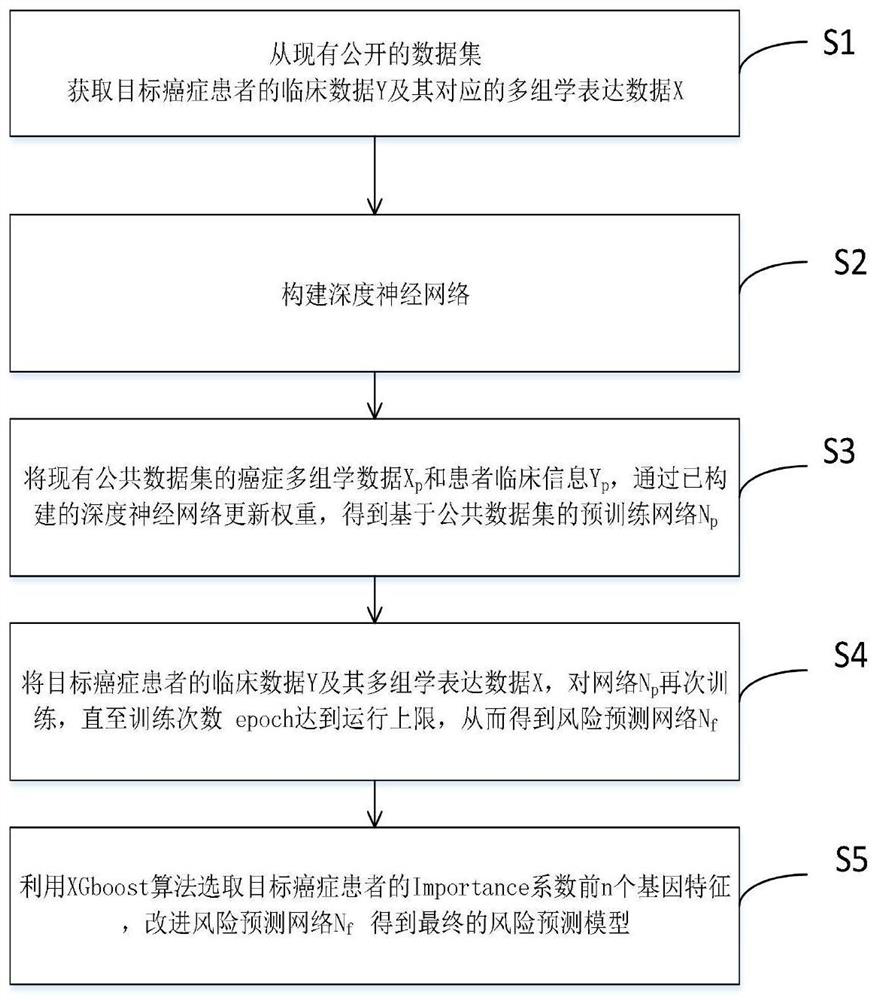
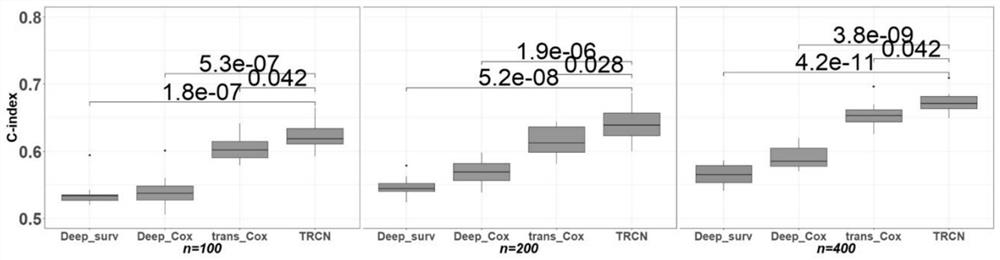
The invention discloses a deep learning method for predicting the prognosis risk of a cancer patient based on multi-omics data, which is used for predicting the prognosis risk of the cancer patient and comprises the following steps: S1, acquiring clinical data Y of a target cancer patient and corresponding multi-omics expression data X from an existing public data set; S2, constructing a deep neural network; S3, updating the weight theta of the cancer multi-omics data Xp and the clinical information Yp of the patient of the existing common data set through the constructed deep neural network to obtain a pre-training network Np based on the common data set; S4, training the network Np again until the training frequency epoch reaches the operation upper limit, thereby obtaining a risk prediction network Nf; and S5, selecting the first n gene features of the Importance coefficient of the target cancer patient by using an XGboost algorithm, and improving the risk prediction network Nf to obtain a final risk prediction model. According to the method, the robustness of the prediction model is improved, and the prognosis risk of the cancer patient is predicted more accurately by using multi-omics data.
Owner:SUN YAT SEN UNIV
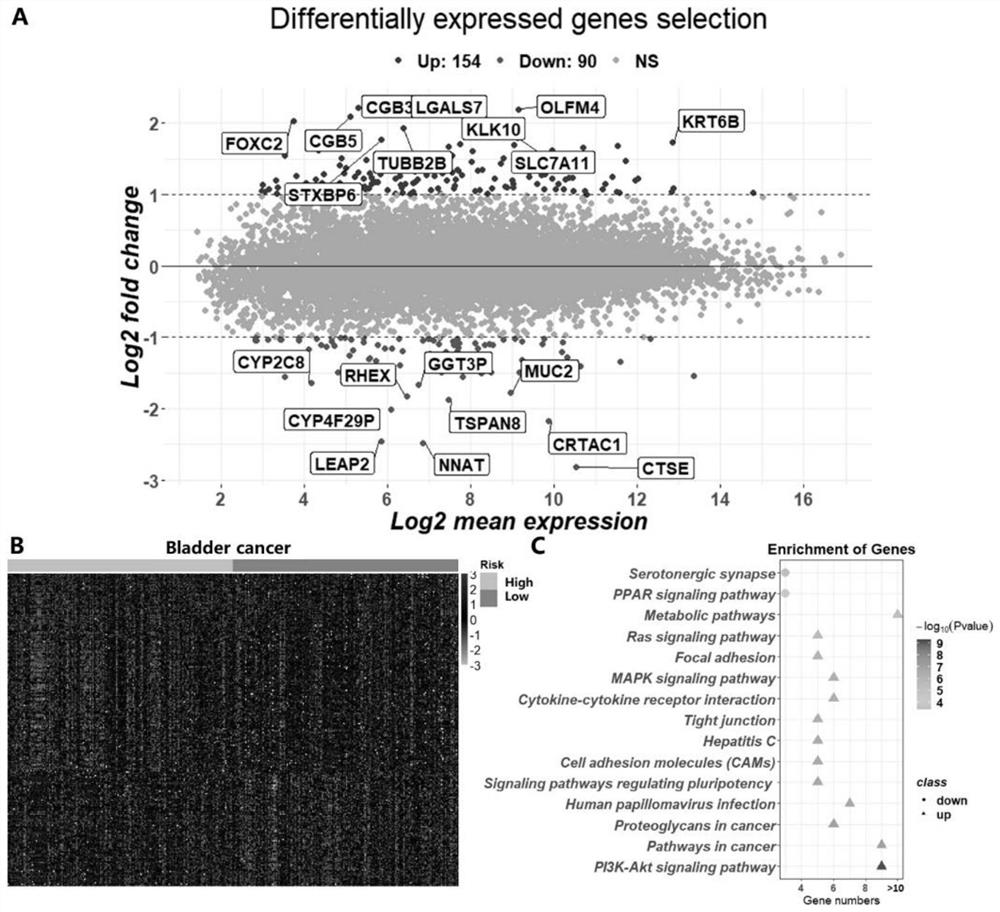
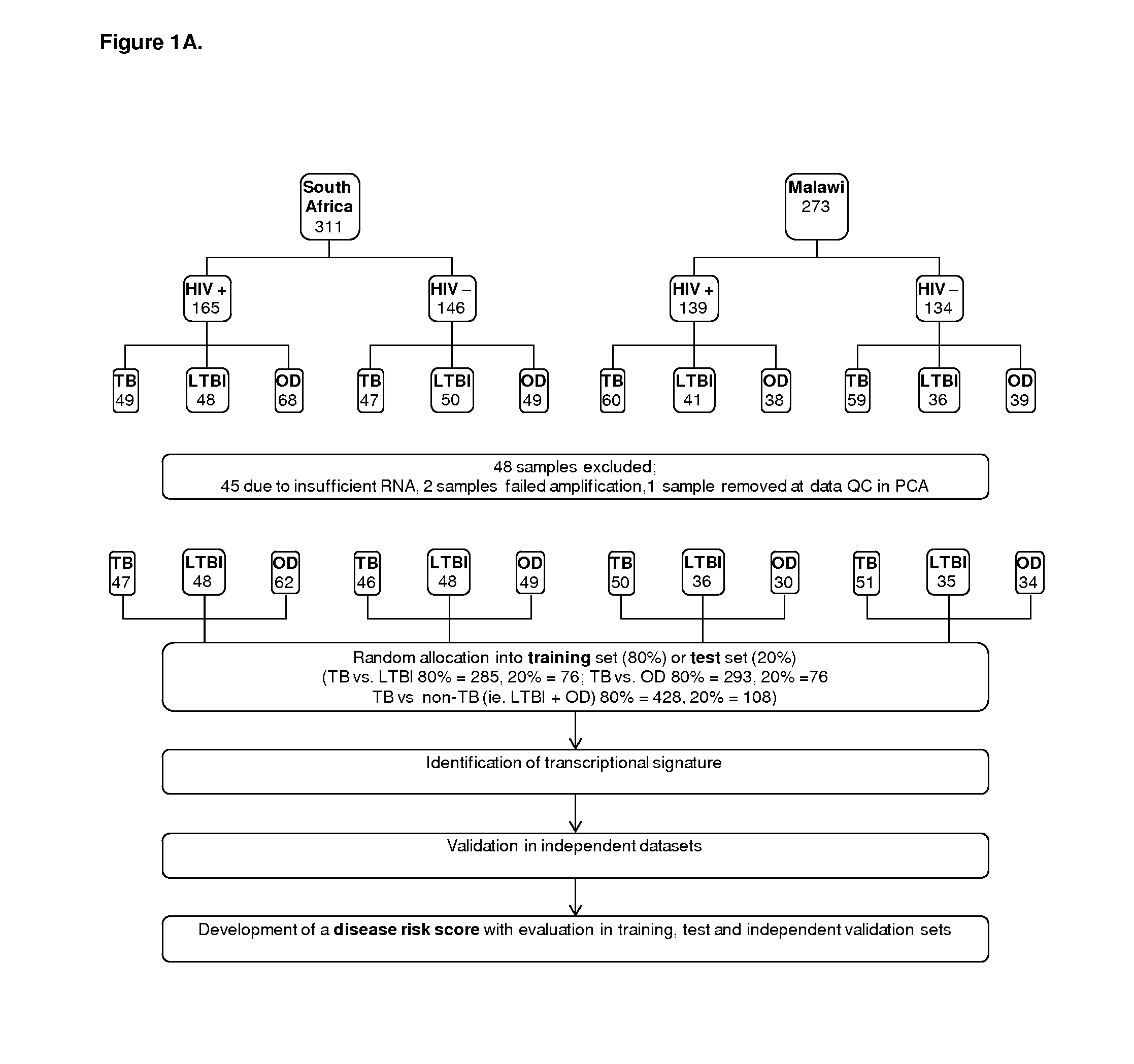
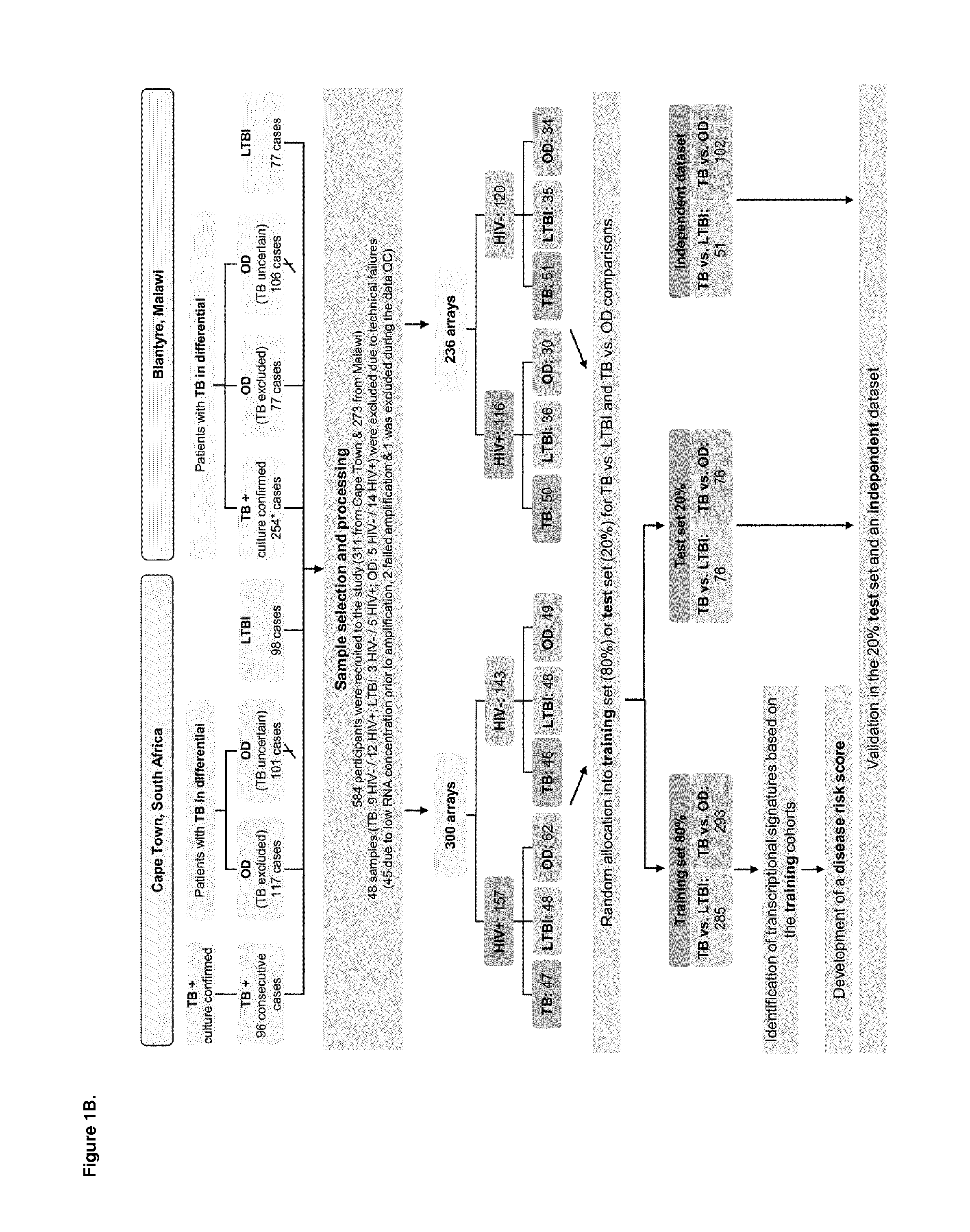
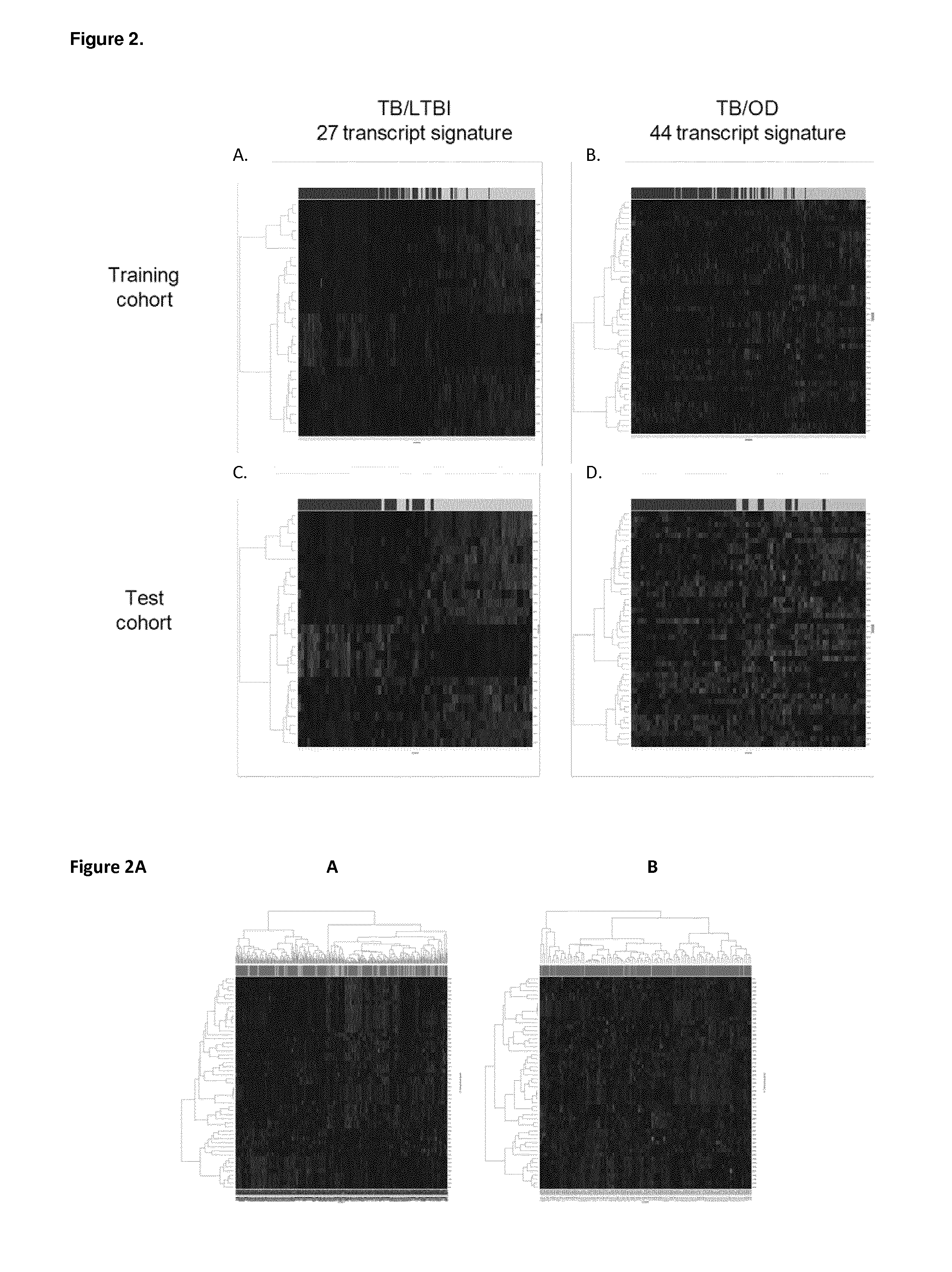
Diagnosis of active tuberculosis by determining the mRNA expression levels of marker genes in blood
InactiveUS20150203899A1Robust and accurate identificationAppropriate treatmentMicrobiological testing/measurementLibrary screeningDiseaseLatent tuberculosis
The present disclosure relates to a method of distinguishing active TB in the presence of a complicating factor, for example, latent TB and / or co-morbidities, such as those that present similar symptoms to TB, such as HIV. The method employs a 27 gene signature to distinguish active tuberculosis from latent TB infection, a 44 gene signature to distinguish active TB from other diseases such as HIV and / or a 53 gene signature to discriminate active TB from latent TB and other diseases. The disclosure also relates to a gene signature employed in the method, a bespoke gene chip for use in the method and a disease risk score obtainable from the method.
Owner:IMPERIAL INNOVATIONS LTD
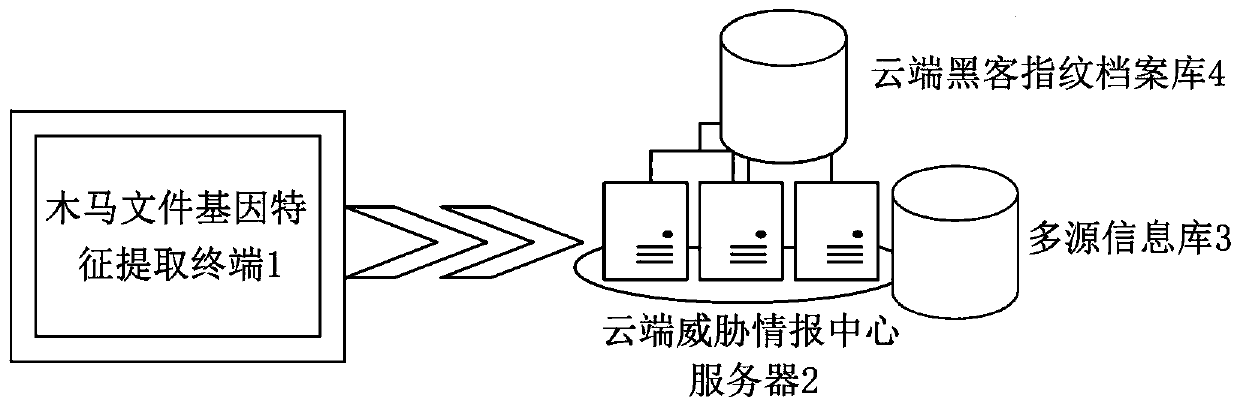
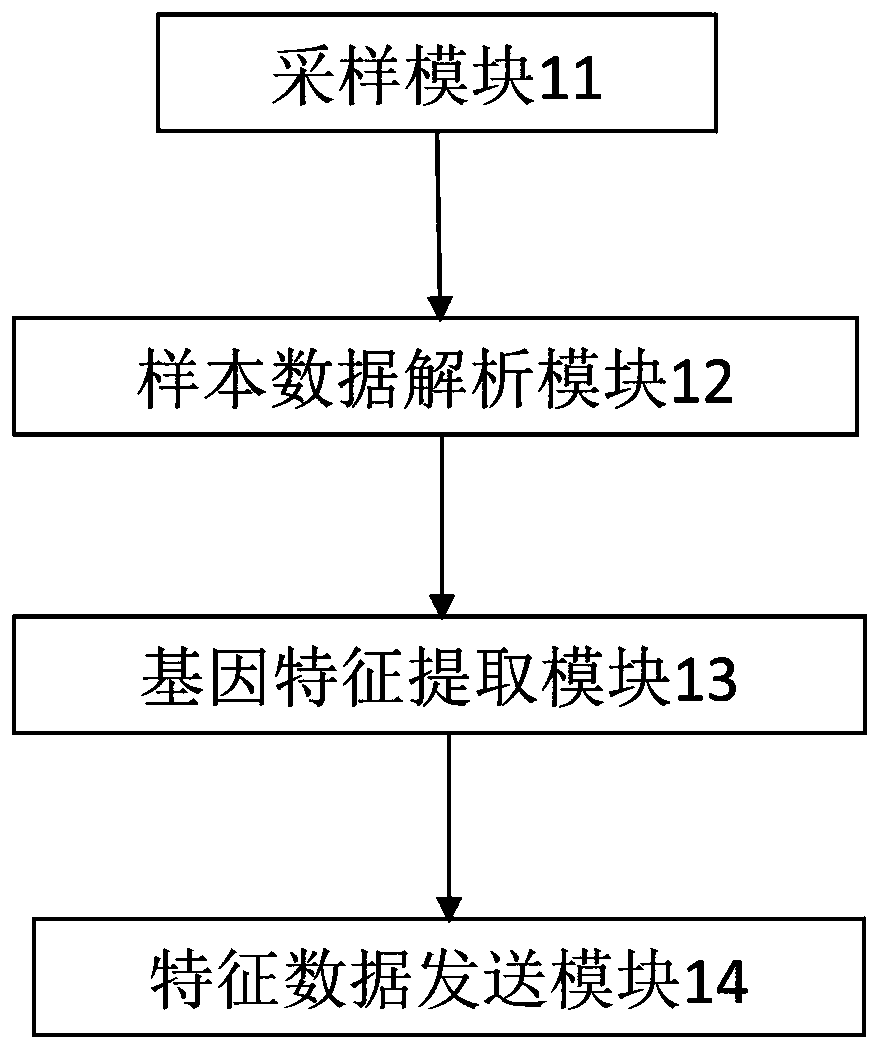
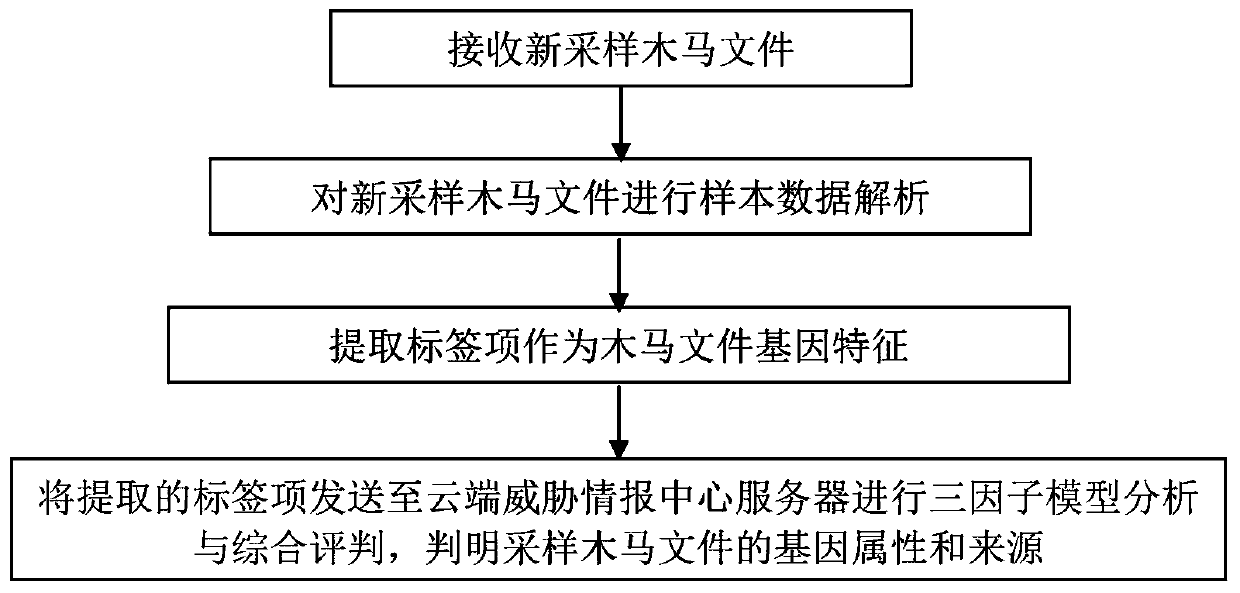
Trojan file tracing method, system and equipment
ActiveCN109784059AExpand the dimension of analysisClever designPlatform integrity maintainanceComplete dataMultiple forms
The invention discloses a Trojan horse file tracing method, system and the equipment. The gene features of the Trojan files are depicted from multiple angles, multiple forms and multiple levels through three-factor model analysis, more comprehensive and more accurate Trojan fingerprint data are provided for Trojan file tracing, the analysis and recognition capability of the Trojan files is improved, and more complete data resources are provided for Trojan file tracing. And the cloud server is linked with the cloud hacker fingerprint archive library, so that the association and traceability ofthe Trojan files can be carried out by combining the behavior habits organized by the hackers, the analysis dimension of the Trojan files is expanded, and the Trojan files which are deeper to hide andmore skillful in design can be identified and traced. Besides, a Trojan file tracing method for multi-source data comprehensive evaluation is adopted, fusion, association and application of multi-source data are truly achieved, high-level and complex Trojan samples can be recognized, and the method has more accurate judgment and tracing capacity.
Owner:北京中睿天下信息技术有限公司
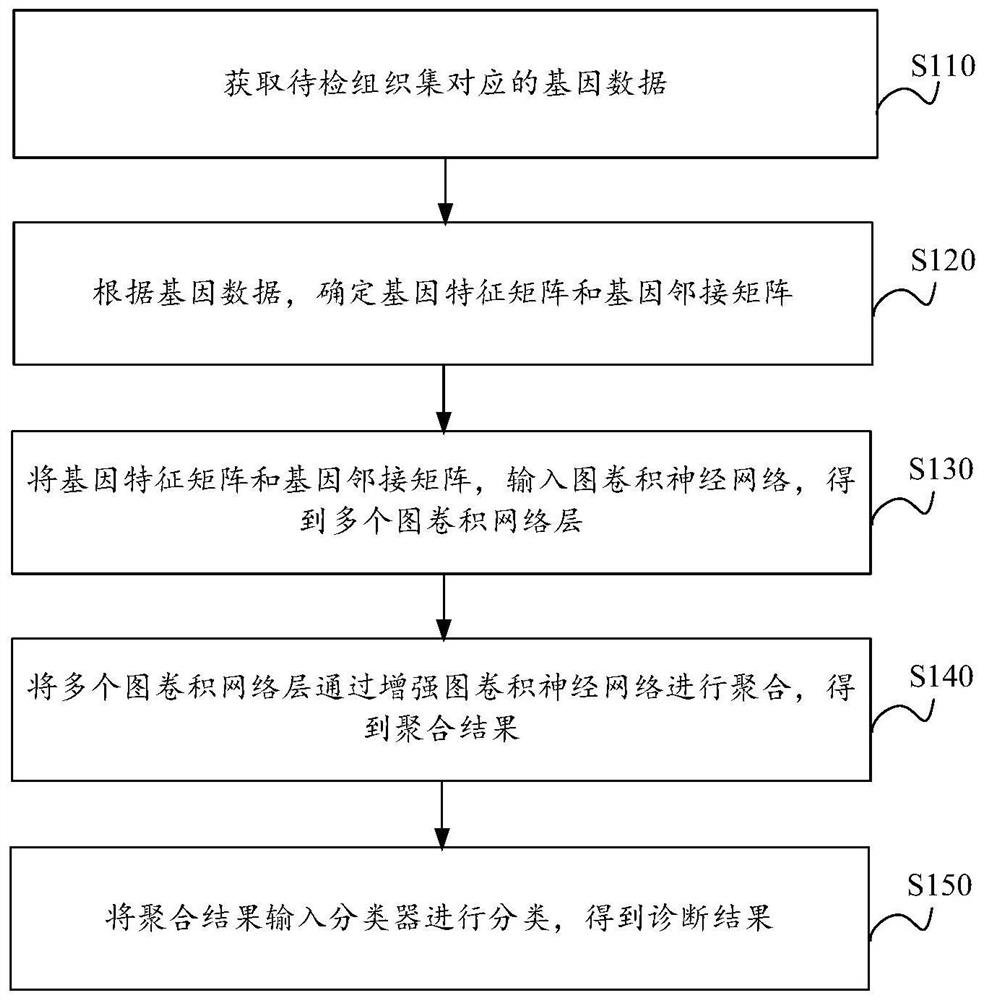
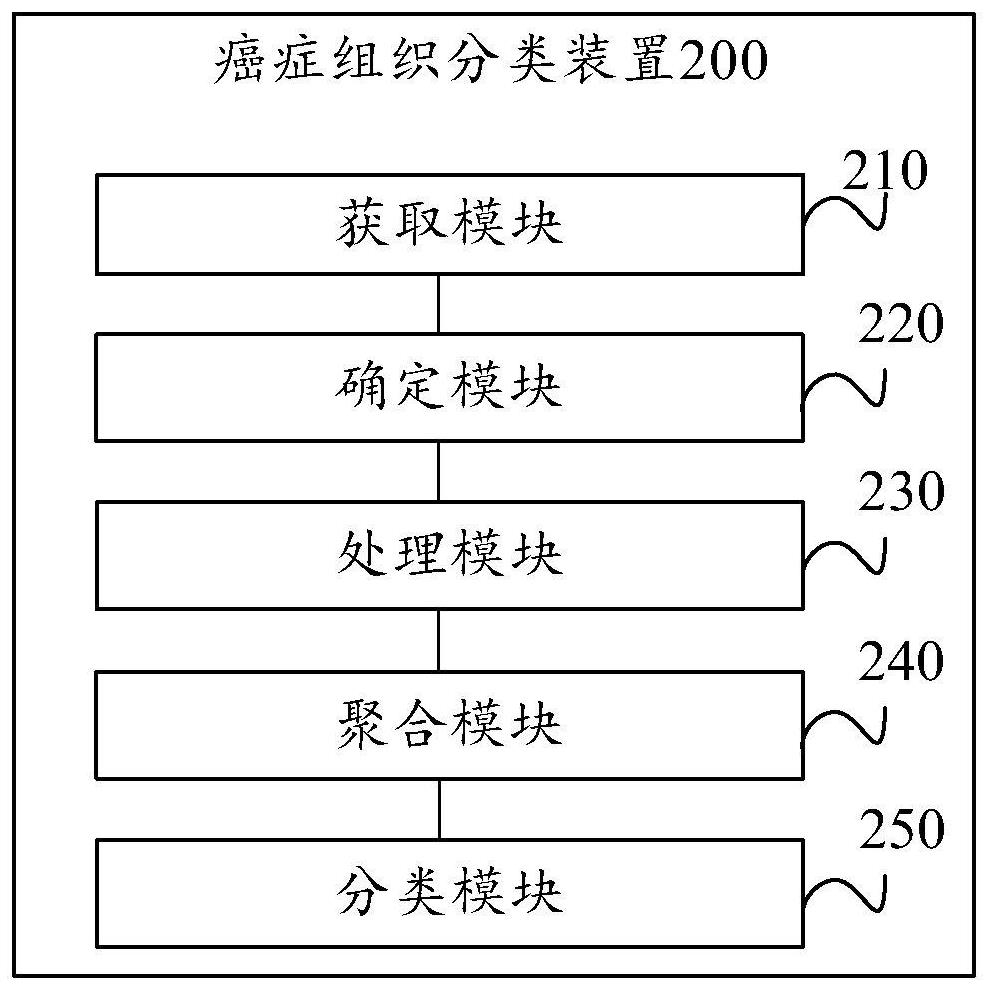
Cancer tissue classification method and device, electronic equipment and storage medium
PendingCN114154557AAddress limitationsTo solve one-sidedness,Character and pattern recognitionNeural architecturesTissue sampleData mining
The invention provides a cancer tissue classification method and device, electronic equipment and a storage medium. The method comprises the following steps: acquiring gene data corresponding to a to-be-detected tissue set; the to-be-detected tissue comprises a plurality of to-be-detected tissue samples; determining a gene feature matrix and a gene adjacent matrix according to the gene data; inputting the gene feature matrix and the gene adjacent matrix into a graph convolutional neural network to obtain a plurality of graph convolutional network layers; aggregating the plurality of graph convolutional network layers through an enhanced graph convolutional neural network to obtain an aggregation result; and inputting an aggregation result into a classifier for classification to obtain a diagnosis result. The scheme is high in cancer tissue classification accuracy.
Owner:CENTRAL UNIVERSITY OF FINANCE AND ECONOMICS
Gene selection method and device
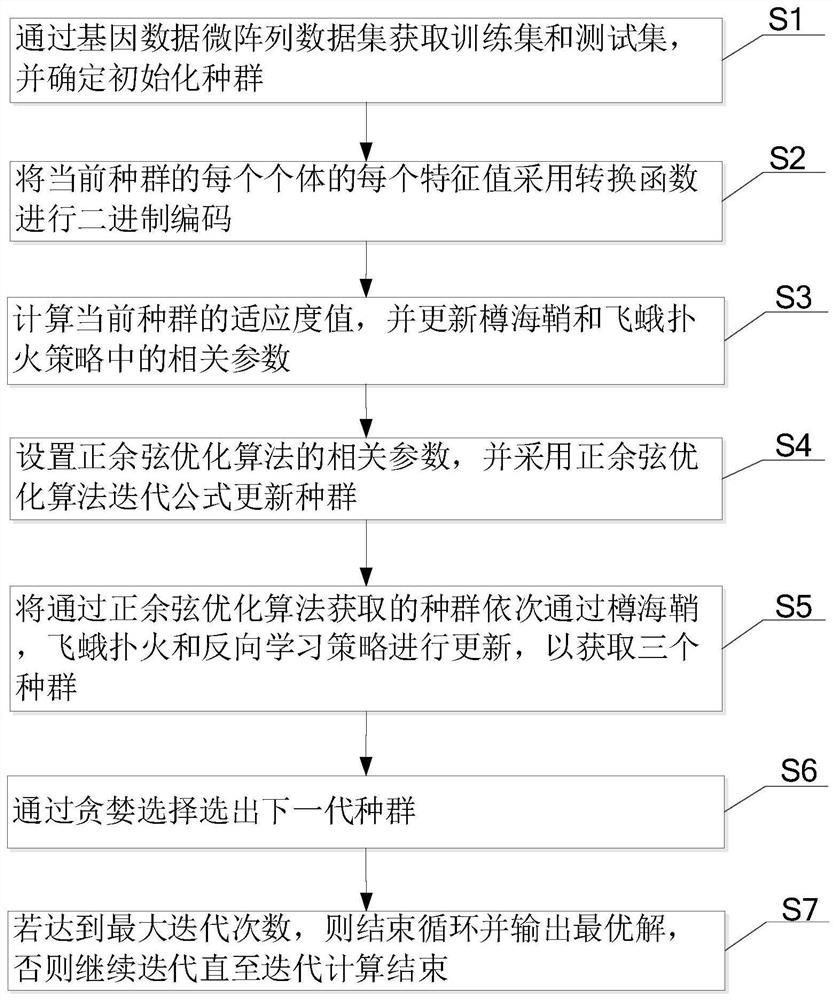
PendingCN112215259ALighten the computational burdenReduce noiseBiostatisticsCharacter and pattern recognitionGene selectionGene Feature
The invention provides a gene selection method which is used for gene characteristic selection. The method comprises the steps of obtaining a training set and a test set through a gene data microarraydata set, and determining an initialized population; carrying out binary coding on each individual of the current population by adopting a conversion function; calculating the fitness value of the current population, and updating related parameters in the dolio-sea squirt and moth fire suppression strategy; setting related parameters of a sine and cosine optimization algorithm, and updating the population by adopting a sine and cosine optimization algorithm iterative formula; updating the populations obtained through the sine and cosine optimization algorithm through a doliola scabra, moth fire suppression and reverse learning strategy in sequence so as to obtain three populations; selecting a next generation of population through greedy selection; and if the maximum number of iterationsis reached, ending the loop and outputting the optimal solution, otherwise, continuing the iteration until the iterative computation is ended. According to the invention, the gene characteristics which contribute most to categories can be screened out from the genes more accurately and more efficiently, and the detection cost is reduced.
Owner:WENZHOU UNIVERSITY
Gene expression profile distance measurement method based on deep learning
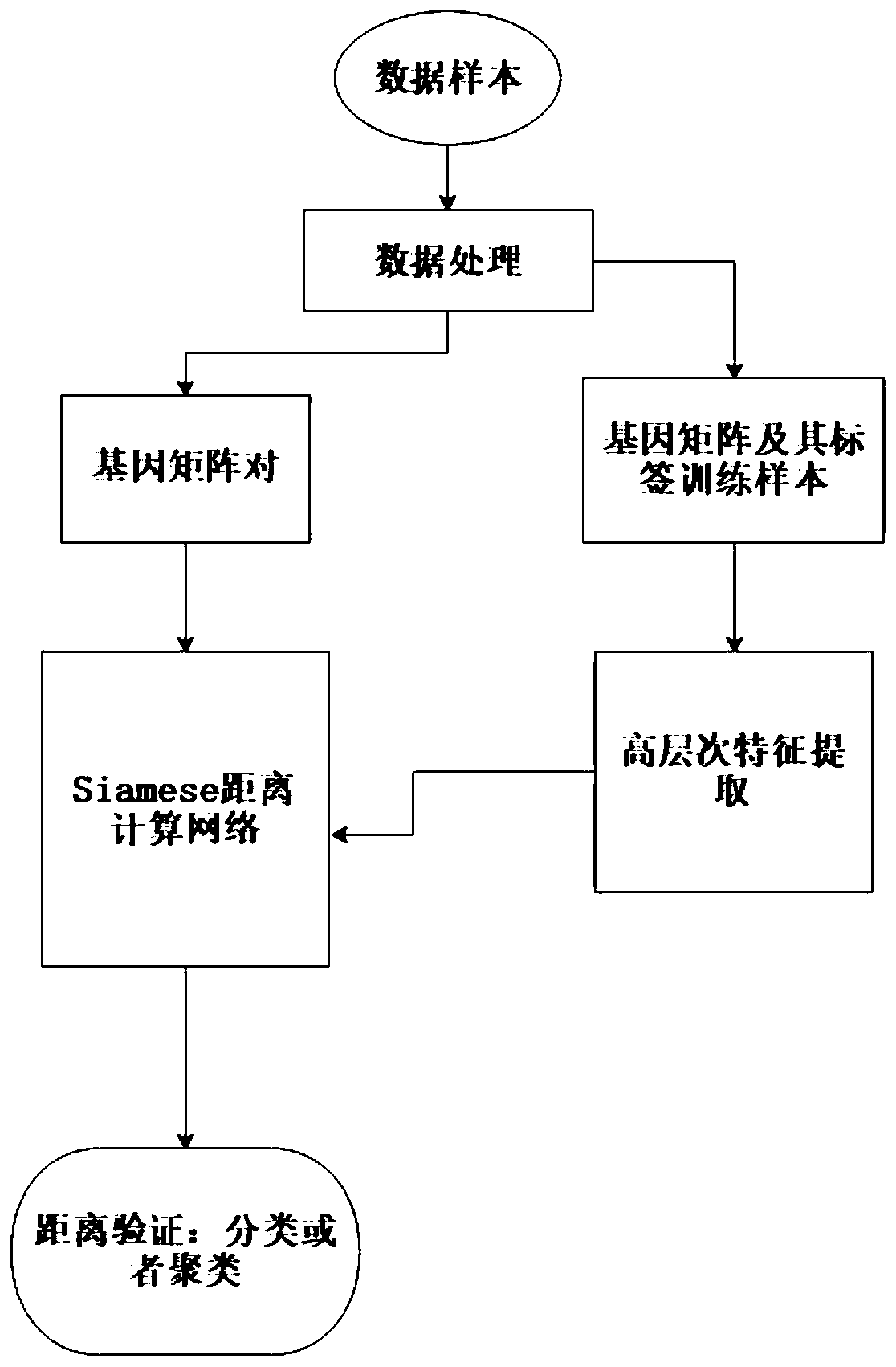
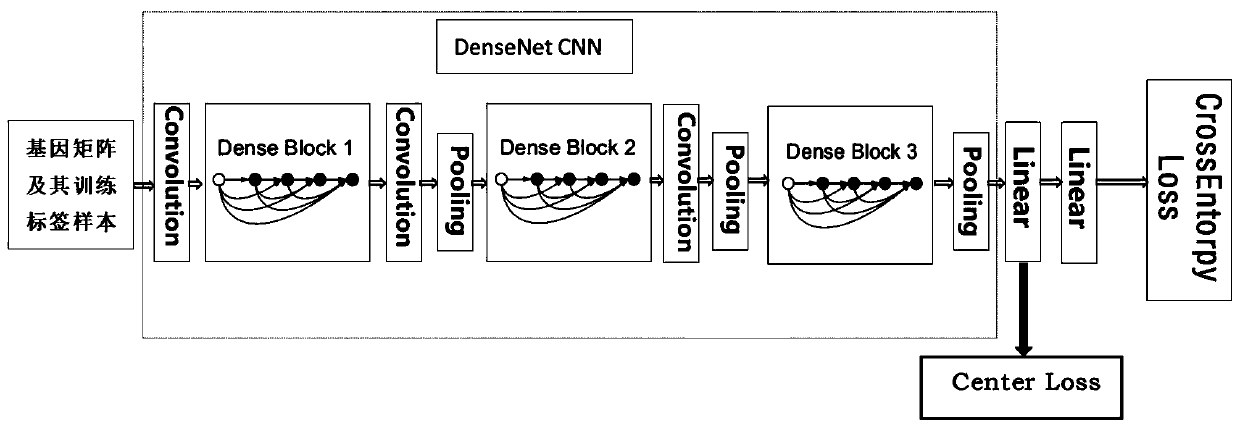
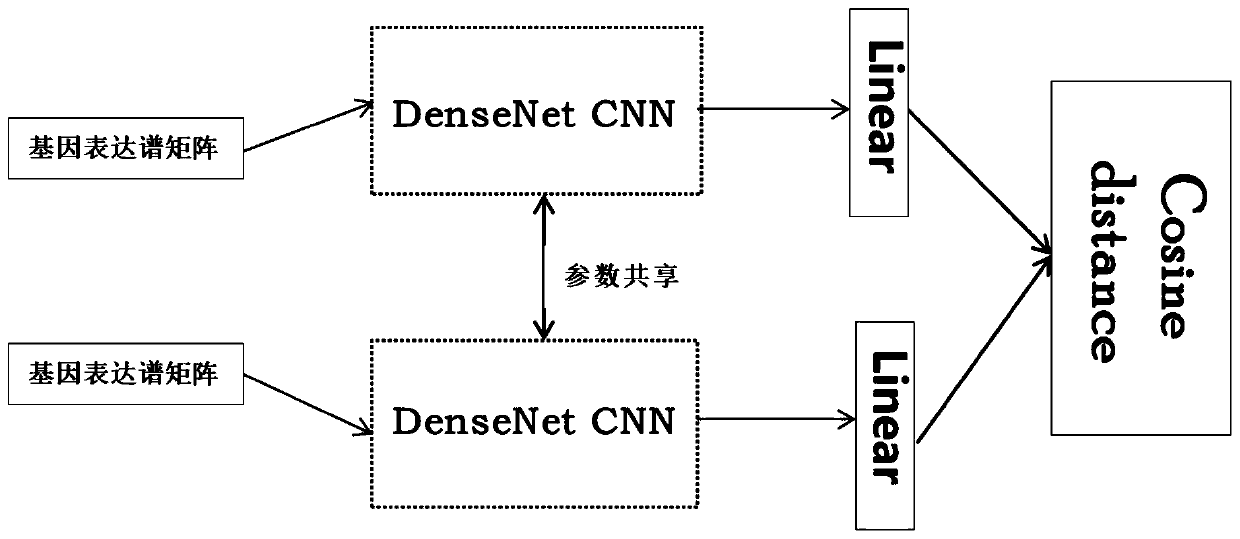
ActiveCN110033041AAccurate and fast calculationCharacter and pattern recognitionNeural architecturesGene EnrichmentProfile analysis
The invention belongs to the field of gene expression profile classification, discloses a gene expression profile distance measurement method based on deep learning, and belongs to mining and application of deep learning on biological big data. Firstly, a convolutional neural network model suitable for gene characteristic metric learning is designed to extract data characteristics, then the distance between the data is calculated by applying an improved cosine distance, and finally, the good performance of the method is measured through the classification effect of a classification algorithm.According to the method, the similarity between different gene expression profiles can be quickly and efficiently measured, and data is provided for subsequent researches such as gene classification,clustering, differential expression analysis and compound screening. Compared with a traditional gene enrichment method, the method has the advantages that the distance measurement effect between thedata is obviously improved, the manual intervention during gene expression profile analysis can be effectively reduced, the overfitting phenomenon easily generated by a conventional deep network is avoided, and the method has relatively high mobility.
Owner:HUNAN UNIV
Biochip for predicting treatment effect of antidepressant and application
InactiveCN102071254AStrong specificityImprove throughputMicrobiological testing/measurementTreatment effectFluorescence
The invention relates to a biochip, in particular to the biochip for predicting the treatment effect of an antidepressant and an application. The biochip for predicting the treatment effect of the antidepressant consists of a carrier and a bioprobe and is characterized in that the bioprobe comprises any one kind or several kinds of materials with the sequence from Seq NO: 1 to 20, and the tail ends of the sequences are modified through fluorescence labels. Through the biochip technology, the treatment effect of the antidepressant on patients can be predicted in an early period according to genetic gene features (single nucleotide polymorphism) of the patients from aspects such as the medicine species, the sexes of the patients receiving the treatment, the putative effect taking time point of medicine, the possibly generated adverse effect and the like, so clinic doctors can be guided to make and implement individualized treatment schemes.
Owner:SOUTHEAST UNIV
Cloning and base sequence determination of novel iturin biosynthesis gene from antagonistic microorganism bacillus subtilis and characteristics of the gene
ActiveUS20130165635A1Easy to useGood antifungal activityBacteriaSugar derivativesBase JBiosynthetic genes
The present invention relates to novel iturin biosynthesis genes, and uses thereof. More specifically, the present invention provides novel iturin biosynthesis genes, wherein the iturin biosynthesis genes were cloned from Bacillus subtilis subsp. krictiensis ATCC 55079, the base sequence was determined after checking whether the cloned genes are iturin biosynthesis genes or not, and it was ascertained that the identified genes are novel genes different from the reported gene by comparing the base sequences with that of the reported gene, and uses thereof.
Owner:KOREA RES INST OF BIOSCI & BIOTECH
Wheat 4B chromosome grain weight QGW4B-CAPS molecular marker and application thereof
ActiveCN104087580AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGrain weight
The invention relates to a wheat 4B chromosome grain weight QGW4B-CAPS molecular marker and an application thereof. A grain weight control genetic locus QGW4B-17 is positioned at 31.3-36.3cM of a short arm of a wheat 4B chromosome and is positioned between a wheat 4B chromosome molecular marker Ex-c101685-705 and a molecular marker RAC875-c27536-611. The invention also discloses a grain weight QGW4B-CAPS molecular marker developed according to the grain weight control genetic locus QGW4B-17. According to the molecular marker QGW4B-CAPS, large grain resources containing the gene feature base sequence can be selected in a purposeful mode and are used for high-yield wheat breeding or molecular designing convergent cross breeding, and molecular marker assisted selective breeding is performed by using the marker, so that breeding of high-yield wheat variety is rapidly and accurately performed.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
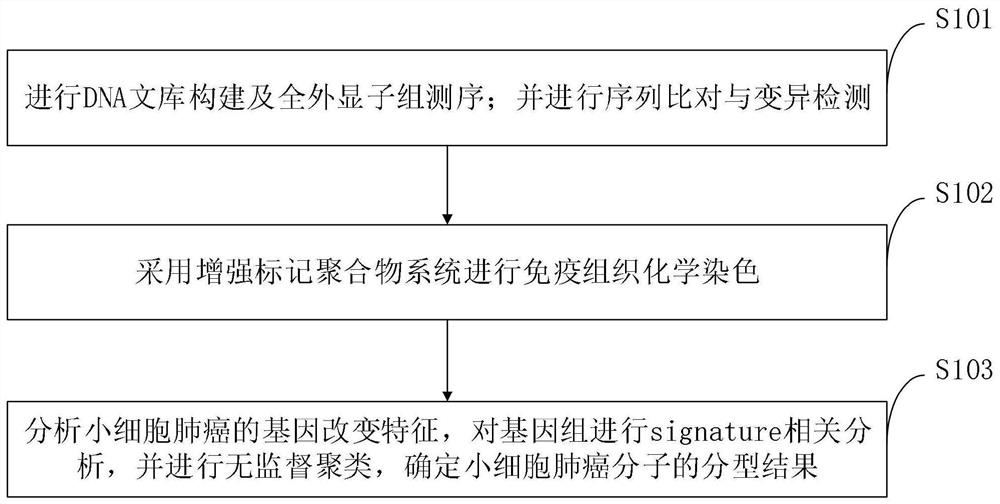
Mutation characteristic-based determination method for small cell lung cancer molecular typing and application
PendingCN113373234AMicrobiological testing/measurementBiological material analysisGenes mutationIndividualized treatment
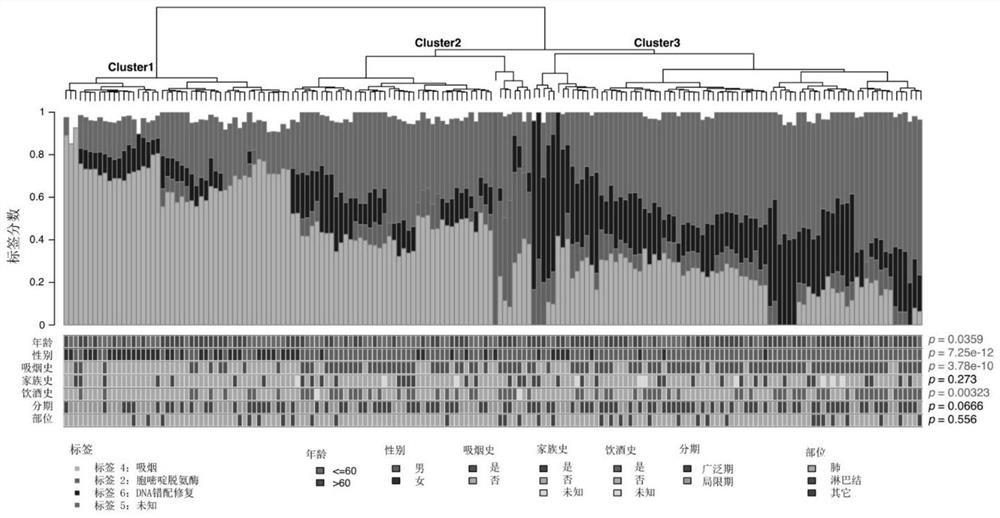
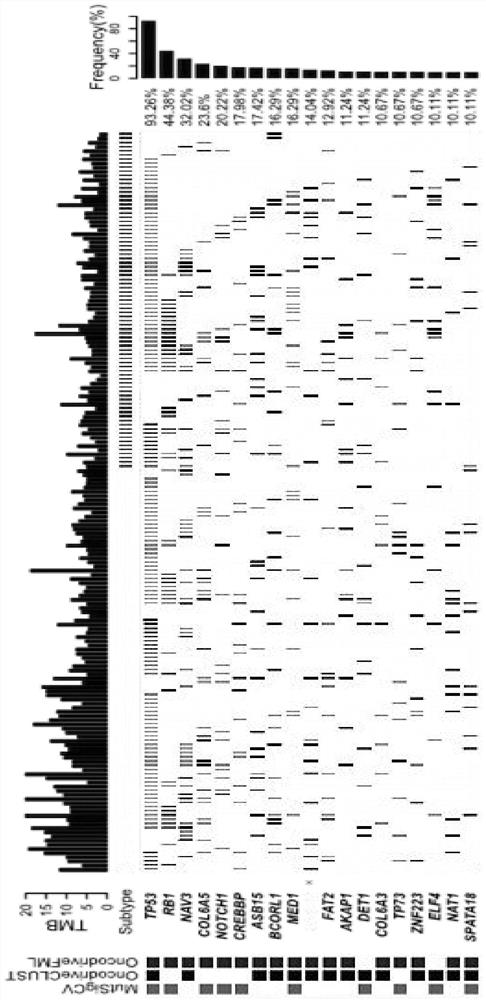
The invention belongs to the technical field of genes, and discloses a mutation characteristic-based determination method for small cell lung cancer molecular typing and an application. The method comprises the following steps: analyzing gene change characteristics of small cell lung cancer, performing signature correlation analysis on a genome, performing unsupervised clustering, and determining a typing result of small cell lung cancer molecules. Through a method of whole exon sequencing (WES), characteristics of gene mutation, copy number change and the like of 178 cases of small cell lung cancer are comprehensively analyzed, and the immune characteristics of TMB, TNB, PDL1, CD8 + T cells and the like are analyzed. According to the invention, gene characteristics and immune characteristics of the small cell lung cancer are integrated. Three gene change molecular types of the small cell lung cancer are provided for the first time. The method is essential for understanding the characteristics of heterogeneity of the small cell lung cancer and developing individualized treatment, and can promote the development of individualized clinical experiments of the small cell lung cancer.
Owner:AFFILIATED CANCER HOSPITAL OF SHANDONG FIRST MEDICAL UNIV SHANDONG CANCER INST (SHANDONG CANCER HOSPITAL)
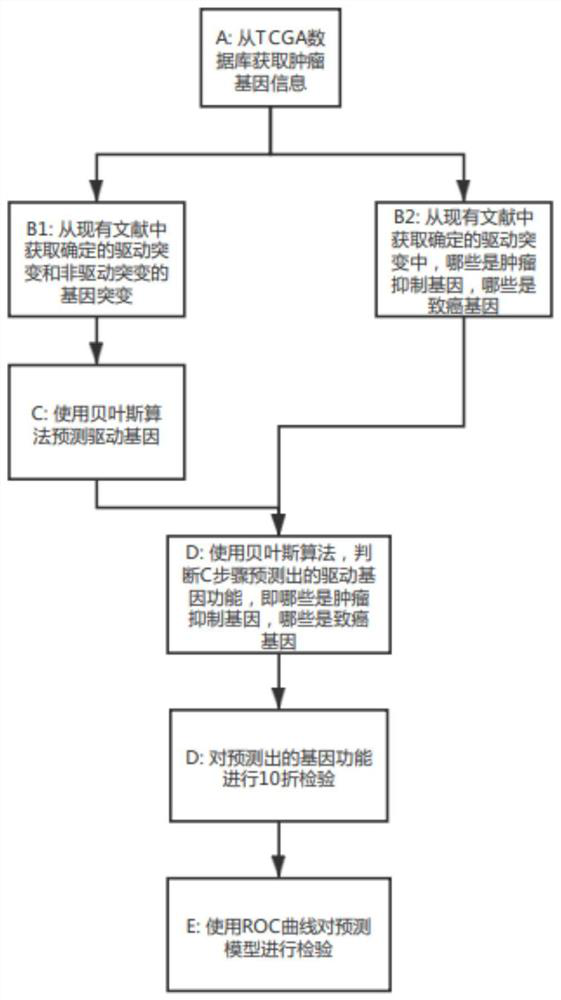
Cancer driver gene prediction method
ActiveCN113517021APromote the development of pre-diagnosisImprove forecast accuracyCharacter and pattern recognitionProteomicsData setData mining
The invention discloses a cancer driver gene prediction method. The method comprises the steps: constructing a first data set and a second data set, wherein the first data set represents the incidence relation between gene features and driving gene mutation types, and the second data set represents the incidence relation between the gene features and driving function types; training a first machine learning classification model by using the first data set, and predicting a new driver gene; confirming data corresponding to the new driver gene predicted by the first machine learning classification model as a second prediction data set; training a second machine learning classification model by using the second data set, and predicting the second prediction data set by using the trained second machine learning classification model to predict the driving function of the new driver gene. According to the invention, the prediction accuracy and the generalization ability of model application can be effectively improved.
Owner:海南精准医疗科技有限公司
Hypoxia-related gene signatures for cancer classification
InactiveUS20140212415A1Nucleotide librariesMicrobiological testing/measurementDiseaseMolecular classification
Biomarkers, particularly hypoxia-related genes, and methods using the biomarkers for molecular classification of disease are provided.
Owner:MYRIAD GENETICS
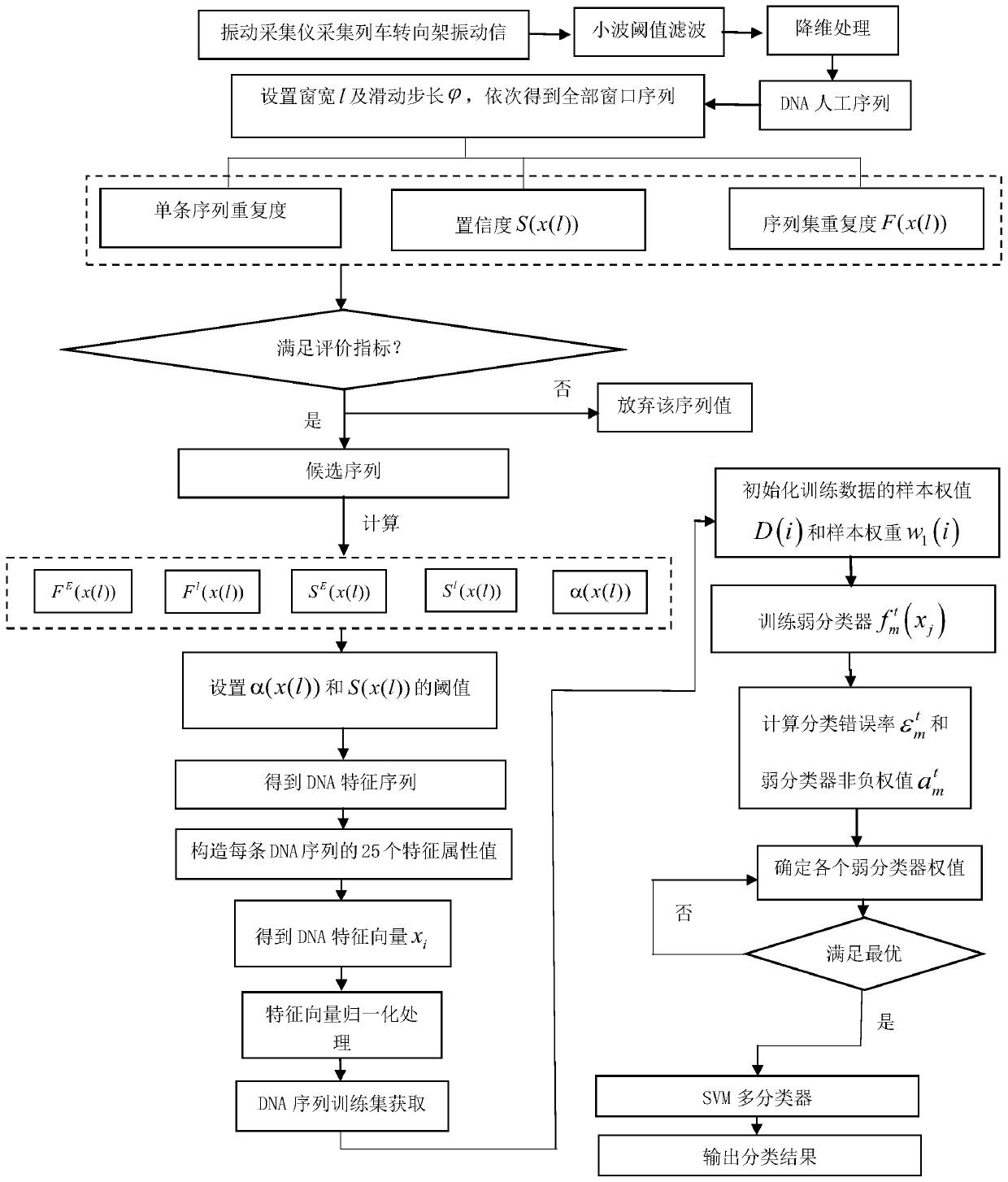
Train bogie fault identification method based on biological information characteristics
ActiveCN110232415AAccurate identificationSpeed up communicationCharacter and pattern recognitionRailway vehicle testingBogieFeature vector
The invention discloses a train bogie fault identification method based on biological information characteristics, which comprises the following steps: obtaining a historical vibration signal of a train bogie, and preprocessing and converting the historical vibration signal into an artificial DNA sequence; obtaining all window sequences through a sliding window method, and selecting a feature sequence meeting a preset requirement from the window sequences; taking the contents of four bases in the artificial DNA sequence, the sequence length and the number of the characteristic sequences as gene characteristic vectors of the artificial DNA sequence; constructing a training sample by using the gene feature vector of the artificial DNA sequence, and training a corresponding LPBoost binary classifier for each fault type; therefore, during fault detection, inputting the corresponding gene feature vector into each LPBoost binary classifier, and determining the fault type of the train bogie to be detected through a voting method. According to the invention, the feature sequence causing each fault type is mined, so that accurate identification and classification of various fault types arerealized.
Owner:CENT SOUTH UNIV
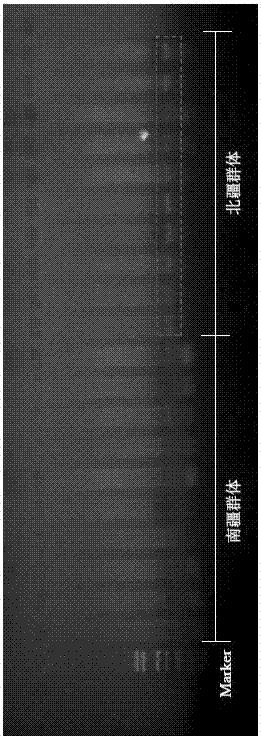
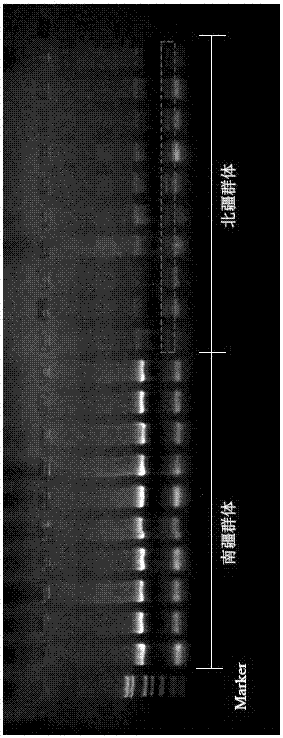
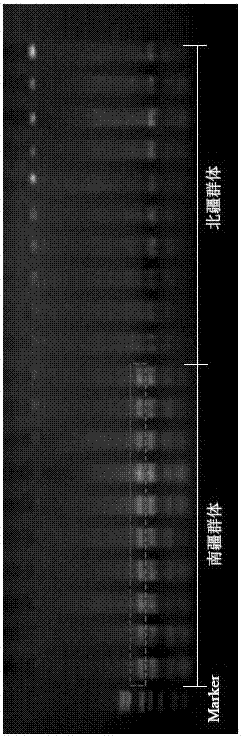
Method for distinguishing gymnodiptychus dybowskii distributed in southern and northern Xinjiang regions by utilizing ISSR molecular marker technology
InactiveCN106967814AMicrobiological testing/measurementDNA/RNA fragmentationGymnodiptychus dybowskiiBiology
The invention discloses a method for distinguishing gymnodiptychus dybowskii distributed in southern and northern Xinjiang regions by utilizing an ISSR molecular marker technology. The method comprises the following steps that 1, a gymnodiptychus dybowskii genome DNA is extracted; 2, the gymnodiptychus dybowskii genome DNA is amplified by utilizing ISSR-PCR reaction; 3, a gymnodiptychus dybowskii DNA electrophoresis mode chart is established; 4, the gymnodiptychus dybowskii distributed in the southern and northern Xinjiang regions is distinguished. The gymnodiptychus dybowskii is distinguished by utilizing the ISSR molecular marker technology, geographical populations distributed in different water systems in the southern and northern Xinjiang regions are distinguished according to genetic characteristics, a reliable basis is provided for species distinguishing, and a technical guarantee is also provided for enhancement and releasing of a parent source and fry quality.
Owner:ZHEJIANG OCEAN UNIV
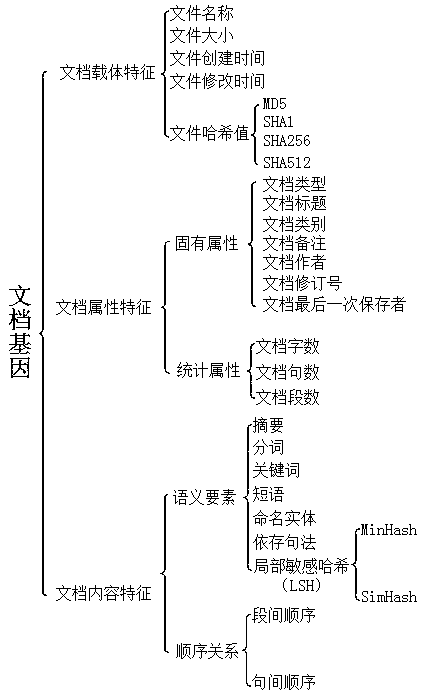
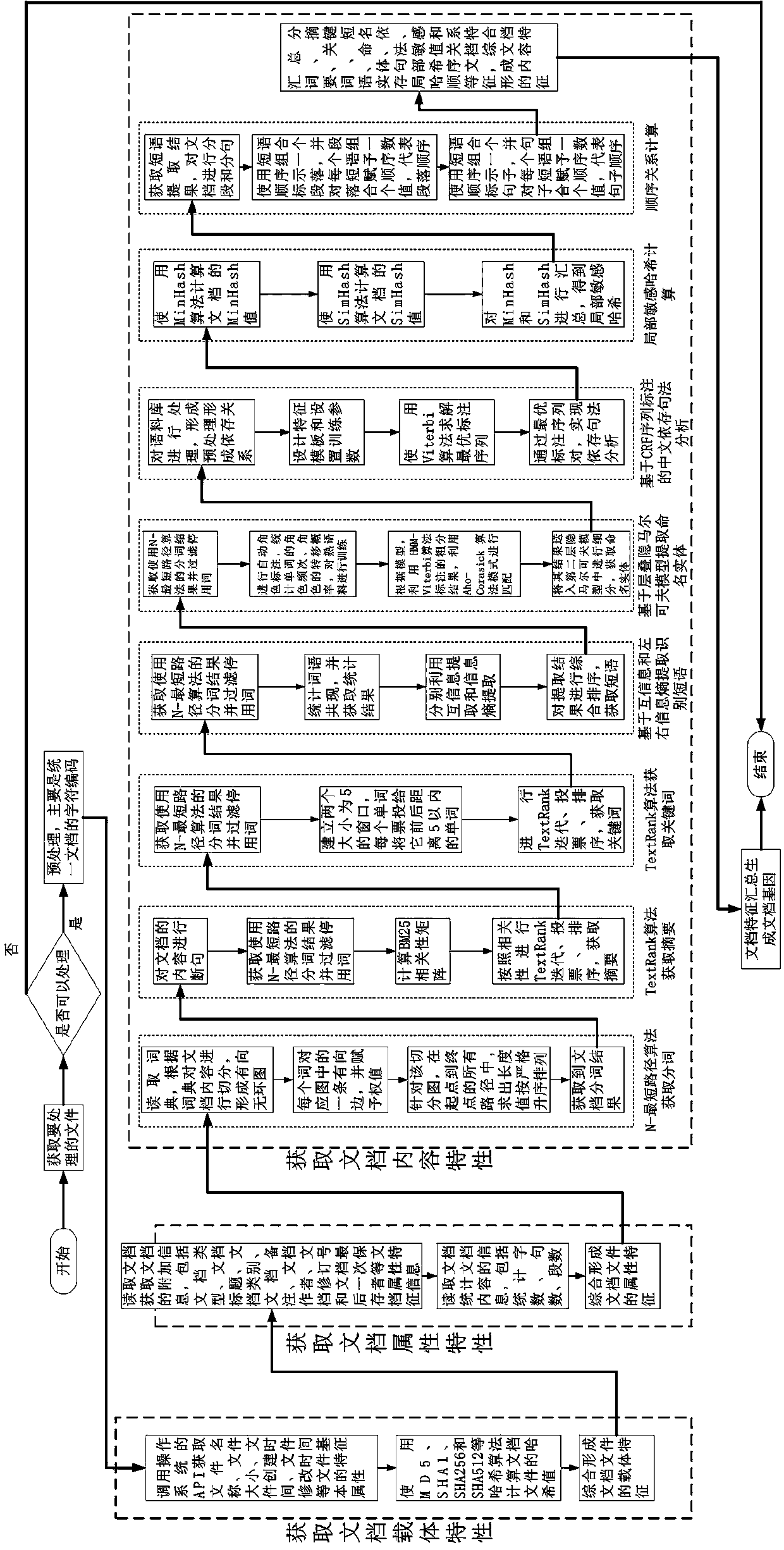
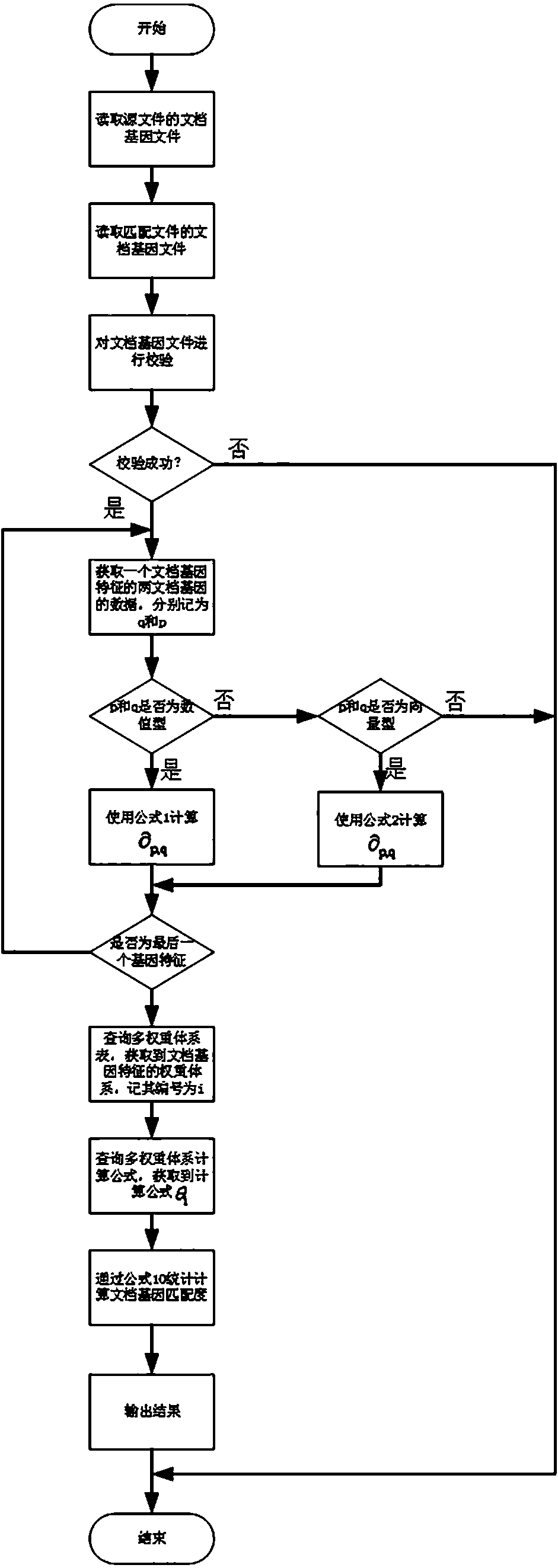
Chinese document gene matching method based on multi-weight system
ActiveCN107491424AReduce jumpingRefined configurationNatural language data processingSpecial data processing applicationsMatching methodsDocumentation
Aiming at the Chinese document gene matching and orienting to document evasion checking scenarios, the present invention proposes a gene feature matching method of 28 hybrid documents. In particular, a multi-weight system is introduced for the first time to reflect the consideration of gene differentiation within and among systems and to form a unified similarity calculation formula. Based on the document gene matching method provided by the present invention, the weight can be finely configured, the process of algorithm condition jump can be reduced, and the achievability and practical applicability are strong.
Owner:北京云量数盟科技有限公司
Gene detection method, feature extraction method, device, equipment and system
PendingCN113517022AGuaranteed accuracyReduce processing costsTherapiesHealth-index calculationProcessed GenesFeature extraction
The embodiment of the invention provides a gene detection method, a feature extraction method, a device, equipment and a system. The gene detection method comprises the following steps: acquiring a to-be-processed gene sequence, wherein the average number of gene segments corresponding to each position in the gene sequence is smaller than or equal to a preset threshold value; performing feature extraction operation on the gene sequence to obtain gene features; enhancing the gene features to obtain enhanced features corresponding to the gene features; and detecting the gene sequence based on the enhanced features to obtain a detection result. According to the technical scheme provided by the embodiment of the invention, the gene sequence is subjected to feature extraction operation to obtain the gene feature, then the gene feature is subjected to enhancement processing to obtain the enhanced feature, and then the gene sequence is detected based on the enhanced feature to obtain the detection result, so the accuracy of gene detection operation is ensured, and the data processing cost and the data processing amount are effectively reduced.
Owner:ALIBABA SINGAPORE HLDG PTE LTD
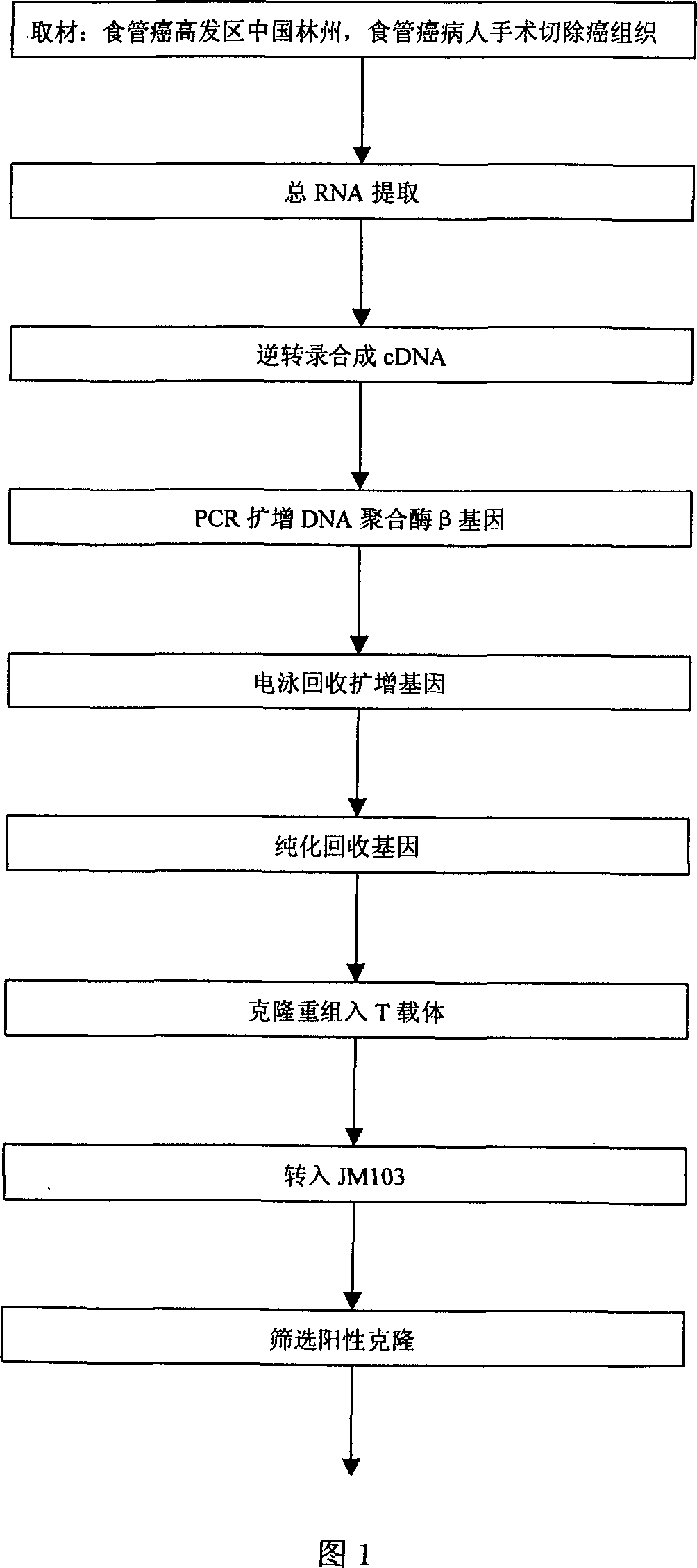
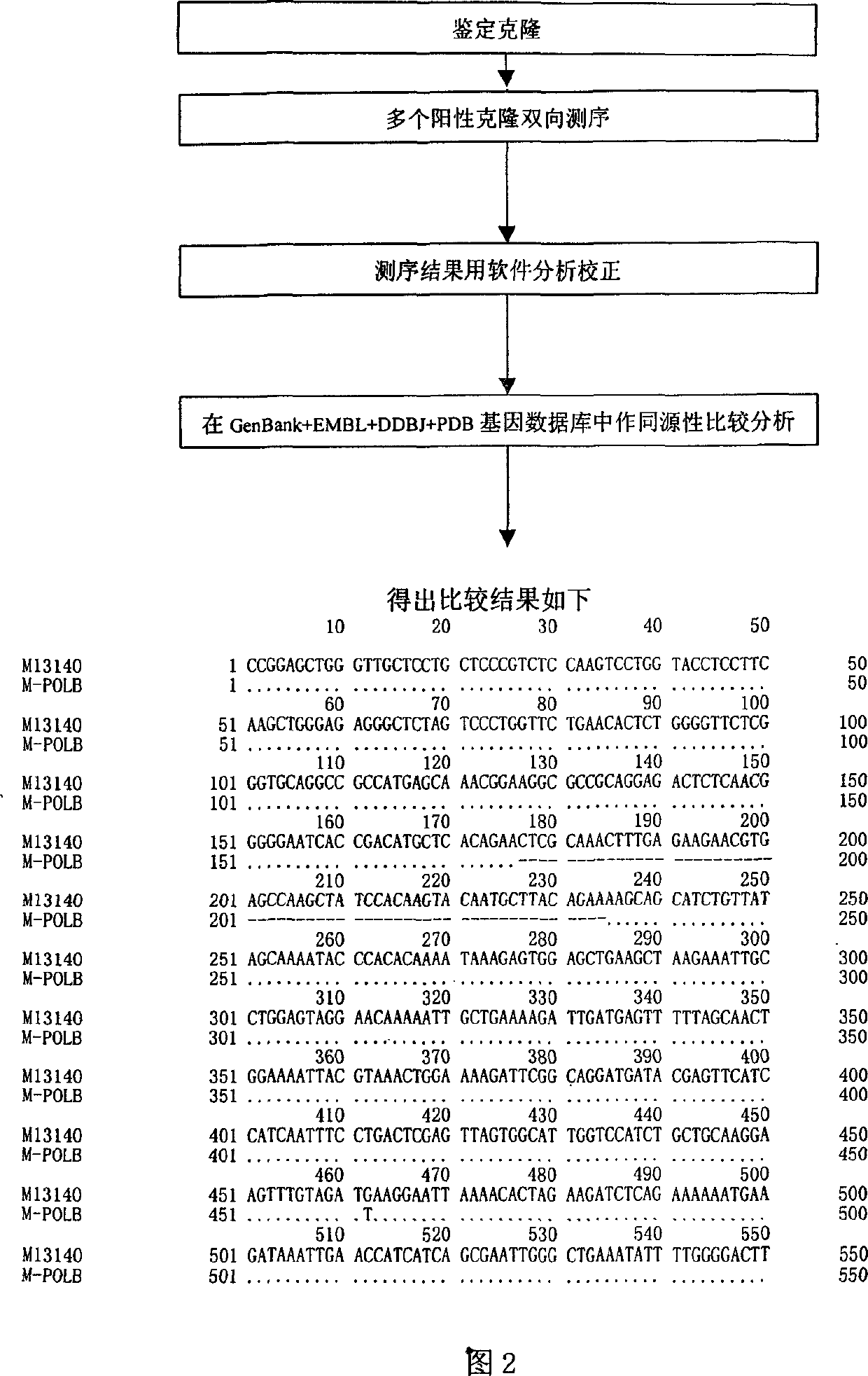
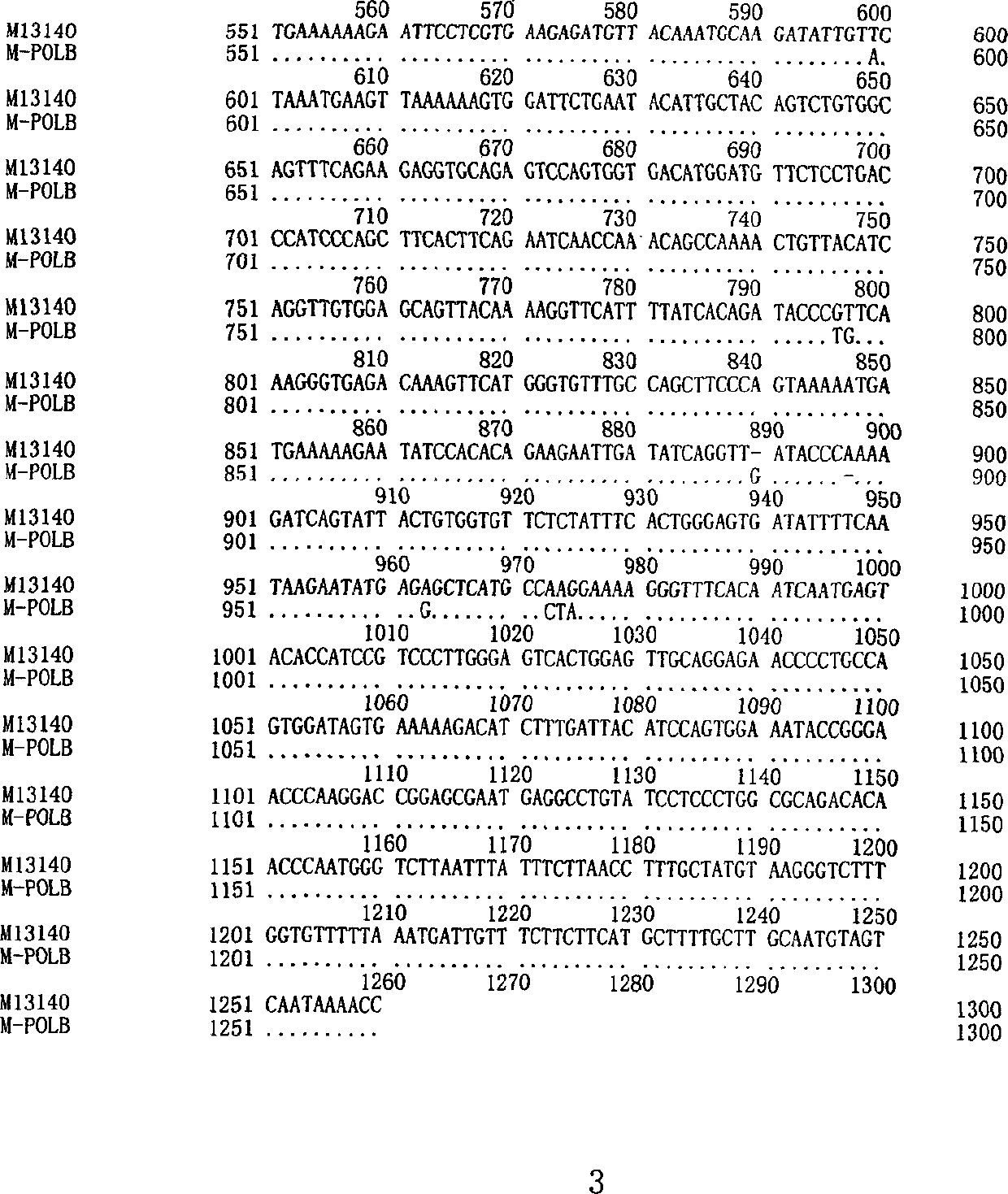
Human DNA polymerase beta mutant gene and its corresponding protein
InactiveCN1366047AEarly treatmentImprove postoperative survivalFermentationAnimals/human peptidesPolymerase LFhit gene
The present invention discloses a cDNA sequence of human repair gene DNA polymerase beta, which is a specific representation of DNA polymerase beta in esophagus cancer. The protein coded by it has fully lost the DNA repair activity of DNA polymerase beta. It can be used for early diagnosis and gene therapy of esophagus cancer.
Owner:ZHENGZHOU UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com