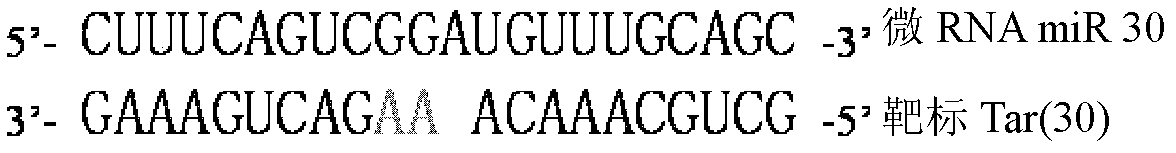Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
56 results about "MCherry" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
MCherry is a member of the mFruits family of monomeric red fluorescent proteins (mRFPs). As a RFP, mCherry was derived from DsRed of Discosoma sea anemones unlike green fluorescent proteins (GFPs) which are often derived from Aequoera victoria jellyfish. Fluorescent proteins are used to tag components in the cell, so they can be studied using fluorescence spectroscopy. mCherry absorbs light between 540-590 nm and emits light in the range of 550-650 nm. mCherry belongs to the group of fluorescent protein chromophores used as vital instruments to visualize genes and analyze their functions in experiments. Genome editing has been improved greatly through the precise insertion of these fluorescent protein tags into the genetic material of many diverse organisms. Most comparisons between the brightness and photostability of different fluorescent proteins have been made in vitro, removed from biological variables that affect protein performance in cells or organisms. It is hard to perfectly simulate cellular environments in vitro, and the difference in environment could have an effect on the brightness and photostability.
Screening method of sgRNA (small guide ribonucleic acid) efficient action target based on CRISPR-Cas13d (clustered red regularly interspaced short palindromic repeat) system and application
ActiveCN110656123AUniversalImprove efficiencyVector-based foreign material introductionDNA/RNA fragmentationVirusBioinformatics
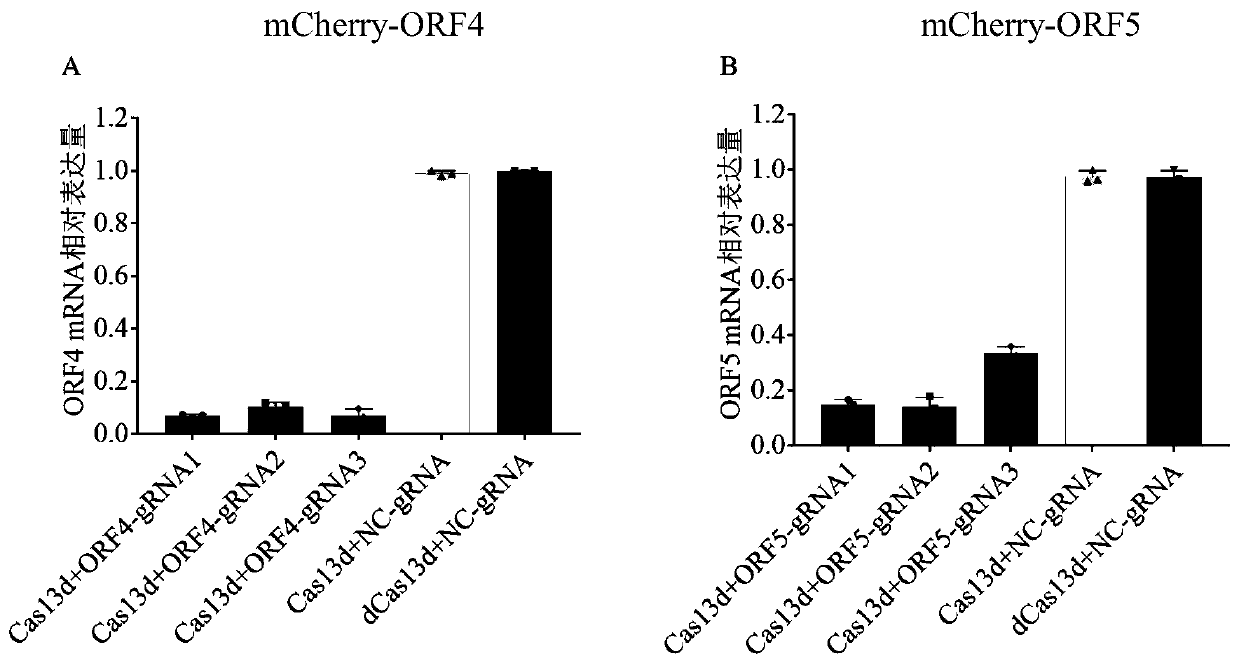
The invention provides a screening method of a sgRNA (small guide ribonucleic acid) efficient action target based on a CRISPR-Cas13d (clustered red regularly interspaced short palindromic repeat) system and application, and particularly application in RNA virus knockdown. According to the screening method, two expression read frame sequences ORF4 and ORF5 of a PRRSV are respectively fused with a carbon end of a mCherry gene through a mCherry fluorescence report gene, a fluorescence report system for screening sgRNA efficient action targets of the CRISPR-Cas13d system is established, and by virtue of the property that mRNA is efficiently cut by CRISPR-Cas13d, rapid screening of sgRNAs with high efficiency is implemented. Additionally, PRRSV-GFP recombinant viruses are efficiently degraded by using the screened efficient targetedly combined sgRNA based on the CRISPR-Cas13d system. The RNA virus knockdown method provided by the invention has the advantages of being high in efficiency, high in precision and low in target missing rate.
Owner:CHINA AGRI UNIV
Preparation method and application for dual-fluorescence reporting system with micro-RNA (ribonucleic acid) function
InactiveCN102443595AJudgment inhibitionMicrobiological testing/measurementFluorescence/phosphorescenceReporting systemEnhanced green fluorescent protein
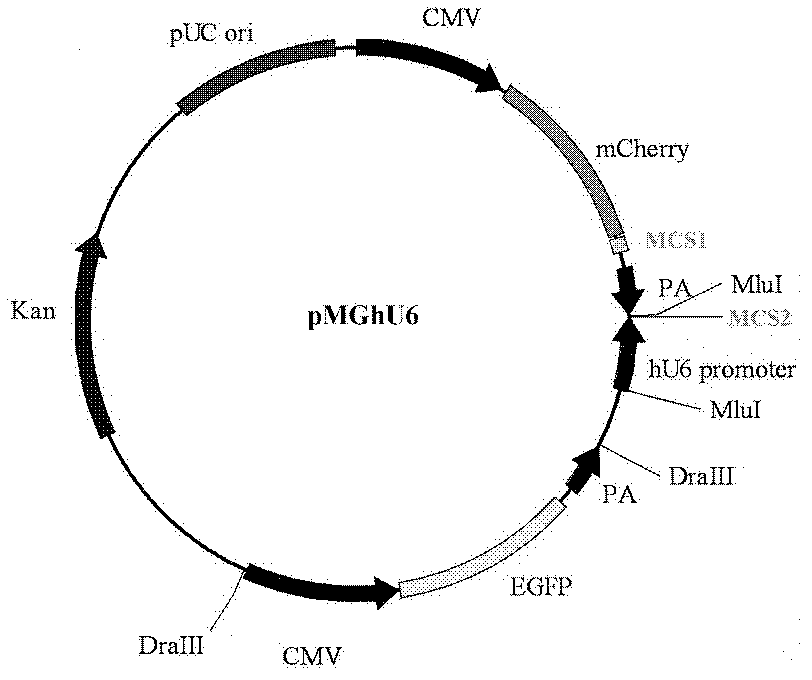
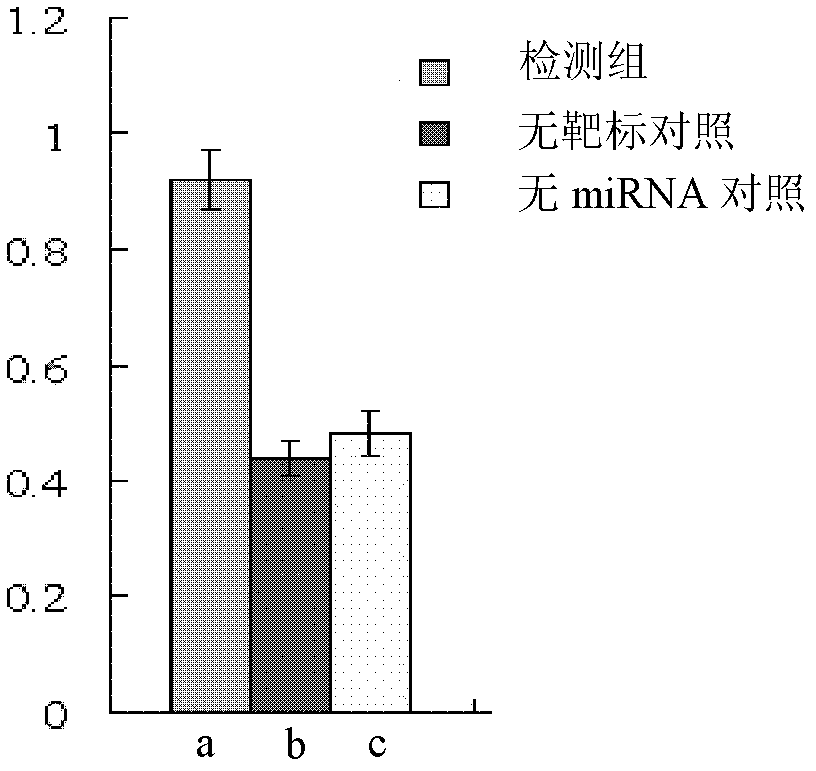
The invention discloses a preparation method and an application for a dual-fluorescence reporting system for detecting a micro-RNA (ribonucleic acid) function, wherein the preparation method comprises the following preparation steps of: A, obtaining a mCherry gene sequence via a PCR (polymerase chain reaction) amplification, inserting a rho EGFP-C1 carrier to replace the EGFP (enhanced green fluorescent protein) sequence of the mCherry gene sequence, and constructing a carrier rho mCherry-C1; B, using a RNAi-Ready rho SIREN-RetroQ plasmid as a template, obtaining a human U6 promoter gene via a PCR amplification, and inserting the carrier rho mCherry-C1 to obtain a plasmid rho hU6-mCherry-C1; and C, obtaining a gene sequence carried with a CMV (cytomegalovirus) promoter and an EGFP expression dialog sequence in an interval of 358 to 1944 on a plasmid rho Adtrack-CMV, and inserting the sequence in the plasmid rho hU6-mCherry-C1 to obtain a plasmid rho MGhU6. The plasmid rho MGhU6 is a dual-fluorescence reporting system simultaneously containing a mCherry reporting gene, an internal reference EGFP, a micro-RNA and the target sequence insertion site thereof. The reporting system can be used for detecting the inhibition function of the micro-RNA to the target sequence expression. The detection method is easy, as well as simple and convenient in operation; and a function detection for the micro-RNA can be realized by transfecting one plasmid only. The system also can be used for visually detecting the inhibition function of a micro-RNA in a single living cell to the target thereof by the aid of fluorescence microscopic imaging.
Owner:WUHAN INST OF VIROLOGY CHINESE ACADEMY OF SCI
Double-tagging microbial cell biosensor for detecting N-acyl homoserine lactone
The invention relates to a double-tagging microbial cell biosensor for detecting N-acyl homoserine lactone (AHL), and the biosensor is suitable for detection of AHL and characterization of host cells in the environment. The cell biosensor comprises: Escherichia coli used as host cells for carrying recombinant plasmids; recombinant plasmid containing an ahlI promoter responding to AHL, a cherry red fluorescent protein reporter gene mCherry and an AHL regulatory protein gene ahlR; and a neomycin phosphotransferase promoter gene nptII for characterization of host bacteria, a green fluorescent protein reporter gene gfp and a T1-4 pUC19 plasmid of four terminator rrnb tandem sequence. According to the biosensor, constitutive type of expression green fluorescent protein is used to characterize the host bacteria; the response time to AHL is 6 h; and the minimum detectable concentration of AHL is 50 nmol / L. The biosensor has the advantages of high sensitivity, fast response, low cost and simple operation, and can be widely applied to detection of AHL and host characterization in the environment.
Owner:RES CENT FOR ECO ENVIRONMENTAL SCI THE CHINESE ACAD OF SCI
African swine fever virus wild strain and vaccine strain identification and detection kit
InactiveCN111876527AHigh utility valueIncreased sensitivityMicrobiological testing/measurementMicroorganism based processesClassical swine fever virus CSFVAfrican swine fever
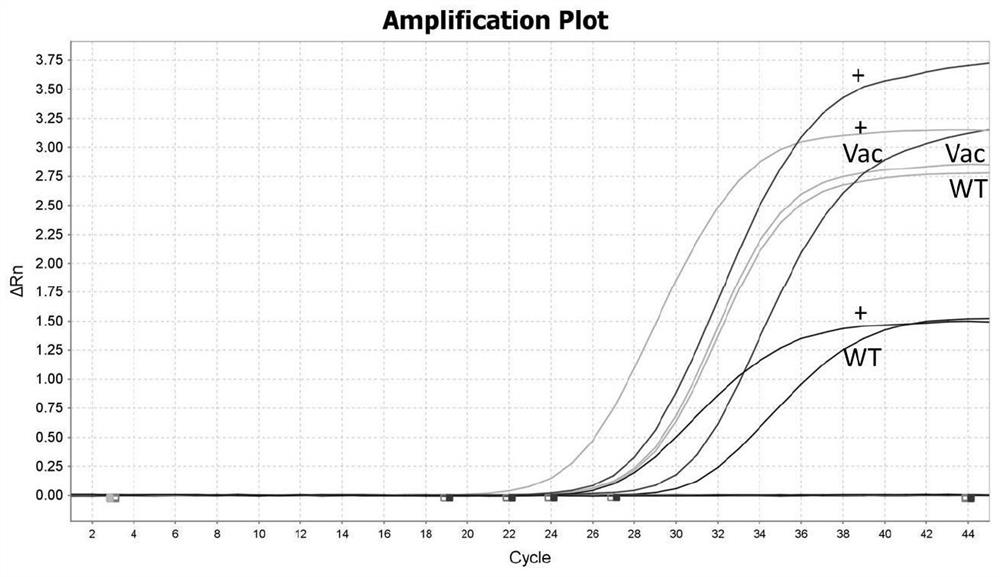
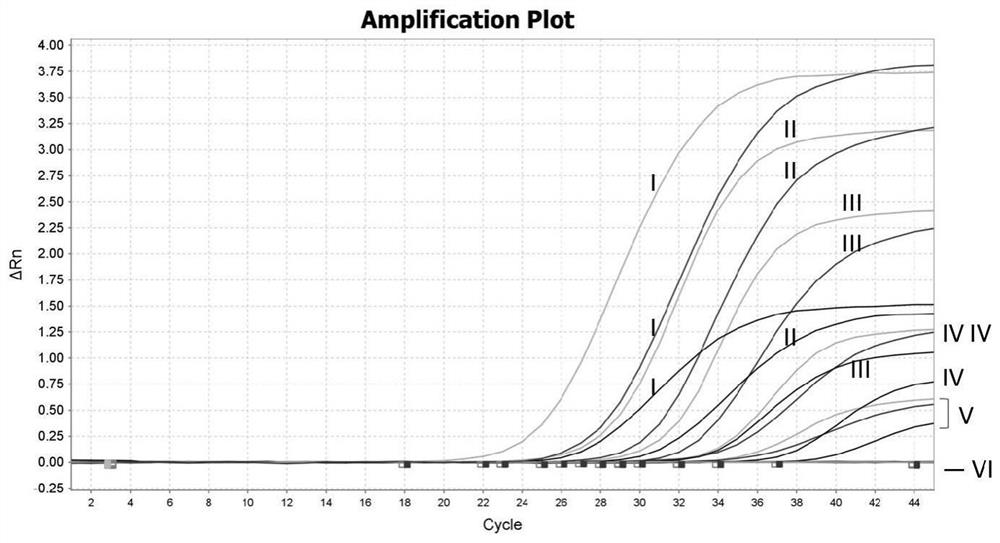
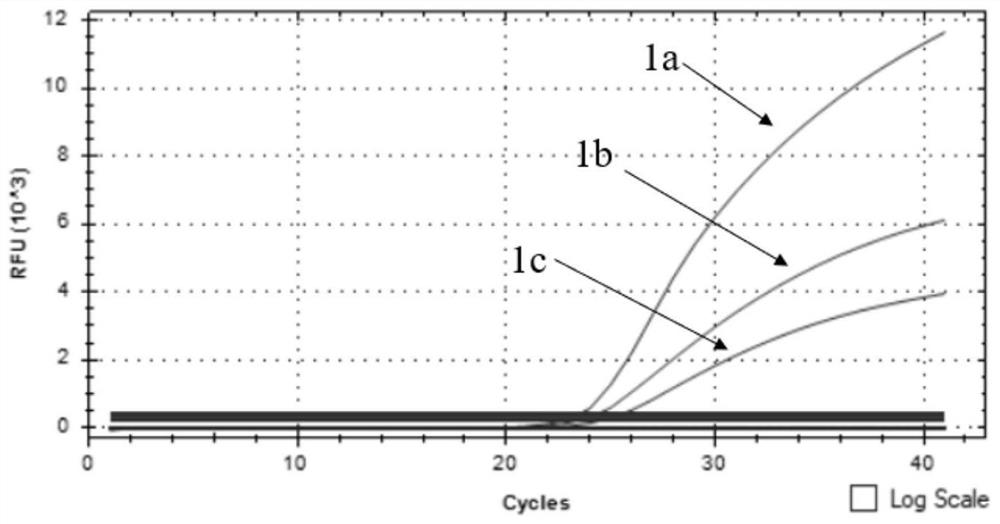
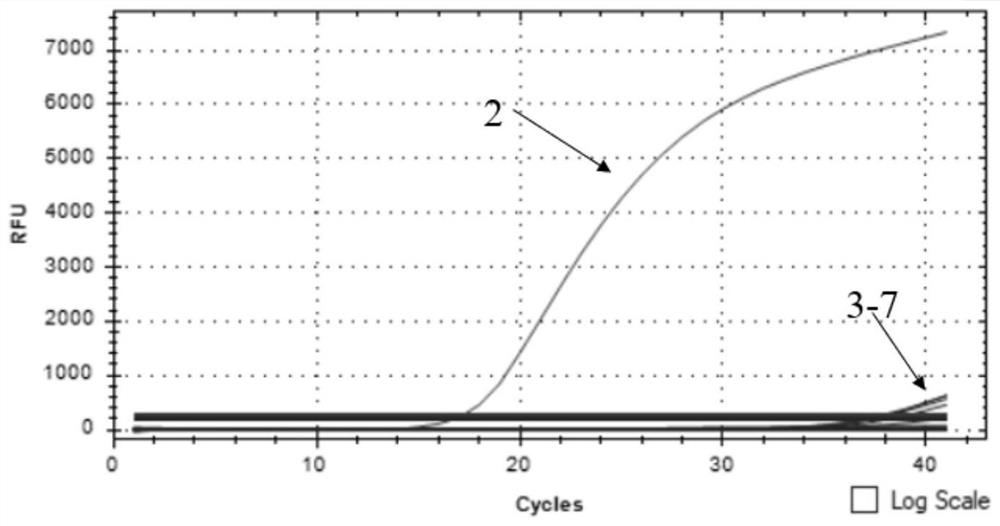
The invention discloses an African swine fever virus wild strain and vaccine strain identification and detection kit and application thereof. The kit provided by the invention comprises three groups of primers and probes, and the first group of primers and probes is completely paired with a specific sequence of an African swine fever virus B646L gene conserved region and is used for detecting theAfrican swine fever virus; the second group of primers and probes is completely paired with a specific sequence of an African swine fever virus EP402R gene conserved region and is used for detecting the African swine fever virus wild strain; and the third group of primers and probes is completely paired with the specific sequence of a red fluorescent protein mCherry gene and is used for detectingthe African swine fever virus vaccine strain. Different fluorescent reporter groups are respectively marked on the three probes. The African swine fever virus wild strain and vaccine strain identification and detection kit is high in detection sensitivity, capable of detecting target DNA with the minimum concentration of 100 copies / [mu] L, good in specificity, easy and convenient to operate and capable of effectively distinguishing African swine fever vaccine immune animals from wild virus infected animals.
Owner:CHINA ANIMAL HEALTH & EPIDEMIOLOGY CENT
Building and application of chronic virus transformation vector with CMV-CBh double promoters
The invention relates to building and application of a chronic virus transformation vector with CMV-CBh double promoters. The chronic virus vector pLenti-CMV-3FLAG-EGFP-tCBh-mCherry-T2A-Puro is obtained by replacing PGK in the pLenti-CMV-3FLAG-EGFP-tCBh-mCherry-T2A-Puro with tCBh fragments obtained by performing PCR (Polymerase Chain Reaction) amplification. The tCBh promoter is used for driving expression of mCherry gene and Puromycin resistance gene, and cells for stabilizing over-expression of target gene can be screened by using Puromycin; the mCherry gene is beneficial for judging the virus packaging efficiency and the infection efficiency, and is also beneficial for stabilizing the screening of the cells. Transfection fluorescence observation proves that the chronic virus vector expression system with the double promoters is successfully built, and a basic vector is provided for further researching target genes of interest.
Owner:OBIO TECH SHANGHAI CORP LTD
Triple fluorescent quantitative PCR detection primers and kit for identifying African swine fever wild strain and gene deletion strain
ActiveCN112646934ARapid identificationRapid epidemic monitoringMicrobiological testing/measurementDNA/RNA fragmentationClassical swine fever virus CSFVAfrican swine fever
The invention discloses triple fluorescent quantitative PCR (Polymerase Chain Reaction) detection primers and a kit for identifying an African swine fever virus wild strain and a gene deletion strain. The invention provides a group of specific primers and probes for detecting the African swine fever virus wild strain and the gene deletion strain, and the specific primers and probes are used for simultaneously detecting the B646L gene of the African swine fever virus wild strain and the EGFP and mCherry genes of the deleted strain through an RT-qPCR method, so that whether a sample is the African swine fever virus wild strain or the gene deletion strain can be identified. The detection method is high in sensitivity and strong in specificity, and the lowest detection concentrations of B646L, EGFP and mCherry gene positive standard plasmids are 5.94*10 <-9> ng / mu L, 9.67*10 <-9> ng / mu L and 5.85*10 <-9> ng / mu L respectively. The method is wide in detection range, can be used for detecting various African swine fever virus gene deletion strains, is rapid and efficient in detection process, is suitable for large-batch detection, provides a new means for identifying African swine fever virus wild strains and gene deletion strains, and can be used for rapidly identifying the African swine fever virus wild strains and gene deletion strains in production practice.
Owner:SOUTH CHINA AGRI UNIV
Method and related biological material for cultivating transgenic plant of fluorescent protein mCherry labeled microtubulin
InactiveCN105018521AEfficient expressionStable expressionBacteriaFluorescence/phosphorescenceNucleotideA-DNA
The invention discloses a method and a related biological material for cultivating a transgenic plant of fluorescent protein mCherry labeled microtubulin. The method comprises the step of importing PTUB6:mCherry-TUB6 to a receiver plant to obtain the transgenic plant of the fluorescent protein mCherry labeled microtubulin, wherein PTUB6:mCherry-TUB6 is formed by connecting mCherry-TUB6 fusion genes with PTUB6 for starting transcription of the mCherry-TUB6 fusion genes, the mCherry-TUB6 fusion genes are genes for encoding fusion protein mCherry-TUB6, the fusion protein mCherry-TUB6 is protein obtained by fusing fluorescent protein mCherry with plant microtubulin, and PTUB6 is a DNA molecule shown in 1st-3004th positions of a nucleotide sequence shown in SEQ ID No.1.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
Optically-controlled expression vector for insect cells and application
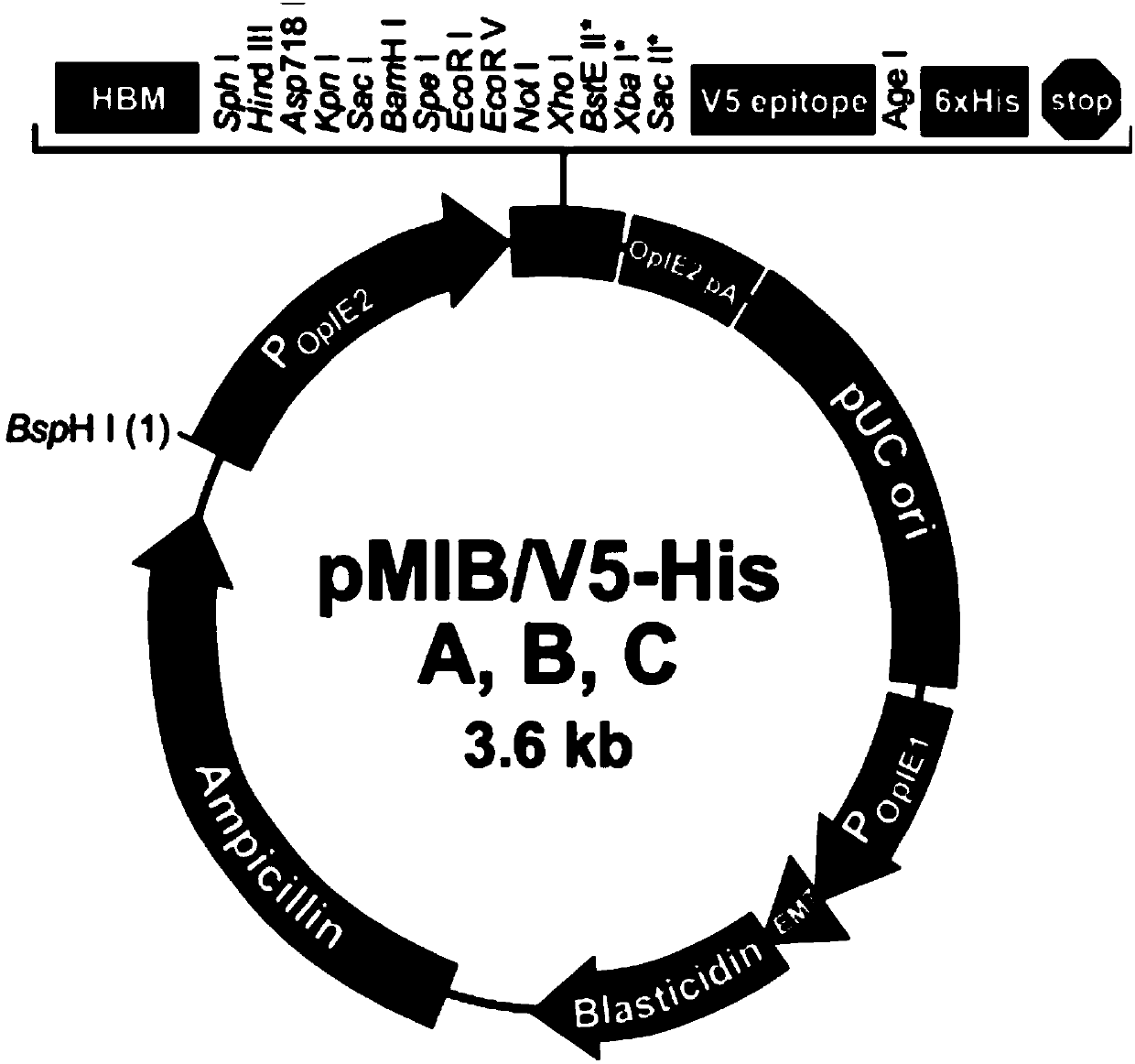
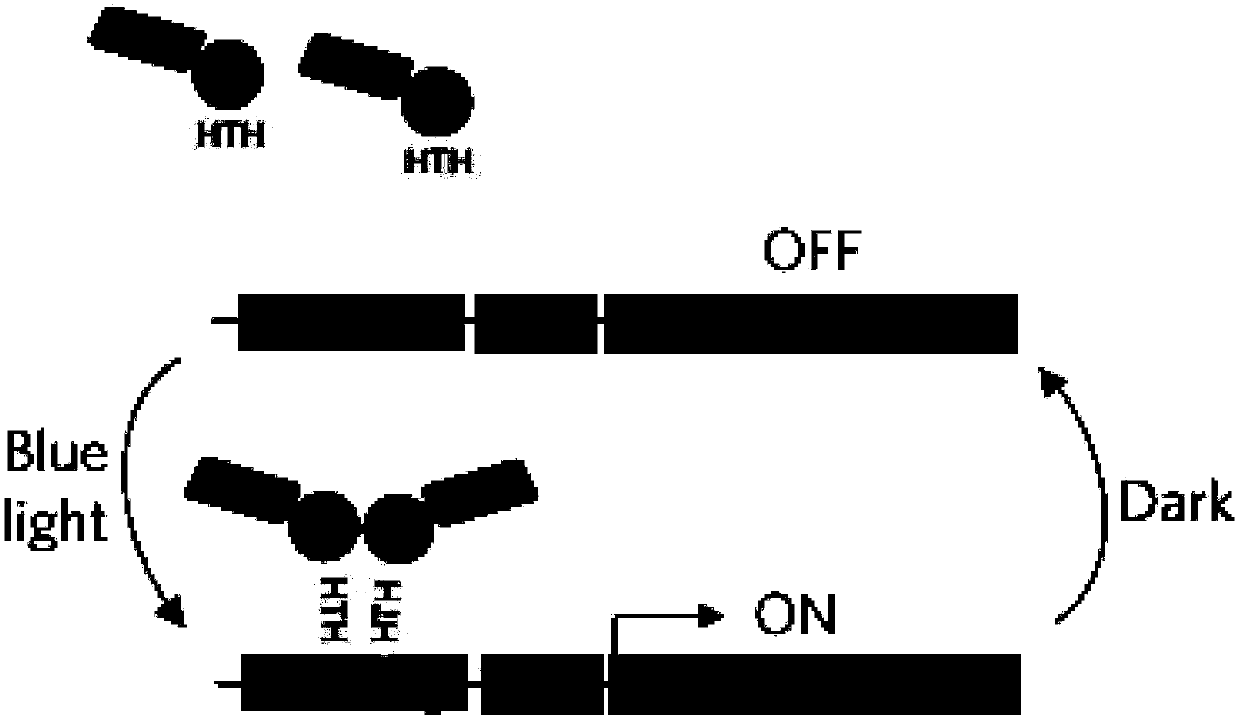
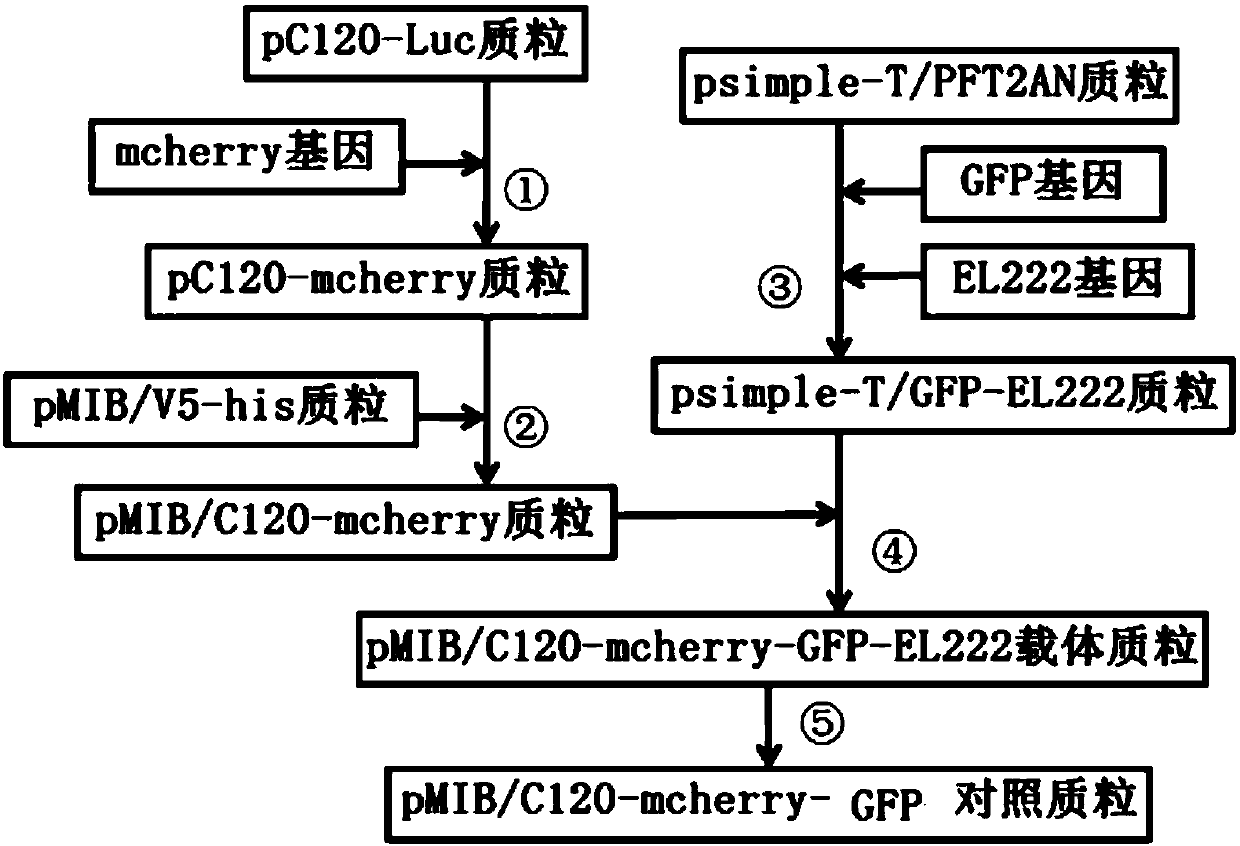
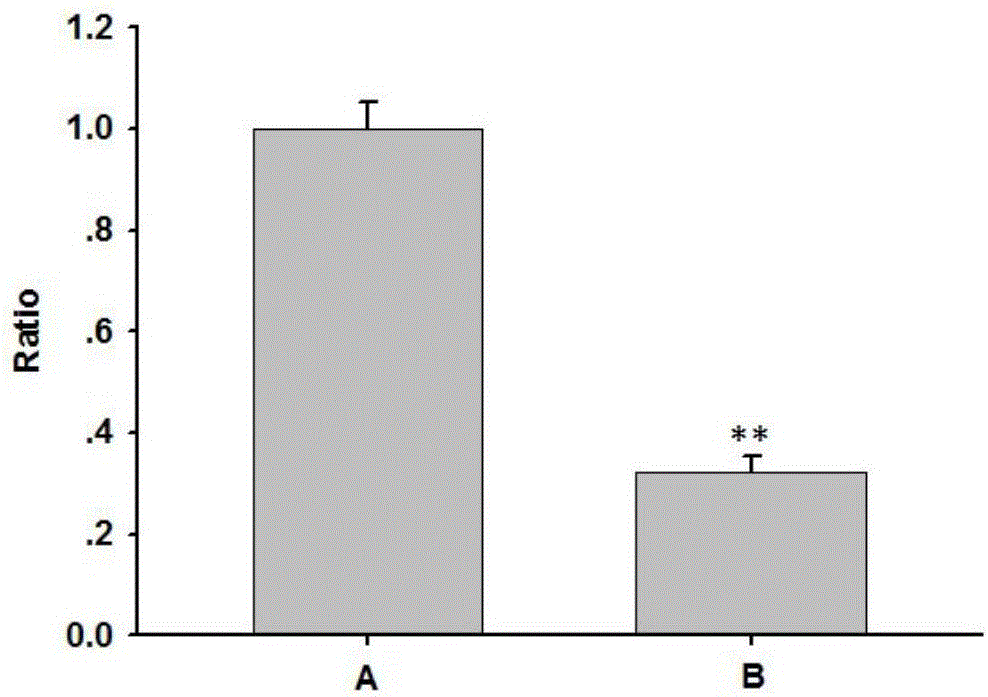
ActiveCN107937437ATest fast and stableIncrease positive rateVector-based foreign material introductionBiologyFluorescent protein
The invention discloses an optically-controlled expression vector for insect cells and application. An applicant successfully constructs a positive plasmid pMIB-C120-mCherry-GFP-EL222 and a control plasmid pMIB-C120-mCherry-GFP on an insect cell expression vector pMIB / V5-HisA based on a VP-EL222 optically-controlled transcription activating system; after the positive plasmid pMIB-C120-mCherry-GFP-EL222 is transfected with Sf9 cells, a green fluorescent protein expresses while a red fluorescent protein does not express; after the positive plasmid pMIB-C120-mCherry-GFP-EL222 is irradiated and stimulated by blue light, the red fluorescent protein expresses; after the control plasmid pMIB-C120-mCherry-GF is transfected with the Sf9 cells, the green fluorescent protein expresses while the red fluorescent protein does not express; after the control plasmid pMIB-C120-mCherry-GF is irradiated and stimulated by the blue light, the red fluorescent protein still does not express due to no EL222 transcription factor. An optically-controlled inducible expression system is successfully applied to the insect cells for the first time, and the application range of the technology is widened. According to a verification method constructed by the invention, the plasmids also provide a flexible and easy-to-use method for using an optically-controlled induced expression technology.
Owner:布林凯斯(深圳)生物技术有限公司
Method for screening activity enhancer by utilizing vector based on random sequence
PendingCN110885845AHigh identification accuracyHigh precisionMicrobiological testing/measurementVector-based foreign material introductionRegulation of gene expressionPromoter
The invention discloses an enhancer screening expression vector and a method for screening an enhancer by utilizing a random sequence. The enhancer screening expression vector is formed by modifying ashuttle plasmid vector, wherein a core component on the enhancer screening expression carrier comprises a mini promoter and a downstream mcherry reporter gene, and the upstream of the mini promoter comprises a recombination site of a random sequence. The invention provides the enhancer screening expression vector and the method for screening the enhancer by using the random sequence. According tothe method, the enhancer sequence is effectively identified by utilizing the enhancer screening expression vector and a screening library; compared with the prior art, the method has the advantages that the identification accuracy of the enhancer in the genome is greatly improved, meanwhile, the enhancer in a dozens of bp sequences can be effectively identified, and the method has important significance in deeply understanding a gene expression regulation mechanism.
Owner:AGRI GENOMICS INST CHINESE ACADEMY OF AGRI SCI
Gene knockout test kit and method for rapidly screening sgRNA
InactiveCN106591366AEasy to operateShorten the experiment cycleNucleic acid vectorFermentationFirefly luciferinFluorescent protein
The invention discloses a gene knockout test kit and method for rapidly screening sgRNA. The test kit includes a kit body, a reagent A and a reagent B. The test kit utilizes pCAG-mCherry-Luc plasmids obtained through fusion of full-length sequences-containing firefly luciferase and green fluorescent protein C terminals, targeted genomic sequences are inserted in the fronts of green fluorescent protein initiation codons, finally the plasmids and expression plasmids of sgRNA and CRISPR-Cas9 transfect HEK293FT cells together, through red fluorescence protein brightness observation and luciferase viability measurement, qualitative or quantitative measurement of sgRNA mediated genome modification activity is quickly conducted, and through a conventional method in the field, the effectiveness of an sgRNA screening system in a slow virus system is verified. The test kit and the method have the advantages that an analog genome of any target gene can be constructed simply and rapidly, and the sgRNA activity is evaluated more quickly and simply and accurately quantified.
Owner:SHANGHAI REPODX BIOTECH CO LTD
Double-fluorescence report recombinant plasmid vector as well as construction method and application thereof
InactiveCN107686846AReduce expressionReduced expression levelMicrobiological testing/measurementVector-based foreign material introductionSequence analysisPlasmid Vector
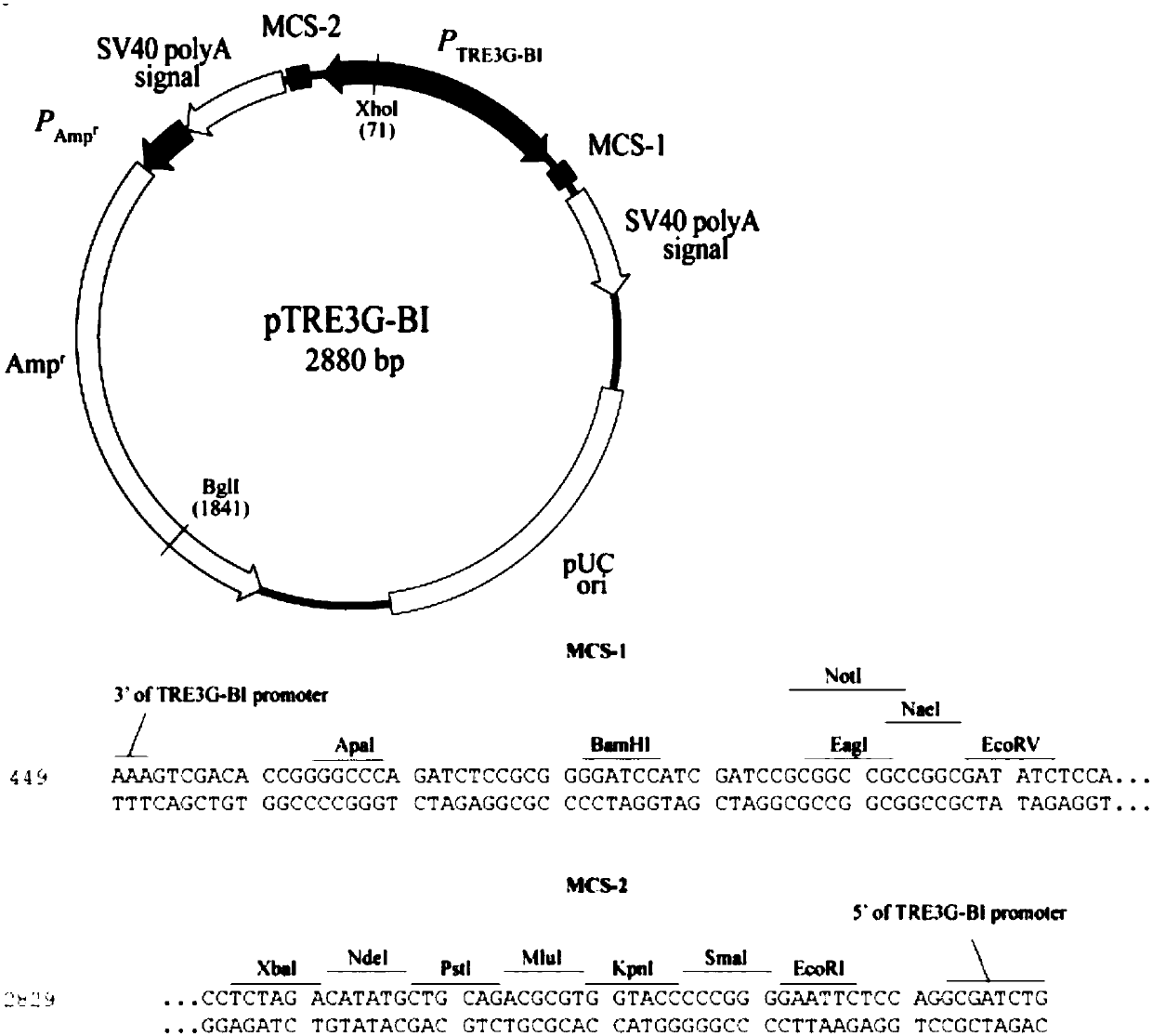
The invention discloses a double-fluorescence report recombinant plasmid vector. The double-fluorescence report recombinant plasmid vector contains a coded green fluorescence protein gene sequence, ared fluorescence protein gene sequence and a 3' non-translation area sequence of a miRNA specific target gene. The construction method comprises the following steps: amplifying a green fluorescence protein gene and a red fluorescence protein gene by a PCR method; inserting the amplified green fluorescence protein gene segment into the 3' direction of a plasmid vector pTRE3G-BI promoter to obtain arecombinant plasmid vector pTRE3G-GFP, and performing sequencing analysis; inserting the amplified red fluorescence protein gene segment into the 5' direction of the plasmid vector pTRE3G-BI promoterto obtain a recombinant plasmid vector pTRE3G-GFP-mCherry, and performing sequencing analysis; amplifying the 3' non-translation area sequence of the target gene by the PCR method and inserting in the mCherry 3'direciton of the plasmid vector pTRE3G-GFP-mCherry; naming the recombinant plasmid vector which is verified to be correct as pTRE3G-GFP-mCherry-3UTR. The double-fluorescence report recombinant plasmid vector can be applied to verify whether miRNA and the target gene thereof have mutual combination relation in the sequence or not.
Owner:SHAANXI NORMAL UNIV
DNA fragment and application thereof in construction of recombinant influenza virus expressing red fluorescent protein gene
ActiveCN110205321ADoes not affect processing modificationDoes not affect assemblySsRNA viruses negative-sensePeptidesFluorescenceA-DNA
The invention discloses a DNA fragment and an application thereof in construction of a recombinant influenza virus expressing a red fluorescent protein gene. An mCherry fluorescent gene is inserted behind an NS1 gene of an H9N2 influenza virus strain, wherein the complete NS gene is formed by sequentially linking an NS1 gene, the mCHerry fluorescent gene and an NEP gene; the gene and the NEP geneare connected by a PTV-1 2A sequence which can be recognized and cut by protease, so a fluorescent protein-labeled NS fusion gene is constructed and has the nucleotide shown in SEQ ID NO:1. The constructed recombinant influenza virus can produce live virus expressing red fluorescent proteins, is relatively stable in the process of virus passage, does not affect the function of the virus itself, has the growth curve and the titer similar to those of a wild virus strain, can be used for subsequent research, and is of great significance on development, screening and production of anti-influenza virus drugs.
Owner:SOUTH CHINA AGRI UNIV
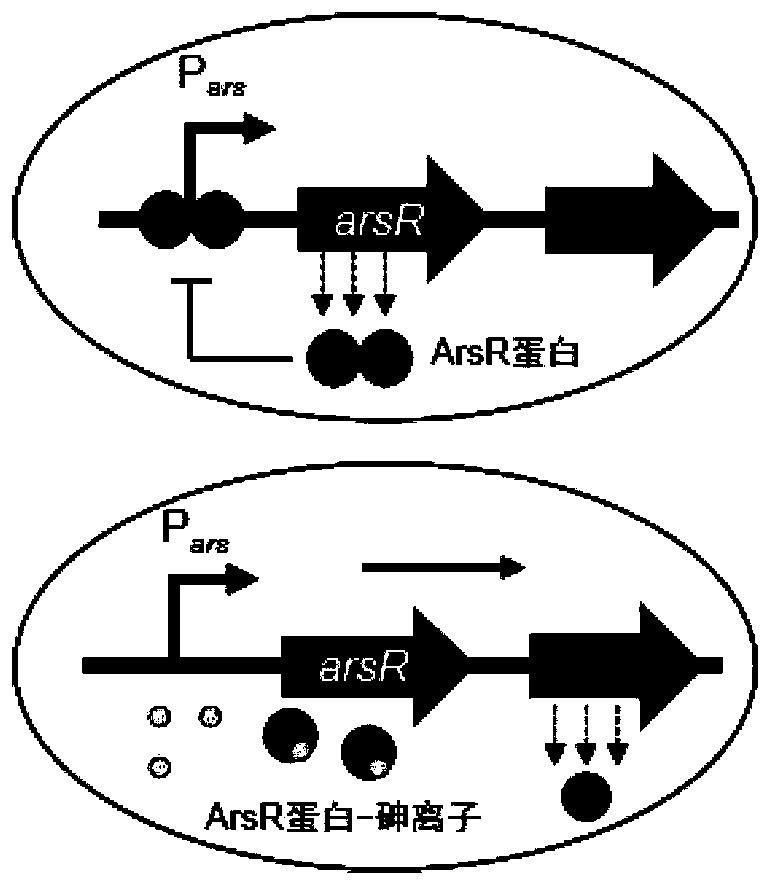
Construction of sensitive arsenic ion whole cell biosensor and arsenic ion concentration detection method
PendingCN111004814AHigh sensitivityUndisturbedMicrobiological testing/measurementBiological material analysisEscherichia coliArsenite ion
The invention relates to construction of a sensitive arsenic ion whole cell biosensor and an arsenic ion concentration detection method. The sensor detection plasmid and the reporter plasmid are recombinant. The detection plasmid consists of an arsenic-specific protein regulatory promoter Pars, an arsenic-specific binding protein ArsR gene, and a self-feedback regulatory protein gene luxR. The reporter plasmid consists of a self-feedback promoter PluxRI, a red fluorescent protein mCherry gene and a self-feedback regulatory protein gene LuxR. By introducing a positive feedback amplification system based on the variant LuxR protein as a regulatory element, the sensitivity of the sensor is increased by 20 times to 0.1 micron, and the specific signal ratio is increased by 1.5 to 7.5 times. Thearsenic ion biosensor takes Escherichia coli DH5 as a host cell, has high sensitivity and specificity for arsenic ion detection, is not interfered by other metal ions, and has wide adaptability.
Owner:TIANJIN UNIV
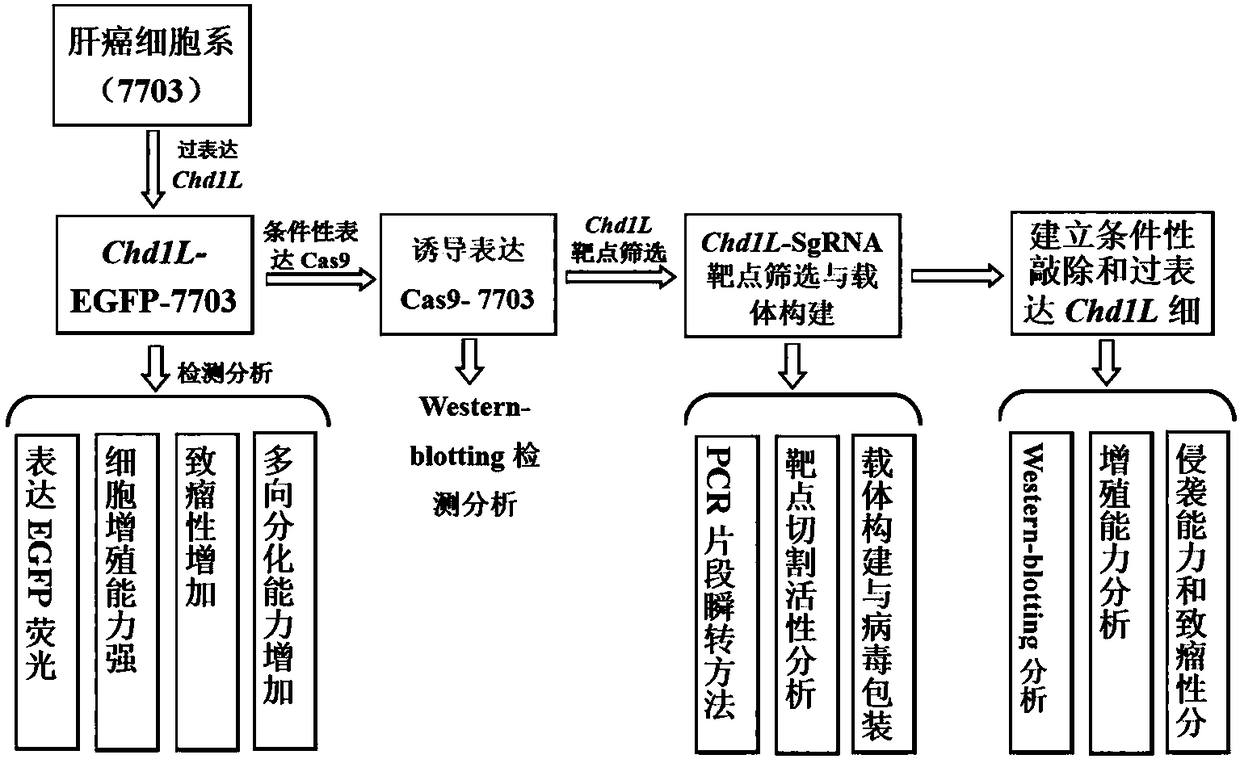
Construction method for hepatoma cell line for conditionally-induced knockout overexpressed Chd1l genes
InactiveCN108342416AReduce the number of culturesReduce experimental errorHydrolasesGenetically modified cellsHepatoma cell lineRed fluorescence
The invention discloses a hepatoma cell line for conditionally-induced knockout overexpressed Chd1l genes and a construction method thereof. The method comprises the steps that firstly, a hepatoma cell line for overexpressed Chd1l and green fluorescent proteins is constructed; secondly, a carrier for conditionally-induced expressed Cas9 is transferred into the hepatoma cell line; finally, a pLVX-mCherry-hU6-Chd1l-SgRNA carrier with red fluorescence and targeted Chd1l cutting activity is constructed and transferred to the cell line, and the hepatoma cell line for conditionally-induced knockoutoverexpressed Chd1l is obtained at last. The cell line has the advantages of being the hepatoma cell line for overexpressed Chd1l before the induction of doxycycline (Dox) and being the hepatoma cellline for expressed knockout Chd1l after induction. The cell line has good contrast, the simultaneous culture of multiple cell lines can be reduced, the scientific research work is simplified, and thecell line is an ideal tool for gene function research.
Owner:GUANGZHOU MEDICAL UNIV
Construction method for coccidia expressing exogenous fluorescent protein
ActiveCN112553079APrecise positioningStrong fluorescent signalProtozoaMicrobiological testing/measurementBiotechnologyCoccidiosis
The invention provides a construction method for coccidia expressing exogenous fluorescent protein, and belongs to the technical field of gene engineering. The construction method for coccidia expressing exogenous fluorescent protein comprises the following steps: designing gRNA for a target gene of the coccidiosis, and constructing a gene editing plasmid containing the gRNA; constructing a homologous recombinant plasmid containing a target gene and an mCherry gene; co-transfecting the gene editing plasmid and the homologous recombinant plasmid into sporozoites of coccidia; and infecting animals with the transfected sporozoites, and performing screening to obtain the coccidia expressing exogenous fluorescent protein. A fluorescent protein mCherry gene and a screening gene are knocked intoa coccidia genome by combining a gene editing technology and a homologous recombination technology, and the constructed coccidia can stably and efficiently express a fluorescent signal. According to the construction method, not only can the coccidia expressing the exogenous fluorescent protein be successfully constructed, but also fluorescent protein expression is stable and high in efficiency, and the construction method is suitable for research on growth, genetic engineering or drug screening of coccidia.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Combining method for optogenetics and neprilysin genes
PendingCN111073906AEfficient degradationCompletely degradedAntibody mimetics/scaffoldsAnimals/human peptidesOptogeneticsPlasmid dna
The invention discloses a combining method for optogenetics and neprilysin (NEP) genes. The method comprises the following main steps: 1) by using mammalian expression plasmid VR1012 as a gene vector,recombining the neprilysin genes and light-activated neuronal channel protein ChR2 genes to a eukaryotic expression plasmid through PCR, digestion, connection and monoclonal picking, and adding a neural specific promoter of hSyn before ChR2 to obtain a recombinant plasmid capable of simultaneously expressing neprilysin and the light-activated neuronal channel protein ChR2 in vivo and in vitro; 2)connecting an EGFP green label after the neprilysin, and connecting an mCherry red label after the ChR2; and 3) performing gene sequencing on the obtained recombinant plasmid DNA, and verifying whether the neprilysin and ChR2 genes are inserted correctly into the VR1012 eukaryotic expression plasmid at the nucleic acid molecular level. The NEP is the most important enzyme for degrading Abeta protein in a brain, so that the NEP can be used to reduce risk.
Owner:TIANJIN UNIV
Construction method for circuit amplification of cadmium ion whole-cell biosensor
ActiveCN112725373AHigh sensitivityImprove featuresVectorsMicrobiological testing/measurementHemt circuitsAmino acid mutation
The invention relates to a construction method for circuit amplification of a cadmium ion whole-cell biosensor. According to the research, the cadmium whole-cell biosensor p2T7RNAPmut-68 is constructed in a mode of adding a T7RNAPmut (amino acid at the 40th site mutates into a termination codon) amplification module for the first time. The sensitivity and specificity of the biosensor are improved by amplifying a circuit through a T7RNAPmut (the 40th amino acid mutates into a termination codon) amplification module, so that the defects of a traditional cadmium ion biosensor in detection are overcome. Plasmid pCDFDuet-2 and pGN68 are used as carriers, and the cadmium ion whole-cell biosensor is obtained by utilizing a cadmium specific binding protein CadR, a reporter element mCherry, a cadO operon and a T7RNAPmut amplification module. The invention provides the construction method of a whole-cell biosensor sensitive to cadmium ions. The whole-cell biosensor sensitive to cadmium ions has wide adaptability.
Owner:TIANJIN UNIV
Dynamic monitoring system for iPSC differentiation
ActiveCN113265427AStable expressionGenetically modified cellsArtificially induced pluripotent cellsCell culture mediaDynamic monitoring
The invention discloses an iPSC differentiation dynamic monitoring system, a dual-fluorescence reporter gene vector system constructed by the invention comprises pCDH-CMV-GFP-EF1a-lift-mCherry plasmids, a hiPSC cell line constructed by transformation of the vector system can stably express red fluorescent protein for a long time and does not express green fluorescent protein, and once differentiation begins, the cells can stably express green fluorescent protein for a long time and also can express red fluorescent protein. The construction of the system provides a powerful research tool for the development of an iPSC culture medium, the detection of iPSC residues, the research of an iPSC stemness maintenance mechanism, the research of cytoskeleton change in an iPSC differentiation process, the tracking of iPSC cell fate and outcome, the optimization of a differentiation system, the detection of an iPSC cell bank, cell transplantation and other downstream researches.
Owner:CHENGNUO REGENERATIVE MEDICINE TECH (ZHUHAI HENGQIN NEW AREA) CO LTD
Method for analyzing protein interactions in prokaryotes based on YESS
The invention belongs to the technical field of gene engineering, and discloses a method for analyzing protein interaction in prokaryotes based on a Yeast Endoplasmic reticulum Sequestration System (YESS). The method includes the steps: constructing an expression plasmid pESD-PPI-GFP-mCherry capable of performing Golden Gate assembling, with gfp and mCherry fluorescent genes, based on pESD-PPI plasmid; selecting 5 mutually interacted protein pairs from the existing data of Zymomonas mobilis on a UniProt database and the protein pairs which are not reported to have interaction as positive and negative contrasts for studying protein interactions of proteins in Zymomonas mobilis; and quantifying the analysis results. The method for analyzing protein interactions in prokaryotes can achieve high-throughput and quantitative study on protein interaction in Zymomonas mobilis. The method for analyzing protein interactions in prokaryotes based on YESS has many advantages of being simple, being easy to operate, being highly efficient, being sensitive and the like.
Owner:武汉睿嘉康生物科技有限公司
CRISPR-Cas13d system for knocking down porcine epidemic diarrhea virus
PendingCN114836418AEfficient degradationHighly inhibitoryHydrolasesNucleic acid vectorPorcine epidemic diarrhoea virusFluorescent reporter
The invention provides a CRISPR-Cas13d (clustered regularly interspaced short palindromic repeats-associated protein 13d) system for knocking down a porcine epidemic diarrhea virus. An mCherry fluorescent reporter gene is utilized, four expression reading frame sequences of PEDV (porcine epidemic diarrhea virus) ORF3, E, M and N are fused to a C terminal of the mCherry gene respectively, a fluorescent reporter system for screening and targeting sgRNA efficient action targets of the PEDV expression reading frames is constructed, and the characteristic that CRISPR-Cas13d efficiently cuts mRNA is utilized to quickly screen efficient sgRNAs. Then, the screened sgRNAs which are combined in a high-efficiency targeting manner are utilized to carry out high-efficiency degradation on a target virus by using a CRISPR-Cas13d (clustered regularly interspaced short palindromic repeats) system. The RNA virus knock-down method provided by the invention has the advantages of high efficiency, high accuracy and low off-target rate.
Owner:CHINA AGRI UNIV
A light-controlled expression vector for insect cells and its application
ActiveCN107937437BTest fast and stableIncrease positive rateVector-based foreign material introductionFluorescent proteinPlasmid transfection
The invention discloses an optically-controlled expression vector for insect cells and application. An applicant successfully constructs a positive plasmid pMIB-C120-mCherry-GFP-EL222 and a control plasmid pMIB-C120-mCherry-GFP on an insect cell expression vector pMIB / V5-HisA based on a VP-EL222 optically-controlled transcription activating system; after the positive plasmid pMIB-C120-mCherry-GFP-EL222 is transfected with Sf9 cells, a green fluorescent protein expresses while a red fluorescent protein does not express; after the positive plasmid pMIB-C120-mCherry-GFP-EL222 is irradiated and stimulated by blue light, the red fluorescent protein expresses; after the control plasmid pMIB-C120-mCherry-GF is transfected with the Sf9 cells, the green fluorescent protein expresses while the red fluorescent protein does not express; after the control plasmid pMIB-C120-mCherry-GF is irradiated and stimulated by the blue light, the red fluorescent protein still does not express due to no EL222 transcription factor. An optically-controlled inducible expression system is successfully applied to the insect cells for the first time, and the application range of the technology is widened. According to a verification method constructed by the invention, the plasmids also provide a flexible and easy-to-use method for using an optically-controlled induced expression technology.
Owner:布林凯斯(深圳)生物技术有限公司
Non-resistance screening double antigen anchor expression vector plq2a and its preparation method
ActiveCN108410901BSolve problemsTo achieve the purpose of non-resistance screeningVector-based foreign material introductionBiotechnologyEscherichia coli
Owner:JILIN AGRICULTURAL UNIV
A method for gene suppression in Gluconobacter oxidans
ActiveCN112094843BImprove gene editing efficiencyFast expressionBacteriaMicroorganism based processesGlucuronateGluconobacter suboxydans
Owner:JIANGNAN UNIV
A method for stably expressing secreted human insulin in mesenchymal stem cells
ActiveCN113201062BSecretion does not affectEasy to purifyGenetically modified cellsSkeletal/connective tissue cellsMesenchymal stem cellPancreatic hormone
The invention discloses a method for stably expressing secretable human insulin in mesenchymal stem cells, comprising the following steps: (1) constructing an mCherry-2A-insulin-His fusion gene lentiviral expression vector; ‑2A‑insulin‑His fusion gene lentiviral expression vector is expressed in human mesenchymal stem cells to obtain extracellular human insulin. After transfection of cells, a stable expression cell line is obtained by screening. The cells express the fusion polypeptide and under the action of self-cleaving polypeptide 2A, human insulin and mCherry red fluorescent gene are separated, and human insulin can be secreted outside the cell. This inventive method solves the current dilemma of expressing human insulin in mesenchymal stem cells, and has great practical significance and potential medical application value.
Owner:WUHAN UNIV OF SCI & TECH
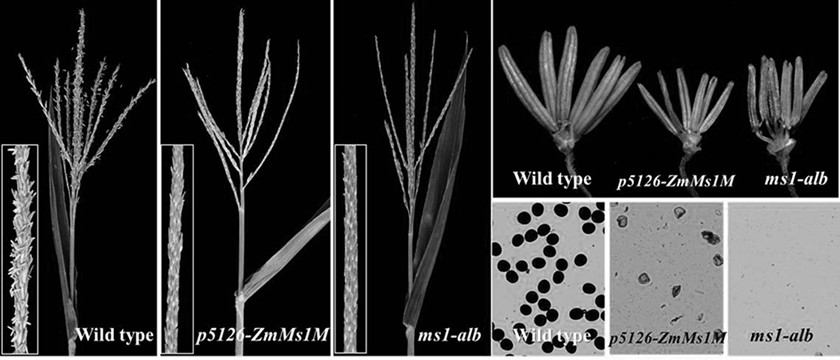
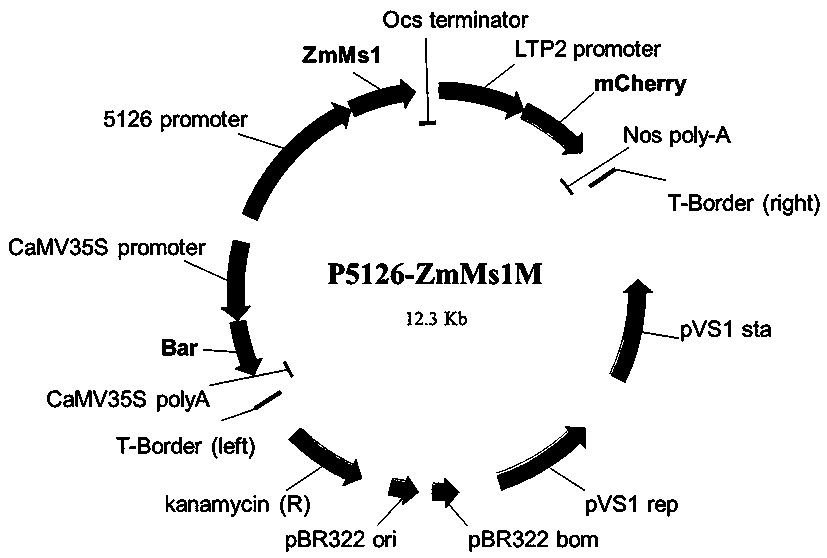
Using the p5126-zmms1m construct to create a maize dominant male sterile line and its application method for breeding and production
ActiveCN109504701BVector-based foreign material introductionPlant genotype modificationBiotechnologyHybrid seed
The invention discloses a technological system for preparing a corn dominant genic male sterility line from a p5126-ZmMs1 and mCherry gene construct and a corn seed cross breeding and producing application method thereof. The construct contains three gene expression cassettes: an expression cassette for causing corn dominant genic male sterility, an expression cassette for marking corn peel colorand a weed killer resistance expression cassette. When the construct is guided into a corn callus tissue, the corn dominant genic male sterility line seeds can be prepared; under normal light, the transgenic sterility line seeds have no obvious difference from the non-transgenic fertile seeds, and goods quality is not affected; however, under green excitation light, the transgenic sterility line seeds show red fluorescence, and the non-transgenic fertile seeds show normal corn color and have no fluorescence. The technological system disclosed by the invention has qualitative difference from acorn recessive male sterility technology (namely a corn multi-control sterility technology system) which has been already applied for authorization by the team and can be efficiently applied to corn sterility cross breeding and hybrid seed production.
Owner:BEIJING SHOU JIA LI HUA SCI TECH CO LTD
Corn dominant nuclear genic sterility line prepared from p5126-ZmMs1M construct and seed breeding and producing application method thereof
ActiveCN109504701AVector-based foreign material introductionPlant genotype modificationTechnology systemCorns callus
The invention discloses a technological system for preparing a corn dominant genic male sterility line from a p5126-ZmMs1 and mCherry gene construct and a corn seed cross breeding and producing application method thereof. The construct contains three gene expression cassettes: an expression cassette for causing corn dominant genic male sterility, an expression cassette for marking corn peel colorand a weed killer resistance expression cassette. When the construct is guided into a corn callus tissue, the corn dominant genic male sterility line seeds can be prepared; under normal light, the transgenic sterility line seeds have no obvious difference from the non-transgenic fertile seeds, and goods quality is not affected; however, under green excitation light, the transgenic sterility line seeds show red fluorescence, and the non-transgenic fertile seeds show normal corn color and have no fluorescence. The technological system disclosed by the invention has qualitative difference from acorn recessive male sterility technology (namely a corn multi-control sterility technology system) which has been already applied for authorization by the team and can be efficiently applied to corn sterility cross breeding and hybrid seed production.
Owner:BEIJING SHOU JIA LI HUA SCI TECH CO LTD
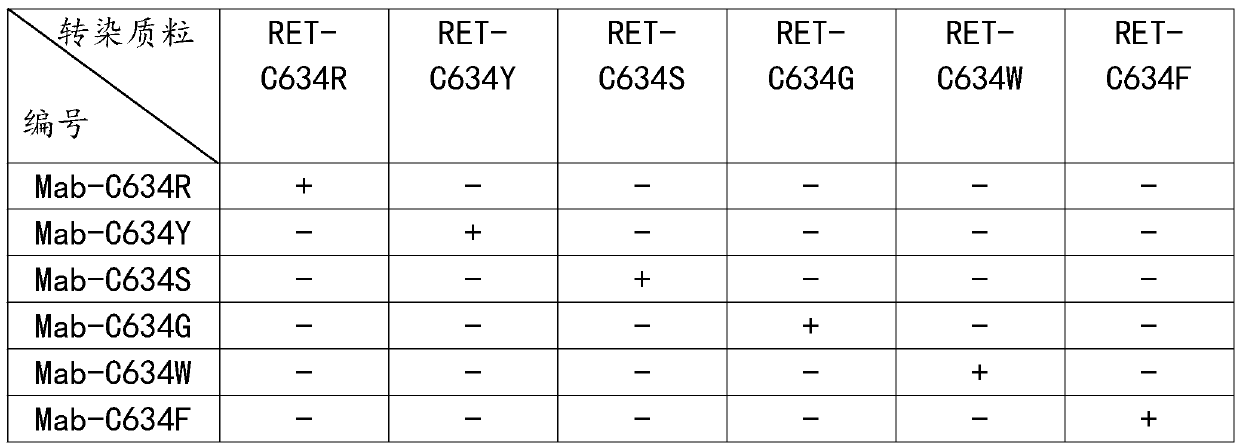
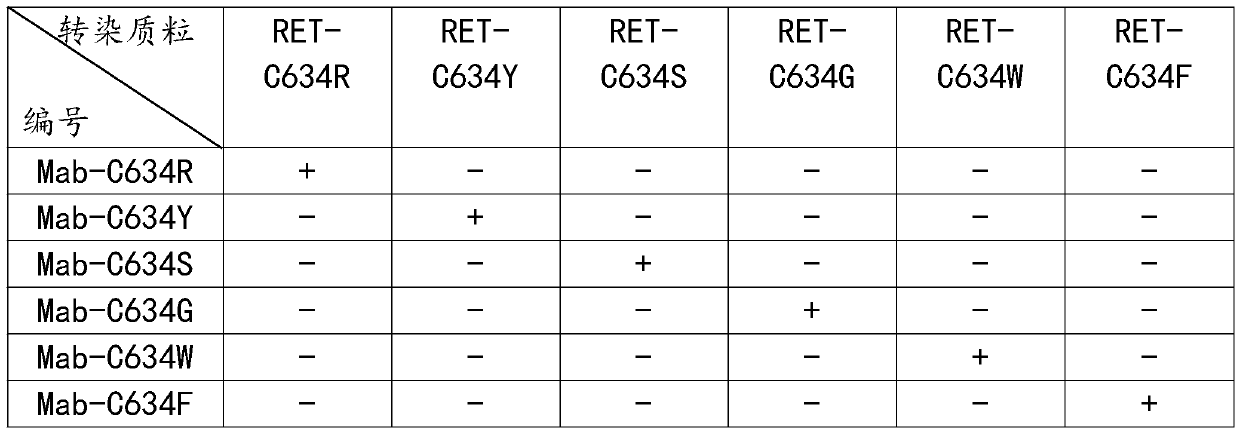
Preparation method of anti-RET mutant protein monoclonal antibody variable region sequence
InactiveCN110938145AQuick checkQuick kindImmunoglobulins against cell receptors/antigens/surface-determinantsAdjuvantMutated protein
The present invention discloses a preparation method of an anti-RET mutant protein monoclonal antibody variable region sequence. Mutation is performed at the C634 position of a RET protein to synthesize a polypeptide sequence, and the polypeptide and a carrier protein KLH are respectively coupled and combined with freund's adjuvant for emulsification to prepare an emulsified antigen, mutants are separately used to construct expression sequences to a purple fluorescent protein mcherry amino terminus to be combined with an expression vector to prepare a detection agent, mice are immunized with the emulsified antigen and mouse myeloma SP2 / 0 cells and spleen cells of the immunized mice are subjected to cell fusion, and are cultured with a semi-solid culture medium and the detection agent, by observing formation of clone balls and color changes of the culture medium, positive clones are selected into new 6-well plates and subjected to expanded culture by a changed liquid culture medium, each cell line is then transferred to a shake flask for culture, a supernatant of the culture medium is collected, antibodies are purified, and the antibodies are sequenced to obtain the variable regionsequence.
Owner:安徽环球基因科技有限公司
Mcherry or mEOS nano antibody as well as preparation method and application of mcherry or mEOS nano antibody
ActiveCN113201069ASmall molecular weightStrong penetrating powerHybrid immunoglobulinsImmunoglobulins against animals/humansMCherryProtide
The invention relates to the technical field of bioengineering, in particular to an mcherry or mEOS nano antibody and a preparation method and application thereof, and relates to the technical field of bioengineering. By constructing an mcherry or mEOS nano antibody library, a specific positive monoclonal nano antibody is screened out, two mcherry nano antibodies capable of being combined with mcherry protein are detected, nucleotide sequences of the two mcherry nano antibodies are shown as SEQ ID NO.1 and SEQ ID NO.3, and seven mEOS nano antibodies capable of being combined with mEOS protein are detected. The nucleotide sequences of the primers are as shown in SEQ ID NO. 5, SEQ ID NO. 7, SEQ ID NO. 9, SEQ ID NO. 11, SEQ ID NO. 13, SEQ ID NO. 15 and SEQ ID NO. 17. Compared with a traditional antibody, the mcherry or mEOS nano antibody constructed by the invention has the advantages that the yield can be increased, the cost can be reduced, and better stability can be obtained.
Owner:NANHUA UNIV
A yess-based method for analyzing protein interactions in prokaryotes
The invention belongs to the technical field of gene engineering, and discloses a method for analyzing protein interaction in prokaryotes based on a Yeast Endoplasmic reticulum Sequestration System (YESS). The method includes the steps: constructing an expression plasmid pESD-PPI-GFP-mCherry capable of performing Golden Gate assembling, with gfp and mCherry fluorescent genes, based on pESD-PPI plasmid; selecting 5 mutually interacted protein pairs from the existing data of Zymomonas mobilis on a UniProt database and the protein pairs which are not reported to have interaction as positive and negative contrasts for studying protein interactions of proteins in Zymomonas mobilis; and quantifying the analysis results. The method for analyzing protein interactions in prokaryotes can achieve high-throughput and quantitative study on protein interaction in Zymomonas mobilis. The method for analyzing protein interactions in prokaryotes based on YESS has many advantages of being simple, being easy to operate, being highly efficient, being sensitive and the like.
Owner:武汉睿嘉康生物科技有限公司
Method for inhibiting genes in gluconobacter oxydans
ActiveCN112094843AImprove gene editing efficiencyFast expressionBacteriaMicroorganism based processesGenetic engineeringMCherry
The invention discloses a method for inhibiting genes in gluconobacter oxydans, and belongs to the technical field of gene engineering and bioengineering. According to the method, a corresponding target gene crRNA is expressed in the gluconobacter oxydans. By verifying the inhibition effect on mCherry fluorescence expression on plasmids, the recombinant gluconobacter oxydans has a gene inhibitioneffect, and by inhibiting other genes in the gluconobacter oxydans genome, a similar inhibition effect is also achieved, and the inhibition function of the system on the genes on the genome is furtherproved. A new method for inhibiting the target genes in the gluconobacter oxydans is provided, the expression of the target genes can be rapidly and effectively inhibited, and the gene editing efficiency of the gluconobacter oxydans is improved.
Owner:JIANGNAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com