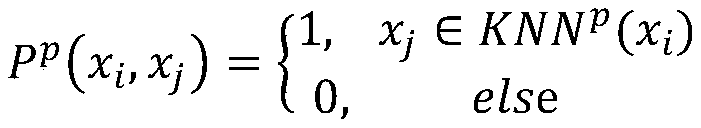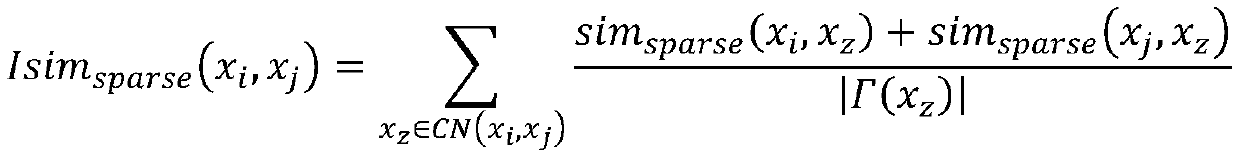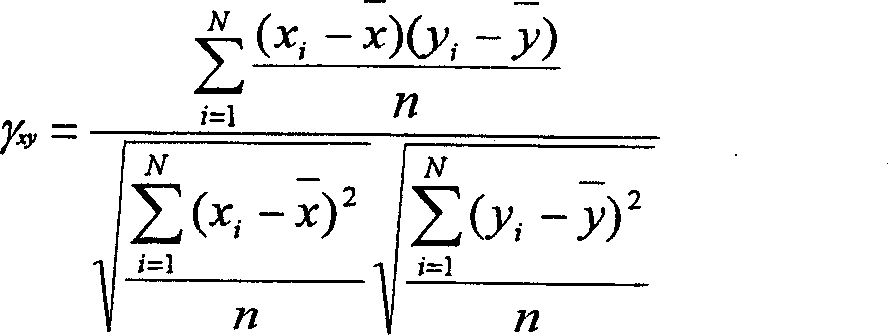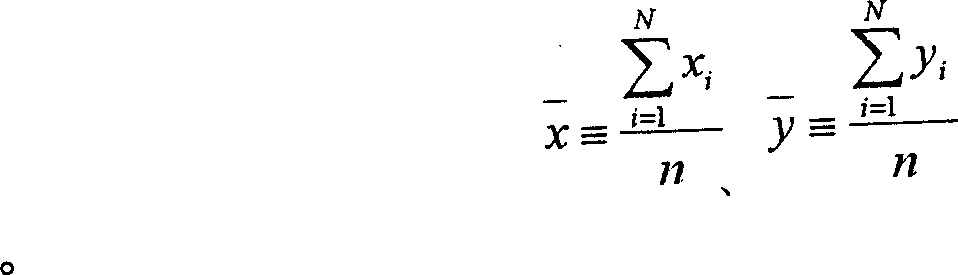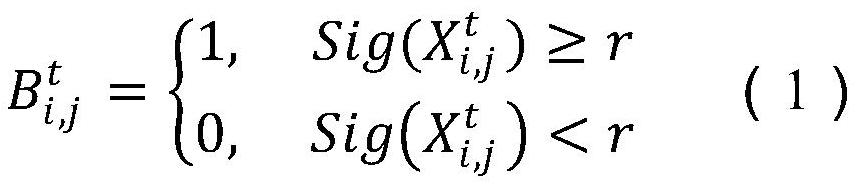Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
87 results about "Gene selection" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Gene selection. An allele that gives most individuals an advantage in survival and reproduction will usually spread through the population. For most of us, that's what "natural selection" really means. But note that it has to be an allele that works well with other genes in the population genome.
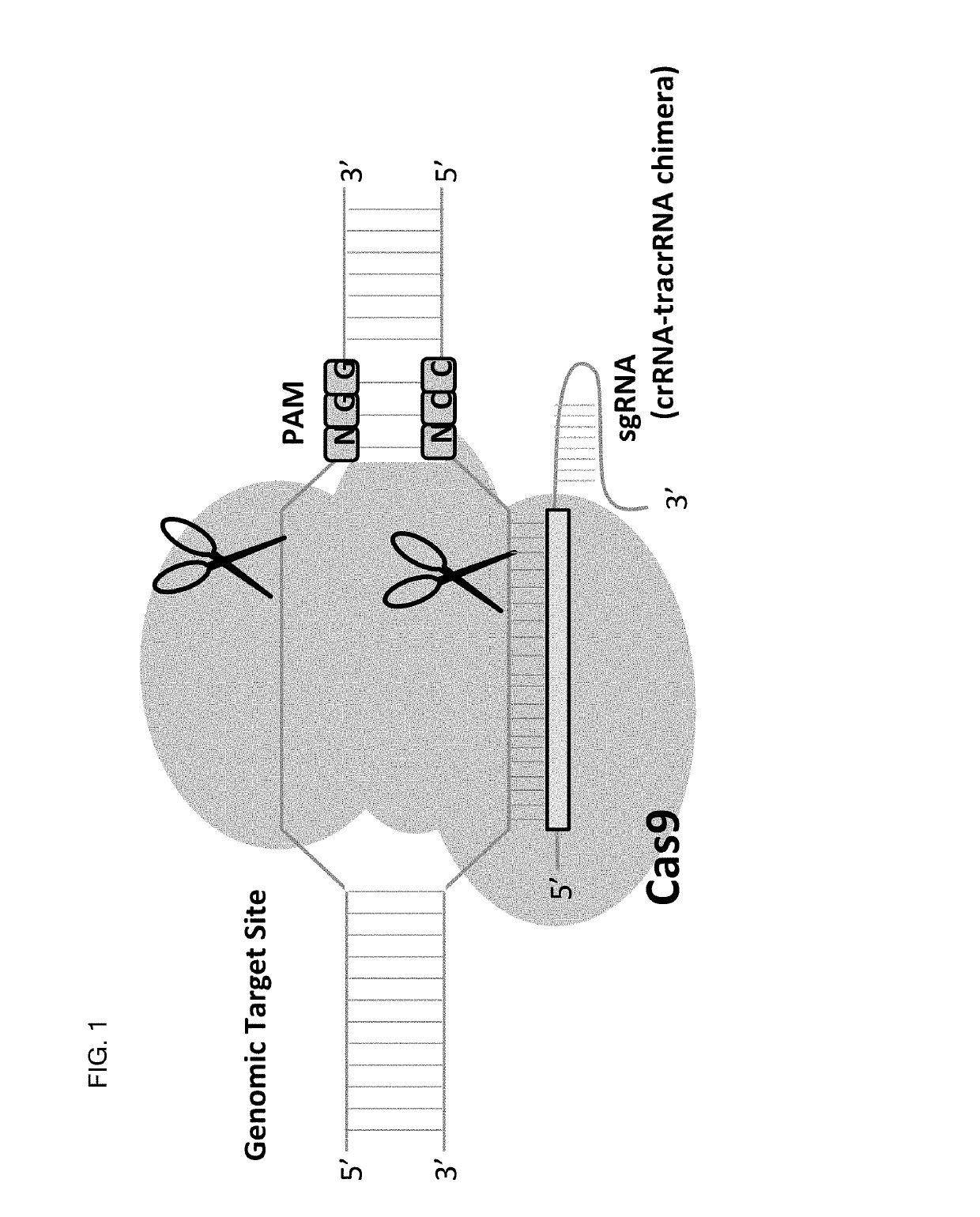
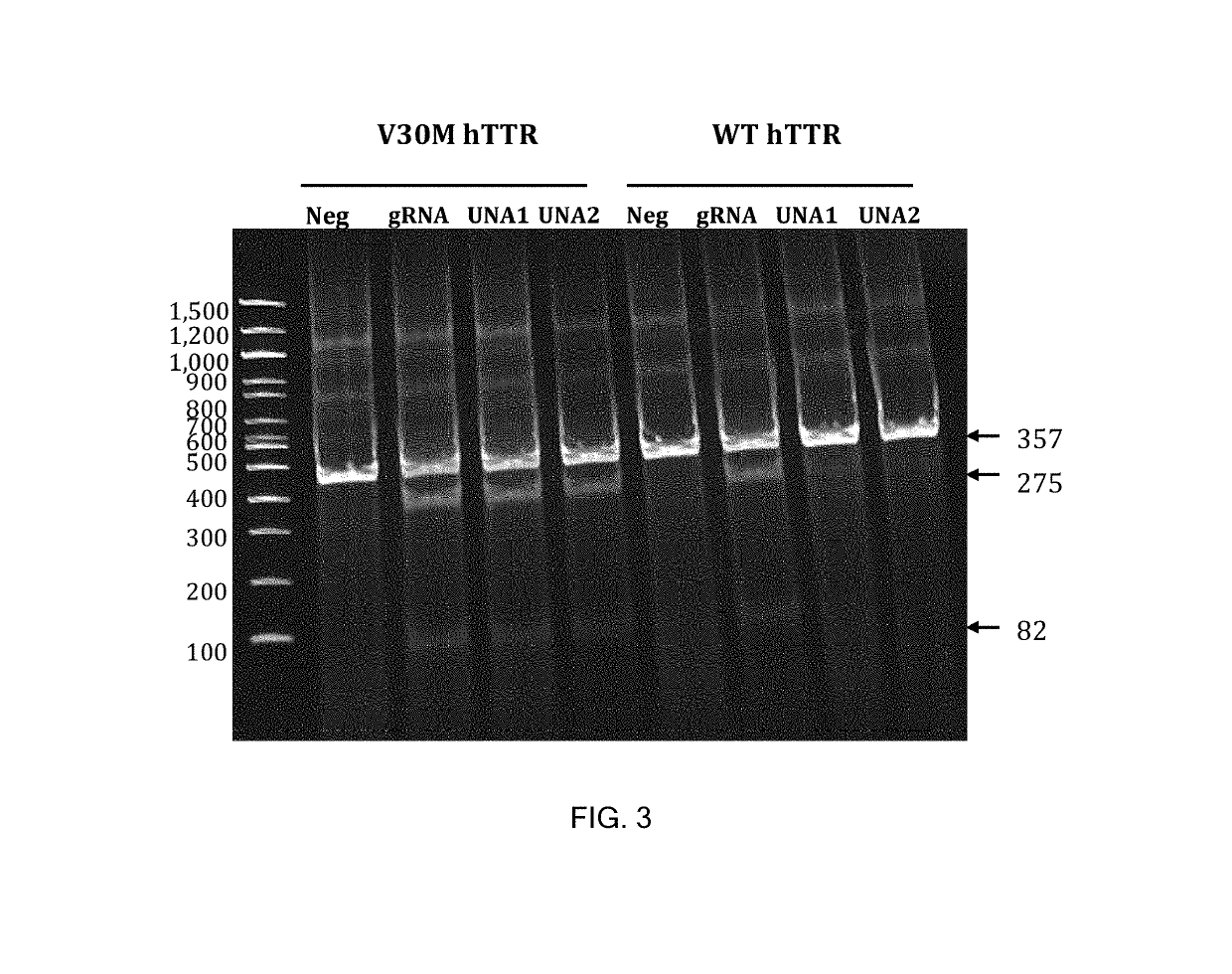
Allele selective gene editing and uses thereof
ActiveUS20170080107A1Improve efficiencyHigh frequencySenses disorderNervous disorderDiseaseGene selection
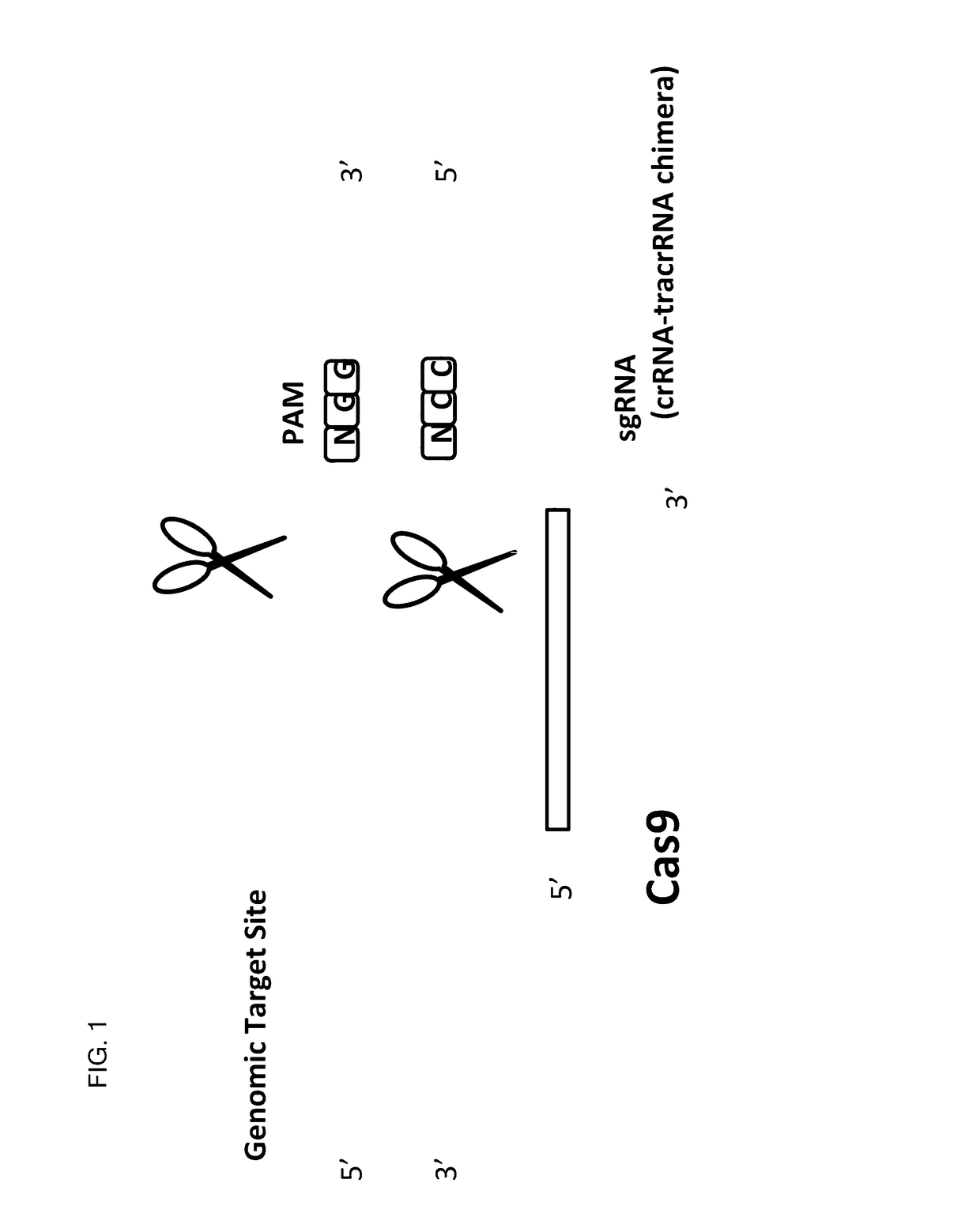
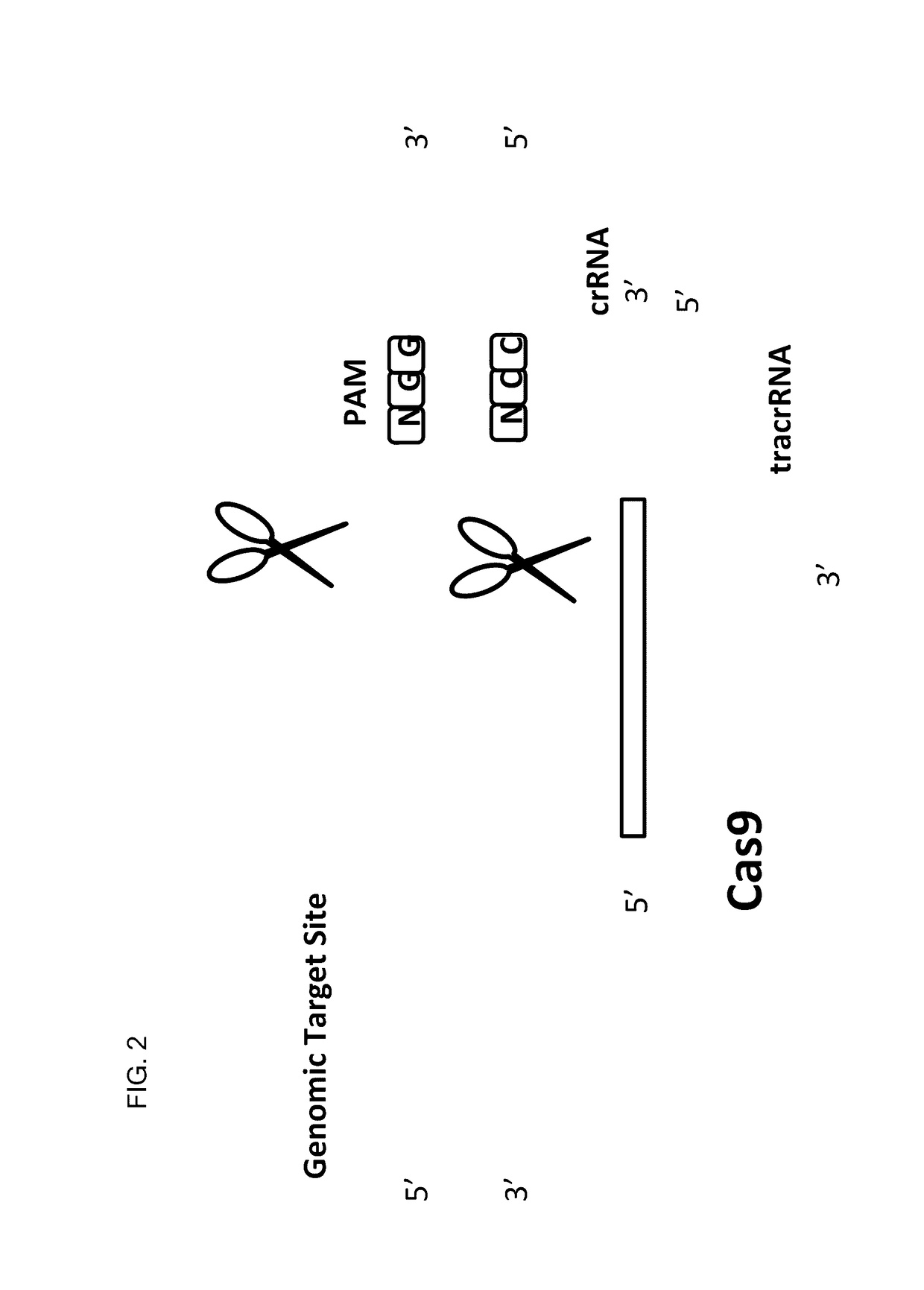
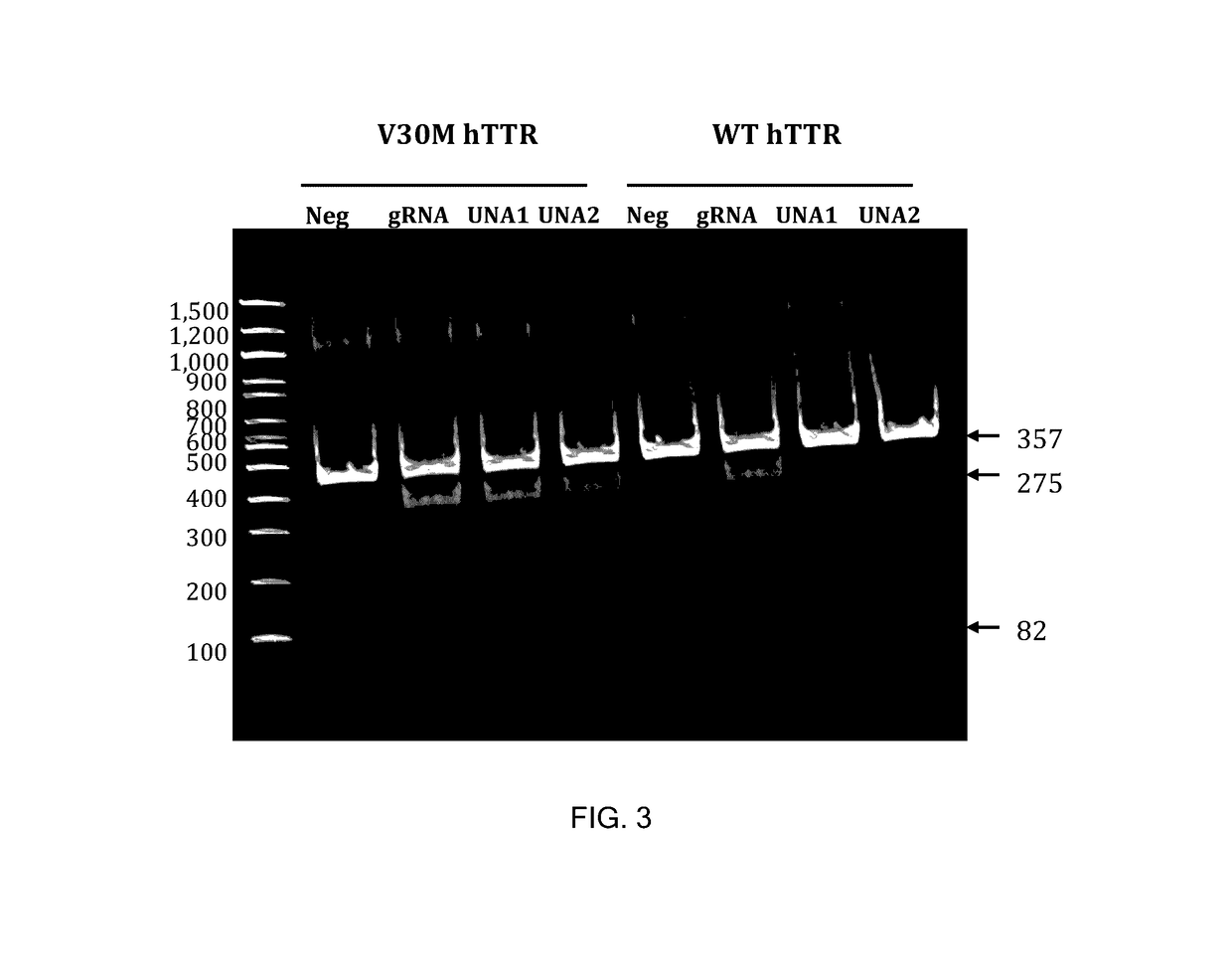
This invention encompasses compounds, structures, compositions and methods for therapeutic guide molecules that direct CRISPR gene editing. A guide molecule for directing gene editing can be allele selective, or disease allele selective, and can exhibit reduced off target activity. A guide molecule can be composed of monomers, including UNA monomers, nucleic acid monomers, and modified nucleotides, wherein the compound is targeted to a genomic DNA. The guide molecules of this invention can be used as active ingredients for editing or disrupting a gene in vitro, ex vivo, or in vivo.
Owner:ARCTURUS THERAPEUTICS
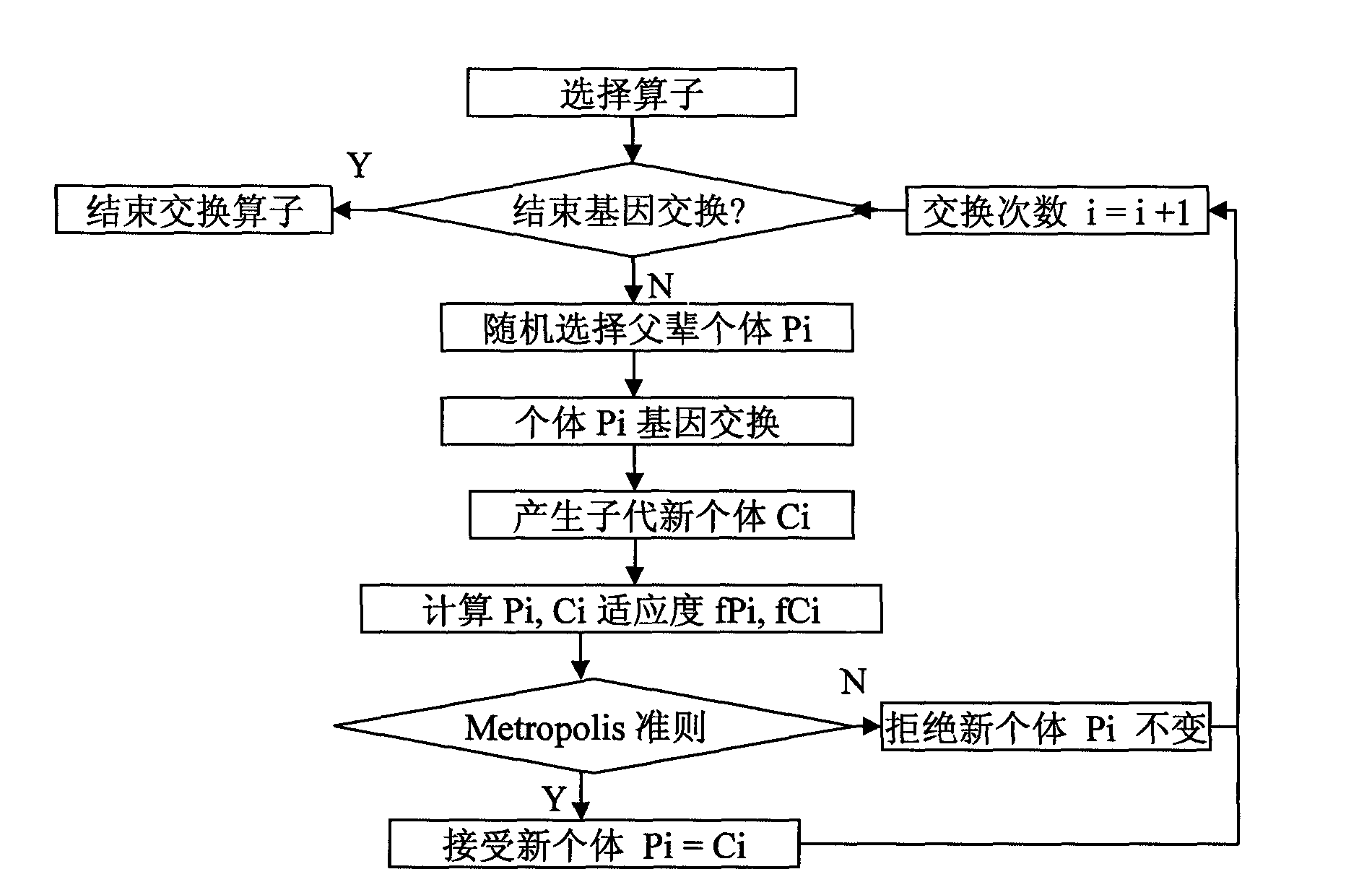
Selection method for subintervals of near infrared spectral characteristics based on simulated annealing-genetic algorithm
InactiveCN101832909ARaise the level of fitnessFix premature convergenceMaterial analysis by optical meansGenetic algorithmsInfraredGene selection
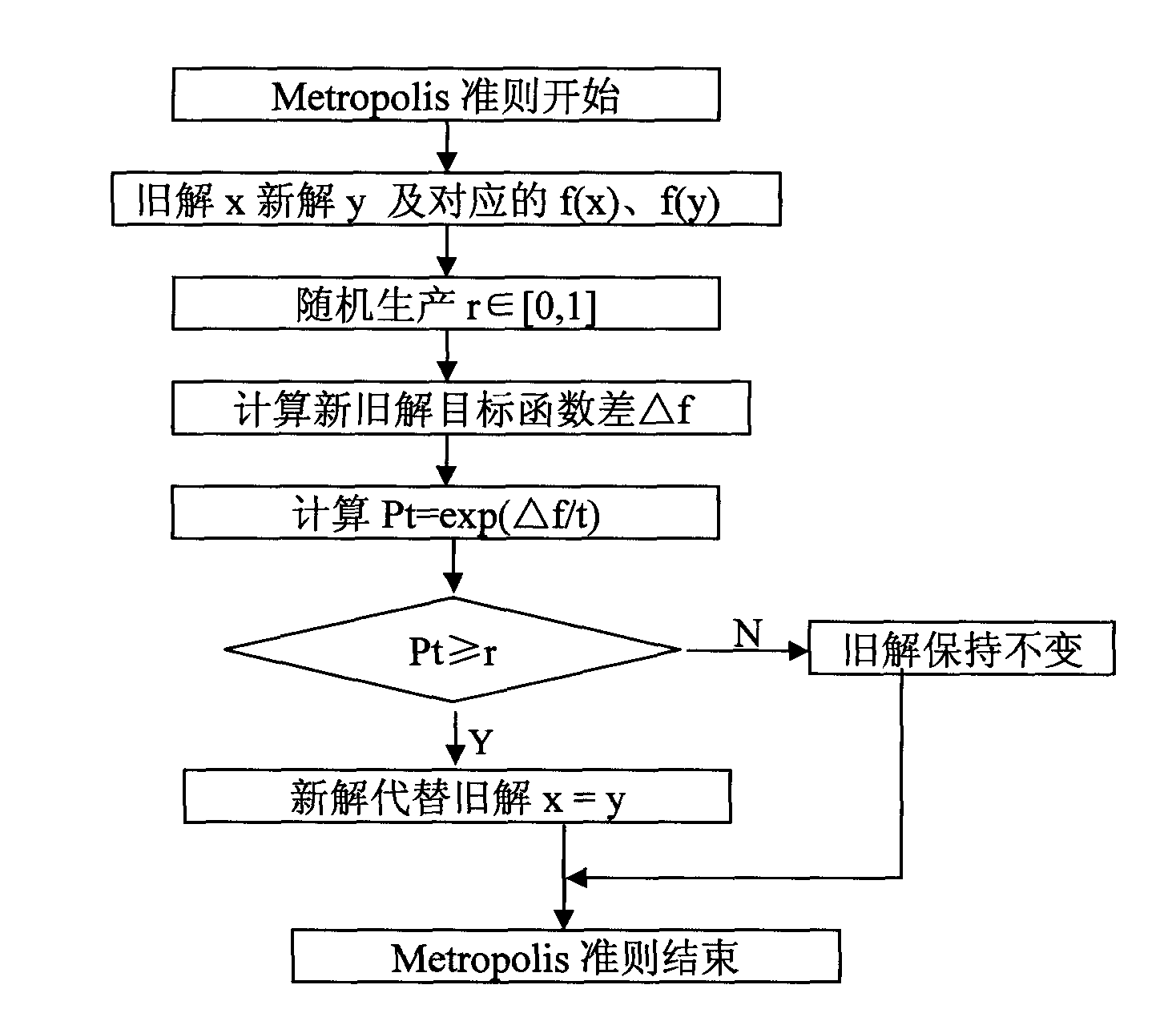
The invention discloses a selection method for subintervals of near infrared spectral characteristics based on a simulated annealing-genetic algorithm. The method comprises the following steps: pretreating a near infrared spectrum; then dynamically dividing subintervals on the pretreated near infrared spectrum, introducing an Metropolis criterion in the simulated annealing algorithm to gene exchange and gene selection operators, and selecting an optimal character subinterval with the simulated annealing-genetic algorithm; and finally judging the best subinterval division method to be combined with the optimal character subinterval and building a PLS model for the selected optimal character subinterval. In the selection method, high-quality offspring individuals can be generated through improved variation and commutating operators, not only adaptability levels of overall populations are improved, but also enough power for population evolution is provided; and deficiency brought by the total number of the spectrum subintervals manually designated according to the experiences in the process of modeling can be avoided, and spectral models with high precision and strong prediction ability can be rapidly obtained.
Owner:JIANGSU UNIV
Systems and methods for parallel processing optimization for an evolutionary algorithm
ActiveUS8255344B2General purpose stored program computerNeural learning methodsAlgorithmGene selection
Owner:THE AEROSPACE CORPORATION
Methods and composition related to in vivo imaging of gene expression
InactiveCN101171337AVectorsCell receptors/surface-antigens/surface-determinantsMethod of imagesIn vivo
Disclosed are nucleic acid sequences encoding a recombinant seven transmembrane G-protein associated receptor (GPCR) amino acid sequence operatively coupled to a tissue-selective promoter sequence. Also disclosed are methods of imaging or treating cells in a subject that involve introducing a nucleic acid encoding a reporter detectable in vivo using non-invasive methods operatively coupled to a tissue-selective promoter or amplified tissue specific promoter to the subject, and subjecting the subject to a non-invasive imaging technique or a therapeutic that selectively interacts with the reporter or gene of interest. For example, the subject may be a subject with cancer, and the therapeutic may be an anti-cancer agent. In particular, the hTERT promoter or amplified hTERT promoter drives expression of a reporter such hSSTR2 and / or hSSTR2 derivative) and these may be linked to a gene of interest. Imaging may be performed by a variety of methods including nuclear medicine.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
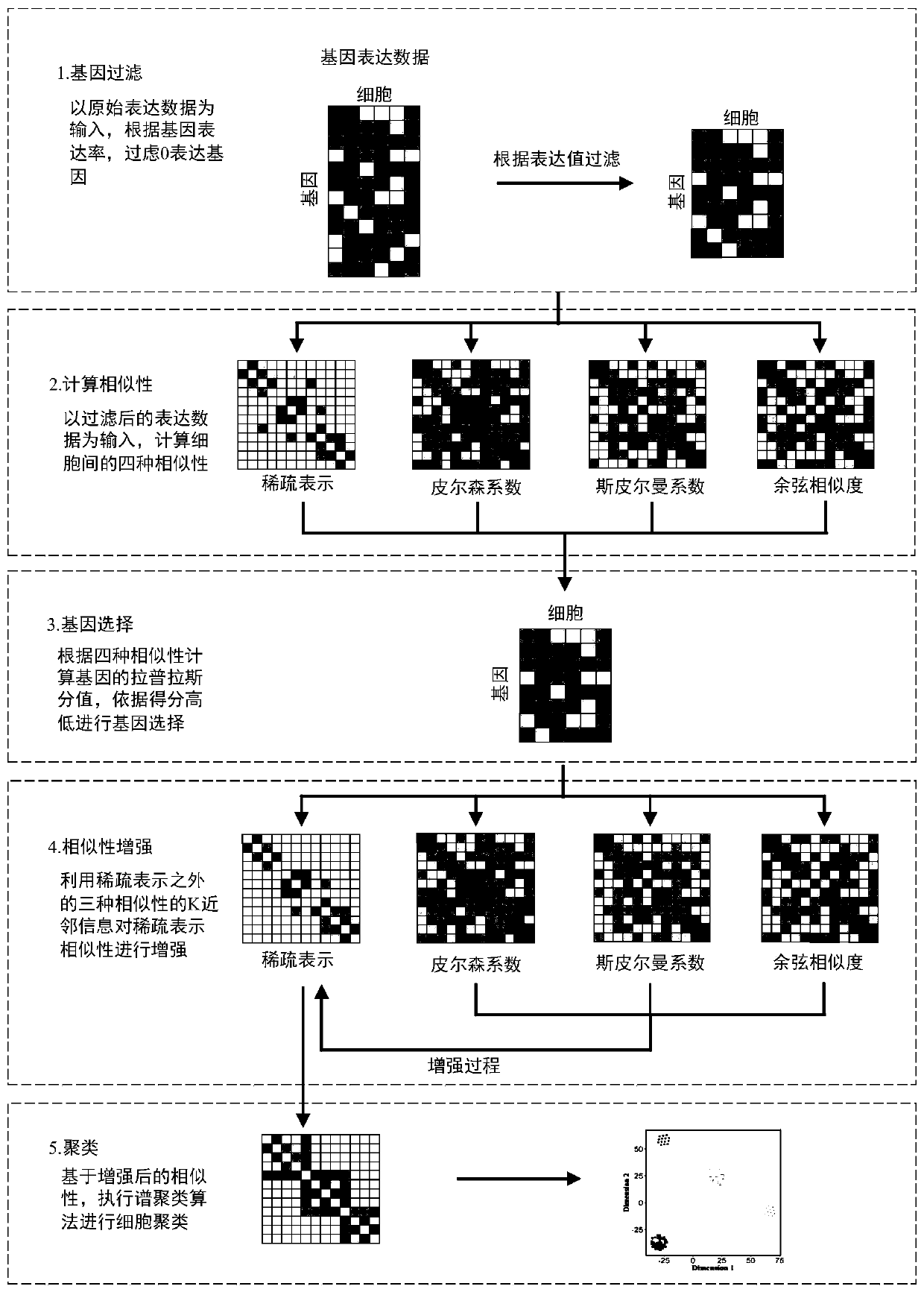
Cell type identification method based on similarity learning and enhancement thereof
ActiveCN110222745AAccurate identificationImprove accuracyBiostatisticsCharacter and pattern recognitionBiological noiseGene selection
The invention discloses a new cell type identification method based on similarity learning and enhancement thereof. The method designs a new global similarity calculation method, combines with other three conventional local similarity information, screens the genes, and carries out enhancement processing on the global similarity with sparse properties. According to the method, a global similaritycalculation method different from the traditional calculation of the local point-to-point similarity is used, the gene selection and the similarity enhancement are carried out by combining multiple different similarities including the global similarity and the local similarity, and a similarity matrix rich in information is obtained. According to the method, the influence of the factors, such as the technical noise, the biological noise, etc., carried by the single cell data can be effectively reduced, and the type of the single cell can be more accurately identified.
Owner:CENT SOUTH UNIV
Tea tree miRNA fluorescent quantitative PCR reference gene under low temperature stress as well as screening method and application of reference gene
ActiveCN105132417AMicrobiological testing/measurementDNA/RNA fragmentationReference genesGene selection
The invention relates to a tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p, which has a nucleotide sequence as shown in SEQ ID NO: 1, as well as a screening method of the reference gene. According to the method, 6 common plant miRNA reference genes and 3 tea tree miRNAs are selected as candidate reference genes, and the expression stabilities of the candidate reference genes under different low temperature stresses and in different tea tree tissues are evaluated by virtue of qRT-PCR technology through two software, namely BestKeeper and geNorm, so as to provide a reference for the selection of reference genes for tea tree miRNA differential expression analysis under low temperature stress. The obtained tea tree miRNA fluorescent quantitative PCR reference gene Pc-222-3p is stable in gene expression under different temperatures of treatment and in different tea tree tissues; therefore, the reference gene Pc-222-3p can be used as a reference gene for tea tree miRNA fluorescent quantitative PCR under low temperature stress.
Owner:ANHUI QIMEN COUNTY QIHONG TEA
Apparatus for forming molecular timetable and apparatus for estimating circadian clock
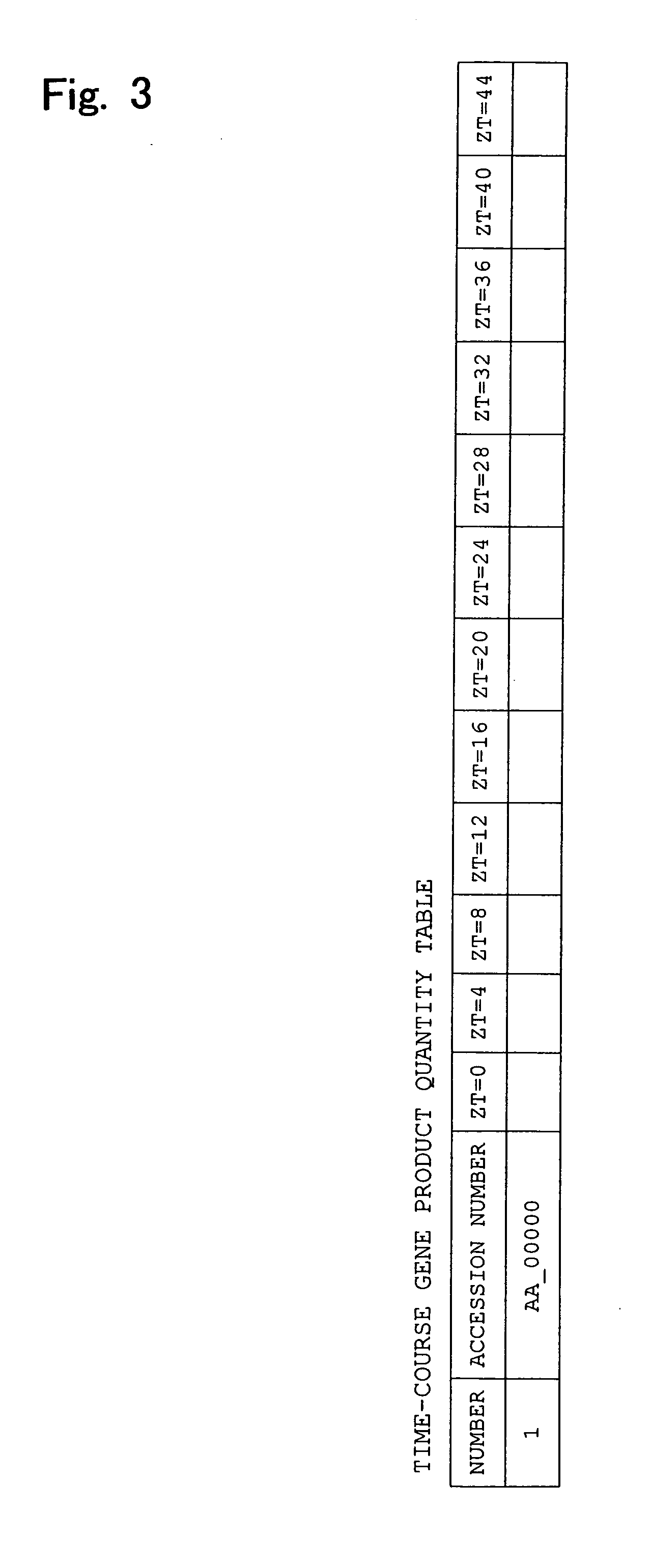
InactiveUS20060078883A1Simple procedureMicrobiological testing/measurementBiological testingCircadian clock geneTime course
An apparatus 11 for estimating internal biological time of a biological individual on the basis of quantity data of a gene product as measured in a specimen sampled from the individual. The apparatus includes circadian oscillatory gene selection means for selecting circadian oscillatory genes whose time-course change in gene product quantity data as measured in standard specimens approximates a cosine curve having a predetermined period; circadian expression curve selection means for selecting, from among a plurality of cosine curves having different phases and sharing a specific period, a circadian expression curve which is similar to the pattern of time-course change in the expression product quantity of each of the above-selected circadian oscillatory genes; and registration means for registering information which identifies the selected circadian expression curve.
Owner:ASTELLAS PHARMA INC
Method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly
The invention relates to a method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly. The method includes the steps that expression values of a large quantity of genes are detected through mRNA and competitive endogenesis RNA expression profile chips, and the correlations of all the genes are ordered; the obtained mRNA feature genes and the obtained competitive endogenesis RNA feature genes are combined; a gene pool is subjected to further gene selection with a genetic algorithm; differential expression genes are selected by comparing the data between different samples of mRNA and competitive endogenesis RNA joint expression profiles of breast cancer patients, breast cancer AIC patients and able-bodied persons; the differential expression genes are predicted, and the genes with the scores larger than 140 and free energy smaller than 20 are selected as reliable target genes; genes with obvious differences are selected from the target genes, and constructed breast-cancer anthracycline cardiotoxicity attacking target genes are verified through the residual RNA. By means of the method, more reliable potential gene markers related to anthracycline cardiotoxicity can be selected as a candidate.
Owner:JINZHOU MEDICAL UNIV
Peanut high-oleic-acid character gene selection method
The invention discloses a peanut high-oleic-acid character gene selection method. The method comprises the steps of extracting leaf gene groups of different peanut breeds, and amplifying AhFAD2A and AhFAD2B genes from the gene groups by using a specificity primer; cutting and recycling specificity amplification stripes and sequencing to obtain the sequence of the gene; translating the obtained sequence, comparing and analyzing, and searching the point mutation or inserting site to judge the oleic acid content and specific value of oleic acid / linoleic acid (O / L) of the breed. The method is simple to operate, variety of peanuts germ plasm can be analyzed, the oleic acid content of single plant of peanut can be determined in the seedling stage, the breeding process can be shortened and the breeding effect of oleic acid can be improved by combining comprehensive agronomic character selection of the single plant.
Owner:SHANDONG PEANUT RES INST
Cancer gene expression profile data identification method based on integration of extreme learning machines
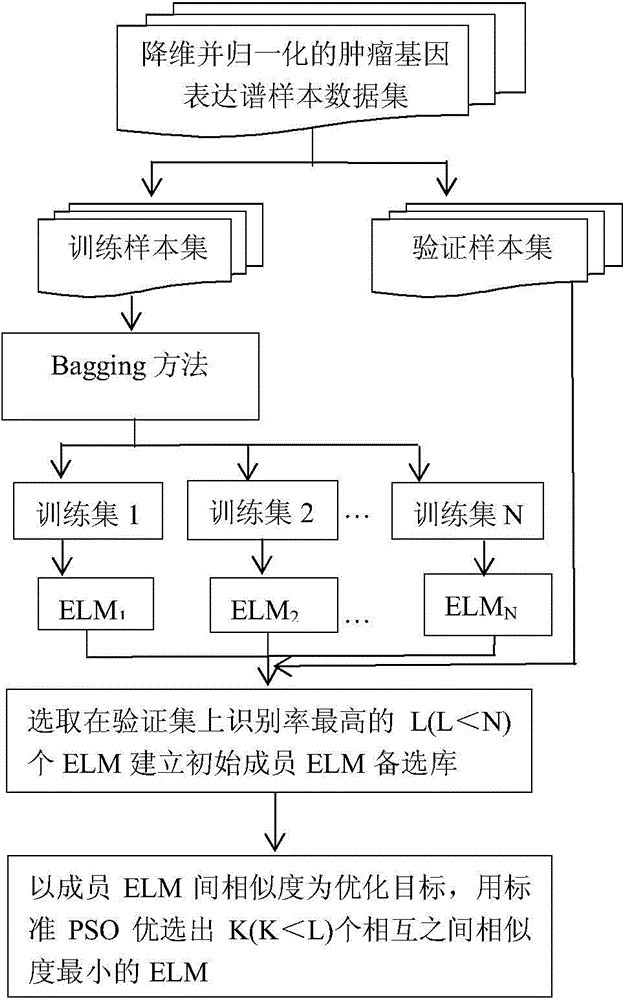
InactiveCN104463251AImprove the accuracy of tumor recognitionReduce learning costsBiostatisticsCharacter and pattern recognitionData setGene selection
The invention discloses a cancer gene expression profile data identification method based on integration of extreme learning machines. The method includes the steps of selection and integration of member extreme learning machines (ELMs). The method concretely includes the steps that preprocessing is carried out on a cancer gene expression profile data set, wherein the preprocessing includes gene selection and normalization of expression profile data; N sample sets are generated through a Bagging method, and each sample set is divided into a training set and a verification set according to a certain proportion; N ELMs are generated on the N training sets in a learning mode, and L ELMs (L&1t; N) with the highest recognition rate on the corresponding verification sets are selected to form an alternative member ELM base; K member ELMs (K&1t; L) forming an integrated system are selected from L ELMs based on the particle swarm optimization algorithm; the integrated vote weight of K member ELMs is worked out by utilizing the minimum-norm least square method; an integrated ELM system is obtained, and the integrated ELM system is used for performing tumor recognition on a newly increased cancer gene expression profile sample. Through the method, the cancer gene expression profile data can be quickly and accurately recognized.
Owner:JIANGSU UNIV OF SCI & TECH
Tumor feature gene selection method combining mRNA and microRNA expression profile chips
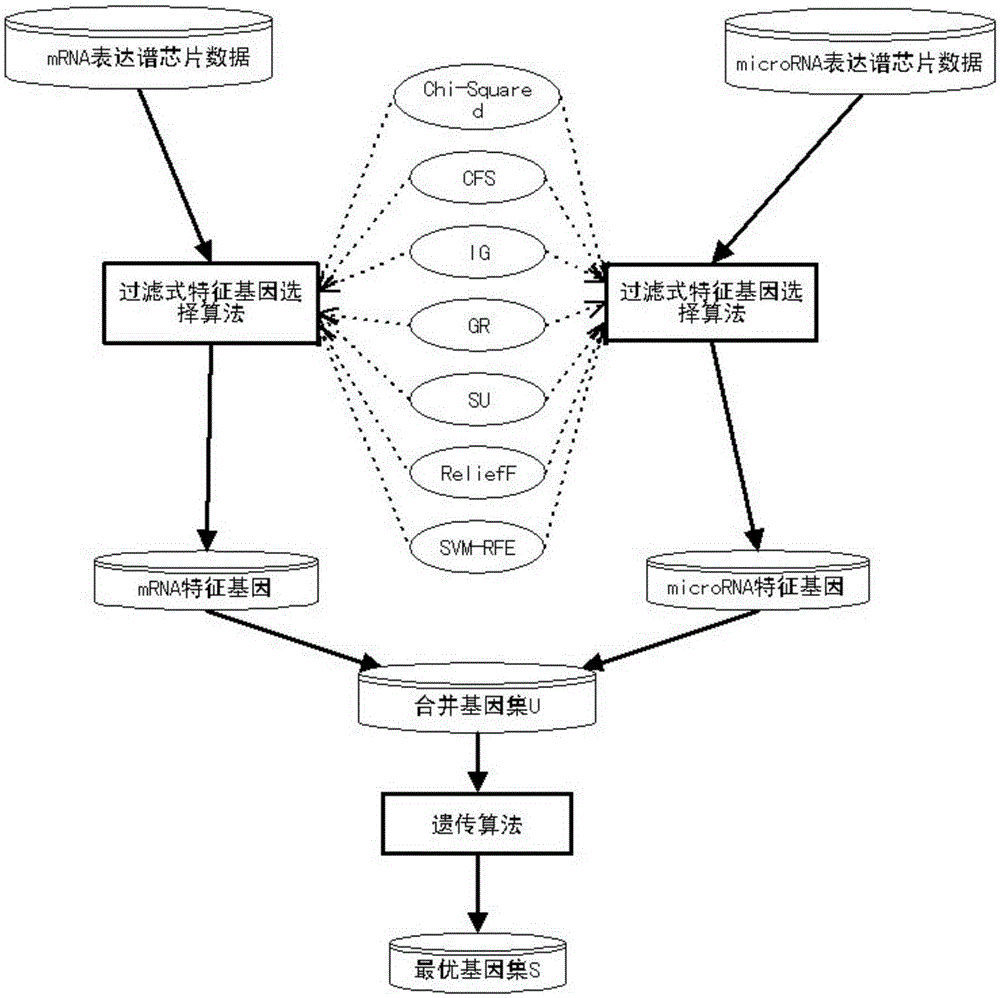
InactiveCN105243296AImprove classification accuracyEasy to analyzeSpecial data processing applicationsGene selectionFiltration
The invention discloses a tumor feature gene selection method combining mRNA and microRNA expression profile chips. The method is implemented by the following steps of step 1, by using the mRNA and microRNA expression profile chips, detecting expression values of a large amount of genes, by adopting a filtration feature gene selection method, ranking relevance of all the genes, removing a large amount of low-relevance genes, and leaving a small amount of genes that are closely related to tumor classification; step 2, combining mRNA and microRNA feature genes obtained by adopting the filtration feature gene selection method to form a gene pool U; and step 3, by adopting a genetic algorithm, further selecting genes for the gene pool, eliminating redundant genes, and performing a search to obtain an optimal gene set S with optimal features. The method is simple in step and high in result accuracy.
Owner:LISHUI UNIV
Lactococcus lactis food-sate secretion expression carrier and its preparing method and application
ActiveCN101168741ABacterial antigen ingredientsGenetic material ingredientsBinding siteGene selection
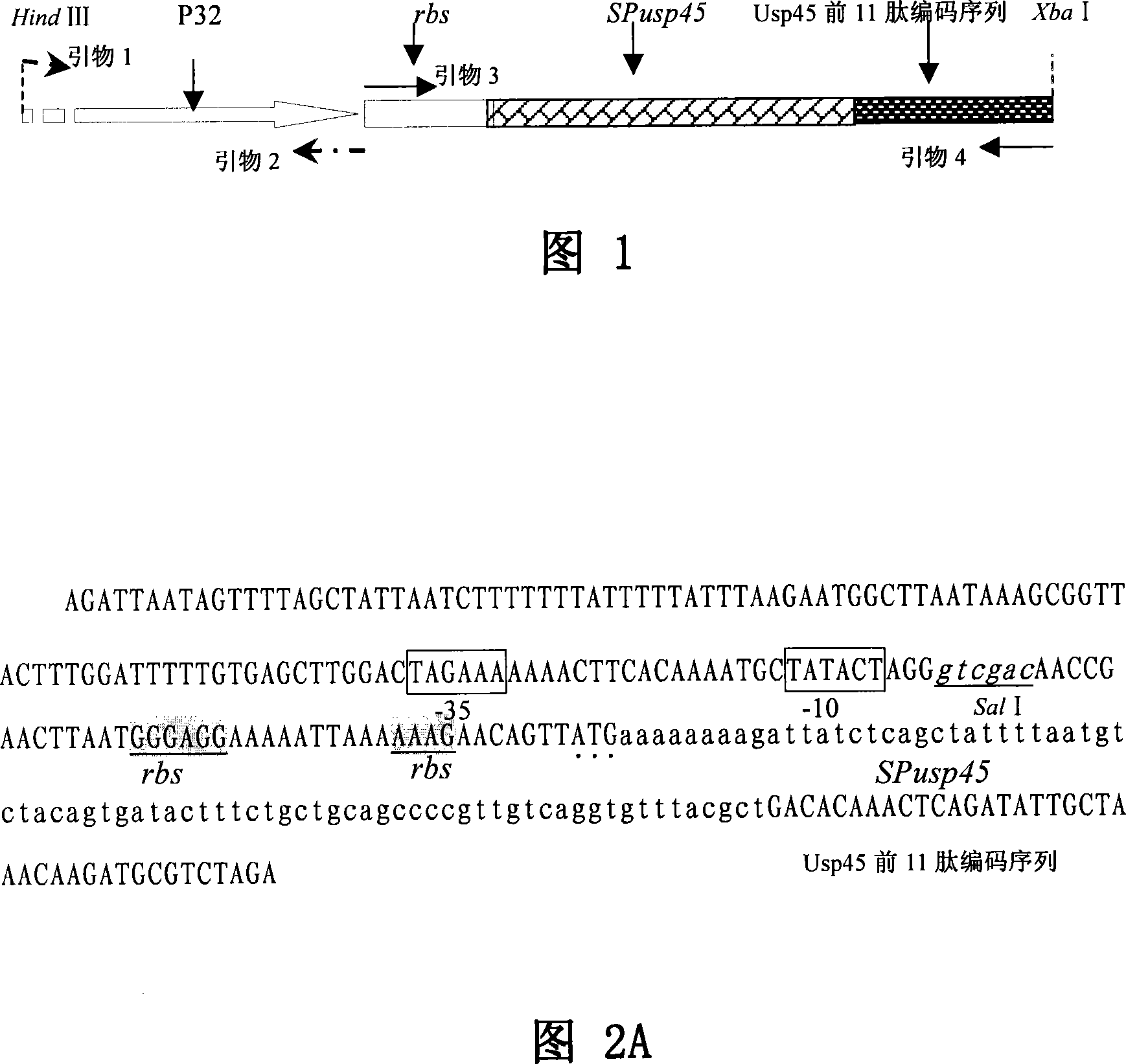
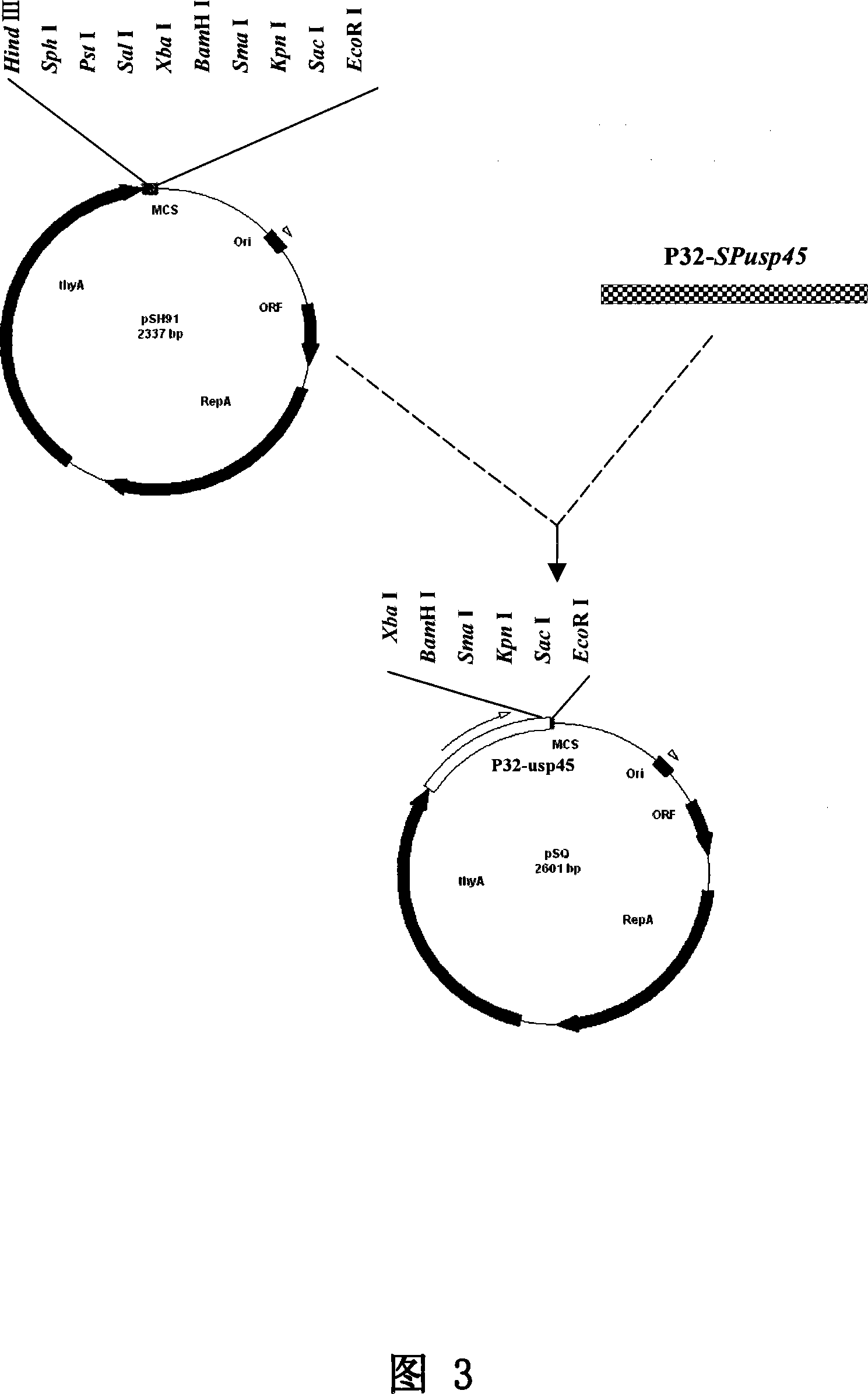
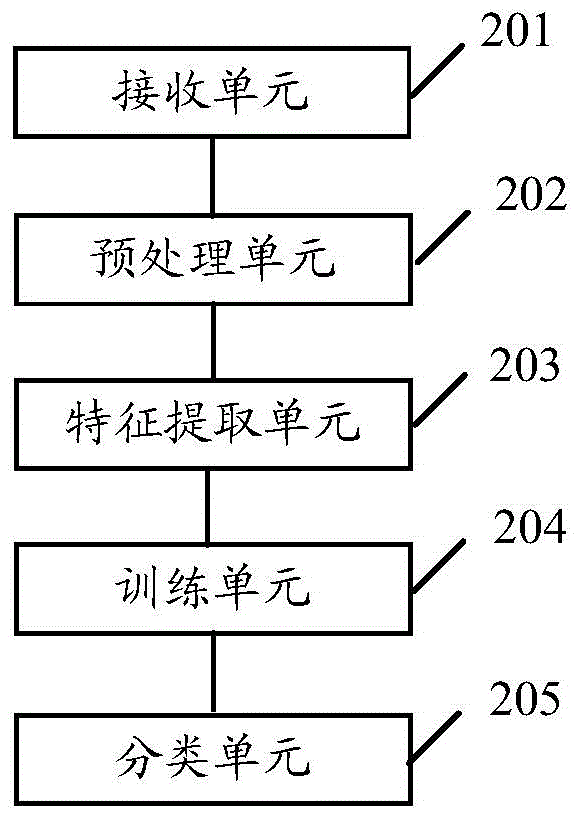
The invention relates to lactic acid galactococcus food-grade secretion expression vector, and the preparation method and the application thereof, in particular to the lactic acid galactococcus food-grade secretion expression vector which comprises the following components: a thyA gene selection marker, replicons of plasmid pWV01, multiple cloning sites, promoter used for secretion expression, a ribosome bind site of Usp45, a signal peptide sequence of Usp45 and a partial Usp45 mature peptides coded sequence.
Owner:ICDC CHINA CDC
Gene data processing method and gene data processing device
InactiveCN104408332AImprove accuracyReduce the impact of classification accuracySpecial data processing applicationsGene selectionTest sample
The embodiment of the invention discloses a gene data processing method and a gene data processing device. The method comprises the following steps: receiving gene data of a specified feature type of a reference population; preprocessing the gene data to obtain standardized gene data; performing feature gene selection on the standardized gene data by a LASSO method to obtain feature gene data; upon a cross validation method, dividing a sample set of the feature gene data into test samples and training samples; injecting the training samples into a classifier to obtain a trained classifier; injecting the test samples into the trained classifier; performing feature classification on the test samples, and performing statistics on the classification accuracy of the classifier. According to the gene data processing method and the gene data processing device provided by the embodiment of the invention, the accuracy of the feature gene selection can be improved, and the influence of the selection of the test samples and the training samples on the classification accuracy is lowered.
Owner:SHENZHEN INST OF ADVANCED TECH
Feature gene selection method based on logistic and relevant information entropy
InactiveCN104598774ASimple structureAvoid lossSpecial data processing applicationsMaximum eigenvalueData set
The invention discloses a novel feature gene selection method based on logistic and relevant information entropy. The method comprises the following steps that a dataset is subjected to logistic regression, a gene variable with great influence on the classification is obtained, a Relief algorithm is used for giving a value on the gene variable, sequencing is carried out, a maximum feature value gene is added to an initial feature gene set, and the relevant information entropy is calculated. The novel feature gene selection method has the advantages that a logistic regression model in the machine study is introduced into the feature gene selection method, and a high-quality gene expression profile is obtained; the correlation between gene variables is measured by the relevant information entropy, redundant genes are deleted, and a feature gene sub set with high classification capability and fewer genes is obtained through searching a feature gene space set.
Owner:HENAN NORMAL UNIV
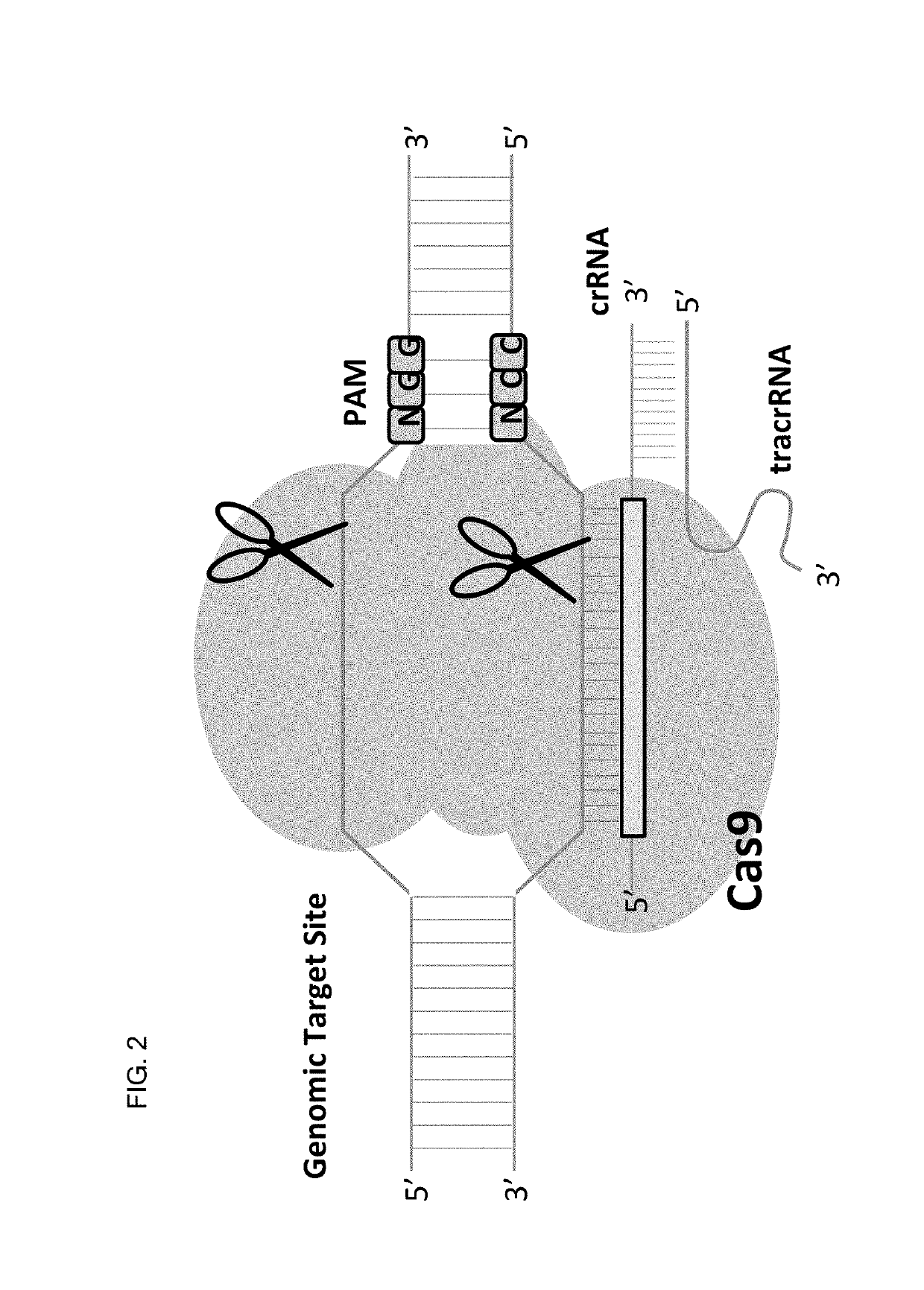
Allele selective gene editing and uses thereof
ActiveUS10369232B2Efficient gene editingReduce off-target effectsSenses disorderNervous disorderNucleotideGenomic DNA
This invention encompasses compounds, structures, compositions and methods for therapeutic guide molecules that direct CRISPR gene editing. A guide molecule for directing gene editing can be allele selective, or disease allele selective, and can exhibit reduced off target activity. A guide molecule can be composed of monomers, including UNA monomers, nucleic acid monomers, and modified nucleotides, wherein the compound is targeted to a genomic DNA. The guide molecules of this invention can be used as active ingredients for editing or disrupting a gene in vitro, ex vivo, or in vivo.
Owner:ARCTURUS THERAPEUTICS
Rice granule shape gene qSS7 as well as preparation method and application
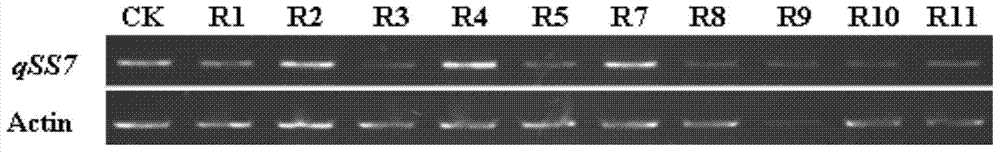
The invention discloses a rice granule shape gene qSS7 as well as a preparation method and application. The sequence of the rice granule shape gene qSS7 is as shown in SEQIDNO.1. Due to the gene, the granule length and the length to width ratio can be increased, the granule width can be reduced, and the chalkiness rate and the chalkiness degree can be also reduced; and when the appearance quality of rice is improved by using the gene, the quality and the yield of rice can be prevented from negative influence; a gene mark is also designed for qSS7, long and short granule allele can be identified in the seedling period, unexpected heterozygous and pure short granule genotype plants can be rejected, the possibility that descendant granule shapes can be separated when being bred is avoided, and breeding procedures for a next season can be reduced. Therefore, the area of a descendant bred material can be reduced by 75% in quality breeding practice, the gene selection accuracy can be improved, and the breeding progress can be accelerated.
Owner:HUAZHONG AGRI UNIV
Breeding method of dedicated silkworm hybrid secreting natural and green sericin with high content
ActiveCN101664019APromote development and utilizationImprove qualityAnimal husbandryFiberHomozygous genotype
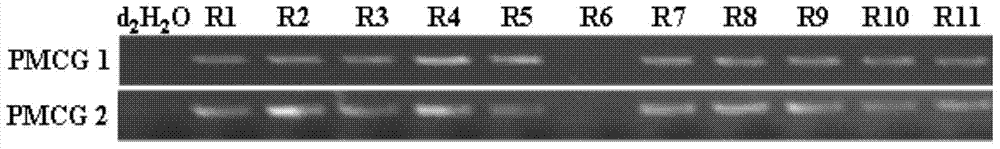
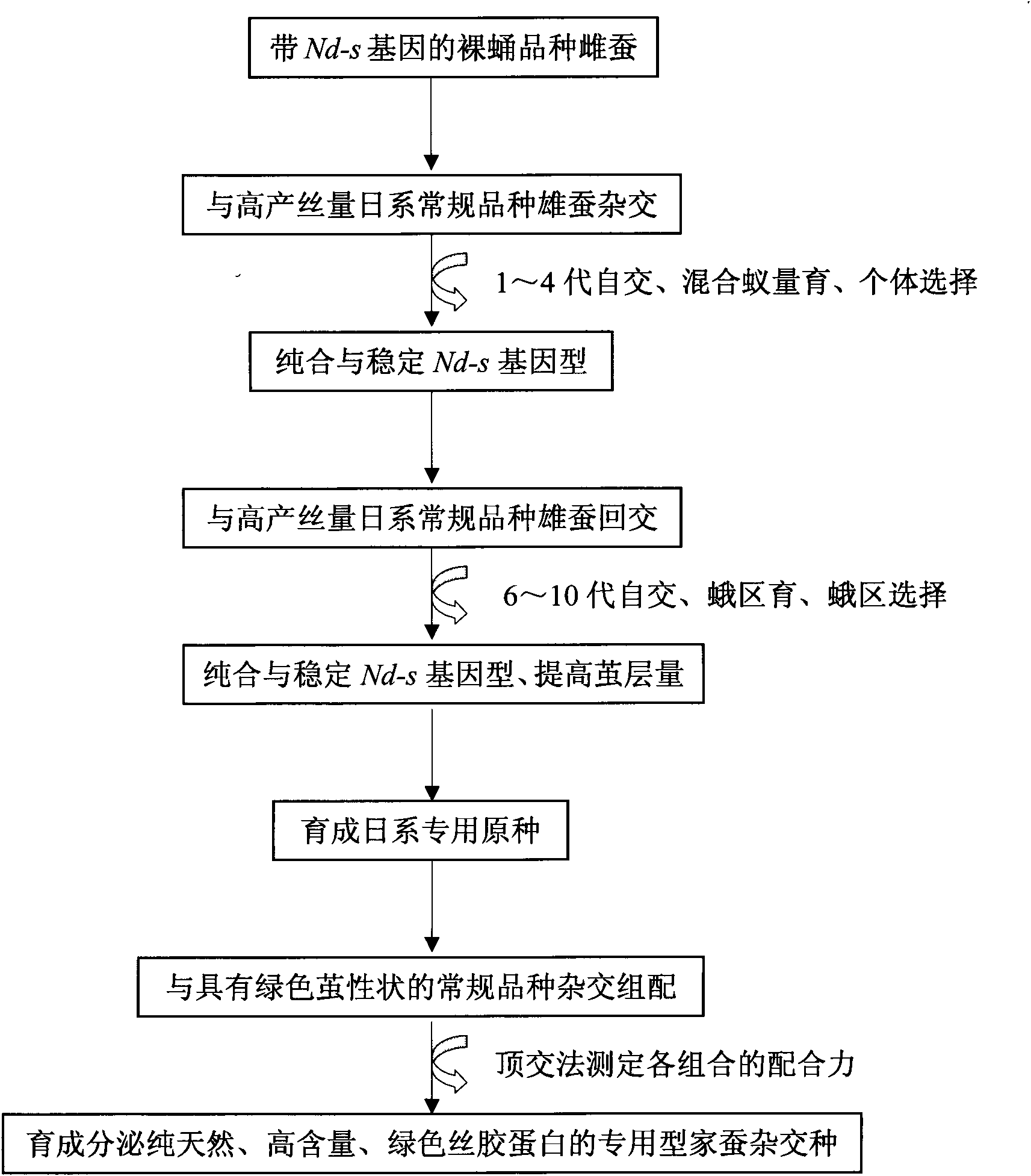
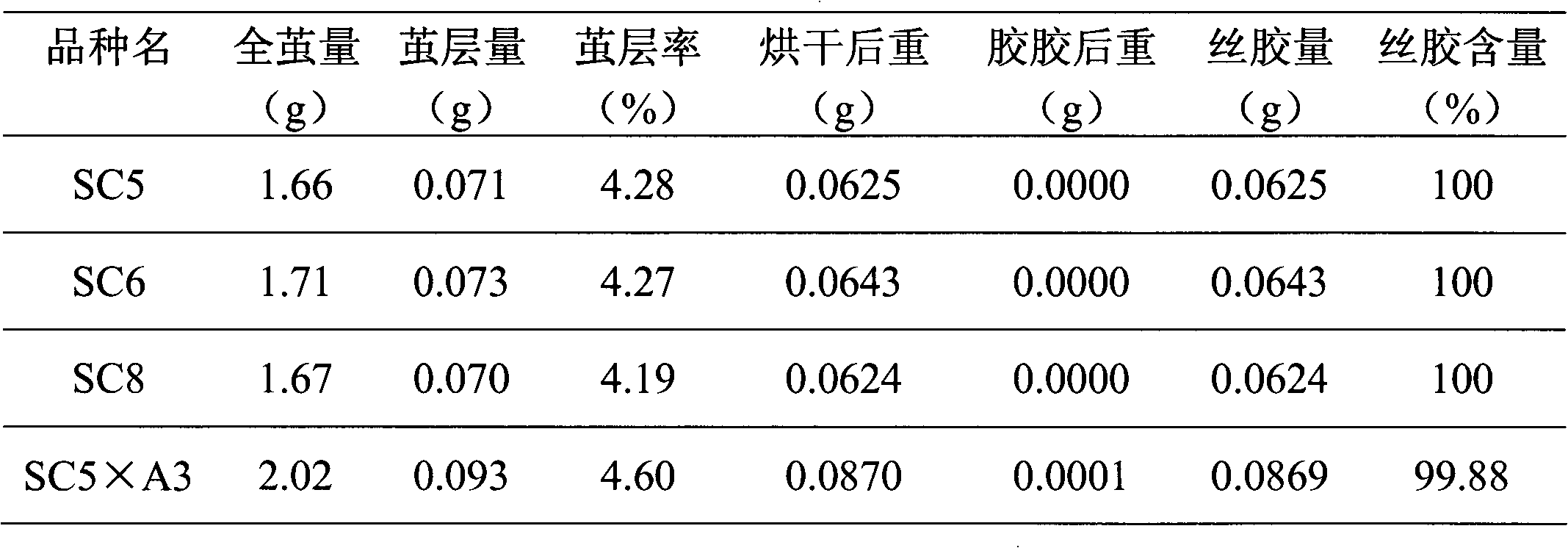
The invention provides a breeding method of dedicated silkworm hybrid secreting natural and green sericin with high content, comprising the following steps: taking exarate pupae breeds (with Nd-s genes) as stuff to be hybridized with the breeds with high amount of silk production, and breeding Japanese special original strains with sericin content being more than 99.0% and homozygous genotypes (Nd-s / Nd-s) by adopting the methods of selfing, backcrossing, marker gene selecting, sericin content tracking and detecting and the like; and hybridizing the Japanese special original strains with the Chinese conventional breeds (Gc / Gc) with green cocoon properties to breed the dedicated silkworm hybrid. The method can provide high quality raw materials to such fields as cosmetics, medicines, food, fiber modified materials and the like and improve the product added value.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Heuristic breadth-first searching method for cancer-related genes
InactiveCN103186717APathogenesis revealedFacilitates personalized treatmentSpecial data processing applicationsGene ordersGene selection
The invention relates to a heuristic breadth-first searching method for cancer-related genes. According to the method, appearance frequencies of genes in a selected gene subset are used for measuring the genes, and genes with higher appearance frequency are considered as the most important cancer-related genes, on the basis, a classifier is designed and a gene ordering method based on HBSA is established. As proved by study, information gene selection plays an important role in improving the classification performance, and the genes can be probably taken as important tumor clinical diagnosis signs, so discovery of the minimum gene subset with the highest classification performance is a very important research objective. As indicated by experimental results, the heuristic breadth-first searching method can not only obtain favorable generalization performance but also discover important tumor genes. And the relationship of the appearance frequencies of the selected genes and the gene number conforms to power-law distribution. The genes in the gene subset with extremely high classification accuracy are in close relationship with specific tumor subtypes, and even the genes are important genes directly related with the tumor.
Owner:HEFEI INSTITUTES OF PHYSICAL SCIENCE - CHINESE ACAD OF SCI
Industrial robot trajectory optimization method based on improved genetic algorithm
ActiveCN112692826AOptimize motion trajectoryMotion track stabilityProgramme-controlled manipulatorAlgorithmGene selection
The invention discloses an industrial robot trajectory optimization method based on an improved genetic algorithm, and aims to provide a trajectory through which an industrial robot completes a given route with the shortest time and the minimum impact based on the given path and a certain constraint condition. Based on a traditional genetic algorithm, the industrial robot trajectory optimization method disclosed by the invention improves a gene selection formula, a crossover formula and a mutation formula, improves crossover and mutation probabilities on the basis of self-adaption PRGA, optimizes the action time of a mechanical arm under the condition that the motion trail of the robot is stable, reduces resonance and shaking, caused by overlarge impact, of the robot, and enables the robot to run stably and smoothly, thereby greatly improving the working efficiency and prolonging the service life of the robot.
Owner:FOSHAN UNIVERSITY
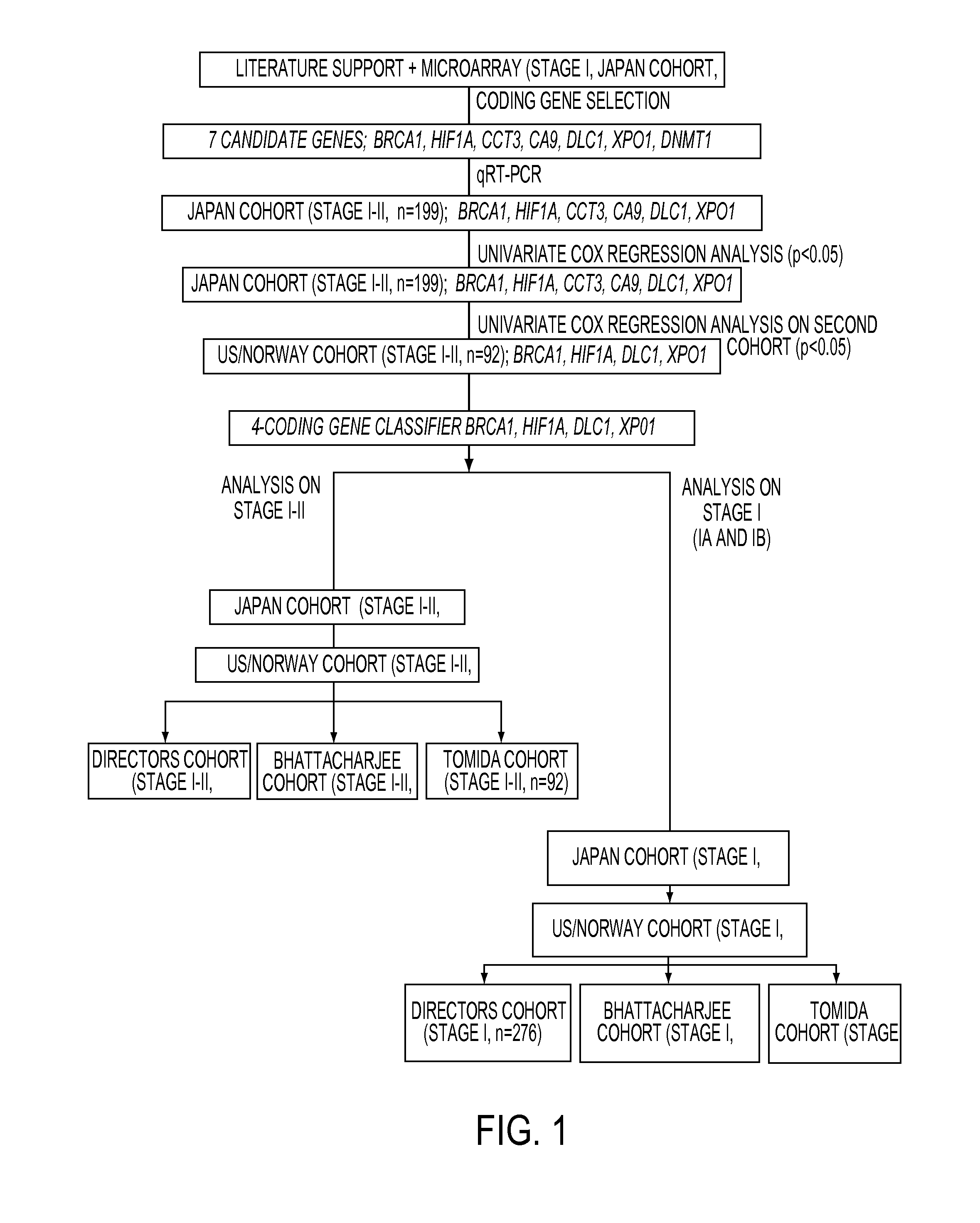
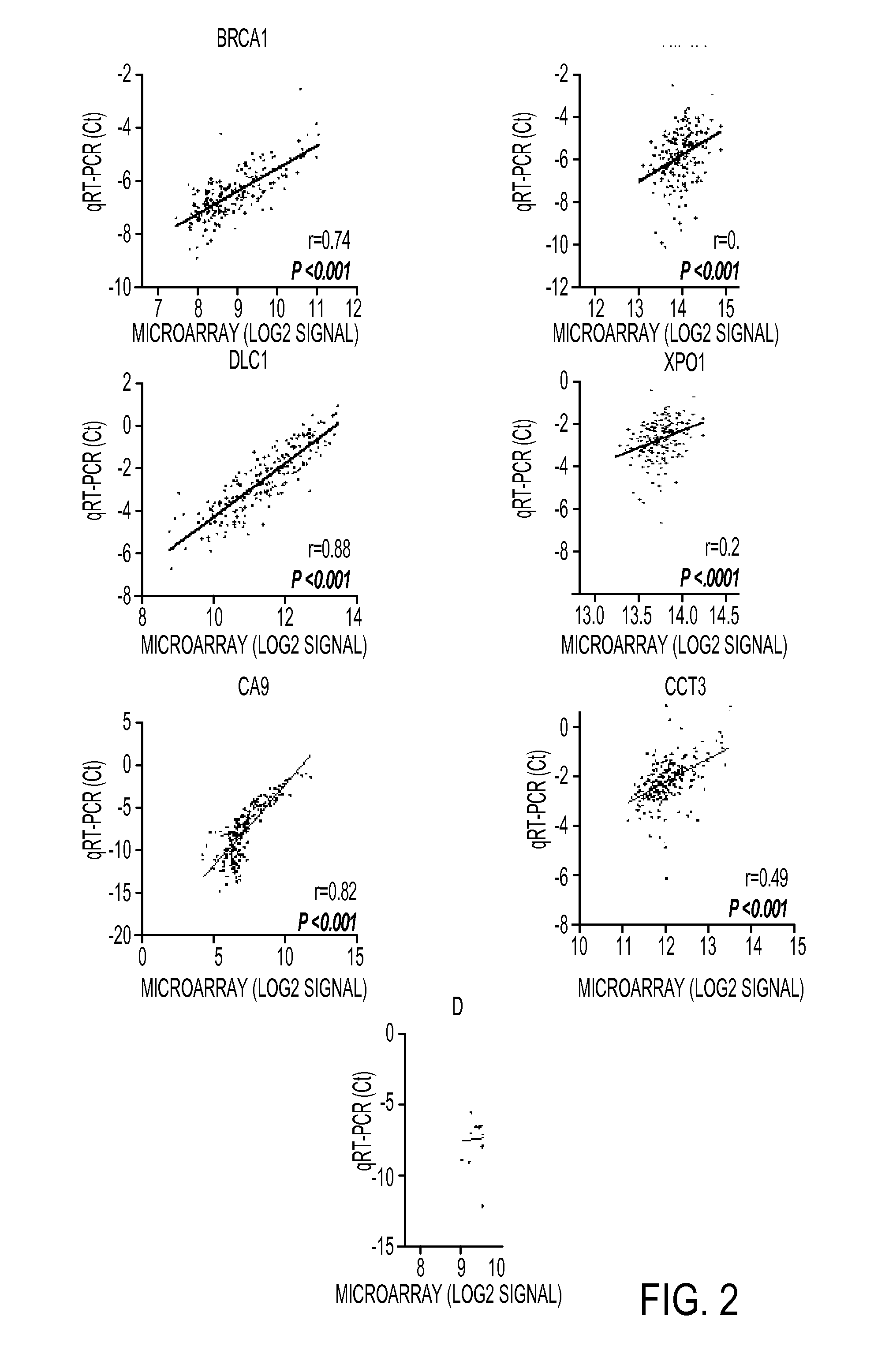
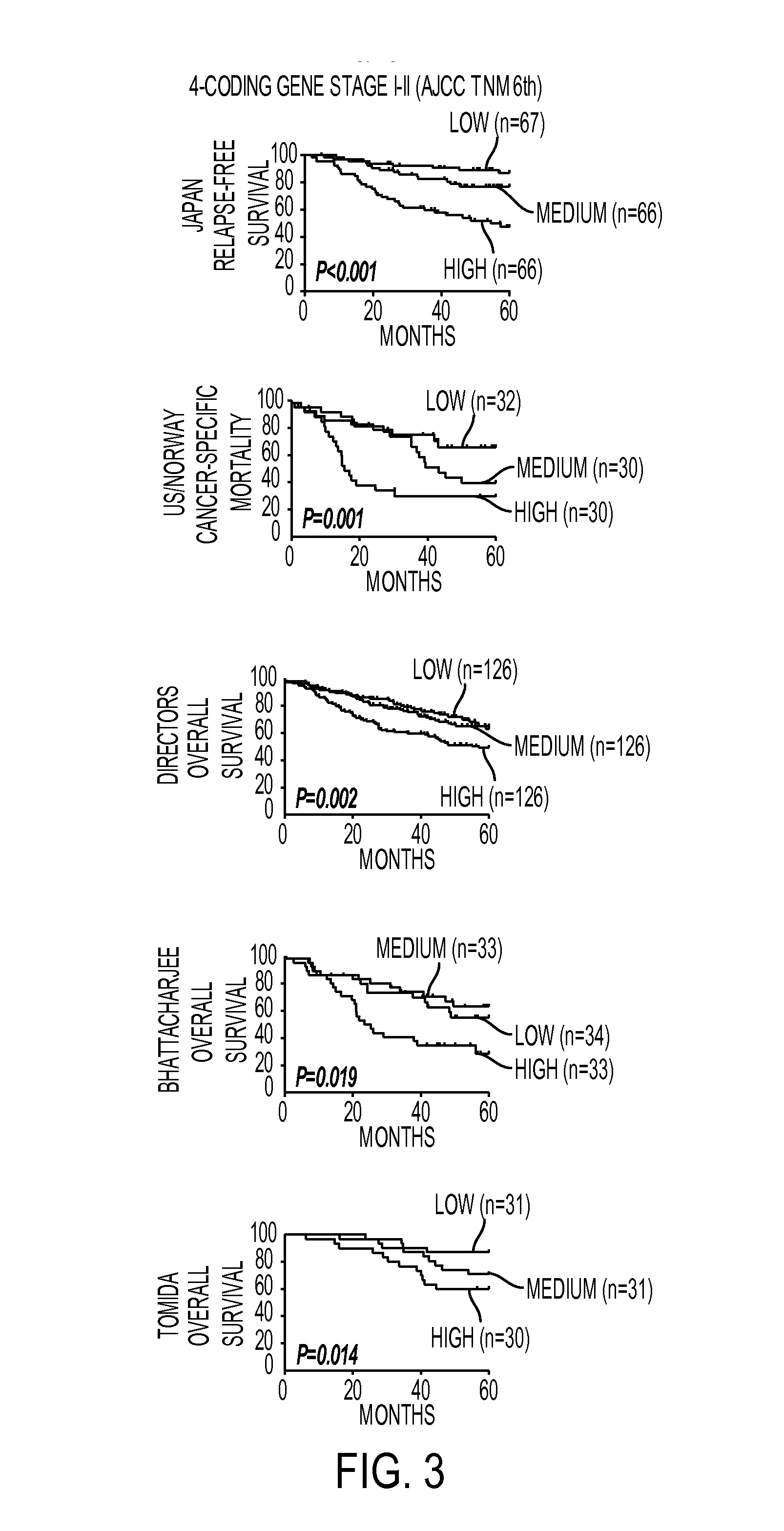
Expression protein-coding and noncoding genes as prognostic classifiers in early stage lung cancer
Owner:UNITED STATES OF AMERICA
Feature gene selection method based on deep learning and evolutionary computation
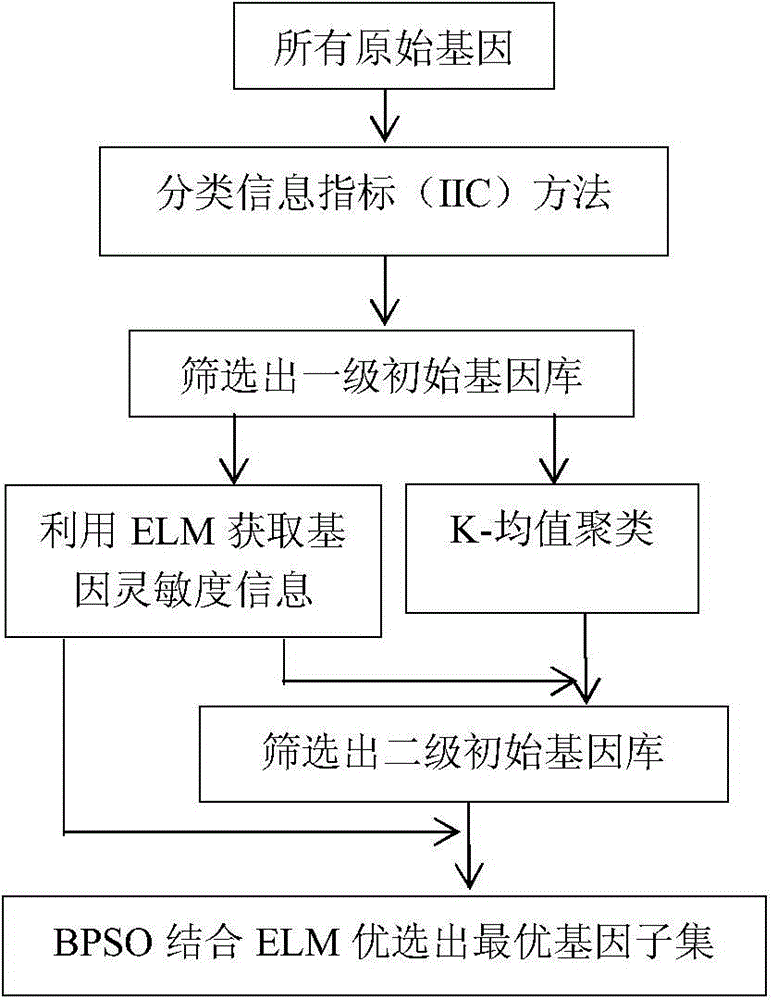
ActiveCN107992945AAccurate removalHigh taxonomic information abundanceCharacter and pattern recognitionNeural architecturesGene selectionEvolutionary computation
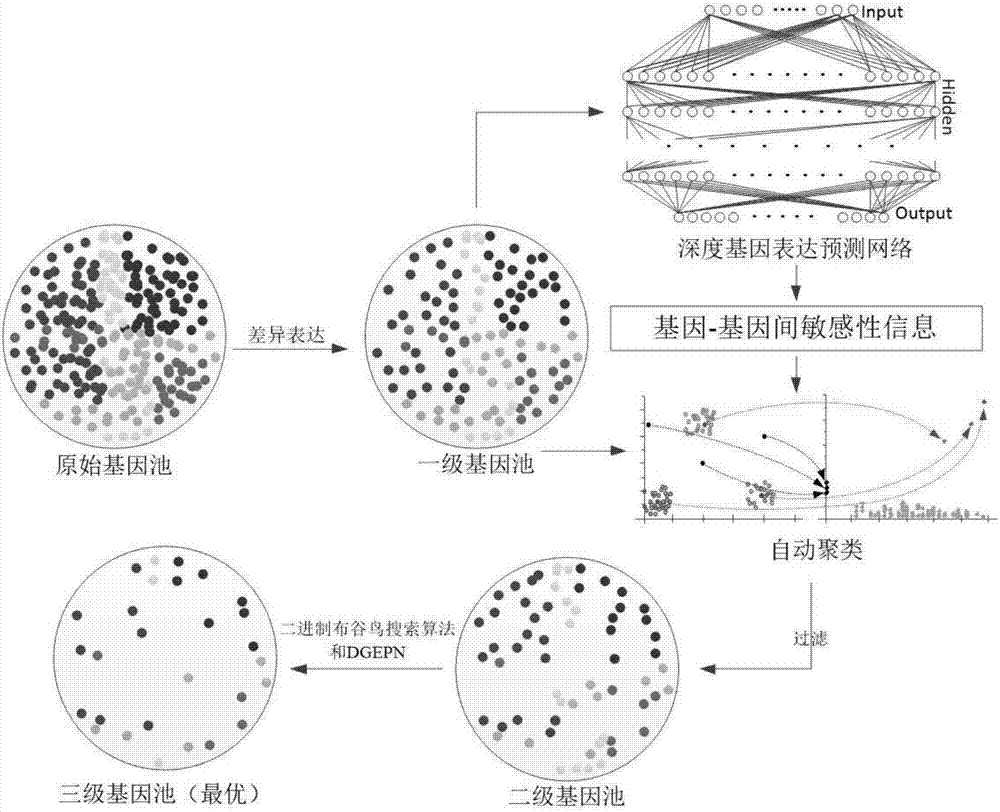
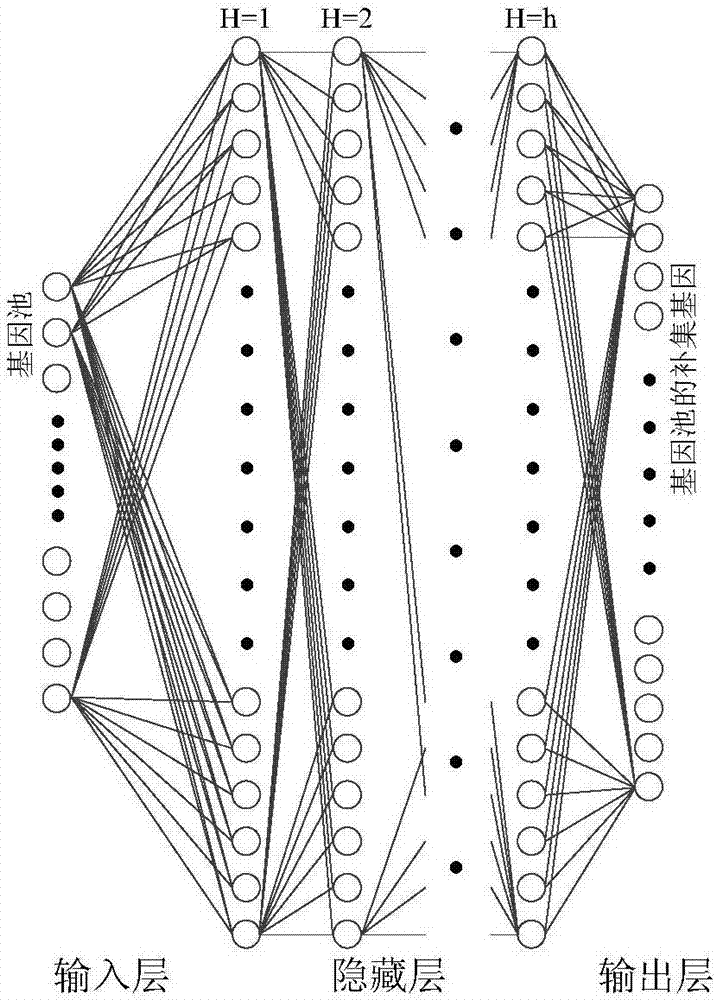
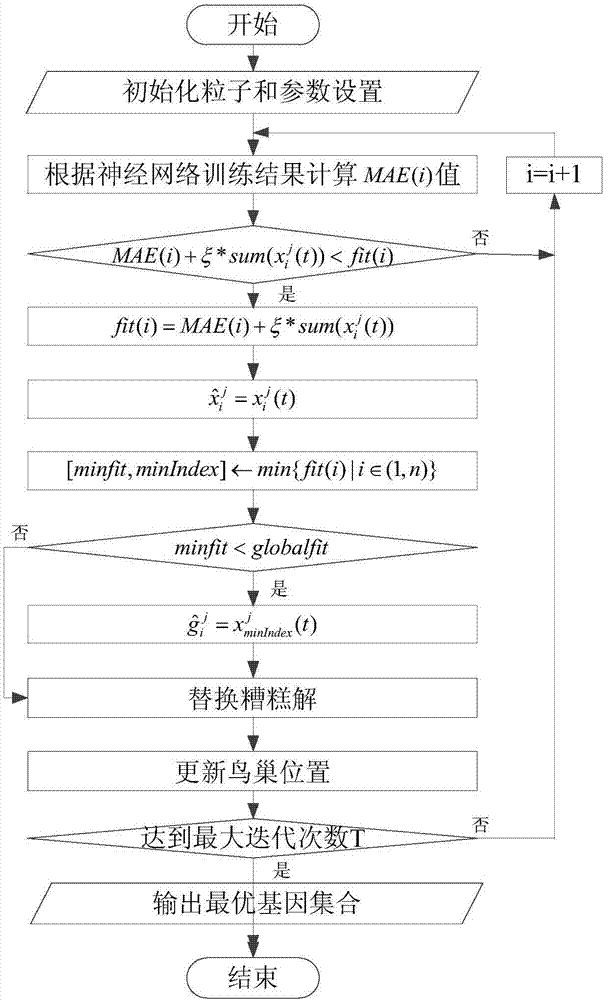
A feature gene search method based on deep learning and evolutionary computation includes the following steps: 1) calculating a differential expression level index and establishing a primary gene poolaccording to a Pareto principle; 2) according to the expression level of each gene, calculating a density matrix and a distance matrix after the mapping of the genes, drawing a decision map, using multiple linear regression analysis to fit a binary plane, and automatically determining a clustering center; 3) constructing a deep gene expression prediction network to calculate gene-genetic susceptibility information (GGSI) of a primary gene pool; 4) removing redundant genes based on GGSI values and creating a secondary gene pool; 5) performing binary coding on a cuckoo search algorithm based onthe GGSI values, selecting the most compact gene sets, and establishing a tertiary gene pool. According to the method, a feature gene selection framework is provided based on a hierarchical structure, which can extract key genes better and use multiple linear regression analysis combined with a deep learning algorithm and an optimization algorithm to select the most compact feature gene sets.
Owner:ZHEJIANG UNIV OF TECH
Orthogonal mirror image filter group realizing method and device based on genetic algorithm
InactiveCN101068108AReduce amplitude distortionReduce storageDigital technique networkGenetic modelsGene selectionGenetic algorithm
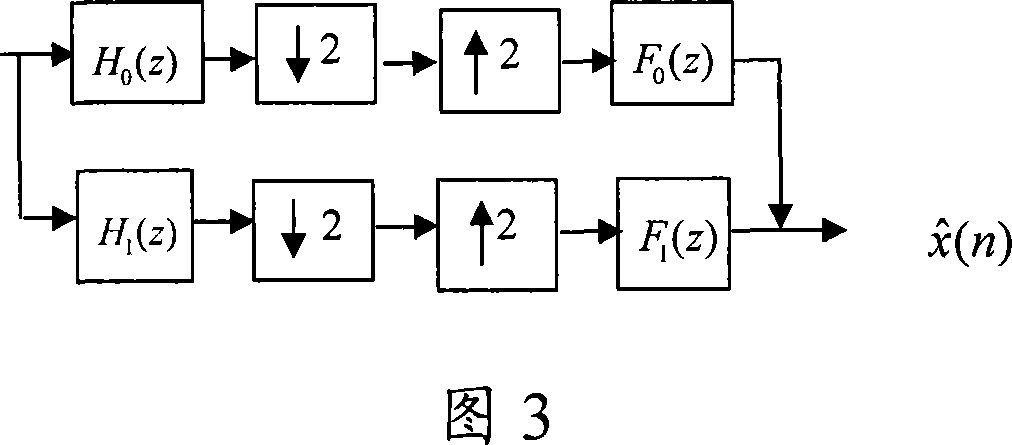
A method for realizing orthogonal mirror image filter set based on genetic algorithm includes generating multi-set of random number to represent parameter of orthogonal mirror image filter set, coding each set of random number to binary system character string in finite length according to current gene selection probability, decoding each character string and calculating out adaptive degree, updating said probability according to gene of character string with highest adaptive degree, repeating said step till ending condition is satisfied, obtaining parameter of said filter set by decoding character string with highest adaptive degree and utilizing obtained parameter to realize said filter set.
Owner:VIMICRO CORP
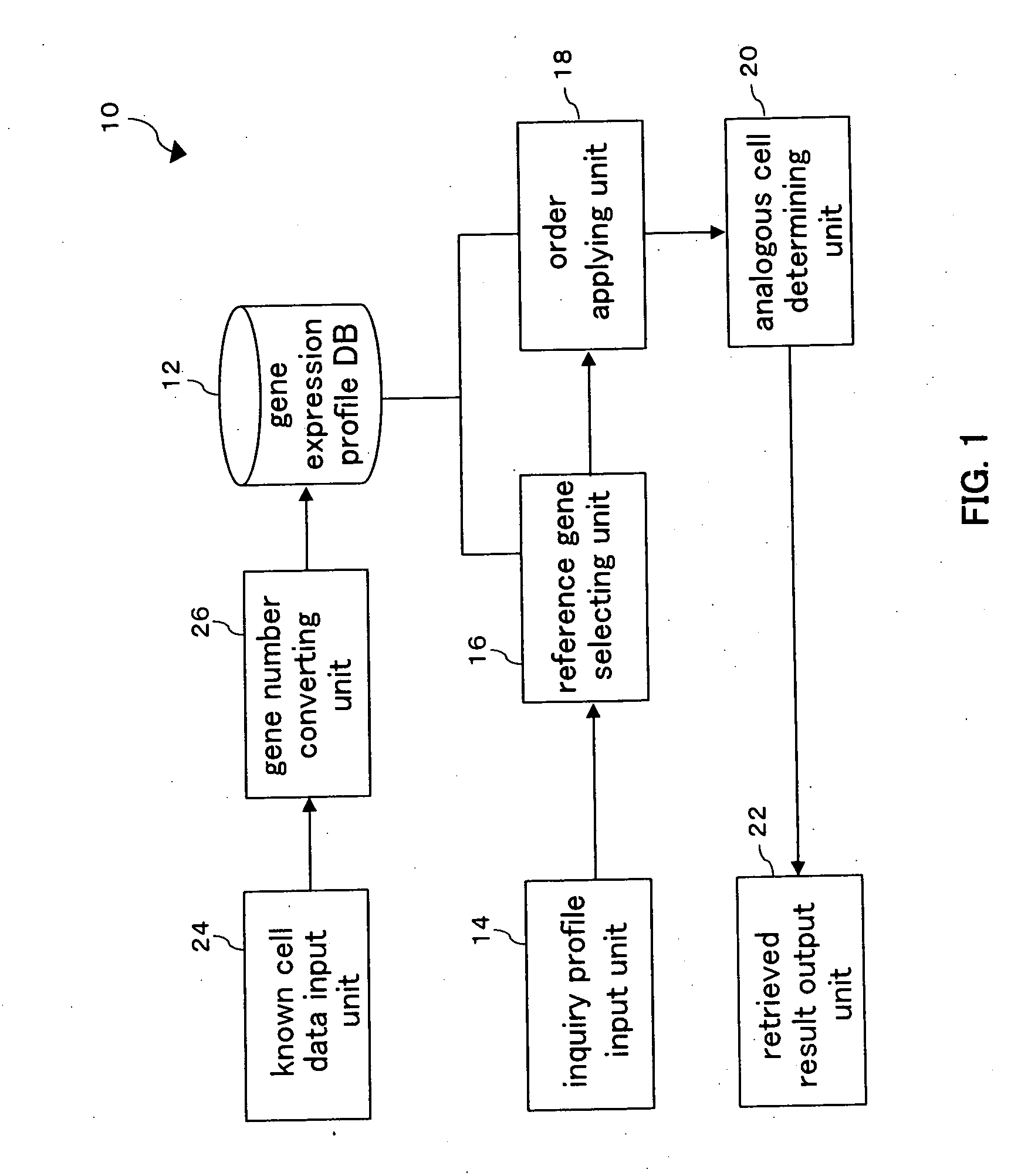
Gene expression profile retrieving apparatus, gene expression profile retrieving method, and program
InactiveUS20070172833A1Microbiological testing/measurementRecombinant DNA-technologyReference genesGene selection
In a gene expression profile retrieving apparatus, while gene expression profile data acquired with different platforms can be used, a cell can be retrieved based upon a gene expression profile. The gene expression profile retrieving apparatus is provided with: a gene expression profile DB stores gene expression profiles of known cells in a plurality of different platforms; a reference gene selecting unit operable to select a plurality of reference genes from genes which are commonly contained in both an inquiry profile and a platform of a gene expression profile stored in the gene expression profile DB; an order applying unit operable to apply orders to the inquiry profile and the reference gene of the known cell stored in the gene expression profile DB according to the expression level; and an analogous cell determining unit operable to acquire from the gene expression profile DB, such a cell which is analogous to the gene expression profile of the inquiry profile in the highest degree based upon the applied order.
Owner:NAT INST OF ADVANCED IND SCI & TECH
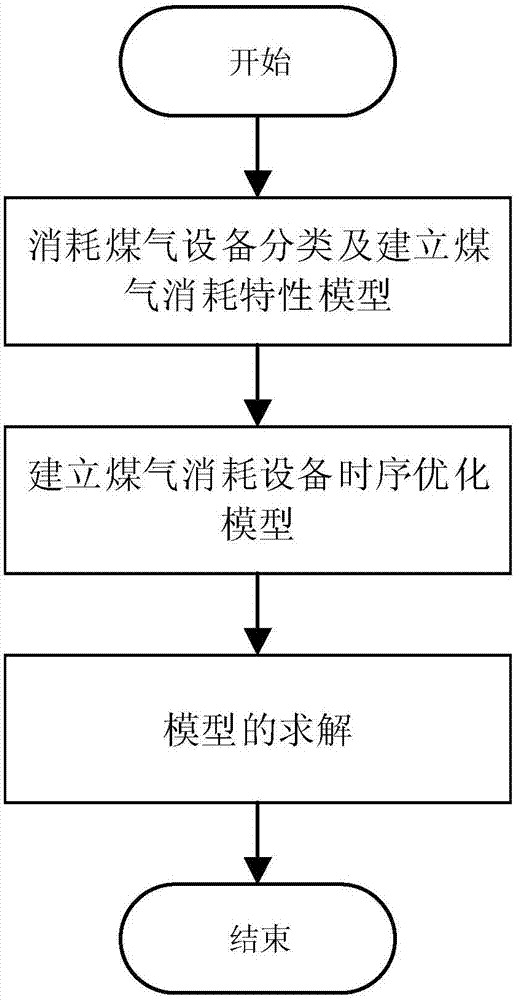
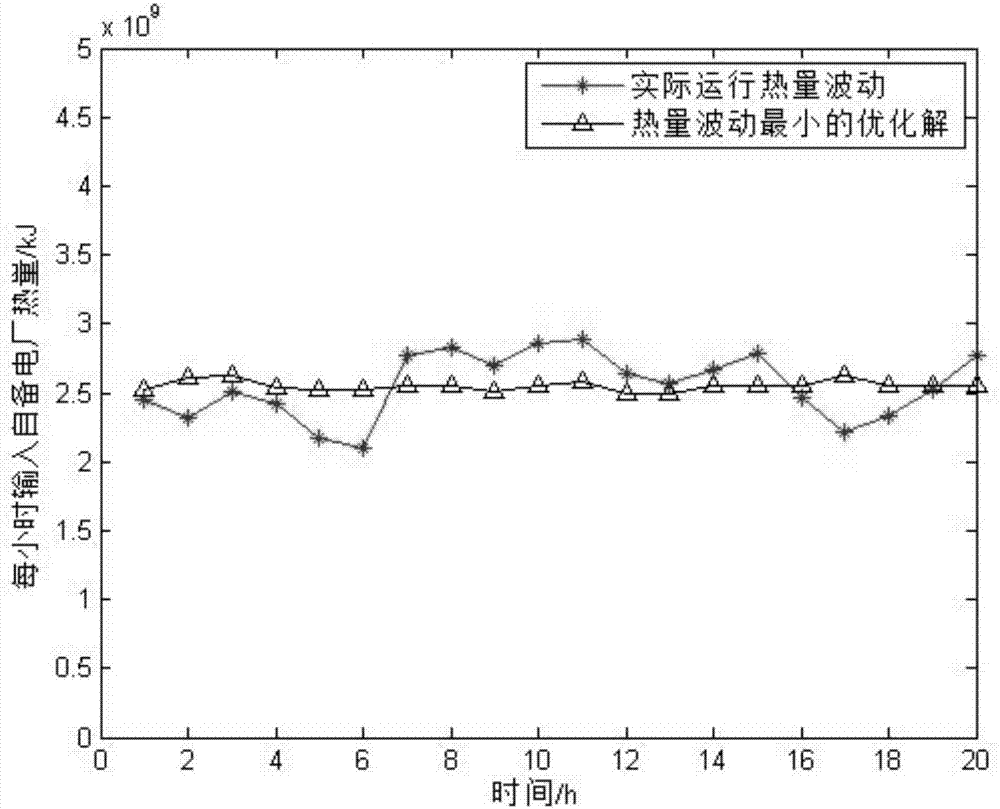
Timing sequence optimization method for gas consumption equipment of steel enterprises
ActiveCN107976976AReduce the difficulty of solvingReduce the numberData processing applicationsTotal factory controlMathematical modelGene selection
The invention discloses a timing sequence optimization method for gas consumption equipment of steel enterprises. Based on the establishment of a gas consumption characteristic model, a timing sequence optimization model for gas consumption equipment is established. The model, by taking the minimum of input heat fluctuation in a self-generation power plant and the minimum number of changes in equipment operating status as an optimization goal, and taking the gas heat, a thermal value, and a flow rate of the power plant as constraints, converts a math problem to 0-1 integer programming problem.The method solves the mathematical problem model by using a multi-objective genetic algorithm, designs the gene selection scheme, sets the position of 0 and 1 in the matrix as the gene of the algorithm, and achieves simple and effective optimization of the equipment timing sequence.
Owner:SOUTHEAST UNIV
Apparatus for forming molecular timetable and appratus for estimating circadian clock
An apparatus (11) for estimating internal biological time of a biological individual on the basis of quantity data of a gene product as measured in a specimen sampled from the individual. The apparatus includes circadian oscillatory gene selection means for selecting circadian oscillatory genes whose time-course change in gene product quantity data as measured in standard specimens approximates a cosine curve having a predetermined period; circadian expression curve selection means for selecting, from among a plurality of cosine curves having different phases and sharing a specific period, a circadian expression curve which is similar to the pattern of time-course change in the expression product quantity of each of the above-selected circadian oscillatory genes; and registration means for registering information which identifies the selected circadian expression curve.
Owner:YAMANOUCHI PHARMA CO LTD
Method for the construction of specific promoters
ActiveUS20130324440A1Increase transcriptionGood choiceSugar derivativesMicroorganismsMethod selectionTissue specific
The present application relates to a system for designing promoters for selective expression of genes. Thereby identified transcription regulatory elements are selected according to a specific methodology and used to create a library of transcription regulatory elements, which are then used to construct specific promoters, especially tissue-specific promoters.
Owner:ASKLEPIOS BIOPHARMACEUTICAL INC
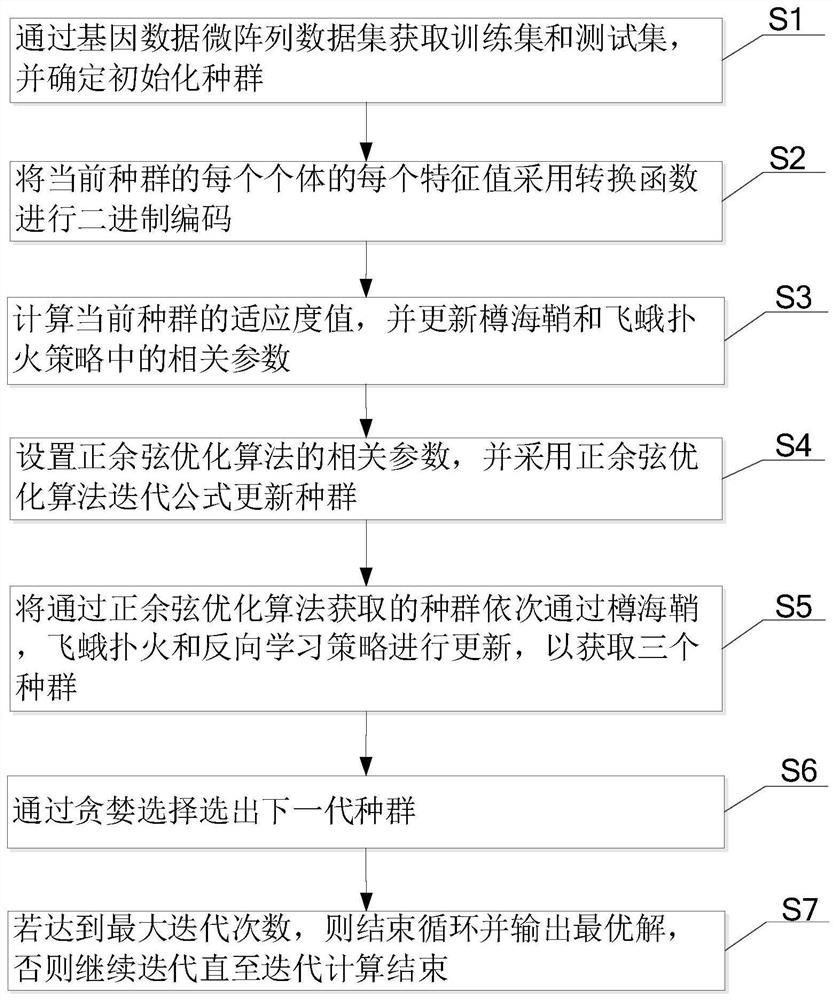
Gene selection method and device
PendingCN112215259ALighten the computational burdenReduce noiseBiostatisticsCharacter and pattern recognitionGene selectionGene Feature
The invention provides a gene selection method which is used for gene characteristic selection. The method comprises the steps of obtaining a training set and a test set through a gene data microarraydata set, and determining an initialized population; carrying out binary coding on each individual of the current population by adopting a conversion function; calculating the fitness value of the current population, and updating related parameters in the dolio-sea squirt and moth fire suppression strategy; setting related parameters of a sine and cosine optimization algorithm, and updating the population by adopting a sine and cosine optimization algorithm iterative formula; updating the populations obtained through the sine and cosine optimization algorithm through a doliola scabra, moth fire suppression and reverse learning strategy in sequence so as to obtain three populations; selecting a next generation of population through greedy selection; and if the maximum number of iterationsis reached, ending the loop and outputting the optimal solution, otherwise, continuing the iteration until the iterative computation is ended. According to the invention, the gene characteristics which contribute most to categories can be screened out from the genes more accurately and more efficiently, and the detection cost is reduced.
Owner:WENZHOU UNIVERSITY
Molecular marker for raphanus sativus L. downy mildew resistant gene close linkage
InactiveCN102978207ARapid identificationMolecular markers are stable and accurateMicrobiological testing/measurementPlant genotype modificationHigh resistanceGene selection
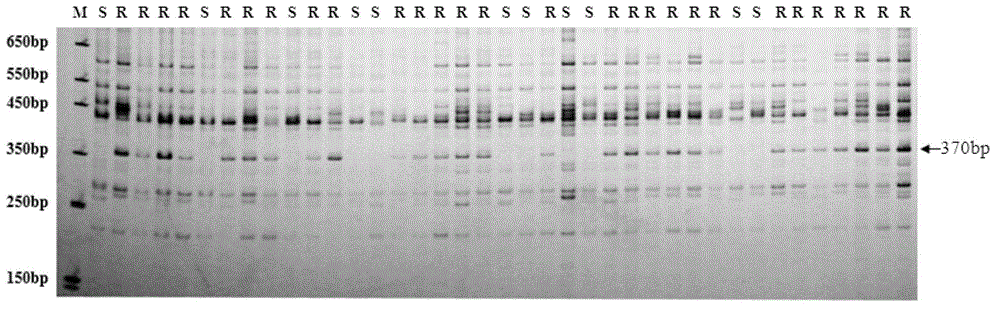
The invention belongs to the field of crop genetic breeding, and discloses a molecular marker for raphanus sativus L. downy mildew resistant gene close linkage. A sequence-related amplified polymorphism (SRAP) molecular marker method is used for carrying out marker genotyping on high-resistance downy mildew raphanus sativus L. parents (NAU-dhp08), high-sensitive downy mildew raphanus sativus L. parents (half green from autumn fields) and a second filial generation (F2) group single plant which is prepared through resistance and sensitive parent hybridization, through the F2 single plant resistance phenotype and marker gene linkage analysis, a resistance marker Em9 / ga24370 of 370-base pair (bp) close linkage is identified, a distance from a resistance locus and a marker genetic distance is 2.3 centimeters. The molecular marker method provided by the invention can be used for the marker assisted selection of raphanus sativus L. downy mildew resistance breeding, quickly and accurately identifies the raphanus sativus L. downy mildew resistance, overcomes the limitations of heavy workload, long period, easily influenced results by environment and the like of the conventional resistance identification method, the high-resistance gene selection efficiency can be obviously improved, so that a breeding process of a new high raphanus sativus L. downy mildew-resistant variety can be accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
Feature gene selecting and cancer classifying method
The invention discloses a feature gene selecting and cancer classifying method which at least comprises the following steps of establishing a logistic regression model according to a super-parameter set and a to-be-processed gene data set; according to maximum likelihood estimation and logarithmic operation, expressing the logistic regression model as a loss function; establishing an SCAD-Net resolving model; according to the loss function and the SCAD-Net resolving model, obtaining an SNL model; calculating an iteration updating operator of the SCAD-Net; according to the iteration updating operator, calculating a gene regression coefficient of the SNL model through a coordinate gradient descent method; and according to the gene regression coefficient, performing feature gene selection andcancer classification. The feature gene selecting and cancer classifying method can effectively improve feature gene selecting and cancer classifying accuracy, thereby facilitating disease researching.
Owner:SHAOGUAN COLLEGE
PCA3 and PSA RNA detection kit and amplification system
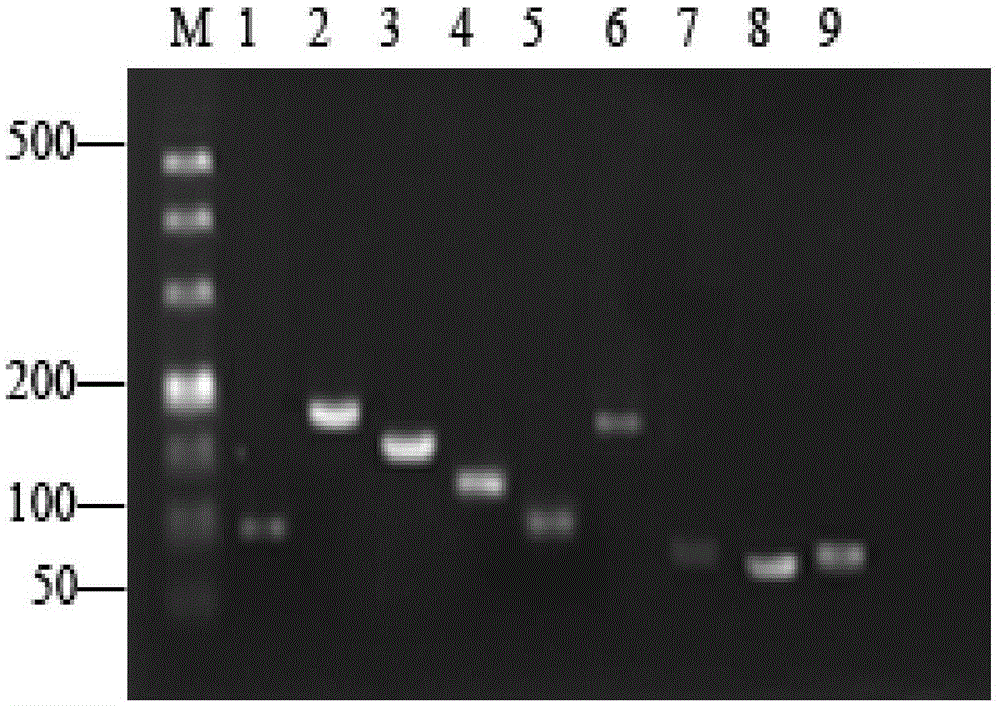
PendingCN110951871AHigh sensitivityImprove featuresMicrobiological testing/measurementFluoProbesGene selection
The invention relates to a PCA3 and PSA RNA detection kit and amplification system. The detection kit includes primers and fluorescent probes aiming at PCA3 and PSA genes, and a Stoffel fragment and Tfl DNA polymerase; the primers aiming at the PCA3 gene are shown as SEQ ID NO. 1 and SEQ ID NO. 2, and the sequence of the fluorescent probe is shown as SEQ ID NO. 3; and the primers aiming at the PSAgene are shown as SEQ ID NO. 10 and SEQ ID NO. 11, and the sequence of the fluorescent probe is shown as SEQ ID NO. 12. Based on existing kits, the detection and amplification system of real-time fluorescence quantitative RT-PCR can be optimized; the PCA3 gene selects the primers and probes which have better detection effects and are designed by the sequence on exon 3; optimized Tris-MOPS-sodiumcitrate buffer which is more suitable for RNA amplification can be introduced into a reaction system, and the Stoffel fragment or the Tfl DNA polymerase is introduced as well; and therefore, the kit can tolerate more templates while detection sensitivity and specificity are enhanced.
Owner:RAYBIOTECH INC GUANGZHOU
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com