Feature gene selection method based on deep learning and evolutionary computation
An eigengene, deep learning technology, applied in the field of bioinformatics, can solve problems such as lack of interpretability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0069] The present invention will be further described below in conjunction with the accompanying drawings.
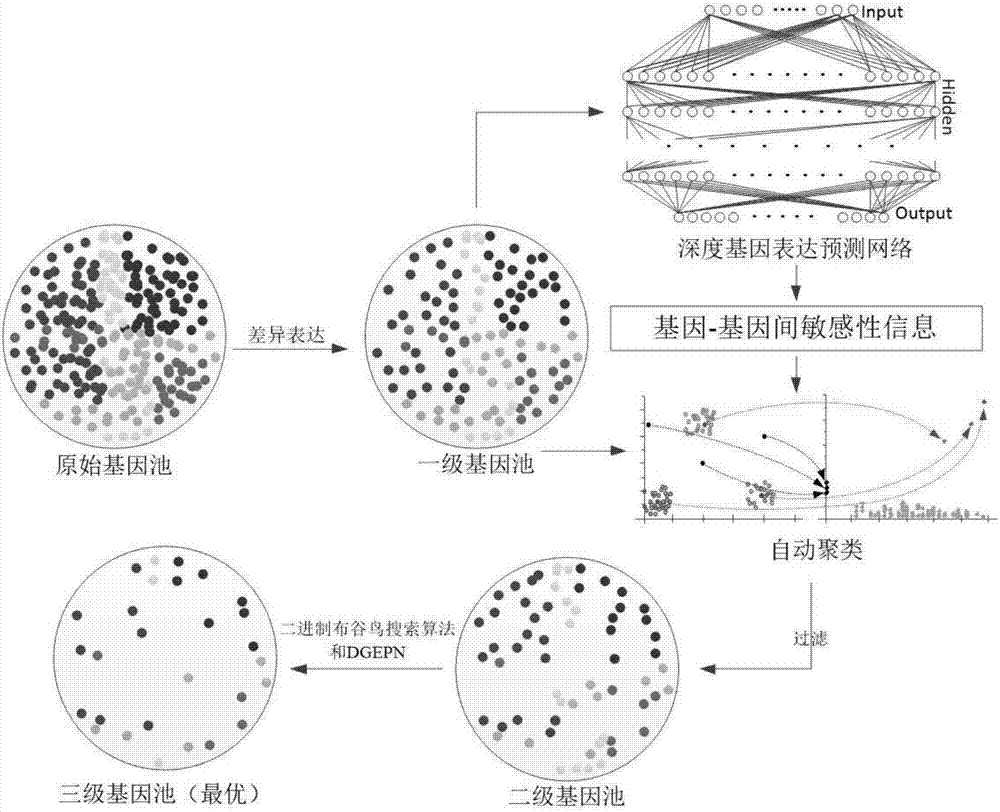
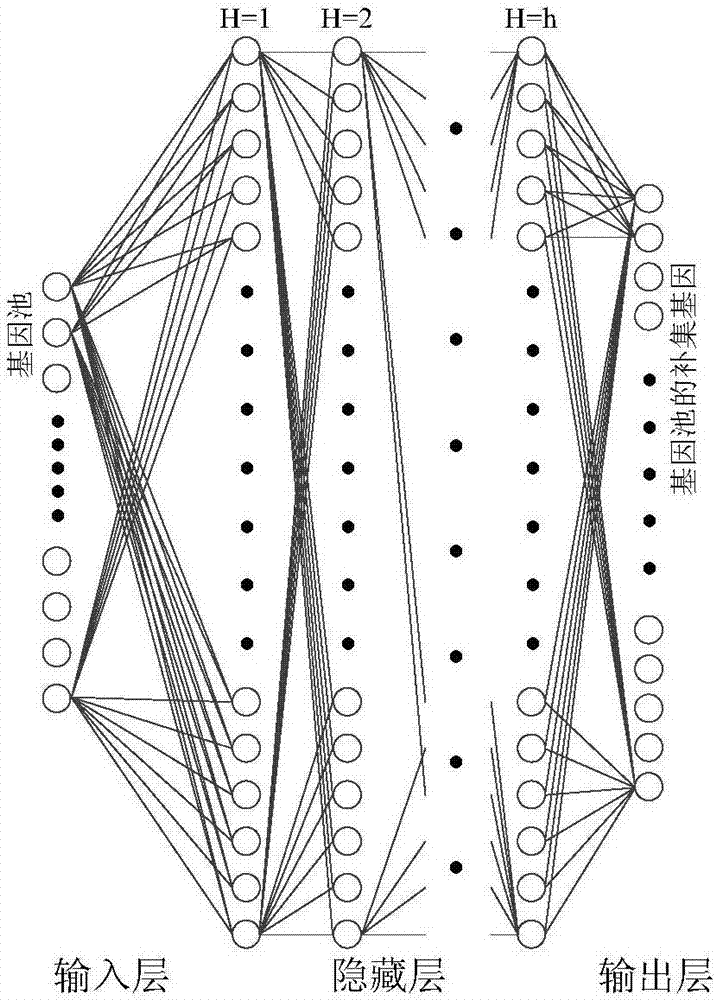
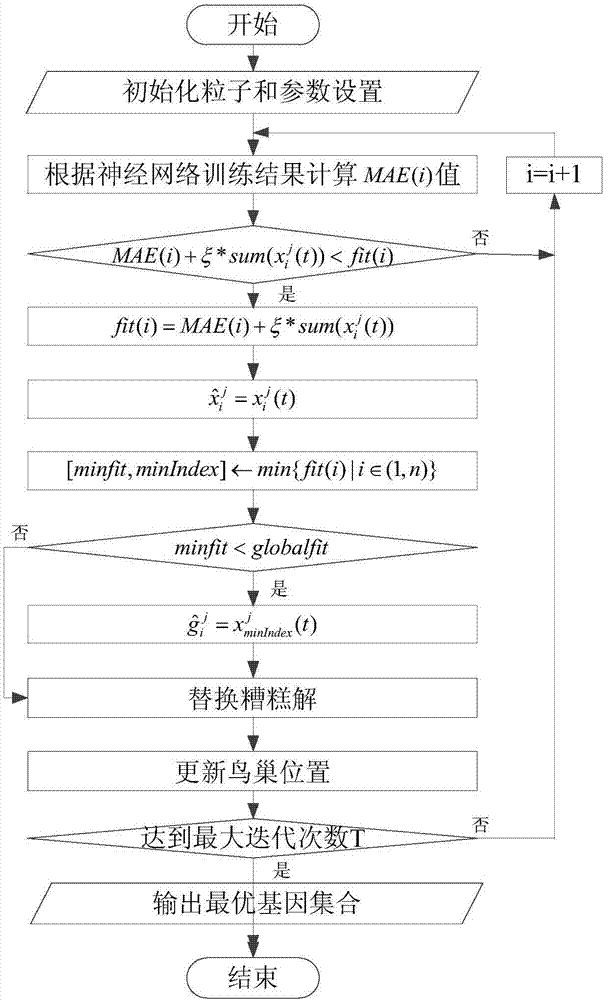
[0070] refer to Figure 1 ~ Figure 3 , a feature gene selection method based on deep learning and evolutionary computation, comprising the following steps:
[0071] 1) Select differentially expressed genes and establish a first-level gene pool. The process is as follows:
[0072] 1.1) Calculate the differential expression level index of each gene in the original gene pool, that is, the IIC-FC index:
[0073]
[0074] Equation (1) is suitable for the calculation of gene differential expression level in multi-classification data sets, where c represents the number of genes in the original gene pool, and represent the average expression levels of gene i and gene j, respectively, and represent the standard deviation of the expression levels of gene i and gene j respectively, the functions max{·,·} and min{·,·} represent the maximum value and minimum value respe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com