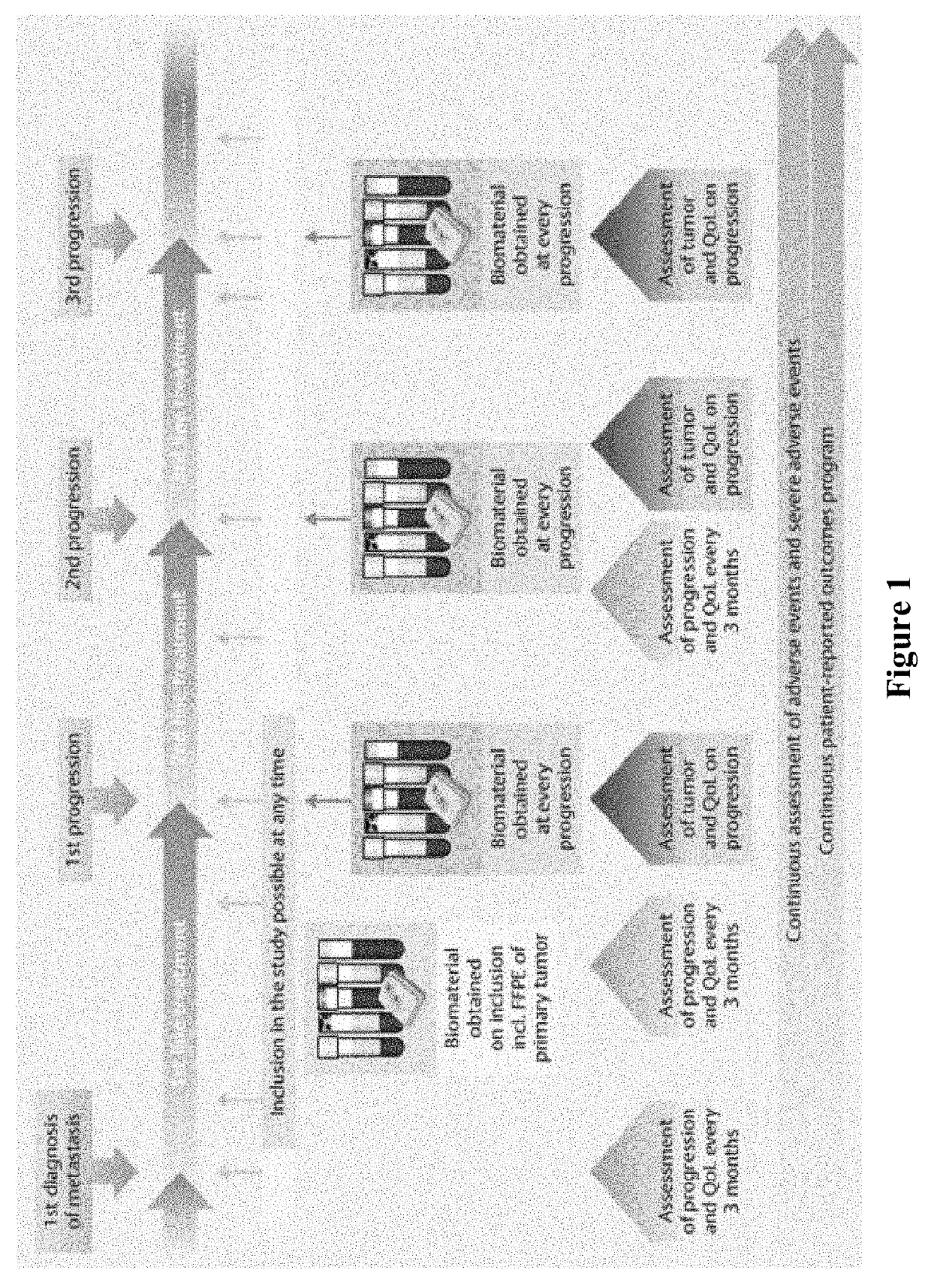
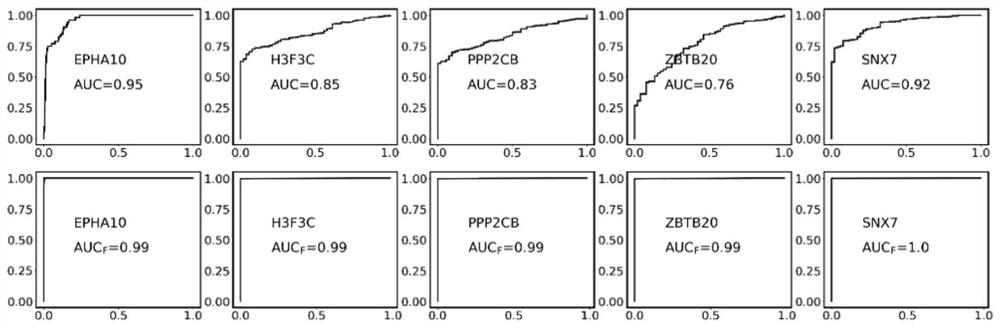
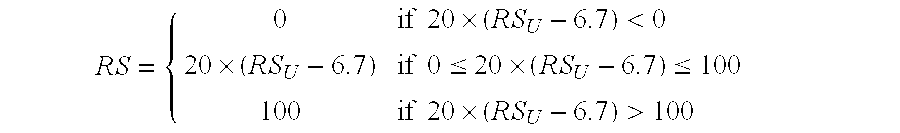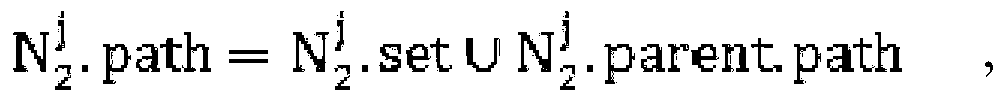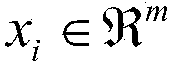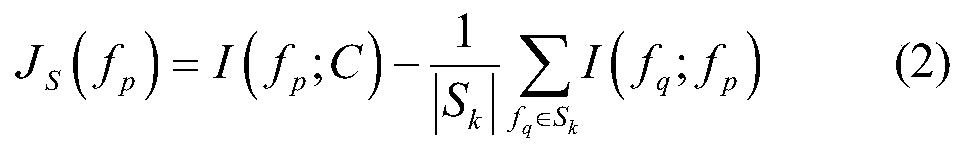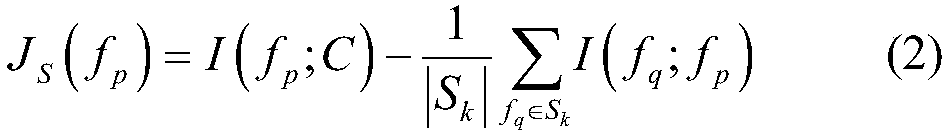Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
31 results about "Gene subset" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
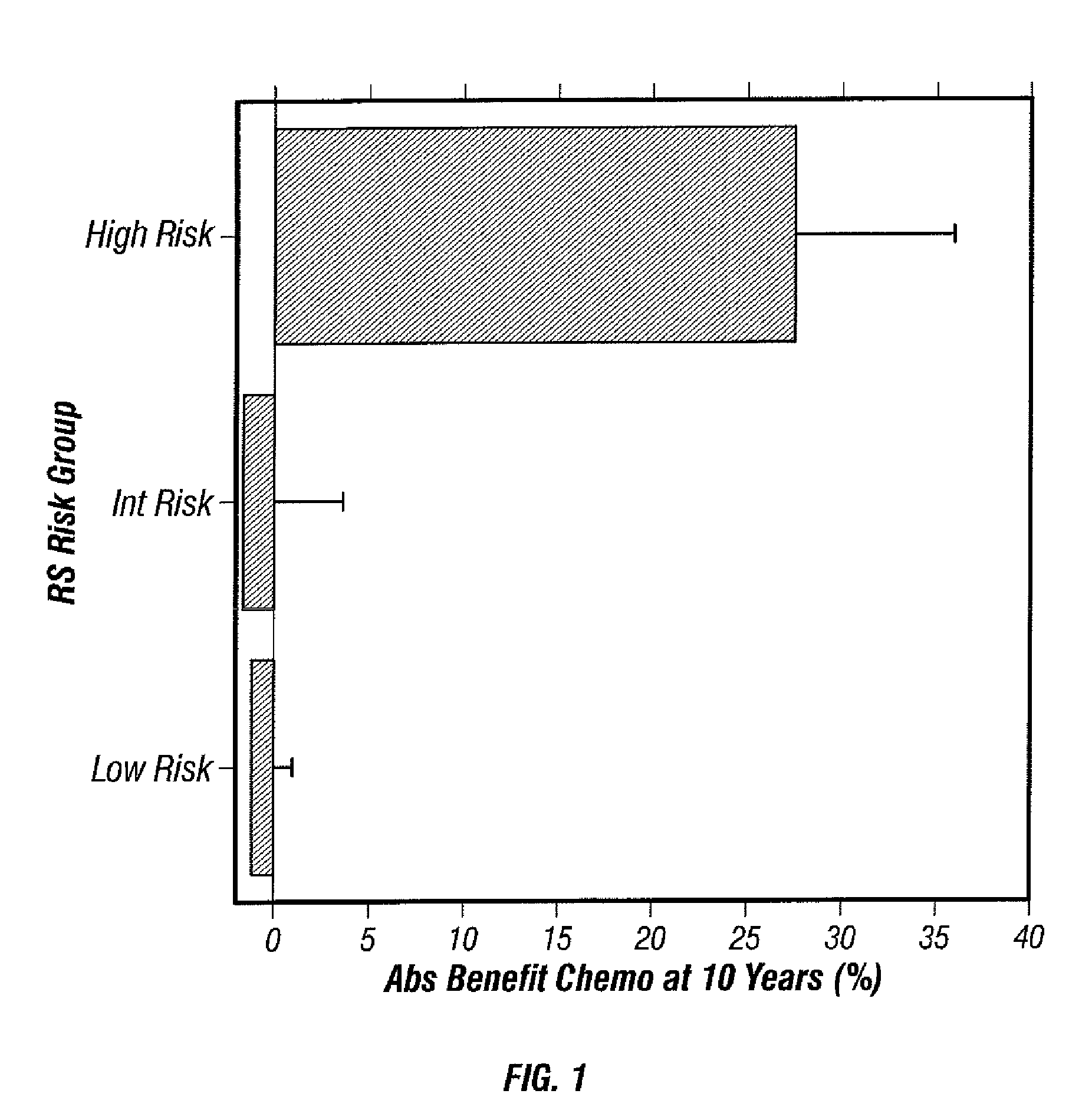
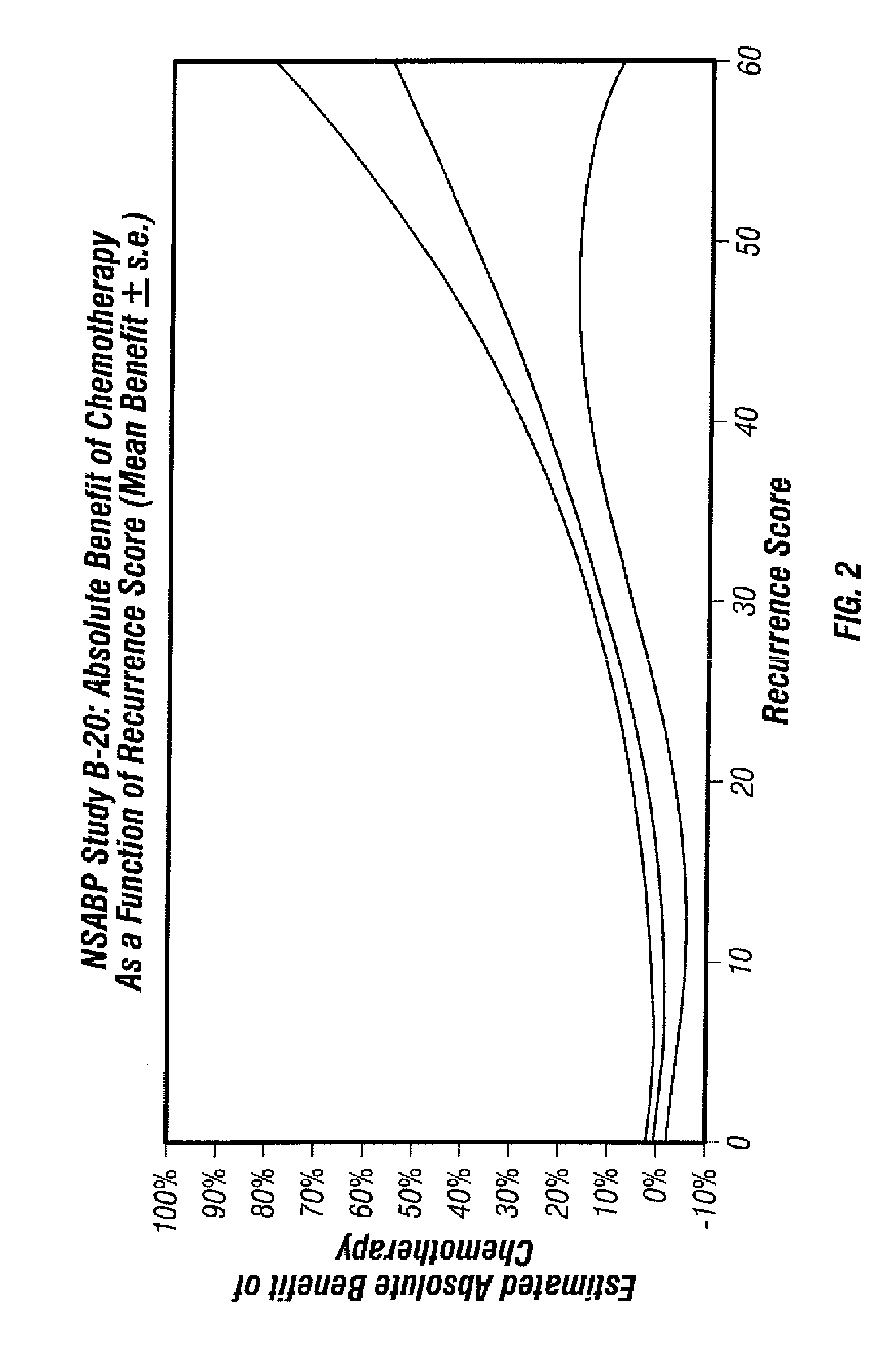
Predicting response to chemotherapy using gene expression markers
ActiveUS7930104B2Raise the possibilityReduce riskMicrobiological testing/measurementAnalogue computers for chemical processesRecurrence scoreOncology
The present invention provides gene expression information useful for predicting whether a cancer patient is likely to have a beneficial response to treatment with chemotherapy, comprising measuring, in a biological sample comprising a breast tumor sample obtained from the patient, the expression levels of gene subsets to obtain a risk score associated with a likelihood of a beneficial response to chemotherapy, wherein the score comprises at least one of the following variables: (i) Recurrence Score, (ii) ESRI Group Score; (iii) Invasion Group Score; (iv) Proliferation Group Score; and (v) the expression level of the RNA transcript of at least one of MYBL2 and SCUBE2, or the corresponding expression product. The invention further comprises a molecular assay-based algorithm to calculate the likelihood that the patient will have a beneficial response to chemotherapy based on the risk score.
Owner:GENOMIC HEALTH INC
Feature selection method and application based on feature identification degree and independence
InactiveCN105938523AValid choiceValid information referenceBiostatisticsHybridisationDiseaseTime complexity
The invention relates to a feature selection method and application based on feature identification degree and independence. The method comprises following steps: calculating the importance degree of each feature by measuring inter-class distinguished ability with feature identification degree and measuring correlational relationship between features with feature independence and sequencing in a descending order; and selecting top k features with the importance higher than those of others to form a feature subset with high class-discrimination performance. Differently-expressed gene subsets selected in application of oncogene expression profile data obtain fine time and class discrimination performance. The feature selection method and application based on feature identification degree and independence have following beneficial effects: easy calculations can be made; time complexity is reduced; selection efficiency runs high; and a good reference is provided for clinical diagnoses and judgments of tumors and other diseases.
Owner:SHAANXI NORMAL UNIV
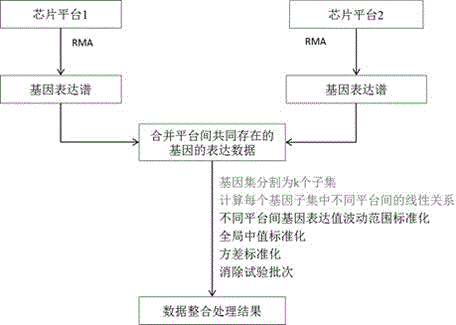
Integration method for gene expression data by crossing chip platforms
ActiveCN103745137AEliminate experimental batch effectsImprove performanceSpecial data processing applicationsGene expression matrixExpression gene
The invention belongs to the technical field of biological information. The invention provides a method for integrating gene expression data by crossing a plurality of different chip platforms; the method comprises the following steps: the standard preprocessing is implemented for the gene expression profile of the plurality of chip platforms; the common gene expression data in the different chip platforms is merged; genes are divided into k subsets according to the expression similarity of the genes on the plurality of chip platforms; the expression linear relation exps1=as*exps2+bs of the different chip platforms in every gene subset can be calculated by the least square method; the gene expression values of the different chip platforms are standardized into the same change range by using the formula exp1=X*A.*exp2+X*B to obtain standard gene expression matrixes, wherein the implications of symbols are defined as the specification.
Owner:艾吉泰康(嘉兴)生物科技有限公司
Heuristic breadth-first searching method for cancer-related genes
InactiveCN103186717APathogenesis revealedFacilitates personalized treatmentSpecial data processing applicationsGene ordersGene selection
The invention relates to a heuristic breadth-first searching method for cancer-related genes. According to the method, appearance frequencies of genes in a selected gene subset are used for measuring the genes, and genes with higher appearance frequency are considered as the most important cancer-related genes, on the basis, a classifier is designed and a gene ordering method based on HBSA is established. As proved by study, information gene selection plays an important role in improving the classification performance, and the genes can be probably taken as important tumor clinical diagnosis signs, so discovery of the minimum gene subset with the highest classification performance is a very important research objective. As indicated by experimental results, the heuristic breadth-first searching method can not only obtain favorable generalization performance but also discover important tumor genes. And the relationship of the appearance frequencies of the selected genes and the gene number conforms to power-law distribution. The genes in the gene subset with extremely high classification accuracy are in close relationship with specific tumor subtypes, and even the genes are important genes directly related with the tumor.
Owner:HEFEI INSTITUTES OF PHYSICAL SCIENCE - CHINESE ACAD OF SCI
Redundancy removal feature selection method LLRFC score+ based on LLRFC and correlation analysis
InactiveCN105740653AOptimal trait gene subsetImprove classification accuracyBiostatisticsSpecial data processing applicationsDiseaseData set
The invention provides a redundancy removal feature selection method LLRFC (Locally Linear Representation Fisher Criterion) score+ based on LLRFC and correlation analysis. A DNA (Deoxyribonucleic Acid) microarray technology provides a new direction for clinic tumor diagnosis. Performance of gene expression data corresponding to different kinds of tumor is different; through the analysis on the tumor gene expression data, study personnel can realize the accurate recognition on the tumor and the tumor subtype in the molecular level; and important biological significance is realized on the diagnosis and the treatment of the tumor. The feature genes in LLRFC judging criterion descending sort gene expression data is used to be combined with the dynamic correlation analysis strategy for further eliminating redundant features; an LLRFC score+ algorithm is provided; and the optimum feature gene subset is selected. The feature selection method LLRFC score+ has the advantages that the classification precision of a classifier can be effectively improved; a sample data set does not need to meet the normal distribution; and the method is applicable to data in various distribution types. The feature selection method LLRFC score+ can help people to find the virulence gene of cancer, and the early-stage diagnosis, tumor staging and typing, prognosis treatment and the like of clinic tumor diseases are facilitated.
Owner:BEIJING UNIV OF TECH
Tumor key gene identification method based on particle swarm optimization and marking criterion
ActiveCN106951728AAccurate identificationEffectively deleteProteomicsGenomicsLocal optimumMulti-swarm optimization
The invention discloses a tumor key gene identification method based on particle swarm optimization and a marking criterion. The method comprises the steps that one gene marking criterion is defined to acquire gene classification ability information so as to filter genes with a low correlation to a tumor category; and the Metropolis criterion is utilized to improve a particle swarm optimization (PSO) algorithm in combination with the gene classification ability information so as to realize identification of tumor key genes. The new method for gene classification information and PSO improvement overcomes the defect that a traditional method for identifying tumor key genes based on PSO is prone to fall into a locally optimal solution, gene subsets in a smaller number and with a high correlation to the tumor category can be selected, and therefore the method is beneficial for improving subsequent tumor identification.
Owner:JIANGSU UNIV
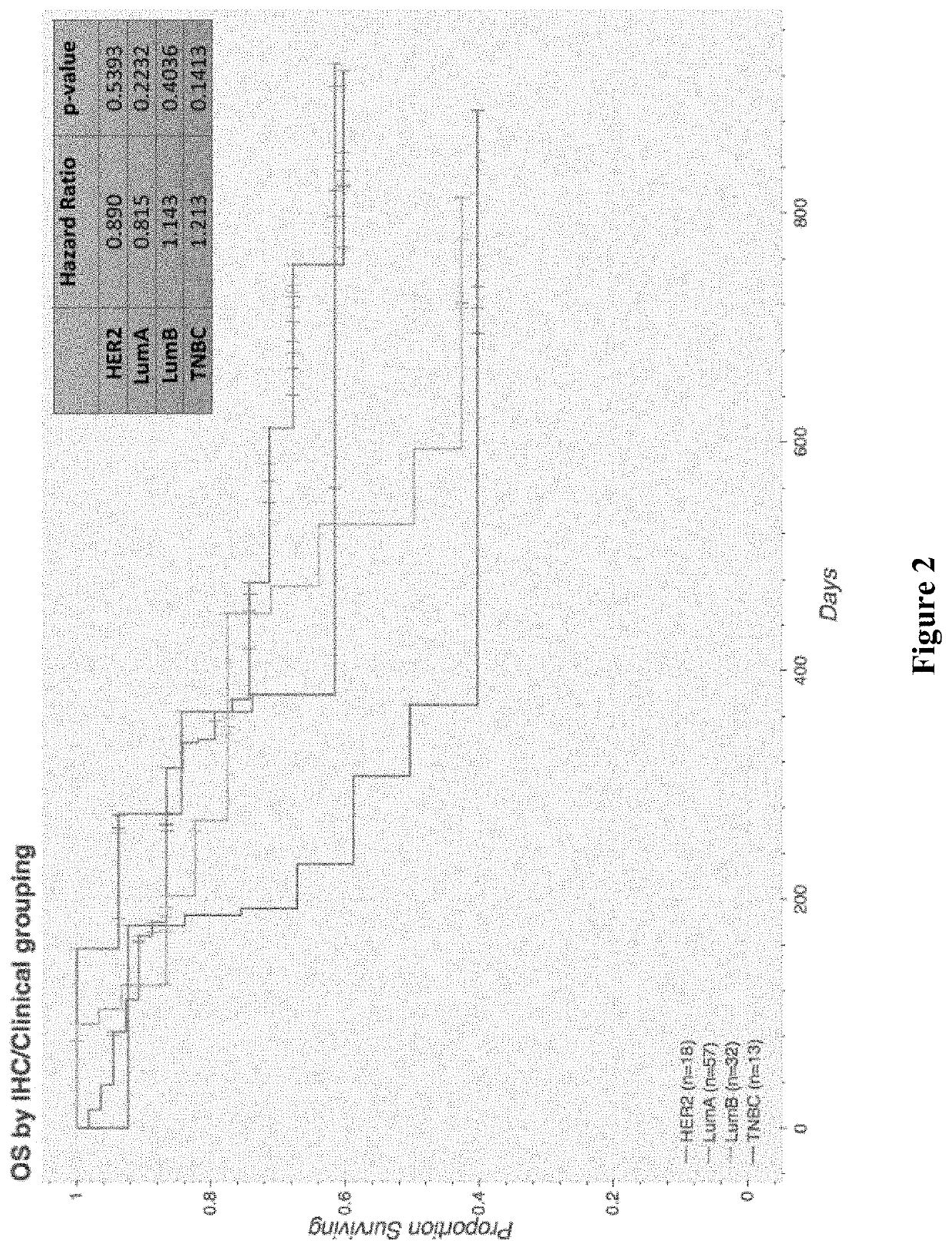
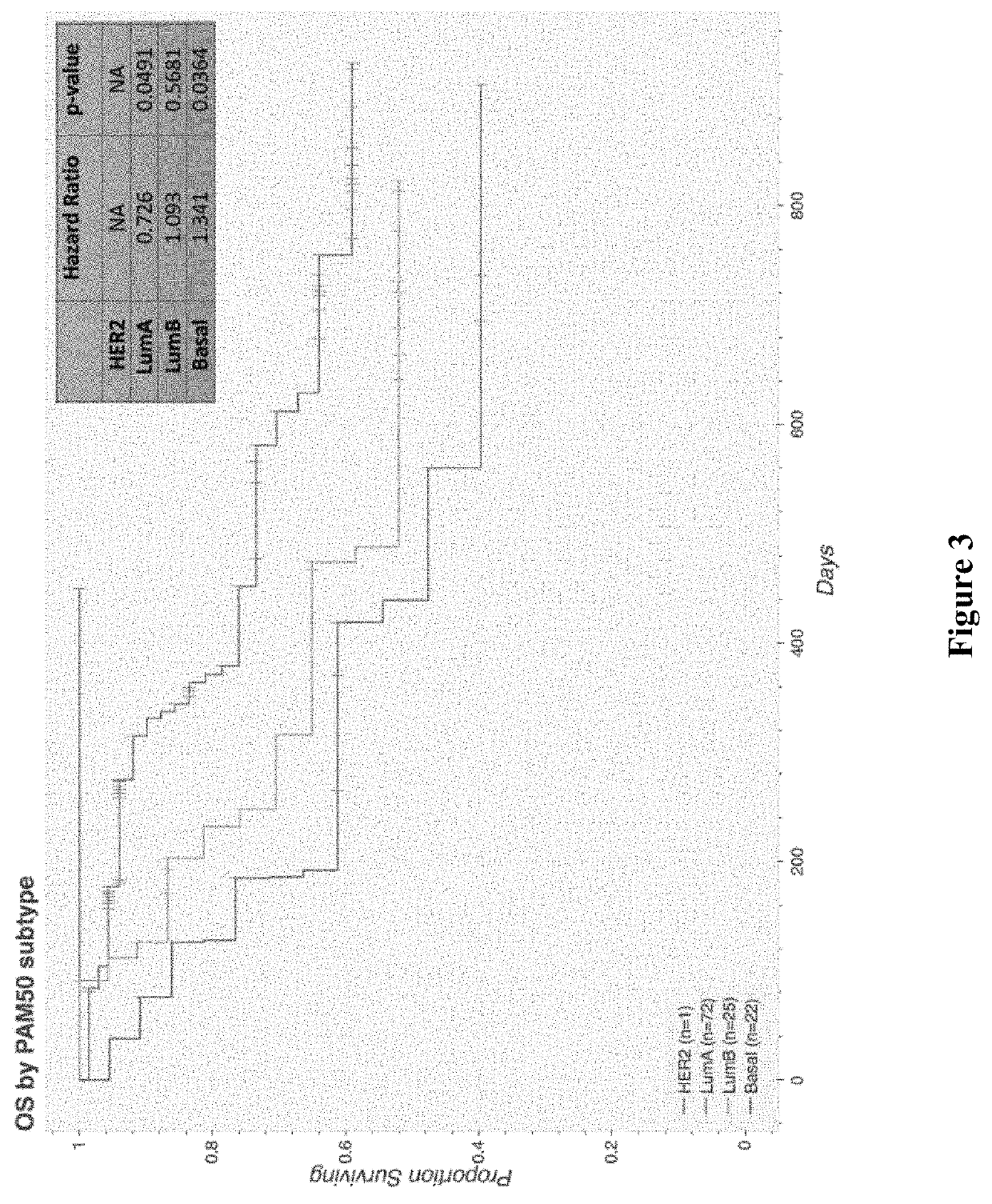
Prognostic indicators of poor outcomes in PRAEGNANT metastatic breast cancer cohort
Transcriptomics data from tumor tissue of patients diagnosed with metastatic breast cancer are clustered and associated with overall survival of the patients. A subset of genes from one of the clusterassociated with poor outcome are used to generate a survival prediction model predicting a survival time based on expression levels of a plurality of genes. Using such generated survival prediction model, a survival time of a patient diagnosed with metastatic breast cancer can be predicted and a treatment regimen can be updated or generated based on the survival time.
Owner:NANTOMICS LLC
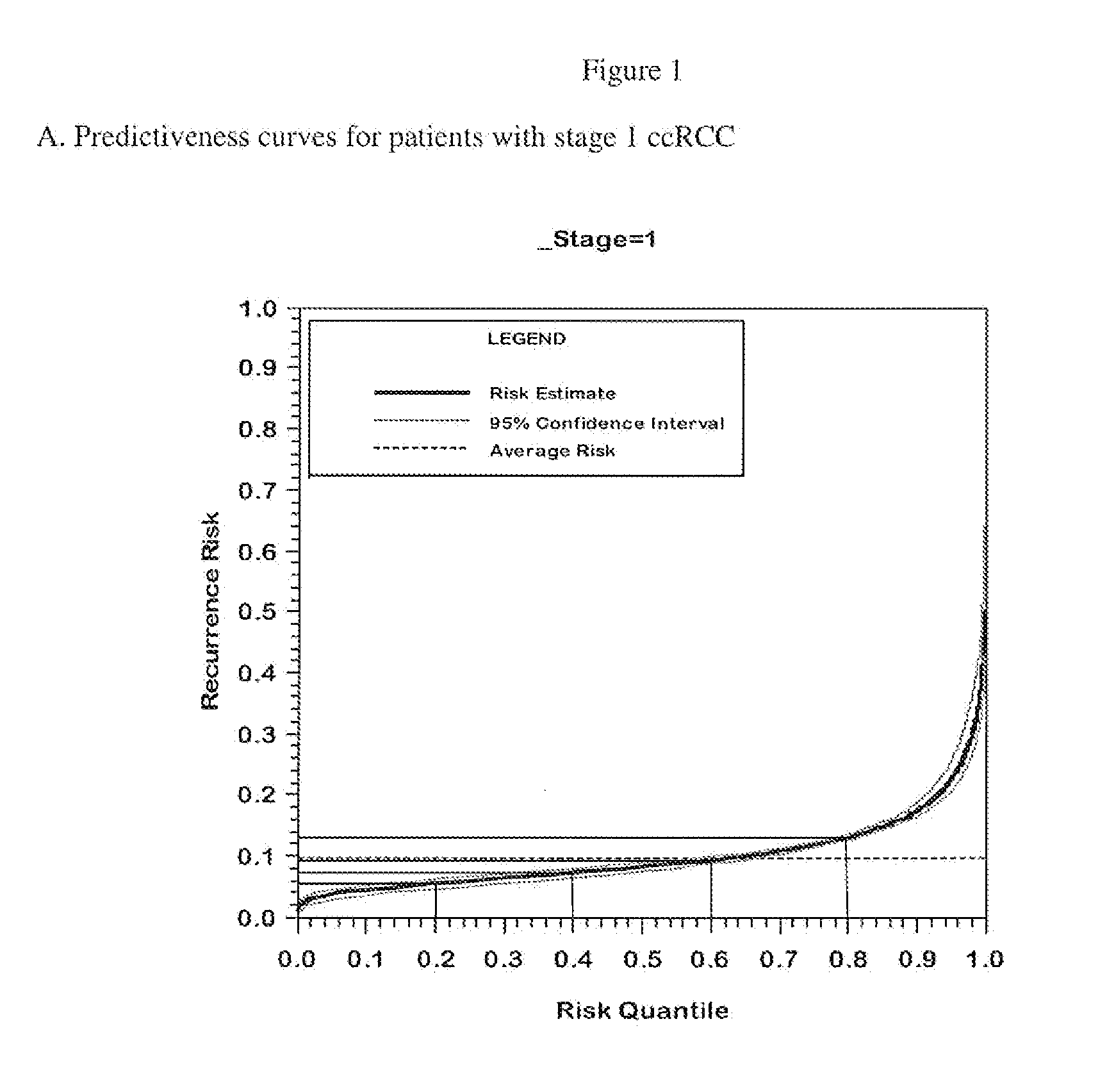
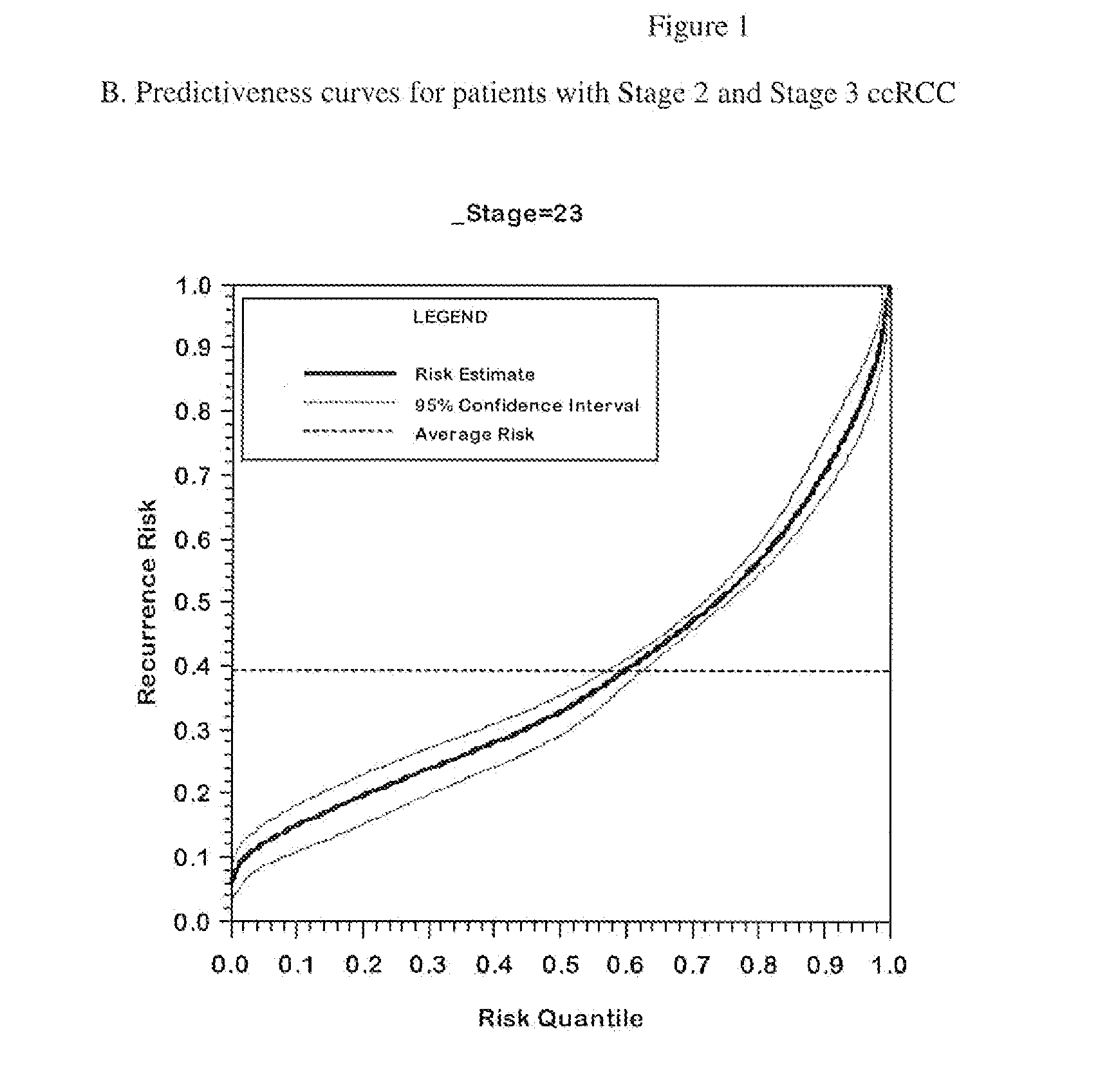
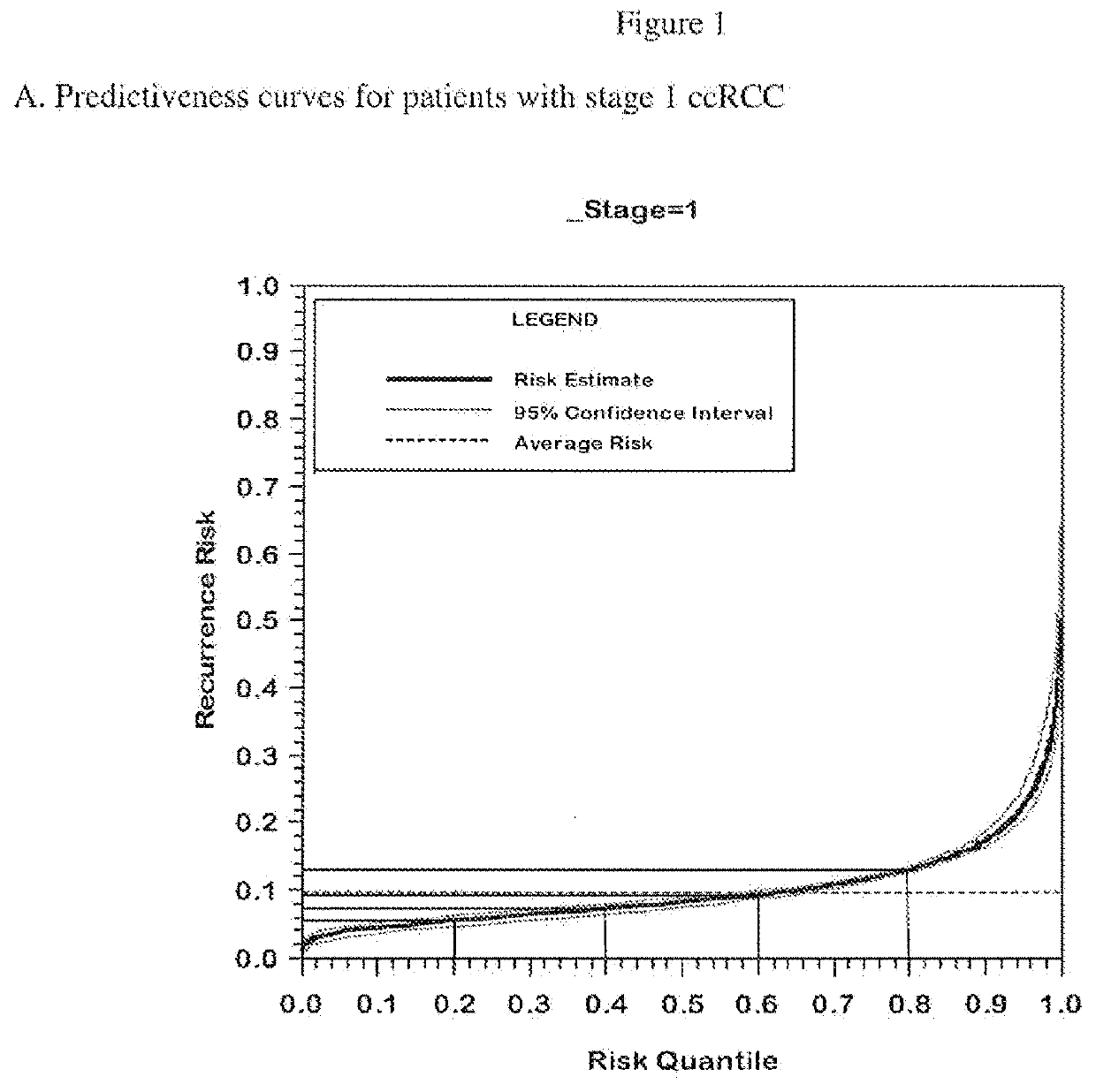
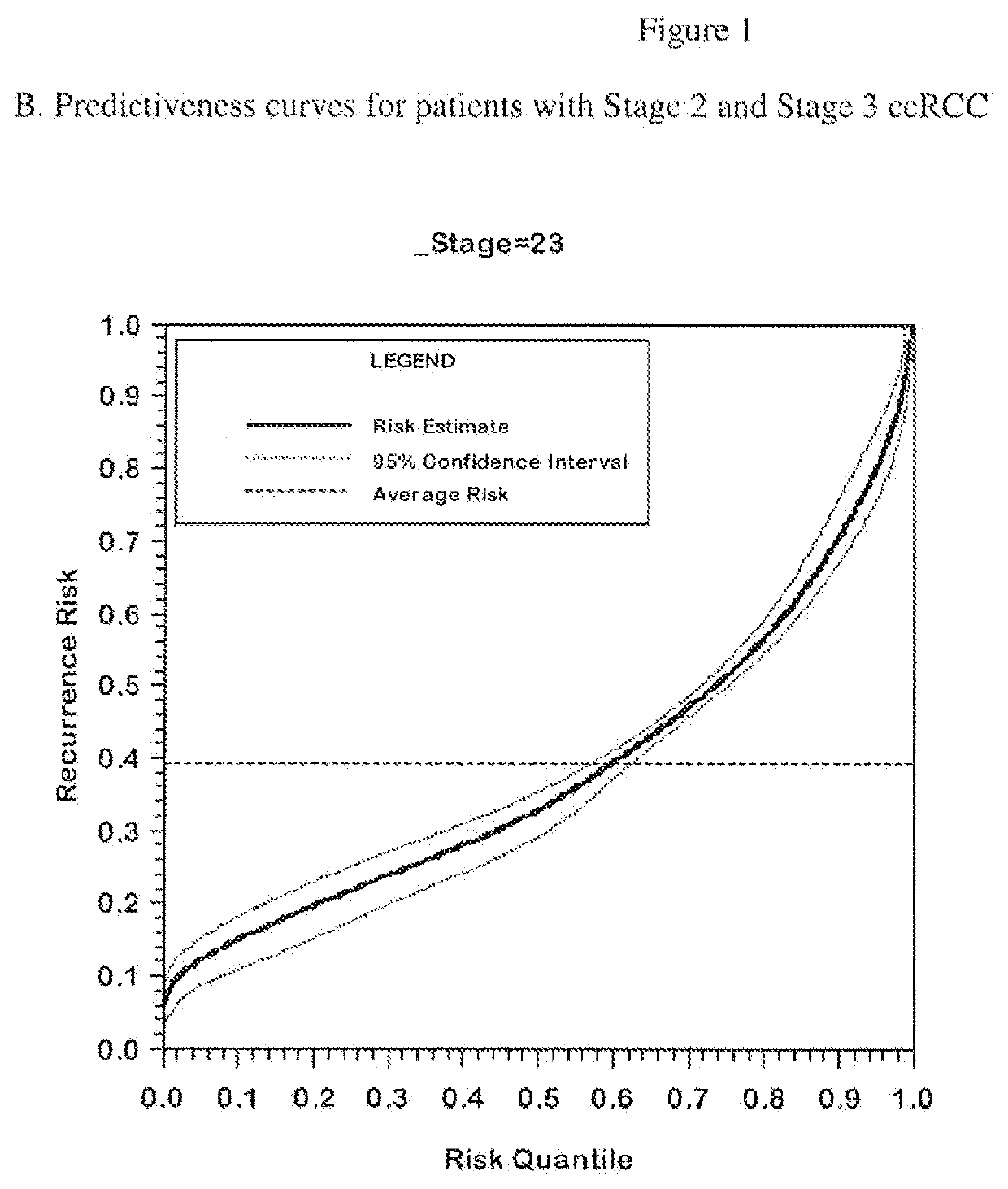
Gene expression profile algorithm for calculating a recurrence score for a patient with kidney cancer
ActiveUS20160048632A1Health-index calculationMicrobiological testing/measurementKidney cancerRecurrence score
Owner:GENOMIC HEALTH INC
Gene expression profile feature selection method based on MFA score and redundancy exclusion
InactiveCN104200135AReduce complexityImprove classification accuracySpecial data processing applicationsCorrelation coefficientScoring algorithm
The invention relates to a gene expression profile feature selection method based on MFA score and redundancy exclusion. Feature selection and classification of oncogene expression profiles facilitate the early diagnosis of tumors, and the causes of the tumors are explained from the angle of gene expression. Firstly, a class inter neighbor matrix Wb and a class inter neighbor matrix Ww are structured through an MFA score algorithm, consequently a class inter Laplacian matrix Lb and a class inter Laplacian matrix Lw are obtained, and lastly genes are ranked. For the feature that gene expression data are high in redundancy, the correlation among the genes is judged through Pearson correlation coefficients, the high correlation genes, namely redundancy genes, are excluded, and finally a gene subset is obtained. The gene expression profile feature selection method based on the MFA score and redundancy exclusion is suitable for training samples distributed in any space, the number of dimensions of features is further reduced by excluding the redundancy genes, the complexity of the algorithm is small, and the high classification accuracy is obtained in experiments.
Owner:BEIJING UNIV OF TECH
A PLS-based multi-disturbance integrated gene selection and tumor-specific gene subset recognition method
ActiveCN109033747AStrong discriminationRich knowledgeSpecial data processing applicationsGene selectionTumor specific
A PLS-based multi-disturbance integrated gene selection and tumor-specific gene subset recognition method features that different disturbance mechanisms are introduced to analyze gene selection of multi-disturbance integrate according to characteristics of tumor microarray data. Based on this framework, a new integrated gene selection method based on PLS is developed by using PLS polygenic metrics. On the one hand, the method of the invention is based on the whole effect of the subset, and can quickly identify the genes with differential expression, and also can identify the genes with weak differential expression signal; on the other hand, the method of the invention is based on a multiple disturbance mechanism, and can identify a series of different subsets of genes with small length andstrong discrimination ability. Therefore, the method of the invention can identify a series of different gene subsets and weakly differentially expressed genes, through which the specific expressionpattern of tumor genes can be more comprehensively recognized.
Owner:FUQING BRANCH OF FUJIAN NORMAL UNIV
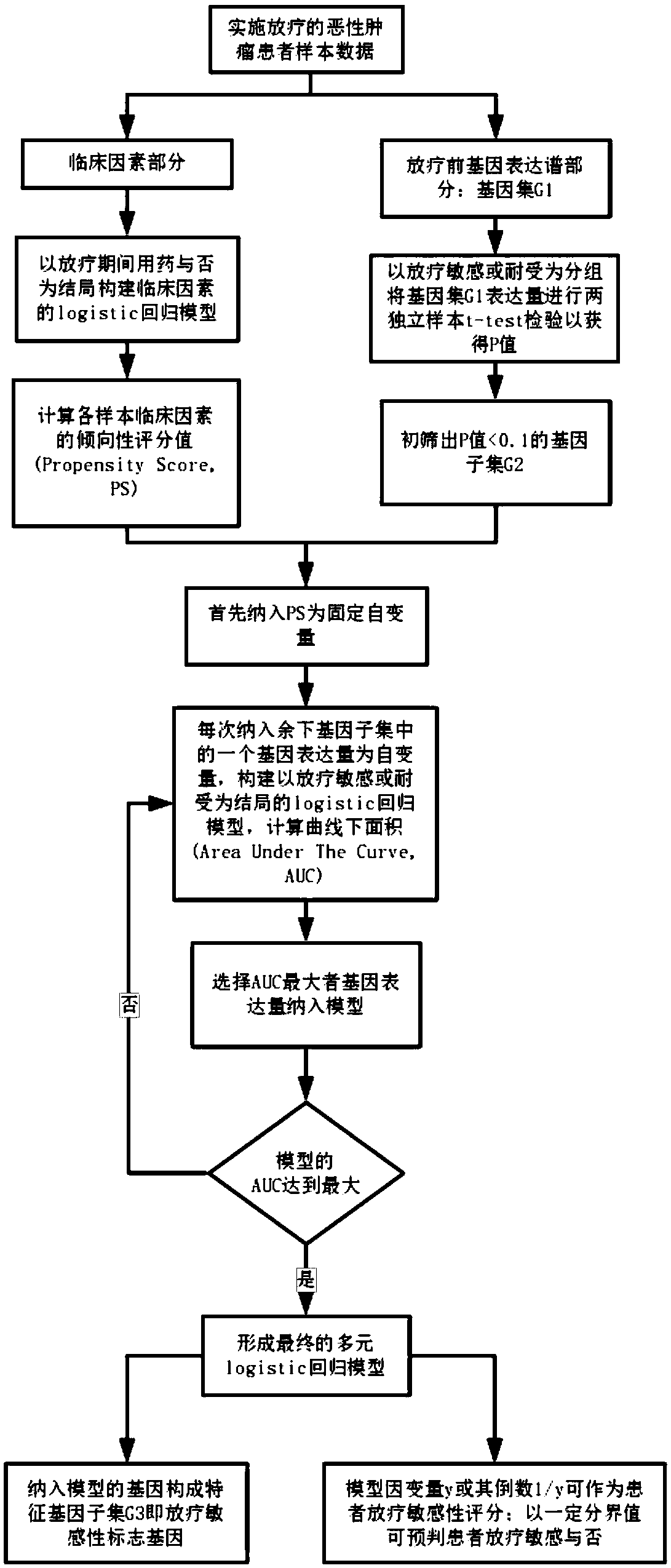
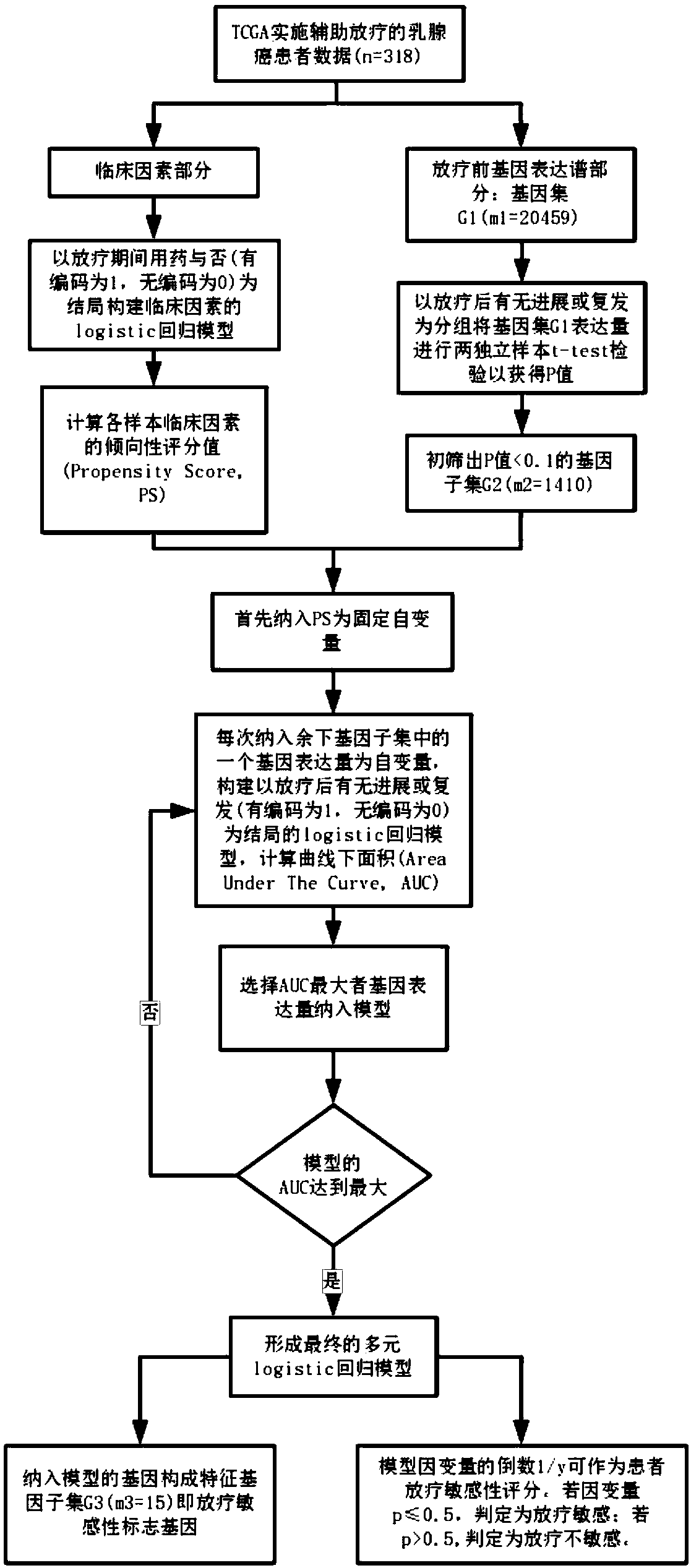
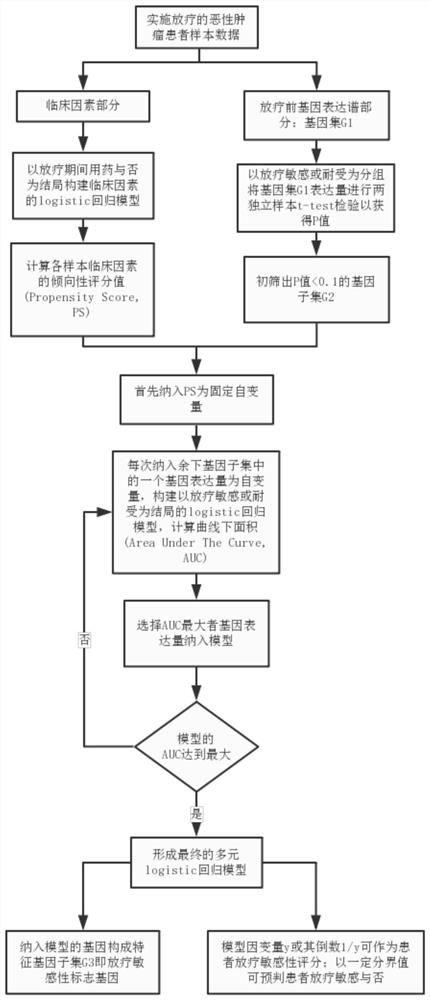
Radiotherapy sensitive marker gene screening method for balancing clinical confounding factors
ActiveCN109346181AImprove predictive performanceReliable methodMedical data miningHealth-index calculationMedicineScreening method
The method is characterized in that data of patients with malignant tumors are divided into clinical factors and pre-radiation gene expression profile data, ie, gene sets; a regression model of the clinical factors is constructed based on whether drugs are used or not for clinical factor data, and the clinical factors PS of each sample are calculated; t-test tests of two independent samples are performed based on radiotherapy sensitivity or tolerance groups, and a subset of differentially expressed genes with P less than 0.1 is screened out; the PS is included as a fixed independent variable,the expression amount of each gene of remaining genes included in the gene subset is taken as an independent variable, a regression model taking the radiotherapy sensitivity or tolerance as the outcome is constructed, the area AUC under a curve is calculated, and the gene expression amount with the largest AUC is selected and included to the regression model till loop termination is realized to form a multi-element regression model when the AUC of the regression model reaches the largest; the genes included in the multi-element regression model constitute radiosensitive sensitivity marker genes; based on the multi-element regression model, a dependent variable or its reciprocal is taken as the patient radiotherapy sensitivity score to predict whether a patient is sensitive to radiotherapywith a certain cutoff value.
Owner:SHANGHAI CHANGHAI HOSPITAL
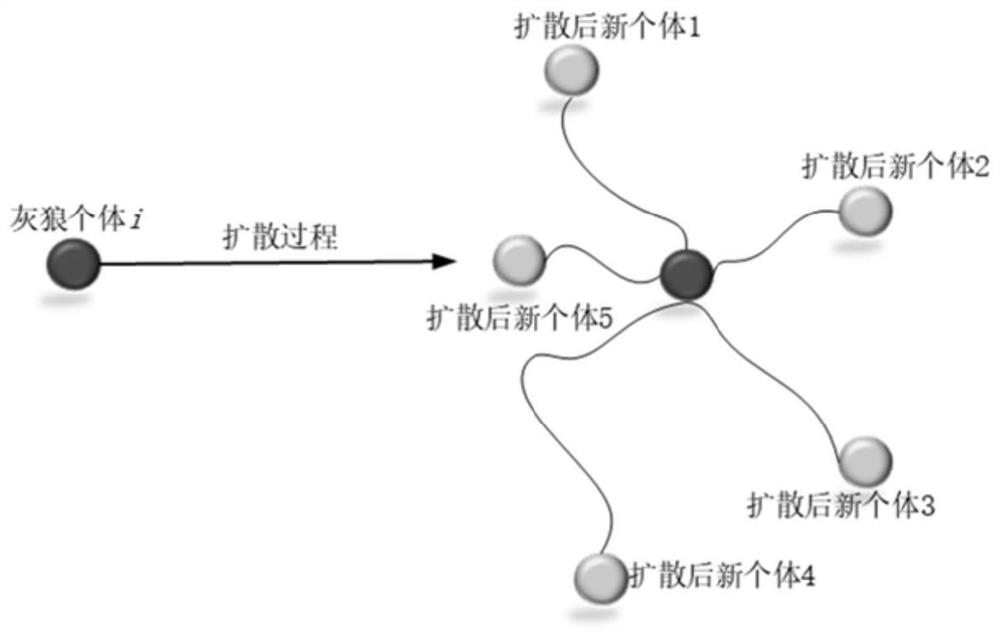
Grey wolf optimization algorithm gene selection method based on diffusion and chaotic local search
PendingCN112116952AOvercoming the problem of low classification accuracyDiversity guaranteedBiostatisticsCharacter and pattern recognitionData setGene selection
The invention provides a grey wolf optimization algorithm gene selection method based on diffusion and chaotic local search, which comprises the following steps of generating a training set and a testset according to a gene data set acquired from a public website, performing global search on the training set and the test set of the gene data set by using a preset grey wolf optimization algorithm,and determining a feature subset of the gene data set by combining fitness functions defined by the training set and the test set in the preset grey wolf optimization algorithm based on a KNN classifier, selecting a globally optimal solution from the determined feature subset of the gene data set by using a preset diffusion strategy, and further performing chaotic local search on the selected globally optimal solution to obtain an optimal training set and an optimal test set of the gene data set as final optimal gene subsets, and outputting the finally obtained optimal gene subset. By implementing the method, the problem of low gene data classification precision of the gene subset obtained in the prior art can be effectively solved, and the optimal gene subset is found.
Owner:WENZHOU UNIVERSITY
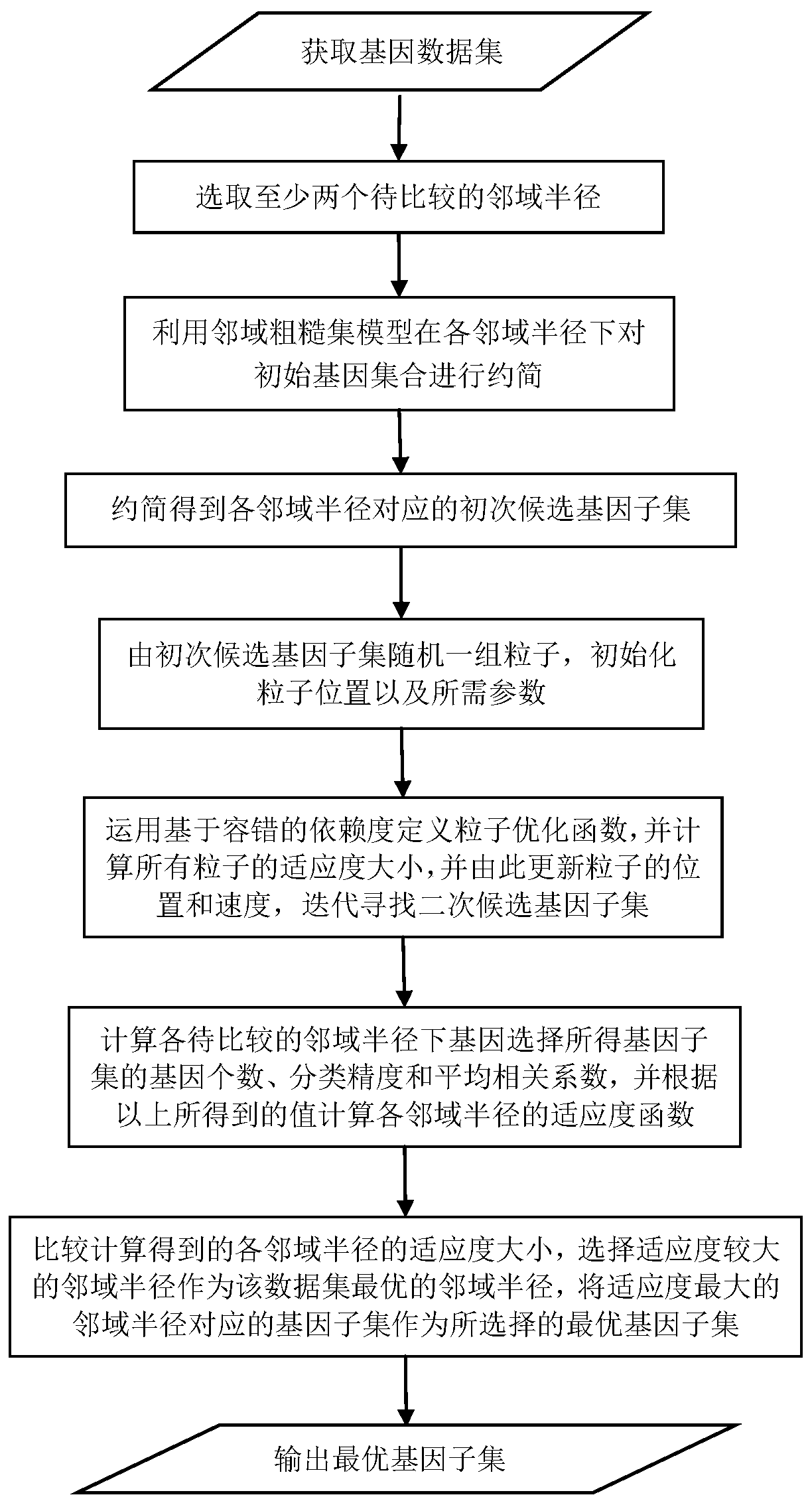
Gene selecting method in consideration of gene correlation and device
ActiveCN110211638AImprove classification accuracyReduce redundancySequence analysisInstrumentsCorrelation coefficientData treatment
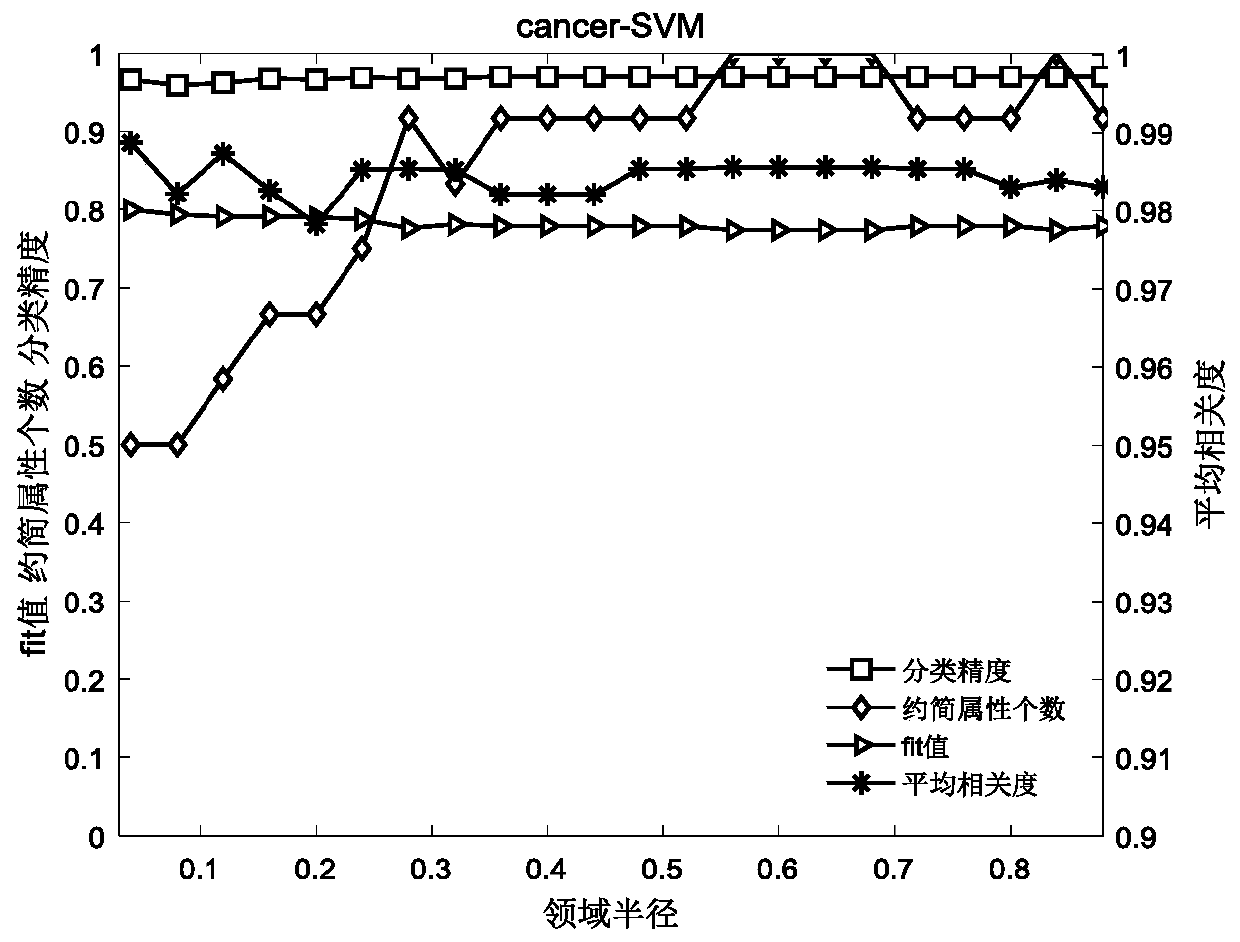
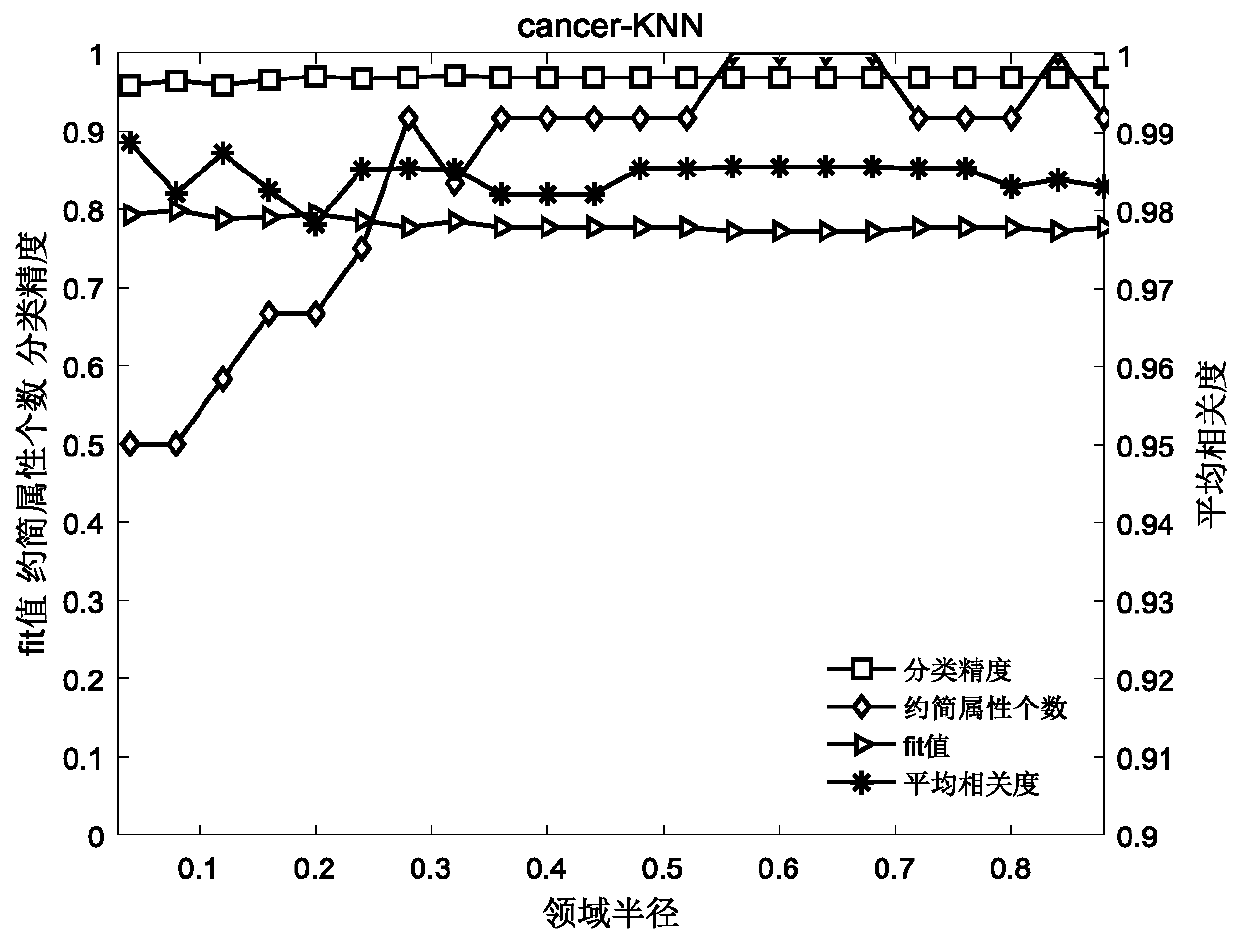
The invention relates to a gene selecting method in consideration of gene correlation and a device, wherein the method and the device belong to the field of data processing technology. Through introducing the correlation between rank correlation coefficient metering genes, an average correlation coefficient between the genes is calculated. By means of the average correlation coefficient, a fitnessfunction for evaluating a neighborhood radius value is improved so that the defined fitness function considers classification precision and the number of genes in result evaluation, and further considers the correlation between the genes. Therefore the gene subset which is selected by the improved fitness function has advantages of low redundancy, low dimension, high precision, etc.
Owner:HENAN NORMAL UNIV
Rapid encapsulation type gene selection method based on maximum correlation and minimum redundancy
InactiveCN110534155AHigh speedReduce the number of times to evaluate a subset of genesBiostatisticsHybridisationGene selectionCandidate Gene Association Study
The invention discloses a rapid encapsulation type gene selection method based on maximum correlation and minimum redundancy, and the method comprises the following steps: 1, searching a gene with themaximum correlation degree with a category label in a gene vector group by using a correlation method, and adding the gene into a candidate gene subset; 2, searching a gene with the maximum correlation redundancy in the gene vector group by utilizing a maximum correlation minimum redundancy method, and adding the gene into the candidate gene subset; 3, judging whether the classification precisionof the two candidate gene subsets before and after updating the candidate gene subsets is reduced or not by utilizing a ten-fold cross validation method; 4, if the classification precision is reduced, outputting the candidate gene subset before updating, otherwise, repeating the step 2. According to the method, the high-quality gene subsets can be obtained, and meanwhile, the time complexity of acommon packaging method is remarkably reduced, so that the gene subsets have good time performance in obtaining, and the obtained gene subsets have good classification performance.
Owner:HEFEI UNIV OF TECH
Gene selection method based on feature discrimination and independence
InactiveCN105938523BImprove efficiencyReduce data dimensionalityBiostatisticsHybridisationDiseaseTime complexity
The invention relates to a feature selection method and application based on feature identification degree and independence. The method comprises following steps: calculating the importance degree of each feature by measuring inter-class distinguished ability with feature identification degree and measuring correlational relationship between features with feature independence and sequencing in a descending order; and selecting top k features with the importance higher than those of others to form a feature subset with high class-discrimination performance. Differently-expressed gene subsets selected in application of oncogene expression profile data obtain fine time and class discrimination performance. The feature selection method and application based on feature identification degree and independence have following beneficial effects: easy calculations can be made; time complexity is reduced; selection efficiency runs high; and a good reference is provided for clinical diagnoses and judgments of tumors and other diseases.
Owner:SHAANXI NORMAL UNIV
Tumor key gene identification method based on multi-objective particle swarm algorithm of preference grid and Levi flight
PendingCN110782950AImprove decision-making efficiencyFast convergenceBiostatisticsProteomicsLearning machineData set
The invention discloses a tumor key gene identification method based on a multi-objective particle swarm algorithm of a preference grid and Levi flight. The method comprises the steps that a classification information index is used to filter an original gene expression profile data set to acquire a primary gene pool; the GCS value of the gene category sensitivity information of each gene in the initial gene pool is calculated, and then particles are encoded by the GCS value; the classification accuracy of gene subsets on an extreme learning machine ELM and the size of the gene subsets are targeted to build a multi-objective optimization model; and the final gene subset is searched through the established multi-objective model, and then the key genes of a tumor are identified. In terms of the multi-objective optimization model, the method provided by the invention can quickly and efficiently identify a small number of key gene subsets with good classification performance in the primarygene pool through the multi-objective model.
Owner:JIANGSU UNIV
Gene expression profile algorithm for calculating a recurrence score for a patient with kidney cancer
ActiveUS10181008B2Health-index calculationMicrobiological testing/measurementRecurrence scoreKidney cancer
The present invention provides algorithm-based molecular assays that involve measurement of expression levels of genes from a biological sample obtained from a kidney cancer patient. The present invention also provides methods of obtaining a quantitative score for a patient with kidney cancer based on measurement of expression levels of genes from a biological sample obtained from a kidney cancer patient. The genes may be grouped into functional gene subsets for calculating the quantitative score and the gene subsets may be weighted according to their contribution to cancer recurrence.
Owner:GENOMIC HEALTH INC
A cross-chip platform gene expression data integration method
ActiveCN103745137BEliminate experimental batch effectsImprove performanceSpecial data processing applicationsGene expression matrixLeast squares
The invention belongs to the technical field of biological information. The invention provides a method for integrating gene expression data by crossing a plurality of different chip platforms; the method comprises the following steps: the standard preprocessing is implemented for the gene expression profile of the plurality of chip platforms; the common gene expression data in the different chip platforms is merged; genes are divided into k subsets according to the expression similarity of the genes on the plurality of chip platforms; the expression linear relation exps1=as*exps2+bs of the different chip platforms in every gene subset can be calculated by the least square method; the gene expression values of the different chip platforms are standardized into the same change range by using the formula exp1=X*A.*exp2+X*B to obtain standard gene expression matrixes, wherein the implications of symbols are defined as the specification.
Owner:艾吉泰康(嘉兴)生物科技有限公司
Gene selection method and device based on fault tolerance
ActiveCN110210552AFault-tolerantFast selectionCharacter and pattern recognitionArtificial lifeFault toleranceData set
The invention relates to a gene selection method and device based on fault tolerance, and belongs to the technical field of data processing, and the method comprises the steps: carrying out the reduction of a gene data set through employing a neighborhood rough set algorithm based on fault tolerance and a discrete particle swarm optimization algorithm, and obtaining an optimal gene subset. In thereduction process, the dependency degree based on the fault tolerance is introduced to evaluate the dependency of the decision attribute set on the condition attribute set, so that a fault tolerance-based dependency degree and neighborhood granularity mixed measurement method is provided, and a fitness function in a discrete particle swarm optimization algorithm, namely a particle fitness functionbased on the dependency degree based on the fault tolerance, is constructed. According to the gene selection method, the attribute set is evaluated from two different perspectives of dependency and neighborhood granularity, the problem of zero fault tolerance of a neighborhood rough set is solved, particles are guided to quickly search an optimal gene subset, and the classification precision of the obtained optimal gene subset is high.
Owner:HENAN NORMAL UNIV
A method for identifying key tumor genes based on particle swarm optimization and scoring criteria
ActiveCN106951728BAccurate identificationEffectively deleteProteomicsGenomicsGene recognitionParticle swarm algorithm
Owner:JIANGSU UNIV
Tumor-specific gene identification method based on pls multi-perturbation integrated gene selection
ActiveCN109033747BRich knowledgeShorten the lengthProteomicsGenomicsGene selectionDifferentially expressed genes
The present invention relates to a tumor-specific gene identification method based on PLS multi-perturbation integrated gene selection. Aiming at the characteristics of tumor microarray data, different perturbation mechanisms are introduced to provide an analysis framework for multi-perturbation integrated gene selection; using PLS multi-gene measurement method, a new PLS-based integrated gene selection method was developed under this framework. On the one hand, the method of the present invention is based on the overall effect of subsets, and can quickly identify genes with differential expression, and at the same time identify genes with weak differential expression signals; on the other hand, the method of the present invention is based on multiple perturbation mechanisms , which can identify a series of different gene subsets with small length and strong discriminative ability. Therefore, the method of the present invention can identify a series of different gene subsets and weakly differentially expressed genes, through which the specific expression patterns of tumor genes can be more comprehensively understood.
Owner:FUQING BRANCH OF FUJIAN NORMAL UNIV
Prognostic indicators of poor outcomes in pregnant metastatic breast cancer cohort
Transcriptomics data from tumor tissue of patients diagnosed with metastatic breast cancer are clustered and associated with overall survival of the patients. A subset of genes from one of the cluster associated with poor outcome are used to generate a survival prediction model predicting a survival time based on expression levels of a plurality of genes. Using such generated survival prediction model, a survival time of a patient diagnosed with metastatic breast cancer can be predicted and a treatment regimen can be updated or generated based on the survival time.
Owner:NANTOMICS LLC
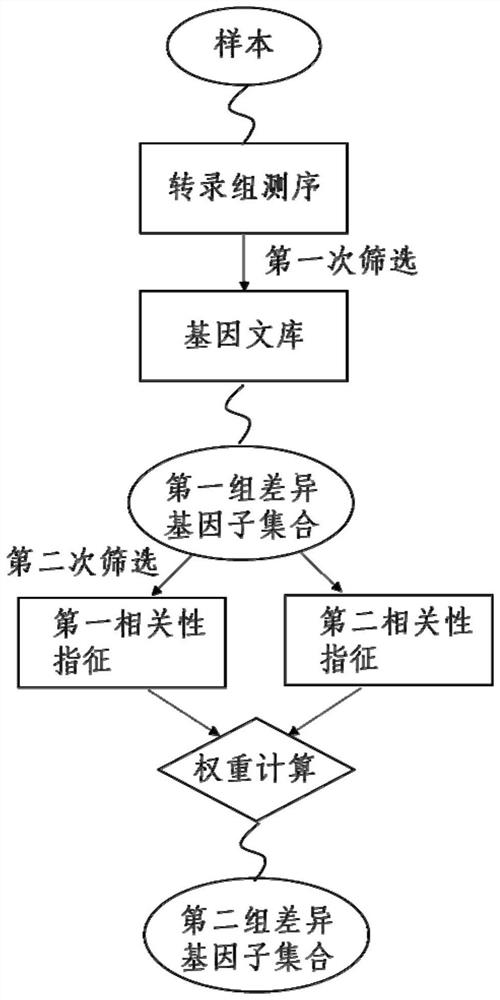
System and method for detecting clinical results of patients with hepatocellular carcinoma by HBV (Hepatitis B Virus)
The invention relates to a method for detecting a clinical result of a hepatocellular carcinoma patient by HBV (Hepatitis B Virus). Qualitative / quantitative evaluation or / and comparison is carried out on the basis of known functional annotation nodes on physiological metabolic pathways for diagnosing liver cancer types and obtained by transcriptome sequencing; a plurality of genes irrelevant to or redundant to the biological function of the diagnosed liver cancer type are excluded after function enrichment clustering to obtain a first group of differential gene subsets after first screening; the voucher used as the comparison opposite direction for excluding the gene or the gene subset in the first screening is the first voucher. The first credential can exist in a first gene library, and is characterized by having no contribution to the generation of a target disease or having weak correlation with physiological pathways involved in the generation of disease phenotypes.
Owner:林根琼
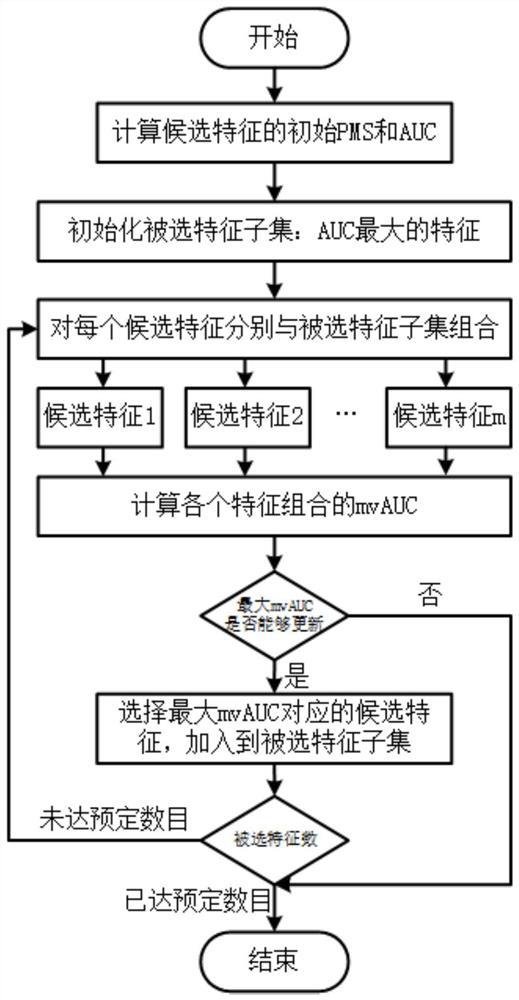
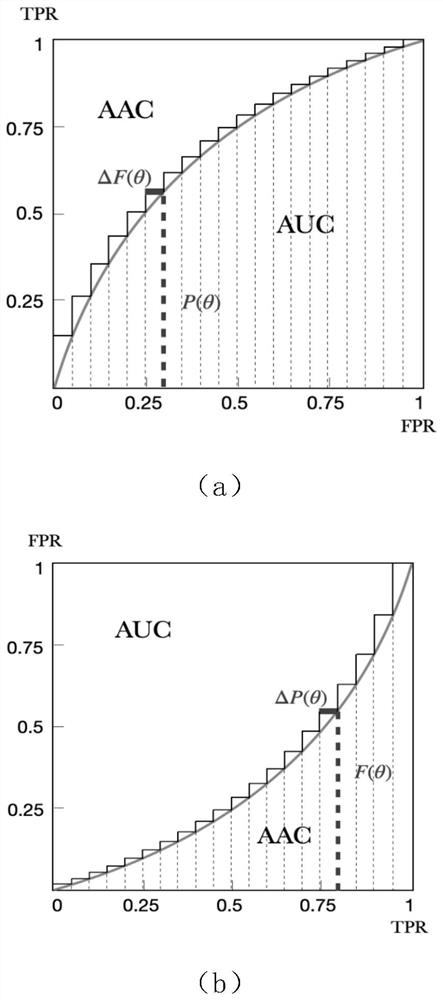
A method for selecting complementary differentially expressed genes based on mvauc
The present invention proposes a feature selection method based on multivariate AUC, which selects the most complementary gene subset from cancer differential expression data to maximize global classification performance. The present invention first proposes a new angle of AUC calculation based on the possible misclassification set of features; then, for a feature set, determine its common possible misclassification set and calculate the new AUC after each feature combination; the new AUC of a feature is compared with the original The difference in AUC shows the complementary effect of other features in the combined feature set on the classification ability of this feature. Finally, mvAUC is calculated based on the new AUC after feature combination, and the candidate features that maximize the current mvAUC are incrementally selected to join the selected feature subset. The method of the present invention has the advantage of being able to directly evaluate the global class discrimination ability of the selected feature subset, and does not need to calculate redundant information between candidate features and each selected feature in pairs.
Owner:NANKAI UNIV
Methods for identifying an increased likelihood of recurrence of breast cancer
Owner:UNIV OF LOUISVILLE RES FOUND INC
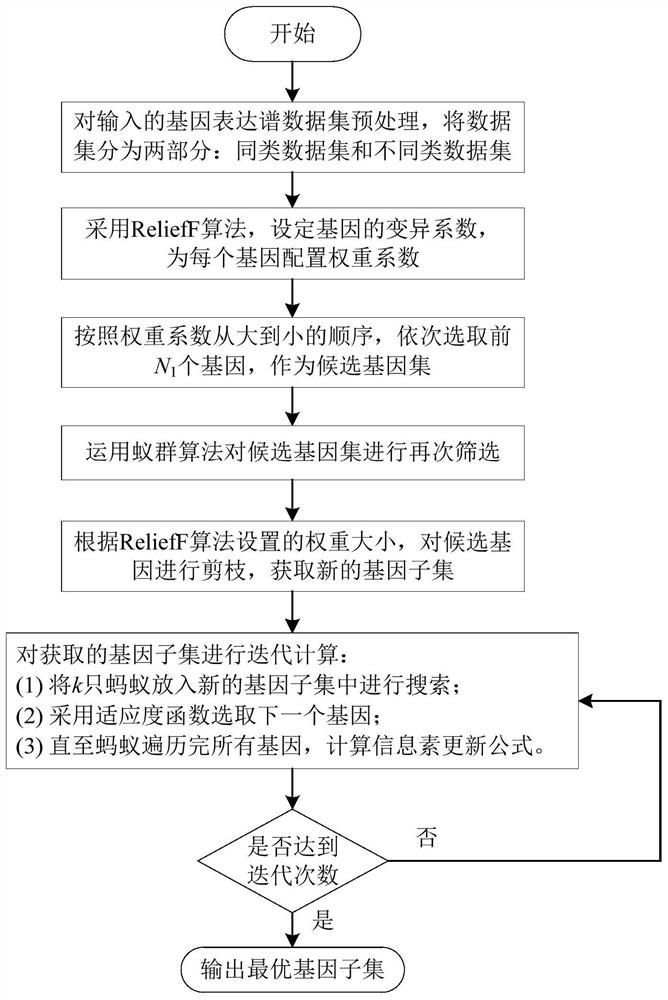
Gene classification method and device
ActiveCN108985010BBiostatisticsCharacter and pattern recognitionCandidate Gene Association StudyComputer science
The invention relates to a gene classification method and device. According to the variation coefficient of the set gene, an attribute weighting algorithm is used to configure a weight coefficient for each gene in the gene sample, and the top N are selected in sequence according to the order of the weight coefficient from large to small. 1 genes as the candidate gene set; use ant colony algorithm to select the optimal gene subset in the candidate gene set, and use the gene subset to classify genes. The present invention first adopts the attribute weighting algorithm to preliminarily screen the candidate gene set, and then screens the candidate gene set again, and uses the ant colony algorithm to select a gene subset in the candidate gene set, effectively removing redundant or invalid genes.
Owner:HENAN NORMAL UNIV
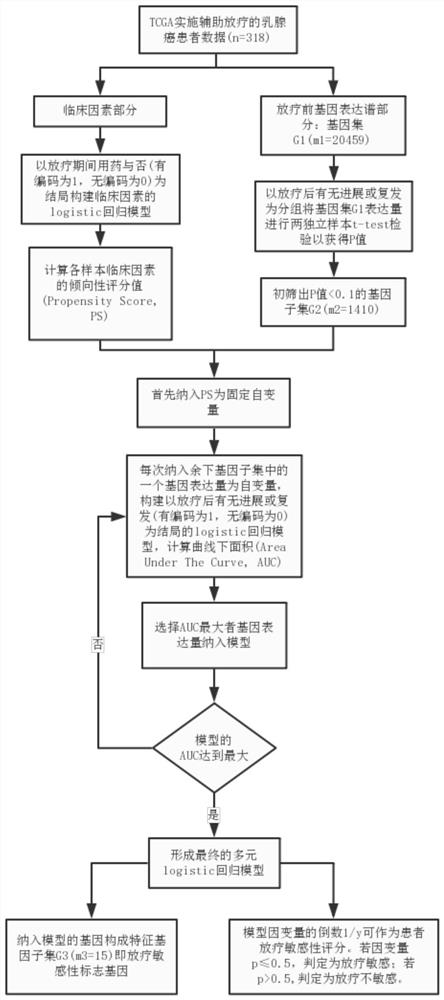
Screening method for radiosensitivity marker genes with balanced clinical confounding factors
ActiveCN109346181BImprove predictive performanceReliable methodMedical data miningHealth-index calculationDifferentially expressed genesIndependent predictor
The present invention divides malignant tumor patient data into clinical factors and pre-radiotherapy gene expression profile data, namely gene sets; constructs a regression model of clinical factors based on whether the clinical factors are used or not, and calculates the clinical factors PS of each sample; Two independent samples t-test test was performed as the group, and the differentially expressed gene subset with P<0.1 was screened out; PS was included as a fixed independent variable, and the expression level of one of the remaining genes in the gene subset was included as an independent variable each time. For the regression model with radiosensitivity or tolerance as the outcome, the area under the curve AUC was calculated, and the gene expression with the largest AUC was selected to be included in the regression model, until the AUC of the regression model reached the maximum and the cycle ended to form a multiple regression model; genes included in the multiple regression model It constitutes a radiosensitivity marker gene; based on the multiple regression model, the dependent variable or its inverse is obtained as the radiotherapy sensitivity score of the patient, and a certain cutoff value is used to predict whether the patient is radiosensitive or not.
Owner:SHANGHAI CHANGHAI HOSPITAL
A tumor identification method based on deterministic particle swarm optimization and support vector machine
ActiveCN106971091BImprove recognition accuracyImprove classification performanceBiostatisticsCharacter and pattern recognitionClassified informationEngineering
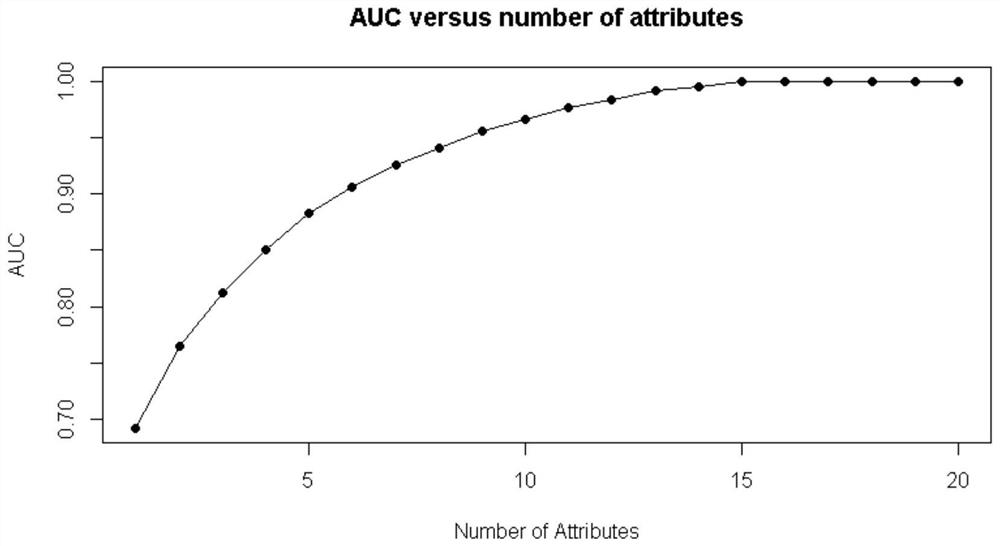
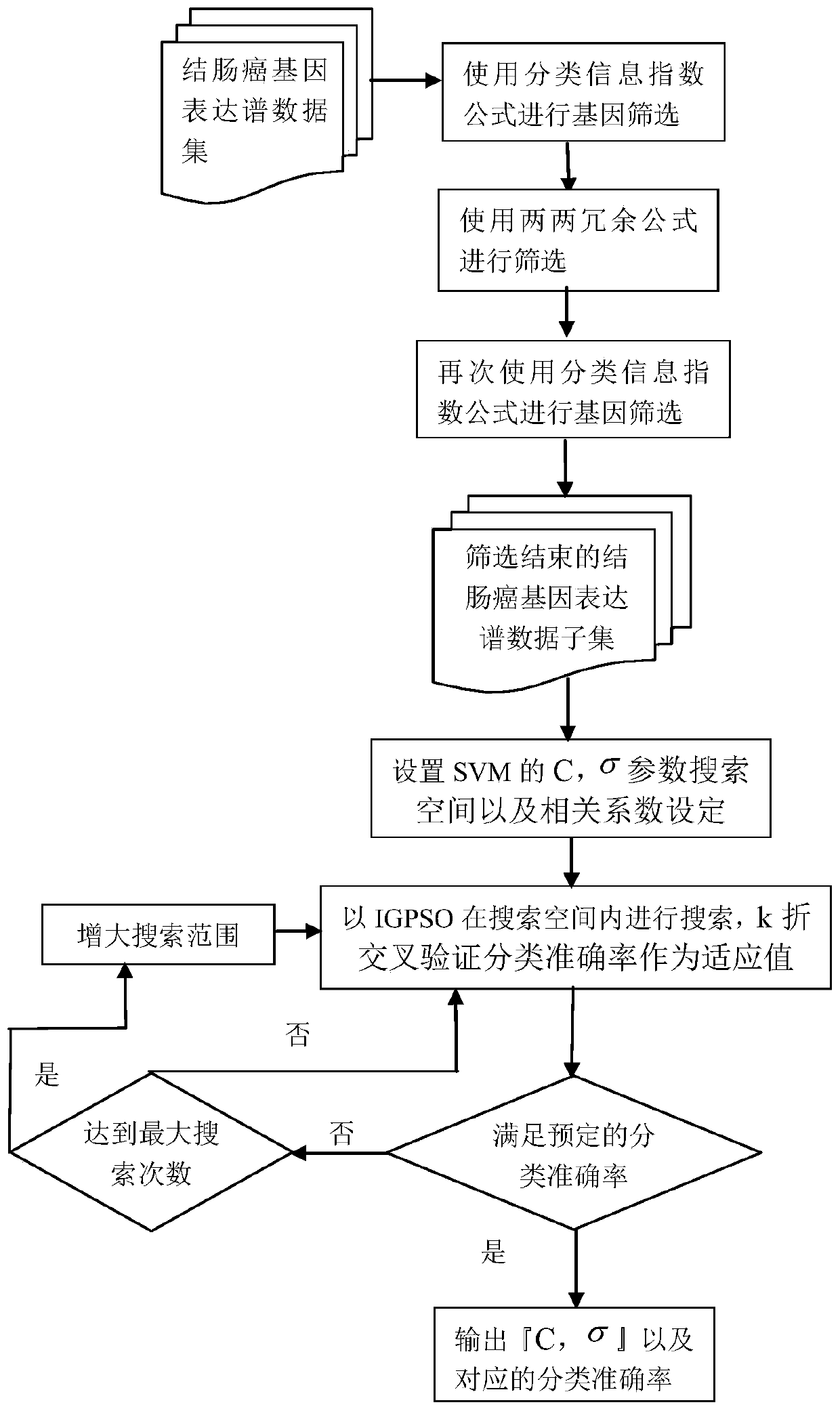
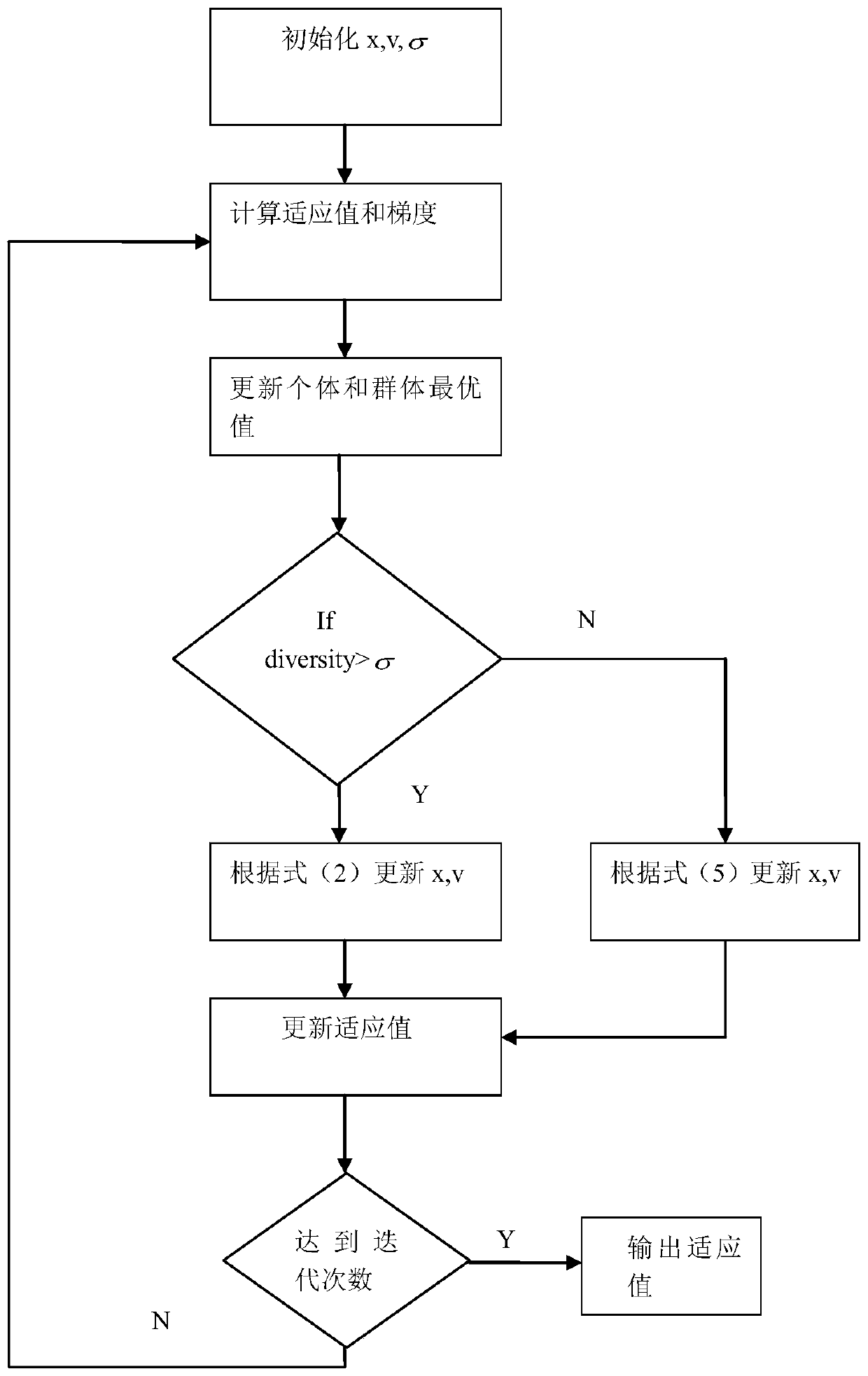
The invention discloses a tumor identification method based on deterministic particle swarm optimization and support vector machine, which includes preprocessing the tumor gene expression spectrum data, using the classification information index method to preliminarily select the information gene on the training set, and then using The pairwise redundancy method removes redundant genes to obtain the candidate gene pool; further uses the classification information index method to obtain key gene subsets on the training set; uses the deterministic particle swarm optimization algorithm on the training set to optimize the parameters of the support vector machine Optimize, and then identify the gene expression profile data of the tumor to be identified. The invention makes full use of the feature that the support vector machine is suitable for small-sample data identification, uses deterministic particle swarm optimization to optimize the support vector machine, further improves the performance of the support vector machine, and thus improves the accuracy of tumor identification.
Owner:JIANGSU UNIV
Gene Expression Profile Algorithm for Calculating a Recurrence Score for a Patient with Kidney Cancer
ActiveUS20210151122A1Health-index calculationMicrobiological testing/measurementKidney cancerRecurrence score
The present invention provides algorithm-based molecular assays that involve measurement of expression levels of genes from a biological sample obtained from a kidney cancer patient. The present invention also provides methods of obtaining a quantitative score for a patient with kidney cancer based on measurement of expression levels of genes from a biological sample obtained from a kidney cancer patient. The genes may be grouped into functional gene subsets for calculating the quantitative score and the gene subsets may be weighted according to their contribution to cancer recurrence.
Owner:GENOMIC HEALTH INC
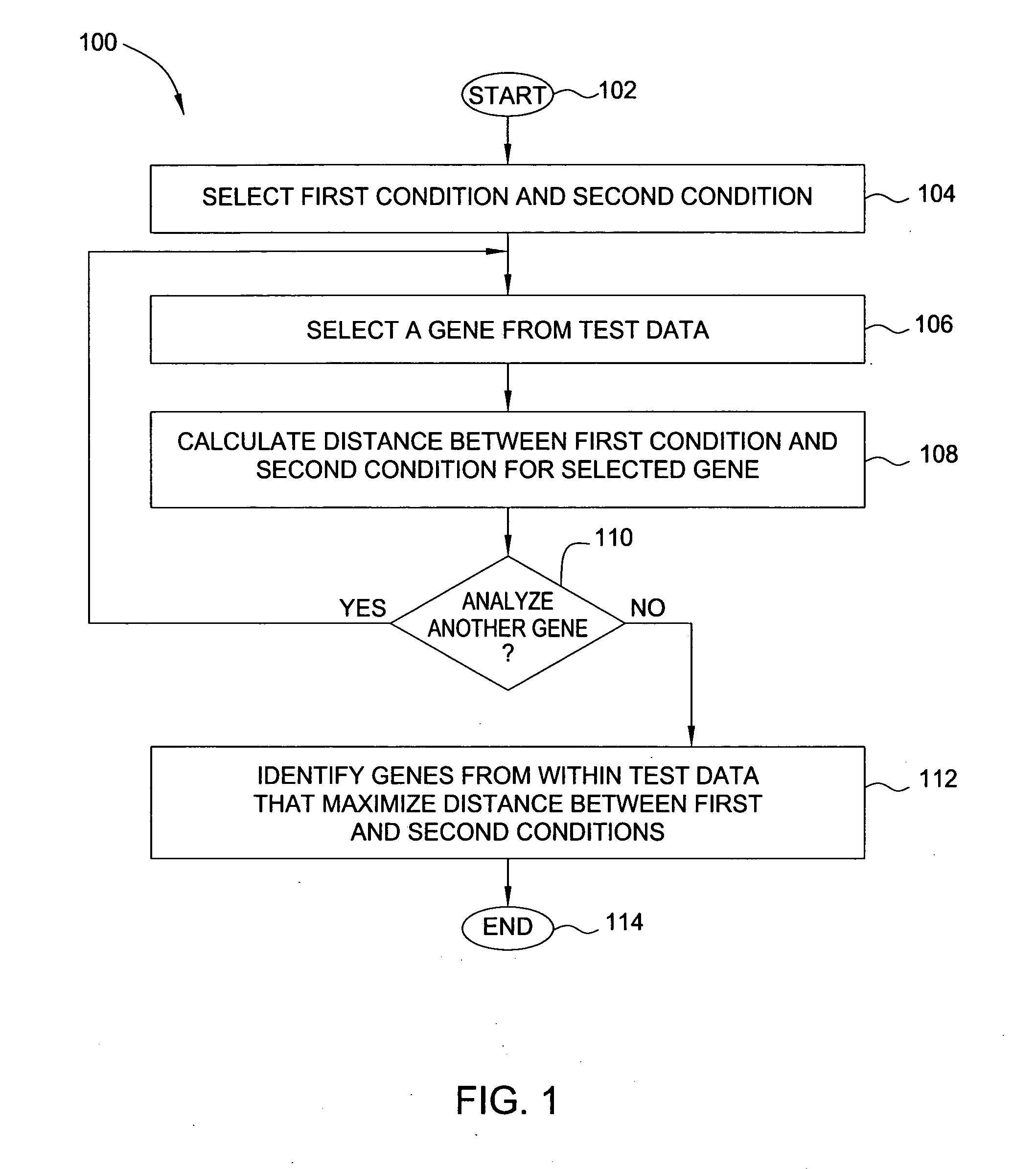
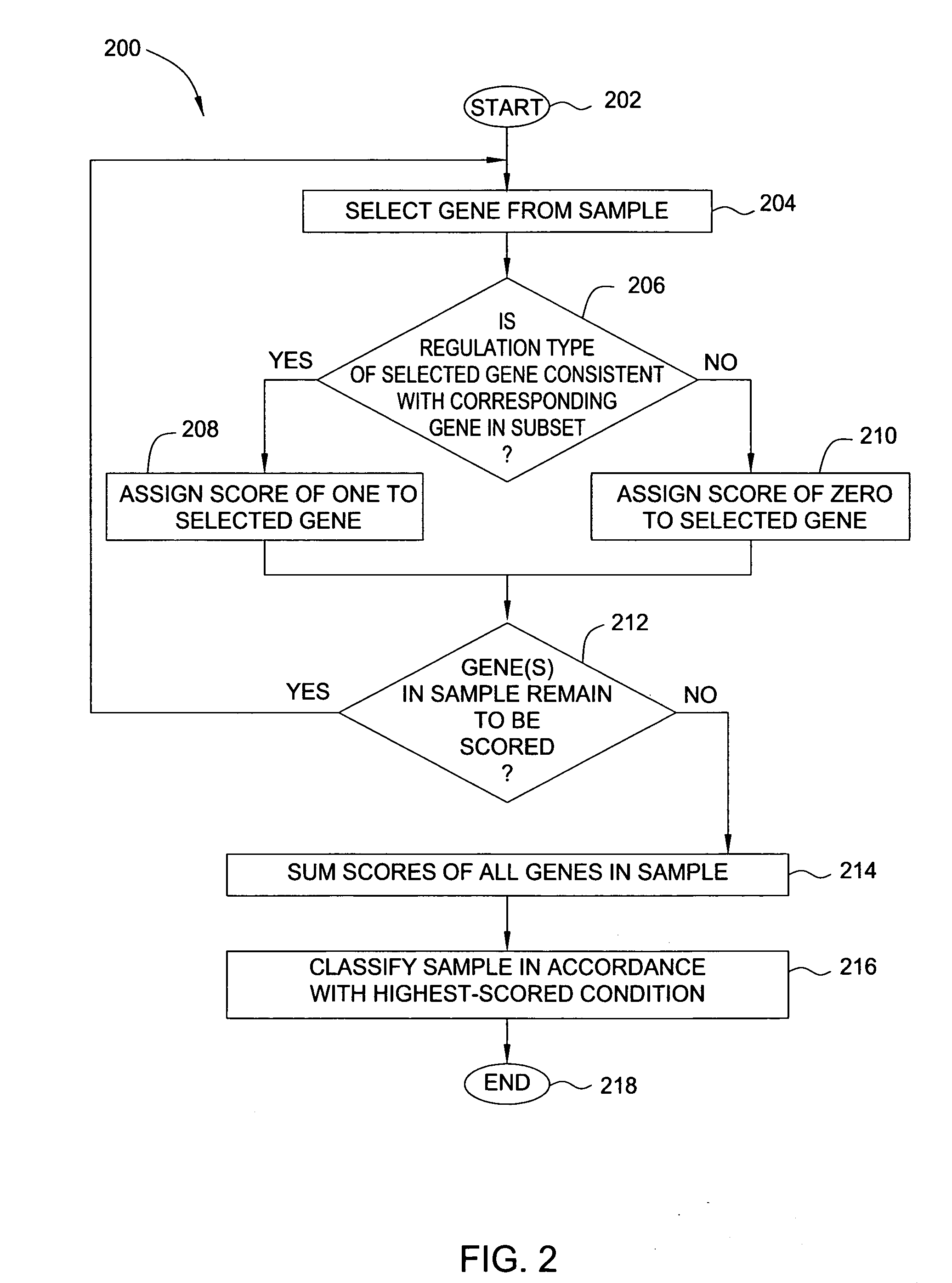
Method and apparatus for classifying nucleic acid responses to infectious agents
InactiveUS20060183143A1DistanceReliable conditionsMicrobiological testing/measurementBiostatisticsDiagnoses diseasesInfectious agent
In one embodiment, the present invention is a method and apparatus for classifying nucleic acid responses to infectious agents. In one embodiment, a method for selecting genes to be analyzed to determine exposure to a condition (from among a plurality of potential conditions) includes determining, for each gene in a set of test data that includes genes and corresponding expression patterns for exposure to given conditions, a distance between each pair of conditions. A subset of genes from within the set of test data is then identified for which the distance between each pair of conditions is maximized. In this way, the number of genes whose expression patterns must be analyzed in order to reliably diagnose a condition is minimized.
Owner:SRI INTERNATIONAL
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com