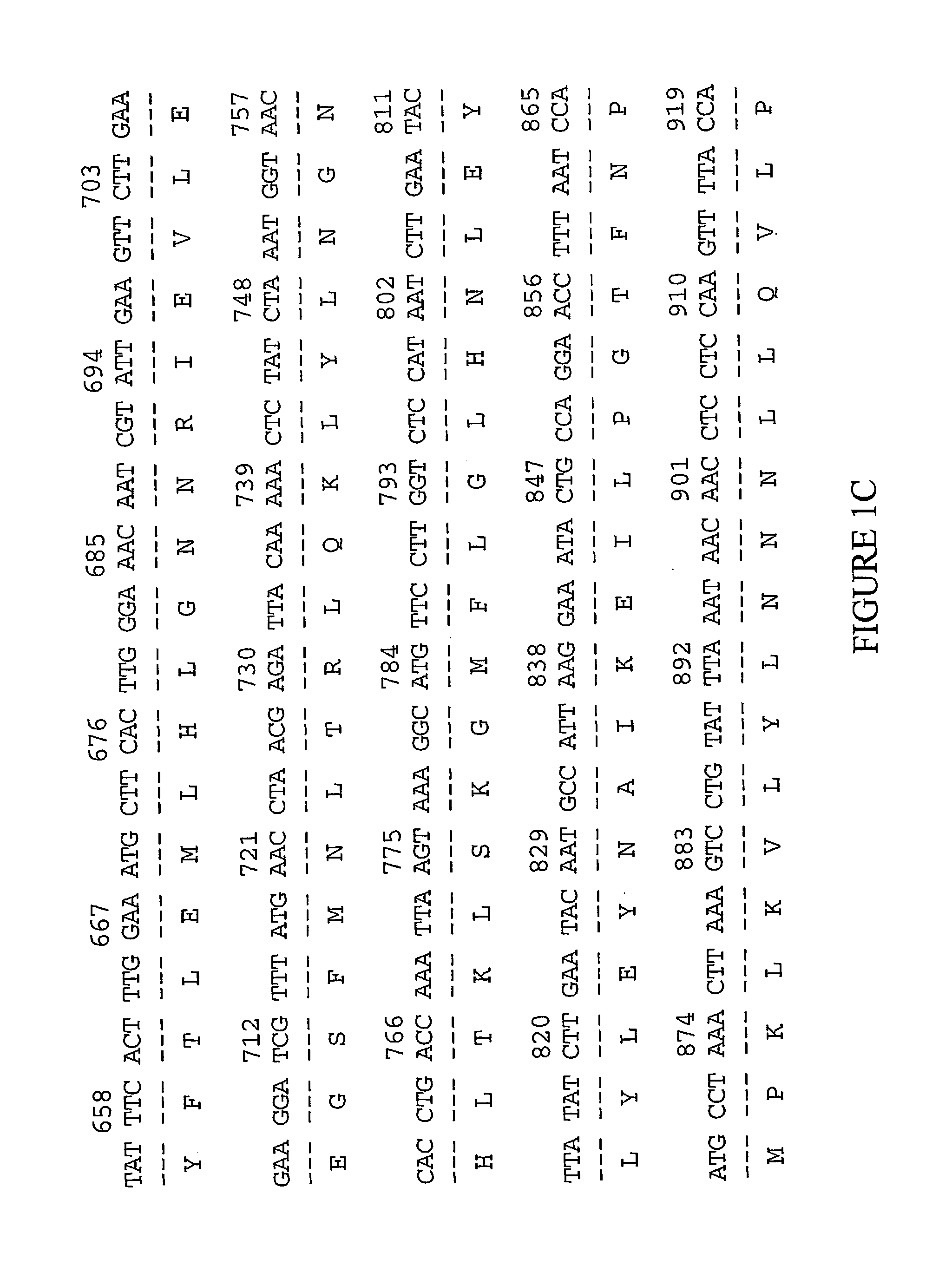
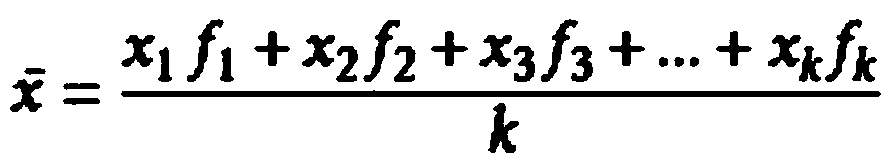Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
212 results about "Gene sets" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Methods for using co-regulated genesets to enhance detection and classification of gene expression patterns
InactiveUS6203987B1Peptide/protein ingredientsMicrobiological testing/measurementCo-regulationIndividual gene
The present invention provides methods for enhanced detection of biological response patterns. In one embodiment of the invention, genes are grouped into basis genesets according to the co-regulation of their expression. Expression of individual genes within a geneset is indicated with a single gene expression value for the geneset by a projection process. The expression values of genesets, rather than the expression of individual genes are then used as the basis for comparison and detection of biological responses with greatly enhanced sensitivity.
Owner:MICROSOFT TECH LICENSING LLC
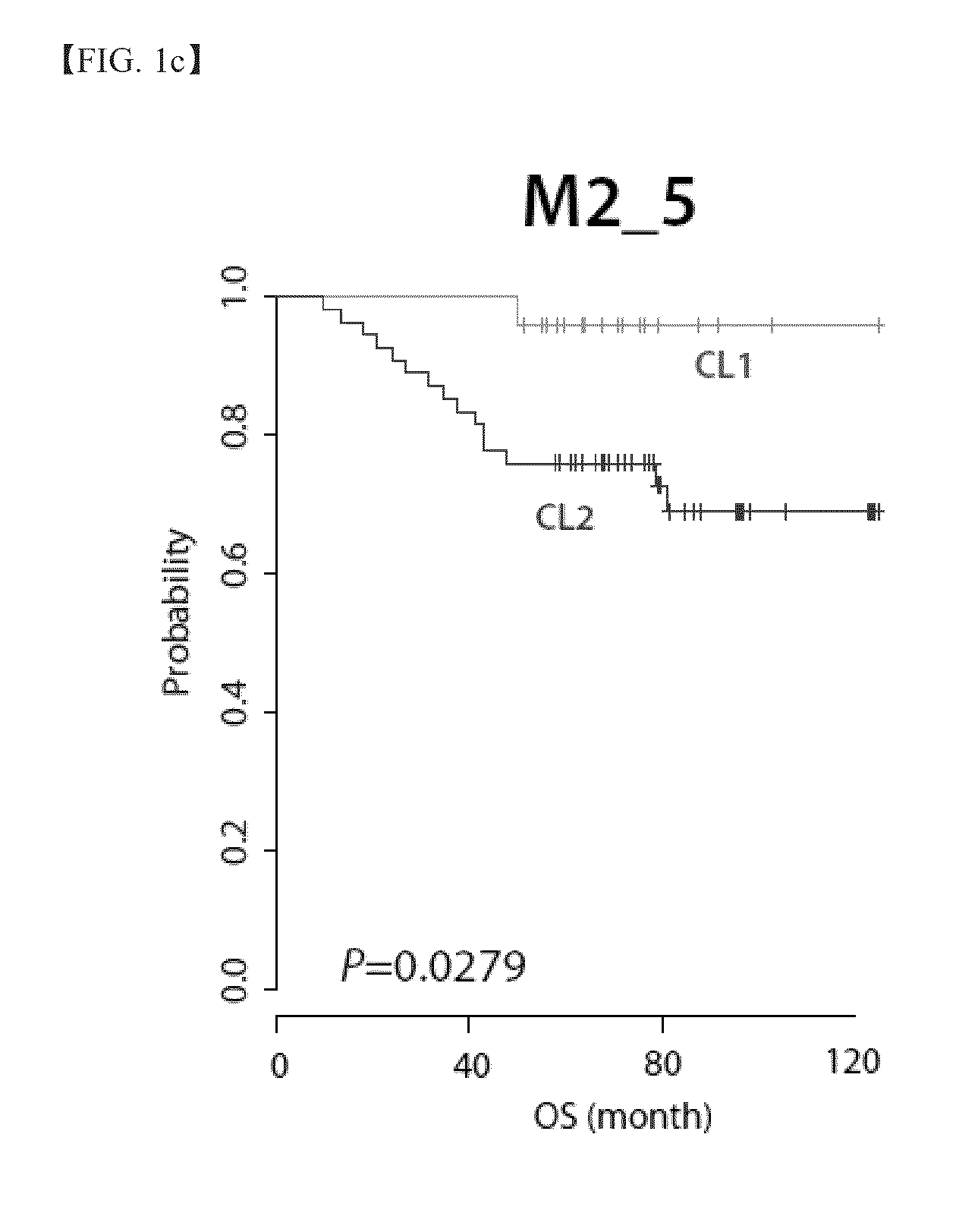
Prediction of likelihood of cancer recurrence
ActiveUS7056674B2Raise the possibilitySugar derivativesMicrobiological testing/measurementOncologyGenome
The present invention provides gene sets the expression of which is important in the diagnosis and / or prognosis of cancer, in particular of breast cancer.
Owner:GENOMIC HEALTH INC
Gene expression markers for breast cancer prognosis
ActiveUS20040209290A1Raise the possibilityMicrobiological testing/measurementResilient suspensionsFhit geneBiology
The present invention provides gene sets the expression of which is important in the diagnosis and / or prognosis of breast cancer.
Owner:GENOMIC HEALTH INC
Predictors Of Patient Response To Treatment With EGFR Inhibitors
InactiveUS20070128636A1Reduce the possibilityReduced responsivenessMicrobiological testing/measurementBiological material analysisGenomeCancer research
The invention concerns genes and gene sets and methods useful in the prediction of the response of a cancer patient to treatment with an epidermal growth factor receptor (EGFR) inhibitor.
Owner:GENOMIC HEALTH INC
Gene expression markers for breast cancer prognosis
The present invention provides gene sets the expression of which is important in the diagnosis and / or prognosis of breast cancer.
Owner:GENOMIC HEALTH INC
Metastasis-associated gene profiling for identification of tumor tissue, subtyping, and prediction of prognosis of patients
InactiveUS20060211036A1Bioreactor/fermenter combinationsBiological substance pretreatmentsAbnormal tissue growthReal-Time PCRs
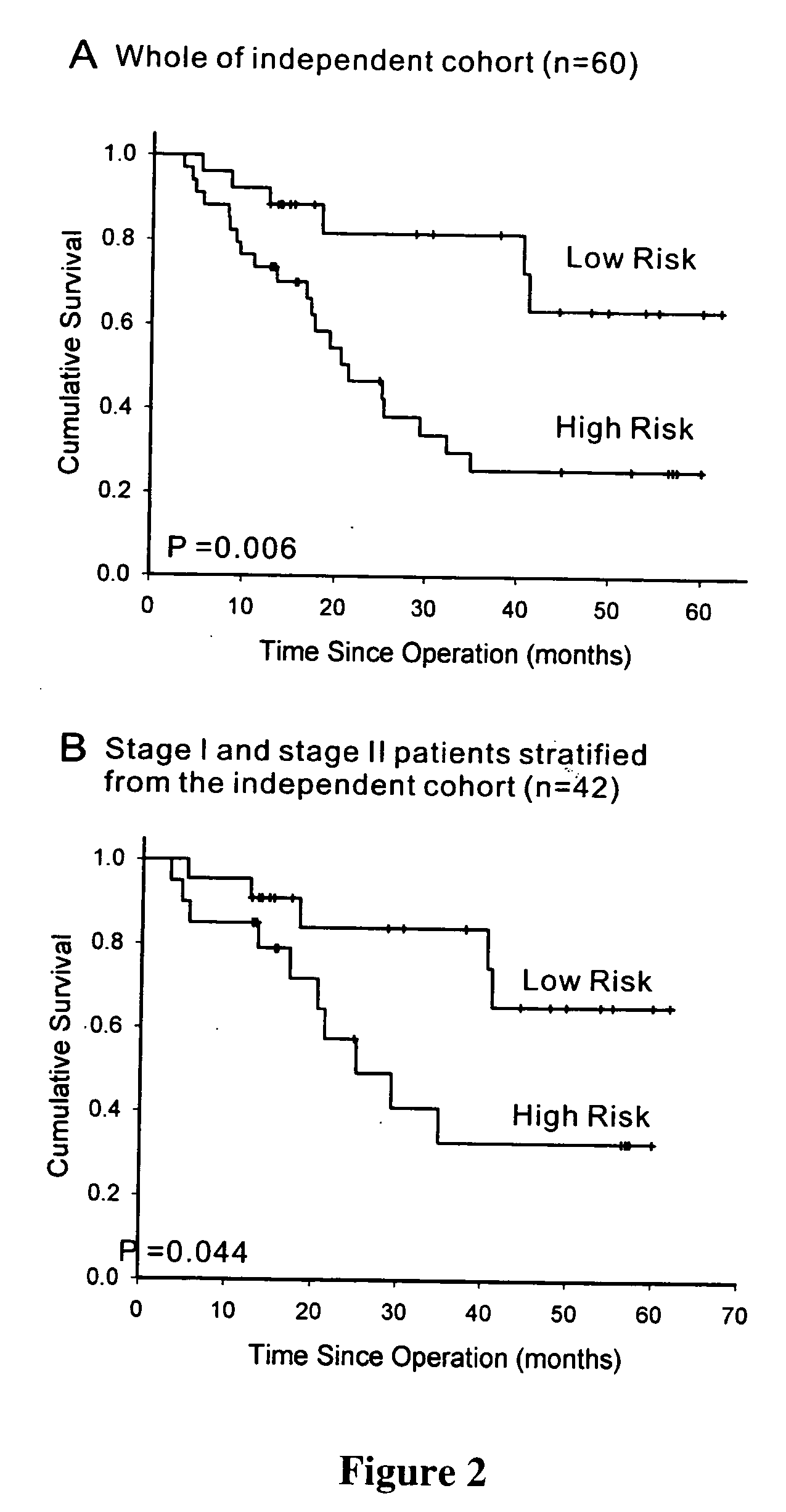
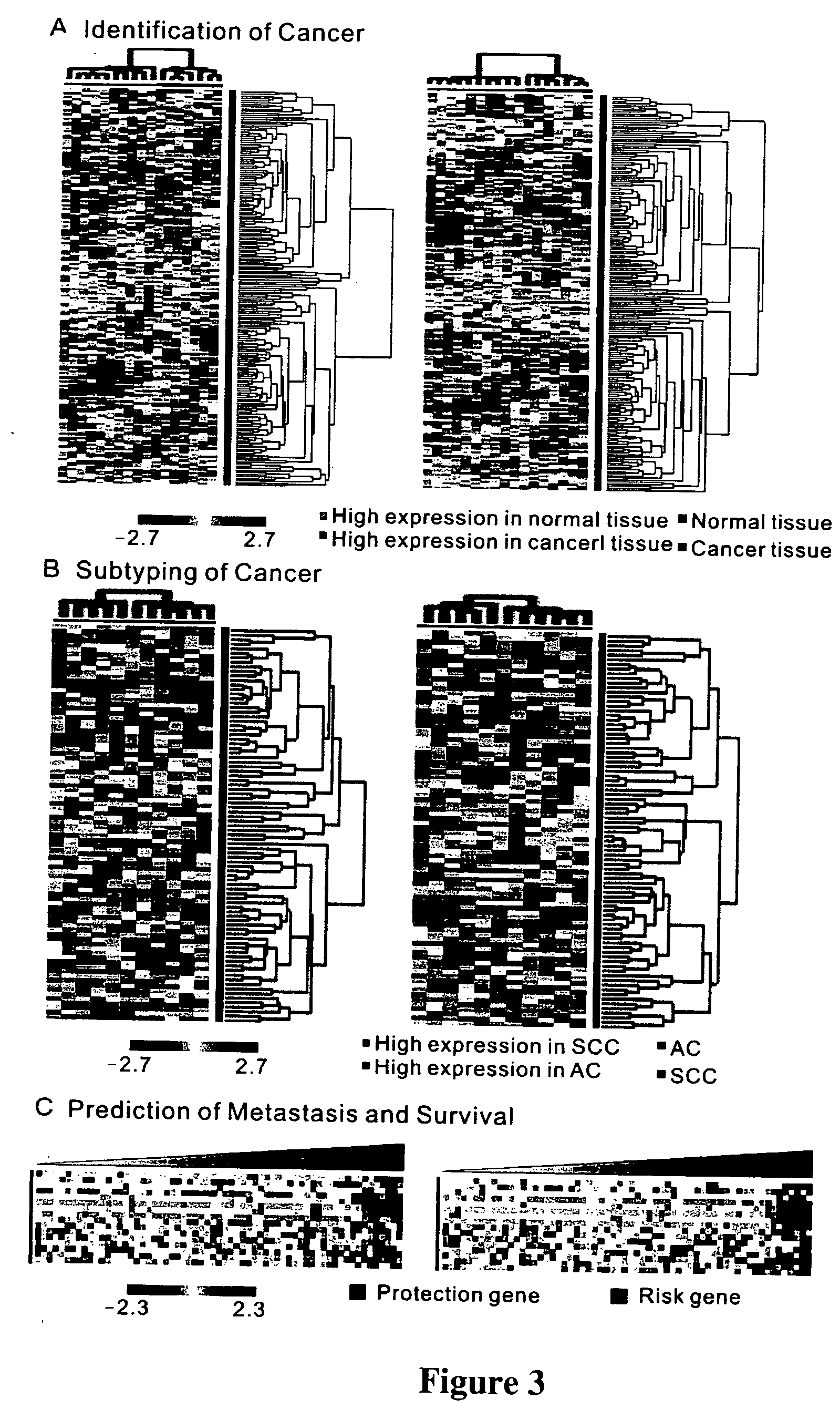
The present invention provides methods using a gene expression profiling analysis (1) to determine whether a human sample is a tumor using a gene set containing nucleic acid sequences of SEQ ID NOS: 1-7, 8-17 or 1-17; (2) to identify whether a tumor tissue is an adenocarcinoma (using a gene set containing nucleic acid sequences of SEQ ID NOS: 15, and 18-21) or a squamous cell carcinoma (using a gene set containing nucleic acid sequences of SEQ ID NOS: 22-27); and (3) to predict the prognosis of survival and metastasis in humans with tumor (using a gene set containing nucleic acid sequences of SEQ ID NOS:19, and 28-42 or SEQ ID NOS: 19, 29, 31, 40, and 42), particularly for those humans who are at the early stage of lung cancer. The gene expression profiling is preferably performed by cDNA microarray-based techniques and / or Real-Time Reverse Transcription-Polymerase Chain Reaction (Real-Time RT-PCR), and analyzed by statistical means.
Owner:ADVPHARMA
Biomarkers for cancer treatment
ActiveUS20080131885A1Reduce healingAnalysis using chemical indicatorsSugar derivativesTumor responseWilms' tumor
The present invention provides identification of a thirty-five gene set that predicts the anticancer activity of inhibitors of the RAF / MEK / MAPK pathway, methods of qualifying cancer status in a subject, methods of identifying an anti-tumor response in a subject, methods of monitoring the efficacy of a therapeutic drug in a subject, and methods of identifying an agent useful in the treatment of a cancer based on expression of the thirty-five gene set.
Owner:MEMORIAL SLOAN KETTERING CANCER CENT
Preparing method for tr-gene products detecting oligonucleotides chip and use thereof
InactiveCN1584049AGuaranteed specificityTo achieve the purpose of mutual verificationMicrobiological testing/measurementHeterologousOligonucleotide chip
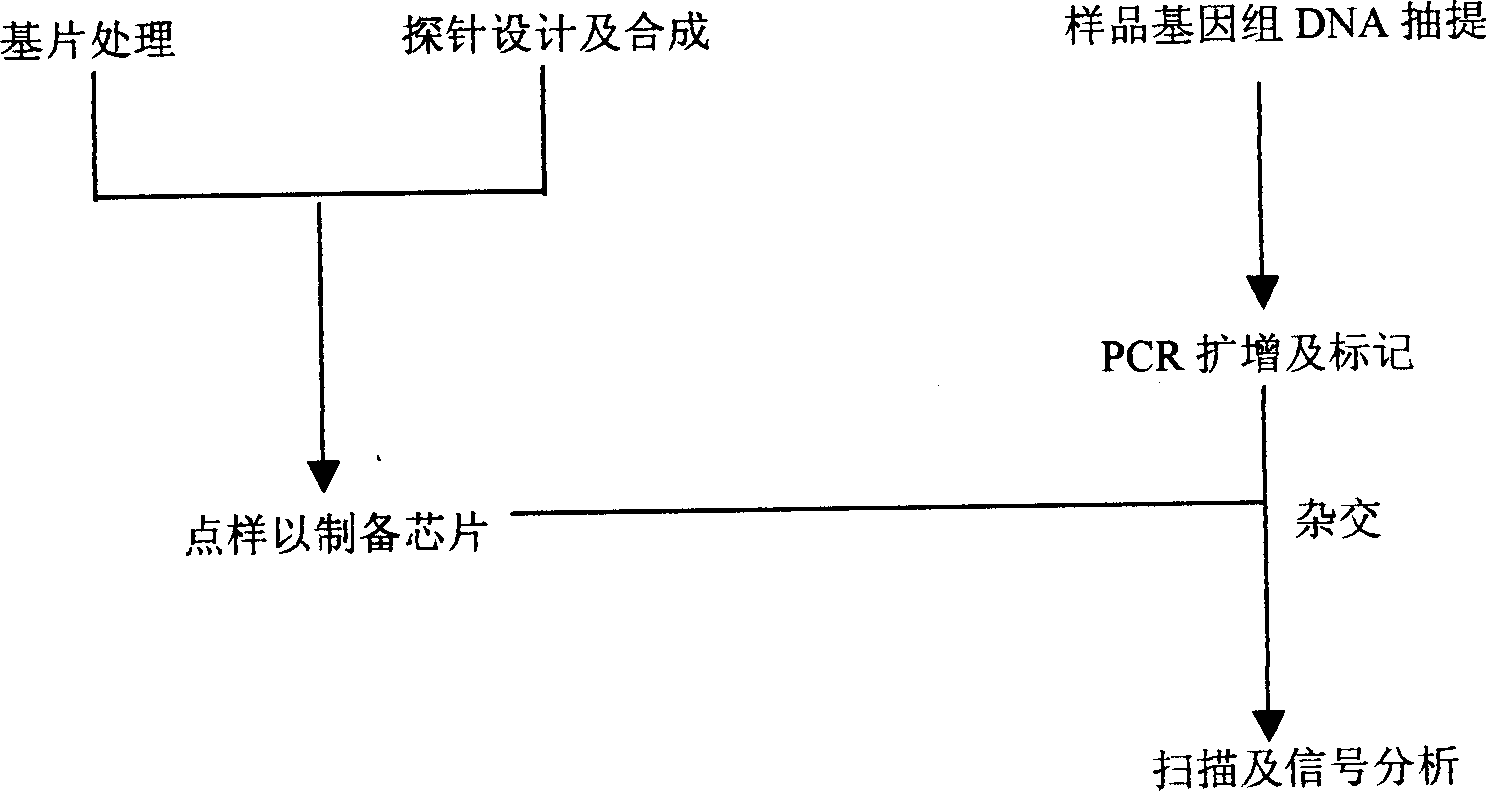
A method for preparing oligonucleotide chip for transgene product detection and use are disclosed. It includes: designing specific oligonucleotide probe and related primer by heterogeneric inserting gene, species internal standard gene, specific boundary sequence in transgene product gene set, fixing probe on glass slide to form transgene product detecting chip, amplifying DNA of plant to be tested by related primmer, marking by fluorescence, and hybridizing with chip. It can be use to acquire variable information.
Owner:国家质量监督检验检疫总局动植物检疫实验所 +2
Gene set for liver cancer detection and detection and design method of panel of gene set
ActiveCN108753967AWide variety of sourcesEasy to detectMicrobiological testing/measurementGene targetsTyping
The invention discloses a preparation method of a liver cancer detection panel. The preparation method comprises the following steps: screening liver cancer information and related genes thereof, thusobtaining a gene set; selecting a gene target zone; designing a panel probe; synthesizing an RNA (Ribonucleic Acid) single-stranded probe, thus obtaining the gene detection panel. The gene detectionpanel designed by the invention can be used for diagnosis, typing and prognosis of liver cancer, has wide gene coverage degree and strong transferability and is suitable for newly-developed detectionplatforms.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV +1
Prediction of likelihood of cancer recurrence
ActiveUS20050079518A1Raise the possibilitySugar derivativesMicrobiological testing/measurementOncologyGenome
The present invention provides gene sets the expression of which is important in the diagnosis and / or prognosis of cancer, in particular of breast cancer.
Owner:GENOMIC HEALTH INC
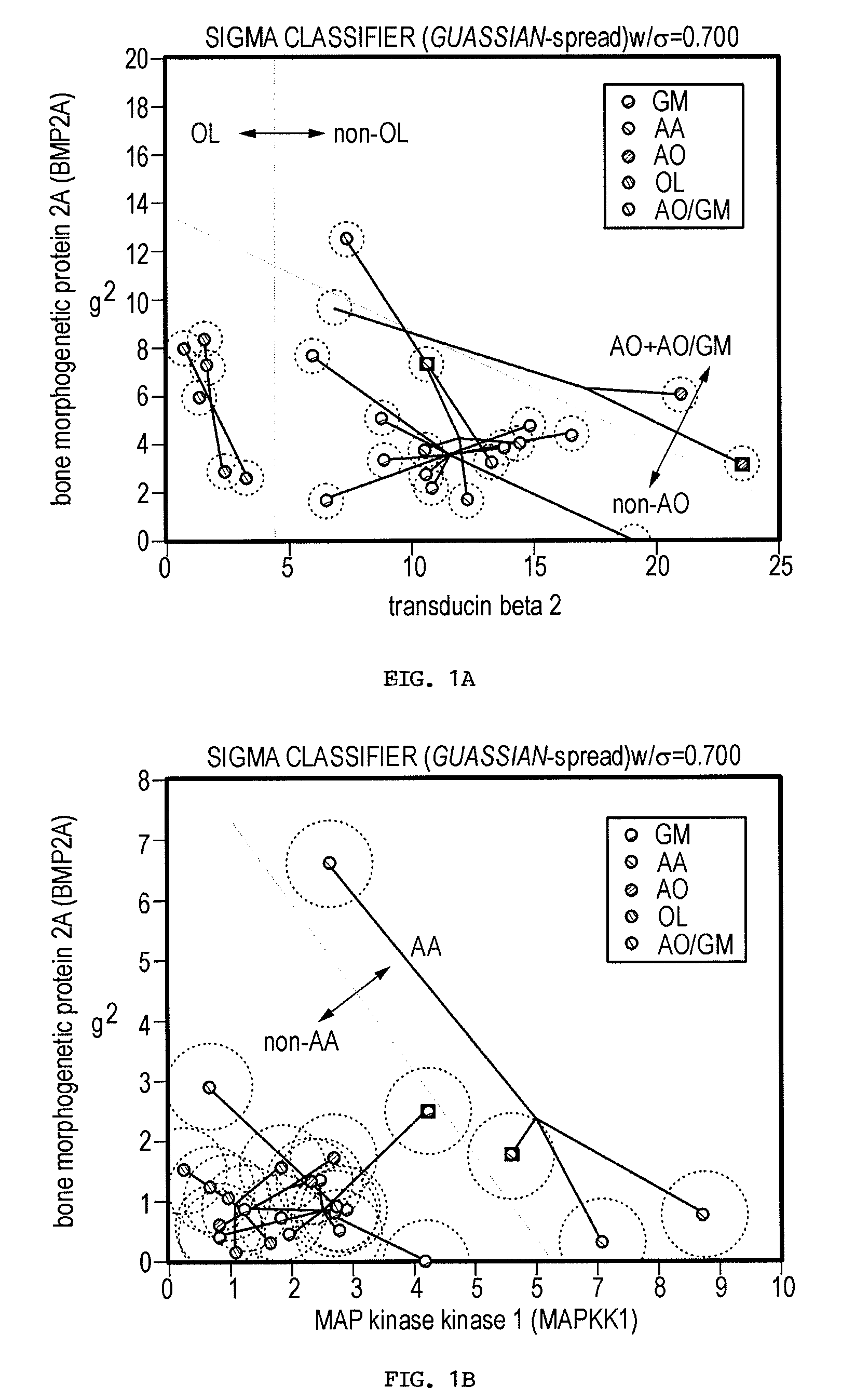
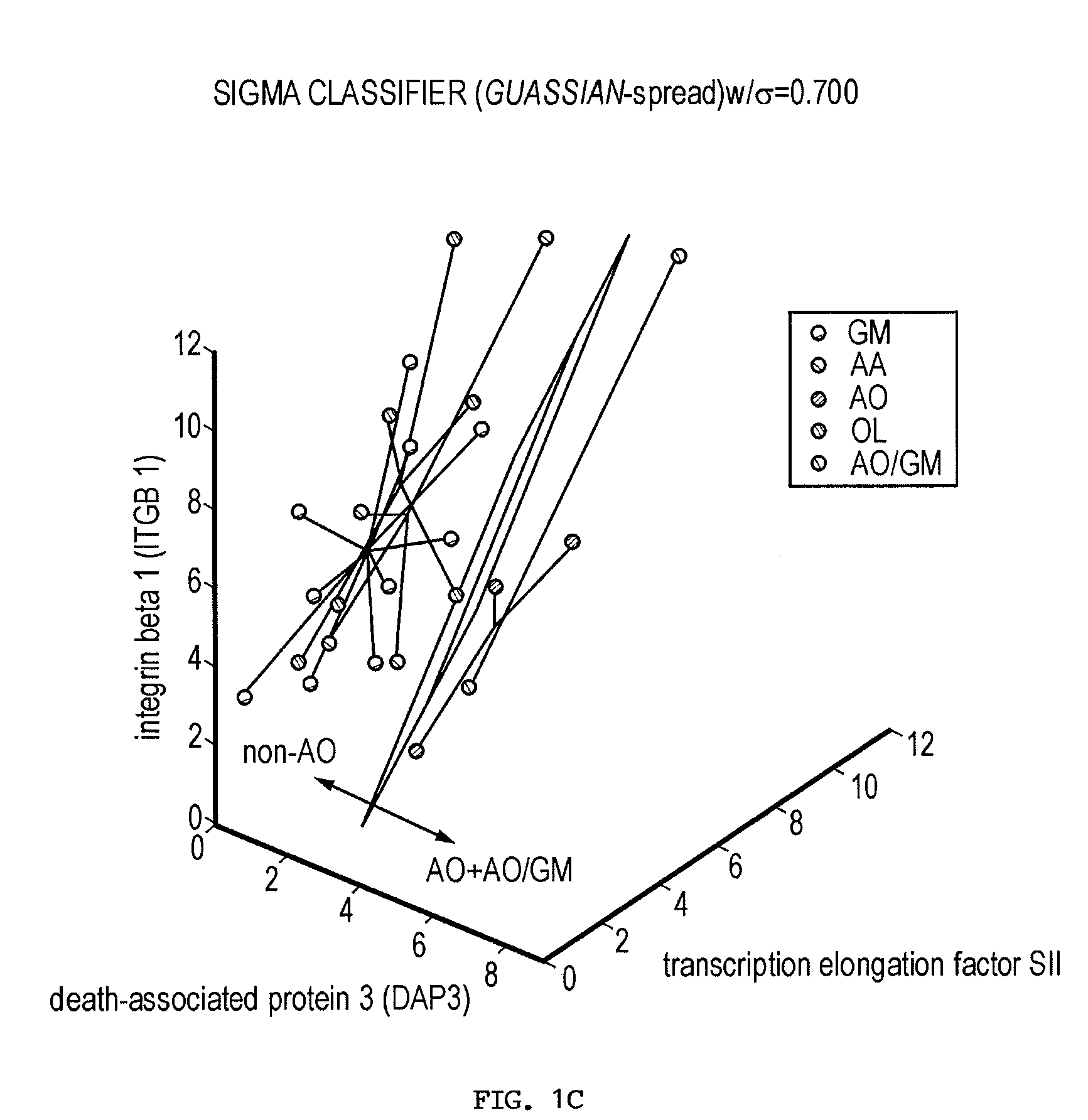
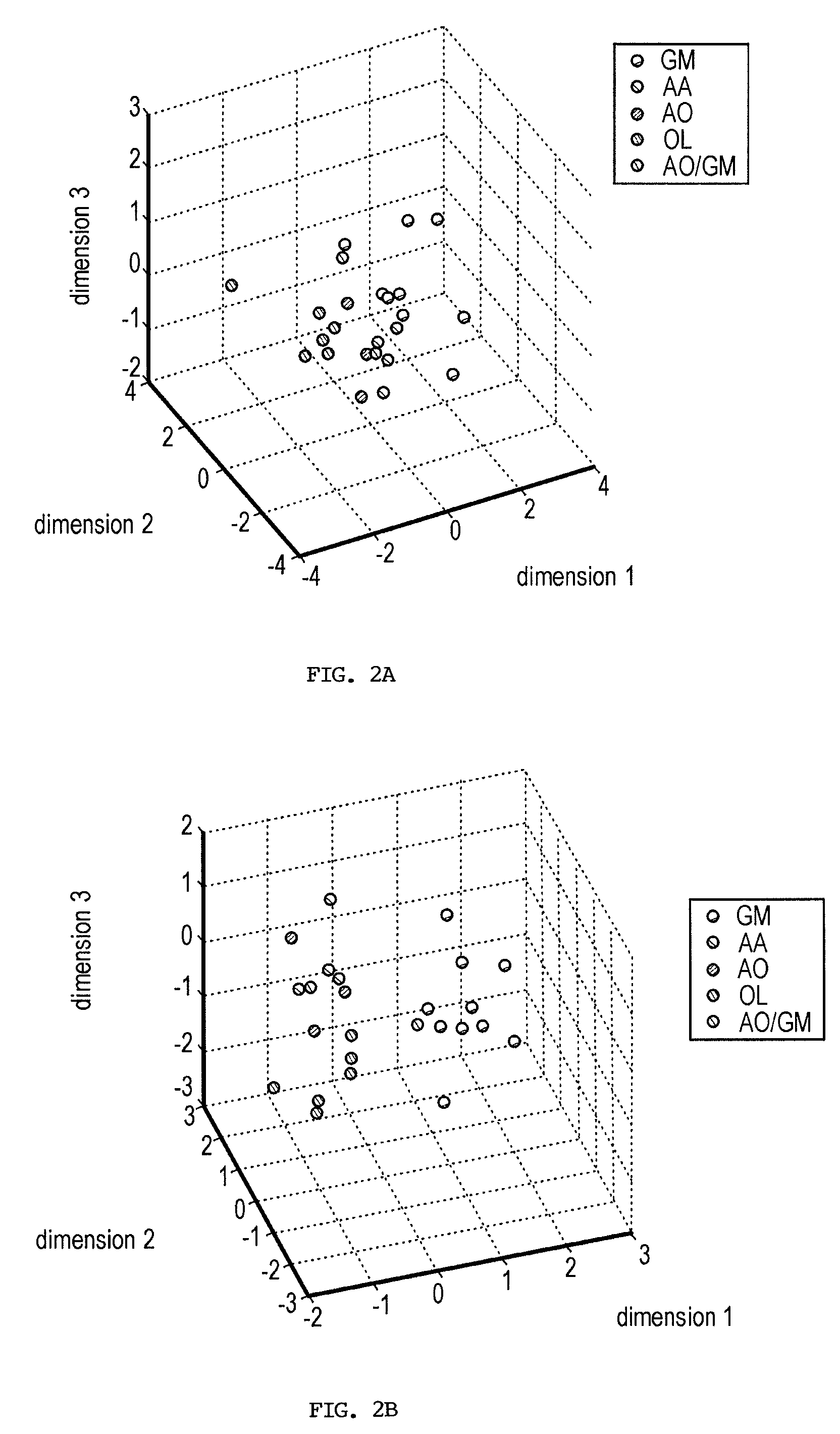
Gene sets for glioma classification
The present invention provides a number of gene markers whose expression is altered in various gliomas. In particular, by examining the expression these markers, one can accurately classify a glioma as glioblastoma multiforme (GM), anaplastic astrocytoma (AA), anaplastic oligodendroglioma (AO) or oligodendroglioma (OL). The diagnosis may be performed on nucleic acids, for example, using a DNA microarray, or on protein, for example, using immunologic means. Also disclosed are methods of therapy.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
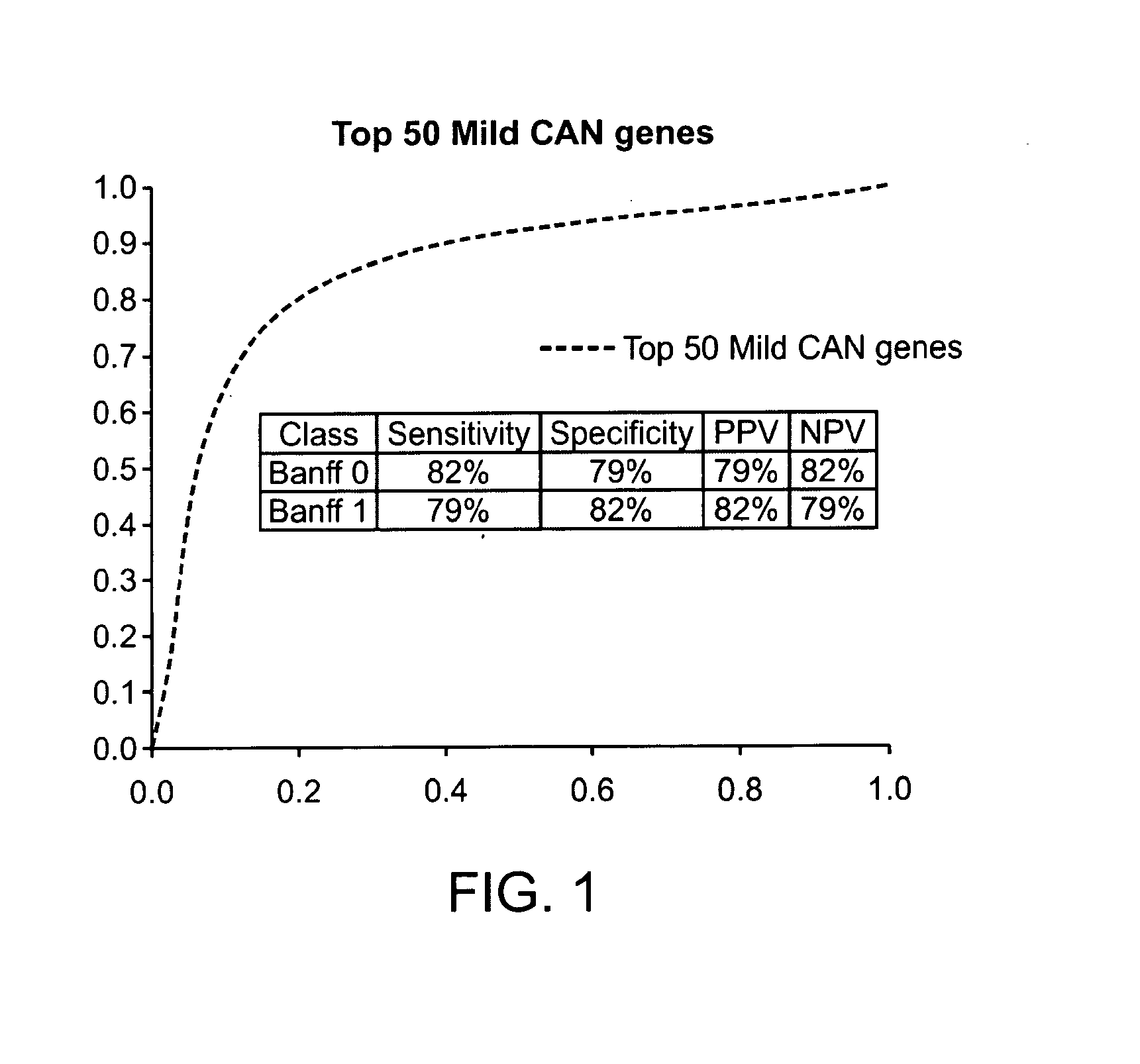
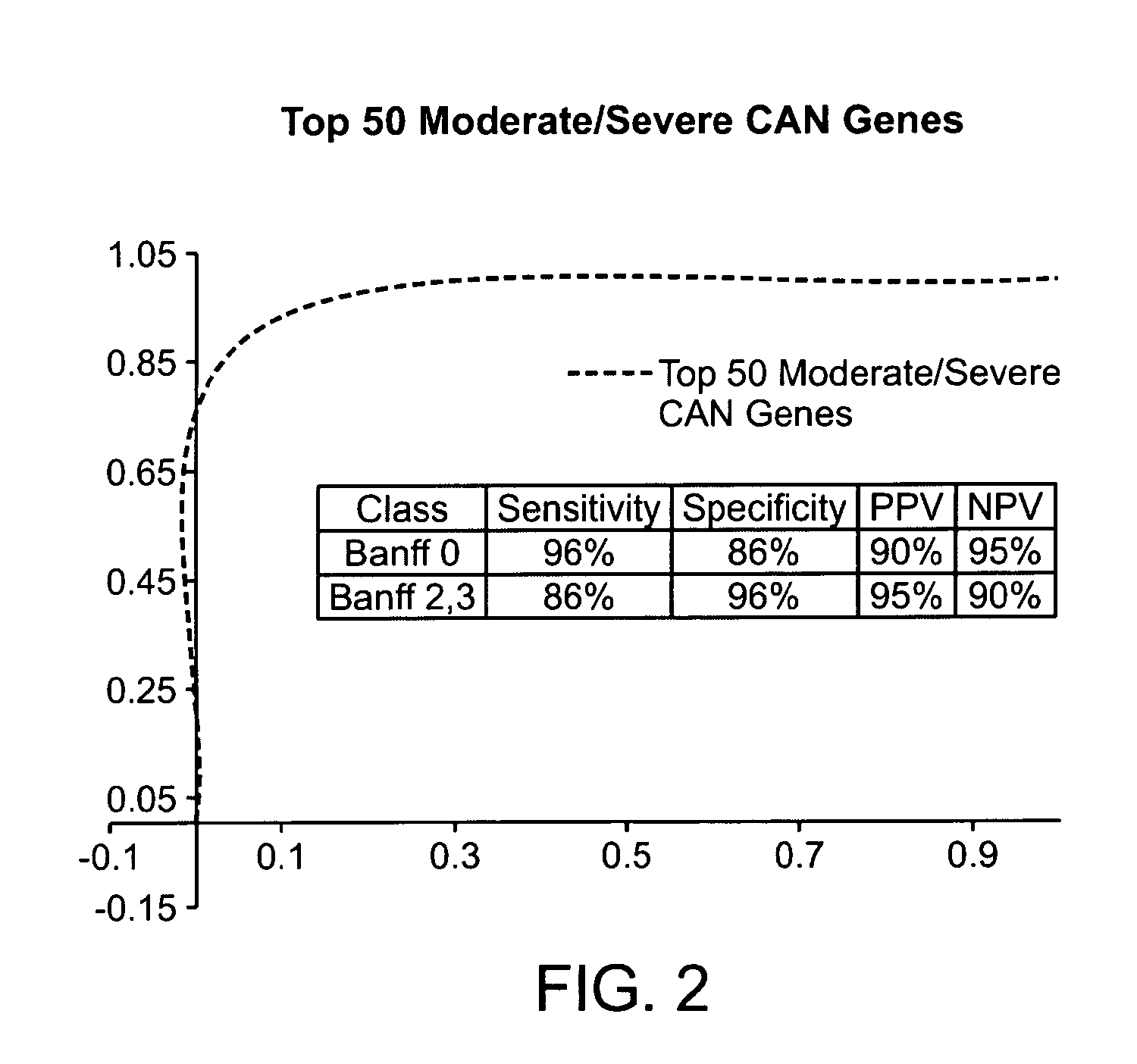
Gene expression profiles associated with chronic allograft nephropathy
ActiveUS20120178642A1Nucleotide librariesMicrobiological testing/measurementChronic allograft nephropathyTubular atrophy
By a genome-wide gene analysis of expression profiles of over 50,000 known or putative gene sequences in peripheral blood, the present inventors have identified a consensus set of gene expression-based molecular biomarkers associated with chronic allograft nephropathy and / or interstitial fibrosis and tubular atrophy CAN / IFTA and subtypes thereof. These genes sets are useful for diagnosis, prognosis, monitoring and / or subtyping of CAN / IFTA.
Owner:THE SCRIPPS RES INST
Diagnostic method and kit for the detection of a lymphocytic variant of hypereosinophilic syndrome
InactiveUS20120289418A1Microbiological testing/measurementLibrary screeningLymphocyteIncreased eosinophils
The present invention is related to a gene set, a kit and a method for diagnosis of lymphocytic variant hypereosinophilic syndrome.
Owner:UNIV LIBRE DE BRUXELIES +1
Gene sets and scoring model for evaluating tumor microenvironment, and application of gene sets
ActiveCN112133365ASignificant prognostic differenceGood forecastMicrobiological testing/measurementBiostatisticsFavorable prognosisPrognosis biomarker
The invention discloses gene sets and a scoring model for evaluating a tumor microenvironment, and application of the gene sets, and belongs to the field of biological information. It is proved for the first time that the gastric cancer has remarkable tumor microenvironment typing, and prognosis difference of gastric cancer patients with different typing is remarkable. According to a constructionmethod of the multi-gene scoring model for evaluating the gastric cancer tumor microenvironment, 44 gene sets capable of representing different microenvironment types are screened out, and the prediction efficiency of the tumor microenvironment multi-gene scoring model corresponding to the gene sets is remarkably better than that of other existing models; and the model is not only a prognostic marker of a gastric cancer patient, but also has a quite remarkable prediction effect in pan-cancer. The model provided by the invention not only can be used as a good prognosis biomarker, but also can be used for predicting checkpoint inhibitor immunotherapy response of various tumors by effectively evaluating the tumor microenvironment, and has important significance and clinical application valuefor better screening immunotherapy benefiting people.
Owner:广东隆宇医疗科技有限公司
Novel compounds
InactiveUS20030211515A1Enhanced interactionHigh frequencyOrganic active ingredientsFungiNucleotidePolynucleotide
Owner:INCYTE
High efficiency transglyco beta galactoside gene
InactiveCN1786170ARemoval of hydrolysis reactionImprove the efficiency of transglycosidationEnzymesFermentationEnzyme GeneReaction temperature
The invention discloses a high efficiency turning glycosyl beta-galactosidase gene. The genes are gained by enterobacter cloacae B5 CGMCC No.1401 gene set DNA under PCR amplification. The whole length of gene nucleotide sequence is 3090 basic groups, and coding 1029 amino acid. The recombinase features are that enzyme is quart-subunit albumen; polymer molecular weight are 391kD; mono subunit are 97.8kD; the enzyme to lactose Km value is 0.32mmol / L, Vmax is 210.70umol / (L.min); and it to onitrophenol-beta-D-galactoside Km value is close to 0; its optimum reaction temperature is 35 centigrade degree; the pH value is from 6.5-9.5; it can be stored at least two months at room temperature; oligomerization galactose yield is 55%; receptor substrate specificity of the enzyme turning glycosyl is very wild; it can used to compose multi galactoside compounds. The gained enzyme gene can be used to modify gene to gain new beta-galactoside synthetase, and improve its combined efficiency fundamentally.
Owner:SHANDONG UNIV
Minimal bacterial genome
The present invention relates, e.g., to a minimal set of protein-coding genes which provides the information required for replication of a free-living organism in a rich bacterial culture medium, wherein (1) the gene set does not comprise the 101 genes listed in Table 2; and / or wherein (2) the gene set comprises the 381 protein-coding genes listed in Table 3 and, optionally, one of more of: a set of three genes encoding ABC transporters for phosphate import (genes MG410, MG411 and MG412; or genes MG289, MG290 and MG291); the lipoprotein-encoding gene MG185 or MG260; and / or the glycerophosphoryl diester phosphodiesterase gene MG293 or MG385.
Owner:SYNTHETIC GENOMICS INC
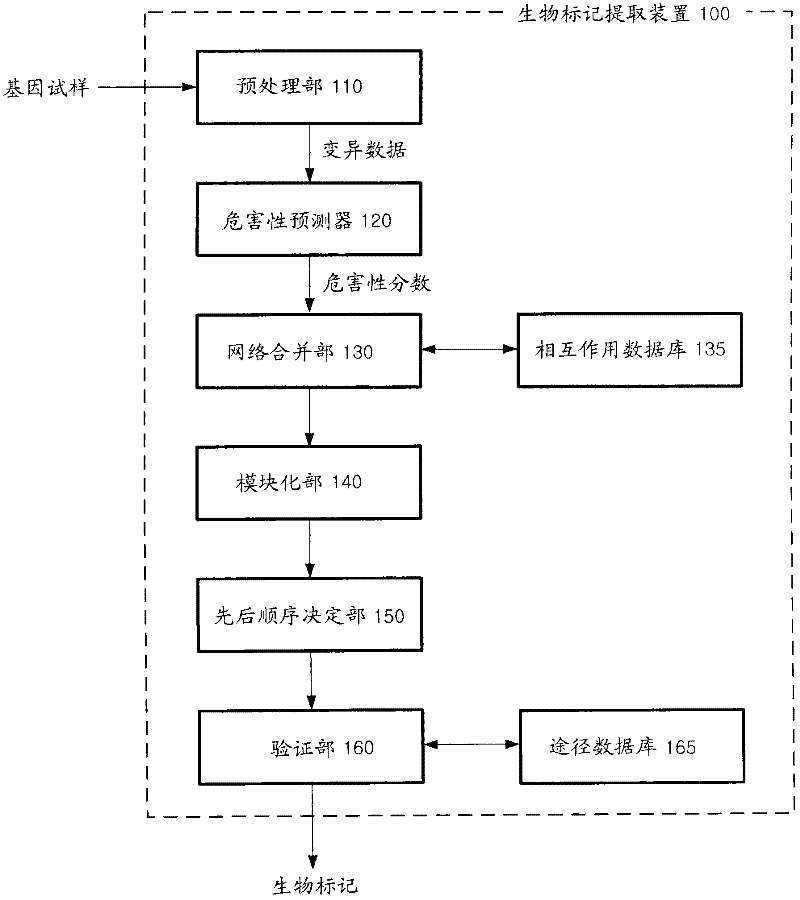
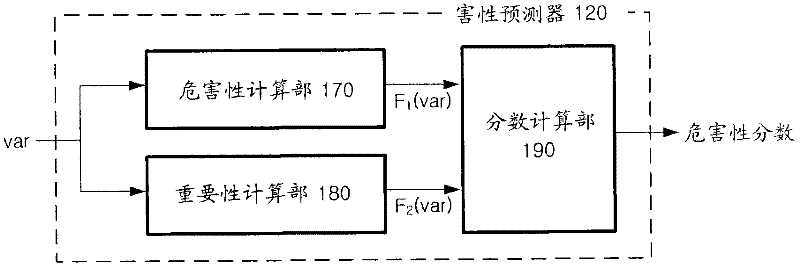
Apparatus and method for extracting biomarkers
An apparatus and method for extracting biomarkers with higher reliability by analyzing toxicity indicating how genetic variants appearing in sequences affect gene functions are provided. The apparatus includes a pre-processor that analyzes sequences of samples of genes and extracts data of genetic variants mapped to the genes, a toxicity prediction unit that obtains toxicity scores obtained by quantifying genetic dysfunctions affected by the data of genetic variants, and a modularization unit that searches for a least one sub-module including a set of genes whose toxicity scores exceed a predetermined critical value from a genetic network.
Owner:SAMSUNG SDS CO LTD
Cynoglossus semilaevis gunther specific molecular label and genetic sex identifying method
This invention relates to specific molecule labeling of the sex of semi-sliding tongue soles and its genetic sex test method including the pick-up of the blood DNA, the AFLP analysis of DNA in the gene set, selection of the AFLP molecular labeling and a method for genetic sex check up of semi-sliding tongue sole, which selects 4 from 64 primer combinations to generate female specific DNA segments and selects 7 female specific AFLP molecular labels of the sole to set up the molecule labeling technology of its genetic sex authentication.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
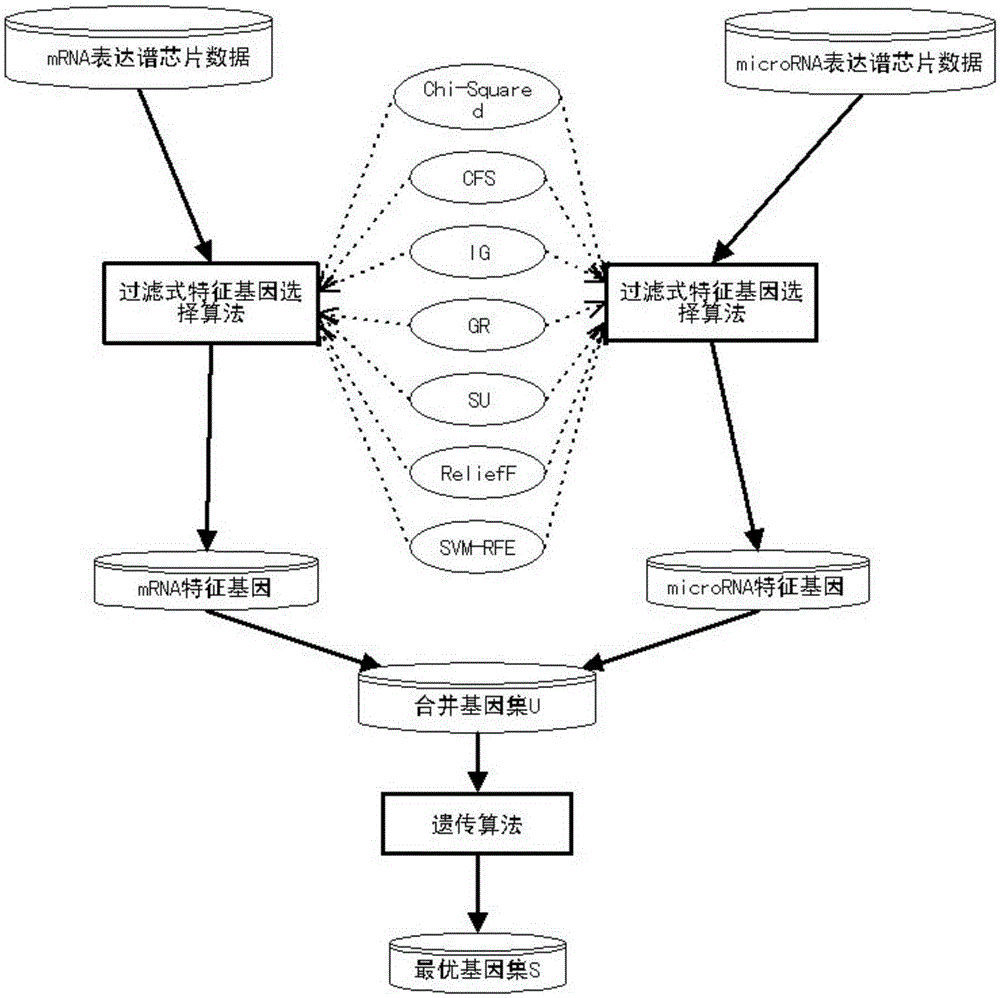
Tumor feature gene selection method combining mRNA and microRNA expression profile chips
InactiveCN105243296AImprove classification accuracyEasy to analyzeSpecial data processing applicationsGene selectionFiltration
The invention discloses a tumor feature gene selection method combining mRNA and microRNA expression profile chips. The method is implemented by the following steps of step 1, by using the mRNA and microRNA expression profile chips, detecting expression values of a large amount of genes, by adopting a filtration feature gene selection method, ranking relevance of all the genes, removing a large amount of low-relevance genes, and leaving a small amount of genes that are closely related to tumor classification; step 2, combining mRNA and microRNA feature genes obtained by adopting the filtration feature gene selection method to form a gene pool U; and step 3, by adopting a genetic algorithm, further selecting genes for the gene pool, eliminating redundant genes, and performing a search to obtain an optimal gene set S with optimal features. The method is simple in step and high in result accuracy.
Owner:LISHUI UNIV
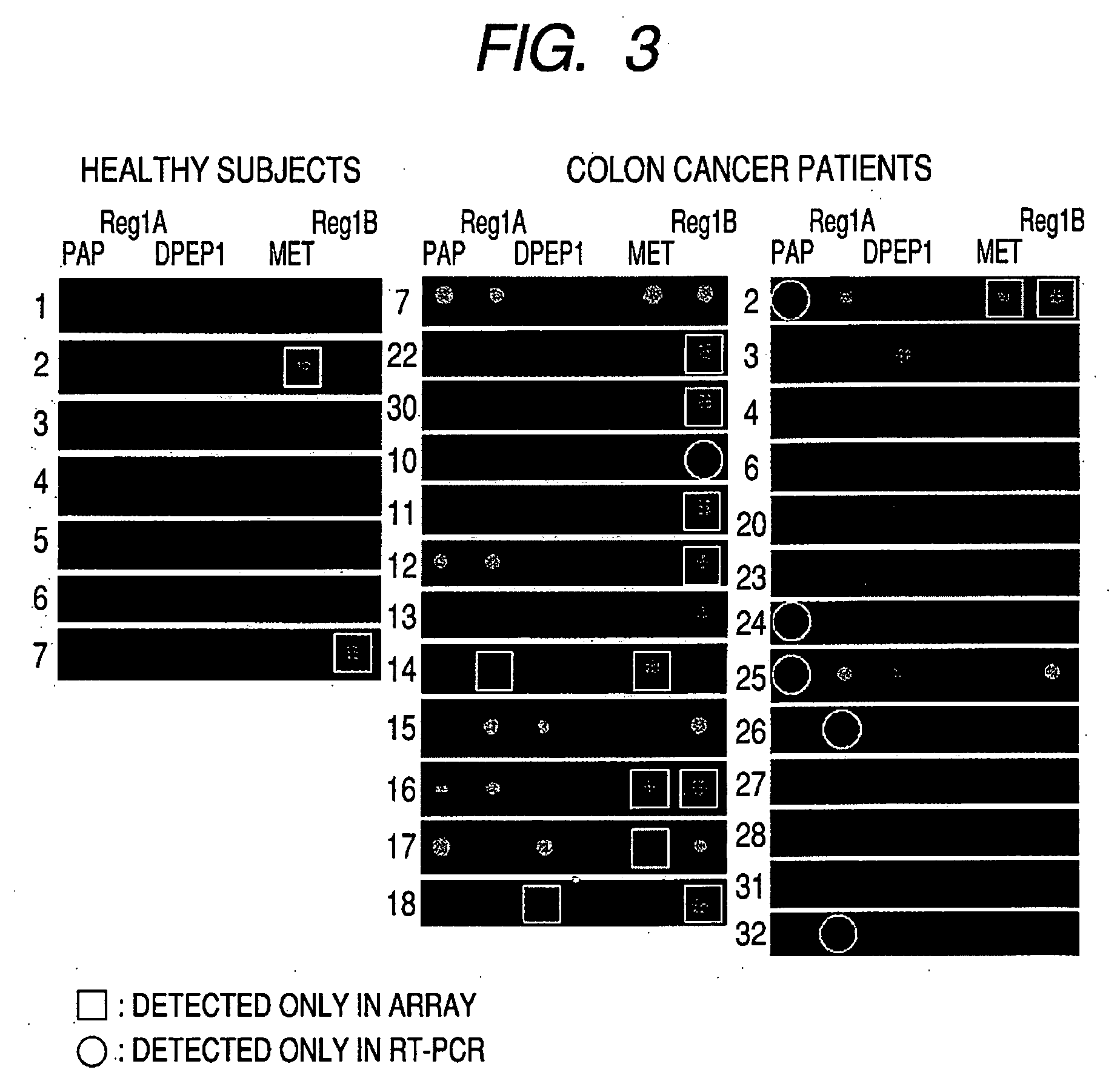
Method for screening colon cancer cells and gene set used for examination of colon cancer
InactiveUS20090011413A1Improve accuracyHigh-precision detectionSugar derivativesMicrobiological testing/measurementGene listColon cancer cell
Colon cancer cells in a sample are screened by analyzing the amount of expression of at least 2 or more genes or products thereof selected from the group of genes listed in Tables 1 and 30. As compared to conventional method, patients having colon cancer can be detected with higher accuracy. Colon cancer cells in stool are also screened by analyzing expression of genes selected from the group of genes listed in Table 37.
Owner:CANON KK +1
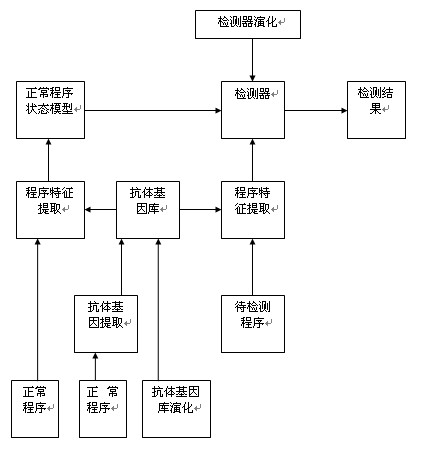
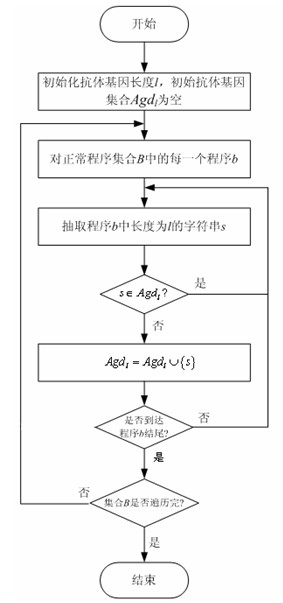
Detection method of bot program
ActiveCN101930517ACan't solveSolve the outstanding contradiction of polymorphic bot synchronizationPlatform integrity maintainanceFeature extractionState model
The invention discloses a detection method of a bot program and belongs to the technical field of information safety, and the method comprises the following steps: extracting antibody genes of a normal program set B, constructing antibody gene sets Agdl, and forming an antibody gene library Agd by the antibody gene sets Agdl of different antibody gene lengths; carrying out feature extraction on a normal program set B' by the antibody gene sets Agdl and constructing a normal program state model; generating detectors by normal program state sets Cb and generating a detector set by the detectors; detecting the bot program by the detector set; and evolving the antibody gene library and the detectors dynamically. The method can not only identify known bot programs but also discover new bot programs or variations of the known bot programs through self-learning and evolvement in a computer environment which changes in real time, thus effectively solving the key issue that a feature code library of computer viruses can not be synchronous with the multistate bot programs.
Owner:四川通信科研规划设计有限责任公司
Feature gene selection method based on logistic and relevant information entropy
InactiveCN104598774ASimple structureAvoid lossSpecial data processing applicationsMaximum eigenvalueData set
The invention discloses a novel feature gene selection method based on logistic and relevant information entropy. The method comprises the following steps that a dataset is subjected to logistic regression, a gene variable with great influence on the classification is obtained, a Relief algorithm is used for giving a value on the gene variable, sequencing is carried out, a maximum feature value gene is added to an initial feature gene set, and the relevant information entropy is calculated. The novel feature gene selection method has the advantages that a logistic regression model in the machine study is introduced into the feature gene selection method, and a high-quality gene expression profile is obtained; the correlation between gene variables is measured by the relevant information entropy, redundant genes are deleted, and a feature gene sub set with high classification capability and fewer genes is obtained through searching a feature gene space set.
Owner:HENAN NORMAL UNIV
Identification of esophageal cancer related characteristic pathways and construction method of early diagnosis model
ActiveCN109841280ALimit random fluctuationsFine molecular mechanismHealth-index calculationMedical automated diagnosisPathway analysisWilms' tumor
The invention belongs to the technical field of tumor diagnosis, and particularly relates to identification of esophageal cancer related characteristic pathways of and a construction method of an early diagnosis model. The identification and constructing method includes the steps: expression spectrum preprocessing, differential expression gene extraction, sample cluster analysis, gene cluster analysis, specific gene set functional pathway analysis, comparison of pathway aberration scores, comparative analysis of functional differences, construction of esophageal cancer specific co-expression networks, characteristic selection of genes, prediction of deep learning models, and the like. The method of the invention divides genes into different groups according to expression similarity and functional consistency of the genes, and analyzes the genes in the form of gene collection, thus being able to avoid the disadvantages of high false positive rate, big random error and unstable result inthe traditional method, and also being able to more specifically identify the function significantly associated with esophageal cancer.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
System for predicting prognosis of locally advanced gastric cancer
The present invention relates to a novel system for predicting a prognosis capable of predicting the prognosis of locally advanced gastric cancer, and more specifically, capable of predicting a clinical result after a resection during gastric cancer surgery through gene set enrichment comparative analysis.
Owner:NOVOMICS CO LTD
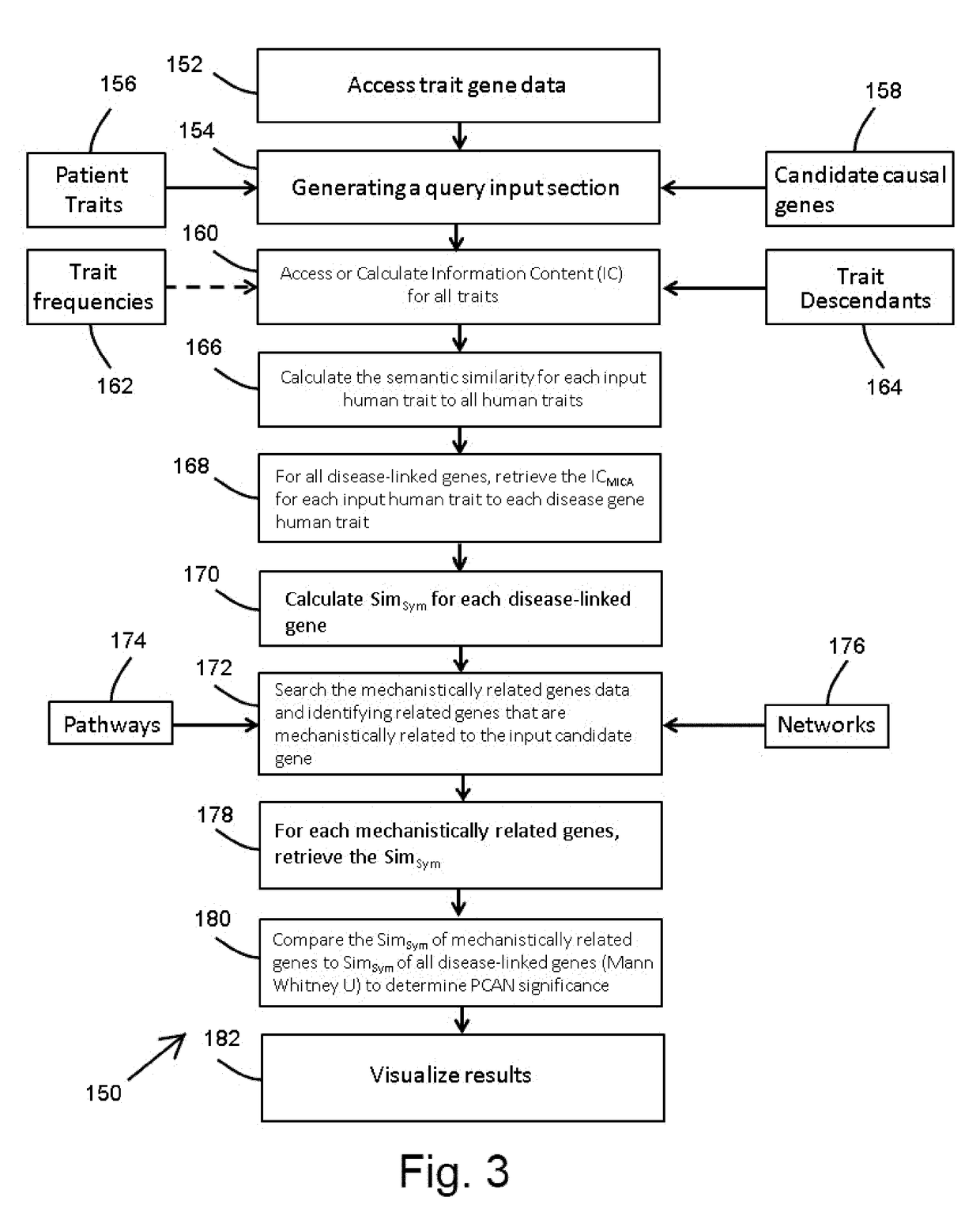
Method and system for quantifying the likelihood that a gene is casually linked to a disease
A computer program product, disposed on a non-transitory computer readable media, for analyzing a biological relevance of a candidate gene to a human phenotype is provided. The product includes computer executable process steps operable to control a computer to receive an input phenotype comprised of a plurality of input human traits and at least one input candidate gene; identify a plurality of disease-linked genes by querying disease-linked gene data and identifying genes causally linked to at least one disease; provide values of a semantic similarity metric for a identified gene set with respect to the input phenotype based on a comparison of human traits linked to each gene of the identified gene set and the input human traits, the identified gene set including genes mechanistically related to the input candidate gene that are included in the identified disease-linked genes; and output a statistical measure indicating whether the values of the semantic similarity metric of the genes of the identified gene set with respect to the input phenotype are greater than the values of the semantic similarity metric of others of the identified disease-linked genes with respect to the input phenotype by a statistically significant amount.
Owner:UCB PHARMA SRL
Markers for responsiveness to an erbB receptor tyrosine kinase inhibitor
InactiveUS20060252056A1Improved prognosisEasy to useSugar derivativesMicrobiological testing/measurementReceptor tyrosine kinase inhibitorGene list
The invention relates to a set of isolated marker genes comprising at least one gene identified as having differential expression as between patients who are responders and non responders to an erbB receptor tyrosine kinase inhibitor; said gene set comprising one or more genes selected from at least the group consisting of the 51 genes listed herein including gene-specific oligonucleotides derived from said genes; and uses of such sets in diagnostic applications.
Owner:ONCOTHERAPY SCI INC
Tumor mutation site screening and mutual exclusion gene mining system
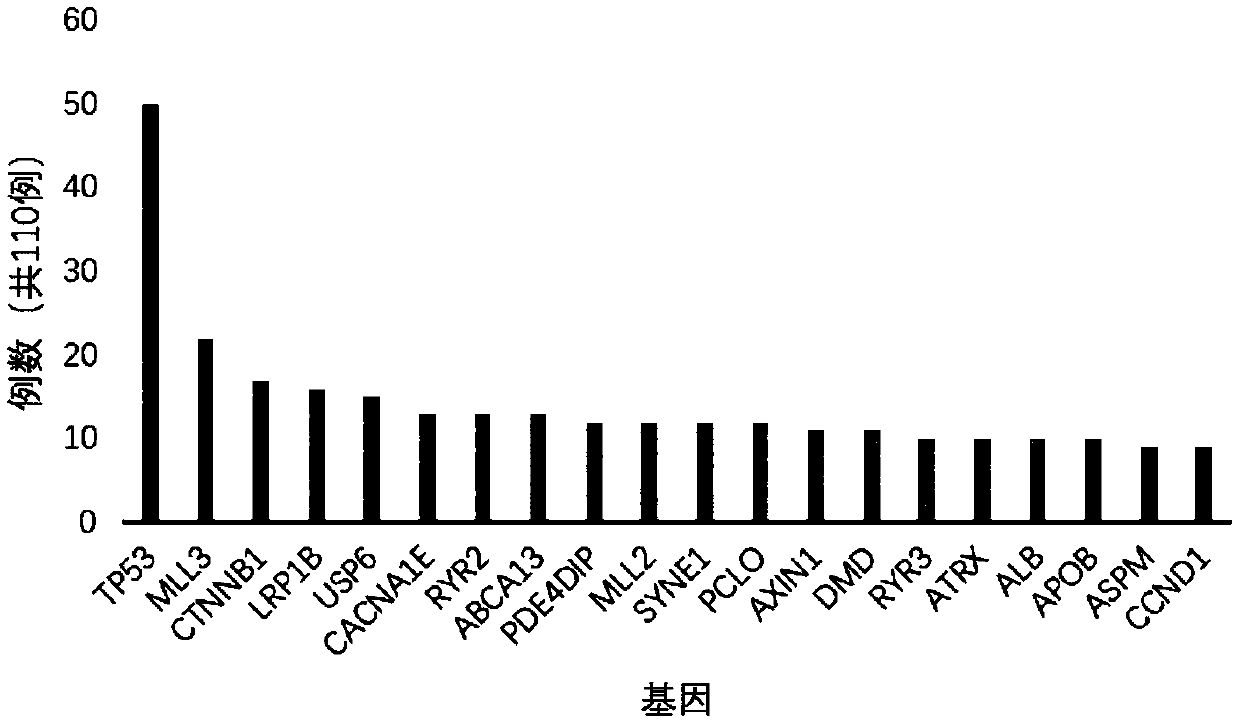
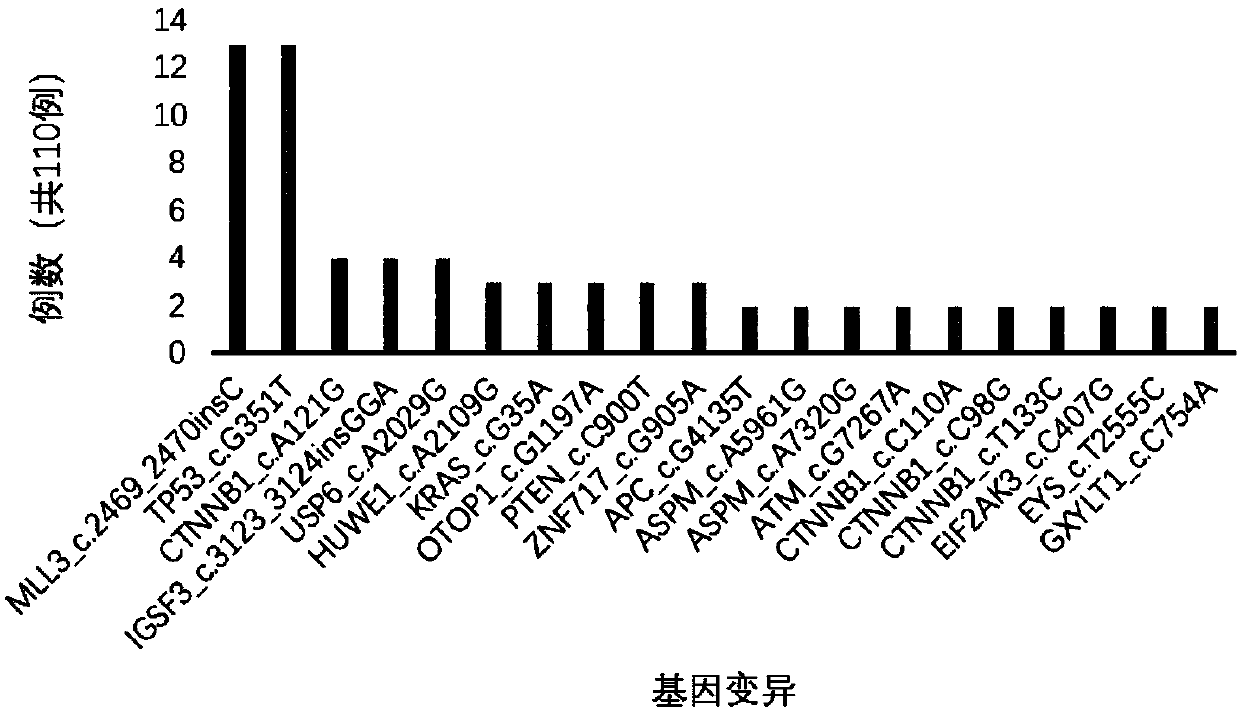
InactiveCN106022001AFacilitate biological interpretationBiostatisticsSpecial data processing applicationsGene miningAnnotation
The invention provides a tumor mutation site screening and mutual exclusion gene mining system. The system comprises a filtering module which is used for filtering a vcf file and an ANNOVAR software output file in an exome processing process; an analysis module which is used for carrying out description analysis on different experiment group mutation sites; a gathering module which is used for gathering mutation genes of each sample and establishing a mutation gene matrix according to an experiment group mutation gene list; and a mining module which is used for carrying out Fisher precise checking based mutual exclusion and co-mutation analysis on the generated mutation gene matrix, thereby determining mutual exclusion and co-mutation genes. According to system, the mutation sites are filtered according to basic parameters such as annotation information of the mutation sites, the sequencing read number and site sequencing depth; and description analysis of different experiment group mutation modes and mining of co-mutation and mutual exclusion gene sets are carried out on the obtained mutation sites.
Owner:WANKANGYUAN TIANJIN GENE TECH CO LTD
Leptin-mediated gene-induction
InactiveUS7291458B2Inducing effectObesity gene productsPeptide/protein ingredientsProtein iThreonine
Methods of activating a signaling cascade comprising, introducing leptin and / or a cytokine to a receptor complex comprising gp 130, optionally in combination with a compound acting on adenylate cyclase or acting on one or more downstream targets of adenylate cyclase, thereby inducing genes in neuro-endocrine cells or cells of neuro-endocrine origin. Two distinct gene-sets are induced, immediate early response genes (STAT-3, SOCS-3, Metallothionein-II, the serine / threonine kinase Fnk and the rat homologue of MRF-1), and late induced target genes (Pancreatitis Associated Protein I, Squalene Epoxidase, Uridinediphosphate Glucuronyl Transferase and Annexin VIII). Strong co-stimulation with the adenylate cyclase activator forskolin was shown with respect to late induced target genes. Transcripts encoding Leptin Induced Protein I (LIP-I) and Leptin Induced Protein II (LIP-II) were identified; however, no forskolin co-stimulatory effect was observed. It is also demonstrated that leptin modulates in vivo expression of MT-II, Fnk and Pancreatitis Associated Protein I genes.
Owner:NEC CORP +1
Method for identifying significantly different expressed genes
InactiveCN101250584AExcellent set recognition effectIncrease valueMicrobiological testing/measurementSpecial data processing applicationsDifferentially expressed genesBioinformatics
The invention discloses a process for identifying significantly differential expression gene sets, which comprises the following steps, firstly setting and inputting data, secondly vesting genes to each gene set, thirdly checking whether the number of genes in each gene is larger than the defined number which is set, if the result is not, then abandoning the gene set, fourthly calculating expression variability index of each gene, fifthly using all genes on a whole chip as background genes, and calculating the expression variability index of the background genes, sixthly randomly sampling from the background genes, and checking the significance of the E value of each gene set, and seventhly outputting the gene set which meets the threshold value requirement as the identification result according to the threshold values of the set E value and p value. The process of the invention has excellent identification effect for large gene sets, guarantees higher accuracy under the condition of fewer detecting times, and greatly increases the value of gene expression values in practical application.
Owner:NANJING UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com