Non-resistance screening double antigen anchor expression vector plq2a and its preparation method
An expression vector and non-resistance technology, which is applied in the field of Lactobacillus plantarum non-resistance screening double antigen-anchored expression vector pLQ2a and its preparation field, can solve the problem of positive recombination in screening of exogenous plasmid electrotransformin host bacteria and Lactobacillus plantarum ligation products Difficulties in fertilization, cell wall thickness of Lactobacillus plantarum, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
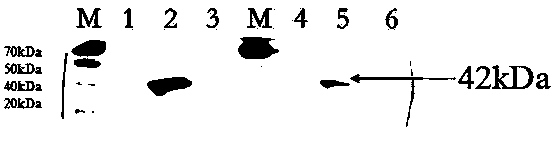
Image
Examples
Embodiment 1
[0043] Example 1 Using alr-EM to double-label the alr gene in the vector 409eatf and host bacteria L. plantarum NC8 / Δalr nutritional complementation screening validation
[0044] (1) pSIP409PgsA'-IL-10 double digestion
[0045] Digest with SalI and PstI, and recover the 4436bp gene fragment containing erythromycin and the replicon on the carrier by agarose gel electrophoresis. The base sequence of the fragment is shown in SEQ ID NO.6.
[0046] (2) Alr t and alr tf gene amplification
[0047] extract L. plantarum The genome of NC8 strain was used as a template to design primers. The primer alr gene contains a SalI restriction site, which is not convenient for vector construction. Therefore, the primers were extended during design, so that alr can be amplified and mutated. The first C in the restriction site was mutated to G, and the amplified gene was named alr t (1149bp). In order to detect the expression of the alr gene after returning to Lactobacillus plantarum, a Fla...
Embodiment 2
[0078] Example 2 Construction of nutritionally complementary asd-alr double-labeled plasmid vector 409ata and its use in E. coli 6212 / Δasd and L. plantarum NC8 / Δalr non-resistance screening verification
[0079] (1) Plasmid 409eat was double digested with BamHI and SalI, and a 4625bp fragment containing the replicon and alr gene was recovered. The base sequence is shown in SEQ ID NO.7.
[0080] (2) Amplify the asd gene, use the PYA4545 plasmid as a template, and use the following primers to amplify a 1758bp fragment containing homologous defects.
[0081] Pasd positive F: CGCTATGCGTGCG GGATCCTCTTCCCTAAATTTAAATATAAAC (underlined is the sequence homologous to the vector, italicized BamHI)
[0082] Pasd R: GGGTTGTCAGGGCTTTTC GTC GAC AACATCAGGTAGTGACA (underlined is the sequence homologous to the vector, italicized to mark Sal I)
[0083] a. The PCR reaction system is as follows:
[0084]
[0085] b. PCR reaction conditions are as follows:
[0086]
[0087] (3...
Embodiment 3
[0090] Example 3 No anti-screening anchor co-anchor expression EGFP and Mcherry verification
[0091] (1) Construction and verification of single-anchor expression EGFP plasmid pSIP409PgsA’-EGFPAT without anti-screening
[0092] 1) Based on the plasmid pSIP409PgsA'-EGFP, replace the EM gene on the vector with asd-alr. Refer to the 409ata gene sequence to design seamless cloning primers containing homologous sequences. The designed primers are versatile. The upstream of the primers is designed with reference to the "256rep" replicon on the vector. Using this primer, the seamless cloning method can be used to clone this series of vectors. The erythromycin was replaced with the asd-alr gene to construct an expression vector for non-resistance screening. The amplified fragment was named 256-asd-alr, the fragment size was 3719bp, and the base sequence was shown in SEQ ID NO.8; The homology arm was added during amplification, so the amplified fragment size was 3765bp.
[0093] Amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com