Tumor key gene identification method based on particle swarm optimization and marking criterion
A particle swarm optimization, key gene technology, applied in the fields of genomics, special data processing applications, instruments, etc., can solve the problems of lack of interpretability, the classification performance needs to be improved, and the subset of key tumor genes is large, so as to improve the tumor subgroup. The effect of type recognition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
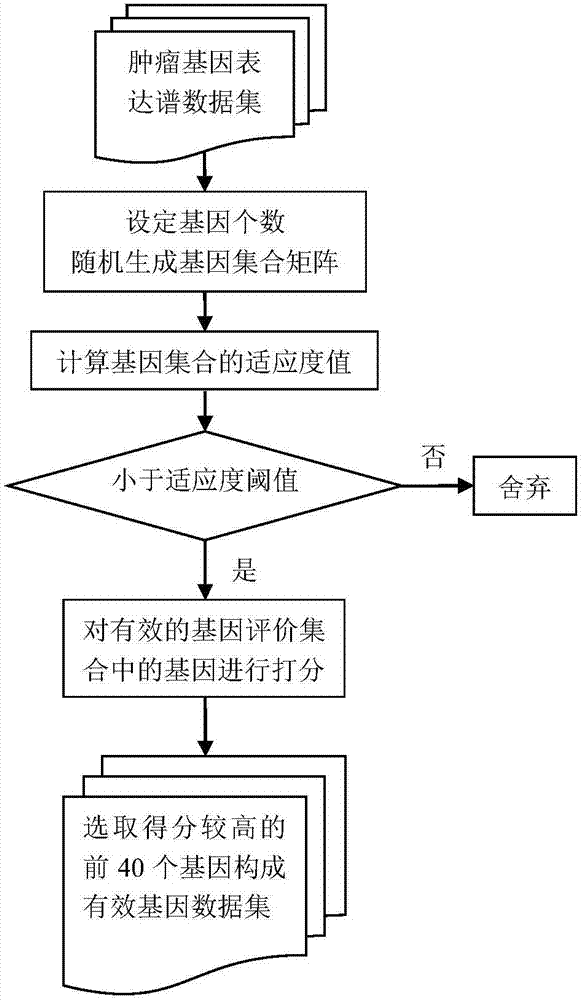
[0055] A key tumor gene identification method based on particle swarm optimization and scoring criteria, including optimizing Particle Swarm Optimization (PSO) through semi-initialization and Metropolis criteria, and using ELM extreme learning machine as an evaluation gene subset for correct classification The classifier of the rate, the step of obtaining the quantitative data of the classification performance of the algorithm comprises the following steps:
[0056] Step 1. Preprocessing of tumor gene expression profile data, including normalization and preliminary dimensionality reduction of tumor gene expression profile data sets, and simultaneously dividing tumor gene expression profile data sets into training sets and test sets;
[0057] Step 2 defines the scoring criteria and evaluates each gene in combination with the extreme learning machine, and screens out the top-scoring genes to establish a candidate gene pool;
[0058] Step 3 Combined with gene scoring information,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com